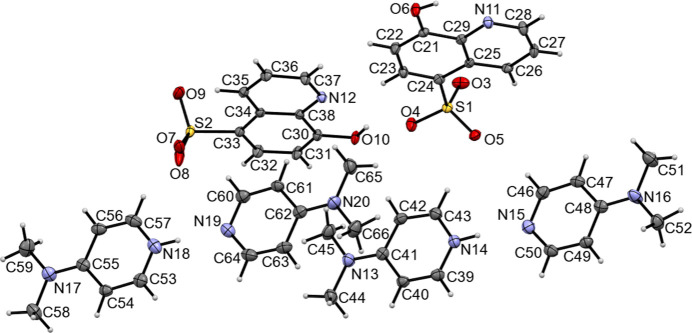
The asymmetric unit of the title compound consists of two independent ion pairs of 4-(dimethylamino)pyridin-1-ium quinolin-8-ol-5-sulfonate (HDMAP+·HqSA−) and neutral N,N-dimethylpyridin-4-amine (DMAP), forming a 1:1:1 cation:anion:neutral molecule co-crystal. The compound has a layered structure, including cation layers of HDMAP+ with DMAP and anion layers of HqSA− in the crystal. The cation and anion layers are linked by intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions.
Keywords: crystal structure, co-crystal, quinolin-8-ol sulfonate, DMAP, N—H⋯N interactions
Abstract
The asymmetric unit of the title compound is composed of two independent ion pairs of 4-(dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate (HDMAP+·HqSA−, C7H11N2+·C9H6NO4S−) and neutral N,N-dimethylpyridin-4-amine molecules (DMAP, C7H10N2), co-crystallized as a 1:1:1 HDMAP+:HqSA−:DMAP adduct in the monoclinic system, space group Pc. The compound has a layered structure, including cation layers of HDMAP+ with DMAP and anion layers of HqSA− in the crystal. In the cation layer, there are intermolecular N—H⋯N hydrogen bonds between the protonated HDMAP+ molecule and the neutral DMAP molecule. In the anion layer, each HqSA− is surrounded by other six HqSA−, where the planar network structure is formed by intermolecular O—H⋯O and C—H⋯O hydrogen bonds. The cation and anion layers are linked by intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions.
1. Chemical context
Ionic co-crystals have much attention in pharmaceuticals for the development of improved drugs based on crystal engineering (Bolla et al., 2022 ▸) and in organic functional materials for achieving rare and multifunctional properties through tunable structures, morphologies, and sizes in co-crystal assemblies (Sun et al., 2019 ▸). In structural chemistry, ionic co-crystals containing pyridine-pyridinium derivatives bridged by an N—H⋯N hydrogen bond have already been proposed (Doring & Jones, 2016 ▸; Fabry et al., 2017 ▸; Zhang et al., 2018 ▸; Vladiskovic et al., 2023 ▸). In addition, the supramolecular synthon preference of pyridinium salts to 8-hydroxyquinoline-5-sulfonate (HqSA−) and various sulfonates has been investigated (Ganie et al., 2021 ▸). On the other hand, quinolin-8-ol and its sulfonated derivative, quinoline-8-ol sulfonic acid (H2qSA), are well-known chelating ligands and analytical reagents (Wiberley et al., 1949 ▸; Kashiwagi et al., 2020 ▸; Kubono et al., 2023 ▸). H2qSA shows higher solubility to water than quionolin-8-ol, especially under basic conditions. We report here the crystal structure of the title compound as an ionic co-crystal composed of the salt of 4-(dimethylamino)pyridin-1-ium (HDMAP+) and quinolin-8-ol-5-sulfonate (HqSA−) with neutral N,N-dimethylpyridin-4-amine (DMAP).
2. Structural commentary
The title compound is composed of two independent HDMAP+·HqSA− ion pairs and neutral DMAP molecules, co-crystallized in the monoclinic system, space group Pc as shown in Fig. 1 ▸. The phenolic H atoms (H6, H10) in the HqSA− moieties are not dissociated.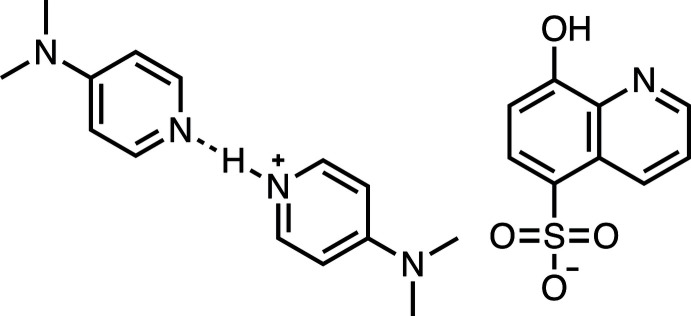
Figure 1.
The molecular structure of the title compound with atom labeling. Displacement ellipsoids are drawn at the 50% probability level. H atoms are represented by spheres of arbitrary radius.
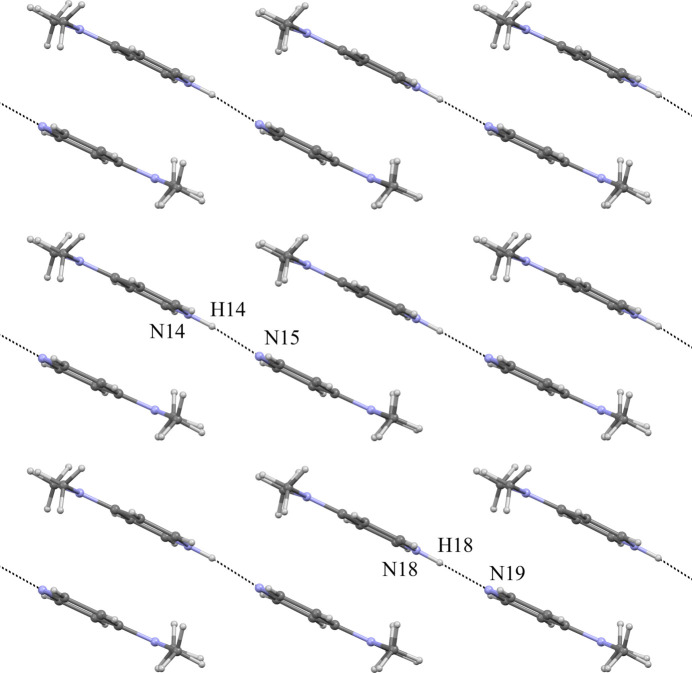
There are intramolecular O—H⋯N hydrogen bonds involving the hydroxy groups and quinoline N atoms (O6—H6⋯N11 and O10—H10⋯N12; Table 1 ▸) generating S(5) ring motifs (Fig. 2 ▸). The proton of the sulfonate group in H2qSA is dissociated and bound to the pyridyl N atom of one DMAP molecule, but there is also another non-protonated DMAP molecule in the crystal. As a result, the co-crystal is formulated as a 1:1:1 HDMAP+:HqSA−:DMAP adduct. The cations of HDMAP+ are formed through intermolecular N14—H14⋯N15 and N18—H18⋯N19 hydrogen bonds in a linear geometry (Fig. 2 ▸, see below). Each H atom attached to the N atom of the pyridine ring in HDMAP+ could be located in a Fourier density map, and the N14—H14 and N18—H18 bond lengths are similar, 0.90 (3) Å. The N atoms of the dimethylamino groups (N13, N16, N17 and N20) show no pyramidalization, with deviations from the plane of the bonded three C atoms of 0.029 (7), 0.031 (3), 0.037 (8) and 0.020 (4) Å, respectively. The quinoline ring systems in HqSA− are essentially planar, the dihedral angles between the mean planes of the pyridine and benzene rings N12/C34–C38 and C30—C34/C38, and N11/C25–C29 and C21–C25/C29 being 0.46 (14) and 0.78 (13)°, respectively.
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2 are the centroids of the N11/C25–C29 and N12/C34–C38 rings, respectively
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O6—H6⋯O8i | 0.84 (5) | 2.00 (5) | 2.679 (3) | 137 (4) |
| O6—H6⋯N11 | 0.84 (5) | 2.24 (5) | 2.728 (3) | 117 (4) |
| O10—H10⋯O4 | 0.89 (5) | 1.90 (5) | 2.674 (3) | 144 (4) |
| O10—H10⋯N12 | 0.89 (5) | 2.31 (5) | 2.730 (3) | 109 (4) |
| N14—H14⋯N15 | 0.90 (3) | 1.91 (3) | 2.814 (4) | 174 (3) |
| N18—H18⋯N19 | 0.90 (3) | 1.92 (4) | 2.816 (4) | 177 (7) |
| C27—H27⋯O6ii | 0.95 | 2.58 | 3.219 (4) | 125 |
| C27—H27⋯O8iii | 0.95 | 2.27 | 3.194 (4) | 164 |
| C36—H36⋯O4iv | 0.95 | 2.33 | 3.244 (4) | 160 |
| C36—H36⋯O10iv | 0.95 | 2.58 | 3.216 (4) | 125 |
| C39—H39⋯O3v | 0.95 | 2.32 | 3.200 (4) | 154 |
| C43—H43⋯O5 | 0.95 | 2.22 | 3.160 (4) | 169 |
| C46—H46⋯O5 | 0.95 | 2.46 | 3.373 (4) | 161 |
| C50—H50⋯O3v | 0.95 | 2.43 | 3.292 (4) | 151 |
| C53—H53⋯O9vi | 0.95 | 2.22 | 3.146 (4) | 164 |
| C54—H54⋯O10vii | 0.95 | 2.55 | 3.450 (4) | 158 |
| C57—H57⋯O7 | 0.95 | 2.22 | 3.143 (4) | 165 |
| C60—H60⋯O7 | 0.95 | 2.40 | 3.325 (4) | 163 |
| C64—H64⋯O9vi | 0.95 | 2.35 | 3.267 (4) | 162 |
| C40—H40⋯Cg1v | 0.95 | 2.63 | 3.498 (3) | 153 |
| C49—H49⋯Cg2viii | 0.95 | 2.87 | 3.754 (3) | 156 |
| C61—H61⋯Cg2 | 0.95 | 2.70 | 3.571 (3) | 152 |
Symmetry codes: (i)  ; (ii)
; (ii) 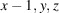 ; (iii)
; (iii) 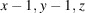 ; (iv)
; (iv) 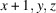 ; (v)
; (v) 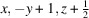 ; (vi)
; (vi) 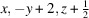 ; (vii)
; (vii) 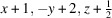 ; (viii)
; (viii) 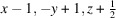 .
.
Figure 2.
The layer structure of the [HDMAP·DMAP]+ cationic unit in the ab plane. The intermolecular N—H⋯N hydrogen bonds are shown as dashed lines.
3. Supramolecular features
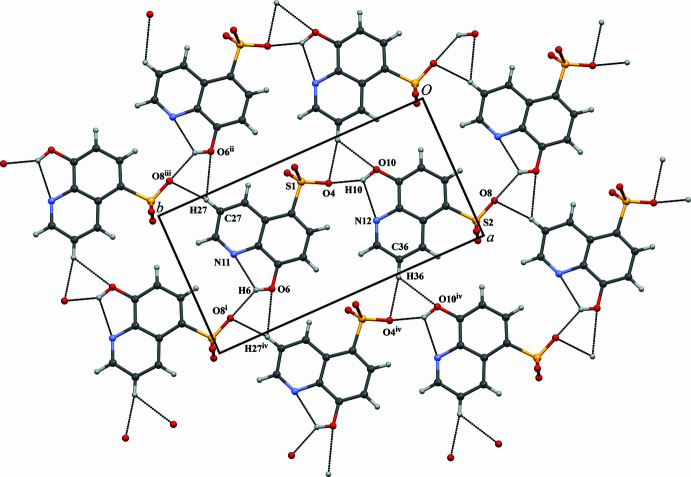
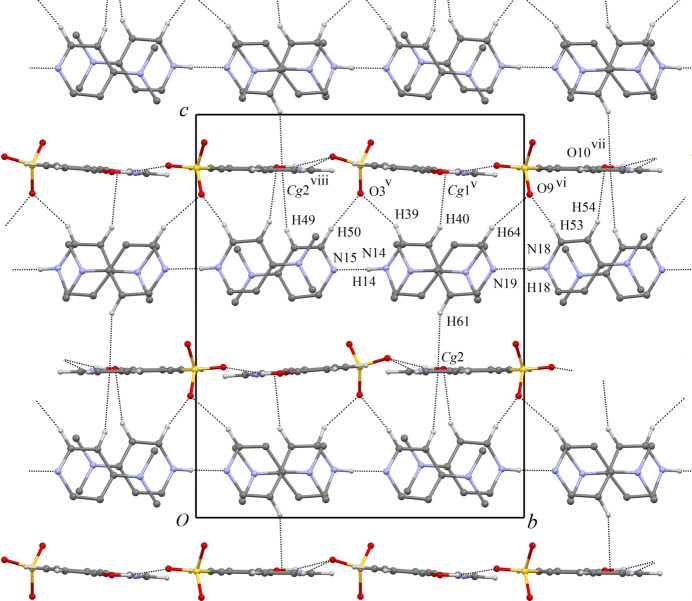
In the title co-crystal, both the cation layers of [HDMAP·DMAP]+ and the anion layers of HqSA− run parallel to the ab plane. The hydrogen-bond geometry is summarized in Table 1 ▸. The pyridine rings in the cation layer are stacked along the ab plane as shown in Fig. 2 ▸. In the cation layer, two independent cation units of [HDMAP·DMAP]+ are formed by intermolecular N—H⋯N hydrogen bonds (N14—H14⋯N15 and N18—H18⋯N19). The N14—H14⋯N15 and N18—H18⋯N19 angles are 174 (3) and 177 (7)°, respectively. The dihedral angles between the two pyridine rings in the [HDMAP·DMAP]+ units are 0.21 (15)° (N14/C39–C43 and N15/C46–C50 rings) and 1.60 (15)° (N18/C53–C57 and N19/C60–C64). The quinoline ring system in the anion layer faces the ab plane as shown in Fig. 3 ▸. In the anion layer, each HqSA− molecule is surrounded by six HqSA− molecules through intermolecular hydrogen bonds, essentially forming an sheet. Each HqSA− molecule binds with two HqSA− molecules having the same molecular orientation through intermolecular C—H⋯O hydrogen bonds [C27—H27⋯O6ii and C36—H36⋯O10iv; symmetry codes: (ii) x − 1, y, z; (iv) x + 1, y, z] and also binds with four HqSA− molecules having the different molecular orientation through intermolecular O—H⋯O and C—H⋯O hydrogen bonds [O6—H6⋯O8i, O10—H10⋯O4, C27—H27⋯O8iii and C36—H36⋯O4iv; symmetry codes: (i) x, y − 1, z; (iii) x − 1, y − 1, z]. The C27—H27⋯O6ii, C36—H36⋯O10iv, O6—H6⋯O8i, O10—H10⋯O4, C27—H27⋯O8iii and C36—H36⋯O4iv angles are 125, 125, 137 (4), 144 (4), 164 and 160°, respectively. The interplanar spacing between adjacent anionic layers (the distance between the closest centroids of the mean planes through N12/C22/C23/C37 within the anionic layers, being across the cationic layer from each other) is 9.562 Å. The interactions between the cationic and anionic layers are attributed to the extended 3D hydrogen-bonding linkages, three C—H⋯π interactions [C40—H40⋯Cg1i, C49—H49⋯Cg2viii, C61—H61⋯Cg2; Cg1 and Cg2 are the centroids of the N11/C25–C29 and N12/C34–C38 rings, respectively; symmetry code: (viii) x − 1, 1 − y, z + 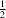 ] and five C—H⋯O interactions [C39—H39⋯O3v, C50—H50⋯O3v, C53—H53⋯O9vi, C54—H54⋯O10vii, C64—H64⋯O9vi; symmetry code: (v) x, 1 − y, z +
] and five C—H⋯O interactions [C39—H39⋯O3v, C50—H50⋯O3v, C53—H53⋯O9vi, C54—H54⋯O10vii, C64—H64⋯O9vi; symmetry code: (v) x, 1 − y, z + 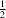 ; (vi) x, 2 − y, z +
; (vi) x, 2 − y, z + 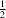 ; (vii) x + 1, 2 − y, z +
; (vii) x + 1, 2 − y, z + 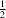 ] as shown in Fig. 4 ▸ and Table 1 ▸. In addition, each independent ion pair forms
] as shown in Fig. 4 ▸ and Table 1 ▸. In addition, each independent ion pair forms 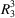 (8) motif by one intermolecular N—H⋯N hydrogen bond and two intermolecular C—H⋯O hydrogen bonds (N14—H14⋯N15, C39—H39⋯O3v and C50—H50⋯O3v; N18—H18⋯N19, C53—H53⋯O9vi and C64—H64⋯O9vi).
(8) motif by one intermolecular N—H⋯N hydrogen bond and two intermolecular C—H⋯O hydrogen bonds (N14—H14⋯N15, C39—H39⋯O3v and C50—H50⋯O3v; N18—H18⋯N19, C53—H53⋯O9vi and C64—H64⋯O9vi).
Figure 3.
The S(5) ring motifs formed by intramolecular O—H⋯N hydrogen bonds involving the hydroxy groups and quinoline N atoms of the HqSA− anionic units. The intramolecular O—H⋯N hydrogen bonds are shown as dashed lines. The sheet structure of the HqSA− anionic units is formed by the planar intermolecular hydrogen-bond networks in the ab plane. The intermolecular O—H⋯O, C—H⋯O, O—H⋯N hydrogen bonds are also shown as dashed lines. [Symmetry codes: (i) x, y − 1, z; (ii) x − 1, y, z; (iii) x − 1, y − 1, z; (iv) x + 1, y, z.].
Figure 4.
The network structure between [HDMAP·DMAP]+ cationic layers and HqSA− anion layers. The intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions are shown as dashed lines. The 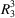 (8) motifs of independent ion pairs formed by an intermolecular N—H⋯N hydrogen bond and two intermolecular C—H⋯O hydrogen bonds are also shown as dashed lines. [Symmetry codes: (v) x, −y + 1, z +
(8) motifs of independent ion pairs formed by an intermolecular N—H⋯N hydrogen bond and two intermolecular C—H⋯O hydrogen bonds are also shown as dashed lines. [Symmetry codes: (v) x, −y + 1, z + 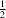 ; (vi) x, −y + 2, z +
; (vi) x, −y + 2, z + 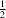 ; (vii) x + 1, −y + 2, z +
; (vii) x + 1, −y + 2, z + 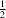 ; (viii) x − 1, −y + 1, z +
; (viii) x − 1, −y + 1, z + 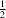 .].
.].
4. Database survey
A search of the Cambridge Structural Database (CSD, Version 2024.1.0, update of March 2024; Groom et al., 2016 ▸) for compounds containing the 4-aminopyridine skeleton with hydrogen atom bound at the 2, 3, 5, 6-positions of the pyridine ring gave 5687 hits. Among those, a search for the containing DMAP molecule gave 1794 hits and for those of protonated DMAP gave 360 hits. A search for compounds containing a pyridine-protonated pyridine skeleton gave 15 hits. In these compounds, the dihedral angles between two pyridine rings are close to 0° in seven structures, which are essentially co-planar due to unique hydrogen-bonding networks stemming from the substituents on the pyridine rings (BAYBIN; Kobayashi et al., 2003 ▸; BECHOG; Glidewell et al., 1982 ▸; KIFBIO; Vladiskovic et al., 2023 ▸; WAZNET; Lackova et al., 2014 ▸; WEVHOX; Zhang et al., 2018 ▸; XACFOW; Mautner & Goher, 1998 ▸; XOHWAT; Santra et al., 2008 ▸). In single crystals of salts of the mellitate anion, which is obtained by deprotonation of mellitic acid (benzene hexacarboxylic acid), with substituted pyridinium derivatives, the triangular hydrogen-bonded unit between the anions induces a two-dimensional sheet self-organizing structure (BAYBIN, Kobayashi et al., 2003 ▸). On the other hand, ferrocene derivatives substituted with pyridine form cationic dimers via a hydrogen bond between two pyridine rings (WOFGII; Braga et al., 2008 ▸). A search for containing both of protonated DMAP and the other neutral DMAP gave 14 hits. There are five hits having the proton between two N-(4-pyridyl)dimethylamine skeletons (2, 3, 5, 6-carbon atoms are bound to hydrogen atoms). In these compounds, the dihedral angles between two pyridine rings are close to 0° in three structures, which are essentially co-planar structures [1.3 (1)° in FETDEO, Aakeroy et al., 2005 ▸; 3.47 (7)° in GOFRUQ, Wagler et al., 2014 ▸; 3.8 (4)° in ZAPNIN, Biradha et al., 1995 ▸]. A fragment search for the 8-hydroxyquinoline-5-sulfonic acid skeleton gave 84 hits, which include two hydrate co-crystals composed of the 8-hydroxyquinoline-5-sulfonicin anion and 4-phenylpyridine (EMEDUY; Ganie et al., 2021 ▸), 4,4′-bipyrydine (INEMAP; Baskar Raj et al., 2003 ▸) cations and three hydrate co-crystals composed of the 8-hydroxy-7-iodoquinoline-5-sulfonic anion and various pyridine derivative cations (EFAQUZ, Smith et al., 2012 ▸; EYIYOA, Smith et al., 2004 ▸; ISUTAR, Hemamalini et al., 2004 ▸). According to the crystal structures of BAYBIN, EFAQUZ, EYIYOA and ISUTAR, these compounds form layered structures by constructing 2D layers of the cationic and anionic moieties with these layers arranged sterically.
5. Synthesis and crystallization
To a solution of DMAP (611 mg, 5.0 mmol) in H2O (5 mL) at 353 K, an ethanol (1 mL) solution of H2qSA (450 mg, 2.0 mmol) was added and then stirred for 30 min. Orange single crystals of the title compound suitable for X-ray diffraction were grown by slow evaporation of the aqueous ethanol solution mentioned above for a week at ambient temperature.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The title compound was refined as an inversion twin in Pc whose twin component mass ratio refined to 0.522 (18):0.478 (18). The hydroxy H atoms, H6 and H10, were located in a difference-Fourier map and freely refined. The N-bound H atoms, H14 and H18, were located in difference-Fourier maps but were refined with a distance restraint of N—H = 0.86 ± 0.02 Å. All H atoms bound to carbon were positioned geometrically and refined using a riding model, with C—H = 0.95 or 0.98 Å and Uiso(H) = 1.2 or 1.5Ueq(C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C7H11N2+·C9H6NO4S−·C7H10N2 |
| M r | 469.55 |
| Crystal system, space group | Monoclinic, Pc |
| Temperature (K) | 100 |
| a, b, c (Å) | 8.00032 (10), 15.14469 (18), 18.9141 (2) |
| β (°) | 100.6050 (12) |
| V (Å3) | 2252.53 (5) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 1.62 |
| Crystal size (mm) | 0.4 × 0.30 × 0.11 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Dualflex, HyPix |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2023 ▸) |
| Tmin, Tmax | 0.731, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 16358, 6813, 6608 |
| R int | 0.030 |
| (sin θ/λ)max (Å−1) | 0.632 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.033, 0.085, 1.04 |
| No. of reflections | 6813 |
| No. of parameters | 620 |
| No. of restraints | 4 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.30, −0.39 |
| Absolute structure | Refined as an inversion twin |
| Absolute structure parameter | 0.478 (18) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698902400642X/ox2006sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902400642X/ox2006Isup3.hkl
Supporting information file. DOI: 10.1107/S205698902400642X/ox2006Isup3.cml
CCDC reference: 2366836
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Crystal data
| C7H11N2+·C9H6NO4S−·C7H10N2 | F(000) = 992 |
| Mr = 469.55 | Dx = 1.385 Mg m−3 |
| Monoclinic, Pc | Cu Kα radiation, λ = 1.54184 Å |
| a = 8.00032 (10) Å | Cell parameters from 11719 reflections |
| b = 15.14469 (18) Å | θ = 3.8–76.8° |
| c = 18.9141 (2) Å | µ = 1.62 mm−1 |
| β = 100.6050 (12)° | T = 100 K |
| V = 2252.53 (5) Å3 | Block, colourless |
| Z = 4 | 0.4 × 0.30 × 0.11 mm |
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Data collection
| XtaLAB Synergy, Dualflex, HyPix diffractometer | 6813 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, PhotonJet (Cu) X-ray Source | 6608 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.030 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 77.2°, θmin = 3.8° |
| ω scans | h = −10→9 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2023) | k = −17→19 |
| Tmin = 0.731, Tmax = 1.000 | l = −22→23 |
| 16358 measured reflections |
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.033 | w = 1/[σ2(Fo2) + (0.0463P)2 + 0.5548P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.085 | (Δ/σ)max < 0.001 |
| S = 1.04 | Δρmax = 0.30 e Å−3 |
| 6813 reflections | Δρmin = −0.39 e Å−3 |
| 620 parameters | Absolute structure: Refined as an inversion twin |
| 4 restraints | Absolute structure parameter: 0.478 (18) |
| Primary atom site location: dual |
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component inversion twin.1. Twinned data refinement Scales: 0.522 (18) 0.478 (18) 2. Fixed Uiso At 1.2 times of: All C(H) groups At 1.5 times of: All C(H,H,H) groups 3. Restrained distances H18-N18 0.86 with sigma of 0.02 H14-N14 0.86 with sigma of 0.02 4.a Aromatic/amide H refined with riding coordinates: C35(H35), C23(H23), C27(H27), C26(H26), C49(H49), C31(H31), C28(H28), C61(H61), C54(H54), C36(H36), C60(H60), C50(H50), C22(H22), C40(H40), C53(H53), C37(H37), C42(H42), C47(H47), C64(H64), C63(H63), C46(H46), C43(H43), C39(H39), C32(H32), C56(H56), C57(H57) 4.b Idealised Me refined as rotating group: C52(H52A,H52B,H52C), C44(H44A,H44B,H44C), C58(H58A,H58B,H58C), C66(H66A,H66B, H66C), C51(H51A,H51B,H51C), C45(H45A,H45B,H45C), C65(H65A,H65B,H65C), C59(H59A, H59B,H59C) |
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.17772 (8) | 0.49921 (4) | 0.37557 (4) | 0.01431 (15) | |
| S2 | 0.88938 (8) | 0.99725 (4) | 0.36763 (4) | 0.01686 (16) | |
| O5 | 0.0868 (3) | 0.46728 (13) | 0.43037 (11) | 0.0198 (4) | |
| O10 | 0.3080 (3) | 0.75413 (13) | 0.36853 (12) | 0.0189 (4) | |
| O4 | 0.2618 (3) | 0.58342 (12) | 0.39552 (12) | 0.0222 (4) | |
| O7 | 1.0147 (3) | 0.97468 (16) | 0.43079 (12) | 0.0274 (5) | |
| O6 | 0.7498 (3) | 0.25632 (14) | 0.35776 (12) | 0.0191 (4) | |
| N11 | 0.4365 (3) | 0.18427 (15) | 0.35601 (13) | 0.0168 (5) | |
| O3 | 0.0728 (3) | 0.49865 (13) | 0.30454 (12) | 0.0251 (5) | |
| N14 | 0.0994 (3) | 0.57921 (17) | 0.61872 (14) | 0.0228 (5) | |
| O9 | 0.9547 (3) | 0.98318 (15) | 0.30206 (12) | 0.0253 (5) | |
| N18 | 1.0806 (4) | 1.06837 (18) | 0.62048 (15) | 0.0283 (6) | |
| N12 | 0.6186 (3) | 0.68257 (14) | 0.36322 (13) | 0.0153 (5) | |
| N17 | 1.3114 (3) | 1.31360 (18) | 0.63022 (14) | 0.0268 (6) | |
| N13 | 0.3275 (3) | 0.82252 (17) | 0.60874 (14) | 0.0248 (6) | |
| C25 | 0.3141 (4) | 0.33136 (18) | 0.36637 (15) | 0.0132 (5) | |
| N16 | −0.3146 (4) | 0.16966 (18) | 0.60144 (14) | 0.0300 (6) | |
| N15 | −0.0827 (3) | 0.41978 (17) | 0.61461 (14) | 0.0258 (6) | |
| C30 | 0.4422 (3) | 0.80852 (18) | 0.36639 (15) | 0.0152 (6) | |
| N20 | 0.6851 (4) | 0.65381 (18) | 0.61189 (15) | 0.0297 (6) | |
| C29 | 0.4524 (4) | 0.27401 (18) | 0.36172 (15) | 0.0141 (5) | |
| N19 | 0.9083 (4) | 0.90581 (18) | 0.61654 (15) | 0.0271 (6) | |
| C35 | 0.9054 (4) | 0.79031 (19) | 0.36300 (15) | 0.0157 (5) | |
| H35 | 1.003375 | 0.825760 | 0.363038 | 0.019* | |
| O8 | 0.8170 (3) | 1.08342 (14) | 0.37345 (18) | 0.0427 (7) | |
| C48 | −0.2420 (4) | 0.25088 (19) | 0.60531 (16) | 0.0222 (6) | |
| C23 | 0.5063 (4) | 0.45581 (18) | 0.37350 (16) | 0.0165 (6) | |
| H23 | 0.525964 | 0.517580 | 0.378068 | 0.020* | |
| C24 | 0.3458 (4) | 0.42408 (18) | 0.37218 (15) | 0.0156 (6) | |
| C27 | 0.1385 (4) | 0.20164 (19) | 0.35746 (16) | 0.0195 (6) | |
| H27 | 0.031056 | 0.173969 | 0.355266 | 0.023* | |
| C26 | 0.1533 (4) | 0.29119 (19) | 0.36372 (15) | 0.0162 (6) | |
| H26 | 0.056312 | 0.326366 | 0.366290 | 0.019* | |
| C34 | 0.7451 (3) | 0.82997 (18) | 0.36388 (15) | 0.0137 (5) | |
| C49 | −0.2102 (4) | 0.29987 (19) | 0.66961 (16) | 0.0224 (6) | |
| H49 | −0.243039 | 0.277078 | 0.711817 | 0.027* | |
| C31 | 0.4187 (4) | 0.89824 (19) | 0.36710 (17) | 0.0192 (6) | |
| H31 | 0.309000 | 0.921934 | 0.367659 | 0.023* | |
| C38 | 0.6054 (4) | 0.77244 (17) | 0.36430 (15) | 0.0130 (5) | |
| C28 | 0.2840 (4) | 0.15080 (18) | 0.35428 (16) | 0.0197 (6) | |
| H28 | 0.271473 | 0.088498 | 0.350666 | 0.024* | |
| C33 | 0.7176 (4) | 0.92240 (18) | 0.36554 (15) | 0.0152 (5) | |
| C61 | 0.8018 (4) | 0.7746 (2) | 0.55202 (16) | 0.0215 (6) | |
| H61 | 0.781003 | 0.744043 | 0.507393 | 0.026* | |
| C54 | 1.1908 (4) | 1.1940 (2) | 0.68862 (16) | 0.0220 (6) | |
| H54 | 1.212304 | 1.223795 | 0.733567 | 0.026* | |
| C36 | 0.9166 (4) | 0.70039 (19) | 0.36211 (16) | 0.0184 (6) | |
| H36 | 1.022730 | 0.672608 | 0.361413 | 0.022* | |
| C62 | 0.7562 (4) | 0.7356 (2) | 0.61344 (17) | 0.0237 (6) | |
| C60 | 0.8763 (4) | 0.8566 (2) | 0.55657 (17) | 0.0257 (7) | |
| H60 | 0.907308 | 0.880142 | 0.514258 | 0.031* | |
| C50 | −0.1315 (4) | 0.3809 (2) | 0.67126 (17) | 0.0247 (6) | |
| H50 | −0.110204 | 0.411581 | 0.715822 | 0.030* | |
| C22 | 0.6432 (4) | 0.39925 (19) | 0.36825 (17) | 0.0193 (6) | |
| H22 | 0.753237 | 0.423009 | 0.368649 | 0.023* | |
| C40 | 0.2142 (4) | 0.7096 (2) | 0.67653 (16) | 0.0209 (6) | |
| H40 | 0.240263 | 0.743534 | 0.719363 | 0.025* | |
| C53 | 1.1159 (4) | 1.1131 (2) | 0.68335 (18) | 0.0257 (7) | |
| H53 | 1.087382 | 1.087116 | 0.725275 | 0.031* | |
| C37 | 0.7696 (4) | 0.64896 (18) | 0.36221 (17) | 0.0183 (6) | |
| H37 | 0.780438 | 0.586507 | 0.361522 | 0.022* | |
| C42 | 0.2165 (4) | 0.6872 (2) | 0.55097 (16) | 0.0229 (6) | |
| H42 | 0.243782 | 0.705189 | 0.506333 | 0.027* | |
| C41 | 0.2560 (4) | 0.74334 (19) | 0.61167 (16) | 0.0200 (6) | |
| C21 | 0.6173 (4) | 0.30979 (18) | 0.36257 (15) | 0.0145 (5) | |
| C47 | −0.1930 (4) | 0.2920 (2) | 0.54526 (16) | 0.0237 (6) | |
| H47 | −0.212812 | 0.263269 | 0.499838 | 0.028* | |
| C64 | 0.8639 (4) | 0.8686 (2) | 0.67528 (18) | 0.0296 (7) | |
| H64 | 0.885457 | 0.901387 | 0.718843 | 0.036* | |
| C55 | 1.2371 (4) | 1.23419 (19) | 0.62685 (16) | 0.0218 (6) | |
| C52 | −0.3516 (5) | 0.1263 (2) | 0.66566 (19) | 0.0318 (7) | |
| H52A | −0.246320 | 0.120190 | 0.701170 | 0.048* | |
| H52B | −0.399916 | 0.067773 | 0.652889 | 0.048* | |
| H52C | −0.433271 | 0.161935 | 0.686096 | 0.048* | |
| C63 | 0.7900 (4) | 0.7873 (2) | 0.67674 (17) | 0.0282 (7) | |
| H63 | 0.761539 | 0.765609 | 0.720146 | 0.034* | |
| C44 | 0.3650 (5) | 0.8790 (2) | 0.67269 (19) | 0.0302 (7) | |
| H44A | 0.258673 | 0.894730 | 0.688314 | 0.045* | |
| H44B | 0.422323 | 0.932840 | 0.661072 | 0.045* | |
| H44C | 0.439097 | 0.847174 | 0.711421 | 0.045* | |
| C46 | −0.1165 (4) | 0.3737 (2) | 0.55256 (17) | 0.0250 (6) | |
| H46 | −0.085302 | 0.399369 | 0.511048 | 0.030* | |
| C43 | 0.1398 (4) | 0.6079 (2) | 0.55592 (17) | 0.0238 (6) | |
| H43 | 0.113795 | 0.571602 | 0.514381 | 0.029* | |
| C39 | 0.1381 (4) | 0.6302 (2) | 0.67832 (17) | 0.0230 (6) | |
| H39 | 0.111083 | 0.609654 | 0.722336 | 0.028* | |
| C32 | 0.5578 (4) | 0.95491 (19) | 0.36698 (17) | 0.0202 (6) | |
| H32 | 0.541065 | 1.016967 | 0.367926 | 0.024* | |
| C56 | 1.2007 (4) | 1.1840 (2) | 0.56237 (17) | 0.0252 (6) | |
| H56 | 1.230849 | 1.206924 | 0.519661 | 0.030* | |
| C58 | 1.3420 (5) | 1.3650 (2) | 0.69703 (19) | 0.0322 (7) | |
| H58A | 1.233475 | 1.377047 | 0.712141 | 0.048* | |
| H58B | 1.397149 | 1.420969 | 0.688945 | 0.048* | |
| H58C | 1.415965 | 1.331447 | 0.734677 | 0.048* | |
| C57 | 1.1241 (4) | 1.1046 (2) | 0.56067 (18) | 0.0292 (7) | |
| H57 | 1.100001 | 1.073059 | 0.516531 | 0.035* | |
| C66 | 0.6549 (5) | 0.6125 (2) | 0.6777 (2) | 0.0355 (8) | |
| H66A | 0.761279 | 0.610761 | 0.712932 | 0.053* | |
| H66B | 0.613111 | 0.552185 | 0.667276 | 0.053* | |
| H66C | 0.569870 | 0.646643 | 0.697307 | 0.053* | |
| C51 | −0.3476 (5) | 0.1193 (2) | 0.53443 (18) | 0.0345 (8) | |
| H51A | −0.429289 | 0.151412 | 0.498551 | 0.052* | |
| H51B | −0.394504 | 0.061445 | 0.543216 | 0.052* | |
| H51C | −0.241090 | 0.111422 | 0.516567 | 0.052* | |
| C45 | 0.3605 (5) | 0.8585 (2) | 0.54079 (18) | 0.0316 (7) | |
| H45A | 0.434078 | 0.817948 | 0.520221 | 0.047* | |
| H45B | 0.416725 | 0.916003 | 0.549571 | 0.047* | |
| H45C | 0.252586 | 0.865738 | 0.507093 | 0.047* | |
| C65 | 0.6452 (5) | 0.6043 (2) | 0.54515 (19) | 0.0320 (7) | |
| H65A | 0.569908 | 0.639435 | 0.509079 | 0.048* | |
| H65B | 0.588361 | 0.549017 | 0.553627 | 0.048* | |
| H65C | 0.750475 | 0.591031 | 0.527717 | 0.048* | |
| C59 | 1.3515 (5) | 1.3552 (2) | 0.5656 (2) | 0.0355 (8) | |
| H59A | 1.429459 | 1.317305 | 0.545016 | 0.053* | |
| H59B | 1.405242 | 1.412637 | 0.578214 | 0.053* | |
| H59C | 1.246532 | 1.363609 | 0.530309 | 0.053* | |
| H14 | 0.047 (5) | 0.5262 (17) | 0.616 (2) | 0.029 (9)* | |
| H10 | 0.336 (6) | 0.697 (3) | 0.372 (2) | 0.037 (11)* | |
| H18 | 1.027 (6) | 1.016 (2) | 0.618 (3) | 0.057 (15)* | |
| H6 | 0.714 (6) | 0.204 (3) | 0.357 (2) | 0.039 (12)* |
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0135 (3) | 0.0121 (3) | 0.0179 (3) | 0.0021 (2) | 0.0043 (2) | 0.0012 (2) |
| S2 | 0.0140 (3) | 0.0113 (3) | 0.0270 (4) | −0.0012 (2) | 0.0083 (3) | −0.0004 (2) |
| O5 | 0.0221 (10) | 0.0170 (9) | 0.0228 (11) | 0.0007 (8) | 0.0104 (8) | −0.0003 (8) |
| O10 | 0.0124 (10) | 0.0129 (10) | 0.0327 (12) | −0.0020 (8) | 0.0079 (8) | 0.0022 (8) |
| O4 | 0.0193 (10) | 0.0115 (9) | 0.0376 (12) | 0.0007 (8) | 0.0100 (9) | −0.0005 (8) |
| O7 | 0.0270 (12) | 0.0331 (11) | 0.0216 (11) | −0.0145 (10) | 0.0027 (9) | −0.0034 (9) |
| O6 | 0.0117 (10) | 0.0142 (10) | 0.0321 (12) | 0.0006 (8) | 0.0054 (8) | 0.0000 (8) |
| N11 | 0.0166 (11) | 0.0136 (11) | 0.0202 (12) | 0.0000 (9) | 0.0030 (9) | 0.0005 (9) |
| O3 | 0.0270 (12) | 0.0286 (12) | 0.0185 (11) | 0.0128 (9) | 0.0012 (9) | 0.0007 (8) |
| N14 | 0.0237 (13) | 0.0207 (12) | 0.0239 (14) | −0.0030 (10) | 0.0041 (10) | −0.0013 (10) |
| O9 | 0.0251 (11) | 0.0316 (11) | 0.0205 (11) | −0.0083 (9) | 0.0076 (9) | 0.0026 (9) |
| N18 | 0.0251 (14) | 0.0240 (14) | 0.0335 (16) | 0.0010 (11) | −0.0006 (11) | −0.0050 (11) |
| N12 | 0.0142 (11) | 0.0127 (11) | 0.0188 (12) | −0.0001 (9) | 0.0029 (9) | −0.0001 (9) |
| N17 | 0.0303 (14) | 0.0248 (13) | 0.0229 (13) | −0.0001 (11) | −0.0010 (11) | 0.0038 (10) |
| N13 | 0.0297 (14) | 0.0218 (13) | 0.0231 (14) | −0.0044 (10) | 0.0052 (11) | 0.0012 (10) |
| C25 | 0.0125 (12) | 0.0147 (13) | 0.0128 (12) | −0.0001 (10) | 0.0035 (10) | 0.0009 (10) |
| N16 | 0.0447 (17) | 0.0249 (13) | 0.0179 (13) | −0.0064 (12) | −0.0008 (12) | −0.0001 (10) |
| N15 | 0.0263 (14) | 0.0231 (13) | 0.0275 (14) | −0.0001 (11) | 0.0036 (11) | −0.0002 (10) |
| C30 | 0.0120 (13) | 0.0159 (13) | 0.0178 (14) | −0.0007 (11) | 0.0029 (10) | 0.0012 (11) |
| N20 | 0.0382 (16) | 0.0294 (14) | 0.0225 (14) | −0.0068 (12) | 0.0081 (12) | 0.0030 (10) |
| C29 | 0.0135 (13) | 0.0158 (13) | 0.0132 (13) | 0.0016 (10) | 0.0029 (10) | 0.0012 (10) |
| N19 | 0.0289 (15) | 0.0246 (13) | 0.0264 (14) | −0.0010 (11) | 0.0014 (11) | −0.0038 (10) |
| C35 | 0.0134 (13) | 0.0174 (13) | 0.0165 (14) | −0.0010 (11) | 0.0035 (10) | 0.0008 (10) |
| O8 | 0.0219 (12) | 0.0115 (10) | 0.100 (2) | −0.0016 (9) | 0.0251 (13) | −0.0010 (12) |
| C48 | 0.0210 (15) | 0.0243 (15) | 0.0199 (15) | 0.0037 (12) | −0.0001 (12) | 0.0005 (12) |
| C23 | 0.0158 (12) | 0.0119 (13) | 0.0220 (14) | −0.0004 (11) | 0.0037 (11) | 0.0004 (10) |
| C24 | 0.0167 (14) | 0.0138 (13) | 0.0167 (14) | 0.0028 (11) | 0.0039 (11) | 0.0014 (10) |
| C27 | 0.0133 (13) | 0.0160 (13) | 0.0295 (16) | −0.0032 (11) | 0.0045 (11) | 0.0022 (11) |
| C26 | 0.0157 (14) | 0.0149 (13) | 0.0190 (14) | 0.0032 (10) | 0.0057 (11) | −0.0009 (10) |
| C34 | 0.0128 (13) | 0.0163 (13) | 0.0119 (12) | −0.0005 (11) | 0.0023 (10) | −0.0003 (10) |
| C49 | 0.0241 (15) | 0.0249 (15) | 0.0182 (14) | 0.0009 (12) | 0.0039 (11) | 0.0013 (11) |
| C31 | 0.0110 (14) | 0.0170 (13) | 0.0308 (17) | 0.0032 (11) | 0.0070 (11) | 0.0025 (11) |
| C38 | 0.0131 (13) | 0.0130 (12) | 0.0129 (12) | −0.0002 (10) | 0.0026 (10) | 0.0004 (10) |
| C28 | 0.0178 (14) | 0.0130 (13) | 0.0274 (16) | −0.0008 (10) | 0.0017 (12) | 0.0014 (11) |
| C33 | 0.0144 (13) | 0.0148 (13) | 0.0172 (14) | −0.0010 (11) | 0.0048 (10) | 0.0006 (10) |
| C61 | 0.0237 (15) | 0.0239 (15) | 0.0160 (14) | −0.0009 (12) | 0.0015 (11) | −0.0010 (11) |
| C54 | 0.0199 (14) | 0.0267 (15) | 0.0185 (14) | 0.0022 (12) | 0.0016 (11) | −0.0023 (11) |
| C36 | 0.0139 (14) | 0.0169 (13) | 0.0248 (15) | 0.0034 (11) | 0.0050 (11) | 0.0003 (11) |
| C62 | 0.0217 (15) | 0.0281 (15) | 0.0212 (15) | 0.0027 (12) | 0.0039 (12) | 0.0021 (12) |
| C60 | 0.0259 (16) | 0.0286 (16) | 0.0219 (15) | 0.0012 (13) | 0.0028 (12) | 0.0012 (13) |
| C50 | 0.0241 (15) | 0.0269 (15) | 0.0218 (15) | 0.0011 (12) | 0.0006 (12) | −0.0047 (12) |
| C22 | 0.0143 (14) | 0.0179 (14) | 0.0272 (16) | −0.0007 (11) | 0.0075 (12) | −0.0005 (11) |
| C40 | 0.0195 (14) | 0.0254 (15) | 0.0172 (14) | 0.0003 (11) | 0.0021 (11) | −0.0011 (11) |
| C53 | 0.0211 (15) | 0.0280 (16) | 0.0270 (17) | 0.0036 (12) | 0.0019 (12) | 0.0001 (12) |
| C37 | 0.0158 (13) | 0.0102 (12) | 0.0295 (16) | 0.0012 (10) | 0.0054 (11) | 0.0003 (11) |
| C42 | 0.0231 (15) | 0.0290 (16) | 0.0176 (14) | 0.0013 (12) | 0.0064 (11) | 0.0005 (12) |
| C41 | 0.0191 (14) | 0.0202 (14) | 0.0206 (15) | 0.0024 (11) | 0.0030 (11) | 0.0029 (11) |
| C21 | 0.0129 (13) | 0.0157 (13) | 0.0154 (13) | 0.0024 (11) | 0.0038 (10) | 0.0006 (11) |
| C47 | 0.0239 (15) | 0.0266 (15) | 0.0198 (15) | 0.0037 (12) | 0.0020 (11) | −0.0034 (11) |
| C64 | 0.0304 (17) | 0.0340 (17) | 0.0237 (16) | 0.0047 (14) | 0.0033 (13) | −0.0076 (13) |
| C55 | 0.0187 (14) | 0.0223 (14) | 0.0229 (16) | 0.0049 (11) | −0.0001 (12) | −0.0012 (11) |
| C52 | 0.0371 (19) | 0.0279 (17) | 0.0296 (18) | −0.0070 (14) | 0.0044 (14) | 0.0041 (13) |
| C63 | 0.0316 (17) | 0.0341 (17) | 0.0196 (16) | 0.0034 (14) | 0.0070 (13) | −0.0004 (13) |
| C44 | 0.0347 (18) | 0.0234 (15) | 0.0319 (18) | −0.0074 (13) | 0.0052 (14) | −0.0007 (13) |
| C46 | 0.0219 (15) | 0.0311 (16) | 0.0218 (15) | 0.0003 (13) | 0.0034 (12) | 0.0040 (12) |
| C43 | 0.0228 (15) | 0.0260 (15) | 0.0222 (15) | −0.0002 (12) | 0.0035 (12) | −0.0037 (12) |
| C39 | 0.0256 (16) | 0.0233 (15) | 0.0205 (15) | 0.0013 (12) | 0.0053 (12) | 0.0007 (12) |
| C32 | 0.0200 (14) | 0.0115 (13) | 0.0309 (17) | 0.0023 (11) | 0.0096 (12) | 0.0016 (11) |
| C56 | 0.0258 (16) | 0.0296 (16) | 0.0194 (15) | 0.0044 (13) | 0.0023 (12) | −0.0015 (12) |
| C58 | 0.0369 (19) | 0.0261 (16) | 0.0303 (18) | −0.0032 (14) | −0.0025 (14) | −0.0027 (13) |
| C57 | 0.0275 (16) | 0.0318 (17) | 0.0254 (16) | 0.0057 (13) | −0.0024 (13) | −0.0090 (13) |
| C66 | 0.038 (2) | 0.0373 (19) | 0.0331 (19) | −0.0040 (15) | 0.0117 (15) | 0.0101 (15) |
| C51 | 0.042 (2) | 0.0318 (18) | 0.0269 (18) | −0.0040 (15) | −0.0022 (15) | −0.0051 (14) |
| C45 | 0.0344 (18) | 0.0333 (17) | 0.0287 (18) | −0.0062 (14) | 0.0100 (14) | 0.0082 (13) |
| C65 | 0.0346 (19) | 0.0261 (16) | 0.0348 (19) | −0.0073 (13) | 0.0054 (15) | −0.0003 (13) |
| C59 | 0.0352 (19) | 0.0381 (19) | 0.0317 (19) | −0.0014 (15) | 0.0023 (14) | 0.0119 (15) |
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Geometric parameters (Å, º)
| S1—O5 | 1.455 (2) | C31—C32 | 1.406 (4) |
| S1—O4 | 1.459 (2) | C28—H28 | 0.9500 |
| S1—O3 | 1.447 (2) | C33—C32 | 1.375 (4) |
| S1—C24 | 1.772 (3) | C61—H61 | 0.9500 |
| S2—O7 | 1.452 (2) | C61—C62 | 1.409 (4) |
| S2—O9 | 1.448 (2) | C61—C60 | 1.373 (4) |
| S2—O8 | 1.440 (2) | C54—H54 | 0.9500 |
| S2—C33 | 1.776 (3) | C54—C53 | 1.359 (4) |
| O10—C30 | 1.359 (3) | C54—C55 | 1.426 (4) |
| O10—H10 | 0.90 (4) | C36—H36 | 0.9500 |
| O6—C21 | 1.350 (3) | C36—C37 | 1.410 (4) |
| O6—H6 | 0.84 (4) | C62—C63 | 1.414 (4) |
| N11—C29 | 1.367 (4) | C60—H60 | 0.9500 |
| N11—C28 | 1.316 (4) | C50—H50 | 0.9500 |
| N14—C43 | 1.358 (4) | C22—H22 | 0.9500 |
| N14—C39 | 1.354 (4) | C22—C21 | 1.372 (4) |
| N14—H14 | 0.90 (2) | C40—H40 | 0.9500 |
| N18—C53 | 1.353 (4) | C40—C41 | 1.425 (4) |
| N18—C57 | 1.359 (5) | C40—C39 | 1.351 (4) |
| N18—H18 | 0.90 (2) | C53—H53 | 0.9500 |
| N12—C38 | 1.366 (3) | C37—H37 | 0.9500 |
| N12—C37 | 1.315 (4) | C42—H42 | 0.9500 |
| N17—C55 | 1.338 (4) | C42—C41 | 1.417 (4) |
| N17—C58 | 1.466 (4) | C42—C43 | 1.360 (4) |
| N17—C59 | 1.462 (4) | C47—H47 | 0.9500 |
| N13—C41 | 1.334 (4) | C47—C46 | 1.375 (4) |
| N13—C44 | 1.467 (4) | C64—H64 | 0.9500 |
| N13—C45 | 1.464 (4) | C64—C63 | 1.369 (5) |
| C25—C29 | 1.422 (4) | C55—C56 | 1.421 (4) |
| C25—C24 | 1.428 (4) | C52—H52A | 0.9800 |
| C25—C26 | 1.415 (4) | C52—H52B | 0.9800 |
| N16—C48 | 1.356 (4) | C52—H52C | 0.9800 |
| N16—C52 | 1.458 (4) | C63—H63 | 0.9500 |
| N16—C51 | 1.461 (4) | C44—H44A | 0.9800 |
| N15—C50 | 1.342 (4) | C44—H44B | 0.9800 |
| N15—C46 | 1.350 (4) | C44—H44C | 0.9800 |
| C30—C31 | 1.372 (4) | C46—H46 | 0.9500 |
| C30—C38 | 1.423 (4) | C43—H43 | 0.9500 |
| N20—C62 | 1.362 (4) | C39—H39 | 0.9500 |
| N20—C66 | 1.453 (4) | C32—H32 | 0.9500 |
| N20—C65 | 1.452 (4) | C56—H56 | 0.9500 |
| C29—C21 | 1.424 (4) | C56—C57 | 1.348 (5) |
| N19—C60 | 1.342 (4) | C58—H58A | 0.9800 |
| N19—C64 | 1.350 (4) | C58—H58B | 0.9800 |
| C35—H35 | 0.9500 | C58—H58C | 0.9800 |
| C35—C34 | 1.419 (4) | C57—H57 | 0.9500 |
| C35—C36 | 1.365 (4) | C66—H66A | 0.9800 |
| C48—C49 | 1.407 (4) | C66—H66B | 0.9800 |
| C48—C47 | 1.413 (4) | C66—H66C | 0.9800 |
| C23—H23 | 0.9500 | C51—H51A | 0.9800 |
| C23—C24 | 1.367 (4) | C51—H51B | 0.9800 |
| C23—C22 | 1.408 (4) | C51—H51C | 0.9800 |
| C27—H27 | 0.9500 | C45—H45A | 0.9800 |
| C27—C26 | 1.365 (4) | C45—H45B | 0.9800 |
| C27—C28 | 1.407 (4) | C45—H45C | 0.9800 |
| C26—H26 | 0.9500 | C65—H65A | 0.9800 |
| C34—C38 | 1.419 (4) | C65—H65B | 0.9800 |
| C34—C33 | 1.418 (4) | C65—H65C | 0.9800 |
| C49—H49 | 0.9500 | C59—H59A | 0.9800 |
| C49—C50 | 1.377 (4) | C59—H59B | 0.9800 |
| C31—H31 | 0.9500 | C59—H59C | 0.9800 |
| N14···H14 | 0.84 | ||
| O5—S1—O4 | 111.94 (13) | C23—C22—H22 | 120.1 |
| O5—S1—C24 | 107.15 (12) | C21—C22—C23 | 119.9 (3) |
| O4—S1—C24 | 104.71 (13) | C21—C22—H22 | 120.1 |
| O3—S1—O5 | 112.26 (14) | C41—C40—H40 | 119.3 |
| O3—S1—O4 | 113.97 (13) | C39—C40—H40 | 119.3 |
| O3—S1—C24 | 106.09 (13) | C39—C40—C41 | 121.3 (3) |
| O7—S2—C33 | 106.42 (13) | N18—C53—C54 | 122.3 (3) |
| O9—S2—O7 | 111.50 (14) | N18—C53—H53 | 118.9 |
| O9—S2—C33 | 106.67 (13) | C54—C53—H53 | 118.9 |
| O8—S2—O7 | 112.15 (17) | N12—C37—C36 | 123.7 (2) |
| O8—S2—O9 | 114.36 (16) | N12—C37—H37 | 118.2 |
| O8—S2—C33 | 105.04 (14) | C36—C37—H37 | 118.2 |
| C30—O10—H10 | 114 (3) | C41—C42—H42 | 119.6 |
| C21—O6—H6 | 107 (3) | C43—C42—H42 | 119.6 |
| C28—N11—C29 | 117.2 (2) | C43—C42—C41 | 120.7 (3) |
| C43—N14—H14 | 115 (2) | N13—C41—C40 | 121.9 (3) |
| C39—N14—C43 | 119.4 (3) | N13—C41—C42 | 122.6 (3) |
| C39—N14—H14 | 125 (2) | C42—C41—C40 | 115.4 (3) |
| C53—N18—C57 | 119.1 (3) | O6—C21—C29 | 120.6 (2) |
| C53—N18—H18 | 121 (3) | O6—C21—C22 | 119.3 (3) |
| C57—N18—H18 | 120 (3) | C22—C21—C29 | 120.2 (3) |
| C37—N12—C38 | 117.4 (2) | C48—C47—H47 | 120.0 |
| C55—N17—C58 | 121.4 (3) | C46—C47—C48 | 120.0 (3) |
| C55—N17—C59 | 120.8 (3) | C46—C47—H47 | 120.0 |
| C59—N17—C58 | 117.6 (3) | N19—C64—H64 | 117.6 |
| C41—N13—C44 | 120.9 (3) | N19—C64—C63 | 124.9 (3) |
| C41—N13—C45 | 121.3 (3) | C63—C64—H64 | 117.6 |
| C45—N13—C44 | 117.7 (3) | N17—C55—C54 | 121.7 (3) |
| C29—C25—C24 | 118.4 (2) | N17—C55—C56 | 122.7 (3) |
| C26—C25—C29 | 116.6 (2) | C56—C55—C54 | 115.6 (3) |
| C26—C25—C24 | 125.0 (2) | N16—C52—H52A | 109.5 |
| C48—N16—C52 | 120.9 (3) | N16—C52—H52B | 109.5 |
| C48—N16—C51 | 121.8 (3) | N16—C52—H52C | 109.5 |
| C52—N16—C51 | 117.1 (3) | H52A—C52—H52B | 109.5 |
| C50—N15—C46 | 115.2 (3) | H52A—C52—H52C | 109.5 |
| O10—C30—C31 | 119.3 (2) | H52B—C52—H52C | 109.5 |
| O10—C30—C38 | 120.1 (2) | C62—C63—H63 | 120.1 |
| C31—C30—C38 | 120.6 (2) | C64—C63—C62 | 119.8 (3) |
| C62—N20—C66 | 120.6 (3) | C64—C63—H63 | 120.1 |
| C62—N20—C65 | 120.8 (3) | N13—C44—H44A | 109.5 |
| C65—N20—C66 | 118.5 (3) | N13—C44—H44B | 109.5 |
| N11—C29—C25 | 123.4 (3) | N13—C44—H44C | 109.5 |
| N11—C29—C21 | 116.8 (2) | H44A—C44—H44B | 109.5 |
| C25—C29—C21 | 119.8 (2) | H44A—C44—H44C | 109.5 |
| C60—N19—C64 | 115.2 (3) | H44B—C44—H44C | 109.5 |
| C34—C35—H35 | 120.5 | N15—C46—C47 | 124.5 (3) |
| C36—C35—H35 | 120.5 | N15—C46—H46 | 117.7 |
| C36—C35—C34 | 118.9 (3) | C47—C46—H46 | 117.7 |
| N16—C48—C49 | 122.2 (3) | N14—C43—C42 | 121.7 (3) |
| N16—C48—C47 | 122.4 (3) | N14—C43—H43 | 119.1 |
| C49—C48—C47 | 115.4 (3) | C42—C43—H43 | 119.1 |
| C24—C23—H23 | 119.2 | N14—C39—H39 | 119.3 |
| C24—C23—C22 | 121.7 (2) | C40—C39—N14 | 121.4 (3) |
| C22—C23—H23 | 119.2 | C40—C39—H39 | 119.3 |
| C25—C24—S1 | 120.6 (2) | C31—C32—H32 | 119.3 |
| C23—C24—S1 | 119.4 (2) | C33—C32—C31 | 121.4 (2) |
| C23—C24—C25 | 120.0 (2) | C33—C32—H32 | 119.3 |
| C26—C27—H27 | 120.4 | C55—C56—H56 | 119.4 |
| C26—C27—C28 | 119.2 (3) | C57—C56—C55 | 121.2 (3) |
| C28—C27—H27 | 120.4 | C57—C56—H56 | 119.4 |
| C25—C26—H26 | 120.2 | N17—C58—H58A | 109.5 |
| C27—C26—C25 | 119.6 (3) | N17—C58—H58B | 109.5 |
| C27—C26—H26 | 120.2 | N17—C58—H58C | 109.5 |
| C35—C34—C38 | 117.1 (2) | H58A—C58—H58B | 109.5 |
| C33—C34—C35 | 124.2 (2) | H58A—C58—H58C | 109.5 |
| C33—C34—C38 | 118.7 (2) | H58B—C58—H58C | 109.5 |
| C48—C49—H49 | 120.0 | N18—C57—H57 | 119.2 |
| C50—C49—C48 | 119.9 (3) | C56—C57—N18 | 121.7 (3) |
| C50—C49—H49 | 120.0 | C56—C57—H57 | 119.2 |
| C30—C31—H31 | 120.2 | N20—C66—H66A | 109.5 |
| C30—C31—C32 | 119.7 (3) | N20—C66—H66B | 109.5 |
| C32—C31—H31 | 120.2 | N20—C66—H66C | 109.5 |
| N12—C38—C30 | 117.2 (2) | H66A—C66—H66B | 109.5 |
| N12—C38—C34 | 123.3 (3) | H66A—C66—H66C | 109.5 |
| C34—C38—C30 | 119.5 (2) | H66B—C66—H66C | 109.5 |
| N11—C28—C27 | 124.0 (3) | N16—C51—H51A | 109.5 |
| N11—C28—H28 | 118.0 | N16—C51—H51B | 109.5 |
| C27—C28—H28 | 118.0 | N16—C51—H51C | 109.5 |
| C34—C33—S2 | 120.5 (2) | H51A—C51—H51B | 109.5 |
| C32—C33—S2 | 119.3 (2) | H51A—C51—H51C | 109.5 |
| C32—C33—C34 | 120.2 (3) | H51B—C51—H51C | 109.5 |
| C62—C61—H61 | 119.9 | N13—C45—H45A | 109.5 |
| C60—C61—H61 | 119.9 | N13—C45—H45B | 109.5 |
| C60—C61—C62 | 120.1 (3) | N13—C45—H45C | 109.5 |
| C53—C54—H54 | 119.9 | H45A—C45—H45B | 109.5 |
| C53—C54—C55 | 120.2 (3) | H45A—C45—H45C | 109.5 |
| C55—C54—H54 | 119.9 | H45B—C45—H45C | 109.5 |
| C35—C36—H36 | 120.2 | N20—C65—H65A | 109.5 |
| C35—C36—C37 | 119.6 (3) | N20—C65—H65B | 109.5 |
| C37—C36—H36 | 120.2 | N20—C65—H65C | 109.5 |
| N20—C62—C61 | 122.2 (3) | H65A—C65—H65B | 109.5 |
| N20—C62—C63 | 122.4 (3) | H65A—C65—H65C | 109.5 |
| C61—C62—C63 | 115.4 (3) | H65B—C65—H65C | 109.5 |
| N19—C60—C61 | 124.6 (3) | N17—C59—H59A | 109.5 |
| N19—C60—H60 | 117.7 | N17—C59—H59B | 109.5 |
| C61—C60—H60 | 117.7 | N17—C59—H59C | 109.5 |
| N15—C50—C49 | 124.9 (3) | H59A—C59—H59B | 109.5 |
| N15—C50—H50 | 117.6 | H59A—C59—H59C | 109.5 |
| C49—C50—H50 | 117.6 | H59B—C59—H59C | 109.5 |
| S2—C33—C32—C31 | −178.5 (2) | C38—C30—C31—C32 | −1.1 (4) |
| O5—S1—C24—C25 | 50.1 (3) | C38—C34—C33—S2 | 178.2 (2) |
| O5—S1—C24—C23 | −131.4 (2) | C38—C34—C33—C32 | −0.5 (4) |
| O10—C30—C31—C32 | 178.2 (3) | C28—N11—C29—C25 | 0.5 (4) |
| O10—C30—C38—N12 | 1.2 (4) | C28—N11—C29—C21 | −179.2 (3) |
| O10—C30—C38—C34 | −178.4 (2) | C28—C27—C26—C25 | 0.5 (4) |
| O4—S1—C24—C25 | 169.1 (2) | C33—C34—C38—N12 | −179.6 (3) |
| O4—S1—C24—C23 | −12.4 (3) | C33—C34—C38—C30 | 0.0 (4) |
| O7—S2—C33—C34 | −57.5 (3) | C61—C62—C63—C64 | −0.7 (5) |
| O7—S2—C33—C32 | 121.2 (3) | C54—C55—C56—C57 | 1.3 (4) |
| N11—C29—C21—O6 | 0.1 (4) | C36—C35—C34—C38 | 0.5 (4) |
| N11—C29—C21—C22 | −179.9 (3) | C36—C35—C34—C33 | 179.4 (3) |
| O3—S1—C24—C25 | −70.0 (3) | C62—C61—C60—N19 | −1.2 (5) |
| O3—S1—C24—C23 | 108.5 (2) | C60—N19—C64—C63 | −0.5 (5) |
| O9—S2—C33—C34 | 61.6 (3) | C60—C61—C62—N20 | −178.7 (3) |
| O9—S2—C33—C32 | −119.6 (3) | C60—C61—C62—C63 | 1.1 (5) |
| N17—C55—C56—C57 | −179.3 (3) | C50—N15—C46—C47 | −0.9 (5) |
| C25—C29—C21—O6 | −179.6 (3) | C22—C23—C24—S1 | −177.6 (2) |
| C25—C29—C21—C22 | 0.4 (4) | C22—C23—C24—C25 | 0.9 (4) |
| N16—C48—C49—C50 | 178.0 (3) | C53—N18—C57—C56 | −0.4 (5) |
| N16—C48—C47—C46 | −178.6 (3) | C53—C54—C55—N17 | −179.8 (3) |
| C30—C31—C32—C33 | 0.6 (5) | C53—C54—C55—C56 | −0.4 (4) |
| N20—C62—C63—C64 | 179.1 (3) | C37—N12—C38—C30 | −179.2 (3) |
| C29—N11—C28—C27 | 0.4 (4) | C37—N12—C38—C34 | 0.5 (4) |
| C29—C25—C24—S1 | 178.2 (2) | C41—C40—C39—N14 | 0.4 (5) |
| C29—C25—C24—C23 | −0.3 (4) | C41—C42—C43—N14 | −0.4 (5) |
| C29—C25—C26—C27 | 0.4 (4) | C47—C48—C49—C50 | −1.9 (4) |
| N19—C64—C63—C62 | 0.4 (5) | C64—N19—C60—C61 | 0.9 (5) |
| C35—C34—C38—N12 | −0.7 (4) | C55—C54—C53—N18 | −0.8 (5) |
| C35—C34—C38—C30 | 179.0 (3) | C55—C56—C57—N18 | −0.9 (5) |
| C35—C34—C33—S2 | −0.7 (4) | C52—N16—C48—C49 | −4.7 (5) |
| C35—C34—C33—C32 | −179.4 (3) | C52—N16—C48—C47 | 175.2 (3) |
| C35—C36—C37—N12 | −0.1 (5) | C44—N13—C41—C40 | −0.5 (5) |
| O8—S2—C33—C34 | −176.6 (2) | C44—N13—C41—C42 | 179.4 (3) |
| O8—S2—C33—C32 | 2.1 (3) | C46—N15—C50—C49 | 0.3 (5) |
| C48—C49—C50—N15 | 1.2 (5) | C43—N14—C39—C40 | 0.8 (5) |
| C48—C47—C46—N15 | 0.1 (5) | C43—C42—C41—N13 | −178.4 (3) |
| C23—C22—C21—O6 | −179.8 (3) | C43—C42—C41—C40 | 1.6 (4) |
| C23—C22—C21—C29 | 0.2 (4) | C39—N14—C43—C42 | −0.8 (5) |
| C24—C25—C29—N11 | 180.0 (3) | C39—C40—C41—N13 | 178.4 (3) |
| C24—C25—C29—C21 | −0.3 (4) | C39—C40—C41—C42 | −1.6 (4) |
| C24—C25—C26—C27 | 179.4 (3) | C58—N17—C55—C54 | −2.7 (4) |
| C24—C23—C22—C21 | −0.8 (4) | C58—N17—C55—C56 | 177.9 (3) |
| C26—C25—C29—N11 | −0.9 (4) | C57—N18—C53—C54 | 1.2 (5) |
| C26—C25—C29—C21 | 178.7 (3) | C66—N20—C62—C61 | 174.1 (3) |
| C26—C25—C24—S1 | −0.8 (4) | C66—N20—C62—C63 | −5.7 (5) |
| C26—C25—C24—C23 | −179.3 (3) | C51—N16—C48—C49 | 179.8 (3) |
| C26—C27—C28—N11 | −0.9 (5) | C51—N16—C48—C47 | −0.3 (5) |
| C34—C35—C36—C37 | −0.1 (4) | C45—N13—C41—C40 | −176.3 (3) |
| C34—C33—C32—C31 | 0.2 (5) | C45—N13—C41—C42 | 3.7 (5) |
| C49—C48—C47—C46 | 1.3 (4) | C65—N20—C62—C61 | −3.0 (5) |
| C31—C30—C38—N12 | −179.6 (3) | C65—N20—C62—C63 | 177.2 (3) |
| C31—C30—C38—C34 | 0.8 (4) | C59—N17—C55—C54 | −177.4 (3) |
| C38—N12—C37—C36 | −0.1 (4) | C59—N17—C55—C56 | 3.2 (5) |
4-(Dimethylamino)pyridin-1-ium 8-hydroxyquinoline-5-sulfonate; N,N-dimethylpyridin-4-amine . Hydrogen-bond geometry (Å, º)
Cg1, Cg2 are the centroids of the N11/C25–C29 and N12/C34–C38 rings, respectively
| D—H···A | D—H | H···A | D···A | D—H···A |
| O6—H6···O8i | 0.84 (5) | 2.00 (5) | 2.679 (3) | 137 (4) |
| O6—H6···N11 | 0.84 (5) | 2.24 (5) | 2.728 (3) | 117 (4) |
| O10—H10···O4 | 0.89 (5) | 1.90 (5) | 2.674 (3) | 144 (4) |
| O10—H10···N12 | 0.89 (5) | 2.31 (5) | 2.730 (3) | 109 (4) |
| N14—H14···N15 | 0.90 (3) | 1.91 (3) | 2.814 (4) | 174 (3) |
| N18—H18···N19 | 0.90 (3) | 1.92 (4) | 2.816 (4) | 177 (7) |
| C27—H27···O6ii | 0.95 | 2.58 | 3.219 (4) | 125 |
| C27—H27···O8iii | 0.95 | 2.27 | 3.194 (4) | 164 |
| C36—H36···O4iv | 0.95 | 2.33 | 3.244 (4) | 160 |
| C36—H36···O10iv | 0.95 | 2.58 | 3.216 (4) | 125 |
| C39—H39···O3v | 0.95 | 2.32 | 3.200 (4) | 154 |
| C43—H43···O5 | 0.95 | 2.22 | 3.160 (4) | 169 |
| C46—H46···O5 | 0.95 | 2.46 | 3.373 (4) | 161 |
| C50—H50···O3v | 0.95 | 2.43 | 3.292 (4) | 151 |
| C53—H53···O9vi | 0.95 | 2.22 | 3.146 (4) | 164 |
| C54—H54···O10vii | 0.95 | 2.55 | 3.450 (4) | 158 |
| C57—H57···O7 | 0.95 | 2.22 | 3.143 (4) | 165 |
| C60—H60···O7 | 0.95 | 2.40 | 3.325 (4) | 163 |
| C64—H64···O9vi | 0.95 | 2.35 | 3.267 (4) | 162 |
| C40—H40···Cg1v | 0.95 | 2.63 | 3.498 (3) | 153 |
| C49—H49···Cg2viii | 0.95 | 2.87 | 3.754 (3) | 156 |
| C61—H61···Cg2 | 0.95 | 2.70 | 3.571 (3) | 152 |
Symmetry codes: (i) x, y−1, z; (ii) x−1, y, z; (iii) x−1, y−1, z; (iv) x+1, y, z; (v) x, −y+1, z+1/2; (vi) x, −y+2, z+1/2; (vii) x+1, −y+2, z+1/2; (viii) x−1, −y+1, z+1/2.
Funding Statement
This work was funded by Japan Society for the Promotion of Science grant JP23 KJ1830 to M. Isobe.
References
- Aakeröy, C. B., Desper, J. & Levin, B. (2005). CrystEngComm, 7, 102–107.
- Baskar Raj, S., Muthiah, P. T., Bocelli, G. & Cantoni, A. (2003). Acta Cryst. E59, o1980–o1983.
- Biradha, H., Edwards, R. E., Foulds, G. J., Robinson, W. T. & Desiraju, G. R. (1995). Chem. Commun. pp. 1705–1707.
- Bolla, G., Sarma, B. & Nangia, A. K. (2022). Chem. Rev.122, 11514–11603. [DOI] [PubMed]
- Braga, D., Giaffreda, S. L., Grepioni, F., Palladino, G. & Polito, M. (2008). New J. Chem.32, 820–828.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst.42, 339–341.
- Döring, C. & Jones, P. G. (2016). Z. Anorg. Allge Chem.642, 930–936.
- Fábry, J. (2017). Acta Cryst. E73, 1344–1347. [DOI] [PMC free article] [PubMed]
- Ganie, A. A., Ismail, T. M., Sajith, P. K. & Dar, A. A. (2021). New J. Chem.45, 4780–4790.
- Glidewell, C. & Holden, H. D. (1982). Acta Cryst. B38, 667–669.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hemamalini, M., Muthiah, P. T., Bocelli, G. & Cantoni, A. (2004). Acta Cryst. C60, o284–o286. [DOI] [PubMed]
- Kashiwagi, Y., Kubono, K. & Tamai, T. (2020). Acta Cryst. E76, 1271–1274. [DOI] [PMC free article] [PubMed]
- Kobayashi, N., Naito, T. & Inabe, T. (2003). Bull. Chem. Soc. Jpn, 76, 1351–1362.
- Kubono, K., Tanaka, R., Kashiwagi, Y., Tani, K. & Yokoi, K. (2023). Acta Cryst. E79, 726–729. [DOI] [PMC free article] [PubMed]
- Lacková, D., Ondrejkovičová, I., Padělková, Z. & Koman, M. (2014). J. Coord. Chem.67, 1652–1663.
- Mautner, F. A. & Goher, M. A. S. (1998). Polyhedron, 18, 553–559.
- Rigaku OD (2023). CrysAlis PRO. Rigaku Oxford Diffraction Ltd, Yarnton, England.
- Santra, R., Ghosh, N. & Biradha, K. (2008). New J. Chem.32, 1673–1676.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Smith, G. (2012). Acta Cryst. E68, o3349. [DOI] [PMC free article] [PubMed]
- Smith, G., Wermuth, U. D. & Healy, P. C. (2004). Acta Cryst. C60, o600–o603. [DOI] [PubMed]
- Sun, L., Wang, Y., Yang, F., Zhang, X. & Hu, W. (2019). Adv. Mater.31, 1902328. [DOI] [PubMed]
- Vladiskovic, C., Mantegazza, S., Razzetti, G. & Masciocchi, N. (2023). Cryst. Growth Des.23, 1119–1126.
- Wagler, J. & Kronstein, M. (2014). CSD Communication.
- Wiberley, S. E. & Bassett, L. G. (1949). Anal. Chem.21, 609–612.
- Zhang, K., Shen, Y., Liu, J., Spingler, B. & Duttwyler, S. (2018). Chem. Commun.54, 1698–1701. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698902400642X/ox2006sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902400642X/ox2006Isup3.hkl
Supporting information file. DOI: 10.1107/S205698902400642X/ox2006Isup3.cml
CCDC reference: 2366836
Additional supporting information: crystallographic information; 3D view; checkCIF report