Abstract
Z-nucleic acid structures play vital roles in cellular processes and have implications in innate immunity due to their recognition by Zα domains containing proteins (Z-DNA/Z-RNA binding proteins, ZBPs). Although Zα domains have been identified in six proteins, including viral E3L, ORF112, and I73R, as well as, cellular ADAR1, ZBP1, and PKZ, their prevalence across living organisms remains largely unexplored. In this study, we introduce a computational approach to predict Zα domains, leading to the revelation of previously unidentified Zα domain-containing proteins in eukaryotic organisms, including non-metazoan species. Our findings encompass the discovery of new ZBPs in previously unexplored giant viruses, members of the Nucleocytoviricota phylum. Through experimental validation, we confirm the Zα functionality of select proteins, establishing their capability to induce the B-to-Z conversion. Additionally, we identify Zα-like domains within bacterial proteins. While these domains share certain features with Zα domains, they lack the ability to bind to Z-nucleic acids or facilitate the B-to-Z DNA conversion. Our findings significantly expand the ZBP family across a wide spectrum of organisms and raise intriguing questions about the evolutionary origins of Zα-containing proteins. Moreover, our study offers fresh perspectives on the functional significance of Zα domains in virus sensing and innate immunity and opens avenues for exploring hitherto undiscovered functions of ZBPs.
Keywords: innate immunity, Zα domain, B-Z conversion, B-to-Z conversion, ADAR1, ZBP1
Nucleic acids adopt a variety of structures other than right-handed double helices. Both DNA and RNA can form triplexes, I-motifs, G-quadruplexes, and left-handed helices referred to as “Z-nucleic acids” (Z-DNA and Z-RNA) (1, 2, 3). These alternative structures are critical components of cellular functions, influencing a myriad of biological processes (4, 5, 6). In particular, it is now well established that the innate immune system exploits Z-nucleic acids as pathogen-associated molecular patterns and damage-associated molecular patterns (7, 8).
Z-DNA/Z-RNA biology owes its now-recognized relevance to several proteins containing Zα domains, which are responsible for recognizing Z-nucleic acids (9). Zα domains are found in metazoan proteins involved in immunity and cancer, as well as in viral proteins. The first crystal structure revealed that the Zα domain of ADAR1 (adenosine deaminase acting on RNA 1) adopts a winged-helix-turn-helix (wHTH) fold. This structure is characterized by a compact α/β architecture encompassing a three-helix bundle (α1 to α3), juxtaposed to a twisted antiparallel β-sheet (β1 to β3) (10). Specific amino acids are important for the specific recognition of Z-nucleic acids, such as N173 and Y177 within helix α3, and P192 and P193; W195 contributes to both protein stability and nucleic acid binding (10). This earlier work demonstrated that the commonly found wHTH domain across proteins only binds to Z-nucleic acids when these amino acids are present.
The only other proteins known to contain Zα domains are ZBP1 in mammals, amphibia, and reptilia (11), PKZ in salmoniform and cypriniform fish (12), E3L in poxviruses (13), ORF112 in cyprinid herpesviruses (CyHV-1-to-3) (14) and I73R in the asfarviridae African swine fever virus (ASFV) (15). Our current understanding of the biology of these proteins points to antagonistic relationships in innate immunity and a potential regulatory role in gene expression (16, 17). Briefly, the recognition of Z-nucleic acids by ADAR1 and ZBP1 is essential for the immune system's ability to detect and respond to viral threats effectively (18). Conversely, viruses have evolved Zα-containing proteins like E3L to subvert and antagonize host immune defenses, thereby enhancing their own virulence and survival within the host organism (19, 20). However, Zα domains have functional roles that need to be nuanced, as they cannot always be swapped, especially across different viruses (20, 21). This level of complexity implies an evolutionary balance between a virus and its host, where the unique characteristics of Zα domains are finely tuned to suit the particular dynamics of each virus-host relationship.
Because Z-nucleic acid biology is now viewed as a determinant for cell fate during infection and auto-immune diseases, one could ask the question of whether more proteins with Zα domains could be discovered, and in what species. Recent computational efforts based on structure similarity search have unveiled the existence of 14 potential Zα domains (22). Additionally, two other potential ZBPs were identified, namely RBP7910 in Trypanosoma brucei (23), and DprA in Riemerella anatipestifer (24). However, these studies did not address the absence of the complete set of required amino acids for binding Z-nucleic acids, and no experimental validations of these putative Zα domains were carried out.
Here, we introduce a computational approach for predicting Zα domains, which combines primary sequence analysis, three-dimensional modeling, and manual examination of the resulting hits through structural analysis. This method led to the discovery of 8 putative Zα domain-containing proteins in eukaryotic organisms, and 68 in giant viruses diverse and mainly unexplored giant viruses in the viral phylum Nucleocytoviricota. We also report Zα-like domains in bacterial proteins, which deviate somewhat from their eukaryotic and viral counterparts in primary amino acid sequence. Finally, we have experimentally validated our predictions by assessing the Z-DNA binding capacity and ability to shift the B-Z equilibrium for two Zα and two Zα-like candidates. This work offers fresh insights into the functional role of Zα domains in virus sensing and innate immunity. Furthermore, it may shed light on previously undiscovered functions of ZBPs, expanding our understanding of their roles in biological processes.
Results and discussion
A combinatory method to predict Zα-domain and Zα-like candidates
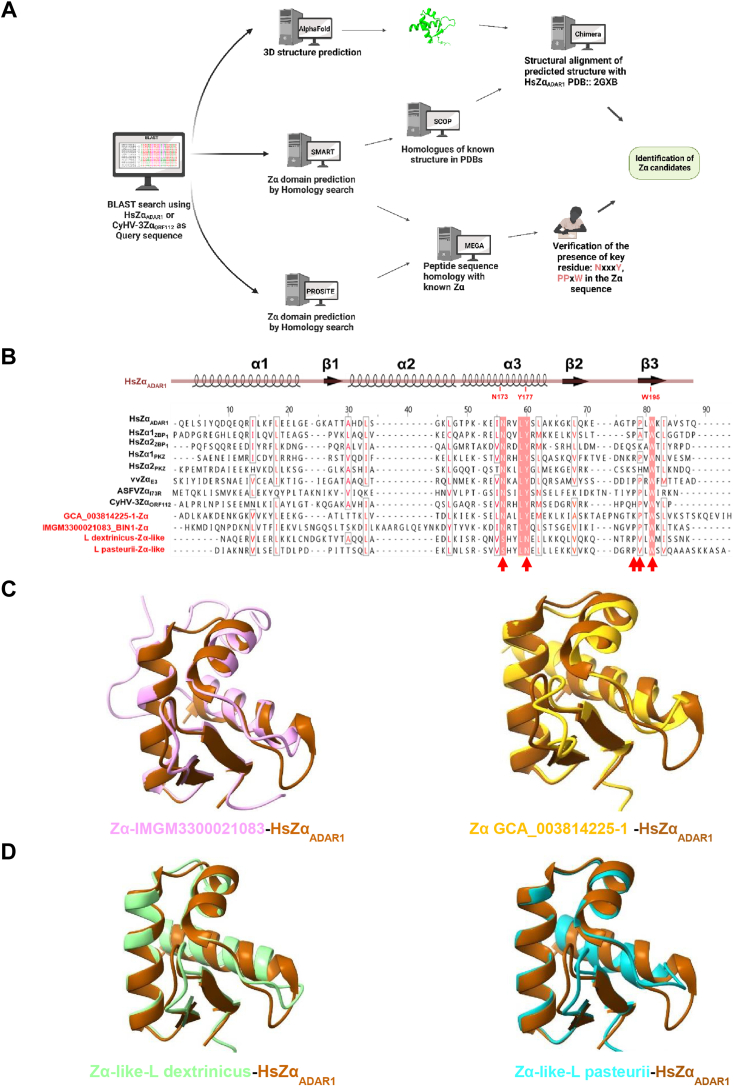
Initially, we employed BLAST search tools in conjunction with primary sequence analysis by domain prediction software SMART and PROSITE to identify the Zα-domain candidates. Subsequently, 3D structure analysis and prediction tools were used to refine our predictions. Leveraging our existing knowledge of the essential attributes of the Zα-domain, we further refined the results derived from our in silico analysis (Fig. 1A). We used multiple sequence alignments to compare the candidate sequences with some representative Zα domains in all 6 known ZBPs. We checked the presence of residues crucial for the functionality of the Zα-domain by primary sequence alignment. Among these, the NxxxY motif within α3 is essential for Z-DNA binding, while the pPxW motif in the β-wing plays a significant role in both Z-DNA (10) binding and A-to-Z conversion (25). In the crystal structure of the HsZαADAR1, tryptophan 195 has been demonstrated to be vital not only for protein stability but also for DNA binding. The double mutation involving asparagine 173 and tyrosine 177 within the α3 helix completely abolishes Zα′s capacity to bind Z-DNA, rendering it functionally inoperative for both mammalian and viral ZBPs. Additionally, proline 192 and proline 193 contribute significantly through van der Waals interactions with DNA (10). Despite the limited overall sequence similarity typical to Zα domains, the aliphatic residues (marked in red) from the three helices, along with Tryptophan 195 in the β-wing, exhibit strong conservation in all candidates. These residues are well-established for their role in supporting the hydrophobic core of the winged helix-turn-helix structure (Figs. 1B, and S1) (10). In line with this, the predicted 3D structures, generated by AlphaFold (26), reveal that all candidates adopt the characteristic winged-HTH conformation. The 3D fold comparisons underscore a remarkable degree of structural similarity between these candidates and the crystallized ADAR1 Zα domain, PDB ID 2GXB (27) (Fig. 1, C and D). Despite these similarities, some candidates found in bacteria lack conserved residues in the corresponding positions N173 and Y177 in HsZαADAR1 displaying instead a serine and an asparagine respectively. As such, we classify them as Zα-like candidates (Figs. 1B and S1).
Figure 1.
Prediction of new Zα candidates. A, the summary of the workflow for identifying Zα candidates. B, alignment of two predicted Zα and two Zα-like with known Zα domains from human and mouse ADAR1 (HsZαADAR1), human ZBP1 (HsZαZBP1), and viral from vaccinia virus and cyprinid-herpesvirus-3 (vvZαE3 ASFV ZαI73R CyHV-3Zα112). The consensus structure Zα domain is shown at the top. The key DNA-interacting residues in ADAR1 (corresponding to Asn174, Tyr177, P192, P193, and Trp195 in HsZαADAR1) are pointed with red arrows. Hydrophobic residues in the hydrophobic core are highlighted in red front. C, structural alignment of AlphaFold predicted structure of Zα candidates with HsZαADAR1 (brown, PDB ID 2GXB). D, structural alignment of AlphaFold predicted structure of Zα-like candidate with HsZαADAR1 (brown, PDB ID 2GXB).
Identification of novel Zα-domain candidates in eukaryotic organisms
All predicted Zα candidates maintain the conserved residues N173 and Y177 in HsZαADAR1 crucial for Z-DNA binding (10). In Anthozoa, a class of marine invertebrates that includes sea anemones, stony corals, and soft corals, we predicted a new Zα containing protein. Interestingly, in some families such as the Poritidae or Actinidea, the novel Zα displays all important residues found in the known Zα. However, some families such as Acroporidae or Caryophylliidae display a Zα domain in which the Y177 is substituted by phenylalanine and W195 is replaced by a tyrosine (Fig. S1). We predicted other potential Zα domain-containing proteins in other eukaryotic organisms out of metazoan and DNA viruses, such as a protein annotated as transcription activator p15 in phytoplankton, and another annotated as acetate-CoA ligase in unicellular microalgae of the Symbiodiniaceae family (Figs. S1 and S3).
Identification of novel Zα-domain candidates in giant viruses
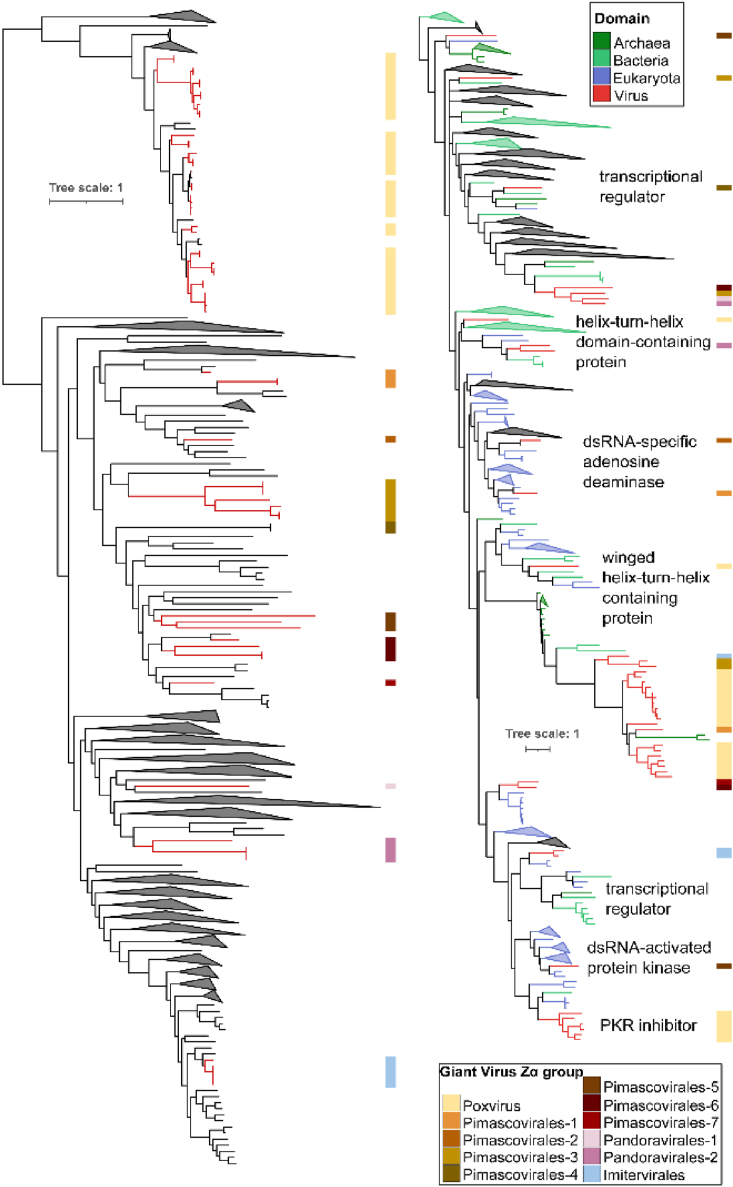
The most substantial contingent of novel ZBP candidates emerged within giant virus metagenome-assembled genomes (GVMAGs), revealing the presence of potential ZBPs in members of the orders pimascovirales, pandoravirales, and imitervirales. These proteins are distinct from previously documented ZBPs in fish-herpesviruses, poxviruses, and African swine fever virus (ASFV) (Fig. 2). These giant viruses were recovered in our previous metagenomic survey of samples from marine and freshwater, soil, and thermal environments (Fig. S2) (28), but also included additional sequences from deep-sea sediments (29) and permafrost (from 49 to 53,000 years old) (30). Our analysis shows that the candidate ZBPs in these giant viruses share similar domain architectures as in previously identified viral-containing proteins (13, 15, 20). The only exception is the viral ZBP candidate in which Zα is associated with tRNA metabolism domains (IMGM3300021083) (Fig. S3).
Figure 2.
Phylogenetic tree of proteins that contain Zα and Zα-like domains. Proteins that were included in the analysis are giant virus ZBPs and homologs identified and extracted from the NCBI nr database.
In our phylogenetic analysis, the clade topology in the ZBPs protein tree does not align with the topology found in the Nucleocytoviricota species tree (Fig. 2). This finding suggests that proteins with Zα domains have been independently acquired, likely multiple times, originating from a specific host via gene transfer. This possibility, as proposed by Kuś et al., suggests that CyHV-3 ORF112 might have been co-opted independently from the host by the common ancestor of the cyprinid herpesviruses, rather than through horizontal transfer from a poxvirus (31). Such scenarios are further supported by the fact that in our tree, proteins from different giant virus lineages group together and are simultaneously intertwined in monophyletic clades with eukaryotic and archaeal homologs.
Our results imply that giant viruses with the Zα domain likely infect a wide range of organisms ranging from dinoflagellates to marine vertebrates and arthropods as well as early diverging metazoans such as cnidarians (32). The presence of ZBP in diverse Nucleocytoviricota lineages raises the question of the role of Zα in the interactions of these viruses with their hosts, and how this fits into our current understanding of the role of ZBPs in animal innate immunity. Furthermore, the manifold presence of ZBPs in divergent giant virus lineages, known to infect protists and algae, sparks curiosity. This observation is particularly intriguing given that virus-protist interactions entail mechanisms distinctly different from the innate immunity found in animals (33). Could the presence of ZBP indicate that these giant viruses infect metazoan hosts? In line with this, the ASFV infects two phylogenetically distant hosts, functioning as an arthropod-borne DNA virus (34), significantly impacting pig farming due to its high virulence (19).
Experimental validation of Zα and Zα-like candidates
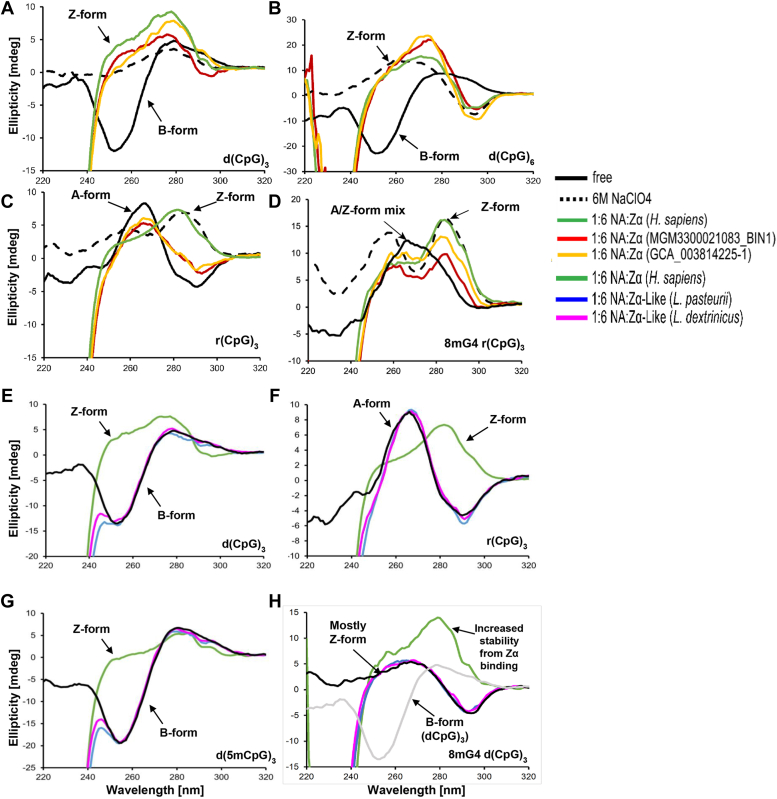
To validate our prediction, we experimentally studied some of these Zα and Zα-like domains by circular dichroism, an established method to probe their ability to induce B(A)-to-Z conversion (35). We found that two predicted Zα domains from two distinct members of the viral order pimascovirales are able to efficiently convert B-DNA duplex of different lengths d(CpG)3 and d(CpG)6 to Z-form (Fig. 3, A and B). Interestingly, we found that these two Zα domains are not able to induce A-to-Z conversion of RNA duplex as seen for the positive control HsZαADAR1 or in the high salt concentration condition (Fig. 3C). Consequently, our study identifies new cases of Zα capable of converting B-DNA to Z-form but not A-RNA. Indeed, the conversion of A-RNA to Z-RNA is a less favorable and energetically costly process compared with B-DNA to Z-DNA conversion (35). To test the binding capacity to sequence contexts that have a lower B-Z energy barrier or preformed Z-form nucleic acids, we used a singly methylated RNA duplex 8mG4 r(CpG)3 which stabilized 50% of the population in Z-form (36, 37) (Fig. 3D). In this case, the Zα domains are capable of flipping the RNA to Z-form. This indicates that Zα domains bind to the pre-stabilized Z-RNA since they cannot flip A-RNA to the Z-form.
Figure 3.
Experimental validation by circular dichroism. A and B, the predicted viral Zα domains induce B-to-Z conversion of DNA. C, viral Zα domains do not convert A-RNA to Z-RNA. D, viral Zα domains do convert an RNA with prestabilized Z-conformation (A:Z form ∼50%/50%) to full Z-RNA. E and F, Zα –like domains are not able to induce B(A)-to-Z conversion. G and H, Zα –like domains do not bind to Z-nucleic acids. For comparison, the profile of a B-form DNA is shown in grey in (H), demonstrating the presence of a large portion of Z-form in 8mG4 d(CpG)3. The profiles overlaid with profiles from second independent measurements are provided in Fig. S4.
It is interesting to speculate on the distinction between competent A/B-to-Z flipping Zα domains, Zα domains that can only flip B-DNA, and the nonfunctional Zα-like domains, and their evolutionary/functional relevance. Numerous studies have tried to identify key residues/regions within the Zα domain that are important for binding and flipping nucleic acids to the Z-conformation. Kim et al. were able to restore the partial function of ADAR1’s Zβ domain by mutating Ile335 back to a tyrosine as is typical for all other ZBDs (38). Additionally, residues within the β-wing play a large role in not only the stabilization of the Z-conformation but also the rate at which it converts nucleic acids to the Z-conformation (39, 40). Furthermore, distantly related wHTH domains with no ability to convert nucleic acids to the Z-conformation can be converted into good Z-binders and converters through directed mutagenesis (41). It is likely that numerous factors contribute to the ability of any given Zα domain’s ability to bind and convert nucleic acids to the Z-conformation, and the loss of one stabilizing interaction may result in the inability to convert A-RNA while still retaining the ability to convert B-DNA.
As expected, we found that for the Zα-like domain, there was no conversion of B-DNA to Z-DNA or A-RNA to Z-RNA, in contrast to our positive control HsZαADAR1 (Fig. 3, E and F). To test the binding capacity to sequence contexts that have a lower B-Z energy barrier or preformed Z-form nucleic acids, we used a methylated DNA duplex d(m5CpG)3 and the singly methylated RNA duplex 8mG4 d(CpG)3 respectively (36, 37). The Zα-like domains show no ability to stabilize or shift the B-Z transition equilibrium of the Z-nucleic acid analogs (Fig. 3, G and H). The Zα-like domain is found in the bacterial protein annotated as sigma 54 (σ54)-interacting transcriptional regulator. σ54 also encompasses an ATPase AAA region, which overlaps with the Holliday junction DNA helicase RuvB. RuvB is a component of the RuvABC revolvasome, crucial for resolving Holliday junctions (42). Remarkably, both Holliday junctions and Z-DNA are found in bacterial biofilms (43, 44). Studies have shown that RuvA protein or DNABII stabilizes Holliday junctions, inducing supercoiling that reduces the energy needed for B-Z transition and promotes Z-DNA formation in the extracellular DNA biofilm matrix (43). Therefore, it would be interesting to test the B-to-Z conversion of the predicted association between the Zα-like domain and the helicase RuvB in σ54. Moreover, knowing that bacteria of the genius of Lactobacillus are biofilm-forming bacteria (45), we hypothesize that σ54 could be involved in the biofilm matrix formation.
Experimental procedures
In silico prediction of Zα domain
To identify new Zα domain candidates, we developed a combinatorial approach. Using the Basic Local Alignment Search Tool, blastp algorithm of NCBI (46), we performed taxid-exclusion blast and taxid-targeted blast using the Zα peptides from CyHV-3 and HsADAR1 as the query sequences. The candidates with a minimum of three crucial residues conserved or featuring less drastic substitutions (such as in A digitifera-Zα, where phenylalanine replaces the tyrosine in helix-α3 and a tyrosine replaces the tryptophane in the β-wing Fig. S1), were considered as the best hits and were selected. Their primary sequences were subjected to Zα domain prediction using SMART (47) and PROSITE (48, 49) software enabling us to determine the peptide length covered by the Zα domain. Leveraging our knowledge of the essential attributes of the Zα-domain, we further refined the results derived from our in silico prediction. We check the presence of certain residues crucial for the functionality of the Zα-domain by primary sequence alignment using MEGA-11 (50) and UGENE (51) tools. The presence or absence of these critical motifs served as the final determinant in our analysis, allowing us to confidently identify Zα candidates. Any candidates lacking tyrosine and asparagine were categorized as Zα-like, given their alignment with other defining characteristics. SMART software also outputs structure primary sequence homologs of known in the protein data bank (PDB) or blast with known SCOPs (structural classification of proteins) of the Zα, for example, d1qbja (52). Finally, to strengthen the Zα prediction, a structure prediction using AlphaFold (26, 53) confirms the winged helix-turn-helix fold superposable with the crystallized Zα available with the PDBs using UCSF ChimeraX (54). With the predicted 3D structures, we performed a 3D-BLAST search (55) to confirm that the best hits are entries corresponding to the Zα domain. Figures 1A and S3 were created with BioRender.com.
Sequence source and annotation
Most of the giant viruses used in our analysis were identified in our metagenomic analysis performed in samples from different biotopes (S2) (28). We also included other giant viruses identified in other studies such as Pithovirus (29) and Marseillesvirus (30). The sequencing of the full viral genome or the viral genome fragment covering the coding sequence of the ZBP allowed the complete identification of the open reading frame corresponding to the ZBP. For the other organisms, the ZBPs are annotated at the PDB genomic level and (protein and mRNA level for Micromonas pusilla (56). Zα domain sequence and the ID numbers of ZBPs are listed in Table S6.
Phylogenetic analysis
Novel Zα and Zα-like domain sequences were clustered at 40% identity using cd-hit (57) and the resulting cluster representative sequences were aligned using mafft-linsi (version 7.505) (58). We used the hmmbuild (version 3.3.2, hmmer.org) to create a profile hidden Markov model from the representative sequence alignment and then used it as a query for the hmmsearch program (version 3.3.2, hmmer.org) on the NCBI nr database (59). Protein sequences that had an overall e-value <0.005 and contained at least one domain with length ≥ 60 amino acids were considered as significant hits to the model. Then we de-replicated the protein sequences with significant hits with cd-hit at 90% identity and an alignment length covering at least 60% the length of the longer sequence (-aL 0.6). The representative protein sequences were aligned together with Zα containing sequences from the viral phylum Nucleocytoviricota using the mafft FFT-NS-2 method (58). The alignment was trimmed with trimal with the -gt 0.1 option (60). A protein tree was built using IQ-Tree (version 1.6.12) (61) with 1000 ultra fast bootstrap replicates (62) and the JTT+R10 model, which was selected with the ModelFinder feature in IQ-Tree (63). The tree was visualised using iToL (64).
To build the giant virus species tree the nsgtree pipeline was used (https://github.com/NeLLi-team/nsgtree) on representative genome of the viral phylum Nucleocytoviricota (65) and additional GVMAGs that contained proteins with a Zα domain. In brief, 7 giant virus orthologous groups (GVOGs) were identified using hmmsearch (version 3.1/b2, hmmer.org), extracted GVOGs were aligned with mafft (version 7.31) (58), trimmed with trimal (-gt 0.1, v1.4) and concatenated. A species tree was built from the supermatrix alignment using IQ-Tree (version 2.03) (66) with LG+F + I + G4 and visualized in iToL (64).
Protein expression and purification
The Zα domains from Homo sapiens ADAR1, L. pasteurii, L. dextrinicus, giant virus (IMGM3300021083 and GCA_003814225–1) were cloned into the pet-28a(+) plasmid (N-terminal 6x His-tag and thrombin cleavage site between His tag and the Zα sequence). The plasmids were transformed and expressed in BL21(DE3) E. coli cells. The cell cultures were grown in Luria Broth (LB) to an OD600 of ∼0.4 and induced with IPTG to a final concentration of 1 mM and allowed to express Zα for 4 h at 37 °C, then centrifuged to collect the cell pellets. Cell pellets were resuspended in lysis buffer (50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole, 1 mM BME) and sonicated. Lysate was centrifuged and the supernatant was applied to a His-trap column, (50 mM Tris-HCl (pH 8.0), 1 M NaCl, 10 mM imidazole, 1 mM BME), and eluted in 20 ml of elution buffer washed with 40 ml of lysis buffer, 80 ml of wash buffer (50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 500 mM imidazole, 1 mM BME). The eluents were concentrated to ∼2 ml and applied to a HiLoad 16/600 Superdex 200 prep grade Gel Filtration Column (GE Healthcare) and the peak corresponding to pure protein (which elutes at ∼80 ml in 20 mM potassium phosphate (pH 6.4), 300 mM NaCl) was collected and concentrated using an Amicon 3 kDa cutoff centrifugal filter (Millipore-Sigma, Burlington, MA). Zα was dialyzed and concentrated into 20 mM potassium phosphate (pH 6.4), 25 mM, 0.5 mM EDTA and concentrated to ∼2 mM using an Amicon 3 kDa cutoff centrifugal filter (Millipore-Sigma). Subsequent dilutions were made in 20 mM potassium phosphate (pH 6.4), 25 mM, 0.5 mM EDTA as needed for the different experiments. We confirmed that all recombinant proteins were free of contaminating nucleic acid (Fig. S5).
DNA and RNA constructs and preparation
The d(CpG)3, d(CpG)6, and d(5mCpG)3 constructs were synthesized by Integrated DNA Technologies. The r(CpG)3 construct was synthesized by Dharmacon (a part of Horizon Discovery). The 8mG4 d(CpG)3 construct was synthesized by the Yale School of Medicine oligo synthesis resource. All nucleic acid constructs in this study were heat annealed prior to use at 95 °C for 5 minutes followed by slow cooling at room temperature for 30 min. For circular dichroism (CD) measurements, all of the constructs were 50 μM in 20 mM potassium phosphate, 25 mM NaCl (pH 6.4), 0.5 mM EDTA, or the same buffer but with 6 M NaClO4 or a 1:6 M ratio of nucleic acid:Zα.
Circular dichroism
All CD measurements were collected using a JASCO J-815 CD spectrometer (run using Spectra Manager version 2 (JASCO)) in a 0.1 cm quartz cuvette. Z-form adoption by high-salt and Zα binding was carried out by incubating the constructs in 6M NaClO4 or with saturating amounts of Zα at a 1:6 M ratio of nucleic acid:Zα for 1 h at 42 °C before cooling to 25 °C and measuring. Spectra were collected in 1-nm steps from 320 to 220 nm with an average of two scans. All measurements with unmodified nucleic acids as well as d(5mCpG)3 and 8mG4 r(CpG)3 were conducted twice, and the profiles shown in Figure 3 overlaid with profiles from second independent measurements are provided in Fig. S4. All duplicate profiles confirmed the high reproducibility of the measurements.
Conclusion
In summary, our study employed a computational combined with an experimental approach that enabled the identification of potential Zα-containing proteins across various organisms. These ZBPs open up new avenues in studies of Zα domains and Z-nucleic acids in virus sensing and innate immunity. The domain architectures within predicted Z-DNA binding proteins imply that they potentially serve functions distinct from those currently attributed to the known ZBPs (Fig. S3). These functions could be conserved in mammals but in the form of synergy between a ZBP and another protein like the cooperative function between ADAR1p150 and DHX9 (67). Furthermore, our experimental validation of two Zα-containing proteins confirms not only their presence in currently uncultivated giant viruses but also their functionality. Our findings open new questions regarding the role of Zα domains in these giant viruses and suggest that the interplay between ZBPs/Z-nucleic acid is an evolutionarily ancient feature as shown for the cGAS/STING pathway (68). It is tempting to speculate that the ZBPs we predicted in giant viruses and early diverging metazoan could be involved in an ancient pathogen detection mechanism that could provide functional and evolutionary insight into innate immune signaling pathways in animals. Our validation results underscore the need for heightened caution in Zα domain prediction. We demonstrate that the mere detection of a Zα domain by prediction tools or the observation of structural similarities between predicted wHTH and crystallized Zα domains fall short of substantiating experimental B(A)-to-Z conversion or Z-nucleic acid-binding. These findings emphasize that the ability to bind to Z-nucleic acid should be the definitive criterion for establishing a wHTH as a true Zα domain.
In our study, we focused on validating the Zα and Zα-like domains found mainly in multi-domain proteins. Although we demonstrate that wHTH domains we classify as Zα-like cannot bind Z-nucleic acid nor convert B-to-Z, we cannot exclude the possibility that the proteins with Zα-like domains may still be involved in Z-nucleic biology. This is because other proteins lacking Zα domains have been found to bind Z-DNA (69) or play a role in regulating Z-RNA sensing (70).
Data availability
The data that support the findings of this study are available from the corresponding authors upon request. The primary sequence and the id-numbers of the new ZBPs and Zα-like containing proteins are summarized in Fig. S3 and Table S6. Other sequences used in this work are available from Frederik Schulz and Mamadou Amadou Diallo upon reasonable request.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The work conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. M. A. D Thanks Dr Anne Silvestre INRA-Nouzilly France, and Dr Anne-Catherine Ahn University of Wageningen Netherlands for their support in this project. We gratefully acknowledge the National Science Foundation (Award #2153787 to Q.V. and B. V.), and the National Institutes of Health (Award #1F31AI167396 to P. N.).
Author contributions
M. A. D., B. V., F. S., Q. V., M. A. H., and E. D. writing–review & editing; M. A. D., B. V., and Q. V. writing–original draft; M. A. D., B. V., F. S., and Q. V. validation; M. A. D., B. V., and Q. V. supervision; M. A. D., F. S., and M. F. R. software; M. A. D., B. V., F. S., and Q. V. resources; M. A. D. project administration; M. A. D., B. V., F. S., Q. V., P. J. N., M. F. R., and J. B. K. formal analysis; M. A. D. and B. V. data curation; M. A. D. conceptualization; M. A. D., B. V., and Q. V. investigation; B. V., F. S., and Q. V. funding acquisition; P. J. N., J. V., M. F. R., and J. B. K. methodology.
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Contributor Information
Frederik Schulz, Email: fschulz@lbl.gov.
Beat Vögeli, Email: beat.vogeli@cuanschutz.edu.
Mamadou Amadou Diallo, Email: mdiallo@uliege.be, mamadouad@irc.vib-ugent.be.
Supporting information
Supplementary tables caption.
References
- 1.Wang A.H.J., Quigley G.J., Kolpak F.J., Crawford J.L., Van Boom J.H., Van Der Marel G., et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 2.Hall K., Cruz P., Tinoco I., Jovin T.M., Van De Sande J.H. Z-RNA’--a left-handed RNA double helix. Nature. 1984;311:584–586. doi: 10.1038/311584a0. [DOI] [PubMed] [Google Scholar]
- 3.Miglietta G., Russo M., Capranico G. G-quadruplex-R-loop interactions and the mechanism of anticancer G-quadruplex binders. Nucleic Acids Res. 2020;48:11942–11957. doi: 10.1093/nar/gkaa944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karki R., Kanneganti T.D. ADAR1 and ZBP1 in innate immunity, cell death, and disease. Trends Immunol. 2023;44:201–216. doi: 10.1016/j.it.2023.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols P.J., Krall J.B., Henen M.A., Vögeli B., Vicens Q. Z-RNA biology: a central role in the innate immune response? RNA. 2023;29:273–281. doi: 10.1261/rna.079429.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song J., Gooding A.R., Hemphill W.O., Love B.D., Robertson A., Yao L., et al. Structural basis for inactivation of PRC2 by G-quadruplex RNA. Science. 2023;381:1331–1337. doi: 10.1126/science.adh0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thapa R.J., Ingram J.P., Ragan K.B., Nogusa S., Boyd D.F., Benitez A.A., et al. DAI senses influenza A virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe. 2016;20:674–681. doi: 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei Y., VanPortfliet J.J., Chen Y.-F., Bryant J.D., Li Y., Fails D., et al. Cooperative sensing of mitochondrial DNA by ZBP1 and cGAS promotes cardiotoxicity. Cell. 2023;186:3013–3032.e22. doi: 10.1016/j.cell.2023.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbert A.G., Spitzner J.R., Lowenhaupt K., Rich A. Z-DNA binding protein from chicken blood nuclei. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3339–3342. doi: 10.1073/pnas.90.8.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz T., Rould M.A., Lowenhaupt K., Herbert A., Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001 doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 12.Rothenburg S., Deigendesch N., Dittmar K., Koch-Nolte F., Haag F., Lowenhaupt K., et al. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1602–1607. doi: 10.1073/pnas.0408714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y.-G., Muralinath M., Brandt T., Pearcy M., Hauns K., Lowenhaupt K., et al. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tome A.R., Kus K., Correia S., Paulo L.M., Zacarias S., de Rosa M., et al. Crystal structure of a poxvirus-like Zalpha domain from cyprinid herpesvirus 3. J. Virol. 2013;87:3998–4004. doi: 10.1128/JVI.03116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L., Miao Y., Wang Z., Chen H., Dong P., Zhang H., et al. Structural insight into African swine fever virus I73R protein reveals it as a Z-DNA binding protein. Transbound. Emerg. Dis. 2022;69:e1923–e1935. doi: 10.1111/tbed.14527. [DOI] [PubMed] [Google Scholar]
- 16.Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh D.-B., Kim Y.-G., Rich A. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc. Nat. Acad. Sci. U. S. A. 2002;99:16666–16671. doi: 10.1073/pnas.262672699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koehler H., Cotsmire S., Zhang T., Balachandran S., Upton J.W., Langland J., et al. Vaccinia virus E3 prevents sensing of Z-RNA to block ZBP1-dependent necroptosis. Cell Host Microbe. 2021;29:1266–1276.e5. doi: 10.1016/j.chom.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Shen Z., Xie Z., Song Y., Li Y., Liang R., et al. African swine fever virus I73R is a critical virulence-related gene: a potential target for attenuation. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2210808120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diallo M.A., Pirotte S., Hu Y., Morvan L., Rakus K., Suárez N.M., et al. A fish herpesvirus highlights functional diversities among Zα domains related to phase separation induction and A-to-Z conversion. Nucleic Acids Res. 2023;51:806–830. doi: 10.1093/nar/gkac761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vijaysri S., Talasela L., Mercer A.A., Mcinnes C.J., Jacobs B.L., Langland J.O. The Orf virus E3L homologue is able to complement deletion of the vaccinia virus E3L gene in vitro but not in vivo. Virology. 2003;314:305–314. doi: 10.1016/s0042-6822(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 22.Bartas M., Slychko K., Brázda V., Červeň J., Beaudoin C.A., Blundell T.L., et al. Searching for new Z-DNA/Z-RNA binding proteins based on structural similarity to experimentally validated Zα domain. Int. J. Mol. Sci. 2022;23:768. doi: 10.3390/ijms23020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikpour N., Salavati R. The RNA binding activity of the first identified trypanosome protein with Z-DNA-binding domains. Sci. Rep. 2019;9:5904. doi: 10.1038/s41598-019-42409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L., Tian X., Liu M., Wang M., Biville F., Cheng A., et al. DprA is essential for natural competence in riemerella anatipestifer and has a conserved evolutionary mechanism. Front. Genet. 2019;10:429. doi: 10.3389/fgene.2019.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langeberg C.J., Nichols P.J., Henen M.A., Vicens Q., Vögeli B. Differential structural features of two mutant ADAR1p150 Zα domains associated with aicardi-goutières syndrome. J. Mol. Biol. 2023;435 doi: 10.1016/j.jmb.2023.168040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Placido D., Brown B.A., Lowenhaupt K., Rich A., Athanasiadis A. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz F., Roux S., Paez-Espino D., Jungbluth S., Walsh D.A., Denef V.J., et al. Giant virus diversity and host interactions through global metagenomics. Nature. 2020;578:432–436. doi: 10.1038/s41586-020-1957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bäckström D., Yutin N., Jørgensen S.L., Dharamshi J., Homa F., Zaremba-Niedwiedzka K., et al. Virus genomes from deep sea sediments expand the ocean megavirome and support independent origins of viral gigantism. mBio. 2019;10 doi: 10.1128/mBio.02497-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rigou S., Christo-Foroux E., Santini S., Goncharov A., Strauss J., Grosse G., et al. Metagenomic survey of the microbiome of ancient Siberian permafrost and modern Kamchatkan cryosols. microLife. 2022;3:uqac003. doi: 10.1093/femsml/uqac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuś K., Rakus K., Boutier M., Tsigkri T., Gabriel L., Vanderplasschen A., et al. The structure of the cyprinid herpesvirus 3 ORF112-Zα·Z-DNA complex reveals a mechanism of nucleic acids recognition conserved with E3L, a poxvirus inhibitor of interferon response. J. Biol. Chem. 2015;290:30713–30725. doi: 10.1074/jbc.M115.679407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz F., Abergel C., Woyke T. Giant virus biology and diversity in the era of genome-resolved metagenomics. Nat. Rev. Microbiol. 2022;20:721–736. doi: 10.1038/s41579-022-00754-5. [DOI] [PubMed] [Google Scholar]
- 33.Queiroz V.F., Tatara J.M., Botelho B.B., Rodrigues R.A.L., Almeida G.M.F., Abrahao J.S. The consequences of viral infection on protists. Commun. Biol. 2024;7:306. doi: 10.1038/s42003-024-06001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basto A.P., Nix R.J., Boinas F., Mendes S., Silva M.J., Cartaxeiro C., et al. Kinetics of African swine fever virus infection in Ornithodoros erraticus ticks. J. Gen. Virol. 2006;87:1863–1871. doi: 10.1099/vir.0.81765-0. [DOI] [PubMed] [Google Scholar]
- 35.Brown B.A., Lowenhaupt K., Wilbert C.M., Hanlon E.B., Rich A. The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13532–13536. doi: 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krall J.B., Nichols P.J., Henen M.A., Vicens Q., Vögeli B. Structure and formation of Z-DNA and Z-RNA. Molecules. 2023;28:843. doi: 10.3390/molecules28020843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichols P.J., Krall J.B., Henen M.A., Welty R., Macfadden A., Vicens Q., et al. Z-form adoption of nucleic acid is a multi-step process which proceeds through a melted intermediate. J. Am. Chem. Soc. 2023;146:694. doi: 10.1021/jacs.3c10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y.G., Lowenhaupt K., Oh D.B., Kim K.K., Rich A. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: implications for development of a therapy for poxvirus infection. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1514–1518. doi: 10.1073/pnas.0308260100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van quyen D., Ha S.C., Lowenhaupt K., Rich A., Kim K.K., Kim Y.G. Characterization of DNA-binding activity of Zα domains from poxviruses and the importance of the β-wing regions in converting B-DNA to Z-DNA. Nucleic Acids Res. 2007;35:7714–7720. doi: 10.1093/nar/gkm748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D., Hur J., Park K., Bae S., Shin D., Ha S.C., et al. Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ) Nucleic Acids Res. 2014;42:5937–5948. doi: 10.1093/nar/gku189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park C., Zheng X., Park C.Y., Kim J., Lee S.K., Won H., et al. Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process. Nucleic Acids Res. 2020;48:12957–12971. doi: 10.1093/nar/gkaa1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ariyoshi M., Nishino T., Iwasaki H., Shinagawa H., Morikawa K. Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8257–8262. doi: 10.1073/pnas.140212997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devaraj A., Buzzo J.R., Mashburn-Warren L., Gloag E.S., Novotny L.A., Stoodley P., et al. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc. Natl. Acad. Sci. U. S. A. 2019;116:25068–25077. doi: 10.1073/pnas.1909017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buzzo J.R., Devaraj A., Gloag E.S., Stoodley P., Bakaletz L.O., Goodman Correspondence S.D., et al. Z-form extracellular DNA is a structural component of the bacterial biofilm matrix. Cell. 2021;184:5740–5758.e17. doi: 10.1016/j.cell.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salas-Jara M.J., Ilabaca A., Vega M., García A. Biofilm forming Lactobacillus: new challenges for the development of probiotics. Microorganisms. 2016;4:35. doi: 10.3390/microorganisms4030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Letunic I., Khedkar S., Bork P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021;49:D458–D460. doi: 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Castro E., Sigrist C.J.A., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sigrist C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary Genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okonechnikov K., Golosova O., Fursov M., Varlamov A., Vaskin Y., Efremov I., et al. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 52.Tung C.H., Yang J.M. FastSCOP: a fast web server for recognizing protein structural domains and SCOP superfamilies. Nucleic Acids Res. 2007;35:W438–W443. doi: 10.1093/nar/gkm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J.M., Tung C.H. Protein structure database search and evolutionary classification. Nucleic Acids Res. 2006;34:3646–3659. doi: 10.1093/nar/gkl395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.TSA: Noctiluca scintillans, MMETSP0253-doi:10.5281-zenodo.249982-Transcript-1000, transcribed RNA Sequence.National Library of Medicine Bethesda, MD 20894
- 57.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50:D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoang D.T., Chernomor O., Von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aylward F.O., Moniruzzaman M., Ha A.D., Koonin E.V. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol. 2021;19:e3001430. doi: 10.1371/journal.pbio.3001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., Von Haeseler A., et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aktaş T., Ilik I.A., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 68.Li Y., Slavik K.M., Toyoda H.C., Morehouse B.R., de Oliveira Mann C.C., Elek A., et al. cGLRs are a diverse family of pattern recognition receptors in innate immunity. Cell. 2023;186:3261–3276.e20. doi: 10.1016/j.cell.2023.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krishna P., Morgan A.R., Van de Sande J.H. Interaction of recA protein with left-handed Z-DNA. Biochem. J. 1991;275:711–719. doi: 10.1042/bj2750711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herbert A., Poptsova M. Z-RNA and the flipside of the SARS Nsp13 helicase: is there a role for flipons in coronavirus-induced pathology? Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.912717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request. The primary sequence and the id-numbers of the new ZBPs and Zα-like containing proteins are summarized in Fig. S3 and Table S6. Other sequences used in this work are available from Frederik Schulz and Mamadou Amadou Diallo upon reasonable request.