Abstract
PARP inhibitor (PARPi) therapy has transformed outcomes for patients with homologous recombination DNA repair (HRR) deficient ovarian cancers, for example those with BRCA1 or BRCA2 gene defects. Unfortunately, PARPi resistance is common. Multiple resistance mechanisms have been described, including secondary mutations that restore the HR gene reading frame. BRCA1 splice isoforms △11 and △11q can contribute to PARPi resistance by splicing out the mutation-containing exon, producing truncated, partially functional proteins. However, the clinical impacts and underlying drivers of BRCA1 exon skipping are not fully understood.
We analyzed nine ovarian and breast cancer patient derived xenografts (PDX) with BRCA1 exon 11 frameshift mutations for exon skipping and therapy response, including a matched PDX pair derived from a patient pre- and post-chemotherapy/PARPi. BRCA1 exon 11 skipping was elevated in PARPi resistant PDX tumors. Two independent PDX models acquired secondary BRCA1 splice site mutations (SSMs) that drive exon skipping, confirmed using qRT-PCR, RNA sequencing, immunoblotting and minigene modelling. CRISPR/Cas9-mediated disruption of splicing functionally validated exon skipping as a mechanism of PARPi resistance. SSMs were also enriched in post-PARPi ovarian cancer patient cohorts from the ARIEL2 and ARIEL4 clinical trials.
Few PARPi resistance mechanisms have been confirmed in the clinical setting. While secondary/reversion mutations typically restore a gene’s reading frame, we have identified secondary mutations in patient cohorts that hijack splice sites to enhance mutation-containing exon skipping, resulting in the overexpression of BRCA1 hypomorphs, which in turn promote PARPi resistance. Thus, BRCA1 SSMs can and should be clinically monitored, along with frame-restoring secondary mutations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02048-1.
Introduction
Defects in the homologous recombination DNA repair (HRR) pathway, including BRCA1 and BRCA2 mutations, are a common feature of high-grade serous ovarian carcinoma (HGSOC) and triple negative breast cancer (TNBC). While BRCA defects are known drivers of malignancy in these cancer types, they also make cancer cells susceptible to DNA-damaging platinum agents and poly ADP ribose polymerase (PARP) inhibitor (PARPi) therapy.
PARPi have prolonged progression-free survival outcomes for many individuals with HRR-deficient (HRD) ovarian cancers; however, drug resistance and disease relapse unfortunately remain common [1]. The most well-established mechanism of platinum/PARPi resistance for patients with HRD HGSOC is secondary somatic “reversion” mutations in HRR genes that restore the open reading frame disrupted by the primary somatic or germline pathogenic variant. The resulting full-length or near full-length protein promotes sufficient HRR to escape PARPi toxicity [1]. Other resistance mechanisms have been characterized in preclinical models, with some clinically observed, including expression of hypomorphic BRCA1 proteins, loss of the 53BP1-Shieldin axis [1], PARP1 mutations [2], loss of HRR gene methylation [1, 3], reduced DNA replication gaps [4] and drug efflux [1].
Overall, HGSOC patients with pathogenic alterations in BRCA1 have a worse prognosis than those with BRCA2 alterations [5]. Among HGSOC patients with BRCA1 alterations, those with frameshift mutations within exon 11 of BRCA1 have a worse cumulative survival, as well as reduced platinum response, compared with individuals with frameshift mutations outside exon 11 [6, 7]. This may be explained, in part, by therapy resistance that can arise from the overexpression of BRCA1 splice isoforms missing most or all of exon 11 (also known as exon 10, but referred to herein as exon 11) [6]. The BRCA1 delta 11q (∆11q) isoform lacks a large portion (c.788–4096) of exon 11 due to splicing at an alternative donor splice site within the exon, resulting in generation of a shorter but partially functional BRCA1 protein. The delta 11 (∆11) isoform of BRCA1 is missing all of exon 11 (c.671–4096), and in human cells is less abundant relative to ∆11q [8]. However, there is evidence that ∆11 can also partially compensate for loss of full length BRCA1, particularly in a TP53-deficient context [9].
Canonical BRCA1 transcripts harboring frameshift exon 11 variants are typically degraded via nonsense-mediated decay (NMD) [10]. However, BRCA1 ∆11 or ∆11q transcripts lack the pathogenic variant-containing exon and are not subject to NMD. Thus, BRCA1-∆11 or -∆11q proteins, although truncated, may cause sufficient levels of HRR to induce PARPi and platinum resistance in cancer cells with BRCA1 exon 11 mutations [6].
Approximately 30% of pathogenic germline BRCA1 variants are estimated to occur in exon 11 [6]. Thus, the cellular mechanisms modulating ∆11 and ∆11q expression are of clinical significance, given BRCA1 isoforms can promote PARPi and platinum resistance.
Using a cohort of nine HGSOC, TNBC and Ovarian Carcinosarcoma (OCS) PDX models, cell lines, and genomic data from circulating tumor DNA (ctDNA) samples from individuals who took part in the ARIEL2 and ARIEL4 PARPi clinical trials, we investigated factors that determine PARPi and platinum response in cancers with primary exon 11 mutations. This included one matched PDX pair from a woman with HGSOC, before and after multiple lines of treatment, including both chemotherapy and PARPi. We discovered that two of five PARPi-resistant PDX harbored secondary BRCA1 splice site mutations (SSMs), and these were shown to drive alternative BRCA1 splicing and PARPi resistance in HGSOC and TNBC models. BRCA1 SSMs, and indeed, an SSM in BRCA2, were identified, with some enriched in ctDNA from women with HGSOC who had received prior PARPi in clinical trials. Herein, we demonstrate that upregulation of alternative BRCA1 isoforms is a mechanism and potential biomarker of PARPi resistance, and identify cases where alternative isoforms are driven by secondary splice site mutation.
Methods
Full details of all materials and methods can be found in the Supplementary Information file. Briefly, our study included the following methods: In vivo PARPi and platinum treatment responses were assessed in PDX. DNA panel sequencing data were used to investigate molecular correlates of PARPi response and resistance. RT-qPCR, RNA sequencing and immunoblotting were used to assess ∆11 and ∆11q expression in our models. BRCA1 minigene modelling was used to evaluate splicing effects of SSMs found in PDX, cell line and patient samples. Targeted CRISPR editing (to generate SSMs) and siRNA of ∆11 and ∆11q isoforms were used to manipulate isoform expression in cell lines, and PARPi responses were assessed using cell viability or colony forming assays. HRR states were measured using the RAD51 foci assay.
Results
PDX models with BRCA1 exon 11 mutations have variable PARPi and platinum responses
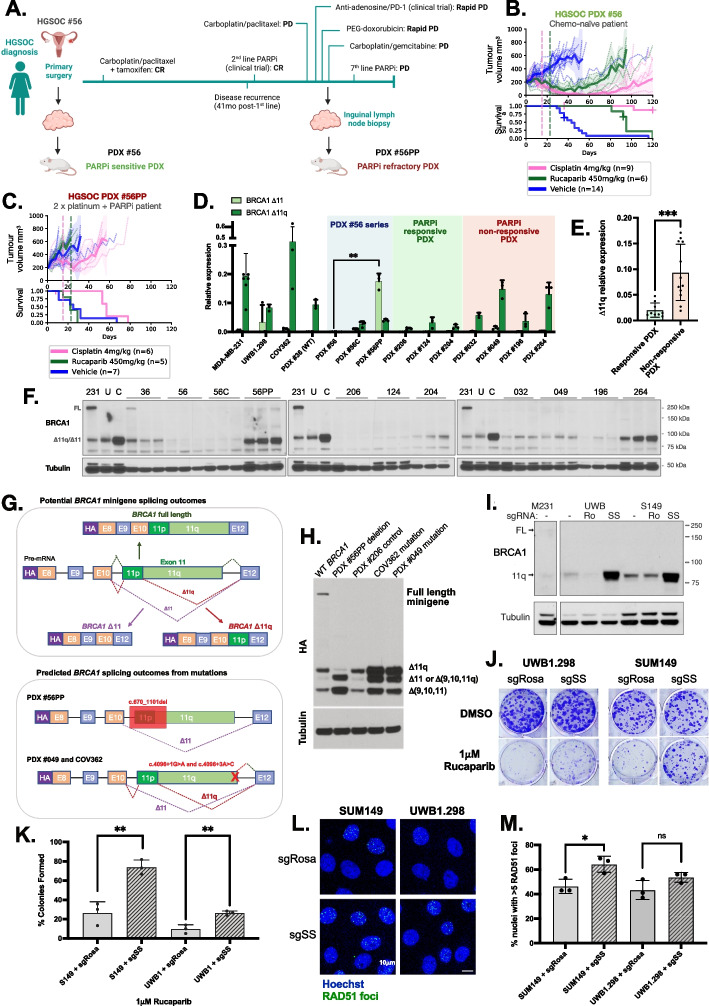
To study therapeutic responses in the setting of BRCA1 exon 11 mutations, we assembled panels of OCS, HGSOC and TNBC PDX models in accordance with Institutional Regulatory Board (IRB) approvals at the Walter and Eliza Hall Institute of Medical Research (WEHI) and Fox Chase Cancer Centers (FCCC). PDX cohorts from each site were generated and treated independently, and treatment responses were classified based on definitions developed independently by each site. A description of PDX, BROCA panel sequencing data and matched patient information is summarized in Supplementary Table S1 (histopathological and immunohistochemical (IHC) review in Supplementary Figure S1; [3]. We were able to generate a matched PDX pair of HGSOC #56: one obtained prior to chemotherapy (PDX #56), and one derived after the patient had received five lines of therapy, including having progressed and relapsed on PARPi (PDX #56PP, post-PARPi) [3] (Fig. 1A-C; Supplementary Figure S2; Supplementary Table S2). The original chemo-naïve PDX #56 lineage was both PARPi- and platinum-responsive (Fig. 1B; Supplementary Figure S2), while PDX #56PP was refractory to both platinum and PARPi, reflecting the patient’s clinical outcomes (Fig. 1C). The same association was observed for other WEHI PDX, where heavily pre-treated post-PARPi patient samples generated PARPi-resistant or refractory PDX (Supplementary Figure S2). FCCC PDX models were treated using slightly different treatment regimens, however, similar trends were observed (Supplementary Figure S2; Supplementary Table S2). In summary, in vivo treatment studies of BRCA1 exon 11 mutated PDX demonstrated a range of PARPi and platinum responses, which correlated with prior clinical platinum/PARPi exposure.
Fig. 1.
Splice-site mutations (SSMs) drive alternative splicing and PARPi resistance in PDX and cell line models of ovarian and breast cancer. A Patient #56 timeline, showing generation of the matched HGSOC PDX #56 (chemo-naïve) and #56PP (post-chemotherapy/PARPi patient). Created with BioRender.com. CR = Complete response; PD = Progressive disease; SD = Stable disease; C6 = cycle 6. B In vivo treatment data for HGSOC PDX #56 (previously published [3]), classified as PARPi responsive (P = 0.005). C HGSOC PDX #56PP was derived from patient #56 following multiple lines of therapy, including PARPi inhibitor, and was refractory to rucaparib (P = 0.375). Mean PDX tumor volume (mm3) ± 95% CI (hashed lines are individual mice) and corresponding Kaplan–Meier survival analysis. Censored events are represented by crosses on Kaplan–Meier plot; n = individual mice. Detailed patient clinical data can be found in Supplementary Table 1. Details of time to harvest, time to progression and Log-Rank test P values for each PDX can be found in Supplementary Table 2. D Relative △11 and △11q expression for each PDX (mean ± SD). PDX #56PP had the highest △11 expression relative to all other PDX (P = 0.0079 compared to matched PDX #56), while PDX #049 and #264 had the highest △11q levels relative to other PDX (classifications in Supplementary Tables 3 and 4). E △11q levels in PARPi responsive PDX models (< 2 prior lines of platinum in patient) were lower than levels in non-responsive PDX models (≥ 2 prior lines of platinum + PARPi) (P = 0.0007). Statistical comparisons of gene expression were made using an unpaired t-test with Welch’s correction. F Lysates from nuclear extracts from 3 independent tumors were probed for BRCA1 expression by immunoblotting. Bands at the anticipated sizes for full length (FL) BRCA1 and the △11/△11q isoforms are marked. Tubulin immunoblotting is included as a loading control. Gels were run simultaneously with cell line lysates included as controls for each gel. MDA-MB-231 [231] cells are a BRCA1 wild-type control with full-length (FL) and △11q expression, UWB1.289 (U) and COV362 (C) cells with exon 11 variants and △11/△11q expression. G Schematic of BRCA1 mini-gene design, and splicing outcomes predicted for each secondary splice site mutation found in PDX and cell line models. H Splicing predictions for each secondary splice-site mutation modelled by the mini-gene were confirmed by immunoblotting for the HA tag. The PDX #56PP deletion was confirmed to drive high ∆11 and potentially also isoform ∆(9,10,11q) expression. COV362 and PDX #049 secondary splice site mutations were confirmed to drive high ∆11q relative to the wild-type (WT) BRCA1 control and the primary deleterious BRCA1 mutation found in PDX #206. I Cells exposed to sgSS (SS) were found to have elevated BRCA1 D11q protein compared to untreated (-) and sgRosa (Ro) cells. J Example image of colony forming assays of UWB1.289 or SUM149 cells exposed to sgSS or sgRosa treated with DMSO control or 1µM Rucaparib. K Quantification of n = 3 colony forming experiments described in part (J). Mean ± SEM plotted; Ns = Not statistically significant; **p < 0.01; using unpaired, two-tailed t-test. L Representative image of RAD51 foci in UWB1.289 or SUM149 cells exposed to sgSS or sgRosa treated with 10 Gy dose of irradiation. M Quantification of nuclei with > 5 RAD51 foci in UWB1.289 (P = 0.11 compared to control) or SUM149 (P = 0.02 compared to control) cells exposed to sgSS or sgRosa and treated with 10 Gy dose of irradiation. Mean ± SEM plotted; Ns = Not statistically significant; *p < 0.05; using unpaired, two-tailed t-test
BRCA1 isoform expression analyses in PDX models
We next sought to quantify BRCA1 exon 11 isoform expression in the PDX models. The mRNA expression of exon 11-deleted ∆11 and ∆11q isoforms was analyzed by qRT-PCR, as well as total BRCA1 transcripts measured using primers specific for exon 14 (Fig. 1D-E; Supplementary Figure S3). PARPi-resistant HGSOC PDX #049 and #264 tumors were found to express high levels of ∆11q (Fig. 1D; Supplementary Tables 1 and 3–4). PARPi-unresponsive HGSOC PDX models #032 and #196 had moderate and low ∆11q expression, respectively, relative to the control cell lines (Fig. 1D; Supplementary Table 1). Interestingly, #56PP expressed an abundance of the ∆11 isoform (P = 0.0079 compared to matched PDX #56). When grouped by treatment responses, platinum- and PARPi-responsive PDX (excluding PDX #56 series) demonstrated low levels of ∆11q transcripts relative to resistant PDX tumors (P = 0.0007; Fig. 1E; Supplementary Table 1).
The mRNA expression patterns observed by qRT-PCR correlated with RNA sequencing (RNAseq) data (Fig. 1D; Supplementary Figure S3), but did not always correspond with protein expression, suggesting additional post-translational regulation [11] (Supplementary Figure S3). Nonetheless, in our cohorts, ∆11 or ∆11q isoform gene expression correlated with PARPi- and, in some cases, platinum-resistance in PDX models derived from patients who had received prior PARPi and multiple lines of platinum therapy.
Secondary exon 11 splice site mutations drive BRCA1 ∆11 and ∆11q expression
To investigate whether ∆11 and ∆11q expression was driving PARPi and platinum resistance in PDX models, we first ruled out other common resistance mechanisms (Supplementary Information; Supplementary Table 2 and 5; Supplementary Figure S4). While no frame-restoring reversion mutations were detected, PDX #049, #56PP and PARPi resistant cell line COV362 all harbored secondary splice site mutations (SSMs) that were predicted in silico to disrupt exon 11 splice junctions and drive exon sipping (Supplementary Information; Supplementary Table S5; Supplementary Figure S5). RNAseq, qRT-PCR and western blotting results supported these in silico predictions (Fig. 1D and Supplementary Figure S3), as did in vitro modelling using a BRCA1 minigene system [6] (Fig. 1G-H). SSMs were not found in matched archival material (Supplementary Figure S6), and all PDX models had high HRD genomic signatures (Supplementary Figure S7-8), suggesting SSMs were acquired. While PARPi-resistant HGSOC PDX #56PP and HGSOC PDX #049 had SSMs that explain the high levels of alternative isoform expression, HGSOC PDX #032 and OCS PDX #264 with high ∆11q expression lacked both driver SSMs in BRCA1 and other possible drivers of ∆11q (Supplementary Information; Supplementary Table S6), suggesting other unknown mechanisms influencing ∆11q expression in these cases.
BRCA1 splice site mutations drive ∆11 and ∆11q isoform expression and PARPi resistance
To measure the effects of BRCA1-∆11q expression in our cell line and PDX models on therapy sensitivity, we first silenced BRCA1-∆11q using siRNA followed by PARPi treatment in COV362 (Supplementary Figure S9). A clear reduction in colony forming capacity was observed in COV362 cells treated with a “broad” exon 9-targeting BRCA1 siRNA and ∆11q siRNA compared to the scrambled control cells (Supplementary Figure S9-S11). Indeed, untreated COV362 cells were found to have some HR activity by RAD51 foci (Supplementary Figure S12-14). Engineered ectopic ∆11q expression in the BRCA1-null PARPi/platinum-sensitive TNBC PDX #1126 also induced platinum and PARPi resistance in vivo [12] (Supplementary Figure S15; Supplementary Table 2), confirming the ability of ∆11q to promote resistance therapy in vitro and in vivo.
While we were able to drive elevated △11q in some PDX models using in vivo PARPi or platinum re-treatment, none of these were found to acquire SSMs (Supplementary Figure S16). We previously reported that the exon 11 frameshift-containing cell lines UWB1.289 and SUM149 expressed the BRCA1-D11q protein, and consequently demonstrated intermediate PARPi resistance amongst a panel of BRCA1 wild-type and mutant cell lines [6]. CRISPR was thus used to test the impacts of exon 11 donor SSMs on PARPi responses. We were able to directly introduce SSMs into cell lines UWB1.289 and SUM149, which harbor exon 11 frameshift mutations and have intermediate D11q expression and PARPi responses. Introduction of exon 11 donor SSMs led to increased BRCA1-D11q protein, PARPi resistance and enhanced HR DNA repair (RAD51 foci) relative to controls (Fig. 1I-M; Supplementary Figure S17). Taken together, these experiments provide direct evidence that SSMs drive high levels of alternative BRCA1 isoform expression and PARPi resistance.
Secondary BRCA1 SSMs enriched post-PARPi in tumors and ctDNA
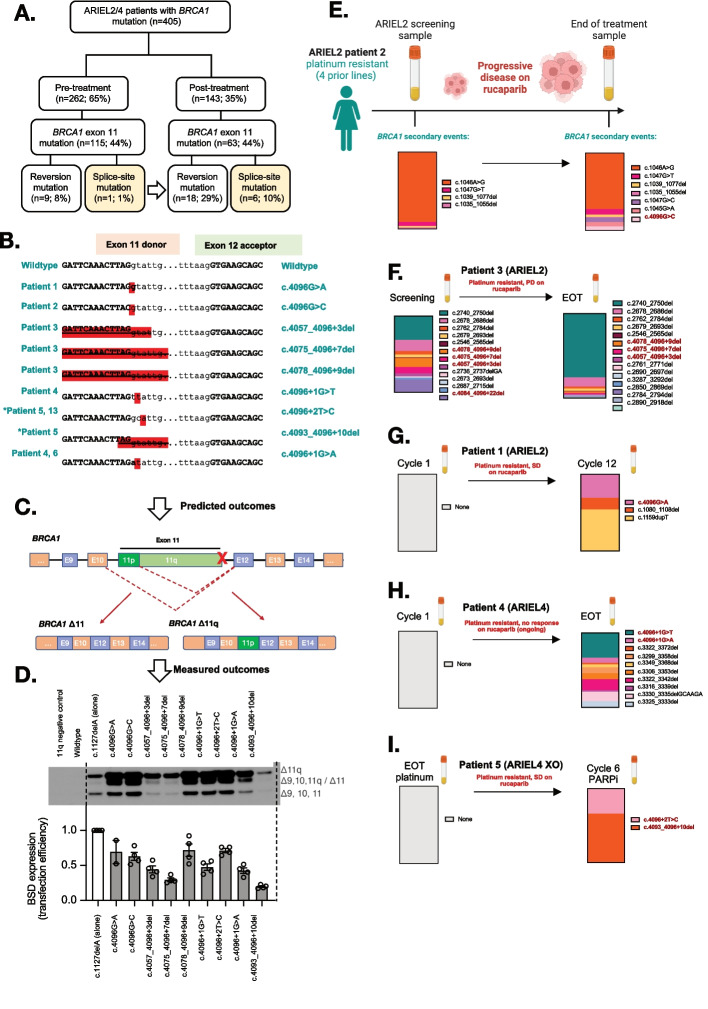
To explore the potential clinical relevance of SSMs, we examined tumor and ctDNA samples from patients enrolled in the clinical trials ARIEL2 (NCT01891344; individuals with platinum-sensitive relapsed HGSOC, Fallopian Tube, or Primary Peritoneal cancer) and ARIEL4 (NCT02855944; individuals with BRCA1/2-mutated relapsed ovarian cancer) before and after treatment with the PARPi rucaparib. We found that, prior to PARPi therapy, 1% (1/115) of patients with BRCA1 exon 11 mutated cancer had an SSM impacting the BRCA1 exon 11 donor splice site (c.4096 region). This rose to 10% (6/63) following PARPi therapy (Fig. 2A; Supplementary Information; Supplementary Table S7-S8). In silico modelling of SSMs predicted that all would drive expression of the ∆11q isoform (Fig. 2B-C; Supplementary Table S8-S9), and this was confirmed using the BRCA1 minigene system (Fig. 2D). Interestingly, in several cases samples with c.4096 SSMs also harbored multiple additional reversion events following PARPi therapy (Fig. 2E-I; Supplementary Table S8). Unlike sequencing of tissue samples (e.g. ARIEL2 patient 6; Supplementary Figure S18) [13], liquid biopsies are not spatially restricted and can sample secondary events across a woman’s entire cancer. Given SSMs and reversion mutations were found to co-exist in some patient samples, we examined whether either the ∆11q or full length BRCA1 proteins provided a selective advantage in an in vivo competition experiment. Equal selection was observed under cisplatin pressure, providing rationale for why these events were detected in the same samples (Supplementary Figure S19).
Fig. 2.
BRCA1 secondary splice-site mutations are enriched in ARIEL2/4 clinical trial patient samples following PARPi treatment. A BRCA1 secondary splice-site mutations increased form 1% (pre-PARPi) to 10% (post-PARPi) in patient tumor/plasma samples from the ARIEL2 and ARIEL4 clinical trials. B The BRCA1 (NM_007294.4) exon 11 donor splice-site mutations identified in these patients and the DNA sequence context are presented. *Patient 5 is an ARIEL4 chemotherapy to PARPi cross-over (XO) arm patient, not included in part A (n = 5). C-D The predicted outcomes on BRCA1 gene splicing based on these disruptions of the exon 11 donor splice site (detailed in Supplementary Table 9) were confirmed for most mutations (D) using the previously described BRCA1 minigene system, with the c.1127delA used as a primary mutation control and included in all SSM minigenes [6]. Mutations driving lower levels of ∆11/∆11q also had a reduced minigene transfection efficiencies relative to other samples (measured as BSD (blasticidin resistance gene) expression). E Summary of BRCA1 secondary events detected in Patient 2 (ARIEL2) before and after PARPi therapy, and their relative proportions in each sample (by colour). F A number of BRCA1 secondary events were also detected in the screening biopsy for platinum and rucaparib resistant Patient 3 (ARIEL2) prior to PARPi, and the number increased at end of rucaparib treatment (EOT). G In contrast, platinum resistant (4 prior lines of platinum) Patient 1 (ARIEL2) had no secondary BRCA1 events detected at first cycle of rucaparib, and had stable disease (SD) on treatment. Three secondary events were detected at cycle 12 of treatment, including a splice-site mutation c.4096G > A. H Patient 4 (ARIEL4) was partially platinum sensitive (2 prior lines) with no secondary BRCA1 events detected at cycle 1 of rucaparib. The EOT plasma sample was positive for multiple reversion events and two splice site mutations (4096 + 1G > T and 4096 + 1G > A) confirmed by minigene to alter splicing (D). I Patient 5 (ARIEL4) was platinum resistant and was enrolled in the chemotherapy arm of ARIEL4. They then crossed over (XO arm) to receive rucaparib treatment where they had stable disease. There were no secondary events detected prior to starting rucaparib, but at cycle 6 there were two BRCA1 splice-site mutations detected (c.4096 + 2 T > C and c.4093_4096 + 10del), without other reversion events. c.4096 + 2 T > C was found to drive alternative BRCA1 splicing (D). Red bold text indicates SSMs detected in patient samples (E-I), while other events presented are exonic secondary/reversion variants predicted to restore the BRCA1 reading frame
In addition to SSMs affecting BRCA1 exon 11, potential SSMs affecting exon 20 were also detected in four individuals enrolled on the ARIEL4 trial (Supplementary Table S8; Supplementary Fig. 18), along with one additional case with a pathogenic BRCA2 variant in exon 8 and an SSM predicted to drive skipping of exon 8 (Supplementary Table S8-9; Supp Figure S18). These cases suggest that this same resistance mechanism may be relevant beyond exon 11 of BRCA1.
Discussion
In the current study we demonstrate that BRCA1 splice site mutations (SSMs) drive high BRCA1 ∆11/∆11q isoform expression, which in turn promotes resistance to PARPi in HGSOC and TNBC. Induction of SSMs in BRCA1 exon 11-mutant HGSOC and TNBC cell lines using CRISPR caused PARPi resistance, demonstrating that SSMs are one avenue to elevated ∆11/∆11q isoform expression and PARPi resistance. BRCA1 SSMs were detected in a proportion of BRCA1 exon 11 mutated cell lines, PDX models and patient samples, with enrichment of SSMs observed following PARPi therapy in patients. Given that SSMs were found in 10% of patients with BRCA1 exon 11 mutation in the ARIEL2 and ARIEL4 clinical trials following PARPi therapy, our work highlights the clinical significance of this drug resistance mechanism. We also illustrate that SSMs can be readily measured in patients using non-invasive liquid biopsies, presenting an opportunity to change clinical practice. This is important given that analysis of ∆11/∆11q isoform expression in patient samples is confounded by normal tissue contamination, while SSMs are readily detectable and could be used as a biomarker for PARPi resistance.
Certain BRCA1 splice site variants can drive elevated ∆11 and ∆11q expression in vitro, for example c.4096 + 1G > A (also called IVS11 + 1G > A) and c.4096 + 3A > G (IVS11 + 3A > G). While these splice site variants have been identified in the germline of multiple families with history of breast and ovarian cancer, evidence of their pathogenicity and impact on inherited cancer susceptibility has been mixed. Indeed, a healthy homozygous carrier of the c.4096 + 3A > G variant has been reported [14]. Interestingly, a c.4096 + 1G > A SSM was previously reported in a patient with a primary germline exon 11 mutation (BRCA1: c.2043dup) following progression on PARPi therapy (ARIEL3 trial) [13]. Thus, the context of these SSMs dictates their impacts on disease outcomes. Primary BRCA1 SSMs might be drivers of disease in some cases, while secondary SSMs in BRCA1 can cause exon skipping and drug resistance.
In addition to SSMs that cause BRCA1 exon 11 skipping, in other cases we found SSMs in ctDNA predicted to drive BRCA1 exon 19 skipping and an SSM previously demonstrated in vitro to remove exon 7 and 8 of BRCA2 [15], in each case to remove a pathogenic germline variant from the transcripts. This suggests that the phenomenon is not restricted to exon 11 of BRCA1, and has relevance for other BRCA1 exons and other HRR pathway genes.
We observed several cases with multiple distinct BRCA1 secondary/reversion events in ctDNA (patients 2, 3 and 4 in Fig. 2). This is unsurprising given the high degree of genetic instability and heterogeneity in HGSOC, and the clonal diversity and selection that can occur under treatment pressure. This further highlights the need for development of more therapeutic options for patients with this heterogeneous and highly adaptable cancer type. This further highlights the need for development of more therapeutic options for patients with this heterogeneous and highly adaptable cancer type. The utility of spliceosome inhibitors combined with PARPi should be explored in cancers where alterative BRCA1 isoforms are identified as a driver of drug resistance. Pladienolide-B has been previously shown to reduce ∆11q expression in vitro [6]. However, the classical small molecule inhibitors of the splicing machinery (e.g. pladienolide-B, spliceostatin A, GEX1A and E1707) have not been clinically useful, due to their high toxicity. Novel spliceosome inhibitors with reduced toxicity profiles, such as the SF3B1-modulator H3B-8800, may provide some hope for patients with ∆11q-driven PARPi resistance [16].
Conclusion
In conclusion, alternative BRCA1 isoform expression is a driver of PARPi resistance across multiple cancer types, and this can be driven by splice site mutations. Previous findings that partially functional △11/△11q isoforms are drivers of platinum and PARPi resistance in preclinical models, have not led to changes in clinical practice. By comprehensively investigating nine new PDX models and relevant cell lines, we have shown that SSMs represent one avenue of alternative isoform expression and PARPi resistance in ovarian and breast cancers. We demonstrated that this mechanism was subject to selection under treatment pressure (whether pre-existing or acquired), and found SSMs were enriched following PARPi in ctDNA from patients. We also identified SSMs in patients with non-exon 11 BRCA1, as well as BRCA2 mutations, providing support for this as a more generalized phenomenon, with wider clinical relevance. Thus, BRCA1 SSMs provide potential for early drug resistance screening and disease management, enabling the development of strategies designed to pre-empt or avert PARPi resistance caused by this mechanism. SSMs should be screened for in people with PARPi-resistant cancers so that approaches can be developed to mitigate the huge challenge we now face, that of PARPi-resistance occurring so widely following recommended first-line PARPi therapy.
Supplementary Information
Acknowledgements
We thank Silvia Stoev, Rachel Hancock, Kathy Barber, Scott Wood and Conrad Leonard for technical assistance. We thank Amanda Spurdle and Michael Parsons (QIMR Berghofer) for their advice regarding BRCA1 SNPs potentially affecting splicing. We thank Clovis Oncology for providing rucaparib for in vivo experiments; Violeta Serra and Judith Balmana for the PDX models #124 and #196; Elgene Lim for #1126, Scott H. Kaufmann for PEO1 and PEO4 cell lines and Dale Garsed and David Bowtell for the UWB1.289 cell line.
Consortia
Australian Ovarian Cancer Study (AOCS), G. Chenevix-Trench, A. Green, P. Webb, D. Gertig, S. Fereday, S. Moore, J. Hung, K. Harrap, T. Sadkowsky, N. Pandeya, M. Malt, A. Mellon, R. Robertson, T. Vanden Bergh, M. Jones, P. Mackenzie, J. Maidens, K. Nattress, Y. E. Chiew, A. Stenlake, H. Sullivan, B. Alexander, P. Ashover, S. Brown, T. Corrish, L. Green, L. Jackman, K. Ferguson, K. Martin, A. Martyn, B. Ranieri, J. White, V. Jayde, P. Mamers, L. Bowes, L. Galletta, D. Giles, J. Hendley, T. Schmidt, H. Shirley, C. Ball, C. Young, S. Viduka, H. Tran, S. Bilic, L. Glavinas, J. Brooks, R. Stuart-Harris, F. Kirsten, J. Rutovitz, P. Clingan, A. Glasgow, A. Proietto, S. Braye, G. Otton, J. Shannon, T. Bonaventura, J. Stewart, S. Begbie, M. Friedlander, D. Bell, S. Baron-Hay, A. Ferrier, G. Gard, D. Nevell, N. Pavlakis, S. Valmadre, B. Young, C. Camaris, R. Crouch, L. Edwards, N. Hacker, D. Marsden, G. Robertson, P. Beale, J. Beith, J. Carter, C. Dalrymple, R. Houghton, P. Russell, M. Links, J. Grygiel, J. Hill, A. Brand, K. Byth, R. Jaworski, P. Harnett, R. Sharma, G. Wain, B. Ward, D. Papadimos, A. Crandon, M. Cummings, K. Horwood, A. Obermair, L. Perrin, D. Wyld, J. Nicklin, M. Davy, M. K. Oehler, C. Hall, T. Dodd, T. Healy, K. Pittman, D. Henderson, J. Miller, J. Pierdes, P. Blomfield, D. Challis, R. McIntosh, A. Parker, B. Brown, R. Rome, D. Allen, P. Grant, S. Hyde, R. Laurie, M. Robbie, D. Healy, T. Jobling, T. Manolitsas, J. McNealage, P. Rogers, B. Susil, E. Sumithran, I. Simpson, K. Phillips, D. Rischin, S. Fox, D. Johnson, S. Lade, M. Loughrey, N. O’Callaghan, W. Murray, P. Waring, V. Billson, J. Pyman, D. Neesham, M. Quinn, C. Underhill, R. Bell, L. F. Ng, R. Blum, V. Ganju, I. Hammond, Y. Leung, A. McCartney, M. Buck, I. Haviv, D. Purdie, D. Whiteman & N. Zeps.
AOCS consortium representative contact:
Nadia Traficante; email: AOCS@petermac.org.
Abbreviations
- WT
Wild-Type
- PARPi
PARP inhibitor
- PDX
Patient-Derived Xenograft
- HGSOC
High Grade Serous Ovarian Carcinoma
- TNBC
Triple Negative Breast Cancer
- OCS
Ovarian Carcinosarcoma
- PCR
Polymerase Chain Reaction
- qRT-PCR
Reverse-Transcription Quantitative Realtime PCR
- IHC
Immunohistochemical
- WEHI
Walter and Eliza Hall Institute of Medical Research
- FCCC
Fox Chase Cancer Centre
- SSM
Splice Site Mutation
- IRB
Institutional Regulatory Board
- ctDNA
Circulating tumor DNA
- HRR
Homologous Recombination DNA Repair
- HRD
HRR-Deficient
- NMD
Nonsense Mediated Decay
- WES
Whole Exome Sequencing
Authors’ contributions
K.N and J.J.K conceived, designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. Y.W. designed and performed experiments, and analyzed/interpreted data. C.V., P.P., K.Q.C., E.L., G.Y.H., H.E.B., A.F., K.S.A., F.G., E.K., R.B. and F.Z. performed experiments and analyzed/interpreted corresponding data. J.B., L.X., and O.K. created analysis pipelines and performed data analysis. A.D., M.R. and J.S.P. performed data analysis/interpretation. G.R. provided pathology review of clinical samples. E.M.S., T.K., S.C., I.O, C.L.S., R.K., A.O., I.M., T.H., K.L., A.D., D.D.L.B., and N.T/ AOCS provided access to clinical samples and interpretation of clinical data. C.L.S., N.J. and M.J.W. conceptualized and led the study. All authors reviewed the manuscript.
Funding
This work was supported by fellowships and grants from the National Health and Medical Research Council (NHMRC Australia; Project grant 1062702 (CLS); Investigator grant 2009783 (CLS); KN, CV, KSA, HEB, EK, MJW); Cancer Council Victoria (Sir Edward Dunlop Fellowship in Cancer Research (CLS) and Grant-in-Aid 1186314 for HEB); the Victorian Cancer Agency (Clinical Fellowships CRF10–20, CRF16014 (CLS) and MCRF23021 fellowship (HEB)), Stafford Fox Medical Research Foundation (to CLS, KN, CV, KSA, HEB, EK, EL, GYH, JB, JSP and MJW), Herman Trust (CLS), University of Melbourne (CLS) and Cooperative Research Centre Cancer Therapeutics (CLS) during the conduct of the study; the National Institute of Health (2P50CA083636) (to EMS), NIH NCI R01 CA251817 and NIH NCI R01 CA237600 (EMS and MR), and the Wendy Feuer Ovarian Cancer Research Fund (to EMS); the Bev Gray Ovarian Cancer Scholarship (PhD top-up scholarship) and Research Training Program Scholarship (PhD Scholarship) (KN); the American Association of Cancer Research (AACR-AstraZeneca Ovarian Cancer Research Fellowship 2022 (KN)); Cancer Research UK Cambridge Institute PhD Studentship, Cambridge Australia Poynton PhD Scholarship, Cambridge Trust International Scholarship, Bryan Tan PhD top-up scholarship (EK). Cancer Council Victoria (HEB); Swiss Cancer Research foundation (KFS-5445–08-2021) (FG); NHMRC Emerging Leader 1 Investigator Grant (APP2008631) (OK), and Stand Up to Cancer (EMS). IM reports grants from Cancer Research UK, Ovarian Cancer Action, National Institute for Health Research and the Imperial NIHR Biomedical Research Centre. MJW reports grants from Stafford Fox Foundation during the conduct of the study. This work was also supported by US National Institutes of Health (NIH) Grants R01CA214799 and R01GM135293 to NJ and JJK. was supported by an American Cancer Society—Tri State CEOs Against Cancer Postdoctoral Fellowship, PF-19–097-01-DMC, Ovarian Cancer Research Alliance and Phil and Judy Messing grant 597484. This work was made possible through the Australian Cancer Research Foundation, the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. All authors, the WEHI Stafford Fox Rare Cancer Program and the AOCS would like to thank all of the patients who participated in these research programs. The AOCS would also like to acknowledge the contribution of the study nurses, research assistants, and all clinical and scientific collaborators to the study. The complete AOCS Study Group can be found at www.aocstudy.org. The Australian Ovarian Cancer Study gratefully acknowledges additional support from Ovarian Cancer Australia and the Peter MacCallum Foundation. The Australian Ovarian Cancer Study Group was supported by the U.S. Army Medical Research and Materiel Command under DAMD17-01–1-0729, The Cancer Council Victoria, Queensland Cancer Fund, The Cancer Council New South Wales, The Cancer Council South Australia, The Cancer Council Tasmania and The Cancer Foundation of Western Australia (Multi-State Applications 191, 211 and 182) and the National Health and Medical Research Council of Australia (NHMRC; ID199600; ID400413 and ID400281).
Availability of data and materials
Sequence data that support the findings of this study have been deposited in the European Genome-Phenome Archive with the primary accession code EGAS50000000022.
Declarations
Ethics approval and consent to participate
For Walter and Eliza Hall Institute of Medical Research (WEHI) PDX models, all experiments involving animals were performed according to the National Health and Medical Research Council Australian Code for the Care and Use of Animals for Scientific Purposes 8th Edition, 2013 (updated 2021), and were approved by the WEHI Animal Ethics Committee (2019.024). Ovarian carcinoma/carcinosarcoma PDX were generated from patients enrolled in the Australian Ovarian Cancer Study (AOCS) or WEHI Stafford Fox Rare Cancer Program (SFRCP). Informed written consent was obtained from all patients, and all experiments were performed according to human ethics guidelines. Additional ethics approval was obtained from the Human Research Ethics Committees at the Royal Women’s Hospital, the WEHI (HREC #10/05 and #G16/02) and QIMR Berghofer (P3456 and P2095). Mouse experiments conducted at FCCC were approved by the Fox Chase Cancer Center Institutional Animal Care and Use Committee (IACUC) and the use of PDX models approved by the Institutional Review Board (IRB).
Competing interests
K. Nesic reports nonfinancial support from Clovis Oncology during the conduct of the study. O. Kondrashova reports personal fees from XING Technologies outside the submitted work. K. Nesic, C. Vandenberg, M.J. Wakefield, C.L. Scott, K. Shield-Artin, A. Farrell, E. Kyran and H.E. Barker all receive research support outside of this study from Eisai Inc, AstraZeneca, Boehringer Ingelheim and Ideaya Biosciences. N. Traficante reports grants from AstraZeneca Pty Ltd. during the conduct of the study and grants from AstraZeneca Pty Ltd. outside the submitted work. D. Bowtell reports grants from AstraZeneca Pty Ltd. during the conduct of the study and research support grants from AstraZeneca, Roche-Genentech and BeiGene (paid to institution) outside the submitted work; and personal consulting fees from Exo Therapeutics, that are outside the submitted work. Australian Ovarian Cancer Study reports grants from AstraZeneca Pty Ltd. during the conduct of the study and grants from AstraZeneca Pty Ltd. outside the submitted work. There are no other conflicts of interest in relation to the work under consideration for publication, nor other relationships / conditions / circumstances that present a potential conflict of interest. A. DeFazio reports grants from AstraZeneca outside the submitted work. A. Dobrovic reports grants from National Breast Cancer Foundation of Australia during the conduct of the study. T.C. Harding, K. Lin and T. Kwan were employees of Clovis Oncology. R. Kristeleit reports funding from Clovis Oncology, AstraZeneca, GSK, Pharma&, and was a Co-ordinating Investigator for the ARIEL4 and ATHENA trials. I. McNeish reports advisory boards for Clovis Oncology, AstraZeneca, GSK, OncoC4, Theolytics, Epsila Bio, Duke St Bio, Scancell, Roche, Takeda, and also institutional grant income from AstraZeneca. E.M. Swisher is on the Scientific Advisory Board for Ideaya Biosciences. C.L. Scott reports research support from AstraZeneca Pty Ltd, Boehringer Ingelheim and Eisai Inc, and other support from Clovis Oncology and Sierra Oncology outside the submitted work; and unpaid advisory boards: AstraZeneca, Clovis Oncology, Roche, Eisai, Sierra Oncology, Takeda, MSD. No disclosures were reported by the other authors.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ksenija Nesic and John J. Krais joint first authors.
Clare L. Scott, Neil Johnson and Matthew J. Wakefield joint senior authors.
Contributor Information
Clare L. Scott, Email: scottc@wehi.edu.au
Neil Johnson, Email: Neil.Johnson@fccc.edu.
Matthew J. Wakefield, Email: matthew.wakefield@unimelb.edu.au
Australian Ovarian Cancer Study:
G. Chenevix-Trench, A. Green, P. Webb, D. Gertig, S. Fereday, S. Moore, J. Hung, K. Harrap, T. Sadkowsky, N. Pandeya, M. Malt, A. Mellon, R. Robertson, T. Vanden Bergh, M. Jones, P. Mackenzie, J. Maidens, K. Nattress, Y. E. Chiew, A. Stenlake, H. Sullivan, B. Alexander, P. Ashover, S. Brown, T. Corrish, L. Green, L. Jackman, K. Ferguson, K. Martin, A. Martyn, B. Ranieri, J. White, V. Jayde, P. Mamers, L. Bowes, L. Galletta, D. Giles, J. Hendley, T. Schmidt, H. Shirley, C. Ball, C. Young, S. Viduka, H. Tran, S. Bilic, L. Glavinas, J. Brooks, R. Stuart-Harris, F. Kirsten, J. Rutovitz, P. Clingan, A. Glasgow, A. Proietto, S. Braye, G. Otton, J. Shannon, T. Bonaventura, J. Stewart, S. Begbie, M. Friedlander, D. Bell, S. Baron-Hay, A. Ferrier, G. Gard, D. Nevell, N. Pavlakis, S. Valmadre, B. Young, C. Camaris, R. Crouch, L. Edwards, N. Hacker, D. Marsden, G. Robertson, P. Beale, J. Beith, J. Carter, C. Dalrymple, R. Houghton, P. Russell, M. Links, J. Grygiel, J. Hill, A. Brand, K. Byth, R. Jaworski, P. Harnett, R. Sharma, G. Wain, B. Ward, D. Papadimos, A. Crandon, M. Cummings, K. Horwood, A. Obermair, L. Perrin, D. Wyld, J. Nicklin, M. Davy, M. K. Oehler, C. Hall, T. Dodd, T. Healy, K. Pittman, D. Henderson, J. Miller, J. Pierdes, P. Blomfield, D. Challis, R. McIntosh, A. Parker, B. Brown, R. Rome, D. Allen, P. Grant, S. Hyde, R. Laurie, M. Robbie, D. Healy, T. Jobling, T. Manolitsas, J. McNealage, P. Rogers, B. Susil, E. Sumithran, I. Simpson, K. Phillips, D. Rischin, S. Fox, D. Johnson, S. Lade, M. Loughrey, N. O’Callaghan, W. Murray, P. Waring, V. Billson, J. Pyman, D. Neesham, M. Quinn, C. Underhill, R. Bell, L. F. Ng, R. Blum, V. Ganju, I. Hammond, Y. Leung, A. McCartney, M. Buck, I. Haviv, D. Purdie, D. Whiteman, and N. Zeps
References
- 1.Veneziani AC, Scott C, Wakefield MJ, Tinker AV, Lheureux S. Fighting resistance: post-PARP inhibitor treatment strategies in ovarian cancer. Ther Adv Med Oncol. 2023;15:17588359231157644. 10.1177/17588359231157644. 10.1177/17588359231157644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pettitt SJ, Krastev DB, Brandsma I, Drean A, Song F, Aleksandrov R, et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun. 2018;9(1):1849. 10.1038/s41467-018-03917-2. 10.1038/s41467-018-03917-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondrashova O, Topp M, Nesic K, Lieschke E, Ho GY, Harrell MI, et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9(1):3970. 10.1038/s41467-018-05564-z. 10.1038/s41467-018-05564-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong K, Peng M, Kousholt AN, Lee WTC, Lee S, Nayak S, et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol Cell. 2021;81(15):3128–44.e7. 10.1016/j.molcel.2021.06.011. 10.1016/j.molcel.2021.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolton KL, Chenevix- Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307(4):382–9. 10.1001/jama.2012.20. 10.1001/jama.2012.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Bernhardy AJ, Cruz C, Krais JJ, Nacson J, Nicolas E, et al. The BRCA1-Delta11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016;76(9):2778–90. 10.1158/0008-5472.CAN-16-0186. 10.1158/0008-5472.CAN-16-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimitrova D, Ruscito I, Olek S, Richter R, Hellwag A, Türbachova I, et al. Germline mutations of BRCA1 gene exon 11 are not associated with platinum response neither with survival advantage in patients with primary ovarian cancer: understanding the clinical importance of one of the biggest human exons. A study of the Tumor Bank Ovarian Cancer (TOC) Consortium. Tumour Biol. 2016;37(9):12329–37. 10.1007/s13277-016-5109-8. 10.1007/s13277-016-5109-8 [DOI] [PubMed] [Google Scholar]
- 8.Tammaro C, Raponi M, Wilson DI, Baralle D. BRCA1 exon 11 alternative splicing, multiple functions and the association with cancer. Biochem Soc Trans. 2012;40(4):768–72. 10.1042/bst20120140. 10.1042/bst20120140 [DOI] [PubMed] [Google Scholar]
- 9.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17(2):201–13. 10.1101/gad.1050003. 10.1101/gad.1050003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11(23):2805–14. 10.1093/hmg/11.23.2805. 10.1093/hmg/11.23.2805 [DOI] [PubMed] [Google Scholar]
- 11.Pettitt SJ, Shao N, Zatreanu D, Frankum J, Bajrami I, Brough R, et al. A HUWE1 defect causes PARP inhibitor resistance by modulating the BRCA1-∆11q splice variant. Oncogene. 2023;42(36):2701–9. 10.1038/s41388-023-02782-8. 10.1038/s41388-023-02782-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson SF, Cruz C, Greifenberg AK, Dust S, Stover DG, Chi D, et al. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep. 2016;17(9):2367–81. 10.1016/j.celrep.2016.10.077. 10.1016/j.celrep.2016.10.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng NNH, Harrell MI, et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017;7(9):984–98. 10.1158/2159-8290.CD-17-0419. 10.1158/2159-8290.CD-17-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrjalsen A, Steffensen AY, Hansen TVO, Wadt K, Gerdes A-M. Classification of the spliceogenic BRCA1 c.4096+3A>G variant as likely benign based on cosegregation data and identification of a healthy homozygous carrier. Clin Case Rep. 2017;5(6):876–9. 10.1002/ccr3.944. 10.1002/ccr3.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraile-Bethencourt E, Valenzuela-Palomo A, Díez-Gómez B, Goina E, Acedo A, Buratti E, et al. Mis-splicing in breast cancer: identification of pathogenic BRCA2 variants by systematic minigene assays. J Pathol. 2019;248(4):409–20. 10.1002/path.5268. 10.1002/path.5268 [DOI] [PubMed] [Google Scholar]
- 16.Steensma DP, Wermke M, Klimek VM, Greenberg PL, Font P, Komrokji RS, et al. Phase I first-in-human dose escalation study of the oral SF3B1 modulator H3B–8800 in myeloid neoplasms. Leukemia. 2021;35(12):3542–50. 10.1038/s41375-021-01328-9. 10.1038/s41375-021-01328-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the European Genome-Phenome Archive with the primary accession code EGAS50000000022.