Abstract
Epstein-Barr virus (EBV) latent membrane protein 1 (LMP1) is essential for EBV-mediated transformation of primary B lymphocytes. LMP1 spontaneously aggregates in the plasma membrane and enables two transformation effector sites (TES1 and TES2) within the 200-amino-acid cytoplasmic carboxyl terminus to constitutively engage the tumor necrosis factor receptor (TNFR)-associated factors TRAF1, TRAF2, TRAF3, and TRAF5 and the TNFR-associated death domain proteins TRADD and RIP, thereby activating NF-κB and c-Jun N-terminal kinase (JNK). To investigate the importance of the 60% of the LMP1 carboxyl terminus that lies between the TES1-TRAF and TES2-TRADD and -RIP binding sites, an EBV recombinant was made that contains a specific deletion of LMP1 codons 232 to 351. Surprisingly, the deletion mutant was similar to wild-type (wt) LMP1 EBV recombinants in its efficiency in transforming primary B lymphocytes into lymphoblastoid cell lines (LCLs). Mutant and wt EBV-transformed LCLs were similarly efficient in long-term outgrowth and in regrowth after endpoint dilution. Mutant and wt LMP1 proteins were also similar in their constitutive association with TRAF1, TRAF2, TRAF3, TRADD, and RIP. Mutant and wt EBV-transformed LCLs were similar in steady-state levels of Bcl2, JNK, and activated JNK proteins. The wt phenotype of recombinants with LMP1 codons 232 to 351 deleted further demarcates TES1 and TES2, underscores their central importance in B-lymphocyte growth transformation, and provides a new perspective on LMP1 sequence variation between TES1 and TES2.
Epstein-Barr virus (EBV) infection of resting primary human B lymphocytes usually does not result in lytic virus infection. Instead, EBV DNA episomes persist in the cell nucleus and express six nuclear proteins (EBNAs) and three integral plasma membrane proteins (latent membrane proteins [LMPs]) (reviewed in reference 31). These proteins mediate the persistence of EBV DNA and the efficient transformation of the infected cells into indefinitely proliferating lymphoblastoid cell lines (LCLs) (reviewed in reference 30). Recombinant EBV-based genetic analyses implicate five EBNAs and LMP1 as the critical proteins for lymphocyte proliferation (5, 14, 26, 35, 39, 58, 71). LMP1 induces many of the phenotypic changes characteristic of EBV-transformed LCLs, and LMP1 has transforming effects on immortalized rodent fibroblasts (1, 52, 74–76). Furthermore, LMP1 is expressed in vivo in EBV-associated lymphoproliferative disease in immunocompromised patients, in nasopharyngeal carcinoma, and in Hodgkin’s disease (57).
LMP1 is a constitutively activated receptor that engages cytoplasmic signal-transducing proteins characteristic of tumor necrosis factor receptors (TNFRs). LMP1 is composed of a 24-amino-acid (aa) cytoplasmic amino terminus, six hydrophobic membrane-spanning segments separated by short turns that function collectively, and a 200-aa cytoplasmic carboxyl terminus (see Fig. 1) (30). The six hydrophobic membrane-spanning segments enable LMP1 molecules to aggregate in the cell plasma membrane independently of an exogenous ligand (23, 26, 37, 75, 76), while the cytoplasmic carboxyl terminus has two sites that constitutively associate with TNFR signal-transducing proteins (24, 53). Site 1 is within the membrane-proximal 45 aa of the cytoplasmic carboxyl terminus and engages TRAF1, TRAF3, TRAF5, and TRAF2 (2, 6, 8, 53, 61). Site 2 is within the distal 35 aa of the carboxyl terminus and engages the TNFR-associated death domain proteins TRADD and RIP (10, 21, 24). In contrast to TNFRs that recruit TRAFs or TRADD and RIP after binding to ligand and receptor aggregation, LMP1 continuously associates with these proteins through these two sites (4, 6, 8, 21, 24, 53, 66, 73). Signaling through site 1 induces NF-κB activation and upregulates expression of TRAF1 and EBI3 in lymphocytes and of the epidermal growth factor receptor in epithelial cells, whereas signaling through site 2 induces NF-κB and c-Jun N-terminal kinase activation, but cannot up-regulate TRAF1, EBI3, or epidermal growth factor receptor expression (7, 8, 10, 16, 20, 32, 44, 46, 48).
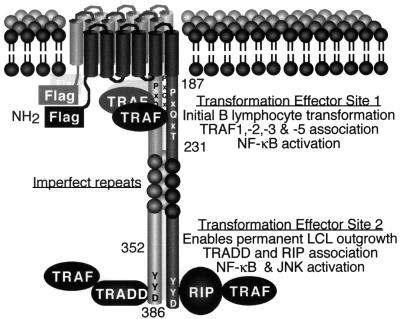
FIG. 1.
Diagram of LMP1. The Flag epitope was introduced at the amino terminus (NH2). The positions of residues 187, 231, 352, and 386 are marked. LMP1 aggregates in the plasma membrane and constitutively associates with TRAFs, TRADD, and RIP. TES1 is located between residues 187 to 231 and aggregates TRAF1, -2, -3, and -5 to mediate NF-κB activation and initial B-lymphocyte growth transformation. TES2 is located between residues 352 and 386; aggregates RIP or TRADD, which associate with TRAFs to mediate high-level NF-κB and c-Jun N-terminal kinase (JNK) activation; and enables permanent LCL outgrowth.
Previously reported recombinant EBV reverse genetic analyses using primary B-lymphocyte transformation assays indicated that the LMP1 transmembrane segments and sites 1 and 2 in the carboxyl terminus are key components for primary B-lymphocyte growth transformation (22, 24, 26, 28). Deletions of specific amino acid sequences from the cytoplasmic amino terminus have little or no effect on transformation by the recombinant EBV as long as arginines and prolines are expressed to tether the first membrane-spanning segment to the cytoplasm (22). Consistent with a stringent requirement for six properly anchored transmembrane segments to achieve aggregation of LMP1 molecules in the plasma membrane, LMP1 lacking the amino terminus and first transmembrane segments accumulates in the plasma membrane, but does not aggregate or support primary B-lymphocyte growth transformation (26). Site 1 is necessary and sufficient for the initiation of lymphocyte transformation (28). The EBV recombinant MS231, which expresses an LMP1 that is carboxy-terminally truncated after site 1, can growth transform B lymphocytes when the transformed cells are cocultivated with fibroblasts, whereas the EBV recombinant MS187, which expresses an LMP1 that is carboxy-terminally truncated before site 1, cannot growth transform B lymphocytes. Furthermore, EBV recombinants with deletion of DNA that encodes the TRAF binding site cannot transform B lymphocytes (22). The importance of site 2 is based on the phenotype of a recombinant with a double point mutation of LMP1 Y384Y385 to isoleucine. This mutation abrogates TRADD binding and cripples transformation, as measured by LCL outgrowth without fibroblast cocultivation (24). Because of genetic and biochemical evidence that sites 1 and 2 are critical for effecting lymphocyte transformation, we use the designation transformation effector sites 1 and 2 (TES1 and -2, respectively).
The experiments reported here are designed to investigate the importance of the 60% of the LMP1 cytoplasmic carboxyl terminus between TES1 and TES2 (aa 232 to 351 [Fig. 1]). These residues are potentially important in signaling and in positioning TES1 or TES2 for interaction with cell proteins. Included in this part of the carboxy terminus are four direct, imperfect repeats of a conserved PQDPDNTDDNG sequence (aa 253 to 301); a PPQLT sequence (aa 320 to 324) that resembles a PxQxT/S TRAF binding core, but does not function as one; a protease cleavage site that has a role in LMP1 catabolism; 19 potential serine or threonine phosphorylation sites, including the major phosphorylated amino acids S313 and T324; and sequences that vary in human isolates and have been reported to affect the ability of LMP1 to transform immortalized rodent fibroblasts (6, 8, 12, 19, 36, 38, 40, 50, 51). To evaluate the importance of aa 232 to 351, an EBV recombinant with these codons deleted has been generated by second site homologous recombination with EBV P3HR-1, and the recombinant phenotype has been characterized in primary B-lymphocyte growth transformation assays (70).
MATERIALS AND METHODS
Cells, virus, growth of infected cells, and assays of effects of cell density on regrowth.
P3HR-1 (42), IB4 (43), BJAB (41), LCLs, and 293 cells were grown as described elsewhere (6, 23). To test the effects of cell density on regrowth, LCLs were serially diluted and seeded into 96-well plates at 30,000 to 1,250 cells/well. Fresh medium was added to cultures after 1 week, and LCL regrowth was monitored for 3 weeks.
LMP1 DNA clones.
EcoRI A and pSVNaeZ DNAs are described elsewhere (67, 70, 71). Plasmid DNA Flag-LMP1 was made by replacing codons 2 to 4 of synthetic wild-type (S-wt) DNA (23) with codons for the Flag antibody epitope (Sigma) between the unique ClaI and XbaI sites, placing a NotI site at a HindIII site at nucleotide (nt) 166480 and a PacI site at a BglII site (nt 169037). Plasmid DNA Flag-LMP1Δ232–351 joins the NaeI site (nt 168627) with a Klenow-filled NcoI site (nt 168758). Expression vectors pSG5 Flag-LMP1 and pSG5 Flag-LMP1Δ232–351 are 2.4- and 2.0-kb MluI DNA fragments from plasmid Flag-LMP1 or plasmid Flag-LMP1Δ232–351 inserted into the Klenow-blunted BamHI site of pSG5 (Stratagene).
NF-κB activation.
A total of 2.5 × 105 293 cells were transformed with LMP1 expression vector, 3×-κB-L luciferase reporter (A gift from Bill Sugden, University of Wisconsin, Madison) (48), and a pGK-β gal transfection control and analyzed as described elsewhere (6).
Recombinant EBV construction, Western blotting, in situ immunofluorescence, and coimmunoprecipitation analyses.
Recombinant EBVΔ232–351 was made by second site homologous recombination as described elsewhere (59, 72). EBV proteins were detected by Western blotting and by in situ immunofluorescence and were coimmunoprecipitated as described elsewhere (23, 28, 59).
RESULTS
An EBV recombinant with LMP1 codons 232 to 351 deleted is competent for primary B-lymphocyte growth transformation.
An EBV LMP genomic clone with an in-frame deletion of codons 232 to 351 and with an insertion of codons for the Flag epitope within the amino-terminal cytoplasmic domain was constructed as described in Materials and Methods. The predicted protein has the Flag epitope and a shorter carboxyl-terminal cytoplasmic tail with TES1 juxtaposed with TES2 (Fig. 1). The deleted sequence comprises the epitope recognized by the LMP1-specific monoclonal antibody S12.
Flag-LMP1Δ232–351 DNA was recombined into the P3HR-1 EBV genome by the second site homologous recombination method. In brief, P3HR-1 cells have an endogenous EBV that is competent for virus replication, but the virus is transformation defective due to a deletion of DNA that codes for EBNA2 and EBNA LP. When 107 P3HR-1 latently infected cells are cotransfected with a cosmid DNA that spans the P3HR-1 deletion, a mutated LMP1 DNA, and an expression vector for the EBV immediate-early transactivator BZLF1, virus replication is induced in the transfected cells and about 107 virions are produced. Almost all are parental P3HR-1 EBV, but about 102 to 103 recombinants with the cosmid DNA are also produced, and these recombinants are able to transform resting human B lymphocytes into proliferating cells. Under optimal conditions, 10% of those recombinants will have undergone homologous recombination with and incorporated the mutated LMP1 DNA. When this mixture of viruses is used to infect about 108 B lymphocytes and the infected cells are plated into 2,000 microwells, as many as one-half of the wells are positive for LCLs.
About one-third of the virus from a P3HR-1 cell cotransfection was used to infect resting primary B lymphocytes from a healthy human donor, and 630 recombinants were recovered, as evidenced by the number of resulting LCLs. A sample of 245 LCLs were further analyzed by PCR for their LMP1 genes. Five of the 245 (2%) LCLs were found to have the Flag-LMP1Δ232–351 DNA, whereas the remaining 240 encoded only wt LMP1 DNA from P3HR-1. All five Flag-LMP1Δ232–351 DNA-containing LCLs were coinfected with P3HR-1 EBV. The smaller than expected percentage of Flag-LMP1Δ232–351 recombinants might be due to recombination constraints imposed by the interruptions of DNA homology at the Flag codon insertion and codon 232-to-351 deletion sites, to a third critical transformation effector site within the deletion, or to a role for the deleted amino acids in protein folding or stability.
To establish the transformation phenotype of the Flag-LMP1Δ232–351 recombinants, one Flag-LMP1Δ232–351 recombinant LCL was transfected with the BZLF1 expression vector to reactivate virus replication, and fresh primary B lymphocytes were infected with the resulting virus progeny. Hundreds of second-generation LCLs resulted, and 140 were examined by PCR for the Flag epitope that is characteristic of the Flag-LMP1Δ232–351 DNA. Two-thirds of the 140 were infected with a Flag-LMP1Δ232–351 EBV recombinant. Twenty-four of these LCLs were selected because the PCR analysis indicated the presence of the Flag codon insertion, but revealed no signal that indicated wt P3HR-1 LMP1 DNA without the Flag codon insertion (data not shown). By another PCR analysis that scores specifically for wt LMP1 DNA, 2 of the 24 LCLs were found to have less than 1 wt LMP1 DNA molecule in 1,000 cells. Of the others, 3 LCLs had no more than 1 wt LMP1 DNA in 100 cells, 9 had no more than 1 wt LMP1 DNA in 10 cells, and 10 had about 1 wt LMP1 DNA per cell. The two LCLs having no more than 1 wt LMP1 DNA in 1,000 cells were cloned by limiting dilution, and two cell lines (Δ24–68 and Δ33–15) were established in which no wt LMP1 DNA could be detected at a level of sensitivity of 1 LMP1 DNA in 104 cells.
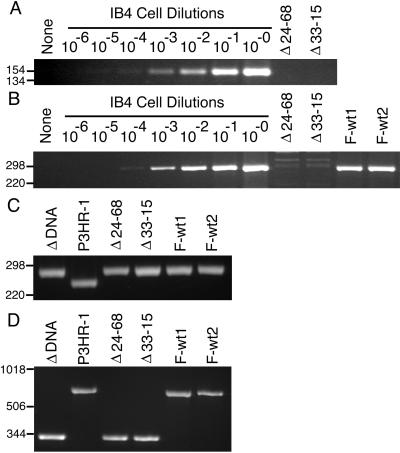
The LMP1 genes in the Δ24–68 and Δ33–15 LCLs were further analyzed by PCR (Fig. 2). Primers that are specific for wt LMP1 amino-terminal codons amplified DNA of the expected size from IB4 cells, which have four integrated EBV genomes per cell. This procedure could detect 1 IB4 cell diluted with 104 EBV-negative BJAB cells (Fig. 2A, lane 4). In lanes 9 and 10, wt LMP1 DNA was not detected in 104 Δ24–68 or Δ33–15 LCLs. This indicates that the Δ24–68 and Δ33–15 LCLs have no wt LMP1 DNA, with a sensitivity of 4 LMP1 genes in 104 cells. In panel B, another set of primers that are identical to codons 232 to 240 and complementary to codons 315 to 323 amplified wt LMP1 DNA of the expected size when at least 1 IB4 cell was diluted with DNA from 104 EBV-negative BJAB cells (lane 4), whereas no DNA was detected in lanes 9 and 10 from 104 Δ24–68 and Δ33–15 LCLs. DNA of the expected wt size was amplified from 104 Flag-wt1 (F-wt1) and F-wt2 LCLs which are infected with Flag-wt LMP1 recombinants (lanes 11 and 12). In panel C, primers were used that amplify LMP1 amino-terminal codons from P3HR-1 cells, which have wt LMP1 DNA (lane 2) or plasmid Flag-LMP1Δ232–351 DNA (lane 1). The PCR product from plasmid Flag-LMP1Δ232–351 DNA is larger due to the Flag codon insertion. Lysates from 104 Δ24–68 and Δ33–15 LCLs (lanes 3 and 4) yielded a PCR product similar in size to DNA amplified from the plasmid Flag-LMP1Δ232–351 DNA that was used in their construction. F-wt1 and F-wt2 LCLs (lanes 5 and 6) have Flag-LMP1 DNA which amplified a DNA of similar size because of the Flag codon insertion. In panel D, primers that flank the coding sequence of the carboxyl-terminal tail amplified a smaller product from plasmid Flag-LMP1Δ232–351 DNA (lane 1) than P3HR-1 cells, which have wt LMP1 DNA (lane 2). The DNA from 104 Δ24–68 and Δ33–15 LCLs (lanes 3 and 4) produced PCR products that were similar in size to the DNA amplified from plasmid Flag-LMP1Δ232–351 DNA used in their construction. DNA from F-wt1 and F-wt2 LCLs which are infected with Flag-LMP1 recombinants (lanes 5 and 6) amplified DNA similar in size to that amplified from P3HR-1 cells which have wt LMP1 DNA (lane 2). Thus, Δ24–68 and Δ33–15 LCLs have no apparent wt LMP1 DNA with a sensitivity of 4 EBV DNA copies in 10,000 cells, and both the codon 232-to-351 deletion and the Flag codon insertion are evident. These results indicate that LMP1 aa 232 to 351 are not essential for EBV-mediated transformation of primary B lymphocytes into indefinitely proliferating lymphoblastoid cell lines.
FIG. 2.
PCR analyses of mutated LMP1 recombinant EBV-infected LCLs for LMP1 DNA. (A) PCR analysis for wt amino-terminal LMP1 DNA with the primers 5′-GAGGATGGAACACGACCTTGAGA-3′ and 5′-CTCCAGTCCAGTCACTCATAACG-3′. Lanes 8 to 2 are 104 IB4 cells (4 EBV DNA per cell) serially 10-fold diluted with 104 EBV-negative BJAB cells. After PCR amplification, DNAs were size separated in 3% agarose gels containing ethidium bromide. The endpoint dilution (lane 4) is the 10−4 dilution for a sensitivity of 4 copies of LMP1 DNA per 104 cells. No wt LMP1 DNA was detected in 104 Δ24–68 LCLs (lane 9) or Δ33–15 LCLs (lane 10) or in water (lane 1). Molecular markers (in base pairs) are indicated to the left of each panel. (B) PCR analysis for wild-type carboxyl-terminal LMP1 DNA as in panel A, except that the primers 5′-GACGGACCCCCACTCTGCTCTC-3′ and 5′-ATTGTGGAGGGCCTCCATCATTTC-3′ were used. (C) PCR analysis for Flag-LMP1 and wt LMP1 amino-terminal DNA as in panel A, except that the primers 5′-CACGCGTTACTCTGACGTAGCCG-3′ and 5′-CTCCAGTCCAGTCACTCATAACG-3′ were used. (D) PCR analysis for Flag-LMP1Δ232–351 deletion mutant and wt LMP1 carboxyl-terminal DNA as in panel A, except that the primers 5′-CTCTATTGGTTGATCTCCTTTGG-3′ and 5′-GCCTATGACATGGTAATGCCTAG-3′ were used.
Flag-LMP1Δ232–351 LMP1 recombinants transform primary B lymphocytes de novo with wt efficiency.
The ability of the Flag-LMP1Δ232–351 EBV recombinants to transform primary B lymphocytes was compared with that of Flag-LMP1 recombinants, which are isogenic except for their LMP1 genes. Δ24–68, Δ33–15, F-wt1, and F-wt2 LCLs were induced to lytic infection, and filtered viruses prepared from the LCLs were serially diluted and then used to infect primary B lymphocytes. In Table 1, the filtered virus DNA titer as determined by endpoint dilution PCR and the number of LCLs growing in microwells 6 weeks after infection at each virus dilution are compared. All LCLs replicated virus, and all viruses were transformation competent. F-wt1 LCL produced the smallest amount of virus, and EBV from this LCL produced the fewest LCLs at every dilution. Δ24–68 LCL produced more virus than F-wt1 LCL, and EBV from Δ24–68 LCL produced more LCLs at every dilution. The virus DNA titer was less than that of F-wt2 or Δ33–15 LCLs, but EBV from the Δ24–68 LCL was similar to EBV from the F-wt2 LCL in the number of transformed cell lines. F-wt2 and Δ33–15 LCLs produced similar amounts of virus, but EBV from Δ33–15 LCLs produced slightly more transformed cell lines. Clearly, Flag-LMP1Δ232–351 recombinants are quite similar to Flag-LMP1 recombinants in replication and in primary B-lymphocyte transformation.
TABLE 1.
Transforming efficiency of recombinant EBVa
| EBV recombinant transformed LCL | Viral DNA titer in cell supernatants | No. of LCLs in 48 wells at dilution:
|
|||
|---|---|---|---|---|---|
| 100 | 10−1 | 10−2 | 10−3 | ||
| F-wt1 | 1 | 17 | 7 | 1 | 0 |
| F-wt2 | 9 | 47 | 32 | 6 | 2 |
| Δ24–68 | 3 | 48 | 36 | 4 | 1 |
| Δ33–15 | 9 | 48 | 47 | 16 | 8 |
F-wt1 and F-wt2 LCLs were infected with Flag-LMP1 recombinant EBV, and Δ24–68 and Δ33–15 LCLs were infected with Flag-LMP1Δ232–351 recombinant EBV. Each LCL was transfected with pSVNael Z, a BZLF1-expressing vector, and treated with phorbol ester to induce lytic infection. After 5 days, cells and media were frozen and thawed, 0.45-μm-pore-diameter filtered, serially 10-fold diluted, and used to infect primary B lymphocytes isolated from peripheral blood. A total of 2.5 × 106 cells were seeded into 48 wells of a 96-well plate. For each dilution, the number of replicating LCLs was scored in columns 3 to 6 at 6 weeks postinfection. To titrate viral DNA (column 2), an aliquot of the virus preparation was serially threefold diluted and PCR analyzed for EBV DNA with the primers 5′-CACGCGTTACTCTGACGTAGCCG-3′ and 5′-CTCCAGTCCAGTCACTCATAACG-3′. The titer is the reciprocal of the maximum dilution at which an EBV-specific, PCR-amplified DNA product is detected after electrophoresis in agarose gels stained with ethidium bromide.
Flag-LMP1Δ232–351 recombinant EBV transformants proliferate into long-term LCLs with wild-type efficiency.
Ten LCLs transformed by each of the four virus stocks from F-wt1, F-wt2, Δ24–68, or Δ33–15 LCLs were continuously subcultivated in vitro to compare the efficiency of long-term LCL outgrowth of cells with that of the second-passage Flag-LMP1Δ232–351 or F-LMP1 EBV recombinants. All of these LCLs continued to expand for the ensuing 6 months in culture. Thus, these cell lines did not differ in their growth rate or ability to expand into long-term LCLs.
Flag-LMP1Δ232–351 LMP1 and Flag-LMP1 recombinant-transformed LCLs are similar in the endpoint dilution from which they can regrow.
LCLs are dependent on cross-feeding for rapid growth. Even after 6 months of continuous subcultivation, LCLs typically require seeding between 104 to 105 cells per ml of complete culture medium in order regrow to 106 cells per ml by 21 days. Subtle growth defects are frequently most evident when cells are challenged by plating at low cell density (15, 28, 77). The inherent sensitivity to dilution of cells transformed by Flag-LMP1Δ232–351 or F-LMP1 EBV recombinants was therefore determined by seeding serial dilutions of cells into 96-well plates at cell concentrations from 30,000 to 1,250 cells per 0.1 ml of medium. LCL outgrowth was monitored over the course of 3 weeks, and the results are presented in Table 2. The progenitor F-wt1, F-wt2, Δ24–68, or Δ33–15 LCLs efficiently regrew when plated at 6,000 to 11,000 cells per 0.1 ml, but not when plated at a lower cell density. Two cell lines transformed by recombinant EBV passaged from each progenitor were tested for regrowth after endpoint dilution. The endpoint dilutions for regrowth of second-passage EBV recombinant-transformed LCLs were 2,500 and ≤5,000 for F-wt1, ≤1,250 and 5,000 for Fwt-2, ≤1,250 and ≤1,250 for Δ24–68, and ≤1,250 and ≤5,000 for Δ33–15. Thus, Flag-LMP1Δ232–351 recombinant-transformed LCLs are similar to Flag-LMP1 recombinant-transformed LCLs in their sensitivity to low-density plating.
TABLE 2.
LCL regrowth after limiting dilutiona
| EBV recombinant | Endpoint for regrowth in EBV recombinant-transformed LCLsb
|
||
|---|---|---|---|
| Progenitorc | 2nd passage
|
||
| Cell line 1 | Cell line 2 | ||
| F-wt1 | 6,667 | 2,500 | ≤5,000 |
| F-wt2 | 10,000 | ≤1,250 | ≤5,000 |
| Δ24–68 | 9,167 | ≤1,250 | ≤1,250 |
| Δ33–15 | 10,833 | ≤1,250 | ≤5,000 |
LCLs were serially diluted with RPMI 1640 medium containing 10% fetal bovine serum, and microwells were seeded with 0.1 ml. Fresh medium was added 1 week later, and regrowth was monitored for 3 weeks total.
The numbers indicate the minimum cell seeding needed to regrow more than 106 cells per ml by 3 weeks.
Mean of three tests.
EBNA and LMP1 expression levels and LMP1 aggregation are similar in Flag-LMP1Δ232–351 and Flag-LMP1 recombinant EBV-transformed LCLs.
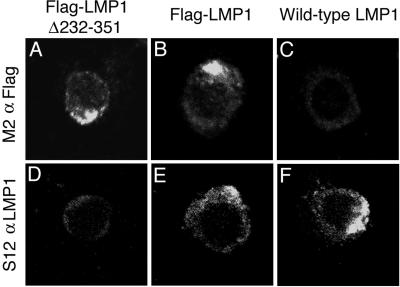
LMP1 characteristically aggregates in a single area in LCL plasma membranes. LMP1 was localized by indirect immunofluorescence on fixed and permeabilized wt, Flag-LMP1, and Flag-LMP1Δ232–351 recombinant-transformed LCLs (Fig. 3). Flag-LMP1 in the F-wt2 LCL was detected in plasma membrane aggregates by using M5 monoclonal antibody to Flag (Fig. 3B) or S12 monoclonal antibody that recognizes an epitope within LMP1 aa 232 to 351 (Fig. 3E). An LCL transformed by an EBV recombinant with a wild-type LMP1 that does not have a Flag epitope tag stained similarly with S12 (Fig. 3F) but not M5 antibody (Fig. 3C). Flag-LMP1Δ232–351 in the Δ33–15 LCL was immunoreactive in similar membrane aggregates with M5 antibody (Fig. 3A), but was not detected with S12 antibody (Fig. 3D). The same results were obtained with the Δ24–68 LCL (data not shown). Thus, deletion of aa 232 to 351 does not alter LMP1 plasma membrane localization or aggregation.
FIG. 3.
Indirect immunofluorescent staining of methanol and acetone-fixed lymphoblastoid cell lines with M2 monoclonal antibody to Flag or S12 monoclonal antibody to the LMP1 carboxyl terminus. LMP1 spontaneously aggregates in the plasma membrane.
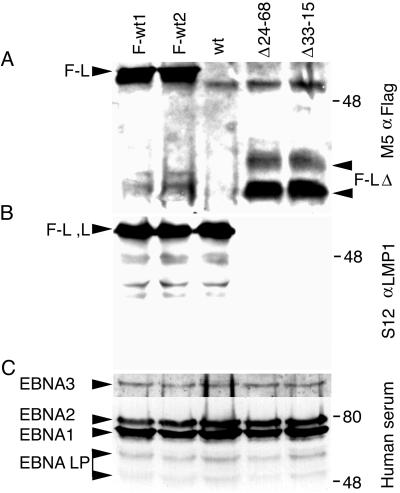
Flag-LMP1 levels were measured by M2 Flag antibody precipitation of Flag-LMP1, size separation in denaturing polyacrylamide gels, and Western immunoblot detection with M5 Flag antibody. In Fig. 4A, a protein of about 60 kDa is present in F-wt1 (lane 1) and F-wt2 (lane 2) LCLs. A prominent, nonspecific band just below Flag-LMP1 is detected in all lanes of the blot. Two proteins of about 40 kDa are present in the Δ24–68 (lane 4) and Δ33–15 (lane 5) LCLs, which express Flag-LMP1Δ232–351. The sum of the two protein band intensities of Flag-LMP1Δ232–351 is similar to that of Flag-LMP1, consistent with the similar level of immunofluorescent staining in situ (Fig. 3). M5 antibody does not detect LMP1 in immunoprecipitates from an LCL that expresses wild-type LMP1 without the Flag epitope tag (Fig. 4, lane 3). In Fig. 4B, F-wt1, F-wt2, and wt LMP1 LCLs have similar quantities of LMP1 in unfractionated cell lysates, as detected by immunoblotting with S12 monoclonal antibody. S12 reactive, full-length LMP1 is absent from the Δ24–68 and Δ33–15 LCLs (lanes 4 and 5). These results confirm that the Δ24–68 and Δ33–15 LCLs do not express the 60-kDa LMP1 but do express the 40-kDa Flag-LMP1Δ232–351. Furthermore, deletion of residues 232 to 351 abolishes the S12 monoclonal antibody epitope, which is likely within the imperfect repeat sequences.
FIG. 4.
Protein expression in recombinant EBV-infected LCLs. (A) Western immunoblot analysis for Flag-LMP1. About 108 cells were Dounce homogenized in buffer containing 0.5% Brij 58, 100 mM NaCl, and 50 mM Tris (pH 7.2) and cleared by centrifugation. Flag-LMP1 was immunoprecipitated with M2 affinity gel (Sigma) for 6 h. Affinity gel was washed once, and precipitated proteins were detached with buffer containing SDS and 2-mercaptoethanol. About 10% of the immunoprecipitates were size separated in denaturing polyacrylamide gels, blotted to nitrocellulose filters, probed with M5 monoclonal antibody to Flag (Sigma) and peroxidase-conjugated secondary antibody to mouse immunoglobulin G (Amersham), and visualized by enhanced chemiluminescence (NEN Life Science). The position of Flag-LMP1 (F-L) is marked on the left, and the position of Flag-LMP1Δ232–351 (F-LΔ) is marked on the right. (B) Western immunoblot analysis for LMP1 carboxyl-terminal amino acids. About 5 × 104 cells were lysed in buffer containing SDS and 2-mercaptoethanol and resolved in denaturing polyacrylamide gels. After Western transfer to nitrocellulose filters, LMP1 was detected as in panel A, except S12 monoclonal antibody was used. The position of Flag-LMP1 (F-L) or LMP1 (L) is marked on the left. (C) Western immunoblot analysis for EBV nuclear antigens (EBNA) leader protein (LP), EBNA1, EBNA2, and EBNA3C. The position of each protein is marked on the left. Analysis was done as in panel B, except that serum from a normal human donor and peroxidase-conjugated secondary antibody to human immunoglobulin G were used. In all panels, molecular mass markers (in kilodaltons) are marked on the right.
Expression of EBV nuclear proteins EBNA LP, -1, -2, and -3C was examined by Western immunoblotting with immune human serum. In unfractionated cell lysates, levels of EBNA1 and EBNA2 were prominent and equivalent in F-wt1, F-wt2, wt, Δ24–68, or Δ33–15 LCLs (Fig. 4C). EBNA LP staining was more diffuse, but the levels were approximately the same in the five LCLs. Detection of EBNA3C required extended exposure to film, but the relative levels were similar in the five LCLs. Thus, Flag-LMP1Δ232–351, Flag-LMP1, and wt LMP1 EBV recombinant-transformed LCLs are not different in latent EBV gene expression.
Flag-LMP1Δ232–351 is similar to Flag-LMP1 in inducing NF-κB activation.
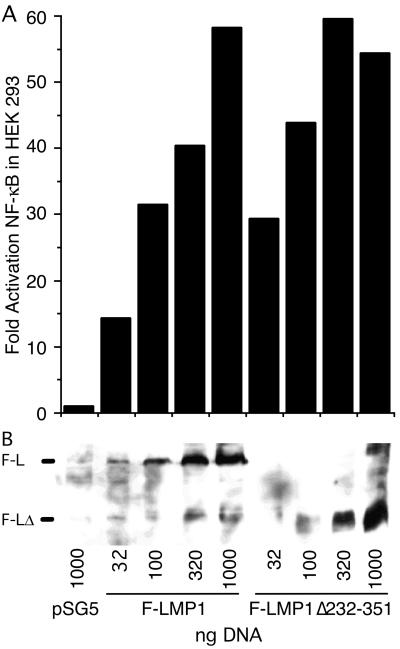
Since NF-κB activation by TES1 and TES2 is genetically linked to B-lymphocyte transformation and likely to be pathophysiologically relevant, we tested the ability of Flag-LMP1Δ232–351 to activate an NF-κB-responsive luciferase reporter in transiently transfected 293 human embryonic kidney cells. Transfection of 32, 100, 320, or 1,000 ng of Flag-LMP1- or Flag-LMP1Δ232–351-expressing vectors activated NF-κB progressively with more DNA (Fig. 5A). Higher levels of NF-κB activation correlated with higher LMP1 expression (Fig. 5B). Flag-LMP1Δ232–351 was consistently as active or somewhat more active than Flag-LMP1.
FIG. 5.
F-LMP1- and F-LMP1Δ232–351-expressing vectors activate NF-κB. (A) HEK293 cells (2.5 × 105) were transfected (Qiagen Superfect) with the indicated amounts of F-LMP1- or F-LMP1Δ232–351-expressing vector or pSG5 vector control and with 350 ng of 3X-κB-L, a luciferase reporter with three NF-κB sites from human major histocompatibility class I and Fos minimal promoter and with 350 ng of pGK-β-gal DNA, a β-galactosidase-expressing vector used to monitor transfection efficiency. After 20 h at 37°C, cells were lysed in reference lysis buffer (Promega) and analyzed for luciferase activity (Promega) and β-galactosidase expression (Tropix) according to the manufacturer’s directions. (B) Western immunoblot analysis for F-LMP1 or F-LMP1Δ232–351. Equivalent amounts of protein from lysates prepared as described above were size separated and analyzed as described in the legend to Fig. 4A. The position of F-LMP1 or F-LMP1Δ232–351 is marked on the left.
Flag-LMP1Δ232–351 and Flag-LMP1 constitutively associate with signaling proteins in LCLs.
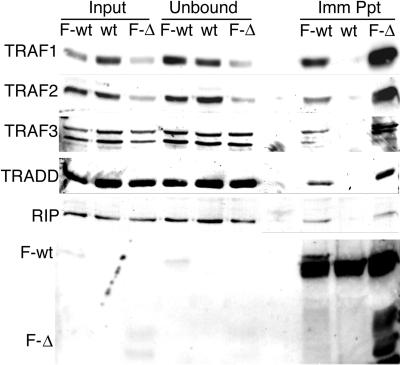
Flag-LMP1 association with signaling proteins was examined by immunoprecipitation with Flag antibody M2 and Western immunoblot analysis. The death domain-containing proteins TRADD and RIP coprecipitated with Flag-LMP1 and Flag-LMP1Δ232–351 with about the same efficiency, whereas neither protein coprecipitated with Flag antibody with extracts from an LCL transformed by an EBV recombinant that expresses a wt LMP1 that lacks Flag (Fig. 6). TRAF1, TRAF2, and TRAF3 also coprecipitated at about the same level with Flag antibody with Flag-LMP1- or Flag-LMP1Δ232–351-transformed LCL extracts, whereas TRAFs did not coprecipitate from extracts of wt LMP1-transformed LCLs. The efficiencies of immunoprecipitation of Flag-LMP1 and Flag-LMP1Δ232–351 were similar, whereas Flag antibody precipitated only a trace amount of wt LMP1. These biochemical results are consistent with the aa 232-to-351 deletion being similar to wild-type EBV in effecting EBV-mediated growth transformation.
FIG. 6.
Coimmunoprecipitation of TRAF1, TRAF2, TRAF3, TRADD, or RIP with F-LMP1 or F-LMP1Δ232–351. Proteins from LCLs (2.0 × 108 cells) infected with F-LMP1, F-LMP1Δ232–351, or a wt LMP1 EBV recombinant were solubilized by Dounce disruption in 0.5% Brij 58, 100 mM NaCl, and 50 mM Tris (pH 7.2) and immunoprecipitated with M2 affinity gel (Sigma) to Flag. Precipitated proteins were Western immunoblot analyzed with antibodies to TRAF1, TRAF2, TRAF3, and TRADD from Santa Cruz Biotechnology, antibody to RIP from Pharmingen, or M5 antibody to Flag from Sigma. Input lanes represent unfractionated cell proteins, unbound lanes represent proteins not precipitated with M2 affinity gel, and Imm Ppt lanes represent immunoprecipitated proteins.
LCLs transformed by Flag-LMP1Δ232–351, Flag-LMP1, and wt LMP1 EBV recombinants are similar for levels of JNK1 and JNK2, phosphorylated JNK1 and JNK2, and Bcl2.
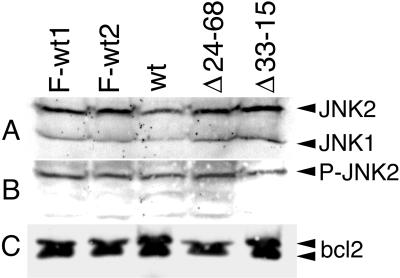
LMP1 activates c-Jun N-terminal kinase (JNK) and upregulates Bcl2 levels (10, 11, 16, 32, 60, 68). Steady-state levels of JNK1 and JNK2, phosphorylated JNK1 and JNK2, and Bcl2 were assayed by Western immunoblotting. Western immunoblot analysis with an antibody that recognizes JNK1 and JNK2 detects similar levels of JNK1 and JNK2 in LCLs transformed by Flag-LMP1Δ232–351, Flag-LMP1, and wt LMP1 EBV recombinants (Fig. 7A). Western immunoblot analysis with an antibody that recognizes the phosphorylated forms of both JNK1 and JNK2 detects phosphorylated JNK2 (P-JNK2) at similar levels in LCLs transformed with EBV recombinants that express Flag-LMP1Δ232–351, Flag-LMP1, and wt LMP1. Although the antibody recognizes both P-JNK1 and P-JNK2, P-JNK1 is not detected in any of these LCLs (Fig. 7B). These results indicate that P-JNK2 is the predominant activated JNK in LCLs and that LCLs transformed by Flag-LMP1Δ232–351 recombinants are similar in JNK activation. Western immunoblot analysis with antibody to Bcl2 reveals that the Bcl2 levels are similar in all of these LCLs (Fig. 7C). These results demonstrate that residues 232 to 351 are not involved with activating JNK2 or with regulating Bcl2, JNK1, or JNK2 levels.
FIG. 7.
Western immunoblot analysis for c-Jun N-terminal kinase 1 and 2 (JNK1 and JNK2), phosphorylated JNK1 and JNK2 (P-JNK1 and P-JNK2), and Bcl2. LCLs (1.5 × 106) transformed with F-LMP1, F-LMP1Δ232–351, or a wt LMP1 EBV recombinant were directly lysed in buffer containing SDS and 2-mercaptoethanol. Cell proteins were resolved in denaturing polyacrylamide gels, Western blotted to nitrocellulose filters, and probed with antibodies. (A) JNK1 and JNK2 are detected at similar levels in all LCLs with an antibody that recognizes JNK1 and JNK2 (New England Biolabs). (B) P-JNK2 but not P-JNK1 is detected at similar levels in all LCLs with an antibody that specifically recognizes P-JNK1 and P-JNK2 (New England Biolabs). (C) Bcl2 protein is detected at similar levels in all LCLs with a Bcl2-specific antibody (Santa Cruz Biotechnology).
DISCUSSION
These data indicate that LMP1 aa 232 to 351 are not critical for primary B-lymphocyte growth transformation in vitro. The assays employed to compare the phenotypic characteristics of Flag-LMP1Δ232–351 with Flag-LMP1 EBV recombinants have previously revealed differences between mutant and wt EBV recombinants in a reduced efficiency of initial lymphocyte growth transformation, in a reduced efficiency of LCL outgrowth at various stages of expansion from the initial transformants, or in an increased cell density dependence (15, 23, 27, 28, 77). The absence of any phenotypic difference between Flag-LMP1Δ232–351 and Flag-LMP1 EBV recombinants in these three assays and the absence of any biochemical difference between Flag-LMP1Δ232–351 and Flag-LMP1 in association with cell signaling proteins, NF-κB activation, JNK activation, or altered cell gene expression indicates that aa 232 to 351 are not important for lymphocyte growth transformation in vitro. These results also underscore the principal positive effector roles of TES1 and TES2 in primary B-lymphocyte growth transformation. This result is consistent with previous recombinant genetic analyses of the role of the LMP1 carboxyl terminus in B-lymphocyte growth transformation. The previous experiments showed that the LMP1 amino terminus, transmembrane segments, and TES1 are sufficient for initial primary B-lymphocyte growth transformation in the absence of the LMP1 carboxyl-terminal 155 aa (27, 28). Cocultivation with fibroblast feeder cells is required for efficient long-term LCL outgrowth after transformation by the MS231 EBV recombinant that expresses LMP1 aa 1 to 231. TES2 appears to be critical for efficient long-term LCL outgrowth, since a double point mutation at the carboxyl terminus of TES2 has a phenotype that is similar to that of MS231 (24). Although there remains the possibility that a point mutant in the TES1 core TRAF binding site can transform when the infected cells are cocultivated with fibroblasts, the data thus far are consistent with a model in which TES1 is the most important effector site overall, and TES2 is the principal effector site in the carboxyl-terminal 155 aa. Certainly, aa 232 to 351 are not important for efficient primary B-lymphocyte growth transformation in vitro.
Either LMP1 residues 232 to 351 are unimportant for primary B-lymphocyte growth transformation, or there are multiple domains within this region, some positive and some negative, such that removal of the entire region has no net effect. Indeed, aa 232 to 351 include four copies of an 11-residue repeat, a site for protease-specific cleavage, sites proposed to interact with Janus kinase 3 (JAK3) and mediate the activation of signal transduction and activation of transcription (STAT) proteins, and sites for specific serine/threonine phosphorylation that are highly conserved among EBV isolates (12, 13, 38, 40, 50, 51). The wt transforming ability of Flag-LMP1Δ232–351 EBV recombinants is compatible with the hypothesis that LMP1’s potential interaction with JAK3 does not mediate signaling that is significant to the growth transformation of primary B lymphocytes. We cannot exclude the possibility that this potential interaction may have a subtle effect on some aspect of latent infection. The absence from Flag-LMP1Δ232–351 of the normal protease cleavage site between asparagine 241 and leucine 242 might have been expected to result in increased accumulation of the deletion mutant LMP1, but Flag-LMP1Δ232–351 was similar in abundance to wt LMP1. No other functional consequences have been attributed to the protease cleavage (52). Furthermore, a previous mutation of the secondary phosphorylation site at T324 to a glutamic acid resulted in an LMP1 that appeared to be unable to transform Rat-1 cells, as measured by a cell contact inhibition assay (51). This finding raised the expectation that deletion of this site would affect B-lymphocyte growth transformation.
The LMP1 aa 232-to-351 deletion includes 9 of the 10 residues (aa 343 to 352) that are deleted from EBV strains that are endemic to southern Chinese ethnic groups that have elevated relative risk for nasopharyngeal carcinoma (19). The same deletion has been noted in some American and European EBV strains, including viral genomes in some Hodgkin’s lymphoma tumor cells and in some patients with lymphoproliferative disease (9, 29, 33, 34, 47, 54–56, 65, 69). Experimental studies comparing the effects of the aa 343-to-352 deletion mutant LMP1 with wild-type LMP1 in BALB/c 3T3 cells indicate that the deletion may be associated with increased transforming effects (36). However, the specific association of this deletion with malignancies has not been documented by formal epidemiological criteria, and recent data indicate this deletion is present in other ethnic groups (17, 18, 64). Furthermore, lymphocytes natively transformed by EBV strains that have the aa 343-to-352 deletion mutant LMP1 are not more tumorigenic in nude or SCID mice, and the deletion has not been associated with poorer outcomes in EBV-associated human lymphoproliferative disease (3, 17, 18, 62, 63). Moreover, LMP1 with aa 343 to 352 deleted did not differ from wild-type LMP1 in NF-κB activation or in induction of CD40 or CD54 surface expression on B-lymphoma cells (25, 45). Thus, the overall significance of this deletion in the pathogenesis of EBV-related malignancies is uncertain.
The absence of a phenotype in B-lymphocyte transformation assays with deletion of aa 232 to 351 may not be fully predictive of the role of these sequences in vivo. In immunocompetent humans, LMP1 is expressed during lytic EBV infection in oropharyngeal epithelial cells, in type III latency that accompanies primary infection in B lymphocytes, and in type II latency in some circulating B lymphocytes. LMP1 is also expressed in EBV-associated nasopharyngeal carcinoma, Hodgkin’s lymphoma, and in some leiomyosarcomas of the intestine (reviewed in reference 57). In these various tissues or stages of EBV infection, aa 232 to 351 may have a role in regulating the effects of TES1 or TES2 or may have other effects that are independent of TES1 or TES2. Amino acids 232 to 351 are highly conserved in all EBV isolates and are therefore likely to have an important role in some aspect of normal EBV infection in vivo. Comparison of the biological properties of mutant versus wt transformed lymphocytes in SCID mouse tumorigenesis models may reveal some difference. However, the difference may be subtle and require study in a primate lymphocryptovirus infection model (49).
The finding that residues 232 to 351 are not important for lymphocyte growth transformation is an important step in defining the amino-terminal boundary of TES2. Previously, TES2 was formally defined by the double point mutation of Y384Y385 to I384 (24). Clearly, aa 352 to 386 are a fully competent TES2 for B-lymphocyte growth transformation in vitro. Since the terminal 11 residues are sufficient to engage TRADD and synergistically activate NF-κB, the amino-terminal boundary of TES2 is likely to be closer to the carboxyl terminus than aa 352. However, RIP also interacts with TES2, and the interaction appears to require more than the terminal 11 residues (21). More precise definition of the boundaries of TES2 and TES1 is important in evaluating whether these sites have additional effector functions.
REFERENCES
- 1.Baichwal V R, Sugden B. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene. 1988;2:461–467. [PubMed] [Google Scholar]
- 2.Brodeur S R, Cheng G, Baltimore D, Thorley L D. Localization of the major NF-κB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J Biol Chem. 1997;272:19777–19784. doi: 10.1074/jbc.272.32.19777. [DOI] [PubMed] [Google Scholar]
- 3.Chen W G, Chen Y Y, Bacchi M M, Bacchi C E, Alvarenga M, Weiss L M. Genotyping of Epstein-Barr virus in Brazilian Burkitt’s lymphoma and reactive lymphoid tissue. Type A with a high prevalence of deletions within the latent membrane protein gene. Am J Pathol. 1996;148:17–23. [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng G, Baltimore D. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-κB activation. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devergne O, Hummel M, Koeppen H, Le Bean M, Nathanson E C, Kieff E, Birkenbach M. A novel interleukin-12 p40-related protein induced by latent Epstein-Barr virus infection in B lymphocytes. J Virol. 1996;70:1143–1153. doi: 10.1128/jvi.70.2.1143-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devergne O, Cahir McFarland E, Mosialos G, Izumi K M, Ware C F, Kieff E. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolcetti R, Zancai P, De Re V, Gloghini A, Bigoni B, Pivetta B, De Vita S, Carbone A, Boiocchi M. Epstein-Barr virus strains with latent membrane protein-1 deletions: prevalence in the Italian population and high association with human immunodeficiency virus-related Hodgkin’s disease. Blood. 1997;89:1723–1731. [PubMed] [Google Scholar]
- 10.Eliopoulos A G, Blake S M S, Floettmann J E, Rowe M, Young L S. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J Virol. 1999;73:1023–1035. doi: 10.1128/jvi.73.2.1023-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliopoulos A G, Young L S. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 12.Fennewald S, van Santen V, Kieff E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J Virol. 1984;51:411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gires O, Kohlhuber F, Kilger E, Baumann M, Kieser A, Kaiser C, Zeidler R, Scheffer B, Ueffing M, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999;18:3064–3073. doi: 10.1093/emboj/18.11.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 15.Harada S, Yalamanchili R, Kieff E. Residues 231 to 280 of the Epstein-Barr virus nuclear protein 2 are not essential for primary B-lymphocyte growth transformation. J Virol. 1998;72:9948–9954. doi: 10.1128/jvi.72.12.9948-9954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzivassiliou E, Miller W E, Raab T N, Kieff E, Mosialos G. A fusion of the EBV latent membrane protein-1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor-kappa B, and stress-activated protein kinase. J Immunol. 1998;160:1116–1121. [PubMed] [Google Scholar]
- 17.Hayashi K, Chen W G, Chen Y Y, Bacchi M M, Bacchi C E, Alvarenga M, Abreu E S, Chang K L, Weiss L M. Deletion of Epstein-Barr virus latent membrane protein 1 gene in United States and Brazilian Hodgkin’s disease and reactive lymphoid tissue: high frequency of a 30-bp deletion. Hum Pathol. 1997;28:1408–1414. doi: 10.1016/s0046-8177(97)90231-8. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi K, Chen W G, Chen Y Y, Murakami I, Chen H L, Ohara N, Nose S, Hamaya K, Matsui S, Bacchi M M, Bacchi C E, Chang K L, Weiss L M. Deletion of Epstein-Barr virus latent membrane protein 1 gene in Japanese and Brazilian gastric carcinomas, metastatic lesions, and reactive lymphocytes. Am J Pathol. 1998;152:191–198. [PMC free article] [PubMed] [Google Scholar]
- 19.Hu L F, Zabarovsky E R, Chen F, Cao S L, Ernberg I, Klein G, Winberg G. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J Gen Virol. 1991;72:2399–2409. doi: 10.1099/0022-1317-72-10-2399. [DOI] [PubMed] [Google Scholar]
- 20.Huen D S, Henderson S A, Croom C D, Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 21.Izumi K M, Cahir McFarland E, Ting A T, Riley E A, Seed B, Kieff E D. The Epstein-Barr virus oncoprotein latent membrane protein 1 (LMP1) engages tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-κB activation. Mol Cell Biol. 1999;19:5759–5767. doi: 10.1128/mcb.19.8.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumi K M, Kaye K M, Kieff E D. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi K M, Kaye K M, Kieff E D. Epstein-Barr virus recombinant molecular genetic analysis of the LMP1 amino-terminal cytoplasmic domain reveals a probable structural role, with no component essential for primary B-lymphocyte growth transformation. J Virol. 1994;68:4369–4376. doi: 10.1128/jvi.68.7.4369-4376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi K M, Kieff E D. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson R J, Stack M, Hazlewood S A, Jones M, Blackmore C G, Hu L F, Rowe M. The 30-base-pair deletion in Chinese variants of the Epstein-Barr virus LMP1 gene is not the major effector of functional differences between variant LMP1 genes in human lymphocytes. J Virol. 1998;72:4038–4048. doi: 10.1128/jvi.72.5.4038-4048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaye K M, Izumi K M, Li H, Johanssen E, Davidson D, Longnecker R, Kieff E. An Epstein-Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J Virol. 1999;73:10525–10530. doi: 10.1128/jvi.73.12.10525-10530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaye K M, Izumi K M, Mosialos G, Kieff E. The Epstein-Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J Virol. 1995;69:675–683. doi: 10.1128/jvi.69.2.675-683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kershaw G R, Berger C, McQuain C, Al Homsi A S, Pihan G, Quesenberry P J, Woda B A, Knecht H. Selective outgrowth of a posttransplant B-immunoblastic lymphoma expressing a latent membrane protein-1 deletion variant. Transplantation. 1997;64:1079–1081. doi: 10.1097/00007890-199710150-00024. [DOI] [PubMed] [Google Scholar]
- 30.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S, editors. Virology. 3rd ed. Philadelphia, Pa: Raven Press, Ltd.; 1996. pp. 2343–2396. [Google Scholar]
- 31.Kieff E, Liebowitz D. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Chanock R M, Hirsh M S, Melnick J L, Monath T P, Roizman B, editors. Virology. 2nd ed. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1889–1920. [Google Scholar]
- 32.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingma D W, Weiss W B, Jaffe E S, Kumar S, Frekko K, Raffeld M. Epstein-Barr virus latent membrane protein-1 oncogene deletions: correlations with malignancy in Epstein-Barr virus-associated lymphoproliferative disorders and malignant lymphomas. Blood. 1996;88:242–251. [PubMed] [Google Scholar]
- 34.Knecht H, Bachmann E, Brousset P, Rothenberger S, Einsele H, Lestou V S, Delsol G, Bachmann F, Ambros P F, Odermatt B F. Mutational hot spots within the carboxy terminal region of the LMP1 oncogene of Epstein-Barr virus are frequent in lymphoproliferative disorders. Oncogene. 1995;10:523–528. [PubMed] [Google Scholar]
- 35.Lee M-A, Diamond M E, Yates J L. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J Virol. 1999;73:2974–2982. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S N, Chang Y S, Liu S T. Effect of a 10-amino acid deletion on the oncogenic activity of latent membrane protein 1 of Epstein-Barr virus. Oncogene. 1996;12:2129–2135. [PubMed] [Google Scholar]
- 37.Liebowitz D, Wang D, Kieff E. Orientation and patching of the latent infection membrane protein encoded by Epstein-Barr virus. J Virol. 1986;58:233–237. doi: 10.1128/jvi.58.1.233-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann K P, Thorley-Lawson D. Posttranslational processing of the Epstein-Barr virus-encoded p63/LMP protein. J Virol. 1987;61:2100–2108. doi: 10.1128/jvi.61.7.2100-2108.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehl A M, Fischer N, Rowe M, Hartmann F, Daus H, Trumper L, Pfreundschuh M, Muller L N, Grasser F A. Isolation and analysis of two strongly transforming isoforms of the Epstein-Barr-virus (EBV)-encoded latent membrane protein-1 (LMP1) from a single Hodgkin’s lymphoma. Int J Cancer. 1998;76:194–200. doi: 10.1002/(sici)1097-0215(19980413)76:2<194::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 41.Menezes J, Liebold W, Klein G, Clements G. Establishment and characterization of an Epstein-Barr virus-(EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- 42.Miller G, Robinson J, Heston L, Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci USA. 1974;71:4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller W E, Mosialos G, Kieff E, Raab-Traub N. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller W E, Cheshire J L, Baldwin A S, Raab-Traub N. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene. 1998;16:1869–1877. doi: 10.1038/sj.onc.1201696. [DOI] [PubMed] [Google Scholar]
- 46.Miller W E, Earp H S, Raab-Traub N. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J Virol. 1995;69:4390–4398. doi: 10.1128/jvi.69.7.4390-4398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller W E, Edwards R H, Walling D M, Raab-Traub N. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J Gen Virol. 1994;75:2729–2740. doi: 10.1099/0022-1317-75-10-2729. . (Erratum, 76:1305, 1995.) [DOI] [PubMed] [Google Scholar]
- 48.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moghaddam A, Rosenzweig M, Lee P D, Annis B, Johnson R P, Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 50.Moorthy R, Thorley-Lawson D A. Processing of the Epstein-Barr virus-encoded latent membrane protein p63/LMP. J Virol. 1990;64:829–837. doi: 10.1128/jvi.64.2.829-837.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moorthy R K, Thorley-Lawson D A. Biochemical, genetic, and functional analyses of the phosphorylation sites on the Epstein-Barr virus-encoded oncogenic latent membrane protein LMP-1. J Virol. 1993;67:2637–2645. doi: 10.1128/jvi.67.5.2637-2645.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moorthy R K, Thorley-Lawson D A. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of Rat-1 fibroblasts. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 54.Natkunam Y, Elenitoba J K, Kingma D W, Kamel O W. Epstein-Barr virus strain type and latent membrane protein 1 gene deletions in lymphomas in patients with rheumatic diseases. Arthritis Rheum. 1997;40:1152–1156. doi: 10.1002/art.1780400621. [DOI] [PubMed] [Google Scholar]
- 55.Odermatt B F, Heitz P U, Bachmann E, Brousset P, Sandvej K, Knecht H. Internal deletions of the latent membrane protein oncogenes of Epstein-Barr virus in Hodgkin’s disease are almost identical with those of Asiatic nasopharyngeal carcinoma. Verh Dtsch Ges Pathol. 1994;78:324–328. [PubMed] [Google Scholar]
- 56.Quintanilla M L, Lome M C, Schwarzmann F, Gredler E, Reyes E, Angeles A A, Fend F. Post-transplantation lymphoproliferative disorders in Mexico: an aggressive clonal disease associated with Epstein-Barr virus type A. Mod Pathol. 1998;11:200–208. [PubMed] [Google Scholar]
- 57.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S, editors. Virology. 3rd ed. Philadelphia, Pa: Raven Press, Ltd.; 1996. pp. 2397–2446. [Google Scholar]
- 58.Robertson E, Kieff E. Reducing the complexity of the transforming Epstein-Barr virus genome to 64 kilobase pairs. J Virol. 1995;69:983–993. doi: 10.1128/jvi.69.2.983-993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robertson E S, Tomkinson B, Kieff E. An Epstein-Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowe M, Peng-Pilon M, Huen D S, Hardy R, Croom-Carter D, Lundgren E, Rickinson A B. Upregulation of bcl-2 by the Epstein-Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NF-κB activation and to induction of cell surface markers. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP-1’s association with TRAF1, TRAF2, and TRAF3. J Virol. 1997;71:4649–4656. doi: 10.1128/jvi.71.6.4649-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sandvej K, Gratama J W, Munch M, Zhou X G, Bolhuis R L, Andresen B S, Gregersen N, Hamilton D S. Sequence analysis of the Epstein-Barr virus (EBV) latent membrane protein-1 gene and promoter region: identification of four variants among wild-type EBV isolates. Blood. 1997;90:323–330. [PubMed] [Google Scholar]
- 63.Sandvej K, Munch M, Hamilton-Dutoit S. Mutations in the Epstein-Barr virus latent membrane protein-1 (BNLF-1) gene in spontaneous lymphoblastoid cell lines: effect on in vitro transformation associated parameters and tumorigenicity in SCID and nude mice. J Clin Pathol. 1996;49:M290–M297. doi: 10.1136/mp.49.5.m290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sandvej K, Peh S C, Andresen B S, Pallesen G. Identification of potential hot spots in the carboxy-terminal part of the Epstein-Barr virus (EBV) BNLF-1 gene in both malignant and benign EBV-associated diseases: high frequency of a 30-bp deletion in Malaysian and Danish peripheral T-cell lymphomas. Blood. 1994;84:4053–4060. [PubMed] [Google Scholar]
- 65.Scheinfeld A G, Nador R G, Cesarman E, Chadburn A, Knowles D M. Epstein-Barr virus latent membrane protein-1 oncogene deletion in post-transplantation lymphoproliferative disorders. Am J Pathol. 1997;151:805–812. [PMC free article] [PubMed] [Google Scholar]
- 66.Shu H B, Takeuchi M, Goeddel D V. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci USA. 1996;93:13973–13978. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takeshita H, Yoshizaki T, Miller W E, Sato H, Furukawa M, Pagano J S, Raab-Traub N. Matrix metalloproteinase 9 expression is induced by Epstein-Barr virus latent membrane protein 1 C-terminal activation regions 1 and 2. J Virol. 1999;73:5548–5555. doi: 10.1128/jvi.73.7.5548-5555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao Q, Robertson K D, Manns A, Hildesheim A, Ambinder R F. Epstein-Barr virus (EBV) in endemic Burkitt’s lymphoma: molecular analysis of primary tumor tissue. Blood. 1998;91:1373–1381. [PubMed] [Google Scholar]
- 70.Tomkinson B, Kieff E. Second-site homologous recombination in Epstein-Barr virus: insertion of type 1 EBNA 3 genes in place of type 2 has no effect on in vitro infection. J Virol. 1992;66:780–789. doi: 10.1128/jvi.66.2.780-789.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. Epstein-Barr virus recombinants from overlapping cosmid fragments. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.VanArsdale T L, VanArsdale S L, Force W R, Walter B N, Mosialos G, Kieff E, Reed J C, Ware C F. Lymphotoxin-beta receptor signaling complex: role of tumor necrosis factor receptor-associated factor 3 recruitment in cell death and activation of nuclear factor κB. Proc Natl Acad Sci USA. 1997;94:2460–2465. doi: 10.1073/pnas.94.6.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 75.Wang D, Liebowitz D, Kieff E. The truncated form of the Epstein-Barr virus latent-infection membrane protein expressed in virus replication does not transform rodent fibroblasts. J Virol. 1988;62:2337–2346. doi: 10.1128/jvi.62.7.2337-2346.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, Springer T, Kieff E. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yalamanchili R, Harada S, Kieff E. The N-terminal half of EBNA2, except for seven prolines, is not essential for primary B-lymphocyte growth transformation. J Virol. 1996;70:2468–2473. doi: 10.1128/jvi.70.4.2468-2473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]