Abstract
Background
Cupriavidus gilardii is an aerobic, gram-negative, motile, glucose-nonfermenting bacillus, first described in 1999. Typically, it exhibits low pathogenicity in humans, causing opportunistic infections primarily in individuals with compromised immune systems. This bacterium has been also found in various environmental sources such as plants and contaminated soils. Notably, there have been no documented cases of C. gilardii infections in animals.
Case presentation
This case report outlines a bovine neonatal diarrhea outbreak that occurred in Northern Greece, during which C. gilardii was isolated. Faecal samples from 5-day-old calves were collected and transported to the laboratory for further examination. Bacterial culture and next generation sequencing techniques were employed to confirm the presence of this bacterium in the samples. Following the isolation and identification of C. gilardii from the samples, an autogenous vaccine was produced and administered to the cows within the farm. Subsequent to vaccination, a progressive reduction in calf diarrhea and deaths was observed, leading to their eventual complete resolution. To the best of our knowledge, this represents the first documentation of C. gilardii isolation from cases of bovine neonatal diarrhea.
Conclusion
This case report presents the first isolation case of C. gilardii from animal samples and more specifically from calf faecal samples. It represents an important observation, providing evidence that this opportunistic human pathogen could contribute to clinical symptoms in animals.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-024-04197-3.
Keywords: Calf diarrhea, Cupriavidus gilardii, Public health
Background
Cupriavidus gilardii is an aerobic, gram-negative, motile bacillus which is characterized by its inability to ferment glucose. Originally identified in 1999 by Coenye et al. [1], this microorganism has been frequently found in diverse environmental sources, including plants and soils contaminated with heavy metals. Previous reports have characterized it as a microorganism with low pathogenicity that primarily causes opportunistic infections, mainly in immunocompromised individuals [2–4]. Reported cases involve patients with conditions such as acute lymphocytic leukemia [2], severe idiopathic aplastic anemia [3], or those who have undergone renal transplantation [4]. Kobayashi et al. [5] described the first case of an infection of C. gilardii in a patient without obvious immunodeficiency. Human clinical samples, such as those from cerebrospinal fluid, bone marrow, wounds, and the respiratory tract, have sporadically revealed the presence of C. gilardii [1, 6, 7]. To date, only six documented cases of clinical infections directly attributed to C. gilardii have been reported [8]. In one of these cases involving a 26-year-old woman undergoing chemotherapy, the patient, among other clinical signs, experienced abdominal pain and diarrhea. An important aspect to note is the intrinsic resistance of C. gilardii to various antimicrobial agents such as aztreonam, meropenem, gentamicin, and tobramycin, as documented in previous reports [3, 9]. So far, C. gilardii has not been associated with any animal diseases, nor has it been detected in clinical samples collected from animals.
Case presentation
In 2022, a severe neonatal diarrhea outbreak was reported at a Limousin-breed beef cattle farm situated in the Central Macedonia Region, near the city of Aridaia, Greece. The herd consisted of 65 animals. The syndrome exhibited clinical signs primarily in 5-day-old calves, characterized by severe diarrhea leading to dehydration, anorexia, and depression. Several of the affected calves succumbed to the illness within an average period of 5 days. Despite treatment attempts using antibiotic therapy (amoxicillin, colistin), the affected animals showed no signs of clinical improvement. During the outbreak the mortality rate was as high as 70% causing substantial economic losses to the farm owner.
Two fresh fecal samples were directly collected from the rectum of diarrheic calves to prevent environmental contamination. These samples were promptly transported under cooling to the laboratory of Microbiology and Infectious Diseases, School of Veterinary Medicine, Aristotle University of Thessaloniki, Greece. Upon arrival at the lab, the samples were diluted in decimal dilutions in Phosphate Buffer Saline (Thermo Fischer Scientific, Massachusetts, USA) and underwent immediate culture onto blood agar (Oxoid, Hampshire, UK) and MacConkey agar plates (Oxoid, Hampshire, UK), which subsequently were incubated at 37 οC overnight. For the isolation of anaerobic bacteria, the same procedure was conducted, and the agar plates were incubated under anaerobic conditions at 37 οC for 48 h.
After the incubation, pale colonies (lactose-nonfermenting) of gram-negative bacilli were grown on MacConkey agar plates while small, white colonies were present on Blood agar plates (Fig. 1). Bacteria had been detected at high dilutions in both samples. More specifically, 1.9 × 109 cfu/ml and 7.8 × 108 cfu/ml of the isolated bacilli were detected in the 2 examined samples, respectively.
Fig. 1.
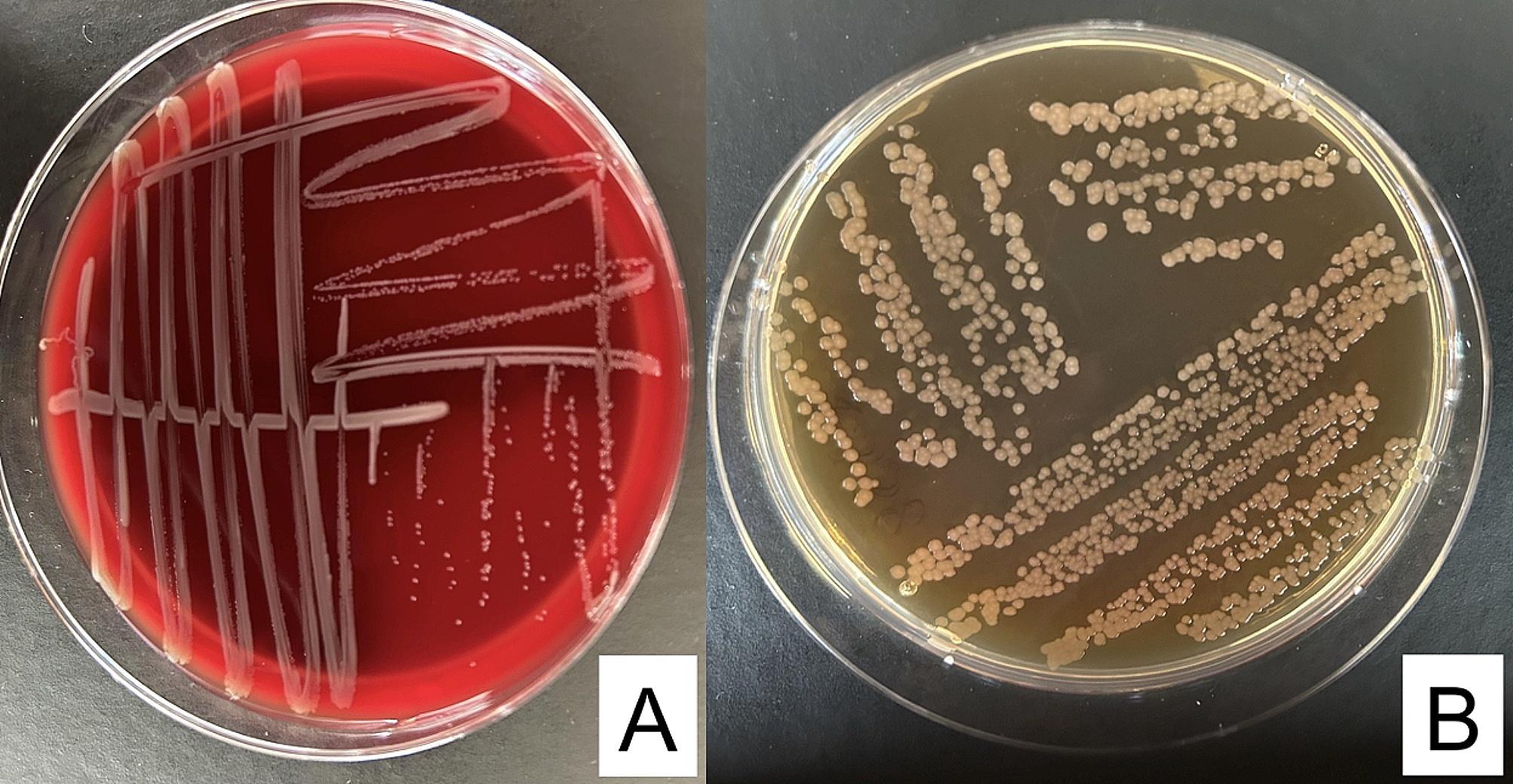
Colonies of C. gilardii on Blood Agar (A) and Mc Conkey Agar (B)
DNA extraction from a single isolated colony at the highest dilution, was performed for both samples using the Nucleospin Microbial DNA kit (Macherey-Nagel, Duren, Germany) following the manufacturer’s protocol. The concentration and purity of the eluted DNA were determined in an Eppendorf Biospectrophotometer (Eppendorf, Wien, Austria). Subsequently, the extracted DNA underwent sequencing using Oxford Nanopore Technology (ONT) sequencing employing the Rapid Barcoding Kit SQK-RBK004 (Oxford Nanopore Technologies, Oxford, UK) for library preparation. Prepared libraries were loaded into a R9.4.1D Flow Cell (Oxford Nanopore Technologies, Oxford, UK) and sequenced using the MinION Mk1C device (Oxford Nanopore Technologies, Oxford, UK). Raw data in FAST5 format were basecalled using Guppy v6.5.7 (Oxford Nanopore Technologies, Oxford, UK) and subsequently demultiplexed into individual sample barcodes. Bioinformatic metagenomic analysis was carried out using the EPI2ME Desktop Agent v3.7.3 (Oxford Nanopore Technologies, Oxford, UK). According to the results of the read analysis, the DNA of C. gilardii was the main taxon in both samples.
Antimicrobial susceptibility testing of the isolated bacteria in both samples was conducted following the guidelines set by the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Given the absence of established veterinary breakpoints for C. gilardii, this study employed clinical breakpoints designated for Pseudomonas aeruginosa, a practice previously used by other researchers [5, 8], and/or Burkholderia pseudomallei, a species within the same family as C. gillardi (Burkholderiaceae). The susceptibility to specific antimicrobial agents was determined by disk diffusion on Mueller-Hinton agar (Oxoid, Hampshire, UK). The susceptibility of the isolated bacteria to various antimicrobial agents is shown in Table 1.
Table 1.
Antimicrobial agent susceptibilities of C. Gilardii
| Antimicrobial agent | Zone diameter (mm) | EUCAST P. aeruginosa |
EUCAST P. aeruginosa |
Result | EUCAST B. pseudomallei |
EUCAST B. pseudomallei |
Result |
|---|---|---|---|---|---|---|---|
| S ≥ | R < | S ≥ | R < | ||||
| AMC | 38 | - | - | 50 | 22 | I | |
| PRL | 38 | 50 | 18 | I | - | - | |
| CAZ | 16 | 50 | 17 | R | 50 | 18 | R |
| FEP | 33 | 50 | 21 | I | - | - | |
| IPM | 40 | 50 | 20 | I | 29 | 29 | S |
| CIP | 36 | 50 | 23 | I | - | - | |
| TE | 25 | - | - | 23 | 23 | S | |
| S | 0 | - | - | - | - | ||
| CN | 0 | - | - | - | - |
* R = Resistant, I = Intermediate, S = Sensitive AMC Amoxicillin-Clavulanic Acid (30 µg), PRL Piperacillin (100 µg), CAZ Ceftazidime (10 µg), FEP Cefepime (30 µg), IPM Imipenem (10 µg), CIP Ciprofloxacin (5 µg), TEΤ Tetracycline (30 µg), S Streptomycin (10 µg), CN Gentamicin (10 µg)
Following successful isolation and identification of C. gilardii from the samples, an autogenous inactivated vaccine was produced and administrated once intramuscularly to the cows, approximately 1 month prior το the expected parturition (Clinical Trial Number 112/2004/18.12.2023 Faculty of Agricultural Sciences, University of Western Macedonia, Florina, Greece). Post-vaccination, a notable decline in incidence of neonatal calf diarrhea was consistently noted, with complete resolution of the symptoms and cessation of deaths over time.
Discussion
Neonatal calf diarrhea is a significant health challenge in both dairy and beef cattle herds worldwide, resulting in economic losses due to reduced calf growth and survival and increased cost of treatment [10]. Calves up to 30 days old is the primary age group.
susceptible to this condition [11]. The disease’s etiology is complex and multifactorial including various infectious agents and predisposing factors (e.g. age, genetics, environment, management, nutrition, immune status, and concurrent illnesses) [11–14]. Among infectious agents, Enteropathogenic Escherichia coli (ETEC) and Salmonella spp. are the predominant bacterial pathogens, Cryptosporidium spp. and Eimeria spp. are the most common isolated protozoa, while bovine rotavirus (BRV), bovine coronavirus (BCoV), and bovine viral diarrhea virus are the most frequent viruses detected in outbreaks of the disease [15–18].
In the specific case of Aridaia beef cattle farm, and despite multiple treatment attempts using antibiotics, neonatal calf diarrhea did not respond. The identification of a specific colony type prevailing in higher dilutions within cultures had revealed the possible involvement of those specific bacteria on the pathogenesis of this situation. The use of novel technologies e.g. next generation sequencing, proved at present to be a very useful tool in verifying a novel potential pathogen e.g. C. gilardii and further directing the next diagnostic and control steps.
The results presented in Table 1, indicate that the isolated bacteria were resistant to ceftazidime, according to the breakpoints of both P. aeruginosa and B. pseudomallei. Moreover, while there are no specific breakpoints defined for streptomycin and gentamicin, the absence of zone diameter in the disk diffusion test for these antibacterial agents (0 mm) signifies resistance against both compounds. Regarding the susceptibility of the isolates to the other tested antimicrobial agents, they demonstrated intermediate susceptibility to piperacillin, cefepime, and ciprofloxacin in line with the breakpoints defined for P. aeruginosa, and also to amoxicillin-clavulanic acid, according to the breakpoints for B. pseudomallei. In addition, both isolates exhibited susceptibility to tetracycline, as per the defined breakpoints for B. pseudomallei. It is noteworthy that although the isolated strain of C. gilardii exhibits susceptibility to imipenem, as per the breakpoints designated for B. pseudomallei, it demonstrates intermediate susceptibility based on the breakpoints specified for P. aeruginosa. To date, investigations on the susceptibility of C. gilardii to different antimicrobial agents in animal populations are missing. However, analogous studies in human contexts provide valuable insights. For instance, Kodayashi et al. [5] demonstrated resistance of the isolated C. gilardii strain to ceftazidime and gentamicin, a finding consistent with our research outcome. Similarly, Kweon et al. [19] reported gentamicin resistance in a C. gilardii strain. On the other hand, the strain was susceptible to ciprofloxacin and cefepime, a finding that contrasts with the results for the strain of the present study. Furthermore, Kweon et al. highlighted C. gilardii’s susceptibility to imipenem and intermediate susceptibility to piperacillin, aligning with our study’s results. In another study, Zhang et al. [8] showed that their isolated strain of C. gilardii was susceptible to ciprofloxacin, piperacillin, and ceftazidime, while our strain displayed intermediate susceptibility (susceptibility-increased exposure) to the first two agents and resistance to the latter. The differences observed in the antimicrobial susceptibility profiles underscore the pressing need to establish specific breakpoints for C. gilardii, both human and veterinary, particularly considering its emerging role as a causative agent in bovine neonatal diarrhea. Such breakpoints would facilitate precise and effective antimicrobial therapy for this bacterium in human and veterinary medicine.
Some important observations emerged during discussions with the farm owner were the following; Firstly, it was noted that the farm owner’s parents, who had frequent contact with the animals, displayed symptoms such as weakness, malaise, and sporadic mild diarrhea. Upon seeking medical attention at the local hospital, physicians attributed their health issue to a bacterial infection; however, the precise etiological agent remained undetermined. Secondly, neighbouring cattle farms in the area were also experiencing similar challenges characterized by calf diarrhea and deaths. These additional observations raise the suspicion that the forementioned specific situations might be linked to the presence of C. gilardii in the farm and the area, reinforcing its substantial involvement in the recorded outbreak of neonatal calf diarrhea.
The fact that after the application of the autogenous vaccine on the cows of the farm, there was a noticeable improvement in the condition, ultimately leading to the cessation of deaths in the calves, confirms the hypothesis that the primary causative agent for this disease may be C. gilardii. This assumption is further supported by the identification of this particular bacterium using a high throughput technique such as ONT sequencing, which eventually supports “personalised” diagnosis and treatment/ control as well as the identification of means for the reduction of antibiotic resistance.
Conclusion
C. gilardii is an opportunistic pathogen that mainly affects the immunocompromised individuals. The present study describes the first case of C. gilardii isolation from a bovine neonatal diarrhea outbreak. The isolation and identification of this specific bacterium was conducted by using a combination of classical microbiology methods and high-throughput sequencing technology. Conducting antimicrobial susceptibility tests on the isolated strain revealed resistance or intermediate susceptibility to various antimicrobial agents. Subsequently, and keeping in mind to reduce antibiotic resistance development, an autogenous vaccine was developed and administered to the cows, leading to a marked enhancement in the farm’s condition regarding the disease in calves. Given the status of C. gilardii as an opportunistic pathogen in humans and its potential implications for public health, the urgency for further investigation of its significance in animal health becomes important. As this being the first reported case of C. gilardii in its involvement in animal disease, the role of novel techniques in directing diagnosis and treatment by specialists in the future for defending unknown or potential pathogens is highlightened.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- C. gilardii
Cupriavidus gilardii
- P. aeruginosa
Pseudomonas aeruginosa
- B. pseudomallei
Burkholderia pseudomallei
Author contributions
K.P. performed the ONT sequencing and wrote the manuscript; A.S. performed the bacteriological investigations and the antibiotic sensitivity test; G.D. contributed to the antibiotic sensitivity test design; I.G. and E.P. produced the autogenous inactivated vaccine; E.S., M.S. and D.P. contributed to the collection and the bacteriological examination of the samples; E.P., S.K.K., N.P. and C.B. designed the experiment; P.P. contributed to the collection of the samples and administered the vaccine to the animals. All authors read and approved the final manuscript.
Funding
This research received no external funding.
Data availability
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The present study had been approved by decision 112/2004/18.12.2023 Faculty of Agricultural Sciences, University of Western Macedonia, Florina, Greece. Informed consent was obtained from the owner of the animals to collect the samples.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coenye T, Falsen E, Vancanneyt M, Hoste B, Govan JR, Kersters K, et al. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int J Syst Bacteriol. 1999;49:405–13. 10.1099/00207713-49-2-405. 10.1099/00207713-49-2-405 [DOI] [PubMed] [Google Scholar]
- 2.Wauters G, Claeys G, Verschraegen G, De Baere T, Vandecruys E, Van Simaey L, et al. Case of catheter sepsis with Ralstonia gilardii in a child with acute lymphoblastic leukemia. J Clin Microbiol. 2001;39(12):4583–4. 10.1128/JCM.39.12.4583-4584.2001. 10.1128/JCM.39.12.4583-4584.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karafin M, Romagnoli M, Fink DL, Howard T, Rau R, Milstone AM, Carroll KC. Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. J Clin Microbiol. 2010;48(3):1005–7. 10.1128/jcm.01482-09. 10.1128/jcm.01482-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tena D, Losa C, Medina MJ, Sáez-Nieto JA. Muscular abscess caused by Cupriavidus gilardii in a renal transplant recipient. Diagn Microbiol Infect Dis. 2014;79(1):108–10. 10.1016/j.diagmicrobio.2014.01.023. 10.1016/j.diagmicrobio.2014.01.023 [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T, Nakamura I, Fujita H, Tsukimori A, Sato A, Fukushima S, et al. First case report of infection due to Cupriavidus gilardii in a patient without immunodeficiency: a case report. BMC Infect Dis. 2016;16:493. 10.1186/s12879-016-1838-y. 10.1186/s12879-016-1838-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coenye T, Vandamme P, LiPuma JJ. Infection by Ralstonia species in cystic fibrosis patients: identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg Infect Dis. 2002;8(7):692–6. 10.3201/eid0807.010472. 10.3201/eid0807.010472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandamme P, Goris J, Coenye T, Hoste B, Janssens D, Kersters K, De Vos P, Falsen E. Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int J Syst Bacteriol. 1999;49(Pt 2):663–9. 10.1099/00207713-49-2-663. 10.1099/00207713-49-2-663 [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Deng W, Wang S, Xu L, Yan L, Liao P. First case report of infection caused by Cupriavidus gilardii in a non-immunocompromised Chinese patient.IDCases. 2017;10:127–9. 10.1016/j.idcr.2017.10.009. [DOI] [PMC free article] [PubMed]
- 9.Wauters G, Claeys G, Verschraegen G, De Baere T, Vandecruys E, Van Simaey L, De Ganck C, Vaneechoutte M. Case of catheter sepsis with Ralstonia gilardii in a child with acute lymphoblastic leukemia. J Clin Microbiol. 2001;39(12):4583–4. 10.1128/jcm.39.12.4583-4584.2001. 10.1128/jcm.39.12.4583-4584.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.USDA. (2018) Health and management practices on U.S. dairy operations, 2014. Accessed 06 Dec 2023.
- 11.Gomez DE, Weese JS. Viral enteritis in calves. Can Vet J. 2017;58(12):1267–74. [PMC free article] [PubMed] [Google Scholar]
- 12.Cho YI, Yoon KJ. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J Vet Sci. 2014;15(1):1–17. 10.4142/jvs.2014.15.1.1. 10.4142/jvs.2014.15.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuirk SM. Disease management of dairy calves and heifers. Vet. Clin. North Am. Food Anim Pract. 2008;24(1):139–53. 10.1016/j.cvfa.2007.10.003. 10.1016/j.cvfa.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Windeyer MC, Leslie KE, Godden SM, Hodgins DC, Lissemore KD, LeBlanc SJ. Factors associated with morbidity, mortal- ity, and growth of dairy heifer calves up to 3 months of age. Prev Vet Med. 2014;113(2):231–40. 10.1016/j.prevetmed.2013.10.019. 10.1016/j.prevetmed.2013.10.019 [DOI] [PubMed] [Google Scholar]
- 15.Bartels CJ, Holzhauer M, Jorritsma R, Swart WA, Lam TJ. Prevalence, prediction and risk factors of enteropathogens in normal and non-normal faeces of young Dutch dairy calves. Prev Vet Med. 2010;93:162–9. 10.1016/j.prevetmed.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izzo MM, Kirkland PD, Mohler VL, Perkins NR, Gunn AA, House JK. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust Vet J. 2011;89(5):167–73. 10.1111/j.1751-0813.2011.00692.x. 10.1111/j.1751-0813.2011.00692.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson S, Hamilton CA, Hope JC, Katzer F, Mabbott NA, Morrison LJ, Innes EA. Bovine cryptosporidiosis: impact, host- parasite interaction and control strategies. Vet Res. 2017;48(1):1–16. 10.1186/s13567-017-0447-0. 10.1186/s13567-017-0447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daugschies A, Najdrowski M. Eimeriosis in cattle: current understanding. J Vet Med. 2005;52(10):417–27. 10.1111/j.1439-0450.2005.00894.x. 10.1111/j.1439-0450.2005.00894.x [DOI] [PubMed] [Google Scholar]
- 19.Kweon OJ, Lim YK, Kim HR, Kim TH, Ha S, Lee MK. Isolation of a novel species in the genus Cupriavidus from a patient with sepsis using whole genome sequencing. 2020;15(5):e0232850. 10.1371/journal.pone.0232850. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.


