Abstract
Long-term recombinant AAV (rAAV) transgene expression in muscle has been associated with the molecular conversion of single-stranded rAAV genomes to high-molecular-weight head-to-tail circular concatamers. However, the mechanisms by which these large multimeric concatamers form remain to be defined. To this end, we tested whether concatamerization of rAAV circular intermediates occurs through intra- or intermolecular mechanisms of amplification. Coinfection of the tibialis muscle of mice with rAAV alkaline phosphatase (Alkphos)- and green fluorescent protein (GFP)-encoding vectors was used to evaluate the frequency of circular concatamer formation by intermolecular recombination of independent viral genomes. The GFP shuttle vector also encoded ampicillin resistance and contained a bacterial origin of replication to allow for bacterial rescue of circular intermediates from Hirt DNA of infected muscle samples. The results demonstrated a time-dependent increase in the abundance of rescued plasmids encoding both GFP and Alkphos, which reached 33% of the total circular intermediates by 120 days postinfection. Furthermore, these large circular concatamers were capable of expressing both GFP- and Alkphos-encoding transgenes following transient transfection in cell lines. These findings demonstrate that concatamerization of AAV genomes in vivo occurs through intermolecular recombination of independent monomer circular viral genomes and suggest new viable strategies for delivering multiple DNA segments at a single locus. Such developments will expand the utility of rAAV for splicing large gene inserts or large promoter-gene combinations carried by two or more independent rAAV vectors.
Adeno-associated virus (AAV) is a unique, nonpathogenic member of the Parvoviridae family of small, single-stranded DNA animal viruses. The linear genome of wild-type AAV (wtAAV) is 4,680 nucleotides in length, may be of either plus or minus polarity, and contains two groups of genes, the Rep and Cap genes (3). Inverted terminal repeats (ITRs), characterized by palindromic sequences producing a high degree of secondary structure, are present at both ends of the viral genome. While other members of the parvovirus group replicate autonomously, AAV requires coinfection with a helper virus (i.e., adenovirus or herpesvirus) for productive replication. In the absence of helper virus, wtAAV establishes a latent, nonproductive infection with long-term persistence by integrating into a specific locus, AAVS1, on chromosome 19 of the host genome. Site-specific integration of AAV has been shown to require expression of the AAV Rep protein (16).
In studies crucial to the development of AAV as a gene therapy vector, methods were developed for propagating recombinant AAV type 2 (rAAV) by inserting therapeutic or reporter genes between two ITRs in place of the AAV Rep and Cap genes (23, 24). rAAV has been shown to be capable of stable, long-term transgene expression both in vitro and in vivo in a variety of tissues, although the transduction efficiency of rAAV is markedly variable in different cell types. For example, rAAV has been reported to transduce lung epithelial cells at low levels (6, 11), while high-level, persistent transgene expression has been demonstrated in muscle cells, neurons, and other nondividing cells (2, 8, 12, 13, 15, 28–30). These tissue-specific differences in rAAV-mediated gene transfer may be due, in part, to variable levels of cellular factors affecting AAV infectivity (i.e., receptors and coreceptors such as heparan sulfate proteoglycan, FGFR-1, and αVβ5 integrin) (21, 25, 26), as well as the latent life cycle (i.e., nuclear trafficking of virus and/or the conversion of single-stranded genomes to expressible forms) (20, 22).
These studies have underscored the potential of rAAV as a gene therapy vector, but they indicate that additional investigation into the mechanisms of transduction and genome persistence in tissues such as muscle may further broaden the utility of this vector for delivering therapeutic genes to other cell types. Although persistence of rAAV has been traditionally attributed to integration, supporting evidence for efficient integration is limited to a recent study with liver (18). Due to deletion of the Rep gene, rAAV does not integrate site specifically at AAVS1. Rather, persistence of rAAV has been attributed to both episomal (1, 5, 10) and randomly integrated (9, 14, 17, 19) AAV genomes. In a recent report, the existence of circular rAAV genomes in muscle was demonstrated, and their episomal persistence correlated with long-term transgene expression (5). These genomes appear to originate through a monomeric circularization process leading to head-to-tail AAV circular genomes. However, over time (between 22 and 80 days), there is a decline in monomer circular intermediates in favor of high-molecular-weight circular concatamers (5, 27). Presently little is known about the mechanisms which lead to formation of these high-molecular-weight concatamers or whether they represent preintegration intermediates. However, one report has suggested that rolling-circle replication may be responsible for the uniformity of head-to-tail concatamers (27).
In the present study, we sought to further characterize potential mechanisms involved in the formation of rAAV circular concatamers associated with long-term episomal persistence and transgene expression in muscle. To address this mechanism, we utilized an approach capable of assessing intermolecular recombination between independently formed circular intermediates by coinfecting muscles with two independent rAAV vectors (green fluorescent protein [GFP] and alkaline phosphatase [Alkphos]) and shuttling circular genomes into bacteria for structural analysis. From these studies we demonstrate a time-dependent increase in coexpression of the two markers genes within single plasmids rescued from muscle Hirt DNA. These studies demonstrate that intermolecular recombinational events between monomer circular intermediates are, at least in part, responsible the formation of high-molecular-weight circular forms of rAAV.
MATERIALS AND METHODS
rAAV vectors.
Two rAAV vector stocks were generated for use in these studies, AV.GFP3ori (5) and AV.Alkphos (also known as CWRAPSP, a gift of Dusty Miller) (11). Virus stocks were generated by cotransfection of 293 cells with either pCisAV.GFP3ori or pCWRAPSP along with pRep/Cap, followed by coinfection with recombinant Ad.CMVlacZ helper virus (5). rAAV was then purified through three rounds of CsCl density gradient centrifugation as previously described (4). Purified viral fractions were heated at 60°C for 1 h to inactivate any residual contaminating helper adenovirus. The yields for AV.GFP3ori and AV.Alkphos were 1 × 1012 and 7 × 1011 particles per ml, respectively, as determined by slot blot hybridization with 32P-labeled GFP or Alkphos probes. Infectious titers determined by infection of 293 cells with rAAVs were 1.1 × 109 (AV.GFP3ori) or 8.6 × 108 (AV.Alkphos) infectious units per ml. Controls testing for contamination of rAAV stocks with wtAAV by anti-Rep immunocytochemical staining in rAAV- and Ad.CMVlacZ-coinfected 293 cells were negative (the limit of sensitivity is less than 1 infectious wtAAV particle per 1010 DNA particles of rAAV). Similarly, histochemical staining for β-galactosidase in rAAV-infected 293 cells showed no detectable contamination with helper adenovirus in 1010 DNA particles of rAAV (limit of sensitivity).
Infection of muscle tissue and evaluation of transgene expression.
The C57BL/6 mice used for these experiments were housed in a virus-free animal care facility and were maintained according to strict University of Iowa and National Institutes of Health guidelines, using a protocol approved by the Animal Care and Use Committee and facility veterinarians. Four- to 5-week-old mice received bilateral 30-μl injections of a mixture of AV.GFP3ori and AV.Alkphos into the tibialis anterior muscle (5 × 109 DNA particles of each virus per muscle). Controls included uninjected muscles and muscles receiving injections of one of the viruses alone. At 14, 35, 80, and 120 days postinfection, animals were euthanized and tissues were harvested for evaluation of transgene expression and preparation of low-molecular-weight Hirt DNA. For each experimental time point, Hirt DNA from at least three independently injected muscles was evaluated and at least two tissue samples from each group were evaluated for expression of GFP and Alkphos in at least 10 sections.
In all experiments, GFP fluorescence was visualized in freshly excised muscle tissue prior to processing. A portion of the same muscle was fixed with 2% paraformaldehyde in phosphate-buffered saline and cryoprotected in graded sucrose solutions before embedding in optimal-cutting-temperature medium. Sections (6 μm) were then evaluated for GFP expression directly and for Alkphos expression following heat inactivation of endogenous Alkphos and histochemical staining for Alkphos activity as previously described (7). To confirm dual localization of GFP and Alkphos expression in the same muscle fibers, either serial sections were evaluated for GFP and Alkphos expression or the same section was first photographed for GFP expression followed by histochemical staining for Alkphos and reimaging of the same field.
Rescue of circular intermediates from muscle Hirt DNA.
Low-molecular-weight Hirt DNA was prepared from 20-mg specimens of injected muscles from three animals at each time point as previously described (5). Yields of Hirt DNA were typically ∼5 μg of DNA/20 mg of tissue. Hirt DNA (4 μl, one-fifth of the total volume; approximately 1 μg) was then used to transform 50 μl of electrocompetent SURE cells (Stratagene) by using a Bio-Rad Escherichia coli electroporater and 0.1-μm cuvettes. Colonies resulting from each bacterial transformation were quantified, and plasmids from 20 colonies from each muscle Hirt DNA sample were purified for analysis. It should be noted that only circular forms carrying the Ampr gene and the bacterial origin of replication from AV.GFP3ori will be rescued by bacterial transformation (5). As previously described, control experiments reconstituting 5 × 1010 viral DNA particles (∼0.14 μg of viral DNA) into uninfected muscle extracts prior to Hirt DNA preparation failed to give rise to replication-competent plasmids in the rescue assay (5). Additional controls in this previous study using AV.GFP3ori virus also demonstrated that linear double-stranded and single-stranded purified viral DNA genomes do not give rise to replication-competent plasmids following transformation into E. coli.
Characterization of genes in rescued circular intermediates.
Several assays were used to characterize the extent of intermolecular recombination between independent circular viral genomes by evaluating the number and type of genes in rescued plasmids from Hirt DNA of muscles coinfected with AV.GFP3ori and AV.Alkphos. Our first analysis involved the bulk evaluation of 60 rescued plasmids (20 from each of three muscle samples for each time point) by dot blot hybridization of miniprep DNA against GFP, Alkphos, and Ampr gene 32P-labeled DNA probes. In these studies, Ampr gene hybridization served as a control to show that there was a sufficient quantity of DNA for the analysis. The percentages of Alkphos- and/or GFP-hybridizing plasmids were calculated by this method for each muscle sample. From this percentage, the total number of plasmids hybridizing to each probe in the Hirt DNA sample was calculated from the total CFU obtained in each transformation. In this analysis, each muscle sample was evaluated independently to determine the mean (± standard error of the mean [SEM]) total Alkphos- and/or GFP-hybridizing plasmids. A second evaluation involved the transfection of rescued plasmids into 293 cells by using Lipofectamine, followed by evaluation of GFP fluorescence and histochemical staining for Alkphos. To confirm that GFP- and Alkphos- coexpressing plasmids were indeed clonal and that both genes were carried on the same plasmid, a selected group of five coexpressing plasmids were retransformed into E. coli, and colonies were reisolated prior to repetition of the transfection studies. In all cases, plasmids coexpressing the two reporter genes remained clonal through this subsequent reisolation.
Structural analysis of concatamer rAAV circular intermediates.
To further characterize the nature of isolated circular intermediates coexpressing both GFP and Alkphos transgenes, we mapped their plasmid structures by Southern blotting and restriction enzyme analysis. The structures of five coexpressing circular intermediate plasmids were determined by digestion with AhdI, HindIII, NotI, HindIII-NotI, ClaI-AseI, and/or SnaBI, and Southern blotting was performed with 32P-labeled GFP, Alkphos, and ITR probes.
RESULTS
Strategy for characterizing mechanisms of rAAV circular intermediate formation.
Efficient circularization of rAAV genomes has been previously demonstrated to occur in muscle in a time-dependent fashion (5). Furthermore, the conversion of monomeric to multimeric circular rAAV intermediates occurred over time and was associated with long-term episomal persistence of AAV genomes. We hypothesized that high-molecular-weight AAV circular genomes might form by either of two mechanisms, one involving the replication of monomer structures and the other involving intermolecular recombination between independent monomers. To evaluate these hypotheses, we developed a rescue assay using two separate rAAV vectors, AV.GFP3ori and AV.Alkphos (Fig. 1A), which allowed for the identification of independent viral genomes through unique transgenes. In this assay, circular-form genomes were rescued in bacteria by virtue of Ampr gene and ori sequences carried in one of the two vectors (AV.GFP3ori). Using these vectors, we proposed a method for characterizing the extent of intermolecular recombination between independent circular rAAV genomes (Fig. 1B).
FIG. 1.
Mechanistic scheme for determining pathways for rAAV circular concatamer formation. (A) The two independent vectors used in these studies, AV.Alkphos and AV.GFP3ori. Restriction sites important in the structural analysis of circular intermediates are also shown. RSV, Rous sarcoma virus promoter; CMV, cytomegalovirus promoter. (B) Schematic representation of two potential models for circular concatamer formation, along with the proposed methods to experimentally differentiate which of these two processes is active in muscle. Following coinfection of the tibialis muscle with AV.Alkphos and AV.GFP3ori, all subsequently rescued plasmids arise solely from circular intermediates containing AV.GFP3ori genomes. If rolling-circle replication is the sole mechanism of concatamerization, only GFP-expressing plasmids should be rescued. In contrast, if intermolecular recombination between independently formed monomer circular intermediates is the mechanism of concatamerization, both GFP-expressing and GFP- and Alkphos-expressing plasmids should be rescued. This figure is diagrammatically sketched and is not drawn to scale.
Coexpression of independently carried rAAV transgenes in muscle myofibers.
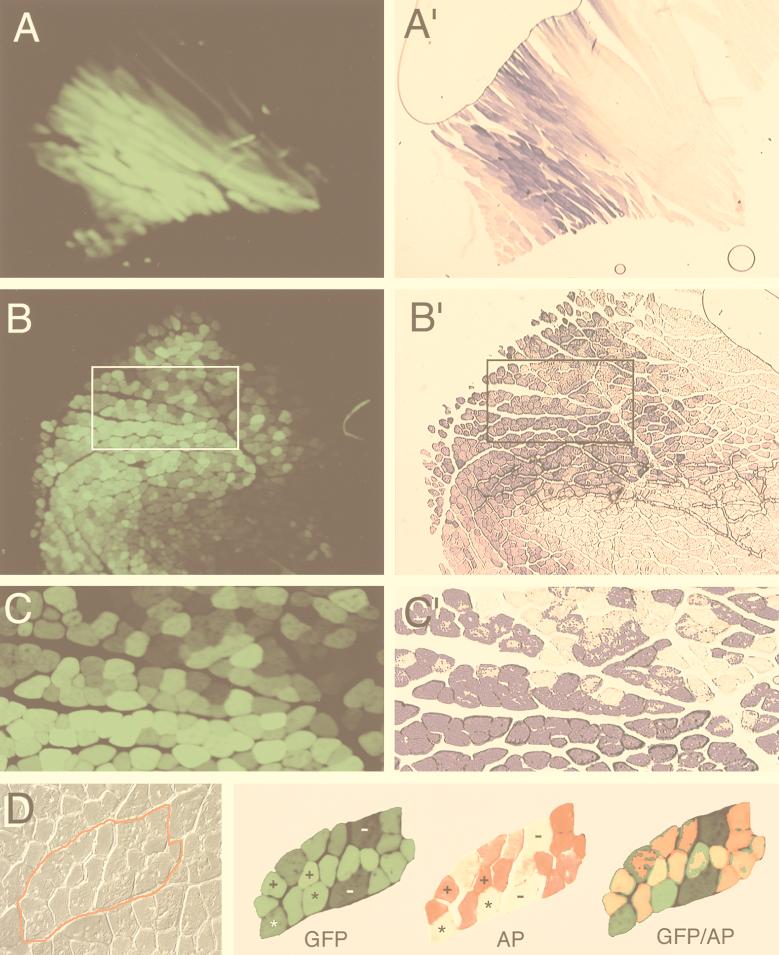
To test our hypotheses regarding intermolecular recombination between circular viral genomes, it was first necessary to confirm that myofibers could be coinfected at a high efficiency with the two rAAV vectors. The tibialis anterior muscles of mice were coinfected with 5 × 109 DNA particles of both AV.GFP3ori and AV.Alkphos. At 14, 35, 80, and 120 days postinfection, muscles were harvested and analyzed for transgene expression. As previously reported, transgene expression from both reporters was weak but clearly visible in 14-day muscle samples (reference 5 and data not shown). By 80 days postinfection, transgene expression was maximal and serial sections demonstrated expression of both Alkphos and GFP transgenes in overlapping regions of the muscle (Fig. 2A to C). At this time point, approximately 50% of the fibers in the tibialis muscle expressed both transgenes. To confirm that coinfection of myofibers occurred with the two independent vectors, colocalization studies were performed on muscle sections by a serial staining procedure. These studies, depicted in Fig. 2D, demonstrate four classes of myofiber transgene expression: (i) GFP positive only, (ii) Alkphos positive only, (iii) GFP and Alkphos positive, and (iv) no transgene expression. The largest fraction of myofibers expressed both GFP and Alkphos transgenes. These results confirm that at the titers of virus used for infection, coinfection occurred in greater than 90% of transgene-expressing myofibers.
FIG. 2.
Coinfection of tibialis muscle of mice with AV.Alkphos and AV.GFP3ori. Transgene expression of rAAV-infected tibialis muscle was determined at 14, 35, 80 (A and A′), and 120 (B to D) days following coinfection with 5 × 109 DNA particles each of AV.Alkphos and AV.GFP3ori. The time course of transgene expression was similar to that previously reported (5), beginning at 14 days and peaking by 35 to 80 days (data not shown). The extent of coinfection of myofibers with both Alkphos and GFP rAAV was determined in serial sections of muscle samples at 80 and 120 days postinfection. (A to C) GFP fluorescence of formalin-fixed, cryoprotected sections; (A′ to C′) histochemical staining for Alkphos in adjacent serial sections. A short staining time (7 min) was necessary to observe variation in staining levels for comparison to GFP. In experiments not shown, it was found that a longer staining times (30 min) saturated the Alkphos signal. The boxed regions in panels B and B′ are enlarged in panels C and C′, respectively. (D) A more precise correlation of GFP and Alkphos staining in myofibers. Colocalization of GFP and Alkphos expression was examined in the same section of a 120-day-postinfection sample. This was performed by photographing the GFP fluorescent image prior to staining for Alkphos activity. The left panel of panel D shows a high-power Nomarski photomicrograph of a group of myofibers (traced in red), while the corresponding GFP and Alkphos (AP) staining patterns are shown in the right panel. Photomicrographs of Alkphos staining were taken with a red filter to allow for superimposition of staining patterns with GFP fluorescence. Coexpression of Alkphos and GFP is shown within myofibers as a yellow-orange color. Myofibers are marked as follows: −, negative for both Alkphos and GFP; ∗, positive for only GFP; and +, positive for both GFP and Alkphos. All tissue samples were evaluated by whole mount for GFP expression prior to Hirt DNA preparation. We evaluated at least two tissue samples from each time point for colocalization studies of Alkphos and GFP, for which at least 10 sections were analyzed.
Rescue of bifunctional rAAV circular intermediates increases over time.
To determine the extent of recombination between circular AAV genomes, circular-form genomes were rescued as plasmids from low-molecular-weight Hirt DNA of muscle tissue coinfected with AV.GFP3ori and AV.Alkphos. Following transformation of E. coli Sure cells with Hirt DNA purified from infected muscles, the total number of GFP- and Alkphos-hybridizing Ampr bacterial plasmids was quantitated for each time point postinfection (Fig. 3A and B). As previously demonstrated (5), the abundance of circular AAV genomes rescued from AV.GFP3ori increased over time. For each muscle sample (three for each time point) 20 plasmid clones were evaluated for hybridization to GFP and Alkphos DNA probes, and the total number of plasmids was back calculated from the total CFU for each individual muscle sample. Figure 3B demonstrates the mean (±SEM; n = 3) total plasmids that hybridized to GFP or GFP and Alkphos probes at each time point. At 14 days postinfection, GFP- and Alkphos-cohybridizing plasmids were never observed. In contrast, at time points after 35 days, the percentage of GFP- and Alkphos-cohybridizing plasmids increased with time, and it reached 33% by 120 days (Fig. 3C). Since bacterial plasmid rescue can occur only through AV.GFP3ori genomes, this data suggests that recombination between independent Alkphos and GFP rAAV genomes takes place over time. Furthermore, no bifunctional plasmids were rescued in bacteria following transformation of mixed Hirt DNA samples prepared from muscles individually infected with AV.Alkphos and AV.GFP3ori, demonstrating that concatamerization did not occur in bacteria but rather had to take place in vivo (data not shown). In summary, these results are consistent with previous studies demonstrating a time-dependent concatamerization of monomer circular rAAV genomes in muscle (5).
FIG. 3.
Rescue of circular intermediates and characterization of DNA hybridization patterns. Using the ampicillin resistance gene and bacterial ori incorporated into the AV.GFP3ori vector, we assessed the extent of circular intermediate formation by rescuing Ampr plasmids following transformation of one-fifth of the isolated Hirt DNA (∼1 μg) into E. coli Sure cells. Twenty plasmids from each muscle sample were prepared and analyzed by slot blot hybridization against GFP, Alkphos, and Ampr gene 32P-labeled DNA probes. Approximately 50 ng of plasmid DNA was analyzed for both rescued plasmids and control plasmids carrying Amp (A8), GFP (A9), or Alkphos (A10) DNA segments. (A) A representative group demonstrating the hybridization patterns. (B) Mean (± SEM) number of rescued bacterial plasmids that hybridized either to GFP alone or to both GFP and Alkphos probes, following transformation of one-fifth of the Hirt DNA. These numbers were calculated from the percentages of plasmids hybridizing to GFP and/or Alkphos and the total CFU plating efficiency derived from the original transformation. In total, three independent muscle samples were analyzed, for a total of 60 plasmids at each time point. (C) Mean (± SEM) percentage of GFP hybridization-positive rescued plasmids that also demonstrated hybridization to Alkphos. These data demonstrate an increase in the abundance of rescued GFP- and Alkphos-coencoding circular intermediates over time.
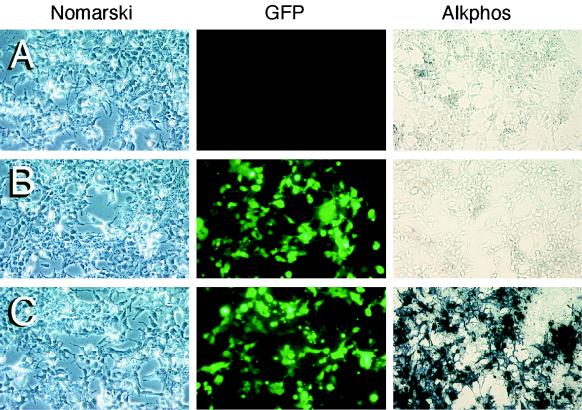
To evaluate the ability of circular intermediates to express the transgenes, we performed transient-transfection studies in 293 cells with rescued circular intermediate plasmids (Fig. 4A to C). Between 85 and 90% of rescued plasmids hybridizing to GFP probes on slot blots also expressed the GFP transgene in this transfection assay (Fig. 4D). The percentage of GFP-expressing plasmids that also expressed Alkphos rose over time in concordance with the hybridization data (Fig. 4D). However, approximately 40 to 50% of plasmids which were hybridization positive for Alkphos did not express the Alkphos transgene. The explanation for this finding is presently unknown, but it may represent recombinational deletion of the Rous sarcoma virus promoter driving Alkphos expression which occurred during concatamerization at sites near the 5′ ITR. These results demonstrate that intermolecular recombination between Alkphos- and GFP-derived circular intermediates occurs as part of the time-dependent concatamerization process of rAAV in muscle. To confirm that amplified plasmid stocks expressing both reporter genes were actually clonal (i.e., one plasmid rather than two independent plasmids resulting from contamination), a select number of bacterial clones expressing both transgenes were reisolated and the transfection assays were repeated. In all cases, plasmids expressing the two reporter genes remained clonal through two rounds of bacterial cloning. Hence, we conclude that dual reporter expression was not due to contamination of independent GFP- and Alkphos-expressing plasmids.
FIG. 4.
Transgene expression from rescued circular intermediates. Rescued circular intermediate plasmids were transfected into 293 cells for assessment of their ability to express transgenes. In these studies all GFP hybridization-positive clones from at least two muscles were tested for each time point and scored for their ability to express GFP and Alkphos. In total, at least 40 clones were evaluated for each time point. Three patterns of transgene expression were observed following transfection of these plasmids: (i) no gene expression (A), (ii) GFP expression only (B), and (iii) GFP and Alkphos expression (C). Panels A to C depict Nomarski photomicrographs (left) of GFP fluorescent fields (center) and Alkphos staining of a different field from the same culture (right). (D) Percentage of GFP hybridization-positive clones that also expressed GFP. Additionally, the percentage of GFP-expressing clones also expressing Alkphos is shown.
Concatamerization of AAV circular intermediates occurs through uniform intermolecular recombination between ITRs of independent viral genomes.
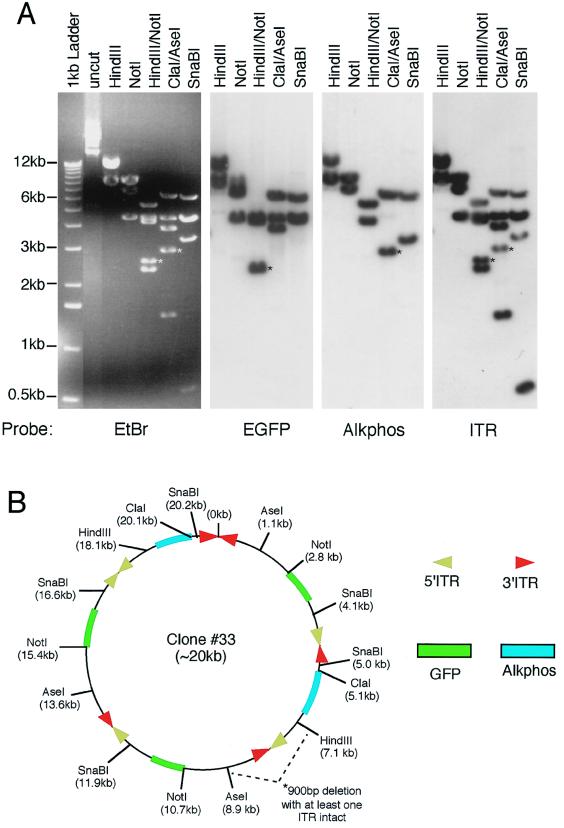
To better understand the mechanisms of circular concatamer formation, we performed detailed structural analysis of five bifunctional circular concatamers isolated from rAAV-infected muscle samples. As previously described for the AV.GFP3ori genome (5), the conversion of monomeric circular AAV genomes to large multimeric circular concatamers with a predominant head-to-tail structure increased with time in muscle (data not shown). To evaluate the structure of bifunctional circular concatamers, we performed restriction enzyme mapping and Southern blotting against 32P-labeled GFP, Alkphos, and ITR probes. The results from five analyzed plasmids demonstrated between three and six genomes within these circular concatamers. Two representative structures from the 35- and 80-day time points are shown in Fig. 5. Several interesting conclusions can be made from this structural analysis. First, as previously reported (5), head-to-tail oriented genomes could be seen in all isolated concatamers. However, several examples of head-to-head and tail-to-tail genome combinations of AV.Alkphos and AV.GFP3ori were also seen. Since head-to-head and tail-to-tail genome concatamers were never seen in muscles infected with AV.GFP3ori alone, we conclude that there must be a selective disadvantage for bacterial replication when ori sequences are in either of these conformations. However, since the AV.Alkphos genomes do not contain a bacterial origin of replication, we feel that this orientation is permitted in chimeric concatamers. Second, noticeable deletions and/or losses of restriction sites close to ITRs were noted in both examples shown in Fig. 5. It is not known whether deletions close to the ITR are a common event in the concatamerization process, but if so, this could account for the fact that only 60% of GFP- and Alkphos-hybridizing circular intermediates also expressed the Alkphos transgene.
FIG. 5.
Structural analysis of bifunctional concatamer circular intermediates. To fully characterize the nature of GFP- and Alkphos-coexpressing circular intermediates, detailed structural analyses were performed by using restriction enzyme mapping and Southern blot hybridization against GFP, Alkphos, and ITR 32P-labeled probes. (A and C) Results from Southern blot analysis of plasmid clones 33 (A) and 5 (C) are given as representative examples of circular intermediates isolated from 80- and 35-day Hirt DNAs of rAAV-infected muscle, respectively. Agarose gels were run in triplicate for each of these clones, and Southern blot filters were hybridized with one of the three DNA probes as indicated below each autoradiogram. Molecular sizes are indicated to the left of the ethidium bromide (EtBr)-stained agarose gel, and restriction enzymes are marked on the top of each gel or filter. (B and D) Deduced structures of plasmid clones 33 and 5, respectively, based on Southern blot analysis. For ease of comparison with the restriction maps of the viral genomes given in Fig. 1A, the positions of restriction enzyme sites are marked with the indicated orientation of intact viral genomes. However, in clone 33 a deletion occurred between the AseI and HindIII site of a head-to-tail array between AV.Alkphos and AV.GFP3ori, as reflected by a 900-bp reduction in the anticipated sizes of HindIII/NotI and ClaI/AseI fragments (marked by asterisks in panel A). Furthermore, the SphI site flanking an ITR was ablated in clone 5 (bands affected by this deletion are marked by asterisks in panel C). These deletions are not reflected in the size markings of the overall concatamer, since the exact region involved and/or the size of the deletion is unclear. Additionally, chemical sequence evidence for rescued circular intermediates suggests that the predominant form of ITR arrays may be in a double-D structure (i.e., one ITR flanked by two D sequences rather than two ITRs) (unpublished data), and hence ITR arrays containing fragments may appear 147 bp shorter than indicated. However, to more easily depict the orientations of viral genomes, we have indicated the positions of 5′ and 3′ ITRs rather than representing a single ITR at these junctions.
DISCUSSION
Concatamerization of rAAV in integrated proviral genomes has been long recognized. Recently, the association of this concatamerization process with the formation of high-molecular-weight circular genomes in muscle has suggested that this process may also be important in episomal persistence. Our findings demonstrating rescue of independent viral genomes within the same circular concatamer suggest that this process of concatamerization occurs through intermolecular recombination. Furthermore, previous findings demonstrating predominantly monomeric circular concatamers in muscle at 14 days (5) correlate with the present results demonstrating only GFP-expressing rescued circular intermediates at this time point. Together with the fact that bifunctional rescued circular concatamers increase with time, these results suggest that large concatamers form, at least in part, by recombination of monomeric circular precursor genomes. Alternative mechanisms involving the generation of circular concatamers from linear single- or double-stranded DNA templates may also contribute to the appearance of bifunctional circular genomes and cannot be ruled out in the present study. However, since an alternative model of concatamerization by rolling-circle replication would be expected to yield only GFP-expressing rescued plasmids in our model system, this mechanism cannot be solely responsible for concatamerization, if it exists at all.
Based on the structural analysis of these bifunctional circular intermediates, recombination between monomeric rAAV genomes undoubtedly is facilitated through ITR sequences. Directionality of this recombinational event appears to play less of a significant role than previously hypothesized (5), since head-to-tail-, head-to-head-, and tail-to-tail-oriented intermolecular concatamers were found. However, the most abundant orientation of ITRs was in a head-to-tail fashion. The extent of various genome orientations in circular concatamers may be affected by the percentages of flip and flop ITR structures within our amplified virus and/or by a selective disadvantage to bacterial replication origins in direct orientation. In addition, the extent to which recombination within ITR regions occurs in bacteria is presently unknown and may account for the deletions and/or restriction site losses near ITR arrays. However, serial passaging of bifunctional circular AAV genomes in bacteria has suggested that the structure of these large concatamers is impressively stable in bacteria.
In summary, these studies have increased our understanding of mechanisms involved in rAAV transduction and genome conversion. Ultimately this knowledge may lead to methods of increasing the utility of rAAV vectors for gene therapy.
ACKNOWLEDGMENTS
The work described here was supported by NIH grant R01 DK/HL58340 (to J.F.E.) and by Gene Therapy Core Center grant P30 DK54759 (to J.F.E.), cofunded by NIDDK and the Cystic Fibrosis Foundation.
We gratefully acknowledge the technical assistance of J. J. Heying and scientific interactions with Dongsheng Duan and Ziying Yan.
REFERENCES
- 1.Afione S A, Conrad C K, Kearns W G, Chunduru S, Adams R, Reynolds T C, Guggino W B, Cutting G R, Carter B J, Flotte T R. In vivo model of adeno-associated virus vector persistence and rescue. J Virol. 1996;70:3235–3241. doi: 10.1128/jvi.70.5.3235-3241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali R R, Reichel M B, Thrasher A J, Levinsky R J, Kinnon C, Kanuga N, Hunt D M, Bhattacharya S S. Gene transfer into the mouse retina mediated by an adeno-associated viral vector. Hum Mol Genet. 1996;5:591–594. doi: 10.1093/hmg/5.5.591. [DOI] [PubMed] [Google Scholar]
- 3.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan D, Fisher K J, Burda J F, Engelhardt J F. Structural and functional heterogeneity of integrated recombinant AAV genomes. Virus Res. 1997;48:41–56. doi: 10.1016/s0168-1702(96)01425-6. [DOI] [PubMed] [Google Scholar]
- 5.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher K J, Engelhardt J F. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan D, Yue Y, Yan Z, McCray P B, Engelhardt J F. Polarity influences the efficiency of recombinant adeno-associated virus infection in differentiated airway epithelia. Hum Gene Ther. 1999;9:2761–2776. doi: 10.1089/hum.1998.9.18-2761. [DOI] [PubMed] [Google Scholar]
- 7.Engelhardt J F, Schlossberg H, Yankaskas J R, Dudus L. Progenitor cells of the adult human airway involved in submucosal gland development. Development. 1995;121:2031–2046. doi: 10.1242/dev.121.7.2031. [DOI] [PubMed] [Google Scholar]
- 8.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 9.Fisher-Adams G, Wong K K, Jr, Podsakoff G, Forman S J, Chatterjee S. Integration of adeno-associated virus vectors in CD34+ human hematopoietic progenitor cells after transduction. Blood. 1996;88:492–504. [PubMed] [Google Scholar]
- 10.Flotte T R, Afione S A, Zeitlin P L. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am J Respir Cell Mol Biol. 1994;11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 11.Halbert C L, Standaert T A, Aitken M L, Alexander I E, Russell D W, Miller A D. Transduction by adeno-associated virus vectors in the rabbit airway: efficiency, persistence, and readministration. J Virol. 1997;71:5932–41. doi: 10.1128/jvi.71.8.5932-5941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzog R W, Hagstrom J N, Kung S H, Tai S J, Wilson J M, Fisher K J, High K A. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplitt M G, Leone P, Samulski R J, Xiao X, Pfaff D W, O’Malley K L, During M J. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 14.Kearns W G, Afione S A, Fulmer S B, Pang M C, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 15.Kessler P D, Podsakoff G M, Chen X, McQuiston S A, Colosi P C, Matelis L A, Kurtzman G J, Byrne B J. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Natl Acad Sci USA. 1996;93:14082–7. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao C H, Snyder R O, Schowalter D B, Patijn G A, Donahue B, Winther B, Kay M A. The kinetics of rAAV integration in the liver. Nat Genet. 1998;19:13–15. doi: 10.1038/ng0598-13. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 19.Ponnazhagan S, Erikson D, Kearns W G, Zhou S Z, Nahreini P, Wang X S, Srivastava A. Lack of site-specific integration of the recombinant adeno-associated virus 2 genomes in human cells. Hum Gene Ther. 1997;8:275–284. doi: 10.1089/hum.1997.8.3-275. [DOI] [PubMed] [Google Scholar]
- 20.Qing K, Khuntirat B, Mah C, Kube D M, Wang X S, Ponnazhagan S, Zhou S, Dwarki V J, Yoder M C, Srivastava A. Adeno-associated virus type 2-mediated gene transfer: correlation of tyrosine phosphorylation of the cellular single-stranded D sequence-binding protein with transgene expression in human cells in vitro and murine tissues in vivo. J Virol. 1998;72:1593–1599. doi: 10.1128/jvi.72.2.1593-1599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 22.Qing K, Wang X S, Kube D M, Ponnazhagan S, Bajpai A, Srivastava A. Role of tyrosine phosphorylation of a cellular protein in adeno-associated virus 2-mediated transgene expression. Proc Natl Acad Sci USA. 1997;94:10879–10884. doi: 10.1073/pnas.94.20.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samulski R J, Chang L S, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summerford C, Bartlett J S, Samulski R J. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 26.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincent-Lacaze N, Snyder R O, Gluzman R, Bohl D, Lagarde C, Danos O. Structure of adeno-associated virus vector DNA following transduction of the skeletal muscle. J Virol. 1999;73:1949–1955. doi: 10.1128/jvi.73.3.1949-1955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westfall T D, Kennedy C, Sneddon P. The ecto-ATPase inhibitor ARL 67156 enhances parasympathetic neurotransmission in the guinea-pig urinary bladder. Eur J Pharmacol. 1997;329:169–173. [PubMed] [Google Scholar]
- 29.Wu P, Phillips M I, Bui J, Terwilliger E F. Adeno-associated virus vector-mediated transgene integration into neurons and other nondividing cell targets. J Virol. 1998;72:5919–5926. doi: 10.1128/jvi.72.7.5919-5926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao X, Li J, Samulski R J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]