Abstract
T-box riboswitches are noncoding RNA elements involved in genetic regulation of most Gram-positive bacteria. They regulate amino acid metabolism by assessing the aminoacylation status of tRNA, subsequently affecting the transcription or translation of downstream amino acid metabolism-related genes. Here we present single-molecule FRET studies of the Mycobacterium tuberculosis IleS T-box riboswitch, a paradigmatic translational T-box. Results support a two-step binding model, where the tRNA anticodon is recognized first, followed by interactions with the NCCA sequence. Furthermore, after anticodon recognition, tRNA can transiently dock into the discriminator domain even in the absence of the tRNA NCCA-discriminator interactions. Establishment of the NCCA-discriminator interactions significantly stabilizes the fully bound state. Collectively, the data suggest high conformational flexibility in translational T-box riboswitches; and supports a conformational selection model for NCCA recognition. These findings provide a kinetic framework to understand how specific RNA elements underpin the binding affinity and specificity required for gene regulation.
Subject terms: Single-molecule biophysics, RNA
smFRET studies of the M. tuberculosis IleS translational T-box riboswitch support a two-step tRNA binding mechanism and the use of conformational selection for tRNA 3’ end recognition, which is distinct from the one used by transcriptional T-boxes.
Introduction
Protein synthesis, or translation, is an essential cellular activity in all known cellular life. The biologically active substrates required for translation are aminoacyl-tRNAs (charged tRNAs), which are synthesized by aminoacyl-tRNA synthetases. To ensure an adequate supply of charged tRNAs, several regulatory mechanisms have evolved1. In Gram-positive bacteria, one such mechanism is represented by T-box riboswitches. Riboswitches are cis-acting RNA elements that undergo structural changes in response to a regulatory signal2,3. While numerous riboswitches regulate gene expression in response to various small molecule metabolites3, T-box riboswitches recognize and respond to the aminoacylation state of tRNAs4. A higher proportion of uncharged tRNAs compared to charged tRNAs indicates a shortage of a specific amino acid or aminoacyl-tRNA synthetase5. By detecting intracellular concentrations of uncharged tRNAs, T-box riboswitches, located at the 5’ leader region of the mRNAs that they regulate, can alter the transcription or translation of aminoacyl tRNA synthetase genes or other amino acid metabolism-related genes6–8. Aside from the ribosome, T-box riboswitches are the only other known RNA capable of both tRNA identity decoding and aminoacylation sensing8, and thus provide a unique paradigm for gene regulation by two interacting, structured non-coding RNAs4,6,9,10.
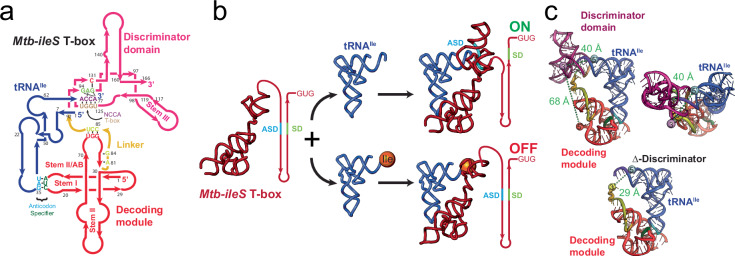
As is the case with other riboswitches; T-box riboswitches are modular and composed of two parts, a ligand binding domain and an expression platform. tRNA decoding and aminoacylation status discrimination reside in two separate structural domains (Fig. 1a). T-box riboswitches have an obligatory Stem I domain that decodes tRNA identity11–13; this process is often facilitated by Stem II and Stem IIA/B domains6,14. In general, Stem I, Stem II, and Stem II A/B comprise the decoding module (Fig. 1a), except for glycine T-box riboswitches, where Stem II and Stem IIA/B are absent. To achieve specificity, the tRNA anticodon pairs with a single codon sequence in the T-box RNA, known as the specifier sequence, which is situated in the specifier loop of Stem I. The discriminator domain examines the aminoacylation status of the tRNA and switches the expression output appropriately14,15(Fig. 1b). The discriminator domain includes Stem III, an antiterminator or antisequestrator region, and the T-box sequence (AGGGUGGNACCGCG), which comprises the 14 most highly conserved nucleotides of the complete T-box RNA (Fig. 1a) and is involved in recognition of the NCCA sequence at the 3’ end of the tRNA.
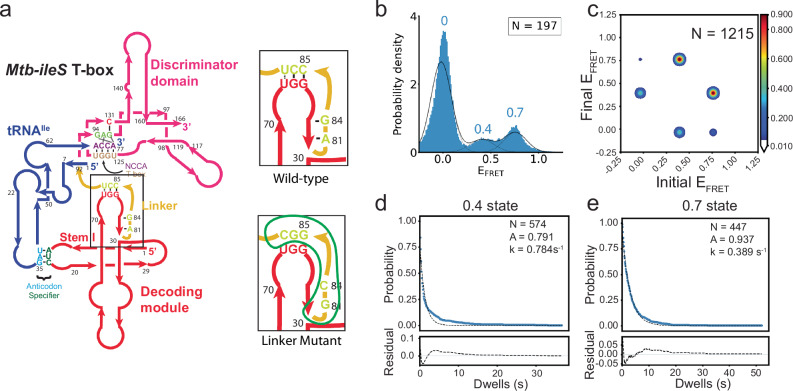
Fig. 1. Structure and mechanism of the Mtb-ileS T-box riboswitch.
a Secondary structure diagram showing the Mtb-ileS T-box riboswitch (pink/red) and tRNAIle (blue). The T-box has two distinct structural domains, the decoding module (red) is involved in recognizing specific tRNAs through anticodon-specifier interactions (cyan, green), whereas the discriminator domain (pink) contains the T-box sequence (brown) involved in recognizing the 3’ NCCA region of the tRNA (purple) and assessing the aminoacylation status. The two domains are joined by a linker region (yellow). The secondary structure diagram is based on the structure of the complex14. b Proposed mechanism for translation-regulating T-box riboswitches. The T-box decodes the tRNA identity of the tRNA and aminoacylation status at the 3’ end. Depending on the aminoacylation status of the tRNA, it exposes or sequesters the Ribosome Binding Site or Shine Dalgarno sequence (SD, green). If the tRNA is uncharged, the 3’ NCCA of the tRNA forms base pairs with the T-box sequence leading to a conformation where the SD region is accessible, and translation is allowed (top). If the tRNA is charged, the interactions with the T-box sequence are precluded, leading to a conformation where the SD and the Anti Shine Dalgarno (ASD, cyan) regions interact, the SD is inaccessible, and translation is not allowed (bottom). c Structure of the Mtb-ileS T-box riboswitch/tRNAIle complex14. The molecules are colored as in (a). The top two pictures show two orientations of the complex. The bottom picture shows a model of a truncated T-box missing the Discriminator domain (Δ-Discriminator mutant). Approximate distances between the 3’ and 5’ ends of the T-box (69 Å), the 3’ end of T-box and the 5’ end of the tRNA (40 Å), and the 3’ end of the Δ-Discriminator Mutant and the 5’ end of the tRNA (29 Å) are shown.
The majority of T-box riboswitches regulate gene expression at the transcription level16–18. In the absence of tRNA, or in the presence of charged tRNA, transcription-regulating T-box riboswitches typically adopt an intrinsic terminator structure that promotes premature termination of transcription. Alternatively, binding of an uncharged cognate tRNA promotes the formation of an antiterminator structure that enables transcription readthrough. The Bacillus subtilis glyQS T-box riboswitch, as a representative transcription-regulating T-box riboswitch, has been the subject of extensive structural and kinetic studies11,13,15,19. Single-molecule characterization of the glyQS T-box riboswitch established a two-step binding model in which the anticodon of tRNAGly is recognized first, leading to a partially bound state, followed by binding of the tRNAGly 3’ NCCA end, leading to a fully bound state19,20. In addition, an intramolecular conformational change between the decoding and the discriminator domains occurs during the second binding step. Notably, in this model, binding of the tRNAGly ligand by the glyQS T-box riboswitch follows the directionality of transcription, with kinetic parameters supporting a cotranscriptional binding and switching mechanism. Specifically, the short lifetime (~4 sec) of the partially bound state allows rapid sampling of the anticodon sequence and ensures that the tRNA remains bound until synthesis of the discriminator domain occurs. On the other hand, the fast transition into the fully bound state when the uncharged NCCA end is present, effectively locks the T-box riboswitch into the antiterminator conformation. Finally, the fully bound state is very stable to allow for transcription readthrough beyond the T-box regulatory sequence.
Translation-regulating T-box riboswitches achieve gene regulation via tRNA-induced conformational changes from a sequestrator conformation, which occludes the Ribosome Binding Site (RBS) sequence, to an antisequestrator conformation, which makes the RBS accessible to the ribosome (Fig. 1b)21. While the core structural domains for anticodon recognition and aminoacylation discrimination are highly similar between the translation and transcription-regulating T-box riboswitches, translation-regulating T-box riboswitches contain distinct structural elements14. Compared to their transcription-regulating counterparts, translation-regulating T-box riboswitches have a shorter Stem I. Stem I in the glyQS T-box riboswitch provides an additional contact with the elbow region of the tRNA that greatly facilitates the first binding step of the tRNA to the riboswitch13,15. The absence of this critical interaction in translation-regulating T-box riboswitches is partially compensated for by additional interactions provided by Stems II and IIA/B at the anticodon binding pocket6,14. However, it is unclear whether these structural differences lead to different binding kinetics in translation-regulating T-box riboswitches. In addition, transcription-regulating riboswitches are limited to cotranscriptional regulation since a decision must be made before transcription of the coding region. In contrast, translation-regulating T-box riboswitches allow for the synthesis of the full transcript, opening the possibility that translation-regulating T-box riboswitches could perform several rounds of regulation throughout the lifetime of the mRNA. Thus, the two types of riboswitches may have evolved to have different structural dynamics that accommodate their functional differences22. Kinetic studies of the tRNA binding mechanisms and associated conformational dynamics of translational T-box riboswitches are necessary to elucidate these differences. While recent kinetic characterizations of translation-regulating T-box riboswitches lacking the discriminator domain have provided mechanistic insights into the conformational landscape of the decoding module and its response to tRNA binding23,24, it remains unknown how conformational dynamics at the decoding module communicate with the rest of the T-box to coordinate their actions, information which is essential to understand the function of these riboswitches.
Here, the interactions between tRNA and a translational T-box riboswitch are investigated using a combination of single-molecule Förster resonance energy transfer (smFRET), ensemble binding assays, and mutagenesis. The use of a translational T-box riboswitch construct including both decoding and discriminator modules enabled kinetic characterization of the full binding process. The results reveal a two-step tRNA binding process with overall similarities to the binding process observed for transcription-regulating T-box riboswitches, but with distinct kinetic features. Particularly, the data presented here supports a conformational selection model for binding of the tRNA NCCA end. In addition, our results highlight critical structural elements at the decoding and discriminator modules that kinetically facilitate tRNA binding efficiency and specificity. This work provides a comprehensive picture of the tRNA binding kinetics of a translational T-box riboswitch, illuminating important biophysical differences between translation- and transcription-regulating T-box riboswitches.
Results
Preparation of fluorophore-labeled Mtb ileS T-box riboswitch
The Mycobacterium tuberculosis ileS T-box riboswitch (Mtb ileS T-box) was chosen as a model system to study translational T-box riboswitches due to the availability of a crystallographic structure of the complex, which encompasses a significant portion of the T-box riboswitch sequence14. The structural information served as a valuable reference to guide the experimental design. Initially, a construct was designed including a 15-nucleotide extension at the 5’ end of the in vitro transcribed T-box RNA, which allowed annealing to a modified DNA probe for immobilization. Purification of this transcript by urea-polyacrylamide gel electrophoresis (urea-PAGE), followed by direct Cy3 labeling at the 3’ end, and in vitro refolding in the presence of a biotinylated oligonucleotide resulted in no measurable tRNAIle binding. This was likely due to the presence of the 5’ extension, which was impeding in vitro refolding of the T-box transcript. To address this issue, a T-box construct was designed employing an alternative labeling method as follows. The 15-nucleotide extension used for immobilization was appended to the 3’ end of the transcript, downstream of the antisequestrator domain. This construct encompasses Stem I, Stem II, Stem II-A/B, the linker sequence, Stem III, and the antisequestrator (nucleotides 1- 166, Fig. 1a). However, it lacks the region spanning the RBS, thereby precluding the transition to the sequestrator conformation. During in vitro transcription, a DNA oligonucleotide complementary to the 3’ extension was added to the reaction. The DNA oligonucleotide carried the donor dye (Cy3) at its 3’ end and a biotin on the 5’ end. The labeled and biotinylated oligonucleotide allows for surface immobilization of the Mtb-ileS T-box via biotin-streptavidin interaction, and also places the donor dye in proximity to the 3’ terminus of the T-box construct for smFRET measurements (Fig. 1c) once hybridized (Fig. 2a). Positioning of the extension at the 3’ end of the riboswitch ensured that the decoding module would have an opportunity for vectorial folding during transcription, and that only full-length transcripts would anneal the oligonucleotide and be observed in smFRET imaging.
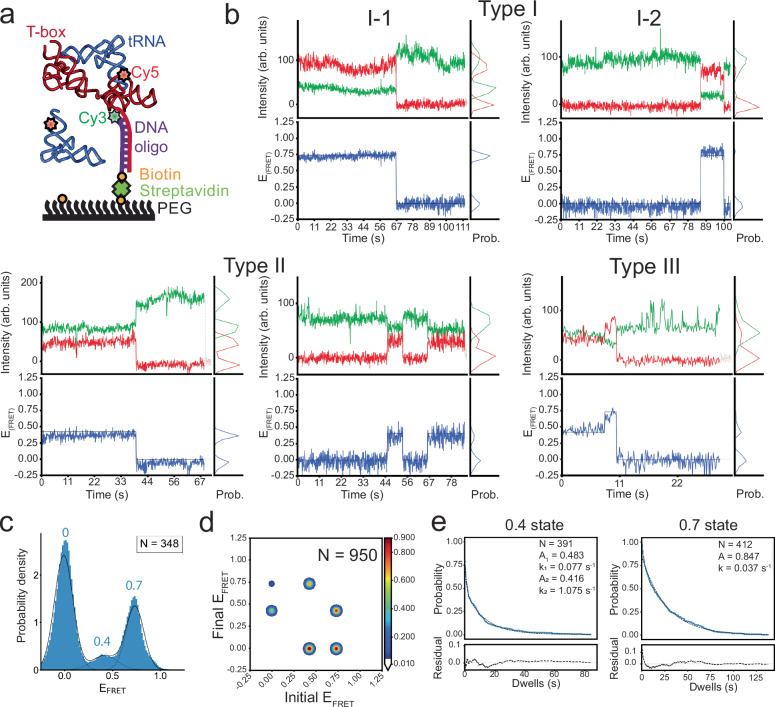
Fig. 2. smFRET experiments show binding of tRNAIle to the wild-type Mtb-ileS T-box riboswitch.
a Schematic diagram illustrating the smFRET experiment. The T-box was anchored to the surface via hybridization to a DNA oligonucleotide (purple) biotinylated on the 5’ end and Cy3-labeled at the 3’ end. The tRNAIle was labeled on the 5’ end with Cy5. Cartoon based on a similar cartoon from19. b Representative Cy3 (green) and Cy5 (red) fluorescence intensity (top) and smFRET trajectories (bottom) of the Mtb-ileS T-box riboswitch/tRNAIle. The smFRET efficiency was calculated as (ICy5/(ICy3-ICy5)). Three types of trajectories were observed: Type I trajectories only show FRET efficiency around 0.7, Type II trajectories only show FRET values around 0.4, and Type III trajectories sample the 0.4 and 0.7 states. c FRET efficiency histogram of the data. Dotted black lines show the individual populations obtained from the consensus HMM modeling, while the model’s population-weighted set of efficiency distributions is plotted with a solid black line. N reports the number of traces included in the histogram. d Transition density plot constructed using the idealized Viterbi paths modelled to the entire dataset using tMAVEN26. The contour level colors report the normalized counts. N reports the total number of transitions. e Dwell time survival plot for the 0.4 and 0.7 FRET states. The data were fitted using either a single (0.7 state) or double (0.4 state) exponential decay function. The number of events (N) used for the analysis and the fitting parameters are shown. Data are shown in blue and the fitting curves are shown in dotted black lines. The fit residuals are shown in the bottom plot. Detailed fitting results are reported in Supplementary Table VI. Source data are provided as a Source Data file.
The Mtb ileS T-box was purified directly from the transcription reaction by size exclusion chromatography (SEC) under conventional conditions25. This process yielded readily distinguishable fractions of both unfolded and folded Mtb-ileS T-box states (Supplementary Fig. 1a). The folded population of Mtb-ileS T-box molecules is consistent with their predicted apparent molecular weight, considering that the substantial negative charge of the molecule due to the phosphate backbone results in a larger hydration shell. The apparent molecular weight of an RNA is 3–5 times larger than a globular protein of comparable molecular weight and hence elutes from a SEC column significantly earlier25. SEC purification also facilitated direct comparison of the overall folding of the wild-type (WT) construct with engineered mutants, ensuring that any binding differences were not due to misfolding (Supplementary Fig. 1a). After transcription and SEC purification, the fraction corresponding to the fully transcribed and correctly folded Mtb ileS T-box riboswitch, already annealed to the biotinylated and Cy3 labeled DNA oligonucleotide, was used for smFRET imaging. The purified material was further analyzed by denaturing PAGE to confirm the integrity and purity of the Mtb ileS T-box for each species (Supplementary Fig. 1b). Additionally, the purified material was analyzed by SEC after one week of storage at the conditions used in this study to ensure the conformational homogeneity and stability of the samples (Supplementary Fig. 1c). The elution profiles of the samples after storage were nearly identical to the initial purification, indicating that the samples maintained their structural integrity during storage. In summary, the design with a 3’ extension, together with direct purification of the T-box construct from the transcription reaction via SEC avoided denaturation and refolding of the T-box riboswitch, which could disrupt its function. For intermolecular smFRET experiments with the cognate tRNA ligand, an acceptor fluorophore (Cy5) was conjugated at the 5’ end of the M. tuberculosis tRNAIle molecule (Methods). The purification of the tRNA was carried out by urea-PAGE, followed by refolding using standard procedures (Methods).
Biolayer Interferometry (BLI) binding assays were used to assess the effect of fluorescent labeling on tRNA binding to the Cy3-labelled Mtb ileS T-box (Supplementary Fig. 2a). For the BLI assays, the Mtb ileS T-box purified as described above was immobilized on the biosensor. tRNA association was measured in a solution containing uncharged tRNAIle. After measuring the tRNAIle association, the biosensor was immersed in a tRNAIle-free solution to enable tRNAIle dissociation detection. To fit the BLI data, three different binding models were considered: one-step binding, parallel one-step binding, and sequential two-step binding (Supplementary Fig. 2b). The binding and dissociation curves were fit most accurately by the two-step sequential binding model (Supplementary Fig. 2c). The apparent equilibrium dissociation constant (KD) was calculated using the two-step sequential binding model as described in the Supplementary Information section. The KD of Mtb ileS T-box binding to tRNAIle obtained in these assays agrees well with previously reported affinities for this T-box class21. KD with Cy5-labeled tRNAIle (0.89 µM) was increased by only 2 to 3-fold compared to unlabeled tRNAIle (0.42 µM), demonstrating that the introduction of a label at the 5’ end of tRNAIle resulted in only a moderate impact on the binding affinity (Supplementary Fig. 2d, Supplementary Table 1). This effect is milder than that previously reported for the B. subtilis glyQS T-box riboswitch19.
Uncharged tRNAIle stably binds to the Mtb ileS T-box
Time-lapsed single-molecule fluorescence images of Mtb ileS T-box were captured in the presence of 100 nM tRNAIle-Cy5 at equilibrium conditions (Fig. 2b). The distribution of FRET efficiency values resulting from tRNAIle-Cy5 binding displays two main populations, with mean values of 0.4 and 0.7 (Fig. 2c, Supplementary Table 2). Furthermore, the presence of a zero FRET state in the histograms can be attributed to both unbinding and acceptor fluorophore photobleaching events. The observed distribution is the result of three types of behaviors (Fig. 2b and Supplementary Table 3). 77.3 % of traces (Type I) sample only the 0.7 FRET state in addition to the zero FRET state. Among Type I traces, the majority of traces (91.8%, Type I-1) display a single, stable signal at 0.7 that undergoes single step photobleaching. A small population (8.2 %, Type I-2) shows transitions from zero to 0.7 FRET efficiency. 17.8% of traces (Type II) sample only the 0.4 state in addition to the zero state, either once or several times before photobleaching. Finally, a very small fraction of traces (Type III traces, 4.9%) sample both the 0.7 and 0.4 states.
In most cases, unless specified otherwise, Global Hidden Markov Modeling (HMM) was used to simultaneously model all trajectories of each particular experimental condition26. Transition frequencies (Supplementary Table 4), transition rates from an initial state to a final state (Supplementary Table 5), and state dwell times (Supplementary Table 6) were revealed by this HMM modeling approach. Throughout this work, the transition rates between two FRET states as described by transition matrices is emphasized (Supplementary Table 5). The lifetime of a state before transitioning out of that state —to any other states— from dwell time analysis is also determined (Supplementary Table 6). In addition, the lifetime of a given state was also estimated using the reciprocal of the sum of transition rates out of that state from the transition matrices (Supplementary Table 5). This calculation is largely consistent with the dwell time analyses in the instances hereby presented.
Type I traces predominately demonstrate transitions from the 0.7 state to the zero FRET state (22.7%, calculated as the number of transitions of this type over the total number of transition events), consistent with the fact that 71.0% of traces (Type I-1) show stable 0.7 FRET before transitioning into the zero state. In contrast, transitions from the zero to the 0.7 state are the least frequent (2.4%, from Type I-2 traces) (Supplementary Fig. 3a). The less frequent Type II traces also contribute to a large number of transition events from 0.4 to zero FRET (25.2%) and from zero to 0.4 FRET (12.7%) (Supplementary Fig. 3b), as a fraction of Type II traces sample the 0.4 state multiple times in each trajectory. 0.4 to zero FRET transitions are more frequent than the reverse transitions as the former also include the photobleaching step. Finally, Type III traces demonstrate inter-state transitions between 0.7 and 0.4 FRET states (20.2% for 0.7 to 0.4 transitions and 16.7% for 0.4 to 0.7 transitions) (Supplementary Fig. 3c, Fig. 2d, Supplementary Table 4).
According to the crystal structure14, there should be a distance of approximately 40 Å between the labeling positions at the 3’ end of the Mtb ileS T-box and the 5’ end of the tRNA (Fig. 1c). The 0.7 FRET value obtained from the measurements is within the predicted range of FRET values, assuming a Förster distance of 40−50 Å for Cy3-Cy5 FRET pair27. Hence, it is plausible to designate the 0.7 FRET state as the fully bound state of tRNAIle to the Mtb ileS T-box. The large population of Type I traces and 0.7 to zero transitions indicate that, preponderantly, the tRNAIle was fully bound to the T-box prior to data collection. This state persisted until the acceptor fluorophore photobleached, thus restricting the observed lifetime of the 0.7 state to 26.9 s (Fig. 2e and Supplementary Table 6). The characteristics of the 0.7 state are strikingly similar to those of the fully bound state of B. subtilis glyQS T-box riboswitch in complex with uncharged tRNAGly19. This similarity is expected due to the structural conservation between antisequestrator and antiterminator domains of translational and transcriptional T-boxes, respectively, in complex with their cognate tRNAs (Supplementary Fig. 3d)14,15.
While the nature of the 0.4 FRET state is not clear from the data above, there are more frequent transitions from the zero to the 0.4 state (12.7%) than from the zero to the 0.7 state (2.4%), suggesting that binding of tRNAIle is more likely to result in this unknown intermediate state (0.4 FRET) than the fully bound state (Fig. 2d, Supplementary Fig. 3a, b, Supplementary Table 4). The lifetime of the 0.4 FRET state is consistently estimated to be 7.0 s, based on a transition rate of 0.144 s−1 out of the 0.4 FRET state in the transition matrix (Supplementary Table 5), and 7.4 s from the dwell time analysis (Fig. 2e, Supplementary Table 6). It was noticed that the dwell time of the 0.4 FRET state is better described by a double-exponential decay, consistent with the presence of two transition pathways and potential populational heterogeneity (see sections below). It should be noted that the binding rate of tRNAIle to either the unknown intermediate or fully bound states from the unbound state is likely to be underestimated by these experiments, given that the majority of the population probed was already in the fully bound state during the data acquisition time window (Supplementary Fig. 8).
Anticodon recognition leads to a partially bound state of the tRNA
The nature of the 0.4 state cannot be deduced from the available structural data. However, it resembles a state observed in studies of the B. subtilis glyQS T-box riboswitch19. In the glyQS T-box, a 0.4 state corresponds to a partially bound state in which only the anticodon/specifier interactions have been established. To test whether the 0.4 FRET state in Mtb ileS T-box represents such partially bound state, two experiments were devised. In the first experiment, smFRET measurements were conducted with the Mtb ileS T-box using a tRNAIle variant lacking the terminal NCCA-3’ sequence (tRNAIle-ΔNCCA-Cy5). It was anticipated that tRNAIle-ΔNCCA-Cy5 would only be able to bind to the decoding module but not to the discriminator domain (Fig. 3a). Using this tRNA substrate, two apparent populations were initially identified in the FRET efficiency distribution: a 0.5 FRET state and a zero FRET state (Fig. 3b, Supplementary Table 2). The largely diminished 0.7 FRET state further supports the assignment of the 0.7 FRET to represent the fully bound state. Hence, the presence of the 0.5 FRET efficiency state was ascribed to a partially bound state in which no interactions with the NCCA sequence had occurred and only the tRNA anticodon region was bound. Symmetrical transitions between the two FRET states were observed (Fig. 3c). The transition rate from the zero state to the 0.5 state was 0.019 s−1, while the transition rate from the 0.5 state to the zero state was 0.053 s−1 (Supplementary Table 5), corresponding to an average lifetime of 18.8 s before tRNA dissociation, which is consistent with a lifetime of 17.3 s determined by dwell time analysis (Fig. 3e, Supplementary Table 6). Again, it was noticed that the dwell time histogram of the 0.5 FRET state is better described by a double exponential decay, similar to the case of the 0.4 FRET state when using tRNAIle (Fig. 3d, e).
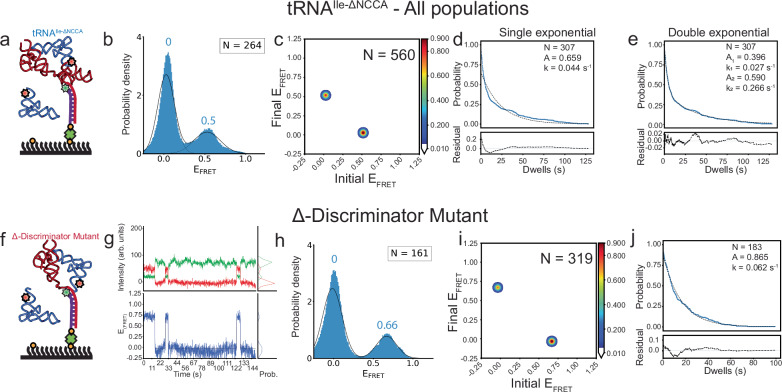
Fig. 3. smFRET experiments show consequences of the absence of the NCCA/specifier interactions.
a Schematic diagram illustrating the smFRET experiment with the tRNAIle-ΔNCCA-Cy5 construct. Cartoon based on a similar cartoon from19. b FRET efficiency histogram for tRNAIle-ΔNCCA-Cy5 mutant. Fitted populations using consensus HMM modeling are shown as in Fig. 2c. c Transition density plot for tRNAIle-ΔNCCA-Cy5 mutant for all data constructed using the idealized Viterbi paths modelled with tMAVEN26, plotted the same way as described in Fig. 2d. d, e Dwell time survival plots for the 0.5 FRET state. The data were fitted using either a single (left) or double (right) exponential decay function. The double exponential decay function better fits the data. Data and fitting results are presented as described in Fig. 2e. f Schematic diagram illustrating the smFRET experiment with the Mtb-ileS T-box riboswitch Δ-Discriminator Mutant and tRNAIle. Cartoon based on a similar cartoon from19. g Representative Cy3 (green) and Cy5 (red) fluorescence intensity (top) and smFRET trajectories (blue, bottom) of the Δ-Discriminator Mutant and tRNAIle complex. h FRET efficiency histogram of the data with the Δ-Discriminator Mutant. Fitted populations using consensus HMM modeling are plotted as in Fig. 2c. i Transition density plot constructed using the idealized Viterbi paths, as described in Fig. 2d. j Dwell time survival plot and fitting results using a single exponential decay function. Data and fitting results are presented as described in Fig. 2e. N in each plot reports the number of traces or events included in the analysis. Source data are provided as a Source Data file.
In a second experiment, a mutant of the Mtb ileS T-box which prevents tRNAIle interactions through the NCCA sequence was constructed by eliminating the discriminator domain (Δ-Discriminator Mutant, nucleotides 1-93). The SEC elution profile of the Δ-Discriminator Mutant resembles that of the wild-type Mtb ileS T-box, with the exception that the peak of interest eluted at a later volume (Supplementary Fig. 1). This shift was anticipated due to the lower molecular weight of the construct. The deletion of the discriminator domain had an impact on the ability of the T-box to bind to tRNAIle, as demonstrated by BLI measurements. The KD increased from 0.42 µM for the wild type, to 4.03 µM for the mutant (Supplementary Table 1), corresponding to a ~10-fold reduction in the binding affinity. smFRET analysis on this mutant using tRNAIle-Cy5 (Fig. 3f) yielded two distinct populations centered at zero and 0.66 FRET respectively (Fig. 3g, h, Supplementary Table 2). Symmetrical transitions between the two FRET states were observed (Fig. 3i and Supplementary Fig. 4f). The transition rate from the zero to the 0.66 state was 0.0134 s−1, whereas the transition rate from the 0.66 state to the zero state was 0.0568 s−1. This corresponds to a 0.66 FRET state average lifetime of 15.5 seconds, consistent with 16.1 seconds as calculated by dwell time analysis (Supplementary Tables 5 and 6). The lifetime of the 0.66 FRET state resembles the lifetime of the 0.5 FRET state in the case of WT T-box with tRNAIle-ΔNCCA. However, interestingly, the dwell time of the 0.66 FRET state can be well described with a single-exponential decay (Fig. 3j).
For the Δ-Discriminator Mutant, the estimated distance between the truncation position and the 5’ end of the tRNA is around 29 Å (Fig. 1c). The observed higher FRET efficiency at 0.66 would be expected for a Förster distance of 40 to 50 Å27. This effect is likely caused by a collapse of the single-stranded linker region in the absence of the discriminator domain, with which it normally establishes contacts (Fig. 1c). However, as the transition rates between the zero and 0.5 states in the WT T-box with tRNAIle-ΔNCCA-Cy5 and between the zero and 0.66 states in the Δ-Discriminator T-box mutant with tRNAIle-Cy5 are similar (Supplementary Table 5), both the 0.5 and the 0.66 FRET states were interpreted to correspond to the same partially bound state. The absence of the NCCA sequence may allow the decoding module to shift the 5’ end of tRNAIle-ΔNCCA-Cy5 closer to the discriminator domain; leading to an apparently higher FRET efficiency state compared to the 0.4 state observed in the case of WT T-box with tRNAIle--Cy5. Taken together, the results presented so far suggest a two-step binding model in which the interactions between the tRNA NCCA and anticodon with the T-box do not happen simultaneously.
tRNA anticodon binding precedes NCCA recognition in Mtb ileS T-box
The above experiments demonstrate that the Mtb ileS T-box can identify the anticodon region of tRNAIle in the absence of the NCCA sequence. Furthermore, two previous studies with translational T-boxes missing the discriminator domain23,24 corroborate this finding. Nevertheless, to determine if interactions between Mtb ileS T-box and the NCCA sequence are possible without anticodon recognition, additional experiments were pursued. First, tRNATrp-Cy5 was introduced into a flow chamber containing pre-immobilized Mtb ileS T-box, but no binding of tRNATrp-Cy5 was detected. Binding by BLI was also not detectable. Second, an additional T-box construct was designed in which the specifier region, responsible for identifying the cognate anticodon, was mutated to recognize tRNATrp instead (AUC mutated to UGG), hereafter referred to as the Specifier Mutant. The Specifier Mutant fraction eluted slightly earlier than the WT Mtb ileS T-box in SEC analysis (Supplementary Fig. 1), likely due to a potential contribution of the specifier sequence in maintaining the native conformational landscape of the decoding module24. No binding was observed when tRNAIle-Cy5 was added to a flow-chamber pre-immobilized with the Specifier Mutant. Consistent with the smFRET measurements, BLI assays measured the KD of tRNAIle binding to the Specifier Mutant to be 120.4 µM, three orders of magnitude larger than the KD of WT Mtb ileS T-box (0.42 µM) (Supplementary Table 1), and well above the expected physiological concentrations of tRNA28.
Overall, the results of these experiments confirm that tRNA binding requires the specifier region to recognize the tRNA anticodon before interactions with the NCCA sequence can be established. The experiments also suggest that the translation-regulating Mtb ileS T-box follows the same sequential binding mechanism as the transcription-regulating B. subtilis glyQS T-box.
The partially bound state in Mtb ileS T-box exhibits conformational heterogeneity
While Mtb ileS T-box follows the same sequential binding mechanism as B. subtilis glyQS T-box, the presence of conformational heterogeneity in the partially bound state of Mtb ileS T-box complex with tRNAIle-ΔNCCA-Cy5 was observed, as indicated by the double exponential fitting of the dwell time data of the 0.5 FRET state (Fig. 3e). This conformational heterogeneity is absent in the B. subtilis glyQS T-box case19. Specifically, two distinct types of traces were observed upon tRNAIle-ΔNCCA-Cy5 binding to the WT Mtb ileS T-box: a population where the 0.5 FRET state is briefly sampled, either once or multiple times (72.7 %) (Fig. 4a and Supplementary Tables 2 and 3), and a second population where the 0.5 FRET state is sampled constantly until photobleaching (27.3 %) (Fig. 4e and Supplementary Tables 2 and 3).
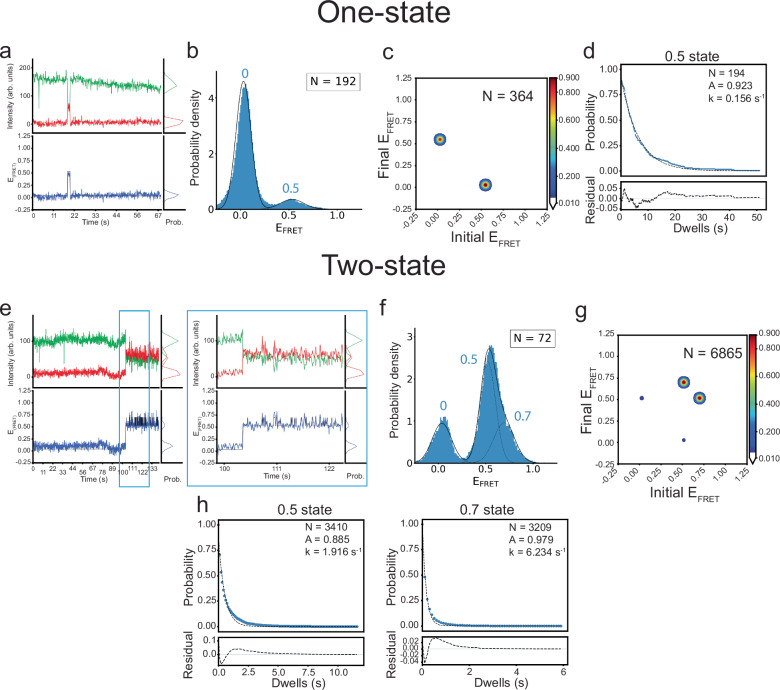
Fig. 4. smFRET experiments with tRNAIle-ΔNCCA reveal conformational sampling in the partially bound state.
a Representative Cy3 (green) and Cy5 (red) fluorescence intensity (top) and smFRET trajectories (blue, bottom) for the One-state population of the Mtb-ileS T-box riboswitch/ tRNAIle-ΔNCCA-Cy5 complex. b FRET efficiency histogram of the One-state population, shown as in Fig. 2c. c Transition density plot for the One-state population constructed using the idealized Viterbi paths, plotted the same way as described in Fig. 2d. d Dwell time survival plots for the 0.5 state in the One-state population, fitted using a single exponential decay function. Data and fitting results are presented as described in Fig. 2e. e Representative fluorescence intensity and smFRET trajectory for the Two-state population of the Mtb-ileS T-box riboswitch/ tRNAIle-ΔNCCA-Cy5 complex. A zoom of a region framed by cyan boxes is displayed on the right, which shows rapid transitions between the 0.5 and 0.7 states. f FRET efficiency histogram of the Two-state population, shown as in Fig. 2c. g Transition density plot constructed using the idealized Viterbi paths for the Two-state population, plotted the same way as described in Fig. 2d. h Dwell time survival plots for the 0.5 and 0.7 states fitted a single exponential decay function. Data and fitting results are presented as described in Fig. 2e. N in each plot reports the number of traces or events included in the analysis. Source data are provided as a Source Data file.
In light of this finding, the two subpopulations were further analyzed separately. For the subpopulation that was briefly sampling the 0.5 state (Fig. 4a, b), transitions were identified via composite HMM, i.e., where individual trajectories are modeled separately26. This approach is more sensitive to FRET states sampled by a few data points per trajectory. Composite HMM analysis revealed homogeneous transitions between 0.5 state and zero state in this population (Fig. 4c, Supplementary Fig. 4c). The dwell time of the 0.5 FRET state in this subpopulation can be fit with a single-exponential decay function, yielding an average lifetime of 6.4 s (Fig. 4d and Supplementary Table 6). This subpopulation, named One-state population, was reminiscent of the case of tRNAGly-ΔNCCA-Cy5 binding to the WT B. subtilis glyQS T-box, in which the dwell time of the partially bound state was described well with single exponential decay function with a similar lifetime19.
Remarkably, molecules that apparently spent a longer time in the 0.5 FRET state actually displayed rapid transitions to a 0.7 FRET state, the latter reminiscent of the FRET efficiency observed in the fully bound state (Fig. 4e, f). In this subpopulation, named Two-state population, the 0.7 FRET state was very transiently sampled by 1-2 time points (<200 ms), which was not initially captured by the global HMM algorithm. In order to model accurately this fast transition behavior, Gaussian mixture modelling (GMM)26 was used (see Methods). Since the short-lived 0.7 FRET state is consistently sampled across the dataset of this subpopulation, this approach, which assumes that all the data points originate from a mixture of a finite number of Gaussian distributions with unknown parameters, is capable of detecting it. The GMM analysis revealed that transition events occurred predominantly between the 0.7 and 0.5 FRET states (92.2% of total transitions) (Fig. 4g and Supplementary Table 4). These FRET states displayed short lifetimes, 0.52 s and 0.16 s for the 0.5 and 0.7 FRET states, respectively (Fig. 4h and Supplementary Table 6).
In summary, these results suggest that, upon anticodon recognition, the Mtb ileS T-box exhibits fast conformational dynamics, unlike the B. subtilis glyQS T-box. In the absence of the 3’ terminal NCCA sequence in tRNAIle, some Mtb ileS T-box molecules undergo futile attempts to achieve the 0.7 FRET state, conformationally resembling the fully bound state. In other words, the 0.7 FRET state can be transiently sampled without establishment of the NCCA-discriminator interaction. The average lifetime of the 0.5 FRET state is 4.8 s considering both the One-state and Two-state populations (Supplementary Figure 8), corresponding to a 0.21 s−1 transition rate out of 0.5 FRET to any other state. This rate is consistent with the total transition rate out of 0.4 FRET (0.14 s−1) in the presence of WT tRNAIle, further confirming that the 0.5 FRET state in the case of tRNAIle-ΔNCCA and the 0.4 FRET state in the case of tRNAIle both represent the partially bound state. Finally, the 0.7 FRET state in the Two-state population is unstable, with a lifetime of 0.16 s. Establishment of the NCCA-discriminator interaction in the presence of uncharged tRNAIle stabilizes the 0.7 FRET by at least 160-fold, leading to a stable fully bound state (Supplementary Fig. 8).
tRNA binding alters the conformational equilibrium of Mtb ileS T-box
To study intramolecular conformational changes of the Mtb ileS T-box upon tRNA binding, a new construct was designed, termed IntraFRET. This construct has a 5’ extension that enables it to bind a 5’-end Cy5-labeled DNA oligonucleotide while retaining the 3’ extension used for immobilization and Cy3 labeling (Fig. 5a). This construct enables probing the distance changes between the 5’ and 3’ ends of the Mtb ileS T-box. The SEC elution profile of the IntraFRET molecule exhibited greater complexity as compared to the T-box construct with only the 3’ oligonucleotide attached. This was evident by the appearance of several peaks in the chromatogram (Supplementary Fig. 1). By measuring both Cy3 and Cy5 absorbance signals from each SEC fraction, only one of the peaks was found to contain both signals. This fraction was used for smFRET analysis. This peak, corresponding to the doubly labeled T-box, eluted earlier compared to T-box with only the 3’ oligonucleotide due to its higher molecular weight.
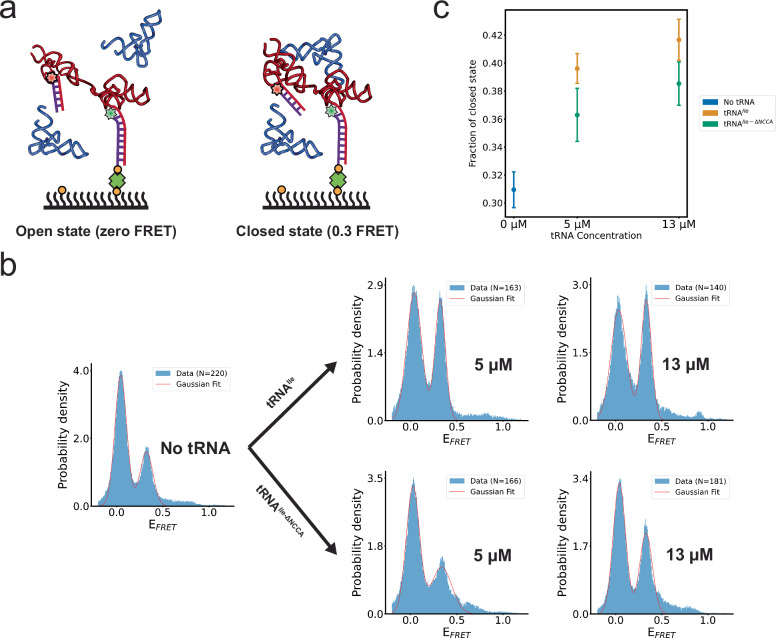
Fig. 5. The Mtb-ileS T-box riboswitch samples open and closed conformations.
a Schematic diagram illustrating the smFRET experiment using an intramolecularly labeled Mtb-ileS T-box construct. Two FRET states were observed, assigned to an open (zero FRET, left) and a closed (0.3 FRET, right) state. Cartoon based on a similar cartoon from19. b FRET efficiency histograms of intramolecularly labeled Mtb-ileS T-box in the absence of tRNA (left), and in the presence of different concentrations of tRNAIle (right, top) and tRNAIle-ΔNCCA (right, bottom). The orange line corresponds to the fits of a Gaussian function to each peak. N reports the number of traces in each histogram. Only the first 40 frames from FRET trajectories are included in the histogram to avoid including data after photobleaching of the fluorophores. c Changes in the fraction of the closed conformation as a function of tRNAIle or tRNAIle-ΔNCCA concentration. The fraction of the closed state was calculated by 1) fitting Gaussian curves to each peak, which provided an estimate of the standard deviation, 2) using these fits to obtain the area under each peak by integration, and 3) calculating the fraction by dividing the area corresponding to the 0.3 FRET state by the sum of the areas corresponding to the zero and the 0.3 FRET states in (b). The error of each fraction was estimated by standard error propagation using the fitting uncertainties as described in the Methods section and correspond to the error bars in the figure. Source data are provided as a Source Data file.
For smFRET measurements, the IntraFRET molecules were immobilized to the imaging slide and time-lapsed fluorescence images were recorded with and without tRNA. To distinguish between molecules in a potential zero FRET state from those with only a Cy3 label, five frames were recorded using direct Cy5 excitation before time-lapsed FRET imaging. In the absence of tRNA, the IntraFRET sample showed two distinct FRET states centered at zero and 0.3, respectively (Fig. 5b). We assigned these states as open (zero FRET) and closed (0.3 FRET) states. The closed state is consistent with the expected distance of around 68 Å between the 3’ and 5’ ends of the T-box in complex with tRNAIle (Fig. 1c)27. Addition of tRNAIle caused a dose-dependent increase in the proportion of the closed state (Fig. 5b, c). Remarkably, addition of tRNAIle-ΔNCCA similarly increased the proportion of the 0.3 FRET efficiency state in a dose-dependent manner, albeit with a reduced efficiency (Fig. 5 b, c). These findings suggest that in the absence of the tRNA ligand, the Mtb ileS T-box samples both the open and closed conformations autonomously. The experiments with tRNAIle-ΔNCCA indicate that anticodon recognition alone increases the population of the closed state. Interactions between the NCCA end of the tRNA and the discriminator of the T-box further enhance the stability of the closed state. These observations are in stark contrast to equivalent experiments with the glyQS T-box, where an intramolecular conformational change is exclusively associated with the binding of both the anticodon and NCCA19.
Stem IIA/B-linker interactions allosterically modulate tRNA binding
The linker region in Mtb ileS T-box (nucleotides 79–92) connects the decoding module and the discriminator domain (Fig. 1a). This linker region is predominantly stabilized by extensive interactions with Stem IIA/B, resulting in limited flexibility. Specifically, nucleotide A81 at the 5’ end of the linker forms an A-minor interaction with the G68-C77 pair in Stem IIA/B. Then, the 5′-half of the linker rests against the interface between Stems II and IIA/B and forms a pseudoknot structure by base pairing with three of the nucleotides in the loop of Stem IIA/B (Fig. 1a). Hence, it has been suggested that this pseudoknot imposes the constraints required to position the discriminator domain in proximity to the tRNA acceptor arm14. Disruption of the aforementioned A-minor interaction (A81G) was shown to significantly decrease the functional efficiency of the B. subtilis tyrS T-box14. To investigate how the Stem II-A/B-linker interactions modulate the tRNA binding kinetics, an Mtb ileS T-box mutant that included the A81G substitution as well as mutations expected to disrupt the three critical base pairs with Stem IIA/B was generated (Fig. 6a). This mutant is hereafter referred to as the “Linker Mutant”. The SEC elution profile and KD (measured by BLI) of the Linker Mutant were similar to those of the WT Mtb ileS T-box (Supplementary Figs. 1, 2d Supplementary Table 1).
Fig. 6. Linker Mutations destabilize the Mtb-ileS T-box/tRNA complex.
a Left, schematic diagram of the Mtb-ileS T-box with the linker region shown inside the black box. Right, schematic diagrams showing the linker region (top) and the mutations introduced in the Linker Mutant (bottom). The region affected is circled in green in the mutant diagram. b FRET efficiency histogram of the data with the Linker Mutant. Fitted populations using consensus HMM modeling are plotted as in Fig. 2c. c Transition density plot constructed using the idealized Viterbi paths, as described in Fig. 2d. d, e Dwell time survival plots and fitting result using a single exponential decay function for the 0.4 FRET state (d) and 0.7 FRET state (e). Data and fitting results are presented as described in Fig. 2e. N in each plot reports the number of traces or events included in the analysis. Source data are provided as a Source Data file.
The FRET values resulting from the binding of tRNAIle-Cy5 to the Linker Mutant form two populations, with mean values of 0.4 and 0.7 (Fig. 6b, Supplementary Table 2), similar to the ones observed using the WT Mtb ileS T-box. However, the 0.7 FRET state is much less populated in this mutant compared to the WT T-box. With the same categorization as the one utilized for the WT construct (Supplementary Fig. 5a), Type I, II, and III traces comprise 20.3%, 46.19%, and 33.5% of the total traces, respectively (Supplementary Table 3). The reduced fraction of Type I traces, and the corresponding increase in the fractions of Type II and III traces suggest that the stability of the fully bound state is compromised in this T-box mutant.
The transitions between 0.7 and 0.4 FRET states in both directions are most frequent, accounting for 63.4% of all transitions (Fig. 6c and Supplementary Table 4). The transition rates between the 0.7 and 0.4 states are 8- to 15-fold faster in the Linker Mutant compared to those in the WT construct (Supplementary Table 5). The transitions between the 0.4 and the zero FRET states contribute to 29.4% of all transitions (Fig. 6c, Supplementary Fig. 5b and Supplementary Table 4), with the rates around 2- to 4-fold faster compared to the WT construct (Supplementary Table 5). Similar to the WT construct, transitions between zero and 0.7 FRET states were the least frequent, accounting for only 7.3% of the total (Fig. 6c, Supplementary Fig. 5b and Supplementary Table 4). Consistent with increased transition rates between the states, the average lifetime of the 0.4 state was 1.3 s, and the lifetime of the 0.7 state was 3.0 s, a ~ 6-fold and ~10-fold decrease compared to the corresponding state in the WT construct, respectively (Figs. 2e, 6d, e and Supplementary Table 6). These results demonstrate the importance of the A-minor interaction and pseudoknot structure in maintaining the stability of both the partially and fully bound states. Overall, these results confirm the significance of the Mtb ileS T-box linker region in tRNA binding and point to allosteric effects on tRNA recognition, since the Stem II A/B-linker interactions are not directly involved in tRNA binding.
The conserved RAG sequence confers specificity and stability to NCCA-Discriminator interactions
Prior structural data had provided insights into aminoacylation state sensing by the T-box discriminator domain. Specifically, a conserved consensus sequence RAG (R = purine) plays a crucial role in this process14. In the Mtb-ileS T-box, the RAG sequence starts with G94, which interacts with nucleotide A77 at the 3’ of the tRNA and with U125 in the antisequestrator. The second nucleotide in the sequence, A95, is crucial for continuous stacking between T-box nucleotides A129 and C131. Lastly, G96 binds C131 in the antisequestrator14. These interactions provide the specificity required to bind an uncharged tRNA containing a terminal adenosine with intact 2′ and 3′ hydroxyl groups. This high level of specificity renders the NCCA binding pocket incompatible with any other chemical moieties, such as an attached amino acid. Indeed, single nucleotide substitutions in the RAG sequence (R94U, A95G, and G96C) significantly decrease the induction of the B. subtilis tyrS T-box14. To investigate how this RAG sequence contributes kinetically to NCCA recognition, a mutant, termed “RAG mutant,” containing substitutions G94C, A95G, and G96C (Fig. 7a) was designed. The RAG mutant has a SEC elution profile that closely resembles that of the WT construct (Supplementary Fig. 1) and exhibits a moderate decrease in binding affinity to tRNAIle, with a KD of 1.2 µM, compared to 0.42 µM in the WT construct (Supplementary Table 1).
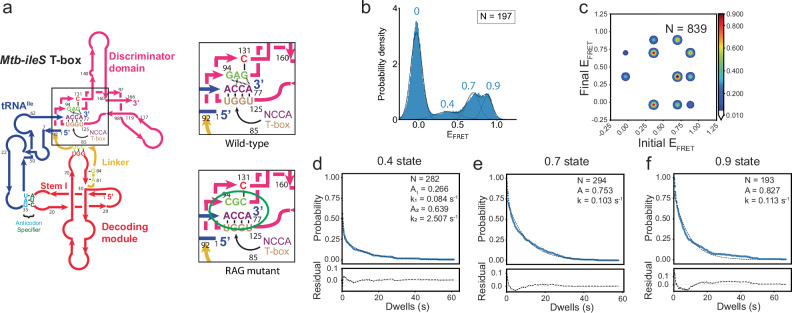
Fig. 7. The RAG sequence is important for aminoacylation state recognition.
a Left: Schematic diagram of the Mtb-ileS T-box with the NCAA/T-box and RAG sequence regions shown inside the black box. Right, schematic diagrams showing the RAG sequence region (GAG) (top) and the mutations introduced in the RAG Sequence mutant (bottom). The interactions affected are circled in green in the mutant diagram. b FRET efficiency histogram of the data with the Linker Mutant. Fitted populations using consensus HMM modeling are plotted as in Fig. 2c. c Transition density plot constructed using the idealized Viterbi paths, as described in Fig. 2d. d–f Dwell time survival plots and fitting result using a single or double exponential decay function for the (d) 0.4 FRET state, (e) 0.7 FRET state and the (f) 0.9 FRET state. Data and fitting results are presented as described in Fig. 2e. N in each plot reports the number of traces or events included in the analysis. Source data are provided as a Source Data file.
Interestingly, smFRET measurements using the RAG mutant and tRNAIle-Cy5 revealed a new FRET state centered at 0.9, in addition to the 0.4 and 0.7 states (Fig. 7b, Supplementary Table 2). Approximately 82.2% of the traces (Type I, II and IV) probe only one FRET state (0.7, 0.4, or 0.9, respectively) either once or a few times before photobleaching, while the remaining 17.8% of traces exhibit transitions between two or three states collectively (Supplementary Fig. 6a). Specifically, the Type II traces, representing traces only sampling of the 0.4 state, represent 17.8% of the data, similar to the WT construct. Traces sampling only the 0.7 (Type I) and only the 0.9 (Type IV) FRET states add up to 64.4%, close to the fraction of the Type I traces in the WT T-box (Supplementary Fig. 6a, b and Supplementary Table 3). This suggests that the fully bound state in the RAG mutant has two conformations corresponding to the 0.7 and 0.9 FRET states respectively. While the 0.7 FRET state corresponds to the same conformation of fully bound state observed in the WT construct, the 0.9 FRET state corresponds a new state only observed in the RAG mutant complex, hereafter referred to as the pseudo fully bound state.
The RAG mutant showed a similar frequency of transitions between 0.7 and 0.4 FRET states compared to the WT complex, albeit with two to three times faster transition rates (Supplementary Tables 4 and 5). In addition, transitions also occurred between the 0.7 and 0.9 FRET states with similar frequency as the 0.7 and 0.4 transitions, suggesting that the RAG mutant complex exhibits additional conformational dynamics between the fully bound state and the pseudo fully bound state. Finally, direct transitions between 0.4 and the 0.9 FRET states were also observed with less frequency (Fig. 7c and Supplementary Table 4). Dwell time analysis determined the average lifetime of the 0.4 state to be 3.8 s (Fig. 7d and Supplementary Table 6), reduced by ~2-fold compared to the 0.4 state of the WT construct (Fig. 2e and Supplementary Table 6), consistent with a faster transition rate from 0.4 FRET to zero FRET in the RAG mutant. The average lifetime of the 0.7 state is 9.7 s (Fig. 7e), reduced by ~2-fold compared to the lifetime of the 0.7 state in WT T-box. The lifetime of the 0.9 state is 8.8 s (Fig. 7f and Supplementary Table 6), comparable to the lifetime of the 0.7 state for this mutant. The lifetime comparison between the RAG mutant and WT construct for each FRET state indicates that: 1) the fully bound states still represent a more energetically stable conformation than the partially bound state, and 2) the mutations make the NCCA-discriminator interactions less structurally constrained by generating an energetically similar pseudo fully bound state. These findings support the notion that the conserved RAG sequence plays a crucial role in regulating the ability of the discriminator domain to sense aminoacylation, likely by restricting potential degrees of freedom of the bound ligand.
Stem II integrity is essential for proper folding of Mtb ileS T-box
In an attempt to investigate its dynamics and involvement in tRNAIle binding, Stem II was systematically shortened to generate three additional constructs, termed Stem II mutants A, B, and C (Supplementary Fig. 7). Amongst the three, only Stem II mutant A produced a distinct peak in the SEC chromatogram (Supplementary Fig. 1). These findings suggest that Stem II mutants B and C are likely to fold into defective conformations. Stem II mutant A did not exhibit any measurable tRNAIle binding, as measured by either smFRET or BLI, hence precluding further study. These results emphasize the importance of the integrity of the decoding module in binding tRNAIle. It was shown in recent single-molecule and Small Angle X-ray Scattering (SAXS) studies of the decoding module that the formation of the anticodon binding groove depends on the conformational dynamics between Stems I, II, and the Stem IIA/B pseudoknot23,24. The mutants presented hereby support that preserving the integrity of all stems is essential for the decoding module to maintain its natural dynamic ensemble. These results highlight the cooperative nature of the conformational landscape of Mtb ileS T-box.
Discussion
There has been a surge in interest in the conformational dynamics and tRNA ligand binding mechanisms of translational T-box riboswitches. The folding landscape of the decoding module (containing Stems I, II, and IIA/B) of the Nocardia farcinica ileS T-box riboswitch and its coupling to tRNA binding have been explored in recent studies23,24. Collectively, these studies showed that the decoding module of a translation-regulating T-box riboswitch exhibits a wide distribution of conformations, which undergo slow interconversion. tRNA binds directly to all conformers, each with unique kinetics. Binding promptly induces rapid rearrangement of the structural elements forming the decoding module, leading to a final, binding competent conformation. Furthermore, following tRNA dissociation, the decoding module returns to its original conformation prior to binding24. In addition, it has been shown that Mg2+ plays an important role in the global folding of the decoding module23. Here, using a larger construct that includes the discriminator domain, the binding kinetics of tRNAIle to both the decoding and discriminator domains of Mtb ileS T-box riboswitch are revealed, significantly expanding our understanding of the tRNA binding mechanism that leads to genetic switching. This work illuminates how the interplay of structural elements in T-box riboswitches leads to unique conformational dynamics in different T-box riboswitches with different roles in biological regulation.
By using smFRET characterization with the same fluorophore pair at equivalent positions in the T-box and tRNA ligand as in our previous studies on the B. subtilis glyQS T-box19, the current study allows comparison of tRNA binding mechanisms between representative members of transcription- and translation-regulating T-box riboswitches. The data supports a similar two-step binding mechanism for the Mtb ileS T-box and B. subtilis glyQS T-box riboswitches, in which anticodon recognition by the decoding module occurs first, leading to the formation of a partially bound state, followed by binding of the tRNA NCCA end to the discriminator module to form the fully bound state (Fig. 8 and Supplementary Fig. 8). In both cases, the fully bound states are very stable, with a lifetime of at least 20 s, to secure sufficient time for transcriptional or translational outputs.
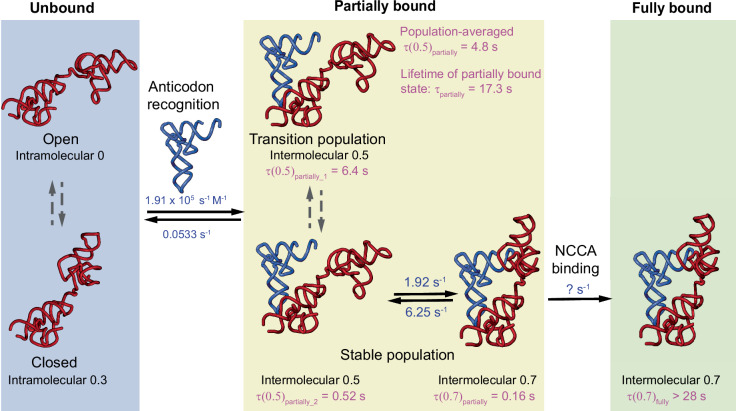
Fig. 8. Proposed kinetic model for the binding process of the Mtb-ileS T-box to tRNA.
In the absence of tRNA, the T-box samples both an open and a closed conformation (left, blue background). In the presence of tRNA, the T-box binds tRNA through a two-step process where interactions with the anticodon are established first, forming a partially bound state (middle, yellow background). The partially bound state exhibits conformational heterogeneity revealed by the WT T-box/ tRNAIle-ΔNCCA complex. In the One-state subpopulation (middle, top), tRNAIle-ΔNCCA transiently binds, leading to a 0.5 FRET state, with a 6.4 s lifetime (τ(0.5)partially_1) before dissociating. In the Two-state subpopulation (middle, bottom), tRNAIle-ΔNCCA remains stably bound, and rapidly fluctuates between a 0.5 FRET state (with a lifetime of τ(0.5)partially_2) and a 0.7 FRET state (with a lifetime of τ(0.7)partially). The 0.7 FRET state in the partially bound state resembles the 0.7 FRET state in the fully bound state (right, green background), but with a much shorter lifetime due to the absence of the NCCA-discriminator interactions. In the presence of an intact, uncharged NCCA end, the NCCA end of tRNA engages with the discriminator domain, leading to a long-lived fully bound state (right, green background, τ(0.7)fully). Rate constants (blue) are shown for the different binding states. Rate constants for the binding and dissociation during anticodon recognition step and for the conformational fluctuations within the Two-state subpopulation of the partially bound state are determined from the experiments using WT T-box riboswitch and tRNAIle-ΔNCCA at a tRNA concentration of 100 nM. The lifetimes of all FRET states are shown in pink and are calculated from dwell time analysis (Supplementary Table 6). The lifetimes of different FRET states in the partially bound state are determined from the experiments using WT T-box riboswitch and tRNAIle-ΔNCCA. The lifetime of the fully bound state is determined from the experiments using WT T-box riboswitch and tRNAIle. A detailed explanation of how the rates and lifetimes were assigned is given in Supplementary Fig. 8.
Besides the general resemblance regarding the two major binding states, the results reveal unique conformational heterogeneity and flexibility in the Mtb ileS T-box riboswitch. The first level of conformational flexibility is observed in the apo state of Mtb ileS T-box riboswitch, evocative of the complex conformational landscape of the decoding domain observed in previous smFRET studies23,24. In contrast to the B. subtilis glyQS T-box, in which the intra-T-box conformational change involves an inward movement of the discriminator module relative to the decoding domain accompanied by the establishment of NCCA-discriminator interactions19, the Mtb ileS T-box displays putatively open (zero FRET) and closed (0.3 state FRET) conformations in the absence of tRNA ligand (Figs. 5 and 8). Binding of either tRNAIle or tRNAIle-ΔNCCA can shift the equilibrium to the closed state from the open state, with tRNAIle being more effective than tRNAIle-ΔNCCA. However, no reversible transitions between the zero and 0.3 FRET states were observed, suggesting that the transition rate between the open and closed conformations may be slower than the imaging time window (~30 s). Therefore, it is not possible to determine the transition rates between the two conformations or conclude whether tRNA binds preferentially to one state over the other. Future 3-color smFRET experiments are needed to fully explore this mechanistic detail. Nevertheless, since binding of tRNA shifts the equilibrium to the closed conformation, it is most likely that the closed conformation corresponds to the conformation after tRNA binds.
The second level of conformational flexibility is observed in the partially bound state of the Mtb ileS T-box riboswitch, where only the anticodon-specifier but not the NCAA-discriminator interactions have been established. The partially bound state in the B. subtilis glyQS T-box exhibits a homogenous population, with a lifetime of ~3.6 s before the tRNA dissociates19. In contrast, the partially bound state of Mtb ileS T-box, represented by the experiments using tRNAIle-ΔNCCA, exhibits two populations (Figs. 4 and 8). Around 73% of the population only samples the 0.5 FRET state (One-state population) with a lifetime of ~6.3 s before the tRNA dissociates. The lifetime of the bound tRNA is noticeably longer in the remaining 27% of the population. In this population, the bound tRNA rapidly transitions between 0.5 and 0.7 FRET states (Two-state population), with the latter resembling the fully bound state when using an uncharged intact tRNA. However, without the possibility to establish the NCCA-discriminator interactions, the lifetime of the 0.7 FRET state in the Two-state population is two orders of magnitude shorter than the 0.7 FRET state representing the fully bound state with an uncharged intact tRNA. Considering both the One-state and Two-state populations, the average lifetime of the partially bound state (17.3 s) in the Mtb ileS T-box complex is 5-fold longer than that for the B. subtilis glyQS T-box. While the exact molecular architectures associated with the conformational heterogeneity observed in the Two-state population remains unclear, this observation suggests that the Mtb ileS T-box riboswitch can sample dynamically the fully bound conformation to attempt interactions between the tRNA NCCA end and the discriminator module, and that the presence of the uncharged tRNA NCCA end greatly stabilizes the fully bound conformation. While it is not possible to directly infer the interconversion between the two populations, it is reasonable to assume that the One-state population is able to transit into the Two-state population. This is due to the fact that the Two-state population represents only 27% of the partially bound state, whereas ultimately 71% of the population is able to reach the fully bound state in the presence of an intact uncharged tRNA. Taken together, these findings support the notion that while binding of the tRNA NCCA to the B. subtilis glyQS T-box may employ an induced-fit model, binding of tRNA NCCA to the Mtb ileS T-box is likely to use a conformational selection model.
Although the B. subtilis glyQS and Mtb ileS T-boxes constructs kinetically examined so far lack, respectively, the terminator and sequestrator sequences, the differences in the kinetic features of these two types of T-box riboswitches may be functionally linked to the temporal constraints of their regulation. In the case of B. subtilis glyQS T-box, the lower minimum free energy (MFE) of the terminator compared to the antiterminator structure (Supplementary Fig. 9b) indicates that terminator is likely to be the preferred conformation in the absence of a tRNA ligand. In order to lock the antiterminator configuration, an uncharged tRNA needs to fully bind cotranscriptionally before transcription and folding of the antiterminator region, otherwise the terminator will form. The time needed for this decision is short. Therefore, a short lifetime of the partially bound state can avoid trapping the glyQS T-box-tRNA in the terminator state and potentially allow multiple sampling events of tRNA ligands. Mtb ileS T-box regulates at the translational level, but the actual timing of this type of regulation is unclear. Interestingly, the difference in MFE between sequestrator and antisequestrator is smaller (Supplementary Fig. 9) compared to that of the B. subtilis glyQS T-box terminator and antiterminator. In addition, the Mtb ileS T-box antisequestrator has lower MFE than the B. subtilis glyQS T-box antiterminator (Supplementary Fig. 9a, b). This suggests that in the absence of a tRNA ligand: 1) The bias of Mtb ileS T-box towards folding into the sequestrator over the antisequestrator conformation cotranscriptionally is less pronounced compared to the terminator over antiterminator conformation in the glyQS T-box; 2) the antisequestrator conformation, if formed during transcription, is more stable throughout transcription; and 3) conformational rearrangements between antisequestrator and sequestrator states may occur post-transcriptionally in the absence of tRNA ligand. Given these possibilities, translational regulation by the Mtb ileS T-box may not be limited to be cotranscriptional but could also occur post-transcriptionally. Indeed, regulation by trans-acting small RNAs has also been observed to occur both co- and post-transcriptionally, with their actual contributions depending on the timing of transcription, transcript folding, and small RNA binding29,30. In this context, a short lifetime of the partially bound state would not provide any functional benefits for Mtb ileS T-box riboswitch. In contrast, a longer lifetime of the partially bound state may allow sufficient time for the establishment of the NCCA-discriminator interactions, through conformational sampling, and downstream conformational changes in the sequestrator/antisequestrator region.
Finally, our results highlight two critical sequence elements that facilitate tRNA binding and NCCA end discrimination. First, previous studies emphasized the central role of the Stem IIA/B-linker pseudoknot as a topological hinge that controls the conformations of Stems I and II14. Consistent with this previous finding, the data from the Linker Mutant further illustrates that the A-minor interaction and the pseudoknot structure are critical for the stability of both the partially bound state and the fully bound state. The critical importance of these interactions helps explain the conservation of the ‘F-box’ sequence in T-boxes (nucleotides 77-81, CGUCA in Mtb ileS T-box)31. Second, the present data elucidate the kinetic impact of the conserved RAG sequence on NCCA discrimination. Structural studies demonstrate that the RAG motif establishes specific interactions with the uncharged tRNA NCCA end. The results here demonstrate that mutating the RAG motif leads to the formation of a pseudo fully bound state, in which the tRNA NCCA end interacts with the discriminator domain with a distinct, likely non-functional configuration. This observation indicates that the RAG motif provides specificity for NCCA binding by constraining the conformational flexibility and reducing sampling of an aberrant NCCA docking state. While no equivalent measurement is available for a transcription-regulating T-box, given the structural similarity of the core discriminator domain and tRNA, it is possible to speculate that the conserved RAG motif may function in the same way in transcription regulating T-boxes as it does in Mtb ileS T-box.
In summary, the current work together with previous kinetic characterization on the decoding domain23,24 provides a more complete picture of tRNA ligand binding process by a representative translational T-box riboswitch (Fig. 8). The differences in the kinetic features of glyQS and ileS T-box riboswitches may reflect a fundamental functional difference between the two types of T-box riboswitches. In addition, the A81:G68:C77 A-minor interaction and the pseudoknot structure formed by Stem IIA/B and the linker region critically coordinate with both binding steps and enhance the stability of the partially bound and fully bound states. Finally, the conserved RAG sequence constrains the binding pocket of the tRNA NCCA end and provides specificity to NCCA recognition by limiting conformational flexibility. Overall, these findings shed light on the sophisticated and dynamic nature of T-box-tRNA interactions, highlighting the complex interplay between structural motifs, conformational flexibility, and ligand binding kinetics. By characterizing the nuanced structural adaptations that enable novel conformational dynamics, these results provide a more comprehensive framework to understand how structured RNA-based regulatory systems adapt to new regulatory niches.
Methods
Template DNA preparation
DNA templates, encompassing the sequence of interest placed downstream of a T7 promoter followed by a BsmI restriction site, were synthesized, and cloned into a pUC19 vector by GenScript (Jiangsu, China). For each plasmid, after transformation into E. coli DH5α cells, a minimum of 1 L of culture was grown overnight at 37 °C shaking at 200 rpm in Lysogeny Broth (LB) supplemented with ampicillin (at a final concentration of 100 µg/ml). Cells were harvested by centrifugation at 5470 x g for 30 min at 4 °C. Each cell pellet from a 1 L culture was resuspended in 10 ml of GTE buffer (50 mM glucose, 25 mM Tris-HCl pH 8.0, 10 mM EDTA, and 0.1 mg/ml RNase A), followed by addition of 20 ml of 0.2 M NaOH and 1% SDS to lyse the cells and left to stand on ice for five minutes. After neutralizing the lysate with 15 ml of 3 M potassium acetate (pH 5.2), the mixture was left on ice for ten min. In order to precipitate the DNA, the lysate supernatant was mixed with 0.6 volume of isopropanol followed by incubation at −20 °C for 15 min. The solution was centrifuged for 30 min at 4 °C at 4000 x g and the supernatant was discarded. The DNA pellet was washed repeatedly with 80% ethanol and then resuspended in 2 ml of TE buffer (10 mM Tris pH 8.0, 0.1 mM EDTA). To eliminate RNA contamination, RNase A (4 mg/ml, final concentration 20 µg/ml) was added and incubated at 37 °C for 20 min. Equal volumes of phenol/chloroform (1:1) were used twice to extract the DNA, followed by addition of 10% (v/v) 3 M sodium acetate pH 5.5 to precipitate the mixture. The precipitated DNA was rinsed twice with 80% ethanol before precipitation by centrifugation at 4000 x g for 30 min at 4 °C. After resuspension in 4 ml of TE buffer, 1.2 ml of 40% PEG 8000 and 516 µl of 5 M NaCl were added, and the mixture was left to precipitate overnight at 4 °C. The precipitated plasmid DNA was recovered by centrifugation at 4000 x g for 30 min at 4 °C, re-suspended in water, and ethanol precipitated again using 3 M sodium acetate pH 5.5 overnight at −20 °C. After three 80% ethanol washes, the pellet was once again resuspended in 1 ml of water. The concentration and purity of the purified plasmid DNA was assessed by measuring the absorbance ratio (A260/A280) using a Nanodrop spectrophotometer (Thermo Scientific) and aiming for a 1.9 ratio. Plasmids DNA were kept at 4 °C until needed. To confirm and validate the plasmid, the entire plasmid was sequenced (Primordium Laboratories).
Fluorophore conjugation of DNA oligonucleotides
For intermolecular smFRET experiments, DNA oligonucleotides containing an amine modification at the 3’ end and a biotin modification at the 5’ end were synthesized by Integrated DNA Technologies (IDT, Coralville IA). For intramolecular smFRET experiments, an additional DNA oligonucleotide containing an amine modification at the 5’ end was synthesized by IDT. To attach either Cy5 or Cy3 to these oligos, 13.5 µl of the 100 µM DNA oligonucleotide was thoroughly mixed with 1.5 µl of 1 M NaHCO3 pH 8.6. Following this, a pre-prepared solution containing 25 µg of N-hydroxysuccinimide (NHS)-conjugated fluorophore (either Cy3 or Cy5 (Lumiprobe, Hunt Valley, MD)) dissolved in 0.5 µl DMSO was added to the DNA oligonucleotide mixture. The mixture was incubated at a constant 37 °C temperature overnight to ensure proper conjugation. To precipitate the conjugated DNA oligonucleotide, 1.67 µl of 3 M sodium acetate pH 5.5 and 50 µl of pure ethanol were added to the mixture, followed by overnight incubation at −20 °C. Post incubation, the mixture was centrifuged at 21000 x g for 30 min. The resulting pellet, containing the conjugated DNA oligonucleotide, was then resuspended in 40 µl of water. To ensure the removal of any residual free dye and salt, the DNA solution was passed through a Micro Bio-Spin 6 column (Bio-Rad, Hercules, CA). The concentrations of oligo and conjugated fluorophore were measured using a Nanodrop spectrophotometer. The labeling efficiency for all labeled oligonucleotides was estimated to be at least 90%.
T-box preparation
The RNA molecules were synthesized by in vitro transcription from a linearized DNA template. To linearize the template, plasmids purified as described above harboring the region of interest were digested with BsmI (NEB, Ipswich MA). The reaction mixture was prepared in a nuclease-free microcentrifuge tube, consisting of 100 µg of plasmid DNA, 500 units of BsmI, the appropriate volume of 10X rCutSmart™ Buffer, and nuclease-free water to bring the final concentration of enzyme to less than 5% (v/v). The tube was then incubated in a constant temperature water bath at 65 °C, for 1 h. Post incubation, the reaction was assessed for completion through gel electrophoresis. The digested product was then purified by two phenol-chloroform extractions, ethanol precipitated, and resuspended in nuclease-free water, and the concentration of the digested product was measured using a Nanodrop spectrophotometer.
For T-box sample preparation the samples were purified from the transcription reaction without any denaturing steps. Transcription reactions were set up in a total volume of 500 μl, containing 50 μg of linearized DNA template, 40 mM Tris-HCl pH 8.0, 10 mM MgCl2, 1 mM spermidine, 50 µg/ml BSA, 20 mM DTT, 5 mM of ATP, CTP, and UTP, 6 mM of GTP, 80 U of Recombinant RNase inhibitor (Promega, Fitchburg, WI), 8 U of inorganic pyrophosphatase (Sigma-Aldrich, St. Louis, MO), and 5 U of T7 RNA polymerase (purified in-house). A Cy3-labelled DNA oligonucleotide was added to a final concentration of 1 µM to the transcription reaction to promote cotranscriptional annealing to the 3’- extension of the T-box sequence. The mixture was gently mixed by pipetting and incubated at 37 °C overnight. The next day, transcription reactions were briefly centrifuged to separate precipitated material before being loaded to a Superdex 200 10/200 GL column (Cytiva) pre-equilibrated with SEC buffer (10 mM Tris pH 7.4, 100 mM KCl, and 20 mM MgCl2). The column was run with an isocratic flow at 0.5 ml/min, and fractions were collected between 11.5 and 14 ml, depending on the mutant (Supplementary Fig. 1). Collected fractions were concentrated to ~40 µl by ultrafiltration using a Vivaspin 500 (Vivaproducts, Littleton, MA) centrifugal concentrator with a 3 kDa MWCO. The concentration of Cy3 in the concentrated fractions was measured using a Nanodrop spectrophotometer. One volume of 50% glycerol was subsequently added to the concentrated fractions before aliquoting, flash-freezing in liquid nitrogen, and storing at −80 °C.
tRNA preparation
For tRNA sample preparation, transcription reactions were set up in a total volume of 5 ml, containing 500 μg of linearized DNA template, 40 mM Tris-HCl pH 8.0, 10 mM MgCl2, 1 mM spermidine, 50 µg/mL BSA, 20 mM DTT, 5 mM of ATP, CTP, and UTP, 6 mM of GTP, 800 U of Recombinant RNase inhibitor (Promega, Fitchburg, WI), 8 U of inorganic pyrophosphatase (Sigma-Aldrich, St. Louis, MO), and 5 U of T7 RNA polymerase (purified in-house). The mixture was gently mixed by pipetting and incubated at 37 °C overnight. One volume of 50% glycerol was subsequently added to the transcription reaction before aliquoting, flash-freezing in liquid nitrogen, and storing at −80 °C. Transcription samples were purified by urea-polyacrylamide gel electrophoresis (urea-PAGE) by cutting out the tRNA band. The excised gel slice was electroeluted using an EluTrap device (Whatman). After electroelution, tRNA was precipitated by the addition of 3 volumes of 100% ethanol and 1/10th volume of 3 M sodium acetate (pH 5.5), followed by incubation at −20 °C for at least 1 hour. The tRNA was then pelleted by centrifugation at 12000 x g for 15 min at 4 °C, and the supernatant was discarded. The tRNA pellet was washed twice with 70% ethanol, centrifuged again at 12000 x g for 5 min at 4 °C. The tRNA pellet was dried using a SpeedVac vacuum concentrator (Savant) for 2–5 min and resuspended in an appropriate volume of nuclease-free water. The concentration of the purified RNA was assessed using a Nanodrop spectrophotometer. The purified tRNA was aliquoted and flash-frozen using liquid nitrogen and stored at −80 °C until further use.
For 5’ end labelling of tRNA, the N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) – NHS coupling method32 was followed, wherein the 5’ monophosphate of the RNA is activated by EDC and imidazole for subsequent labelling. First the 5’ triphosphate of RNA was converted into a 5’ monophosphate by incubating 100 µg RNA with 100 units of RNA 5’ Pyrophosphohydrolase (NEB, Ipswich MA) at 37 °C for 1 hour within a 100 µl reaction volume. Following this, the enzyme was removed by phenol chloroform extraction followed by passing the supernatant through a Micro Bio-Spin 6 column to exchange to a 10 mM HEPES pH 7.0, 150 mM NaCl, and 10 mM EDTA buffer. After the buffer exchange, the RNA solution was treated with 12.5 mg of EDC, 50 µl of ethylene diamine, and 200 µl of a 0.1 M imidazole pH 6.0 buffer. This mixture was then incubated for 3 hours at constant 37 °C temperature. Post incubation, the RNA was ethanol precipitated and the pellet was resuspended in 0.1 M sodium carbonate pH 8.7 buffer. Any residual EDC was removed by passing the solution through a Bio-Spin 6 column. For the final labelling step, the RNA solution in 0.1 M sodium carbonate pH 8.7 buffer was incubated with Cy5 NHS (Lumiprobe, Hunt Valley, MD) dye for 45 min, using an approximate RNA to dye molar ratio of 1:200 followed by ethanol precipitation and RNA resuspension in water. To remove any residual free dye, the final RNA solution was passed through a Micro Bio-Spin 6 column. The labeling efficiency of tRNAs was consistently around 90%-100%.
Refolding tRNA for smFRET or BLI experiments was done by initially diluting it to the required final concentration in a solution containing 10 mM HEPES pH 7.5 and 50 mM NaCl. The sample was then heated to 95 °C using a thermocycler, incubated for 3 min, and then transferred to ice to cool it rapidly for 3 min. After cooling, MgCl2 was added to a final 15 mM concentration. The tRNA solution was heated to 50 °C in a thermocycler and incubated for 10 min at this temperature, followed by a 30 min incubation at 37 °C. Lastly, the sample was removed from the thermocycler and allowed to equilibrate to room temperature. For BLI binding assays, the folded tRNAs were dialyzed into 10 mM Tris pH 7.4, 100 mM KCl, and 20 mM MgCl2. For smFRET studies, one volume of 50% glycerol was added to the tRNA solution, aliquoted and flash frozen in liquid nitrogen for long-term storage prior to use.
Biolayer interferometry assays
Biolayer Interferometry (BLI) was used to determine the equilibrium dissociation constant (KD) of Mtb-ileS T-box and tRNAIle using streptavidin biosensors (SAX) in an Octet K2 system (FortéBio Inc. San Jose, CA). The assays were performed on black, flat-bottom 96-well microplates (Greiner Bio-One 655209) in a total volume of 200 μl with orbital shaking at 1000 rpm. The instrument was controlled with the Data Acquisition 11.1 (FortéBio Inc. San Jose, CA) software package. Before each experiment, biosensors were hydrated in SEC buffer (10 mM Tris pH 7.4, 100 mM KCl and 20 mM MgCl2). At the start of each experiment, a baseline was established using SEC buffer for 30 seconds. Subsequently the T-box at a 7.5 ng/µL concentration in SEC buffer was bound to the streptavidin sensor for 250 seconds, followed by washing for 30 seconds with the same buffer to eliminate non-specific binding and establish a pre-association baseline. Next, the purified and refolded tRNA was introduced into the biosensor and the association signal was measured for up to 500 seconds, until apparent equilibrium was reached at the highest concentration. Finally, the dissociation curves were obtained by moving the biosensors back into wells with SEC buffer (Supplementary Fig.1b). Each BLI experiment was done at several different tRNA concentrations (measured before each experiment) in the 1.6 to 55.3 μM range. For each set of experiments, a sensor that was not loaded with any T-box was used as a negative control and reference well for later data subtraction. New streptavidin biosensors were used for each experiment.
The raw data were preprocessed using the Octet Data Analysis Software version 11.1 (FortéBio Inc. San Jose, CA). Preprocessing included Y-axis alignment, reference subtraction, and interstep correction to account for noise while sensors are moved between solutions. Three binding models were considered to describe the binding kinetics: a one-step binding model, a parallel one-step binding model, and a sequential two-step binding model. Data were fit to the three considered models using either the Octet Data Analysis Software version 11.1 (FortéBio Inc. San Jose, CA) for the one-step and parallel one-step binding models, or a custom software for the two-step binding model. To prepare the data for our custom model fitting software, preprocessed data files were exported from the Octet Data Analysis software and converted to plain text (.txt). These data were then processed using Python scripts in Jupyter notebooks to fit the data to the sequential two-step binding model. Due to the high multicollinearity of the model fitted to the data, the standard method to estimate parameter errors by extracting the diagonal elements of the covariance matrix of the regression model led to artificially inflated variances and, in turn, standard errors. Thus, the estimation of parameter errors was performed using a residuals bootstrapping approach. For this purpose, initial fitting was done using the curve_fit function from the scipy.optimize module in Python, which provided the best-fit values for nonlinear fitting of all parameters for each dataset. The residuals between the original data and the fitted values were then calculated. Bootstrapping was performed for 10000 cycles. In each cycle, a new set of bootstrap data were generated by randomly sampling the residuals with replacement and adding them to the original values. Nonlinear fitting was then repeated on these bootstrap data using the same initial guesses and parameter bounds as the original fit. The resulting best-fit parameters and R-squared values from each bootstrap cycle were stored. The means and standard deviations of the bootstrap parameter distributions were then calculated to estimate the parameter uncertainties. Jupyter notebooks containing the code used to fit and plot these data are provided in the Source Data. A detailed description of the mathematical model used to fit these data is provided in the Supplementary Information and the Jupyter Notebooks.
smFRET measurement
Quartz slides and glass coverslips for total internal reflection fluorescence microscopy were prepared according to previously published protocols33,34. The surface of the slides and coverslips were coated with a mixture of poly-ethylene glycol (PEG, Mw = 500,000) and biotin-PEG (Mw = 500,000). Microfluidic channels were constructed between the slides and coverslips as previously described in refs. 33,34. The channels were further passivated with a solution of 10 µM BSA to prevent non-specific binding, followed by incubation with a solution containing 10 µM BSA and 1 μM streptavidin. An aliquot of folded T-box riboswitch, carrying the labeled and biotinylated DNA oligo, was diluted to a concentration of 0.5-1 nM, and immobilized to the surface of the chamber via biotin-streptavidin interactions. tRNA was diluted to a concentration of 100 nM in an imaging buffer comprising of 50 mM HEPES pH 7.0, 100 mM KCl, 15 mM MgCl2, 5 mM protocatechuic acid (PCA) (Sigma), 1 U/mL protocatechuate-3,4-dioxygenase (PCD) (MP Biomedical), and 2 mM Trolox (Sigma), and was flowed into the microfluidic channel. For the intramolecular smFRET experiments, the concentration of the dual labeled T-box RNA was 1 nM and the unlabeled tRNA concentration was either 5 or 13 μM. An objective based total internal reflection fluorescence (TIRF) microscope based on a Nikon Ti-E with 100X NA 1.49 CFI HP TIRF objective (Nikon) was used to perform the smFRET measurements. A 561 nm laser (Coherent Obis at a power density of 4.07 × 105 W/cm2) and a 647 nm laser (Cobolt MLD at a power density of 5.88 × 105 W/cm2) were used for Cy3 and Cy5 excitation respectively. Emissions from both donor and acceptor were passed through an emission splitter (OptoSplit III, Cairn), and collected at different locations on an EMCCD (iXon Ultra 888, Andor). For intermolecular FRET measurement, 1500 frames of time-lapse images were taken with 100 ms exposure time and 561 nm laser excitation. For the intramolecular FRET measurements, 5 frames of time-lapse images with 100 ms exposure time and 647 nm laser excitation were taken before the FRET recording using the 561 nm laser excitation. The biological samples (i.e. the in vitro transcribed tRNAs and T-boxes) were generated at least twice, each considered as a biological replicate. For each biological replicate, at least three technical replicates were done. In each measurement, T-box and tRNA were diluted, smFRET images were recorded, and analysis were performed independently. Before each smFRET experiment, Tetraspeck beads (ThermoFisher) were used to align the emission channels. NIS Elements software was used for all smFRET data acquisition.
Transition rates and lifetime analyses
NIS Elements software was used to pick individual spots and generate intensity trajectories from the time-lapse images. For intermolecular FRET experiments, maximum intensity projection of Cy5 emission signal was generated, and signals above an intensity threshold were selected to generate regions of interest (ROIs). For the intramolecular FRET measurements, maximum intensity projection of Cy5 emission signal from the 5 frames with Cy5 direct excitation was generated, and signals above an intensity threshold were selected to generate ROIs. ROIs were projected into the Cy3 and Cy5 channels in the time-lapse images to generate Cy3 and Cy5 intensity trajectories. Fluorescence intensity trajectories were corrected for baseline and bleed-through in MATLAB as previously described in ref. 33. FRET traces were generated by calculating ICy5 / (ICy5 + ICy3) at each time point from the intensity trajectories.
Idealization of FRET trajectories was achieved using the global modeling algorithm in tMAVEN26 unless otherwise specified. The following settings for the parameters of the priors were used: α: 1.0, β: 0.25, π: 1.0, a: 2.5, b: 0.01. The prior b was set to 0.4 for the Linker Mutant and 0.2 for the RAG Mutant. When a global modeling approach was used, transition rates were directly extracted from the transition matrix output of tMAVEN (Supplementary Table 5). Mean lifetime values calculated from these transition rates are also described in Supplementary Table 5. To calculate errors in transition rate matrices (Supplementary Table 5), variances of the un-normalized counts matrix output by tMAVEN were calculated using the following Dirichlet variance formula:
| 1 |
where , is the i entry of the counts matrix, and is the sum of the row is in. Upper and lower bounds on the rates were calculated using the following relation between transition probabilities and rate constants:
| 2 |
where Δt is the frame rate of data acquisition. To get upper and lower bounds for lifetimes calculated by either transition rate analysis or dwell time analysis, the error propagation formula was used:
| 3 |
where a function F depends on variables x1, x2,…, each with their respective errors x1, x2,…. Jupyter notebook files with error propagation calculations are provided in the Source Data files.
Since global modeling is equivalent to concatenating the ensemble of trajectories into a single, long trajectory, which is then analyzed with an HMM, this approach may be insensitive to infrequently and transiently sampled states. Therefore, in the analysis of two subpopulations in the partially bound state using WT Mtb ileS T-box and tRNAIle-ΔNCCA-Cy5, a composite approach was used. Traces of the One-state population were analyzed by a variational Bayesian algorithm clustered by K-means (composite HMM + K-means)26. This approach infers an HMM for each trace and then clusters the inferred FRET means for each HMM using K-means, allowing it to capture more accurately the 0.5 FRET events of each trace. Modeling of the Two-state population was achieved using the variational Bayesian Gaussian mixture modelling (vbGMM) algorithm in tMAVEN26. This approach assumes that all the data points are generated from a mixture of a finite number of Gaussian distributions with unknown parameters and best identified the short-lived 0.7 FRET state.
In all cases, lifetimes were also calculated using either single or double exponential fitting to the dwell time histogram for each FRET state before transitioning out of that state using tMAVEN26. In most of the cases, the single-exponential decay fitting is sufficient to describe the dwell time data (when A > 0.75). For the cases where A < 0.75, the double-exponential decay fitting was selected (Supplementary Table 6).
IntraFRET data analysis
The FRET state population analysis for the IntraFRET sample was analyzed using custom software provided in the Source Data as a Jupyter notebook. Fluorescence intensity trajectories were read from files, each containing paired columns of donor and acceptor fluorescence intensities. To ensure consistent sampling before photobleaching, we considered the first 400 data points from each file. The FRET efficiency for each pair of donor and acceptor intensities was calculated using the formula:
| 4 |
Efficiencies were computed for all paired columns in the data set, and the results saved for subsequent analysis. The histogram data were then fit to a double Gaussian model defined as:
| 5 |
where and are the amplitudes, and are the means, and and are the standard deviations of the two Gaussian components. The parameters were estimated using non-linear least squares fitting. The area under each Gaussian curve was computed to determine the contributions of each FRET population. The area under each Gaussian component is given by:
| 6 |
where is the fitted amplitude, and is the fitted standard deviation. The errors of these areas were propagated from the fitting uncertainties using the following formula:
| 7 |
where and are the standard errors of the fitted parameters. The total area under both Gaussians was calculated as the sum of the individual areas, and its error as the root sum square of the individual errors. The ratio of the areas under the two Gaussian components was computed to quantify the relative contributions of the two populations. The error in this ratio was propagated using standard error propagation formulas:
| 8 |
| 9 |
Diagrams of the Mtb ileS T-box were made using PyMOL35 based on the coordinates of the structure of the complex14 (PDB ID 6ufg, 6ufh).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Dr. Jiacheng Zhang for help with the initial experiments, Arabela Grigorescu for help with the BLI experiments, as well as other members of the Fei and Mondragón laboratory for help and suggestions. We thank Dr. Ruben Gonzalez (Columbia University) for sharing tMAVEN. Research was supported by the NIH (R35-GM118108 to A.M., NIH Director’s New Innovator Award 1DP2GM128185 to J.F.) and the Yen Fellowship from The University of Chicago to S.P. We acknowledge the help from the Northwestern University Keck Biophysics Facility. Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Keck Biophysics Facility is acknowledged.
Author contributions
E.C.C., J.F., and A.M. designed the research. S.P. and Z.Z. conducted smFRET experiments and S.P., Z.Z., E.C.C., and J.F. analyzed the smFRET data. E.C.C. and D.A. prepared all the samples. E.C.C. performed BLI experiments and E.C.C. and A.M. analyzed the BLI results. A.R.V. contributed software and helped with data analysis. E.C.C., J.F., and A.M. wrote the manuscript; all authors edited and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw data supporting the findings of this study are available from the corresponding authors upon request. Source data for the figures and Supplementary Figs. are provided as a Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Eduardo Campos-Chavez, Sneha Paul.
Contributor Information
Jingyi Fei, Email: jingyifei@uchicago.edu.
Alfonso Mondragón, Email: a-mondragon@northwestern.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50885-x.
References
- 1.Chantranupong, L., Wolfson, R. L. & Sabatini, D. M. Nutrient-sensing mechanisms across evolution. Cell161, 67–83 (2015). 10.1016/j.cell.2015.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henkin, T. M. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev.22, 3383–3390 (2008). 10.1101/gad.1747308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salvail, H. & Breaker, R. R. Riboswitches. Curr. Biol.33, R343–R348 (2023). 10.1016/j.cub.2023.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundy, F. J. & Henkin, T. M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell74, 475–482 (1993). 10.1016/0092-8674(93)80049-K [DOI] [PubMed] [Google Scholar]
- 5.Bullwinkle, T. J. & Ibba, M. Translation quality control is critical for bacterial responses to amino acid stress. Proc. Natl Acad. Sci. USA113, 2252–2257 (2016). 10.1073/pnas.1525206113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suddala, K. C. & Zhang, J. An evolving tale of two interacting RNAs-themes and variations of the T-box riboswitch mechanism. IUBMB Life71, 1167–1180 (2019). 10.1002/iub.2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver, J. W. & Serganov, A. T-box RNA gets boxed. Nat. Struct. Mol. Biol.26, 1081–1083 (2019). 10.1038/s41594-019-0340-9 [DOI] [PubMed] [Google Scholar]
- 8.Zhang, J. Unboxing the T-box riboswitches-A glimpse into multivalent and multimodal RNA-RNA interactions. Wiley Interdiscip. Rev. RNA11, e1600 (2020). 10.1002/wrna.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putzer, H., Condon, C., Brechemier-Baey, D., Brito, R. & Grunberg-Manago, M. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res30, 3026–3033 (2002). 10.1093/nar/gkf415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, J. & Ferre-D’Amare, A.R. Structure and mechanism of the T-box riboswitches. Wiley Interdiscip. Rev. RNA6, 419–433 (2015). 10.1002/wrna.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigg, J. C. & Ke, A. Structural determinants for geometry and information decoding of tRNA by T box leader RNA. Structure21, 2025–2032 (2013). 10.1016/j.str.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann, J., Jossinet, F. & Gautheret, D. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders. Nucleic Acids Res41, 5494–5502 (2013). 10.1093/nar/gkt219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, J. & Ferre-D’Amare, A.R. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature500, 363–366 (2013). 10.1038/nature12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battaglia, R. A., Grigg, J. C. & Ke, A. Structural basis for tRNA decoding and aminoacylation sensing by T-box riboregulators. Nat. Struct. Mol. Biol.26, 1106–1113 (2019). 10.1038/s41594-019-0327-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, S. et al. Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions. Nat. Struct. Mol. Biol.26, 1094–1105 (2019). 10.1038/s41594-019-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez-Preciado, A., Henkin, T. M., Grundy, F. J., Yanofsky, C. & Merino, E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol. Biol. Rev.73, 36–61 (2009). 10.1128/MMBR.00026-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchand, J. A., Smela, Pierson, Jordan, M. D., Narasimhan, T. H. H. & Church, K. G.M. TBDB: a database of structurally annotated T-box riboswitch:tRNA pairs. Nucleic Acids Res49, D229–D235 (2021). 10.1093/nar/gkaa721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitreschak, A. G., Mironov, A. A., Lyubetsky, V. A. & Gelfand, M. S. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA14, 717–735 (2008). 10.1261/rna.819308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, J. et al. Specific structural elements of the T-box riboswitch drive the two-step binding of the tRNA ligand. Elife7, e39518 (2018). [DOI] [PMC free article] [PubMed]
- 20.Suddala, K. C. et al. Hierarchical mechanism of amino acid sensing by the T-box riboswitch. Nat. Commun.9, 1896 (2018). 10.1038/s41467-018-04305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherwood, A. V., Grundy, F. J. & Henkin, T. M. T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions. Proc. Natl Acad. Sci. USA112, 1113–1118 (2015). 10.1073/pnas.1424175112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breaker, R. R. Riboswitches and Translation Control. Cold Spring Harb. Perspect. Biol.10, a032797, (2018). [DOI] [PMC free article] [PubMed]
- 23.Niu, X. et al. Structural and dynamic mechanisms for coupled folding and tRNA recognition of a translational T-box riboswitch. Nat. Commun.14, 7394 (2023). 10.1038/s41467-023-43232-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suddala, K. C. et al. Direct observation of tRNA-chaperoned folding of a dynamic mRNA ensemble. Nat. Commun.14, 5438 (2023). 10.1038/s41467-023-41155-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapkota, K. P., Li, S. & Zhang, J. Cotranscriptional Assembly and Native Purification of Large RNA-RNA Complexes for Structural Analyses. Methods Mol. Biol.2568, 1–12 (2023). 10.1007/978-1-0716-2687-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verma, A. R., et al. Increasing the accuracy of single-molecule data analysis using tMAVEN. Biophys J, 10.1016/j.bpj.2024.01.022 (2024). [DOI] [PubMed]
- 27.Son, H., Mo, W., Park, J., Lee, J. W. & Lee, S. Single-Molecule FRET Detection of Sub-Nanometer Distance Changes in the Range below a 3-Nanometer Scale. Biosensors (Basel), 10, 168 (2020). [DOI] [PMC free article] [PubMed]
- 28.Dong, H., Nilsson, L. & Kurland, C. G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol.260, 649–663 (1996). 10.1006/jmbi.1996.0428 [DOI] [PubMed] [Google Scholar]
- 29.Fei, J., Kosuri, P., MacDougall, D. D. & Gonzalez, R. L. Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell30, 348–359 (2008). 10.1016/j.molcel.2008.03.012 [DOI] [PubMed] [Google Scholar]
- 30.Rodgers, M. L., O’Brien, B. & Woodson, S. A. Small RNAs and Hfq capture unfolded RNA target sites during transcription. Mol. Cell83, 1489–1501.e5 (2023). 10.1016/j.molcel.2023.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rollins, S. M., Grundy, F. J. & Henkin, T. M. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol25, 411–421 (1997). 10.1046/j.1365-2958.1997.4851839.x [DOI] [PubMed] [Google Scholar]
- 32.Rinaldi, A. J., Suddala, K. C. & Walter, N. G. Native purification and labeling of RNA for single molecule fluorescence studies. Methods Mol. Biol.1240, 63–95 (2015). 10.1007/978-1-4939-1896-6_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchard, S. C. et al. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA101, 12893–12898 (2004). 10.1073/pnas.0403884101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ha, T. et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature419, 638–641 (2002). 10.1038/nature01083 [DOI] [PubMed] [Google Scholar]
- 35.Schrodinger, L. L. C. The PyMOL Molecular Graphics System. Version1, 8 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the findings of this study are available from the corresponding authors upon request. Source data for the figures and Supplementary Figs. are provided as a Source Data file. Source data are provided with this paper.