Abstract
The resistance of cancer cells to treatment significantly impedes the success of therapy, leading to the recurrence of various types of cancers. Understanding the specific mechanisms of therapy resistance may offer novel approaches for alleviating drug resistance in cancer. Recent research has shown a reciprocal relationship between circular RNAs (circRNAs) and N6-methyladenosine (m6A) modification, and their interaction can affect the resistance and sensitivity of cancer therapy. This review aims to summarize the latest developments in the m6A modification of circRNAs and their importance in regulating therapy resistance in cancer. Furthermore, we explore their mutual interaction and exact mechanisms and provide insights into potential future approaches for reversing cancer resistance.
Keywords: Circular RNAs, N6-methyladenosine, Crosstalk, Cancer, Therapeutic resistance
Introduction
Up to now, more than 160 varieties of chemical alterations have been identified in RNA molecules, with methylation being the most prevalent form [1]. These RNA methylation modifications include N6-methyladenosine (m6A), 5-methylcytosine (m5C), 7-methylguanine (m7G), 2'-O-dimethyladenosine (m6Am), N1-methyladenosine (m1A), and 5-hydroxymethylcytosine (5hmC) [2]. Among these modifications, m6A is the most prevalent modification in eukaryotic cells [3]. Also known as N6-methyladenosine modification, m6A is a type of post-transcriptional RNA modification that plays a crucial role in regulating gene expression [4, 5]. The discovery of m6A modification in RNA was first reported in the 1970s [6], and it has essential functions in various processes, such as RNA stability, splicing, translation, and RNA–protein interactions [7].
Circular RNAs (circRNAs) are a type of RNA molecule that forms a single-stranded closed loop structure, lacking the typical 5' to 3' ends [8]. Researchers now widely accept that circRNAs can be translated into functional peptides through internal ribosome entry site (IRES)-mediated translation. The absence of a 5' cap in circRNAs suggests that they must rely on cap-independent mechanisms, such as m6A modification, for translation initiation. Unlike linear RNA molecules, circRNAs resist degradation by exonucleases and exhibit more excellent stability [9]. The topic of m6A modification in circRNAs is an area of ongoing research and still needs to be fully understood. Initially, it was believed that circRNAs, due to their circular structure, were not susceptible to m6A modification [10]. However, recent studies have challenged this notion by providing evidence of m6A modifications in circRNAs.
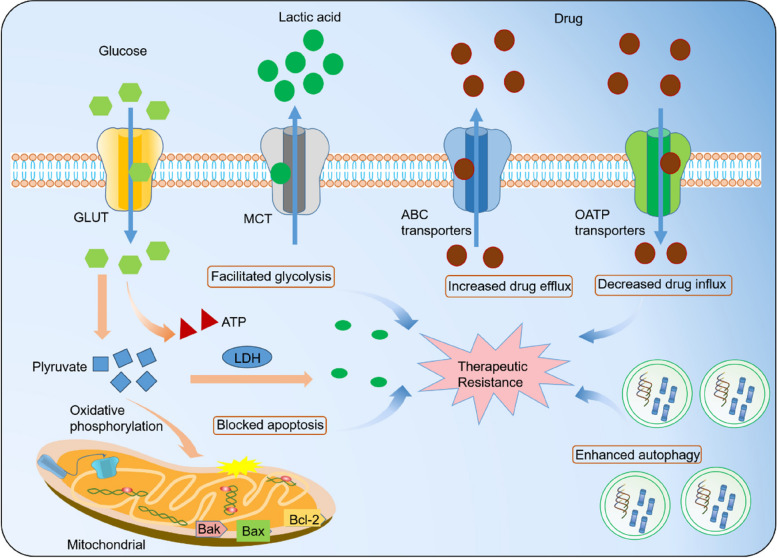
A growing body of experimental data has elucidated that m6A modification can also occur in circRNAs [11]. This modification can potentially influence the stability, localization, and function of circRNAs [12]. It has been suggested that m6A modification in circRNAs can regulate their interactions with RNA-binding proteins (RBPs), miRNAs, and specific molecules, thereby impacting their overall cellular functions [13]. However, our understanding of m6A modification in circRNAs is still relatively limited, and further studies are urgently needed to investigate the underlying mechanisms and functional significance [14]. Nevertheless, the study of m6A modification in circRNAs has great potential for uncovering novel regulatory mechanisms in RNA biology [15]. Under treatment pressure, cancer cells can develop resistance to therapy, allowing them to evade death. On one hand, changes in drug transport and metabolism can impair the efficacy of various anticancer drugs. On the other hand, cancer cells can acquire survival advantages through mechanisms like apoptosis resistance, DNA damage repair, and induction of epithelial-to-mesenchymal transition (EMT) (Fig. 1).
Fig. 1.
Mechanisms of therapeutic resistance in cancer. Enhanced drug efflux and diminished drug influx result in a reduced accumulation of drugs within cancer cells. In addition, inhibiting programmed cell death, facilitating DNA damage repair, and augmenting cellular self-digestion processes also contribute to the survival of tumor cells under treatment-induced stress. Furthermore, cancer cells prefer glucose metabolism, which produces lactic acid through glycolysis, promoting rapid cell proliferation in response to therapeutic interventions. Lastly, an imbalanced tumor microenvironment (TME), promotion of the transition from epithelial to EMT, and heightened properties of cancer stem cells (CSCs) also impede the effectiveness of cancer treatment
Herein, this review comprehensively analyzes the latest developments and mechanisms of m6A modification of circRNA in tumor treatment, the crosstalk between m6A modifications and circRNAs, and their interactions in modulating treatment resistance. This highlights the potential of targeting these modifications and RNAs to overcome resistance to cancer treatment.
m6A modification and circRNAs
m6A modification regulators
Readers are crucial in various aspects of RNA processing, including RNA splicing, transport, degradation, and translation [16]. Some well-known reader proteins include the YTH domain-containing family (YTHDF) proteins and YTH domain-containing family (YTHDC) proteins [17]. The interaction among writers, erasers, and readers of m6A modification regulates RNA processing and function [18]. The addition and removal of the methyl group can affect RNA stability, splicing, translation efficiency, and RNA–protein interactions [10]. The reader proteins determine the fate of the modified RNA, directing it toward degradation or translation [19]. A concise overview is depicted in Fig. 2.
Fig. 2.
Overview of m6A modification. A complex of multicomponent m6A methyltransferases installs the writers of the m6A modification, while the erasers remove it through demethylases. In the nucleus, nuclear readers recognize m6A and regulate RNA transcription, splicing, and structure. In the cytoplasm, cytoplasmic readers detect m6A and regulate RNA stability, translation, and binding capacity
The YTH domain functions as a reader molecule, specifically recognizing m6A modifications on RNA in a methylation-dependent manner. Humans have five known YTH domain-containing proteins: YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3. These proteins play a critical role in post-transcriptional regulation, influencing splicing, translation, RNA localization, and overall RNA lifespan [20]. A comparison of the crystal structures of YTHDC2 and YTHDC1 domains reveals that both a conserved hydrophobic pocket and a positively charged surface are crucial for recognizing m6A-modified RNA [21]. Similarly, YTHDF1, YTHDF2, and YTHDF3 also possess aromatic cages, specific residues for m6A recognition, and a basic patch that facilitates RNA backbone binding [22]. Scutenaire et al. conducted a comprehensive evolutionary analysis of YTH domains in Viridiplantae [23]. Their study revealed that vascular plants possess YTHDF- and YTHDC-type motifs, which contain the essential amino acids for RNA binding and m6A accommodation. These motifs are predicted to adopt a similar structural fold as YTH domains found in animals and yeast.
Within the cell nucleus, the dynamic regulation of RNA methylation is carried out by m6A writers and erasers. For instance, YTHDC1 plays a crucial role in splicing and controlling the export of m6A-modified mRNAs. This is achieved by recruiting SRSF3 while simultaneously blocking the binding of SRSF10 to the mRNA. YTHDF1 recognizes m6A-containing mRNAs and promotes both initiation and elongation of translation in the cytoplasm. Additionally, YTHDF2 recognizes the same mRNAs and targets them for degradation through deadenylation mediated by the CCR4-NOT complex and endoribonucleolytic cleavage mediated by HRSP12. Notably, YTHDF3 interacts with YTHDF1 and YTHDF2, accelerating the overall metabolism of m6A-modified mRNAs. YTHDC2, on the other hand, plays a distinct role in regulating the switch from mitosis to meiosis through its interaction with MEIOC. Additionally, YTHDC2 destabilizes target RNAs by interacting with proteins such as XRN1, UPF1, and MOV10. Interestingly, YTHDC2 also binds to the 18S ribosomal RNA and exhibits 3'-5' RNA helicase activity, both of which contribute to the translation of its target RNAs.
Writers, erasers, and readers actively interact in the process of m6A modification
The m6A modification process is controlled by a trio of proteins known as writers, erasers, and readers. These proteins, including METTL3/14/16, VIRMA, and WTAP, add a methyl group to the RNA, effectively turning the switch on. Conversely, erasers, such as FTO and ALKBH proteins, remove the methyl group, acting as molecular "off switches". Additionally, reader proteins like YTHDC1/2 and IGF2BP1/2/3 act as cellular sensors, detecting the presence or absence of the m6A mark and triggering downstream events accordingly. This dynamic interplay allows for precise control over the fate and function of RNA.
The intricate process of RNA m6A modification is dependent on the dynamic interaction between writers, readers, and erasers within eukaryotic cells [24]. These specialized proteins, including methyltransferases (writers) such as METTL3, METTL14, and WTAP, work together to form the m6A methyltransferase complex (MTC) responsible for adding m6A to target mRNA [25, 26]. Reader proteins in both the nucleus and cytoplasm act as chemical regulators. They can directly influence RNA processing by recruiting specific partners such as YTHDC1, YTHDF2, and YTHDF3, which can alter RNA base-pairing, secondary structures, and protein-RNA interactions, ultimately determining the fate of the RNA [27].
Erasers, also known as enzymes, play a crucial role in this dynamic dance by removing the m6A modification and creating a reversible system alongside writers and readers [28, 29]. This dynamic regulation of m6A has become a critical area of cancer research, as it is closely linked to how environmental pollutants can trigger carcinogenesis.
Mutual regulation between circRNAs and m6A modification
CircRNAs can be classified into four categories: circular intronic RNAs (ciRNAs), exonic circRNAs (ecRNAs), tRNA intronic circRNAs (tricRNAs), and exon–intron circRNAs (EIciRNAs), each with distinct formation mechanisms. Researchers have elucidated several biogenesis mechanisms, including lariat-driven circularization and circularization associated with RBPs.
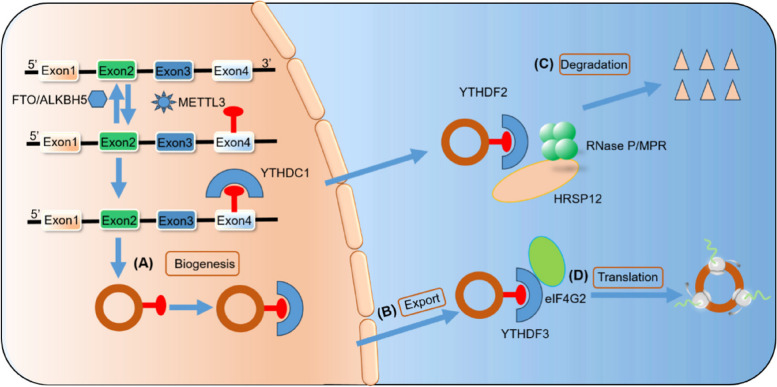
Surprisingly, emerging research has revealed that m6A fulfills an essential role in various aspects of circRNAs. These aspects are illustrated in Fig. 3 and include: (1) Biosynthesis of circRNAs. Previous studies have shown that m6A modulates the biosynthesis of circZNF609 in a YTHDC1/METTL3-dependent manner. Similarly, m6A modification is strongly related to the biogenesis of circ1662, circARL3, and circMETTL3 in various tumors. (2) Export of circRNAs from the nucleus to the cytoplasm. Exporting circRNAs from the nucleus to the cytoplasm is essential for their function. m6A-modified circNSUN2 can interact with YTHDC1 in the nucleus, thereby facilitating its export from the nucleus to the cytoplasm. (3) Degradation of circRNAs. There is limited research on the degradation of circRNAs, and the potential mechanisms are still unclear. However, m6A modification has been found to suppress the degradation of circRNAs, thereby increasing their stability. For example, a study showed that the t1/2 of m6A-modified circCUX1 was extended when treated with Actinomycin D, indicating that m6A modification enhances the stability of circCUX1 [30]. (4) Translation of circRNAs. CircRNAs can be translated through two cap-independent pathways: m6A-dependent translation and the IRES. The translation of m6A-modified circRNAs is dependent on eIF4G2 and the involvement of METTL3/14.
Fig. 3.
Impact of m6A modification on biosynthesis, export, translation, and degradation of circRNAs. A CircRNA biosynthesis: METTL3, an m6A writer, adds m6A sites to pre-mRNAs. These m6A sites can then be detected by YTHDC1, an m6A reader, which facilitates the process of back-splicing. B CircRNA export: YTHDC1 also promotes the export of circRNAs to the cytoplasm by binding to m6A residues. C CircRNA degradation: YTHDF2, another m6A reader, recognizes m6A-modified circRNAs and forms a complex with RNase P/MRP and HRSP12, leading to the degradation of circRNAs. D CircRNA translation: YTHDF3 and eIF4G2 play vital roles in initiating the translation of m6A-modified circRNAs
m6A modulates circRNA subcellular localization
The m6A modification plays a crucial role in regulating the subcellular localization of RNA. This modification affects the binding of RBPs to RNA molecules, which influences their localization within the cell [31]. RBPs recognize the m6A modification on RNA and bind to the modified RNA molecules. These RBPs then have the ability to identify and transport the RNA to specific subcellular compartments, such as the nucleus or cytoplasm [32]. For example, the YTH domain-containing proteins, which are RBPs that specifically recognize m6A, can shuttle between the nucleus and cytoplasm, facilitating the transport of m6A-modified RNA [33].
CircRNAs localize to various cellular compartments, such as the nucleus or cytoplasm, interacting with other molecules to influence gene expression. However, disruptions in circRNA localization can hinder these interactions, potentially affecting protein partners and leading to cellular dysfunction or disease. Therefore, understanding the location of a circRNA is crucial in comprehending its regulatory mechanisms, particularly in the cytoplasmic competitive endogenous RNA (ceRNA) model. For example, in colorectal cancer (CRC), the m6A reader YTHDC1 promotes the export of circNSUN2 to the cytoplasm, where it forms a circNSUN2/IGF2BP2/HMGA2 RNA–protein complex that stabilizes HMGA2 mRNA. This complex has been shown to drive cancer metastasis by promoting the epithelial-to-mesenchymal transition (EMT) process [34].
A previous study found a weak correlation between the cytoplasmic localization and stability of circRNAs [35]. This suggests that long-lived circRNAs accumulate in the cytoplasm, likely during mitosis when the nucleus disassembles. The subcellular localization of circRNAs can also be affected by m6A modification, which can target them to specific cellular compartments [10]. Similarly, another study showed that m6A modification of the circRNA circDENND4C suppressed its export from the nucleus to the cytoplasm. In addition to regulating the export of circRNAs to the cytoplasm, m6A modification may also impact the localization of circRNAs within the cytoplasm [36]. For example, m6A modification of the circSRY promotes its localization to stress granules [37].
m6A modulates the expression of circRNAs
Previous studies have demonstrated that METTL3 is responsible for installing m6A modification on the reverse complementary sequences of circ1662, which is essential for the production of circ1662 through a process called intron pairing-driven circularization [38]. Additionally, circRNAs modified with m6A can be recognized by specific reader proteins, potentially leading to changes in their stability and subsequent alterations in their expression levels [30, 39]. Some circRNAs with m6A modification may undergo endoribonucleolytic degradation mediated by the RNase P/MRP complex and requires the cooperative binding of HRSP12 and YTHDF2 proteins [40].
m6A regulates the function of circRNAs
Due to their unique structure, the translation process of m6A-modified circRNAs differs from that of their parental genes. This mechanism involves the reader protein YTHDF3, which identifies m6A-circRNAs and recruits translation initiation factors, eIF3A and eIF4G2, ultimately triggering cap-independent translation [41].
The m6A modification also affects the role of circRNAs in regulating immunity. CircRNAs without m6A modification can directly activate the Retinoic acid-inducible gene I (RIG-I) signaling pathway to facilitate immune response [42]. In bladder cancer, YTHDF2, an m6A reader, targets DDX58 mRNA, which encodes the RNA helicase RIG-I. The interaction between YTHDF2 and DDX58 mRNA promotes the degradation of RIG-I, a tumor suppressor protein. Elevated levels of YTHDF2 promote bladder cancer progression by inhibiting RIG-I-mediated innate immune signaling. This discovery sheds light on the role of m6A modifications in bladder cancer and highlights the potential impact of targeting YTHDF2 as a therapeutic strategy for improving patient outcomes [43].
CircRNAs regulate m6A modification
CircRNAs have the ability to bind to m6A writers and erasers, preventing them from modifying other RNAs. This can lead to changes in the m6A modification status of different RNAs, ultimately affecting their expression and function. For example, circ-METTL3 functions as a sponge for the m6A eraser FTO, leading to increased m6A modification and expression of target mRNAs [44]. Besides, circRNAs can interact with proteins that bind to m6A-modified RNAs, influencing the fate of these RNAs in terms of stability, translation, and localization. One notable example is circ-YTHDF1, which interacts with the m6A reader YTHDF1, preventing it from binding to and degrading m6A-modified mRNAs [45]. Furthermore, circRNAs can regulate the expression of genes that encode m6A writers, erasers, and readers, ultimately impacting the overall level of m6A modification in cells. For example, circ-IGF2BP2 controls the expression of the m6A writer METTL3, which eventually affects the overall level of m6A modification in cells [46]. This ability to regulate m6A modification adds another layer of complexity to the regulatory network.
m6A modification of circRNAs and its role in regulating cancer therapy resistance
In the context of cancer therapeutic resistance, the m6A modification of circRNAs influences the expression and function of genes involved in drug response [47]. Furthermore, m6A modification can affect the interaction between circRNAs and RBPs, leading to changes in their stability and function [48, 49]. The m6A modification of circRNAs has been found to significantly impact the signaling pathways involved in cancer therapy resistance [50], providing new insights into the mechanisms underlying cancer therapeutic resistance [51]. Further investigation is necessary to completely comprehend the precise mechanisms by which m6A modification of circRNAs contributes to this resistance and to examine its potential as a therapeutic target [52]. The increasing knowledge about circRNAs has revealed their crucial functions in the growth, movement, and infiltration of different types of tumor cells. Emerging evidence suggests that circRNAs can impact the sensitivity of cancer treatment through various mechanisms, including regulating drug transportation, DNA repair, cell death, the tumor microenvironment, cellular self-degradation, EMT, cancer stem cells, and glucose metabolism (Table 1). This subsection primarily provides an overview of how m6A modification influences the tumor response to treatment through these various mechanisms (Fig. 4).
Table 1.
CircRNAs involved in cancer therapy resistance
| Therapeutic resistance | Mechanisms | Cancer types | CircRNAs | Roles | Functions | Ref |
|---|---|---|---|---|---|---|
| Chemoresistance | Drug transport | LUAD | circPVT1↑ | miR-145-5p↓ → ABCC1↑ |
Cisplatin/pemetrexed resistance |
[53] |
| CRC | circ_0007031↑ | miR-133b↓ → ABCC5↑ | 5-FU resistance | [54] | ||
| GC | circMTHFD2↑ | miR-124↓ → ABCB1↑ | Pemetrexed resistance | [55] | ||
| DNA damage repair | GC | circAKT3↑ | PI3K/AKT pathway↑ → BRCA1↑ | Cisplatin resistance | [56] | |
| breast cancer | circSMARCA5↓ | SMARCA5↑ | Cisplatin resistance | [57] | ||
| Apoptosis | GC | circCCDC66↑ | miR-618↓ → BCL-2↑ | Cisplatin resistance | [58] | |
| EC | cDOPEY2↓ | CPEB4↑ → MLC-1↑ | Cisplatin resistance | [59] | ||
| NSCLC | circ_0002874↑ |
miR- 1273f↓ → MDM2↑ → P53↓ |
Paclitaxel resistance | [60] | ||
| Radioresistance | TME | GC | circNRIP1↑ | miR-138-5p↓ → HIF-1α↑ | 5-FU resistance | [61] |
| NSCLC | circASXL1↑ | miR-206↓ → HIF-1α↑ | Cisplatin resistance | [62] | ||
| Autophagy | laryngocarcinoma | circPGAM1↑ | miR-376a↓ → ATG2A↑ | Cisplatin resistance | [63] | |
| GC | circCUL2↓ | miR-138-5p↑ → ROCK2↓ | Cisplatin resistance | [64] | ||
| breast cancer | circ_0092276↑ | miR-384↓ → ATG7↑ | Doxorubicin resistance | [65] | ||
| EMT and CSCs | prostate cancer | circ_0092367↓ | miR-1206↑ → ESRP1↓ | Gemcitabine resistance | [66] | |
| NSCLC | circ_0000079↓ | FXR1/PRCKI complex↑ | Cisplatin resistance | [67] | ||
| NSCLC | circRNA CDR1as↑ | HOXA9↓ → miR-641↑ | Cisplatin resistance | [68] | ||
| CRC | circ_001680↑ | miR-340↓ → BMI1↑ | Irinotecan resistance | [69] | ||
| Glycolysis | ESCC | circGOT1↑ | miR-606↓ → GOT1↑ | Cisplatin resistance | [70] | |
| prostate cancer | circARHGAP29↑ | c-Myc↑ → LDHA↑ | Docetaxel resistance | [71] | ||
| neuroblastoma | circDLGAP4↑ | miR-143↓ → HK2↑ | Doxorubicin resistance | [72] | ||
| NSCLC | circ_0008928↑ | miR-488↓ → HK2↑ | Cisplatin resistance | [73] | ||
| Radioresistance | TME | HCC | cZNF292↑ |
SOX9 nuclear translocation↑ → Wnt/β-catenin pathway↑ |
Radioresistance | [74] |
| Glycolysis | breast cancer | circABCB10↑ | miR-223-3p↓ → PFN↑ | Radioresistance | [75] | |
| Targeted therapy resistance | Drug transport | NSCLC | circSETD3↑ | miR-520 h↓ → ABCG2↑ | Gefitinib resistance | [76] |
| Autophagy | CML | circ_0009910↑ | miR-34a-5p↓ → ULK1↑ | Imatinib resistance | [77] | |
| EMT and CSCs | PC | circ_0013587↓ | miR-1227↑ → E-cadherin↓ | Erlotinib resistance | [78] | |
| Immunotherapy resistance | TME | ICC | circHMGCS1–016↑ |
miR-1236–3↓ → CD73/ GAL-8↑ |
Anti-PD-1 therapy resistance | [79] |
| HNSCC | circFAT1↑ |
STAT3↑ → CD8 + T cells infiltration↓ |
Anti-PD-1 therapy resistance | [80] |
Fig. 4.
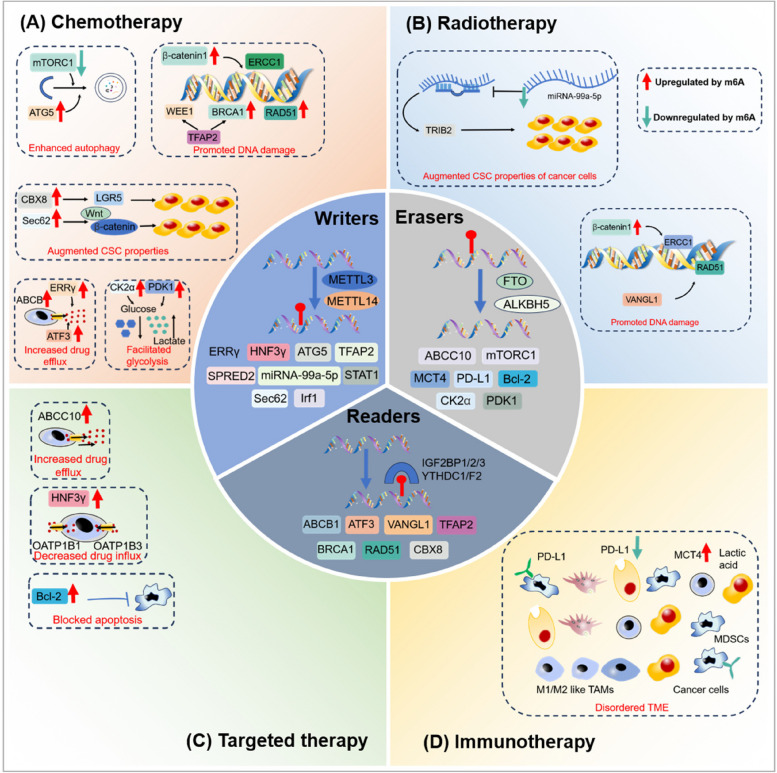
The role of m6A regulators in mediating therapy resistance in cancer. A During chemotherapy: m6A modification can hinder the effectiveness of treatment by promoting processes such as glycolysis, DNA damage repair, CSC properties, autophagy, and drug efflux. B In radiotherapy: m6A modification can enhance radioresistance by improving DNA damage repair mechanisms and promoting characteristics associated with CSCs. C In targeted therapy: m6A modification can increase drug efflux, inhibit cell apoptosis, and impede drug influx, ultimately reducing the efficacy of targeted therapy. D In immunotherapy: m6A modification can remodel the tumor microenvironment (TME), thereby contributing to resistance against immunotherapy
Effects of m6A modification on glycolysis, DNA damage repair, CSC properties, autophagy, and drug efflux during chemotherapy
m6A-induced alterations in glycolysis
Researchers have now identified disrupted energy metabolism as a defining characteristic of cancer. In 1924, Otto Warburg discovered that tumor cells primarily rely on glycolysis for energy production, even in the presence of oxygen [81]. This phenomenon, known as "aerobic glycolysis" or the "Warburg effect", involves rapid glucose consumption by tumor cells to generate lactic acid and ATP. While glycolysis yields less ATP than oxidative phosphorylation, it is crucial for the rapid proliferation of cancer cells due to its faster energy production rate [82]. Additionally, the abundant lactic acid produced by tumor cells contributes to an acidic TME. Furthermore, glycolytic intermediates can act as building blocks for synthesizing essential biomolecules, which play a critical role in driving tumor growth, metastasis, and treatment resistance [83]. Recent research conducted by Yu's group has revealed that the depletion of ALKBH5 results in an increase in the expression of casein kinase 2α (CK2α). This finding suggests that the m6A modification promotes cisplatin resistance in bladder cancer (BC) by upregulating the CK2α-mediated glycolysis pathway [84].
m6A-induced alterations in DNA damage repair
The field of m6A modification of circRNAs in DNA damage repair is rapidly expanding. The m6A modification regulates the expression and functions of circRNAs in DNA damage repair processes [85]. This modification regulates glycolysis efficiency and promotes oncogenesis [86]. Furthermore, m6A modification in circRNAs is linked to various physiological processes, such as DNA damage repair [87]. However, more research is needed to fully elucidate the role of m6A modification of circRNAs in DNA damage repair [88].
Several proteins involved in repairing DNA damage, such as ERCC1, have been identified as crucial factors in the resistance to chemoradiotherapy. These proteins are closely linked to m6A modification. ERCC1, located on chromosome 19, plays a vital role in the nucleotide excision repair (NER) pathway [89]. Higher levels of ERCC1 correlate with cisplatin resistance in various cancers, including gastric (GC) and cervical cancers [90]. To better understand the role of FTO in chemoradiotherapy resistance, Zhou et al. investigated its expression and discovered its regulatory function in both cisplatin and radiotherapy resistance. Their research showed that FTO increases the expression of β-catenin, which in turn upregulates ERCC1. This upregulation of ERCC1, through NER activation, leads to treatment failure in cervical squamous cell carcinoma (CSCC) [91]. Double-strand breaks (DSBs) are the most lethal form of DNA damage, but fortunately, cells have the ability to repair them through homologous recombination (HRR) [92]. However, abnormal expression of essential genes involved in HRR can alter the sensitivity of tumor cells to cancer therapy. This concept has been illustrated in several studies. For example, reduced expression of epithelial membrane protein 3 (EMP3) leads to increased levels of YTHDC1, which promotes DNA repair in breast cancer cells by upregulating BRCA1 and RAD51, ultimately resulting in chemoresistance [93]. Interestingly, IGF2BP1-mediated stabilization of TFAP2C in cisplatin-resistant seminoma cells contributes to the activation of WEE1 and BRCA1, further promoting DNA repair [94].
A previous study has shown that the m6A modification of circRNA circNSUN2 increases its stability and enhances the recruitment of DNA damage repair proteins to sites of DNA damage. This circRNA acts as a scaffold to promote the assembly of DNA repair complexes, ultimately enhancing efficient DNA damage repair [95]. Another study has revealed that the m6A modification of circRNA circAmotl1a regulates the expression of DNA damage response genes by interacting with specific RBPs [96]. Additionally, the m6A modification of circHIPK3 promotes its interaction with the DNA damage repair protein PARP1, which is involved in single-strand break and base excision repair [97]. Furthermore, the m6A modification of circDENND4C suppresses its interaction with the DNA damage repair protein ATM [98], thereby affecting the recruitment of DNA repair factors to DNA damage sites and altering the efficiency of DNA damage repair [99].
These findings indicate that m6A modification of circRNAs plays a crucial role in regulating DNA damage repair processes [93]. By modulating the expressions and functions of circRNAs, m6A modification can influence the efficiency and accuracy of DNA damage repair, thereby maintaining genomic stability [100]. Additionally, more research is necessary to identify the precise mechanisms by which m6A modification influences circRNA-mediated DNA damage repair. Furthermore, it is important to explore the potential implications of DNA damage in diseases such as cancer and neurodegenerative disorders [101, 102].
The impact of circRNA m6A on EMT and CSCs
Recent studies have revealed that the modification of circRNAs through m6A can play a crucial role in regulating EMT and CSCs [103], ultimately influencing cancer progression [104]. EMT is a process closely associated with increased invasiveness and metastasis in cancer [105]. The impact of m6A modification on EMT-related circRNAs and their effect on cancer progression is currently a topic of active research.
Firstly, m6A modification can increase the stability of circRNAs, resulting in higher levels of expression [106]. These increased expressions of certain circRNAs are associated with EMT and CSC properties [107]. Furthermore, circRNAs are identified as promoting EMT by serving as miRNA sponges or binding to specific proteins, thereby regulating the expression of genes related to EMT [108].
Secondly, m6A modification can potentially regulate the function of circRNAs in EMT and CSCs. Studies have shown that m6A modification can influence the binding ability of circRNAs to specific proteins or miRNAs, thereby affecting their regulatory roles in EMT and CSCs [109]. For example, m6A modification of circRNA can alter its interaction with RBPs, resulting in changes to downstream signaling pathways involved in EMT and CSC maintenance [110]. Furthermore, circRNAs play a significant role in EMT-related processes such as cell invasion, metastasis, and the regulation of EMT-related transcription factors and signaling pathways [111]. Additionally, m6A modification of the circRNA circZEB1 promotes EMT in GC cells by increasing the expression of the EMT transcription factor ZEB1 [112]. Moreover, m6A modification encourages the acquisition of CSC properties by activating the Wnt/β-catenin signaling pathway, leading to chemoresistance in colorectal cancer (CRC) [113].
In the context of CSCs, circRNAs have been implicated in their functions and potential applications. Exosome-associated circRNAs have also been identified as critical regulators of EMT in cancer [114]. Additionally, m6A modification affects the expression of genes involved in EMT and CSCs [115]. m6A modification on mRNA transcripts can affect their stability, translation, and splicing, ultimately regulating the genes related to EMT and CSCs [116]. m6A modification of circSOX2 promotes CSC self-renewal and tumorigenicity in colorectal cancer by increasing SOX2 expression [117]. Consistently, m6A modification of the circCD44 promotes CSC self-renewal and tumorigenicity in glioblastoma by upregulating the expression of the stem cell marker CD44 [118]. This modulation of gene expression has also been linked to regulating the EMT process and maintaining CSC properties [119].
In summary, the m6A modification of circRNAs is identified as a crucial regulatory mechanism in the processes of EMT and CSCs [120]. Understanding the impact of circRNA m6A modification on EMT and CSCs can offer valuable insights for developing new therapeutic approaches in tumor treatment [121]. As research in this area progresses, we can expect to gain a deeper understanding of the specific mechanisms by which m6A modification regulates EMT and CSCs, and how this regulation can be utilized for therapeutic purposes [122].
m6A modification of circRNAs regulates autophagy
Recent research has shown that m6A modification is crucial in activating autophagy and forming autophagosomes. A study conducted by Li et al. demonstrated that overexpression of YTHDF1 under hypoxia facilitated the translation of ATG2A and ATG14, resulting in the activation of autophagy [123]. Moreover, the activation of autophagy during hypoxia can lead to drug resistance in various types of tumor cells. This is attributed to the function of FTO in decreasing levels of pro-survival autophagy, rendering gastric cancer cells more susceptible to chemotherapy through the mTORC1 pathway [124]. METTL3 also plays a role in promoting autophagy, resulting in decreased sensitivity of seminoma cells to cisplatin [125].
Furthermore, m6A modification of circRNAs impacts the interaction between circRNAs and miRNAs. miRNAs are small RNA molecules responsible for regulating gene expression at the post-transcriptional level. CircRNAs can bind to miRNAs and inhibit their activity, acting as sponges for miRNAs. The m6A modification of circRNAs can be altered in their binding affinity to miRNAs, thereby modulating the regulation of miRNA targets and ultimately impacting autophagy-related pathways [126]. In summary, the m6A modification of circRNAs represents a novel mechanism by which circRNAs can regulate autophagy [127]. Additional experimental studies are needed to identify the specific circRNAs, uncover the mechanisms of m6A modification in regulating autophagy, and determine the implications for various cellular processes and diseases [11, 128].
m6A-induced alterations in drug transport
The m6A modification significantly affects the resistance of cancer therapy through various mechanisms, such as the regulation of gene expression and the alteration of protein function [129]. One of these mechanisms is the alteration of drug transport [130]. Additionally, the m6A modification plays an essential role in the expression of the ABC transporter family, which strongly contributes to multidrug resistance [131].
Resistance to cancer drugs is often caused by enhanced drug efflux and reduced drug influx. A recent study has identified a new m6A binder, insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3), which regulates gene expression by improving the stability and nuclear export of specific mRNA targets [132]. Overexpression of IGF2BP3 interacts with the m6A site of P-gp, leading to increased expression of P-gp and reduced sensitivity of colorectal cancer cells to therapy [133]. Moreover, m6A modification-induced upregulation of ERRγ promotes chemoresistance by increasing P-gp levels. Interestingly, Liu et al. found that YTHDF2 regulates the expression of ATF3, a transcription factor that interacts with the enhancer region of ABCB1 and promotes its expression, resulting in tamoxifen resistance [134].
The impact of m6A modification on DNA damage repair and CSC characteristics in radiotherapy
m6A modification improves DNA damage repair in radiotherapy
Cells have the ability to repair DNA damage caused by radiotherapy. A study on lung adenocarcinoma (LUAD) found that increased levels of m6A and decreased levels of miR-29b-3p resulted in an upregulation of VANGL1. This activation of VANGL1 triggers the BRAF/TP53BP1/RAD51 pathway, promoting DNA repair and protecting cells from radiation damage [135].
m6A modification augments CSC characteristics in radiotherapy
Recent studies have shown that the modification of circRNAs through m6A has an impact on radiotherapy, thereby influencing cancer progression. For example, a study conducted by Liu et al. identified the METTL14/miR-99a-5p/TRIB2 axis as a significant contributor to the characteristics of CSCs and their resistance to radiotherapy in esophageal squamous cell carcinoma (ESCC). The positive correlation between this axis and these aggressive features suggests potential therapeutic targets for ESCC treatment [136].
The potential of m6A modification to affect drug efflux, cell apoptosis, and drug influx in targeted therapy
m6A modification increases drug efflux in targeted therapy
Apart from chemotherapy, m6A modification also occurs in targeted therapy. For example, Gefitinib, a drug commonly used to treat non-small cell lung cancer (NSCLC), can be effluxed by the transporter protein ABCC10, thereby impacting its intracellular concentration inside the cell [137]. Interestingly, research has shown that FTO, an enzyme enriched in serum exosomes from patients resistant to gefitinib, can increase the expression of ABCC10 through m6A modification. This suggests that overexpression of FTO may contribute to the development of gefitinib resistance in NSCLC [138].
m6A modification induces cell apoptosis alterations in targeted therapy
Apoptosis, a programmed cell death process, is crucial in normal development and disease progression [139]. Evading apoptosis is a hallmark of cancer and a major cause of treatment failure. Key regulators of apoptosis include the BCL-2 family proteins. The BCL-2 family consists of both pro-apoptotic members, such as BAX and BAK, which trigger cell death, and anti-apoptotic members, such as BCL-2 and MCL-1 [140]. m6A modifications can influence the expression of these BCL-2 family proteins. For example, METTL3, an m6A writer enzyme, increases BCL-2 expression in breast cancer, thereby suppressing apoptosis [141]. This finding is particularly relevant because high levels of BCL-2 are linked to resistance to multiple drugs, including paclitaxel, tamoxifen, and trastuzumab [142, 143].
m6A modification impedes drug influx in targeted therapy
In addition to the ABC efflux transporter family, the m6A modification also affects the expression of specific transporters, such as organic anion-transporting polypeptides (OATP) [144]. The OATP transporter family consists of 11 members. Notably, OATP1B1 and OATP1B3 are located on the basement membrane of human liver cells (hepatocytes) and responsible for transporting drugs within the liver. Sorafenib, a targeted therapy for inoperable or advanced hepatocellular carcinoma (HCC) and clear cell renal cell carcinoma (ccRCC), is known to be a substrate for both OATP1B1 and OATP1B3. A recent study revealed that HCC tissues have lower levels of hepatocyte nuclear factor 3γ (HNF3γ) compared to healthy neighboring tissues. This decrease was linked to reduced METTL14 expression, contributing to sorafenib resistance by suppressing OATP1B1 and OATP1B3 expression [145].
The impact of circRNA m6A regulators on the tumor microenvironment in immunotherapy
The TME comprises various cell types, including immune cells, tumor cells, and stromal cells [146]. These cells interact with each other, creating a microenvironment that promotes tumor growth and survival [31]. The m6A modification of circRNAs is crucial in remodeling the TME by regulating gene expression involved in tumor progression and metastasis [147, 148]. By targeting specific circRNAs, m6A regulators can influence the expression of genes involved in the TME, such as immune checkpoint molecules, cytokines, and chemokines [32, 149]. This can result in either an enhanced immune response against tumors or immune evasion by tumor cells [150]. Consistently, circRNA m6A regulators can also influence the interaction between cancer cells and stromal cells in the TME [151]. Furthermore, m6A modification can regulate the expression of extracellular matrix components and signaling molecules involved in cell–cell communication, ultimately impacting tumor cell invasion, angiogenesis, and metastasis [152]. Apart from regulating gene expression, m6A modification of circRNAs can also influence the interaction between circRNAs and proteins that regulate the TME [153]. The potential for using circRNA m6A regulators to remodel the TME as a target for cancer treatment has been identified [154]. However, research on the role of m6A modification of circRNAs in remodeling the TME is still in its early stages [155], but it has shown potential for regulating immune responses in cancer.
The relationship between m6A modification and circRNAs in cancer therapy resistance
In this review, we provide a concise overview of the recent studies that have examined the regulatory role of m6A-modified circRNAs in the development of treatment resistance in various types of cancers, including hepatocellular carcinoma (HCC), hypopharyngeal squamous cell carcinoma (HSCC), and non-small cell lung carcinoma (NSCLC) (Table 2, Fig. 5).
Table 2.
m6A-modified circRNAs in cancer therapy resistance
| Cancer types | M6A regulators | Roles of m6A in circRNAs | Functions | Mechanisms | Ref |
|---|---|---|---|---|---|
| HSCC | METTL3 | Promotes m6A modification of circCUX1 to stabilizes its expression | Radiotherapy resistance | Decreases the release of inflammatory factors in TME | [30] |
| HCC | m6A |
Elevates circRNA-SORE expression via increasing RNA stability |
Sorafenib resistance | Activates the Wnt/β-catenin pathway | [156] |
| IGF2BP1 |
Promotes m6A-modified circMAP3K4 translation |
Cisplatin resistance | Inhibits apoptosis | [157] | |
| NSCLC | YTHDC1 | Facilitates the biogenesis of m6A-modified circIGF2BP3 | Anti-PD-L1 therapy resistance | Promotes tumor immune evasion | [158] |
| YTHDF2 |
Increases m6A-modified circASK1 degradation |
Gefitinib resistance | Represses apoptosis | [159] |
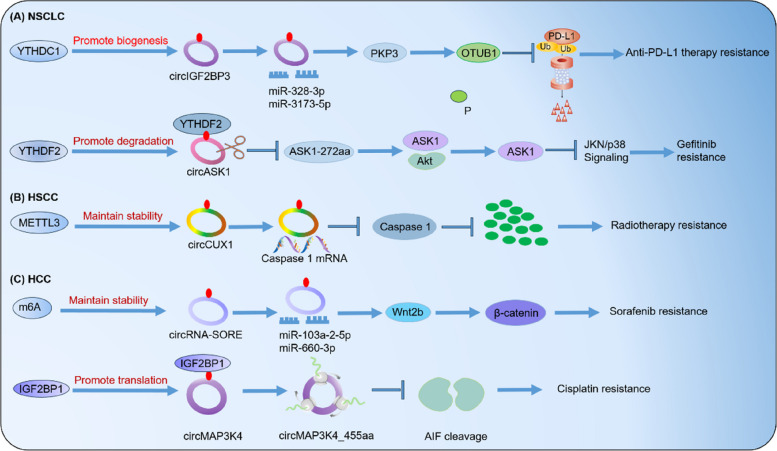
Fig. 5.
The role of m6A-modified circRNAs in cancer therapy resistance. m6A modification significantly impacts the biogenesis, translation, and degradation of circRNAs, ultimately affecting their role in therapeutic resistance in various types of cancer. A In non-small cell lung cancer (NSCLC), YTHDC1 and YTHDF2 play a crucial role in facilitating anti-PD-L1 therapy and gefitinib resistance by increasing the biogenesis of circIGF2BP3 and degrading circASK1, respectively. B In head and neck squamous cell carcinoma (HSCC), METTL3 increases the stability of circCUX1, leading to radioresistance. C In hepatocellular carcinoma (HCC), m6A regulators promotes the stability of circRNA-SORE and the translation of circMAP3K4, resulting in resistance to sorafenib and cisplatin, respectively
Hypopharyngeal squamous cell carcinoma (HSCC)
Improving the diagnosis of HSCC requires identifying the molecular mechanisms that promote therapy resistance. A previous study found that the expression of circ_0058106 is upregulated in HSCC tissues. Circ_0058106 is reported to regulate the Wnt2b/β-catenin/c-Myc pathway, thereby promoting tumorigenesis and EMT in HSCC [160]. Similarly, another study showed that upregulated circMATR3 enhances proliferation and invasion, while inhibiting apoptosis [161]. It is plausible that m6A modification plays a regulatory role in the expression levels of circRNAs. This hypothesis is supported by a recent study by Wu et al., which found that METTL3-mediated m6A methylation stabilizes the expression of circCUX1. CircCUX1, in turn, interacts with caspase 1, leading to a decrease in its expression and a subsequent reduction in IL-1β levels in the tumor microenvironment. This may potentially contribute to the development of tolerance in HSCC [30].
Hepatocellular carcinoma (HCC)
Recently, several studies have highlighted the crucial role of the interaction between circRNAs and m6A modification in various aspects of HCC [162, 163]. For example, it is found that METTL3 promotes the production of circHPS5 in HCC. The m6A-regulated circHPS5 functions as a sponge for miR-370, increasing HMGA2 expression and promoting EMT, ultimately facilitating the progression of HCC [164]. Sorafenib, a commonly prescribed targeted drug for HCC, is hindered in its therapeutic effectiveness by the development of acquired resistance. In sorafenib-resistant HCC, the elevated levels of circRNA-SORE, transported through exosomes, impede the degradation of YBX1 by interacting with it, thereby reducing the drug's therapeutic effectiveness [165].
Non‑small‑cell lung cancer (NSCLC)
Lung cancer is a significant threat to human well-being and is one of the leading causes of cancer-related deaths worldwide [157]. In recent years, targeted therapy and immunotherapy have made promising advancements, providing hope for patients with lung cancer. However, a major challenge remains in the development of acquired resistance to these treatments. Mettl3 plays a crucial role in promoting the circularization of circIGF2BP3 through its m6A modification. This circular RNA acts as a competitive endogenous RNA (ceRNA), sequestering miR-328-3p and miR-3173-5p to increase the expression of PKP3. PKP3 interacts with the RNA-binding protein FXR1, stabilizing OTUB1 mRNA and promoting the abundance of PD-L1 through deubiquitination. Importantly, deletion of tumor PD-L1 abolishes the effect of the circIGF2BP3/PKP3 axis on CD8+ T cell response. In a Lewis lung carcinoma mouse model, inhibition of the circIGF2BP3/PKP3 pathway enhanced the efficacy of anti-PD-1 therapy. Furthermore, the PKP3/PD-L1 signature and the level of infiltrating CD8+ T cells can be used to classify NSCLC patients into distinct risk groups [166]. It has been confirmed that IGF1R promotes proliferation by activating the PI3K/AKT pathway, leading to osimertinib resistance [158]. Overall, understanding the mechanisms behind acquired resistance in NSCLC and exploring the role of circRNAs, such as circIGF2BP3 and hsa_circ_0005576 [167, 168], can provide valuable insights for developing more targeted and effective treatment strategies [169].
Furthermore, studies have demonstrated that increased levels of circNDUFB2 can effectively impede the proliferation and dissemination of NSCLC cells. This is achieved through circNDUFB2 as a scaffold, facilitating the interaction between TRIM25 and IGF2BPs, a protein known to drive tumor progression and metastasis. The formation of the TRIM25/circNDUFB2/IGF2BPs complex results in the tagging and degradation of IGF2BPs, a process further enhanced by the m6A modification of circNDUFB2. Additionally, circNDUFB2 can activate the RIG-I-MAVS signaling pathway by binding to RIG-I, attracting immune cells to the TME [170].
CircRNA influences m6A modification in cancer therapy resistance
Notably, abnormal circRNA expression affects m6A modification in cancer. We have compiled a summary of studies on m6A regulation by circRNAs, including CRC, BC, HCC and glioma, as shown in Table 3 and Fig. 6.
Table 3.
CircRNAs related to m6A modification in different types of cancer
| Cancer | circRNAs | Roles of circRNAs in m6A | Functions | Mechanisms | Ref |
|---|---|---|---|---|---|
| Glioma | circ_0072083 | Promotes ALKBH5 expression via sponging miR-1252-5p | Temozolomide resistance | Maintains glioma stem cells | [171] |
| CRC | circPTK2 | Elevates YTHDF1 level by targeting miR-136-5p | 5-FU/oxaliplatin resistance | – | [172] |
| HCC | circRHBDD1 | Recruits YTHDF1 | Anti-PD-1 therapy resistance | Elevates glycolysis | [173] |
| Bladder cancer | circ0008399 | Facilitates the formation of MTC through combining with WTAP | Cisplatin resistance | Boosts anti-apoptosis | [174] |
| circMORC3 |
Interacts with VIRMA and elevates global m6A level |
Cisplatin resistance |
Promotes DNA repair and suppresses DNA damage |
[175] | |
| Prostate cancer | circARHGAP29 | Interacts with IGF2BP2 | Docetaxel resistanc | Promotes glycolysis | [71] |
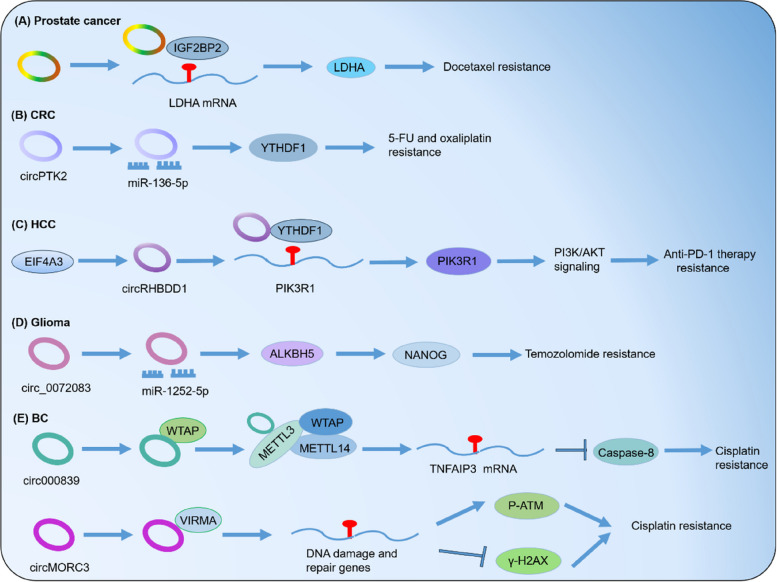
Fig. 6.
CircRNAs influence m6A modification in cancer therapy resistance. CircRNAs can either modulate the expression of m6A regulators or interact with them, significantly impacting m6A functions in various types of cancer. A In prostate cancer, circARHGAP29 interacts with IGF2BP2, leading to an increase in docetaxel resistance. B In colorectal cancer (CRC), circPTK2 increases the level of YTHDF1, resulting in resistance to 5-FU and oxaliplatin. C In hepatocellular carcinoma (HCC), circRHBDD1 recruits YTHDF1, leading to resistance to anti-PD-1 therapy. D In glioma, circ_0072083 promotes the expression of ALKBH5, resulting in resistance to temozolomide. E In breast cancer (BC), circ0008399 facilitates the formation of MTC by interacting with WTAP, leading to an increase in cisplatin resistance. Additionally, circMORC3 promotes cisplatin resistance by interacting with VIRMA
Potential challenges associated with targeting m6A writers, readers, and erasers for therapeutic benefit
Small molecules are crucial tools for understanding the roles of specific RNA modifications in certain types of cancer. Table 4 summarizes small molecule inhibitors that target writers, readers, and erasers of m6A modifications.
Table 4.
Small-molecule inhibitors targeting RNA modifications
| Target gene | Inhibitor name | Cancer | Anticancer potency |
|---|---|---|---|
| METTL3/METTL14 | STM2457 | AML | 0.6–10.3 μM |
| STC-15 | Advanced tumors | ||
| Multiple small molecules |
AML, ovarian adenocarcinoma |
80 nM–1.39 μM | |
| Multiple small molecules | AML | < 1 μM, < 10 μM | |
| UZH2 | AML | 12 μM, 70 μM | |
| UZH1a | prostate cancer | Not tested | |
| CDIBA | AML | 13–22 μM | |
| Eltrombopag | osteosarcoma | 8.28 μM | |
| Elvitegravir | AML | Reduced metastasis | |
| YTHDF | Ebselen | Prostate cancer | 26.83 μM |
| YTHDF1 | Salvianolic acid (SAC) | Neuronal tissue | N/A |
| YTHDF2 | CpG-siRNAYTHDF2 | Mouse melanoma | Reduced tumor growth |
| FTO | CS1 | AML | 22–753 nM |
| FB23-2 | AML | 1.9–5.2 μM | |
| MO-I-500 |
Triple-negative Breast cancer |
Inhibited survival and colony formation | |
| MA | Hela | N/A | |
| MA2 | GSCs | Inhibited growth and self-renewal of GSCs | |
| FTO-04 | GSCs, glioblastoma | Impaired self-renewal properties | |
| Dac51 | Melanoma | Inhibited tumor growth | |
| R-2HG | Glioma | Decreased cell proliferation | |
| ALKBH5 |
2-[(1-hydroxy-2-oxo-2-phenylethyl)sulfanyl]acetic acid, 4-{[(furan-2-yl)methyl] amino}-1,2-diazinane-3,6-dione |
AML | 1.38–47.8 μM |
| PUS7 | C17 | Glioblastoma | 56.77–177.4 nM |
Abbreviations: ALKBH5 AlkB homolog 5, AML Acute myeloid leukemia, FTO Fat mass and obesity-associated protein, GSCs Glioma stem-like cells, IDH Isocitrate dehydrogenases, MA Meclofenamic acid, METTL3 Methyltransferase-like 3, METTL14 Methyltransferase-like 14, N/A Not available, PUS Pseudouridine synthase, YTHDF YTH domain-containing family protein
Despite promising preclinical data, targeting dysregulated m6A modifiers with small-molecule inhibitors remains challenging. A deeper understanding of the role and underlying mechanisms of m6A machinery in cancer is crucial for selecting effective therapeutic targets. In addition to small molecules, protein degraders offer an alternative strategy for targeting m6A readers. Combinatorial targeting of multiple oncogenic m6A modifiers shows promise for achieving optimal therapeutic outcomes. For example, simultaneously targeting writers and erasers could have a synergistic effect, considering their distinct tumorigenic mechanisms. Functional studies using genetic depletion models and combinatorial genetic deletions can validate synergy and guide researchers in developing novel combination therapies. Due to the context-dependent nature of m6A modifiers, different cancer types or subtypes may require unique combination therapies. Future possibilities may include CRISPR-based manipulation of specific m6A modifications critical for cancer development. The complex and interactive nature of cancer and the TME suggests that targeting oncogenic m6A modifiers will likely require a combination with other therapies to achieve curative effects.
Developing effective RNA-based therapies and targeting RNA-modifying proteins requires optimizing drug discovery pipelines. Current drugs targeting these enzymes often cause off-target effects, hindering their clinical potential. Overcoming the challenge of off-target effects and ushering in a new era of targeted RNA-based interventions can be achieved through advancements in computational models, high-throughput enzymatic assays, and exploration of interconnected pathways.
Naturally, RNA modifications can enhance the stability, efficacy, and specificity of therapeutic RNAs. We can optimize these benefits, such as increased stability, improved efficacy, and enhanced specificity, through strategic natural and synthetic modifications targeting specific sequence contexts and locations. Integrating comparative single-cell analyses with advanced molecular, biochemical, and cellular studies in healthy and diseased human cells can fully harness the potential of RNA modifications for clinical applications.
Exosomal circRNA functions as clinical biomarker
A compilation of the most recent discoveries exploring circRNAs and exosomal circRNAs as potential diagnostic or prognostic indicators for different cancer types can be found in Table 5. As more experimental data is gathered, we are optimistic that exosomal circRNAs will prove to be highly promising cancer biomarkers and eventually have practical applications in clinical settings.
Table 5.
The clinical applications of circRNAs and exosomal circRNAs as biomarkers for cancer diagnosis or prognosis
| Registration number/NCT number |
Study type | Study phase | Recruiting status | Tumor type | Sample name | Sample size |
|---|---|---|---|---|---|---|
| ChiCTR2300069863 | Observational study | 0 | Recruiting | Cholangiocarcinoma | Bile, serum | 320 |
| ChiCTR1900027419 | Basic science | 0 | Not yet recruiting | Lung cancer | Blood | 58 |
| ChiCTR1900024188 | Diagnostic test | 0 | Recruiting | Prostate cancer | Urine | 300 |
| ChiCTR1800019529 | Diagnostic test |
Diagnostic new technique clinical study |
Recruiting | Prostate cancer | Plasma | 200 |
| ChiCTR1800018038 | Diagnostic test |
Diagnostic new technique clinical study |
Not yet recruiting | Pancreatic cancer | Blood | 20 |
| NCT05771337 | Observational study | Not available | Not yet recruiting | Breast cancer | Blood | 80 |
| NCT04584996 | Observational study | Not available | Recruiting | Pancreatic cancer, biliary tract cancer | Blood, bile | 186 |
| NCT04464122 | Observational study | Not available | Recruiting | Neuroendocrine neoplasm | Not available | 60 |
| NCT03334708 | Observational study | Not available | Recruiting | Pancreatic cancer | Blood | 700 |
CircRNAs boast exceptional stability due to their unique covalent closed-loop structure, which protects them from exonuclease degradation. This results in a significantly longer lifespan compared to their parent mRNAs, while mRNAs typically degrade within 10 h, circRNAs can persist for up to 48 h [176]. These advantages make circRNAs a promising tool for cancer diagnosis and prognosis prediction. However, the low abundance of exosomal circRNAs poses a challenge in accurately detecting and identifying them in body fluids using current methods [177]. Furthermore, the cost of detecting circRNAs in tissue or exosomes is higher than that of existing tests. Therefore, advanced techniques are needed to improve the accuracy and affordability of detecting exosomal circRNAs before they can be effectively used in lipid biopsies for clinical applications. Combining exosomal circRNAs with other exosomal cargoes or current biomarkers may provide a more precise and comprehensive assessment for early cancer diagnosis and predicting clinical outcomes.
Although ultrasensitive flow cytometry has been widely used to identify the origin of external vesicles by staining cell-specific markers, the classification of exosomes needs to be clarified and consistent across different studies [178]. Furthermore, the accuracy of using exosomal circRNAs as diagnostic and prognostic biomarkers for human malignancies may be affected by the choice of circRNA profiling method. Techniques such as RNA-seq, microarray, and qRT-PCR can each impact the results [179]. Therefore, standardized protocols for exosome isolation, purification, characterization, and profiling methods are urgently needed in exosomal circRNA research.
Conclusions and future prospects
The development of resistance to cancer treatment presents significant challenges, such as higher mortality rates and unfavorable prognoses for cancer patients. Therefore, it is imperative to thoroughly investigate the potential molecular mechanisms that contribute to therapeutic resistance in cancer. While many studies have proposed various factors that can impact the responsiveness of tumor cells to treatment, the exact mechanisms responsible for therapeutic resistance still need to be fully understood. In this comprehensive review, we have synthesized the latest findings on the interplay between m6A modification and circRNAs, and their implications for therapeutic resistance. This review offers valuable insights for future research on overcoming therapeutic resistance in cancer.
Epigenetic regulation, specifically modifications in RNA, can significantly impact post-transcriptional gene expression. This dysregulation includes alterations in ABC transporters, OATP transporters, genes involved in autophagy, DNA damage and repair genes, all of which contribute to cancer relapse. However, the extent to which these epigenetic mechanisms contribute to the dysregulation of circRNAs. Our research has revealed that m6A modifications significantly influence the biosynthesis, localization, translation, and degradation, potentially leading to abnormal circRNA expression. However, only a few m6A-modified circRNAs have been identified as being associated with the response to tumor therapy.
Improving the detection of m6A modification presents a significant technical challenge. While several techniques have been developed in recent years, such as methylated RNA immunoprecipitation sequencing (MeRIP-seq), m6A labeling-based sequencing (m6A-label-seq), and methylation-iCLIP (miCLIP), they all have limitations and require further refinement. For instance, MeRIP-seq relies on antibodies, which can lead to false-positive results due to their lack of specificity. Similarly, although m6A-label-seq can recognize m6A sites with single-base resolution, it is limited in its ability to detect a large number of m6A residues [180–183]. Furthermore, there is growing evidence to suggest that other RNA modifications, such as pseudouridine (Ψ) and 5-methylcytosine (m5C), are prevalent in non-coding RNAs, including ribosomal RNAs, miRNAs, and lncRNAs [184]. Therefore, it is hypothesized that circRNA levels may be regulated not only by m6A modifications but also by other types of RNA modifications.
Currently, scientists have discovered various natural compounds that target m6A readers, writers, and erasers. For example, Saikosaponin D (SsD) increases the overall m6A modification level by inhibiting the m6A demethylase FTO. This, in turn, can alleviate leukemia resistance to TKIs therapy by reducing the stability of BCL-2 [174]. In addition to SsD, other substances such as fusaric acid, curcumin, STM2457, and chidamide can modulate m6A regulatory proteins [185]. The discovery of these modulators that target m6A offers new possibilities for overcoming therapeutic resistance.
Recently, numerous studies have revealed that circRNAs are highly abundant in exosomes. These exosomes facilitate the progression of cancer by acting as vehicles for transporting molecules, particularly circRNAs. These exosomes are present in various bodily fluids, including urine, blood, and saliva. Consequently, the potential of exosomal circRNAs as valuable markers for cancer prognosis in liquid biopsies should not be disregarded [186]. To fully realize this potential, the development of highly accurate detection techniques is crucial [42]. Certain circRNAs can be to be translated through IRES and m6A-dependent pathways [187]. Additionally, certain circRNAs possess the capacity to regulate the expression of m6A regulators through their ability to sponge miRNAs and interact with m6A writers/readers/erasers. However, it is still unclear whether circRNAs can impact m6A modification through other biological functions, such as gene transcription regulation or translation, remains unclear.
In summary, m6A modification, circRNAs, and their interactions are crucial in regulating resistance to cancer treatment. In the future, targeting m6A modification and circRNAs in drug development may help researchers conquer treatment resistance.
Acknowledgements
We would like to thank all our colleagues, both local, national, and worldwide, with whom we have had many discussions, exchanges of ideas, and collaborations in the field of circRNA over the years.
Abbreviations
- circRNAs
Circular RNAs
- CSCs
Cancer stem cells
- ecRNAs
Exonic circRNAs
- EIciRNAs
Exon-intron circRNAs
- EMT
Epithelial-to-mesenchymal transition
- HCC
Hepatocellular carcinoma
- HSCC
Hypopharyngeal squamous cell carcinoma
- IGF2BP3
Insulin-like growth factor 2 mRNA-binding protein 3
- IRES
Internal ribosome entry site
- m1A
N1-methyladenosine
- m6A
N6-methyladenosine
- m5C
5-Methylcytosine
- m7G
7-Methylguanine
- m6Am
2'-O-dimethyladenosine
- MeRIP-seq
Methylated RNA immunoprecipitation sequencing
- m6A-label-seq
M6A labeling-based sequencing
- miCLIP
Methylation-iCLIP
- NSCLC
Non-small cell lung carcinoma
- RBPs
RNA-binding proteins
- TME
Tumor microenvironment
- tricRNAs
TRNA intronic circRNAs
Authors’ contributions
All authors listed have made a substantial, direct and intellectual contribution to the work. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China 82274000 (to Y.Y.) and 82173849 (to C.G.); a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (Traditional Chinese Medicine).
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have agreed to publish this manuscript.
Competing interests
No potential conflicts of interest were disclosed.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chunyan Gu, Email: guchunyan@njucm.edu.cn.
Ye Yang, Email: yangye876@sina.com, Email: 290422@njucm.edu.cn.
References
- 1.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A,: MODOMICS: a database of RNA modification pathways., et al. update. Nucleic Acids Res. 2017;2018(46):D303–d307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Yang X, Qi Z, Sang Y, Liu Y, Xu B, Liu W, Xu Z, Deng Y. The role of mRNA m(6)A methylation in the nervous system. Cell Biosci. 2019;9:66. 10.1186/s13578-019-0330-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, Liu Q, Shi H, Wang F, Wang Y. m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221–35. 10.1080/15548627.2019.1659617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang Z, Ye H, Ma J, Wei Z, Wang Y, Zhang Y, Huang D, Song B, Meng J, Rigden D, Chen K. m6A-Atlas v2.0: updated resources for unraveling the N6-methyladenosine (m6A) epitranscriptome among multiple species. Nucleic Acids Res. 2024;52(D1):D194-202. 10.1093/nar/gkad691. [DOI] [PMC free article] [PubMed]
- 5.Jain S, Koziej L, Poulis P, Kaczmarczyk I, Gaik M, Rawski M, Ranjan N, Glatt S, Rodnina M. Modulation of translational decoding by mA modification of mRNA. Nat Commun. 2023;14:4784. 10.1038/s41467-023-40422-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su H, Cheung H, Lau H, Chen H, Zhang X, Qin N, Wang Y, Chan M, Wu W, Chen H: Crosstalk between gut microbiota and RNA N6-methyladenosine modification in cancer. FEMS Microbiol Rev. 2023;47(4):fuad036. 10.1093/femsre/fuad036. [DOI] [PubMed]
- 7.Liu Y, Yang D, Liu T, Chen J, Yu J, Yi P. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol Med. 2023;29:454–67. 10.1016/j.molmed.2023.03.005 [DOI] [PubMed] [Google Scholar]
- 8.He T, Zhang Q, Xu P, Tao W, Lin F, Liu R, Li M, Duan X, Cai C, Gu D, et al. Extracellular vesicle-circEHD2 promotes the progression of renal cell carcinoma by activating cancer-associated fibroblasts. Mol Cancer. 2023;22:117. 10.1186/s12943-023-01824-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Circular RNA. detection pipelines yield divergent sets of circular RNAs. Nat Methods. 2023;20:1135–6. 10.1038/s41592-023-01945-5 [DOI] [PubMed] [Google Scholar]
- 10.Liao Y, Qiu X, Liu J, Zhang Z, Liu B, Jin C: The role of m6A-modified CircEPHB4 in glioma pathogenesis: Insights into cancer stemness metastasis. Ann Clin Translat Neurol. 2023;10(10):1749-67. 10.1002/acn3.51864. [DOI] [PMC free article] [PubMed]
- 11.Li K, Peng Z, Wang R, Li X, Du N, Liu D, Zhang J, Zhang Y, Ma L, Sun Y, et al. Enhancement of TKI sensitivity in lung adenocarcinoma through m6A-dependent translational repression of Wnt signaling by circ-FBXW7. Mol Cancer. 2023;22:103. 10.1186/s12943-023-01811-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, Jiang J, Qian H, Yan Y, Xu W. Exosomal circRNA: emerging insights into cancer progression and clinical application potential. J Hematol Oncol. 2023;16:67. 10.1186/s13045-023-01452-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, Tang T, Lu X, Fu X, Yang Y, Peng L: MPCLCDA: predicting circRNA-disease associations by using automatically selected meta-path and contrastive learning. Brief Bioinform. 2023;24(4):bbad227. 10.1093/bib/bbad227. [DOI] [PubMed]
- 14.Wu P, Hou X, Peng M, Deng X, Yan Q, Fan C, Mo Y, Wang Y, Li Z, Wang F, et al. Circular RNA circRILPL1 promotes nasopharyngeal carcinoma malignant progression by activating the Hippo-YAP signaling pathway. Cell Death Differ. 2023;30:1679–94. 10.1038/s41418-023-01171-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma C, Wang X, Zhang L, Zhu X, Bai J, He S, Mei J, Jiang J, Guan X, Zheng X, et al. Super enhancer-associated circular RNA-CircKrt4 regulates hypoxic pulmonary artery endothelial cell dysfunction in mice. Arterioscler Thromb Vasc Biol. 2023;43:1179–98. 10.1161/ATVBAHA.122.318842 [DOI] [PubMed] [Google Scholar]
- 16.Fang Z, Mei W, Qu C, Lu J, Shang L, Cao F, Li F. Role of m6A writers, erasers and readers in cancer. Exp Hematol Oncol. 2022;11:45. 10.1186/s40164-022-00298-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816–31. 10.1093/nar/gkaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang J, Cai H, Hou C, Song F, Jiang Y, Wang Z, Qiu D, Zhu Y, Wang F, Yu D, Hou J: METTL14 inhibits malignant progression of oral squamous cell carcinoma by targeting the autophagy-related gene RB1CC1 in an m6A-IGF2BP2-dependent manner. Clin Sci. (London, England: 1979). 2023;137(17):1373-89. 10.1042/CS20230219. [DOI] [PMC free article] [PubMed]
- 19.Qiu L, Jing Q, Li Y, Han J. RNA modification: mechanisms and therapeutic targets. Molecular biomedicine. 2023;4:25. 10.1186/s43556-023-00139-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507–19. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 21.Ma C, Liao S, Zhu Z. Crystal structure of human YTHDC2 YTH domain. Biochem Biophys Res Commun. 2019;518:678–84. 10.1016/j.bbrc.2019.08.107 [DOI] [PubMed] [Google Scholar]
- 22.Shi R, Ying S, Li Y, Zhu L, Wang X, Jin H. Linking the YTH domain to cancer: the importance of YTH family proteins in epigenetics. Cell Death Dis. 2021;12:346. 10.1038/s41419-021-03625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scutenaire J, Deragon JM, Jean V, Benhamed M, Raynaud C, Favory JJ, Merret R, Bousquet-Antonelli C. The YTH Domain Protein ECT2 Is an m(6)A Reader Required for Normal Trichome Branching in Arabidopsis. Plant Cell. 2018;30:986–1005. 10.1105/tpc.17.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–24. 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Q, Mo J, Liao Z, Chen X, Zhang B. The RNA m(6)A writer WTAP in diseases: structure, roles, and mechanisms. Cell Death Dis. 2022;13:852. 10.1038/s41419-022-05268-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer KD, Jaffrey SR. Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319–42. 10.1146/annurev-cellbio-100616-060758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flamand MN, Tegowski M, Meyer KD. The proteins of mrna modification: writers, readers, and erasers. Annu Rev Biochem. 2023;92:145–73. 10.1146/annurev-biochem-052521-035330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. 10.1038/ncomms2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–9. 10.1016/j.febslet.2008.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, Fan Y. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12:298. 10.1038/s41419-021-03558-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Pi H, Zheng W, Guo X, Shi C, Wang Z, Zhang J, Qu X, Liu L, Shen H, et al: The 3' Non-Coding Sequence Negatively Regulates PD-L1 Expression, and Its Regulators Are Systematically Identified in Pan-Cancer. Genes. 2023;14(8):1620. 10.3390/genes14081620. [DOI] [PMC free article] [PubMed]
- 32.Deng X, Sun X, Hu Z, Wu Y, Zhou C, Sun J, Gao X, Huang Y. Exploring the role of m6A methylation regulators in glioblastoma multiforme and their impact on the tumor immune microenvironment. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2023;37: e23155. 10.1096/fj.202301343 [DOI] [PubMed] [Google Scholar]
- 33.Bai R, Sun M, Chen Y, Zhuo S, Song G, Wang T, Zhang Z. H19 recruited N 6 -methyladenosine (m 6 A) reader YTHDF1 to promote SCARB1 translation and facilitate angiogenesis in gastric cancer. Chin Med J. 2023;136:1719–31. 10.1097/CM9.0000000000002722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. 10.1038/s41467-019-12651-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Guo Y, Guo Y, Wu X, Si C, Xu Y, Kang Q, Sun Z. m6A Modification in Non-Coding RNA: The Role in Cancer Drug Resistance. Front Oncol. 2021;11: 746789. 10.3389/fonc.2021.746789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Sun M, Tao Y, Ren J, Peng M, Jing Y, Xiao Q, Yang J, Lin C, Lei L, et al: Cytoplasmic Expression of TP53INP2 Modulated by Demethylase FTO and Mutant NPM1 Promotes Autophagy in Leukemia Cells. Int J Mole Sci. 2023;24(2):1624. 10.3390/ijms24021624. [DOI] [PMC free article] [PubMed]
- 37.Liu S, Guo X, Shang Q, Gao P. The biogenesis, biological functions and modification of Circular RNAs. Exp Mol Pathol. 2023;131:104861. 10.1016/j.yexmp.2023.104861 [DOI] [PubMed] [Google Scholar]
- 38.Chen C, Yuan W, Zhou Q, Shao B, Guo Y, Wang W, Yang S, Guo Y, Zhao L, Dang Q, et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021;11:4298–315. 10.7150/thno.51342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, Gorshkov K, Mao Q, Xia S, Cen D, et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. 2020;19:163. 10.1186/s12943-020-01281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74:494–507.e498. 10.1016/j.molcel.2019.02.034 [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell. 2019;76:96–109.e109. 10.1016/j.molcel.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Li Y, Zhou L, Zhou H, Ye L, Ou T, Hong H, Zheng S, Zhou Z, Wu K, et al. The m6A Reader YTHDF2 Promotes Bladder Cancer Progression by Suppressing RIG-I-Mediated Immune Response. Cancer Res. 2023;83:1834–50. 10.1158/0008-5472.CAN-22-2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang ZX, Wang YN, Li ZY, Dai ZH, He Y, Chu K, Gu JY, Ji YX, Sun NX, Yang F, Li W. The m6A mRNA demethylase FTO in granulosa cells retards FOS-dependent ovarian aging. Cell Death Dis. 2021;12:744. 10.1038/s41419-021-04016-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bao Y, Zhai J, Chen H, Wong CC, Liang C, Ding Y, Huang D, Gou H, Chen D, Pan Y, et al. Targeting m(6)A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut. 2023;72:1497–509. 10.1136/gutjnl-2022-328845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y, Zhou K, Li W, Hu J, Fu C, et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. 2022;40:1566–1582.e1510. 10.1016/j.ccell.2022.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ju G, Lei J, Cai S, Liu S, Yin X, Peng C: The Emerging, Multifaceted Role of WTAP in Cancer and Cancer Therapeutics. Cancers. 2023;15(11):3053. 10.3390/cancers15113053. [DOI] [PMC free article] [PubMed]
- 48.Sun X, Fu S, Yuan X, Pu X, Wang R, Wang X, Lu H: RNA N6-methyladenosine (m6A) modification in HNSCC: molecular mechanism and therapeutic potential. Cancer Gene Therapy. 2023;30(9):1209-14. 10.1038/s41417-023-00628-9. [DOI] [PubMed]
- 49.Wang L, Tang Y. N6-methyladenosine (m6A) in cancer stem cell: From molecular mechanisms to therapeutic implications. Biomed Pharmacother. 2023;163:114846. 10.1016/j.biopha.2023.114846 [DOI] [PubMed] [Google Scholar]
- 50.Dong H, Zeng L, Chen W, Zhang Q, Wang F, Wu Y, Cui B, Qi J, Zhang X, Liu C, et al. N6-methyladenine-mediated aberrant activation of the lncRNA SOX2OT-GLI1 loop promotes non-small-cell lung cancer stemness. Cell death discovery. 2023;9:149. 10.1038/s41420-023-01442-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, Tan X, Wu R, Deng T, Wang H, Jiang X, Zeng P, Tang J. m6A-mediated upregulation of lncRNA-AC0263561 promotes cancer stem cell maintenance in lung adenocarcinoma via activating Wnt signaling pathway. Aging. 2023;15:3538–48. 10.18632/aging.204689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Ye M, Bai J, Hu C, Lu F, Gu D, Yu P, Tang Q. el insights into the interplay between m6A modification and programmed cell death in cancer. Int J Biol Sci. 2023;19:1748–63. 10.7150/ijbs.81000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng F, Xu R. CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR-145-5p/ABCC1 axis. Biomed Pharmacother. 2020;124:109828. 10.1016/j.biopha.2020.109828 [DOI] [PubMed] [Google Scholar]
- 54.He X, Ma J, Zhang M, Cui J, Yang H. Circ_0007031 enhances tumor progression and promotes 5-fluorouracil resistance in colorectal cancer through regulating miR-133b/ABCC5 axis. Cancer Biomark. 2020;29:531–42. 10.3233/CBM-200023 [DOI] [PubMed] [Google Scholar]
- 55.Xu QY, Xie MJ, Huang J, Wang ZW. Effect of circ MTHFD2 on resistance to pemetrexed in gastric cancer through regulating expression of miR-124. Eur Rev Med Pharmacol Sci. 2019;23:10290–9. [DOI] [PubMed] [Google Scholar]
- 56.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, Xu Z, Zeng A, Zhang X, Zhang X, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. 10.1186/s12943-019-0969-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, Yuan X, Yin W, Xu J, Chen K, et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer. 2020;19:128. 10.1186/s12943-020-01246-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q, Miao Y, Fu Q, Hu H, Chen H, Zeng A, Jin Y, Jiang Y, Qian L, Wu L, et al. CircRNACCDC66 regulates cisplatin resistance in gastric cancer via the miR-618/BCL2 axis. Biochem Biophys Res Commun. 2020;526:713–20. 10.1016/j.bbrc.2020.03.156 [DOI] [PubMed] [Google Scholar]
- 59.Liu Z, Gu S, Wu K, Li L, Dong C, Wang W, Zhou Y. CircRNA-DOPEY2 enhances the chemosensitivity of esophageal cancer cells by inhibiting CPEB4-mediated Mcl-1 translation. J Exp Clin Cancer Res. 2021;40:361. 10.1186/s13046-021-02149-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu J, Ni L, Zhao F, Dai X, Tao J, Pan J, Shi A, Shen Z, Su C, Zhang Y. Overexpression of hsa_circ_0002874 promotes resistance of non-small cell lung cancer to paclitaxel by modulating miR-1273f/MDM2/p53 pathway. Aging (Albany NY). 2021;13:5986–6009. 10.18632/aging.202521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu G, Li M, Wu J, Qin C, Tao Y, He H. Circular RNA circNRIP1 Sponges microRNA-138-5p to Maintain Hypoxia-Induced Resistance to 5-Fluorouracil Through HIF-1α-Dependent Glucose Metabolism in Gastric Carcinoma. Cancer Manag Res. 2020;12:2789–802. 10.2147/CMAR.S246272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W, He Y, Zhang Y. CircRNA in ocular neovascular diseases: Fundamental mechanism and clinical potential. Pharmacol Res. 2023;197:106946. 10.1016/j.phrs.2023.106946 [DOI] [PubMed] [Google Scholar]
- 63.Feng B, Chen K, Zhang W, Zheng Q, He Y. circPGAM1 enhances autophagy signaling during laryngocarcinoma drug resistance by regulating miR-376a. Biochem Biophys Res Commun. 2021;534:966–72. 10.1016/j.bbrc.2020.10.063 [DOI] [PubMed] [Google Scholar]
- 64.Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, Li X, Dang Y, Zhang G. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19:156. 10.1186/s12943-020-01270-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Q, Liang D, Shen P, Yu Y, Yan Y, You W. Hsa_circ_0092276 promotes doxorubicin resistance in breast cancer cells by regulating autophagy via miR-348/ATG7 axis. Transl Oncol. 2021;14: 101045. 10.1016/j.tranon.2021.101045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu S, Wang M, Zhang H, Guo X, Qin R: Circ_0092367 Inhibits EMT and Gemcitabine Resistance in Pancreatic Cancer via Regulating the miR-1206/ESRP1 Axis. Genes (Basel). 2021;12(11):1701. 10.3390/genes12111701. [DOI] [PMC free article] [PubMed]
- 67.Chen C, Zhang M, Zhang Y. Circ_0000079 Decoys the RNA-Binding Protein FXR1 to Interrupt Formation of the FXR1/PRCKI Complex and Decline Their Mediated Cell Invasion and Drug Resistance in NSCLC. Cell Transplant. 2020;29:963689720961070. 10.1177/0963689720961070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Y, Zheng R, Chen J, Ning D. CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC). Cancer Cell Int. 2020;20:289. 10.1186/s12935-020-01390-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X, Chen L, Yang M, Peng K, Zhang Z, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19:20. 10.1186/s12943-020-1134-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou S, Guo Z, Lv X, Zhang X. CircGOT1 promotes cell proliferation, mobility, and glycolysis-mediated cisplatin resistance via inhibiting its host gene GOT1 in esophageal squamous cell cancer. Cell Cycle. 2022;21:247–60. 10.1080/15384101.2021.2015671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang X, Guo S, Wang S, Zhang Y, Chen H, Wang Y, Liu R, Niu Y, Xu Y. EIF4A3-Induced circARHGAP29 Promotes Aerobic Glycolysis in Docetaxel-Resistant Prostate Cancer through IGF2BP2/c-Myc/LDHA Signaling. Cancer Res. 2022;82:831–45. 10.1158/0008-5472.CAN-21-2988 [DOI] [PubMed] [Google Scholar]
- 72.Tan WQ, Yuan L, Wu XY, He CG, Zhu SC, Ye M. Exosome-delivered circular RNA DLGAP4 induces chemoresistance via miR-143-HK2 axis in neuroblastoma. Cancer Biomark. 2022;34:375–84. 10.3233/CBM-210272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi Q, Ji T, Ma Z, Tan Q, Liang J: Serum Exosomes-Based Biomarker circ_0008928 Regulates Cisplatin Sensitivity, Tumor Progression, and Glycolysis Metabolism by miR-488/HK2 Axis in Cisplatin-Resistant Nonsmall Cell Lung Carcinoma. Cancer Biother Radiopharm 2021. [DOI] [PubMed]
- 74.Yang W, Liu Y, Gao R, Xiu Z, Sun T. Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance. Cell Signal. 2019;60:122–35. 10.1016/j.cellsig.2019.04.011 [DOI] [PubMed] [Google Scholar]
- 75.Zhao Y, Zhong R, Deng C, Zhou Z. Circle RNA circABCB10 Modulates PFN2 to Promote Breast Cancer Progression, as Well as Aggravate Radioresistance Through Facilitating Glycolytic Metabolism Via miR-223-3p. Cancer Biother Radiopharm. 2021;36:477–90. [DOI] [PubMed] [Google Scholar]
- 76.Huang Y, Dai Y, Wen C, He S, Shi J, Zhao D, Wu L, Zhou H. circSETD3 Contributes to Acquired Resistance to Gefitinib in Non-Small-Cell Lung Cancer by Targeting the miR-520h/ABCG2 Pathway. Mol Ther Nucleic Acids. 2020;21:885–99. 10.1016/j.omtn.2020.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao HX, Miao CF, Sang LN, Huang YM, Zhang R, Sun L, Jiang ZX. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 2020;243: 117255. 10.1016/j.lfs.2020.117255 [DOI] [PubMed] [Google Scholar]
- 78.Xu H, Chen R, Shen Q, Yang D, Peng H, Tong J, Fu Q. Overexpression of Circular RNA circ_0013587 Reverses Erlotinib Resistance in Pancreatic Cancer Cells Through Regulating the miR-1227/E-Cadherin Pathway. Front Oncol. 2021;11: 754146. 10.3389/fonc.2021.754146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu YP, Dong ZN, Wang SW, Zheng YM, Zhang C, Zhou YQ, Zhao YJ, Zhao Y, Wang F, Peng R, et al. circHMGCS1-016 reshapes immune environment by sponging miR-1236-3p to regulate CD73 and GAL-8 expression in intrahepatic cholangiocarcinoma. J Exp Clin Cancer Res. 2021;40:290. 10.1186/s13046-021-02095-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jia L, Wang Y, Wang CY. circFAT1 Promotes Cancer Stemness and Immune Evasion by Promoting STAT3 Activation. Adv Sci (Weinh). 2021;8:2003376. 10.1002/advs.202003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. 10.1038/nrc3038 [DOI] [PubMed] [Google Scholar]
- 82.Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. 10.1186/1476-4598-12-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marcucci F, Rumio C. Glycolysis-induced drug resistance in tumors-A response to danger signals? Neoplasia. 2021;23:234–45. 10.1016/j.neo.2020.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu H, Yang X, Tang J, Si S, Zhou Z, Lu J, Han J, Yuan B, Wu Q, Lu Q, Yang H. ALKBH5 Inhibited Cell Proliferation and Sensitized Bladder Cancer Cells to Cisplatin by m6A-CK2α-Mediated Glycolysis. Mol Ther Nucleic Acids. 2021;23:27–41. 10.1016/j.omtn.2020.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tian Y, Xiao H, Yang Y, Zhang P, Yuan J, Zhang W, Chen L, Fan Y, Zhang J, Cheng H, et al. Crosstalk between 5-methylcytosine and N-methyladenosine machinery defines disease progression, therapeutic response and pharmacogenomic landscape in hepatocellular carcinoma. Mol Cancer. 2023;22:5. 10.1186/s12943-022-01706-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu X, Zhang P, Huang Y, Shi W, Mao J, Ma N, Kong L, Guo L, Liu J, Chen J, Lu R. METTL3-mediated m6A mRNA contributes to the resistance of carbon-ion radiotherapy in non-small-cell lung cancer. Cancer Sci. 2023;114:105–14. 10.1111/cas.15590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Du H, Zou N, Zuo H, Zhang X, Zhu S. YTHDF3 mediates HNF1α regulation of cervical cancer radio-resistance by promoting RAD51D translation in an m6A-dependent manner. FEBS J. 2023;290:1920–35. 10.1111/febs.16681 [DOI] [PubMed] [Google Scholar]
- 88.Li E, Xia M, Du Y, Long K, Ji F, Pan F, He L, Hu Z, Guo Z: METTL3 promotes homologous recombination repair and modulates chemotherapeutic response in breast cancer by regulating the EGF/RAD51 axis. eLife. 2022:11:e75231. 10.7554/eLife.75231. [DOI] [PMC free article] [PubMed]
- 89.Torii Y, Kato R, Minami Y, Hasegawa K, Fujii T, Udagawa Y. ERCC1 expression and chemosensitivity in uterine cervical adenocarcinoma cells. Anticancer Res. 2014;34:107–15. [PubMed] [Google Scholar]
- 90.Li W, Jie Z, Li Z, Liu Y, Gan Q, Mao Y, Wang X. ERCC1 siRNA ameliorates drug resistance to cisplatin in gastric carcinoma cell lines. Mol Med Rep. 2014;9:2423–8. 10.3892/mmr.2014.2112 [DOI] [PubMed] [Google Scholar]
- 91.Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590–7. 10.1002/mc.22782 [DOI] [PubMed] [Google Scholar]
- 92.Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem. 2018;293:10524–35. 10.1074/jbc.TM118.000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou K, Sun Y, Dong D, Zhao C, Wang W. EMP3 negatively modulates breast cancer cell DNA replication, DNA damage repair, and stem-like properties. Cell Death Dis. 2021;12:844. 10.1038/s41419-021-04140-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei J, Yin Y, Zhou J, Chen H, Peng J, Yang J, Tang Y. METTL3 potentiates resistance to cisplatin through m(6) A modification of TFAP2C in seminoma. J Cell Mol Med. 2020;24:11366–80. 10.1111/jcmm.15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou M, Liu W, Zhang J, Sun N. RNA mA Modification in Immunocytes and DNA Repair: The Biological Functions and Prospects in Clinical Application. Frontiers in cell and developmental biology. 2021;9: 794754. 10.3389/fcell.2021.794754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang C, Zhou S, Zhang C, Jin Y, Xu G, Zhou L, Ding G, Pang T, Jia S, Cao L. ZC3H13-mediated N6-methyladenosine modification of PHF10 is impaired by fisetin which inhibits the DNA damage response in pancreatic cancer. Cancer Lett. 2022;530:16–28. 10.1016/j.canlet.2022.01.013 [DOI] [PubMed] [Google Scholar]
- 97.Li S, Li N, Li B, Zhu L, Xu T, Wang L, Zhang J, Kong F: CircHIPK3 promotes proliferation and metastasis of villous trophoblasts through miR-30a-3p/Wnt2 axis. J Genet. 2022;101:55. [PubMed]
- 98.Ren S, Liu J, Feng Y, Li Z, He L, Li L, Cao X, Wang Z, Zhang Y. Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J Exp Clin Cancer Res. 2019;38:388. 10.1186/s13046-019-1398-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hayashi M, Sugahara K, Yamamura A, Iida Y. Evaluation of the Properties of the DNA Methyltransferase from Aeropyrum pernix K1. Microbiology spectrum. 2021;9:e0018621. 10.1128/Spectrum.00186-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miranda-Gonçalves V, Lobo J, Guimarães-Teixeira C, Barros-Silva D, Guimarães R, Cantante M, Braga I, Maurício J, Oing C, Honecker F, et al. The component of the mA writer complex VIRMA is implicated in aggressive tumor phenotype, DNA damage response and cisplatin resistance in germ cell tumors. Journal of experimental & clinical cancer research : CR. 2021;40:268. 10.1186/s13046-021-02072-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu F, Wei J, Cui X, Yu C, Ni W, Bungert J, Wu L, He C, Qian Z. Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 2021;49:5779–97. 10.1093/nar/gkab415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qu F, Tsegay P, Liu Y. N-Methyladenosine, DNA Repair, and Genome Stability. Front Mol Biosci. 2021;8:645823. 10.3389/fmolb.2021.645823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li R, Chen H, Li C, Qi Y, Zhao K, Wang J, You C, Huang H. The prognostic value and immune landscaps of m6A/m5C-related lncRNAs signature in the low grade glioma. BMC Bioinformatics. 2023;24:274. 10.1186/s12859-023-05386-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, Peng J. METTL5 serves as a diagnostic and prognostic biomarker in hepatocellular carcinoma by influencing the immune microenvironment. Sci Rep. 2023;13:10755. 10.1038/s41598-023-37807-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang Y, Guan Y, Zhang X: METTL3-Mediated Maturation of miR-99a-5p Promotes Cell Migration and Invasion in Oral Squamous Cell Carcinoma by Targeting ZBTB7A. Mol Biotechnol. 2023 Jul 27. 10.1007/s12033-023-00815-x. [DOI] [PubMed]
- 106.Hu B, Gao J, Shi J, Wen P, Guo W, Zhang S: m6A reader YTHDF3 triggers the progression of hepatocellular carcinoma through the YTHDF3/m A-EGFR/STAT3 axis and EMT. Mole Carcinogenesis 2023;62(10):1599-1614. 10.1002/mc.23602. [DOI] [PubMed]
- 107.Shen S, Jin H, Zhang X, Zhang Y, Li X, Yan W, Xie S, Yu B, Hu J, Liu H, et al. LINC00426, a novel mA-regulated long non-coding RNA, induces EMT in cervical cancer by binding to ZEB1. Cell Signal. 2023;109:110788. 10.1016/j.cellsig.2023.110788 [DOI] [PubMed] [Google Scholar]
- 108.Chen M, Tian B, Hu G, Guo Y: circUHRF2METTL3-Modulated Promotes Colorectal Cancer Stemness and Metastasis through Increasing mRNA Stability by Recruiting IGF2BP1. Cancers. 2023;15(12):3148. 10.3390/cancers15123148. [DOI] [PMC free article] [PubMed]
- 109.Xi S, Ming D, Zhang J, Guo M, Wang S, Cai Y, Liu M, Wang D, Zhang Y, Li Y, Yuan S. Downregulation of N6-methyladenosine-modified LINC00641 promotes EMT, but provides a ferroptotic vulnerability in lung cancer. Cell Death Dis. 2023;14:359. 10.1038/s41419-023-05880-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang X, Bai Q, Chen W, Liang J, Wang F, Gu W, Liu L, Li Q, Chen Z, Zhou A, et al: m A-Dependent Modulation via IGF2BP3/MCM5/Notch Axis Promotes Partial EMT and LUAD Metastasis. Advanced science (Weinheim, Baden-Wurttemberg, Germany) 2023;10:e2206744. [DOI] [PMC free article] [PubMed]
- 111.Zhang Y, Wang L, Yan F, Yang M, Gao H, Zeng Y. Mettl3 Mediated m6A Methylation Involved in Epithelial-Mesenchymal Transition by Targeting SOCS3/STAT3/SNAI1 in Cigarette Smoking-Induced COPD. Int J Chron Obstruct Pulmon Dis. 2023;18:1007–17. 10.2147/COPD.S398289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen H, Zhang J, Yang L, Li Y, Wang Z, Ye C. circ-ZEB1 regulates epithelial-mesenchymal transition and chemotherapy resistance of colorectal cancer through acting on miR-200c-5p. Transl Oncol. 2023;28: 101604. 10.1016/j.tranon.2022.101604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu X, Su K, Sun X, Jiang Y, Wang L, Hu C, Zhang C, Lu M, Du X, Xing B. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2021;40:132. 10.1186/s13046-021-01934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hu W, Klümper N, Schmidt D, Ritter M, Ellinger J, Hauser S. Depletion of the mA demethylases FTO and ALKBH5 impairs growth and metastatic capacity through EMT phenotype change in clear cell renal cell carcinoma. American journal of translational research. 2023;15:1744–55. [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Y, Chen Y, Liang J, Jiang M, Zhang T, Wan X, Wu J, Li X, Chen J, Sun J, et al: METTL3-mediated m6A modification of HMGA2 mRNA promotes subretinal fibrosis and epithelial-mesenchymal transition. J Mole Cell Biol. 2023;15(3):mjad005. 10.1093/jmcb/mjad005. [DOI] [PMC free article] [PubMed]
- 116.Wang C. Danli Ma, Yu H, Zhuo Z, Ye Z: N6-methyladenosine (m6A) as a regulator of carcinogenesis and drug resistance by targeting epithelial-mesenchymal transition and cancer stem cells. Heliyon. 2023;9: e14001. 10.1016/j.heliyon.2023.e14001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang Y, Wang N, Zhang Y, Jiang W, Fang C, Feng Y, Ma H, Jiang F, Dong G. Self-restricted circular RNA circSOX2 suppressed the malignant progression in SOX2-amplified LUSC. Cell Death Dis. 2022;13:873. 10.1038/s41419-022-05288-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao S, Xu F, Ji Y, Wang Y, Wei M, Zhang L. Circular RNA circ-CD44 regulates chemotherapy resistance by targeting the miR-330-5p/ABCC1 axis in colorectal cancer cells. Histol Histopathol. 2023;38:209–21. [DOI] [PubMed] [Google Scholar]
- 119.Zhao F, Zhao P, Chang J, Sun X, Ma X, Shi B, Yin M, Wang Y, Yang Y. Identification and vitro verification of the potential drug targets of active ingredients of Chonglou in the treatment of lung adenocarcinoma based on EMT-related genes. Front Genet. 2023;14:1112671. 10.3389/fgene.2023.1112671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen C, Huang J, Huang J, Deng J, Shangguan X, Chen A, Chen L, Wu W. Metformin attenuates multiple myeloma cell proliferation and encourages apoptosis by suppressing METTL3-mediated m6A methylation of THRAP3, RBM25, and USP4. Cell cycle (Georgetown, Tex). 2023;22:986–1004. 10.1080/15384101.2023.2170521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Duan Y, Du Y, Mu Y, Gu Z, Wang C. Prognostic value, immune signature and molecular mechanisms of the SUMO family in pancreatic adenocarcinoma. Front Mol Biosci. 2022;9:1096679. 10.3389/fmolb.2022.1096679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Garg R, Melstrom L, Chen J, He C, Goel A: Targeting FTO Suppresses Pancreatic Carcinogenesis via Regulating Stem Cell Maintenance and EMT Pathway. Cancers. 2022;14(23):5919. 10.3390/cancers14235919. [DOI] [PMC free article] [PubMed]
- 123.Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, Hu Y, Qiu J, Pu L, Tang J, Wang X. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021;6:76. 10.1038/s41392-020-00453-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feng S, Qiu G, Yang L, Feng L, Fan X, Ren F, Huang K, Chen Y: Omeprazole improves chemosensitivity of gastric cancer cells by m6A demethylase FTO-mediated activation of mTORC1 and DDIT3 up-regulation. Biosci Rep. 2021;41(1):BSR20200842. 10.1042/BSR20200842. [DOI] [PMC free article] [PubMed]
- 125.Chen H, Xiang Y, Yin Y, Peng J, Peng D, Li D, Kitazawa R, Tang Y, Yang J. The m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting ATG5 in seminoma. Transl Androl Urol. 2021;10:1711–22. 10.21037/tau-20-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li J, Cao H, Yang J, Wang B. CircCDK1 blocking IGF2BP2-mediated m6A modification of CPPED1 promotes laryngeal squamous cell carcinoma metastasis via the PI3K/AKT signal pathway. Gene. 2023;884:147686. 10.1016/j.gene.2023.147686 [DOI] [PubMed] [Google Scholar]
- 127.Chen X, Zhu S, Li H, Wang J, Sun L, Xu J, Hui Y, Li X, Li L, Zhao Y, et al. N-methyladenosine-modified circIRF2, identified by YTHDF2, suppresses liver fibrosis via facilitating FOXO3 nuclear translocation. Int J Biol Macromol. 2023;248:125811 10.1016/j.ijbiomac.2023.125811 [DOI] [PubMed] [Google Scholar]
- 128.Qi K, Dou Y, Zhang Z, Wei Y, Song C, Qiao R, Li X, Yang F, Wang K, Li X, Han X: Expression Profile and Regulatory Properties of m6A-Modified circRNAs in the Longissimus Dorsi of Queshan Black and Large White Pigs. Animals. 2023;13(13):2190. 10.3390/ani13132190. [DOI] [PMC free article] [PubMed]
- 129.Lin Z, Li J, Zhang J, Feng W, Lu J, Ma X, Ding W, Ouyang S, Lu J, Yue P, et al. Metabolic Reprogramming Driven by IGF2BP3 Promotes Acquired Resistance to EGFR Inhibitors in Non-Small Cell Lung Cancer. Can Res. 2023;83:2187–207. 10.1158/0008-5472.CAN-22-3059 [DOI] [PubMed] [Google Scholar]
- 130.Yang L, Yan B, Qu L, Ren J, Li Q, Wang J, Kan X, Liu M, Wang Y, Sun Y, et al. IGF2BP3 Regulates TMA7-mediated Autophagy and Cisplatin Resistance in Laryngeal Cancer via m6A RNA Methylation. Int J Biol Sci. 2023;19:1382–400. 10.7150/ijbs.80921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun Y, Shen W, Hu S, Lyu Q, Wang Q, Wei T, Zhu W, Zhang J. METTL3 promotes chemoresistance in small cell lung cancer by inducing mitophagy. Journal of experimental & clinical cancer research : CR. 2023;42:65. 10.1186/s13046-023-02638-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang Z, Zhao F, Gu X, Feng L, Xu M, Li T, Liu X, Zhang X. Binding of RNA m6A by IGF2BP3 triggers chemoresistance of HCT8 cells via upregulation of ABCB1. Am J Cancer Res. 2021;11:1428–45. [PMC free article] [PubMed] [Google Scholar]
- 134.Liu X, Yuan J, Zhang X, Li L, Dai X, Chen Q, Wang Y. ATF3 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an N(6)-Methyladenosine-Based Epitranscriptomic Mechanism. Chem Res Toxicol. 2021;34:1814–21. 10.1021/acs.chemrestox.1c00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hao CC, Xu CY, Zhao XY, Luo JN, Wang G, Zhao LH, Ge X, Ge XF. Up-regulation of VANGL1 by IGF2BPs and miR-29b-3p attenuates the detrimental effect of irradiation on lung adenocarcinoma. J Exp Clin Cancer Res. 2020;39:256. 10.1186/s13046-020-01772-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu Z, Wu K, Gu S, Wang W, Xie S, Lu T, Li L, Dong C, Wang X, Zhou Y. A methyltransferase-like 14/miR-99a-5p/tribble 2 positive feedback circuit promotes cancer stem cell persistence and radioresistance via histone deacetylase 2-mediated epigenetic modulation in esophageal squamous cell carcinoma. Clin Transl Med. 2021;11: e545. 10.1002/ctm2.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao H, Huang Y, Shi J, Dai Y, Wu L, Zhou H. ABCC10 Plays a Significant Role in the Transport of Gefitinib and Contributes to Acquired Resistance to Gefitinib in NSCLC. Front Pharmacol. 2018;9:1312. 10.3389/fphar.2018.01312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiao P, Liu YK, Han W, Hu Y, Zhang BY, Liu WL. Exosomal Delivery of FTO Confers Gefitinib Resistance to Recipient Cells through ABCC10 Regulation in an m6A-dependent Manner. Mol Cancer Res. 2021;19:726–38. 10.1158/1541-7786.MCR-20-0541 [DOI] [PubMed] [Google Scholar]
- 139.Fulda S. Tumor resistance to apoptosis. Int J Cancer. 2009;124:511–5. 10.1002/ijc.24064 [DOI] [PubMed] [Google Scholar]
- 140.Villanova L, Careccia S, De Maria R, Fiori ME: Micro-Economics of Apoptosis in Cancer: ncRNAs Modulation of BCL-2 Family Members. Int J Mol Sci. 2018;19(4):958. 10.3390/ijms19040958. [DOI] [PMC free article] [PubMed]
- 141.Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020;722: 144076. 10.1016/j.gene.2019.144076 [DOI] [PubMed] [Google Scholar]
- 142.Gu M, Zheng W, Zhang M, Dong X, Zhao Y, Wang S, Jiang H, Zheng X. LncRNA NONHSAT141924 promotes paclitaxel chemotherapy resistance through p-CREB/Bcl-2 apoptosis signaling pathway in breast cancer. J Cancer. 2020;11:3645–54. 10.7150/jca.39463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049–57. 10.1093/carcin/bgq192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. 10.1111/j.1476-5381.2009.00430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou T, Li S, Xiang D, Liu J, Sun W, Cui X, Ning B, Li X, Cheng Z, Jiang W, et al. m6A RNA methylation-mediated HNF3γ reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct Target Ther. 2020;5:296. 10.1038/s41392-020-00299-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022;21:14. 10.1186/s12943-022-01500-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhu Y, Tan J, Goon J: Cuproptosis- and m6A-Related lncRNAs for Prognosis of Hepatocellular Carcinoma. Biology. 2023;12(8):1101. 10.3390/biology12081101. [DOI] [PMC free article] [PubMed]
- 148.Yuan F, Cai X, Wang Y, Du C, Cong Z, Zeng X, Tang C, Ma C. Comprehensive analysis of mA subtype classification for immune microenvironment of pituitary adenomas. Int Immunopharmacol. 2023;124: 110784. 10.1016/j.intimp.2023.110784 [DOI] [PubMed] [Google Scholar]
- 149.Ma C, Zheng Q, Wang Y, Li G, Zhao M, Sun Z: Pan-cancer analysis and experimental validation revealed the m6A methyltransferase KIAA1429 as a potential biomarker for diagnosis, prognosis, and immunotherapy. Aging. 2023;15(17):8664-8691. 10.18632/aging.204968. [DOI] [PMC free article] [PubMed]
- 150.Yue S, Liu H, Su H, Luo C, Liang H, Zhang B, Zhang W. m6A-regulated tumor glycolysis: new advances in epigenetics and metabolism. Mol Cancer. 2023;22:137. 10.1186/s12943-023-01841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu W, Li H, Hameed Y, Abdel-Maksoud M, Almutairi S, Mubarak A, Aufy M, Alturaiki W, Alshalani A, Mahmoud A, Li C. Elucidating the clinical and immunological value of m6A regulator-mediated methylation modification patterns in adrenocortical carcinoma. Oncol Res. 2023;31:819–31. 10.32604/or.2023.029414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lian B, Yan S, Li J, Bai Z, Li J. HNRNPC promotes collagen fiber alignment and immune evasion in breast cancer via activation of the VIRMA-mediated TFAP2A/DDR1 axis. Molecular medicine (Cambridge, Mass). 2023;29:103. 10.1186/s10020-023-00696-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xiao Y, Li J, Wu J. Development and validation of a novel prognostic signature based on m6A/m5C/m1A-related genes in hepatocellular carcinoma. BMC Med Genomics. 2023;16:177. 10.1186/s12920-023-01611-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Z, Gao W, Liu Z, Yu S, Jian H, Hou Z, Zeng P. Comprehensive analysis of m6A regulators associated with immune infiltration in Hepatitis B virus-related hepatocellular carcinoma. BMC Gastroenterol. 2023;23:259. 10.1186/s12876-023-02873-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hu J, Xue C, Wang Q. N-methyladenosine modification: an important player in the tumor immune microenvironment. Biomed Pharmacother. 2023;165:15171. 10.1016/j.biopha.2023.115171 [DOI] [PubMed] [Google Scholar]
- 156.Najafi M, Mortezaee K, Majidpoor J. Cancer stem cell (CSC) resistance drivers. Life Sci. 2019;234:116781. 10.1016/j.lfs.2019.116781 [DOI] [PubMed] [Google Scholar]
- 157.Khan RIN, Malla WA. m(6)A modification of RNA and its role in cancer, with a special focus on lung cancer. Genomics. 2021;113:2860–9. 10.1016/j.ygeno.2021.06.013 [DOI] [PubMed] [Google Scholar]
- 158.Liu S, Jiang Z, Xiao P, Li X, Chen Y, Tang H, Chai Y, Liu Y, Zhu Z, Xie Q, et al. Hsa_circ_0005576 promotes osimertinib resistance through the miR-512-5p/IGF1R axis in lung adenocarcinoma cells. Cancer Sci. 2022;113:79–90. 10.1111/cas.15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. 10.1186/s12943-020-1135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li C, Li W, Cao S, Xu J, Qian Y, Pan X, Lei D, Wei D. Circ_0058106 promotes proliferation, metastasis and EMT process by regulating Wnt2b/β-catenin/c-Myc pathway through miR-185-3p in hypopharyngeal squamous cell carcinoma. Cell Death Dis. 2021;12:1063. 10.1038/s41419-021-04346-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang Z, Wei P, Wei D, Cao S, Liu H, Chen L, Han X, Zhao X, Liu C, Li G, et al. Effect of up-regulation of circMATR3 on the proliferation, metastasis, progression and survival of hypopharyngeal carcinoma. J Cell Mol Med. 2020;24:4687–97. 10.1111/jcmm.15134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rao X, Lai L, Li X, Wang L, Li A, Yang Q. N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life. 2021;73:408–17. 10.1002/iub.2438 [DOI] [PubMed] [Google Scholar]
- 163.Qin S, Mao Y, Chen X, Xiao J, Qin Y, Zhao L. The functional roles, cross-talk and clinical implications of m6A modification and circRNA in hepatocellular carcinoma. Int J Biol Sci. 2021;17:3059–79. 10.7150/ijbs.62767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rong D, Wu F, Lu C, Sun G, Shi X, Chen X, Dai Y, Zhong W, Hao X, Zhou J, et al. m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids. 2021;26:637–48. 10.1016/j.omtn.2021.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, Gorshkov K, Sun Q, Lin H, Zheng X, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5:298. 10.1038/s41392-020-00375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu Z, Wang T, She Y, Wu K, Gu S, Li L, Dong C, Chen C, Zhou Y. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20:105. 10.1186/s12943-021-01398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tobiume K, Matsuzawa A, Takahashi T, Nishitoh H, Morita K, Takeda K, Minowa O, Miyazono K, Noda T, Ichijo H. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001;2:222–8. 10.1093/embo-reports/kve046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen X, Ma W, Yao Y, Zhang Q, Li J, Wu X, Mei C, Jiang X, Chen Y, Wang G, et al. Serum deprivation-response protein induces apoptosis in hepatocellular carcinoma through ASK1-JNK/p38 MAPK pathways. Cell Death Dis. 2021;12:425. 10.1038/s41419-021-03711-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M, Li S, Xie D, Sun X, Hu X, Chen C. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 2021;520:321–31. 10.1016/j.canlet.2021.08.007 [DOI] [PubMed] [Google Scholar]
- 170.Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, Ma X, Cheng Z, Yu C, Wang S, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12:295. 10.1038/s41467-020-20527-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer. 2020;19:167. 10.1186/s12943-020-01287-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cai J, Chen Z, Zhang Y, Wang J, Zhang Z, Wu J, Mao J, Zuo X. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol Ther Oncolytics. 2022;24:755–71. 10.1016/j.omto.2022.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, Zhong H, Jiang Y, Zhang X, Jiang G. Circ0008399 Interaction with WTAP Promotes Assembly and Activity of the m(6)A Methyltransferase Complex and Promotes Cisplatin Resistance in Bladder Cancer. Cancer Res. 2021;81:6142–56. 10.1158/0008-5472.CAN-21-1518 [DOI] [PubMed] [Google Scholar]
- 174.Sun K, Du Y, Hou Y, Zhao M, Li J, Du Y, Zhang L, Chen C, Yang H, Yan F, Su R. Saikosaponin D exhibits anti-leukemic activity by targeting FTO/m(6)A signaling. Theranostics. 2021;11:5831–46. 10.7150/thno.55574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Koukourakis MI, Giatromanolaki A, Panteliadou M, Pouliliou SE, Chondrou PS, Mavropoulou S, Sivridis E. Lactate dehydrogenase 5 isoenzyme overexpression defines resistance of prostate cancer to radiotherapy. Br J Cancer. 2014;110:2217–23. 10.1038/bjc.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sulaiman SA, Abdul Murad NA, Mohamad Hanif EA, Abu N, Jamal R. Prospective Advances in Circular RNA Investigation. Adv Exp Med Biol. 2018;1087:357–70. 10.1007/978-981-13-1426-1_28 [DOI] [PubMed] [Google Scholar]
- 178.Saenz-Pipaon G, San Martín P, Planell N, Maillo A, Ravassa S, Vilas-Zornoza A, Martinez-Aguilar E, Rodriguez JA, Alameda D, Lara-Astiaso D, et al. Functional and transcriptomic analysis of extracellular vesicles identifies calprotectin as a new prognostic marker in peripheral arterial disease (PAD). J Extracell Vesicles. 2020;9:1729646. 10.1080/20013078.2020.1729646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2021;12:911–46. 10.1007/s13238-020-00799-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang Y, Jia G. Detection methods of epitranscriptomic mark N6-methyladenosine. Essays Biochem. 2020;64:967–79. 10.1042/EBC20200039 [DOI] [PubMed] [Google Scholar]
- 181.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shu X, Cao J, Cheng M, Xiang S, Gao M, Li T, Ying X, Wang F, Yue Y, Lu Z, et al. A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat Chem Biol. 2020;16:887–95. 10.1038/s41589-020-0526-9 [DOI] [PubMed] [Google Scholar]
- 183.Werner S, Galliot A, Pichot F, Kemmer T, Marchand V, Sednev MV, Lence T, Roignant JY, König J, Höbartner C, et al. NOseq: amplicon sequencing evaluation method for RNA m6A sites after chemical deamination. Nucleic Acids Res. 2021;49:e23. 10.1093/nar/gkaa1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nombela P, Miguel-López B, Blanco S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. 2021;20:18. 10.1186/s12943-020-01263-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M, Lyu WY, Qi Q, Tiwari AK, Chen JX, Zhang DM, Chen ZS. m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer. 2022;21:52. 10.1186/s12943-022-01510-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. 10.1186/s12943-019-1041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Qu L, Yi Z, Shen Y, Lin L, Chen F, Xu Y, Wu Z, Tang H, Zhang X, Tian F, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–1744.e1716. 10.1016/j.cell.2022.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.