Abstract
Glioblastoma (GBM) is the most prevalent and aggressive malignant primary brain tumor. GBM proximal to the lateral ventricles (LVs) is more aggressive, potentially because of subventricular zone contact. Despite this, cross-talk between GBM and neural stem/progenitor cells (NSC/NPCs) is not well understood. Using cell-specific proteomics, we show that LV-proximal GBM prevents neuronal maturation of NSCs through induction of senescence. In addition, GBM brain tumor–initiating cells (BTICs) increase expression of cathepsin B (CTSB) upon interaction with NPCs. Lentiviral knockdown and recombinant protein experiments reveal that both cell-intrinsic and soluble CTSB promote malignancy-associated phenotypes in BTICs. Soluble CTSB stalls neuronal maturation in NPCs while promoting senescence, providing a link between LV-tumor proximity and neurogenesis disruption. Last, we show LV-proximal CTSB up-regulation in patients, showing the relevance of this cross-talk in human GBM biology. These results demonstrate the value of proteomic analysis in tumor microenvironment research and provide direction for new therapeutic strategies in GBM.
Cell-specific proteomics identifies GBM-derived cathepsin B promoting tumor growth and senescence in SVZ neural progenitor cells.
INTRODUCTION
Glioblastoma (GBM) is the most common and aggressive primary brain tumor in adults (1). The location of GBM contributes to patient outcomes, where tumors in contact with the lateral ventricles (LVs) result in increased tumor expression of stem cell genes, a functionally distinct immune tumor microenvironment, increased incidence of distal recurrence, and decreased median overall survival independent of age and extent of resection (2–7). This may be due to the presence of the subventricular zone (SVZ), the largest neurogenic niche in mammals, which is located along the lateral wall of the LVs (8–10).
The SVZ contains populations of neural stem and progenitor cells (NSC and NPCs, respectively) throughout life, though the human SVZ lacks the migrating neuroblast chains observed in rodent models (9, 10). NSCs extracted from the adult SVZ have been characterized in vitro by their ability to proliferate, self-renew, and differentiate into multiple lineages (11). There is a subpopulation of stem-like GBM cells located throughout the tumor, termed brain tumor–initiating cells (BTICs), with very similar properties to NSCs. BTICs are able to form neurospheres, differentiate into multiple progeny, and share several migratory behaviors and markers with NSCs, with the additional ability to form tumors in vivo (12). The presence of these stem-like cells contributes to tumor progression, therapeutic resistance, and worse patient outcome (13). The biological similarity of these cell types to NSCs supports the idea that stem cell–promoting factors in the SVZ may promote the progression of nearby GBM tumors and that NSCs may play a role in GBM malignancy. Previous work has shown that cell types within the SVZ, both neurogenic and non-neurogenic, are altered in response to LV-associated GBM in rodents (14–16).
Gene expression in both GBM and the SVZ have been well characterized at a transcriptomic level using combinations of bulk and single-cell RNA sequencing (17–24). Despite this, proteomic characterization is still in earlier stages for these niches (25–28), and cell-specific proteomics has not yet been performed to differentiate neurogenic and non-neurogenic proteomes. The proteomic evaluation of NPCs as well as GBM BTICs is incredibly important, as studies have shown that there is considerable disconnect between the transcriptome and translatome in both the neurogenic cell population of the adult SVZ and the neoplastic cells of GBM tumors. In the SVZ, this is due, in large part, to differentiation being posttranscriptionally controlled in NPC and their progeny (29). In GBM, it was concluded that proteomic and phosphoproteomic analysis in addition to transcriptomics is crucial to fully understand malignant pathways at work (28), again pointing to the necessity of advancing proteomic knowledge of this disease.
In a recent study using a rodent model, we have demonstrated that LV-proximal GBM tumors exhibit faster progression compared to their LV-distal counterparts, mirroring the findings observed in patients. In addition, we found that these tumors disrupt SVZ neurogenesis (16). However, it is still unknown what GBM- and NPC-specific mechanisms are at play in this bidirectional cellular interaction. Here, we use a nascent proteomic labeling system via the L274G mutation of methionine tRNA synthetase (MetRS L274G, MetRS*) to investigate cell-specific proteomic changes in BTICs and SVZ NPCs when GBM is located in proximity to the LV. We demonstrate that SVZ NSC/NPC neuronal maturation is suppressed, and senescence is promoted in the presence of nearby GBM tumors, which is partially driven by increased cathepsin B (CTSB) expression in tumor cells, implicating the contribution of this signaling axis to the worse prognosis of LV-proximal GBM.
RESULTS
GBM proximity to the LVs increases tumor malignancy and disrupts normal neurogenic processes
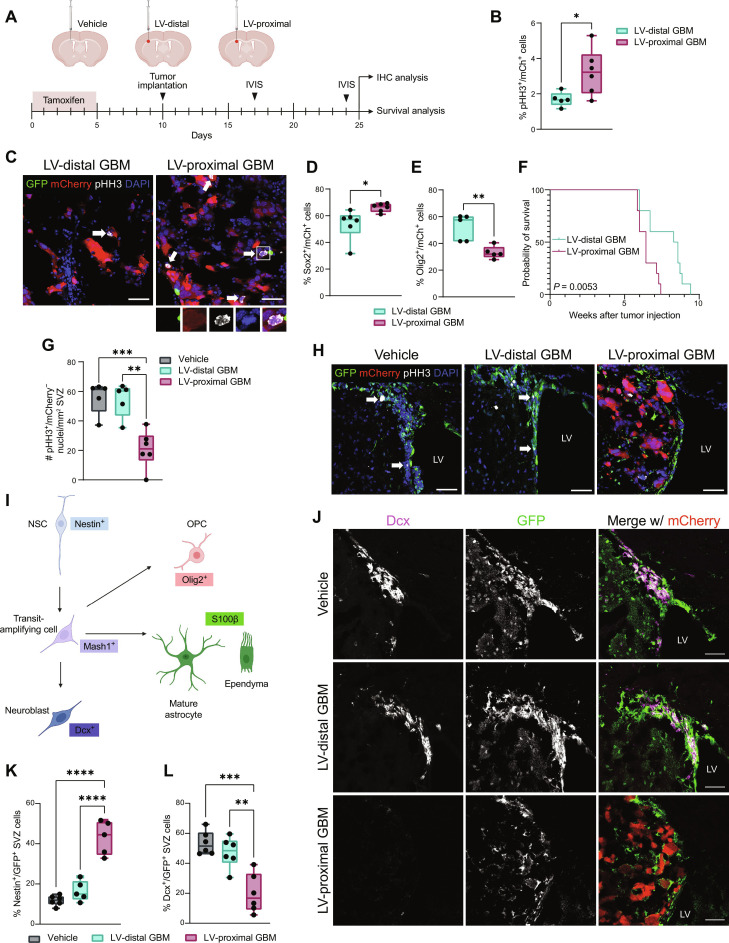
Previous studies have determined that GBM in contact with the LVs is more aggressive than LV-distant counterparts in patients and animal models, potentially because of contact with components of the SVZ (2, 3, 15, 16, 30–34). We first investigated whether GBM tumor malignancy was increased and resulted in altered neurogenesis when tumors were close to the LVs in an immunocompetent rodent model. To this end, we generated transgenic mice to express green fluorescent protein (GFP) and MetRS* under the Nestin promoter with tamoxifen (TAM)–inducible activation of Cre (Nestin-CreERT2; STOPflox R26-GFP MetRS L274G) (Nes-MetRS* mice), allowing for fate tracking of NSCs. Following TAM administration, mCherry and luciferase (mCh-luc)–positive Gl261 murine glioma cells were implanted at locations proximal and distal to the LV for immunohistochemical and survival analysis. An LV-proximal vehicle [phosphate-buffered saline (PBS)]–injected group was included to account for the effects of intracranial injection on the SVZ (Fig. 1A). Within the tumor, there was an increase in the percent of proliferating phospho-histone H3–positive (pHH3+)/mCh+ cells in LV-proximal GBM compared to LV-distal GBM (Fig. 1, B and C). There was also an increased percentage of Sox2+/mCh+ cells (Fig. 1D) and a decrease in the percentage of Olig2+/mCh+ cells (Fig. 1E) in LV-proximal tumors, displaying increased stem cell marker expression. In addition, there was significantly decreased survival in LV-proximal GBM animals compared to LV-distal GBM counterparts (Fig. 1F), indicating that this model captures the worse prognosis of LV-proximal GBM observed in patients.
Fig. 1. Tumor malignancy is increased and SVZ neurogenesis is decreased in LV-proximal GBM-bearing animals.
(A) Experimental groups and timeline. Created with Biorender.com. (B) Percentage quantification of proliferation in murine GBM cells in vivo (n = 5 to 6 biological replicates). Data compared using two-tailed unpaired t test. (C) Representative immunohistochemistry (IHC) showing proliferation in GL261 mCh+ tumors. White arrows and inset indicate proliferating tumor cells. Scale bar, 50 μm. DAPI, 4′,6-diamidino-2-phenylindole. (D) Percentage quantification of Sox2+/mCh+ tumor cells (n = 6 biological replicates). Data compared using two-tailed unpaired t test. (E) Percentage quantification of Olig2+/mCh+ tumor cells (n = 5 biological replicates). Data compared using Mann-Whitney test. (F) Kaplan-Meier curves of survival outcomes for LV-distal GBM and LV-proximal GBM-bearing animals (n = 10 biological replicates). Data compared using Mantel-Cox log-rank test. (G) Quantification of proliferative (pHH3+) cells/mm2 SVZ (n = 5 to 6 biological replicates). Data compared using ordinary one-way analysis of variance (ANOVA) with Tukey multiple comparisons. (H) Representative IHC showing proliferation in the SVZ. White arrows indicate proliferating GFP+ cells. Scale bar, 50 μm. (I) Schematic describing neurogenesis markers used and cell types labeled. Created with Biorender.com. (J) Representative IHC showing Dcx+/GFP+ cells in the SVZ. Scale bar, 50 μm. (K) Percentage quantifications of GFP+ cells in the SVZ that are Nestin+ (n = 5 to 6 biological replicates). Data tested with ordinary one-way ANOVA with Tukey multiple comparisons. (L) Percentage quantifications of GFP+ cells in the SVZ that are Dcx+ (n = 5 to 6 biological replicates). Data tested with ordinary one-way ANOVA with Tukey’s multiple comparison. Data represented as median ± minimum/maximum; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Conversely, proliferation in the SVZ (pHH3+/mCh−) was decreased in the LV-proximal GBM animals compared to LV-distal GBM animals and to LV-proximal vehicle controls (Fig. 1, G and H). We then examined the phenotype of SVZ GFP+ cells across our conditions using well-studied markers of neurogenesis and glial differentiation to determine cell fate when exiting the Nestin+ stem cell state (Fig. 1I). We identified a significant decrease in the percentage of GFP+ cells forming neuroblasts [doublecortin–positive (Dcx+)/GFP+] and a significant increase in the percentage of Nestin+/GFP+ cells in the SVZ of the LV-proximal GBM group (Fig. 1, J to L). There was additionally a decreased percentage of S100b+/GFP+ SVZ cells, although there was no change in Olig2+ or Mash1+ cell populations (fig. S1, E to G). These results indicate reduced SVZ proliferation and arrested NPC as a result of LV-proximal GBM.
To confirm these effects are due to the interaction between GBM and NSC/NPCs rather than an effect from other factors in the tumor microenvironment, we performed treatment of primary human brain tumor-initiating cells (BTICs) with conditioned medium from primary human fetal NPCs (hfNPCs). We identified that treatment with hfNPC-conditioned medium significantly increased GBM BTIC viability over time compared to nontreated (NT) or BTIC-conditioned medium (fig. S1A). For further exploration of the BTIC-hfNPC interaction, BTICs were cocultured with hfNPCs using semipermeable Transwell chamber inserts. Through Ki67 staining, we found that coculture with hfNPCs increased the proliferation percentage of multiple BTIC lines compared to both NT lines and BTIC-BTIC coculture controls (fig. S1B). In addition, hfNPCs induced a significant increase in BTIC migration compared to control conditions via Transwell migration assay (fig. S1C). In the opposite conditions, coculture with BTICs resulted in significantly decreased proliferation of hfNPCs compared to self-coculture or NT groups (fig. S1D). These results indicate that there is a direct interaction between the neurogenic cell population of the SVZ and GBM BTICs that contributes to increased tumor malignancy and decreased neurogenesis.
NSC/NPC-specific proteomics of the SVZ reveals decreased neuronal maturation in the presence of LV-proximal GBM
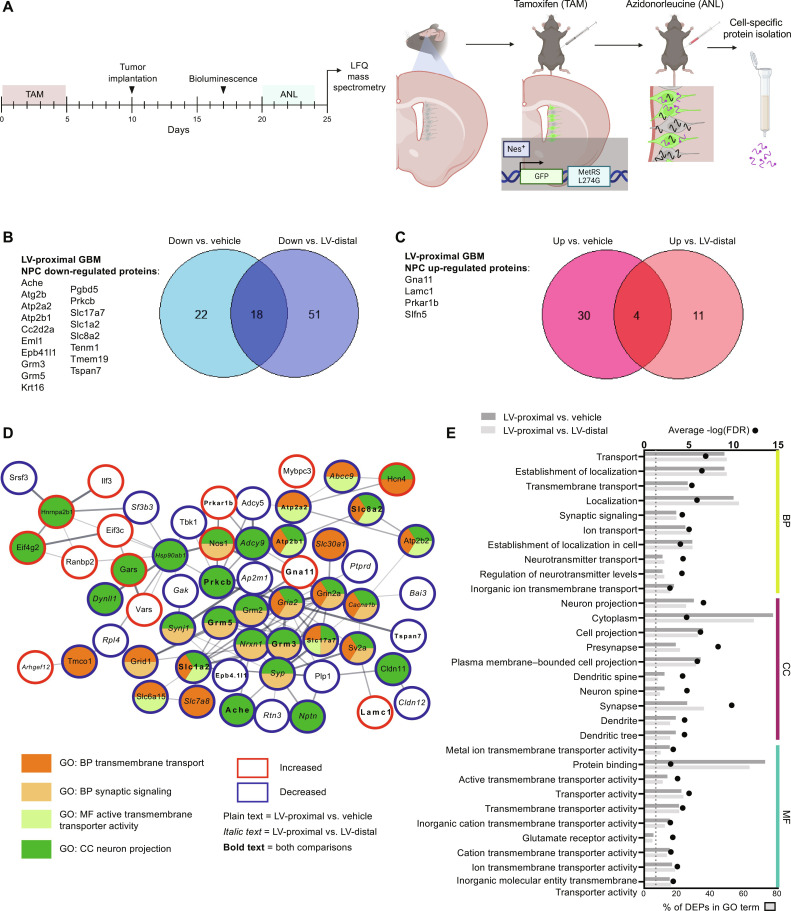
Because of the observed alterations in neurogenesis and NSC cell fate via immunohistochemistry (IHC), we next wanted to examine changes in the NSC/NPC proteome dependent on GBM proximity to the LV using the MetRS L274G nascent protein labeling system. In Nes-MetRS* mice, we first ensured that our Cre driver was correctly inducing GFP expression in Nestin+ cells only following TAM administration. We injected Nes-MetRS* mice with TAM or corn oil intraperitoneal for 5 days and examined brain tissue for GFP expression 2 weeks later. No GFP+ cells were found within corn oil–injected animals, while Nestin+/GFP+ cells were identified at high levels above background within the SVZ upon TAM administration (fig. S2A), as well as in the rostral migratory stream and sparsely in the dentate gyrus of the hippocampus (fig. S2, B and C). We next confirmed that the noncanonical methionine analog, azidonorleucine (ANL), was being incorporated into proteins in NSC/NPC-specific manner. Two weeks after TAM administration in Nes-MetRS* mice, ANL was provided intraperitoneally over 5 days, followed by fluorescent noncanonical amino acid tagging (FUNCAT) analysis of the SVZ. Only ANL-injected animals had substantial FUNCAT signal, which was mainly limited to GFP+ NPCs of the SVZ and the surrounding area (fig. S2D). In addition, via bio-orthogonal noncanonical amino acid tagging (BONCAT) we found that ANL-injected animals with the mutated MetRS enzyme had increased cell-specific metabolic protein labeling over background (fig. S2E) and resulted in successful pulldown of higher levels of clicked, biotinylated proteins in animals induced with TAM and injected with ANL (fig. S2F).
We then activated NSC labeling with TAM and injected animals intracranially with either LV-proximal vehicle, LV-distal GBM, or LV-proximal GBM. ANL was then delivered to animals, and the ipsilateral SVZ was extracted for NSC/NPC-specific label-free quantitative (LFQ) proteomic analysis (Fig. 2A). Through an analysis of five replicates per group, we identified a total of 2669 NPC-specific proteins from the SVZ. After performing enrichment analysis with an analysis of variance (ANOVA) P value cutoff of 0.05, there were 74 differentially expressed proteins (DEPs) in the SVZ of LV-proximal GBM animals compared to LV-proximal vehicle animals and 84 DEPs in LV-proximal GBM SVZ compared to LV-distal GBM SVZ. Of these proteins, there were a total of 22 overlapping DEPs (18 down-regulated and 4 up-regulated) in LV-proximal GBM between the two analyses (Fig. 2, B and C). Of the 18 down-regulated proteins, 12 are related to nervous system development, neuronal signaling, and synaptic function, such as Ache, Grm3, Grm5, Slc1a2 (EAAT2, glutamate transporter 1), and Slc17a7 (VGLUT1). Search Tool for Retrieval of Interacting Genes/Proteins (STRING) analysis and analysis of gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were performed for all DEPs in the two analyses (Fig. 2, D and E, and table S1). Commonly altered terms were largely related to transmembrane transport, synaptic transmission, neural cell projection, and calcium signaling (Fig. 2E), all of which are important for the function of mature neurons (35). This confirms histological findings of decreased neurogenesis and neuroblast differentiation in the presence of an LV-proximal GBM and implicates the identified shared altered proteins in regulating neuronal maturation of NPCs.
Fig. 2. Cell-specific quantitative proteomics of SVZ NPCs reveals decreased expression of neuronal maturation proteins in the presence of LV-proximal glioma.
(A) Schematic illustrating experimental timeline and methodology for Fig. 2. Created with Biorender.com. (B) Venn diagram and list of down-regulated proteins in the SVZ of LV-proximal GBM animals compared to vehicle and LV-distal GBM controls. (C) Venn diagram and list of up-regulated proteins in the SVZ of LV-proximal GBM animals compared to vehicle and LV-distal GBM controls. (D) STRING interaction network for a subset of the LV-proximal GBM-associated DEPs that belong to the indicated GO or KEGG terms or directly interact with proteins belonging to these terms. Node colors indicate protein affiliation with the indicated terms. Plain text indicates a DEP in the LV-proximal GBM versus vehicle interaction. Italic text indicates a DEP in the LV-proximal GBM group versus LV-distal GBM group. Bold text indicates the DEP overlaps between the two interactions. (E) Overlapping GO terms between the two interaction DEPs. BP, biological process; CC, cellular compartment; MF, molecular function.
LV-proximal GBM induces DNA damage–mediated senescence in SVZ NPCs
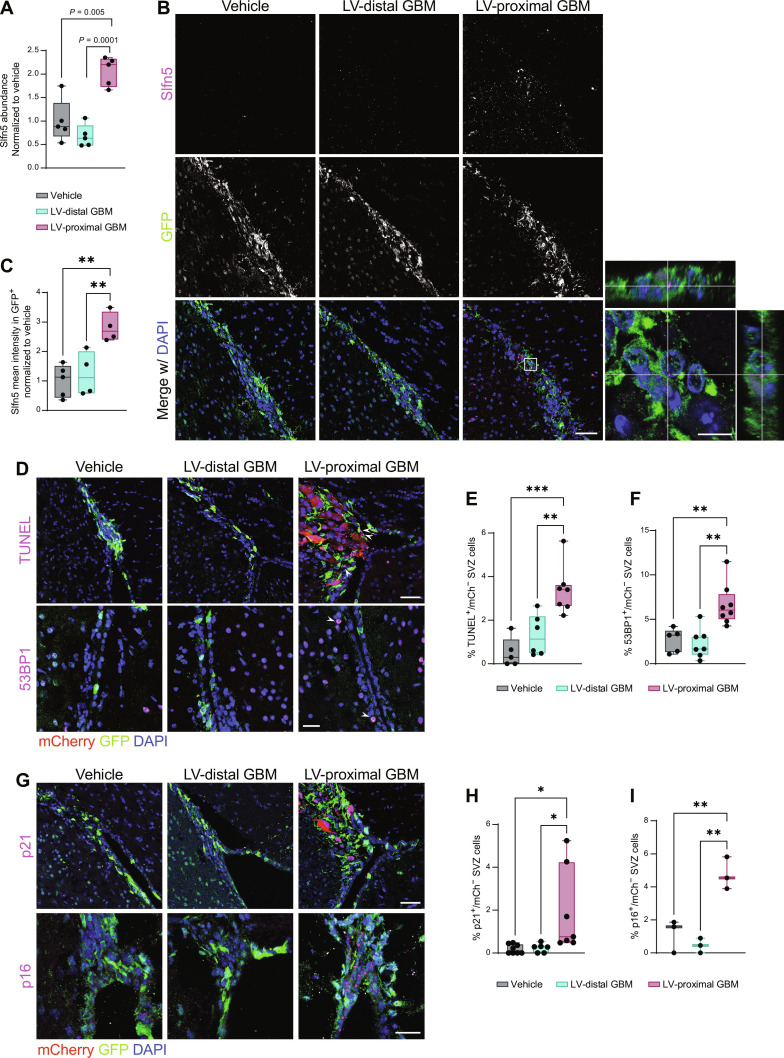
One of the significantly increased proteins in the NPC proteome in response to LV-proximal GBM is schlafen family member 5 (Slfn5; Fig. 3A). To confirm the increased expression of this protein in NSCs/NPCs in the presence of LV-proximal tumors, we probed tissue from Nes-MetRS* mice for Slfn5 expression. Slfn5 was found to be increased in NPCs in the presence of LV-proximal GBM at the tissue level via IHC (Fig. 3, B and C), confirming a cell-specific Slfn5 increase in NPCs and their progeny. Recently, Slfn5 has been identified to participate in reorganizing chromatin to facilitate nonhomologous end joining of double-strand breaks in DNA (36). Slfn5+ puncta were confirmed to lie within nuclei of GFP+ cells of the SVZ (Fig. 3B, inset), suggesting the presence of DNA damage and repair mechanisms at work in the SVZ of LV-proximal GBM-bearing animals. To examine whether LV-proximal GBM induces DNA breakage in the SVZ, we performed a terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay on the Nes-MetRS* brain tissue. We identified that compared to LV-distal GBM or LV-proximal vehicle controls, LV-proximal GBM induced a significant increase in the percent of TUNEL+ cells in the SVZ (Fig. 3, D and E), confirming an accumulation of DNA damage. In addition, cells of the SVZ in LV-proximal GBM-bearing mice had an increased percentage of cells positive for p53 binding protein 1 (53BP1), an additional marker of double-strand DNA breakage (Fig. 3, D to F) (37). The DNA damage response results in halted proliferation while promoting either apoptosis or cellular senescence (38), depending on the severity of the damage and activation of downstream pathways. Our previous work identified no significant changes in apoptosis of SVZ cells in the presence of nearby GBM, despite decreased stem cell number and decreased proliferation (16). We therefore decided to investigate alterations in senescence using the markers p21 and p16, important regulators of the DNA damage response in senescent cells (39, 40). In the Nes-MetRS* mouse model, we identified an increased percentage of p21+ and p16+ SVZ nuclei in the presence of LV-proximal glioma (Fig. 3, G to I). Together, these data indicate that LV-proximal GBM induces increased DNA damage and senescence in NPCs of the SVZ.
Fig. 3. LV-proximal GBM induces increased DNA damage and senescence in SVZ NPCs.
(A) LFQ proteomic results for Slfn5 quantification (n = 5 biological replicates). Normalized to vehicle group. Data were compared with one-way ANOVA with false discovery rate (FDR) correction. (B) Representative immunofluorescent images of increased Slfn5+ in GFP+ cells of the LV-proximal GBM SVZ. Scale bar, 25 μm. White box indicates where inset on right is taken, showing Slfn5+ puncta within GFP+ cells. Scale bar for inset, 10 μm. (C) Slfn5+ mean intensity quantification in GFP+ cells via immunofluorescence (n = 4 to 5 biological replicates). Normalized to vehicle group. Data were compared via ordinary one-way ANOVA with Tukey multiple comparisons. (D) Representative immunofluorescence images of increased TUNEL+ cells (top) and increased 53BP1+ cells (bottom) in LV-proximal GBM SVZ. Scale bar, 50 μm for TUNEL images and 25 μm for 53BP1 images. White arrows indicate DNA damage+/mCh− nuclei. (E) Quantification of percentage of TUNEL+ cells in SVZ (n = 5 to 7 biological replicates). Data were compared via ordinary one-way ANOVA with Tukey multiple comparisons. (F) Quantification of percentage of 53BP1+ cells in SVZ (n = 5 to 7 biological replicates). Data were compared via ordinary one-way ANOVA with Tukey multiple comparisons. (G) Representative immunofluorescence images of increased p21+ cells (top) and p16+ cells (bottom) in LV-proximal GBM SVZ. Scale bar, 50 μm for p21 images and 25 μm for p16 images. (H) Quantification of percentage of p21+ cells in SVZ (n = 5 to 7 biological replicates). Data were compared via ordinary one-way ANOVA with Tukey multiple comparisons. (I) Quantification of percentage of p16+ cells in SVZ (n = 3 biological replicates). Data were compared via ordinary one-way ANOVA with Tukey multiple comparisons. Data represented as median ± minimum/maximum; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
BTICs increase expression of promalignancy proteins, including CTSB, upon coculture with NPCs
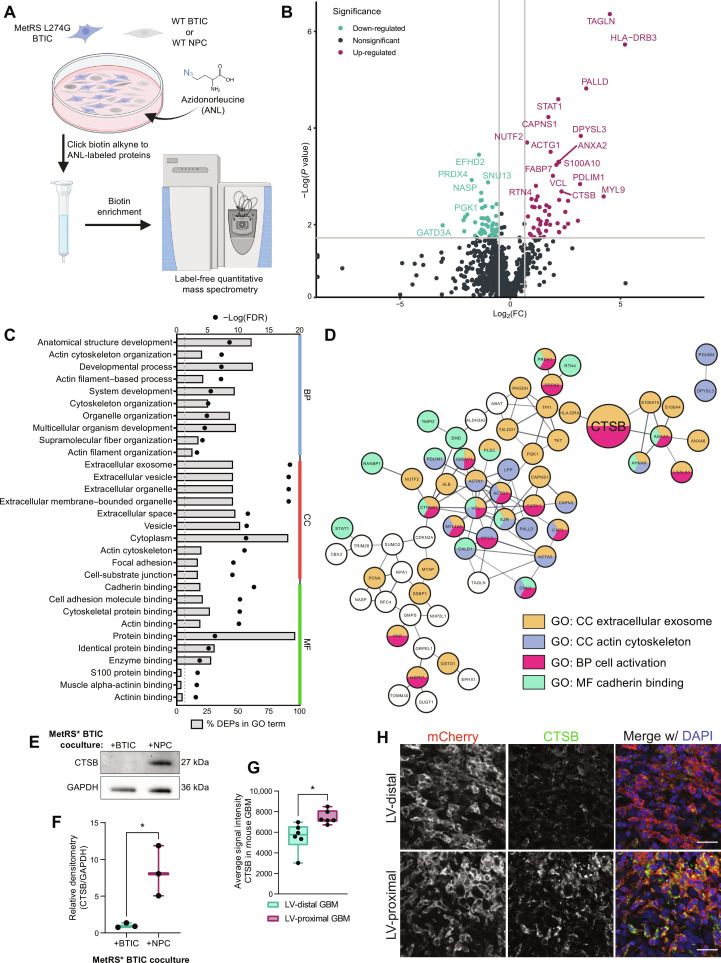
To analyze the direct influence of NPCs on GBM BTIC protein expression in the absence of other brain microenvironmental factors such as cerebrospinal fluid (CSF), we performed cell-specific proteomics of BTICs using the MetRS* labeling system in coculture. Patient-derived BTICs were transduced with lentivirus encoding the mutant MetRS and cocultured with wild-type BTICs or hfNPCs (Fig. 4A). LFQ proteomics of three independent replicates identified 9452 peptides corresponding to 1058 different BTIC-specific labeled proteins. Following enrichment analysis with a cutoff threshold of 1.5 fold change, we identified 54 proteins down-regulated and 52 proteins up-regulated by NPC coculture (Fig. 4B). GO and KEGG analysis of BTIC-specific DEPs revealed strong biological process signatures related to developmental biology and cell migration, including actin cytoskeleton organization, developmental process, actin filament–based process, and system development (Fig. 4, C and D, and table S2). In addition, several of the proteins are related to extracellular cellular compartment terms such as extracellular exosome, extracellular vesicle, and extracellular space (Fig. 4, C and D), implicating changes in intercellular signaling due to coculture with NPCs. The DEP most increased by NPC coculture and that can be both cell-contained and secreted extracellularly in GBM is the lysosomal cysteine protease CTSB. We confirmed proteomic findings of increased CTSB in coculture by performing Western blot on BTIC-specific proteins (Fig. 4, E and F). We also identified increased expression of CTSB in LV-proximal GBM compared to LV-distal counterparts in our rodent model (Fig. 4, G and H). Together, these analyses identify several malignancy- and migration-related proteins up-regulated in BTICs upon exposure to NPC and among these implicates CTSB as a potential target for LV-proximal GBM.
Fig. 4. BTIC coculture with hfNPCs results in GBM-specific increase in the expression of promalignancy proteins including CTSB.
(A) Schematic illustrating coculture setup. Created with Biorender.com. WT, wild-type. (B) Volcano plot of BTIC-specific proteins up-regulated and down-regulated by coculture with NPCs. Gray lines indicate fold change = 1.5 (x axis) and adjusted P value at 0.05 (y axis). (C) Top 10 GO terms in each GO category associated with DEPs from BTIC coculture proteomics. Both the percentage of DEPs (bars) and FDR (points) of terms are included. (D) STRING interaction network for a subset of the NPC coculture associated DEPs that belong to the indicated GO or KEGG terms or directly interact with proteins belonging to these terms. Node colors indicate protein affiliation with the indicated terms. CTSB highlighted by larger size. (E) Representative Western blot for CTSB in BTIC-specific proteins from control coculture and NPC coculture. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is shown as a loading control. (F) Quantification of Western blot densitometry for GBM1A BTIC-specific CTSB normalized to GAPDH (n = 3 biological replicates). Data were compared with two-tailed unpaired t test. (G) Quantification of IHC mean intensity of CTSB within murine GBM (n = 6 biological replicates). Data were compared with two-tailed unpaired t test. (H) Representative IHC of mouse tumors showing increased CTSB in LV-proximal GBM. Scale bar, 25 μm. Data represented as median ± minimum/maximum; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Both cell-intrinsic and soluble CTSB contribute to GBM malignancy
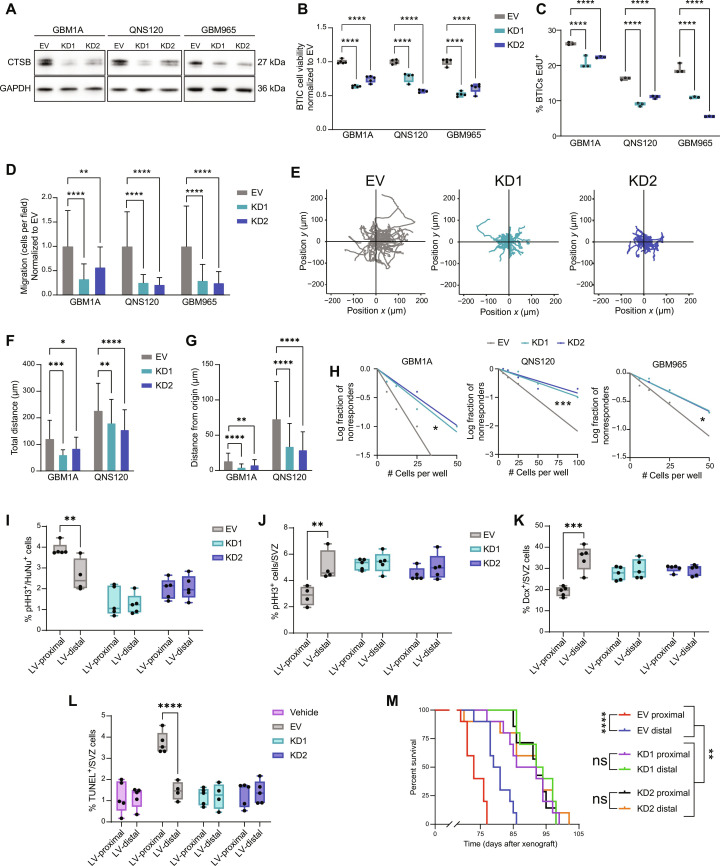
Because of the increase in CTSB expression in BTICs upon coculture with NPCs, we next explored the effects of both cell-intrinsic and secreted CTSB on BTIC biology. To test the effect of endogenous CTSB on patient-derived BTICs, we performed lentiviral knockdown (KD) of CTSB in three BTIC lines using two short hairpin RNA (shRNA) constructs. An empty vector (EV) construct was also used as an experimental control. Following transduction, we confirmed CTSB KD using quantitative polymerase chain reaction (qPCR) and Western blot (Fig. 5A and fig. S3A). We then investigated how CTSB intrinsic expression affects the malignant behavior of BTICs in culture. KD of CTSB decreased viability (Fig. 5B) and proliferation rate (Fig. 5C) compared to EV controls, indicating that CTSB strongly contributes to the cellular growth of human GBM BTICs. In addition, CTSB KDs exhibited decreased Transwell migration (Fig. 5D), total distance migrated, and distance traveled from origin (Fig. 5, E to G) in time-lapse migration studies. In examining stem cell frequency using limiting dilution assay (LDA) to test for self-renewal capacity, we found that CTSB-silenced BTICs had reduced stem cell frequency compared to EV controls (Fig. 5H). These results demonstrate that silencing the expression of endogenous CTSB reduces the stem cell fraction and the proliferation, viability, and migration capacity of GBM cells.
Fig. 5. Cell-intrinsic CTSB contributes to GBM malignancy and LV-proximal GBM phenotypes.
(A) Western blot confirming CTSB KD. (B) BTICs cell viability at 48 hours (n = 5 technical replicates). Normalized to EV. (C) EdU proliferation assay in EV and CTSB KD (n = 3 technical replicates). (D) Transwell migration of EV and CTSB KD. (E) Representative plots of migration from origin in QNS120 EV and CTSB KD. (F) Total distance traveled by EV and CTSB KD BTICs in time-lapse migration. (G) Distance from origin traveled by EV and CTSB KD BTICs in time-lapse migration. (H) LDA self-renewal for EV and CTSB KD. (I) Proliferation in EV and CTSB KD GBM1A tumors proximal or distal to LV (n = 4 to 5 biological replicates). (J) Proliferation in the SVZ in the presence of EV and CTSB KD GBM1A tumors proximal or distal to LV (n = 4 to 5 biological replicates). (K) Quantified Dcx+ neuroblasts in the SVZ in the presence of EV and CTSB KD GBM1A tumors proximal or distal to LV (n = 5 biological replicates). (L) Quantified TUNEL+ cells in the SVZ in the presence of PBS vehicle injection or EV and CTSB KD GBM1A tumors proximal or distal to LV (n = 4 to 5 biological replicates). (M) Kaplan-Meier curves of mice bearing EV or CTSB shRNA GBM1A tumors proximal or distal to LV (n = 7 to 10 biological replicates). Data represented as median ± maximum/minimum (box and whisker) or mean ± SD (bar charts). Data compared with two-way ANOVA, Dunnett multiple comparisons for (B), (C), (D), (F), and (G); Pearson’s chi-square test for (H); two-way ANOVA, Šídák multiple comparisons for (I) to (L); and Mantel-Cox log-rank test accounting for multiple comparisons for (M). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
To examine how tumor-derived CTSB contributes to GBM prognosis, either the EV or CTSB KD BTIC lines were injected at LV-distal and LV-proximal locations in immunocompromised athymic nude mice. While EV control GBM tumors resulted in the previously observed increased tumor proliferation and decreased SVZ proliferation when injected in LV-proximal locations, CTSB KDs did not induce decreased SVZ proliferation and had significantly lower GBM cell proliferation irrespective of injection location (Fig. 5, I and J). CTSB KD in tumors also prevented the loss of Sox2+ stem cells in the SVZ and increase in Sox2+ tumor cells seen in control LV-proximal GBM (fig. S3, B and C). In addition, while LV-proximal EV GBMs resulted in significantly decreased Dcx+ neuroblasts in the SVZ compared to LV-distal EV GBMs, CTSB KD tumors did not significantly alter neuroblast production (Fig. 5K), implicating GBM-derived CTSB in decreased neuronal differentiation. SVZ cell DNA damage was also prevented by CTSB KD, where LV-proximal GBM no longer induced increased TUNEL+ cells in the SVZ (Fig. 5L), suggesting that GBM CTSB may play a role in mediating microenvironmental damage. Within the GBM, CTSB KD tumors had significantly increased TUNEL+/HuNu+ cells, showing increased DNA damage within the tumors themselves (fig. S3D). The silencing of CTSB in BTICs prolonged median survival overall in comparison to EV controls and also abolished the malignancy-promoting effect of LV proximity (Fig. 5M). These results indicate that cell-intrinsic CTSB plays a major role in GBM progression and that the effect of the SVZ on GBM malignancy is mediated in large part by GBM CTSB.
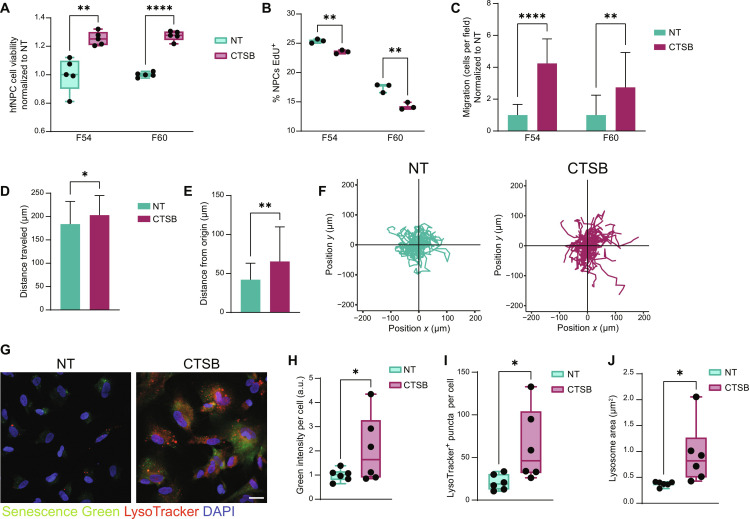
To determine how soluble CTSB affects GBM BTIC biology, we added recombinant human CTSB to BTIC culture media. We determined that the addition of soluble CTSB increased both the viability (fig. S4A) and proliferation rate (fig. S4B) of BTICs. BTIC migration was also increased by soluble CTSB in Transwell (fig. S4C) and time-lapse migration assays (fig. S4, D to F). These results show that secreted CTSB in the microenvironment also contributes to increased BTIC malignancy via autocrine and paracrine signaling.
Soluble CTSB promotes NPC senescence
To evaluate how BTIC-secreted CTSB may contribute to the disruption in neurogenesis, we treated hfNPCs with recombinant CTSB in the culture medium. Although cell viability increased in hfNPCs with the addition of soluble CTSB (Fig. 6A), proliferation rate was decreased (Fig. 6B), matching the observed in vivo phenotype. The migration activity of hfNPCs was also increased in the presence of soluble CTSB as observed in Transwell (Fig. 6C) and time-lapse migration (Fig. 6, D to F), implicating a promigratory role of soluble CTSB on NPCs. Previous studies have identified increased CTSB expression and lysosome number in low-proliferating NSCs compared to high-proliferating progenitor cells (41). In addition, lysosomes are increased in size upon SVZ aging (41), suggesting a potential role of CTSB and lysosomes in increased senescence. Therefore, we next tested whether soluble CTSB contributes to senescence or lysosomal phenotypes in vitro. Treatment with recombinant CTSB resulted in increased β-galactosidase expression in NPCs, identified via Senescence Green assay (Fig. 6, G and H). In addition, we used LysoTracker to quantify the number and size of lysosomes in treated NPCs. Recombinant CTSB resulted in increased lysosome number and size in NPCs (Fig. 6, G, I, and J). These results together suggest that soluble CTSB pushes NPCs toward a more senescent and less proliferative phenotype in vitro, strongly suggesting that GBM CTSB drives this phenotype in the SVZ of LV-proximal GBM-bearing animals.
Fig. 6. Soluble CTSB contributes to decreased proliferation and increased senescence of hfNPCs.
(A) hfNPC cell viability measurements for NT and +CTSB at 48 hours (n = 5 technical replicates). Data were tested with multiple unpaired t tests with Benjamini FDR. (B) Proliferation assay indicating percentage of EdU+ hfNPCs in NT and +CTSB conditions (n = 3 technical replicates). Data were tested with multiple unpaired t tests with Benjamini FDR. (C) Transwell migration of hfNPCs NT and +CTSB. Data were tested with Data were tested with multiple unpaired t tests with Benjamini FDR. (D) Total distance traveled by hfNPCs F54 NT and +CTSB in time-lapse migration. Data were compared with two-tailed unpaired t test. (E) Distance from origin traveled by hfNPCs F54 NT and +CTSB in time-lapse migration. Data were tested with two-tailed unpaired t test. (F) Representative plots of hfNPC migration from origin when NT or +CTSB in line F54. (G) Representative immunofluorescent images of F60 with Senescence Green (β-galactosidase) and LysoTracker labeling in NT or +CTSB conditions. Scale bar, 25 μm. (H) Quantification of Senescence Green levels in F60 NT or +CTSB (n = 6 technical replicates). Normalized to NT. Data were tested with unpaired t test. (I) Quantification of number of LysoTracker+ lysosomes per cell (n = 6 technical replicates). Data were tested with two-tailed unpaired t test. (J) Quantification of LysoTracker puncta area per cell (n = 6 technical replicates). Data were tested with two-tailed unpaired t test. Data represented as median ± maximum/minimum (box and whisker) or mean ± SD (bar charts); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
CTSB plays a role in human patient LV-contacting GBM
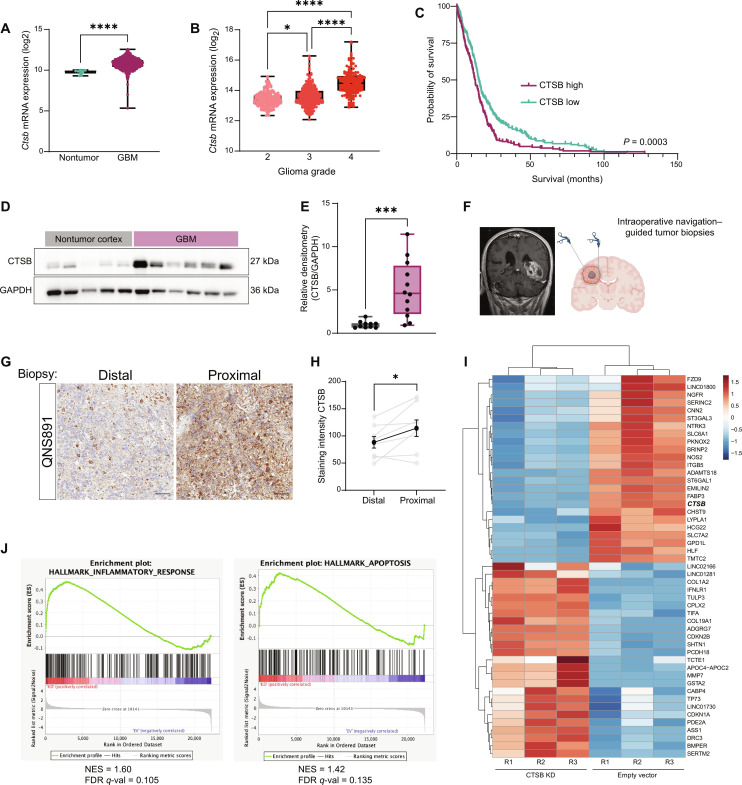
We have thus far shown that the interaction between BTICs and NPCs results in a malignancy-promoting up-regulation of CTSB in culture and in our rodent model. However, because of the decreased frequency of neurogenesis in the adult human SVZ compared to rodents (9, 10), it is unclear whether these findings apply to human cases. To evaluate the contribution of CTSB to glioma patient outcome, we analyzed the The Cancer Genome Atlas (TCGA) GBM and LGG databases using the GlioVis data visualization tool (42). We found that CTSB transcript is increased in GBM tissue compared to normal brain (Fig. 7A). CTSB is also increased by glioma grade (Fig. 7B) and is associated with decreased overall survival in the TCGA GBM cohort (Fig. 7C) at the mRNA level. In addition, we confirmed that CTSB protein is up-regulated in GBM tissues compared to nontumor cortex via Western blot (Fig. 7, D and E). Previous studies have not identified any changes in GBM gene expression related to LV contact at a bulk sequencing level (43), but the heterogeneity inherent to GBM may require analysis of more precise biopsied samples from areas close to the LV or examination at a protein level to identify molecular changes. To determine whether CTSB protein is up-regulated by proximity to the LV, we obtained intraoperative patient-matched surgical navigation–guided biopsies of GBM at locations close and far from the LV in patients with SVZ-contacting tumors (n = 8 patients) (Fig. 7F and table S3). We found that CTSB protein is increased in LV-proximal GBM biopsies compared to the patient-matched LV-distal biopsies (Fig. 7, G and H), confirming that the LV or SVZ microenvironment up-regulates CTSB protein expression in patients with GBM.
Fig. 7. CTSB plays a role in the malignancy of patient LV-contacting GBM.
(A) CTSB gene expression in nontumor and GBM tissues from TCGA database. Data were tested with Mann-Whitney test. (B) CTSB gene expression among different glioma grades from TCGA database. Data were tested with one-way ANOVA with Tukey multiple comparisons. (C) Kaplan-Meier curves of survival outcomes for CTSB-high and CTSB-low GBM. Groups separated at median expression. Data were tested with Mantel-Cox log-rank test. (D) Representative Western blot for CTSB in patient-derived nontumor cortex and GBM tissues. (E) Quantification of Western blot densitometry data from patient tissues (n = 10 to 12 biological replicates). Normalized to nontumor cortex. Data were tested with two-tailed unpaired t test. (F) Schematic outlining collection of matched patient-derived biopsies from LV-contacting tumors. Created in Biorender.com. (G) Representative staining for CTSB in matched patient-derived LV-distal and LV-proximal biopsies for GBM patient QNS891. (H) Quantification of CSTB staining intensity in matched LV-distal and LV-proximal GBM biopsies (n = 8 biological replicates). Data were tested with two-tailed paired t test. (I) Gene heatmap of the 25 most down-regulated and up-regulated genes in GBM1A BTIC CTSB KD compared to EV controls (n = 3 technical replicates). CTSB gene indicated in bold text. (J) GSEA plots for inflammatory response and apoptosis hallmark gene sets indicating increase in CTSB KD. Data represented as median ± maximum/minimum (box and whisker) or mean ± SD (line graph), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine how CTSB gene expression contributes to gene signatures, we performed bulk RNA sequencing on EV versus CTSB KD1 GBM BTICs. A variety of genes were found to be regulated by CTSB expression in BTICs (Fig. 7I), including several previously identified to contribute to BTIC biology. Upon performing gene set enrichment analysis (GSEA) for hallmark gene sets in the Broad Institute Molecular Signatures Database, we observed a strong up-regulation of the inflammatory response and apoptosis gene signatures in CTSB KD compared to control (Fig. 7J).
DISCUSSION
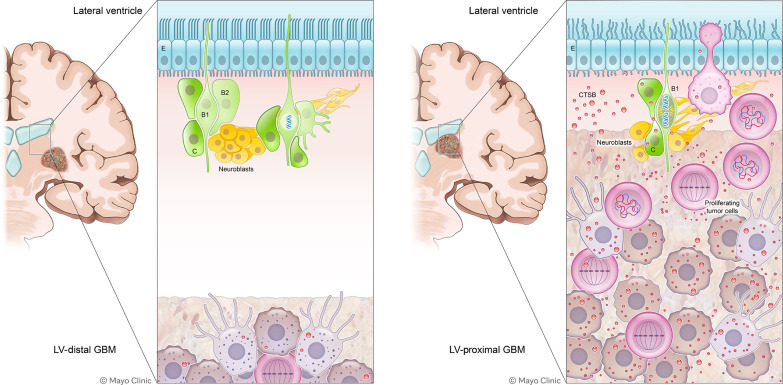
The distinct pathobiology and specific molecular mechanisms driving LV-contacting GBM is of great clinical importance, because of the increased aggressiveness of these tumors resulting in worse patient outcomes. In this study, we determined that proximity of GBM to the SVZ contributes to both increased GBM malignancy and decreased neuronal differentiation in an immunocompetent rodent model. Through cell-specific proteomics, we determined that LV-proximal GBM reduced expression of several proteins related to neuronal maturation and synaptic function in SVZ NPCs while increasing DNA damage and senescence. On the GBM side, coculture with NPCs results in cell-specific up-regulation of CTSB. We have shown that cell-intrinsic CTSB contributes to the malignant phenotype of GBM BTICs in vitro and in vivo, while soluble CTSB also plays a role in promoting BTIC proliferation and migration. Last, we show that soluble CTSB may be one of the molecules stalling NPC differentiation via up-regulating senescence and that CTSB is also an important molecule in LV-contacting GBM in the patient population. Together, these data implicate the bidirectional interaction between NPCs and GBM as an important contributor to tumor malignancy and decreased brain health (Fig. 8).
Fig. 8. A schematic summarizing these findings.
B1 and B2, SVZ neurogenic astrocytes; C, transit-amplifying progenitor; E, ependymal layer.
Our data indicate a contribution of the SVZ to tumor growth and readouts of tumor malignancy. Thus, we believe that the presence of factors from the SVZ contribute to tumor malignancy. However, it may also be possible that the proximity of tumor cells to the SVZ and its contained neurotrophic factors may preferentially select for the growth and expansion of stem-like glioma cells over a more differentiated phenotype. Our current experiments are unable to separate these two phenomena. Discerning between these two scenarios should be a priority for ongoing research on LV-proximal GBMs.
Our data show that exogenous CTSB induces senescence in NPCs, shifting them to a less proliferative and more aged phenotype. Previous work has shown that the CTSB gene is up-regulated in quiescent NSCs compared to activated transit-amplifying cells and their progeny (41). This is likely due to lysosomal CTSB; it was found that quiescent NSCs contained larger lysosomes with protein aggregates compared to activated NSCs, which were even further increased in size upon aging (41). The expression of lysosomal biosynthesis regulators contributes to maintaining the stemness of NPCs, where KD of these genes or lysosomal transporters results in premature neuronal differentiation (44). When we treated NPCs with soluble CTSB, we observed an increase in the number and size of the lysosomes alongside increasing senescence. This is an interesting area for further study, as it is still unknown how exposure to soluble CTSB affects lysosomal dynamics in NPCs, or whether soluble CTSB is internalized in these cells. Furthermore, it remains to be studied how senescence in NPCs may alter functional neurogenesis outcomes, such as neuroblast migration down the rostral migratory stream and neuronal maturation in the olfactory bulb. These areas are important areas of future study to fully understand the effect of CTSB on NPC biology.
Our data show that glioma tumor proximity to the SVZ results in the down-regulation of several proteins involved in neuronal maturation and synaptic function. This is interesting, as it shows that the endogenous NPCs of the SVZ likely do not contribute to the newly found neuron-glioma synapses in the field of cancer neuroscience (45, 46). This could be due to two potential explanations; either these neuron-glioma synapses are less important in LV-proximal GBM or these tumors receive neuronal input from other pre-existing neurons in the nearby striatum while maintaining a growth factor–rich niche in the presence of undifferentiated NSCs. Future work on the influence of cancer neuroscience on LV-proximal GBM will elucidate which of these hypotheses is true in these tumors.
Our work identifies CTSB as an important malignancy-promoting factor in LV-proximal GBM and that it is increased in tumor cells upon exposure to NPCs. It currently remains unknown what factor from NPCs induces the up-regulation of CTSB in GBM cells and whether this is dependent on cell-cell contact or is modulated by a secreted factor. Furthermore, the downstream effect of soluble CTSB on NPCs and the pathway through which it acts needs further exploration. The identification of the NPC-derived factor and how CTSB regulates NPC biological pathways need additional studies to fully understand.
Previously, CTSB expression in GBM cell lines has been closely linked to tumorigenic potential, invasiveness, and radioresistance (47–51). These studies, however, have largely been performed using commercial cell lines and do not explore these findings in the context of patient heterogeneity. Here, we confirm the importance of CTSB in several low-passage patient-derived BTICs and show that the cell-intrinsic expression of this molecule is important in the proliferation, invasiveness, and stemness properties of these cells. The contribution of CTSB to radioresistance in patient-derived lines should be further explored in future studies, as this would make this molecule an excellent target for new therapeutics. In addition to the cell-intrinsic contribution, we found that soluble CTSB also contributes to GBM cell growth and migration, suggesting additional malignancy-promoting properties of autocrine and paracrine signaling. CTSB can be released into the tumor microenvironment by a variety of cell types including endothelial cells, fibroblasts, and tumor-associated macrophages and myeloid-derived suppressor cells (52, 53). Additional cell-specific proteomic studies on other cell types of the tumor microenvironment will be essential for elucidating the role of soluble CTSB, as well as other molecules, on GBM progression.
Though this study focuses on the cross-talk between neoplastic cells and their microenvironment, there is no perfect in vitro or in vivo model to study this aspect of GBM. Therefore, we use several systems to confirm our findings. Our in vitro studies use coculture between human BTICs and hfNPCs, but the interaction between these two cell types may be further mediated by a third cell type such as tumor-associated macrophages, or a noncellular source of factors such as the CSF. In our transgenic model for MetRS* studies, we use an immunocompetent rodent with a syngeneic murine glioma line. Although Gl261 recapitulates several aspects of human GBM (54), it is a commercial cell line and lacks the classic heterogeneity that is characteristic of the human disease. Therefore, in our KD experiments, we implant patient-derived BTICs in immunosuppressed mice. This allows for a more heterogeneous human glioma model, but these animals lack the functional immune system that further contributes to disease progression (55–57). Currently, studying the tumor microenvironment in GBM necessitates the use of multiple models and cross-checks, as well as including patient samples to verify implicated pathways. Here, we have combined the use of in vitro culture systems, immunodeficient and immunocompetent murine models, and patient samples to fully implicate the CTSB and senescence pathways in LV-proximal GBM. Hopefully, with the technological and methodological growth of the field, such as the development of organoid coculture (58, 59) and three-dimensional biomimetic culture systems (60, 61), the simulated interaction of tumor cells with their microenvironment will more closely model the human condition.
Here, we use a nascent proteomic labeling system, MetRS L274G in the presence of ANL, to explore the proteomic contributions in the cross-talk between GBM and NPCs. Although this system has been used in other areas of neuroscience (62–64), cancer biology (65), and stem cell biology (66–68), neurogenesis and brain tumor cell–specific nascent proteomics has been relatively unexplored. Our approach to this system can now be used to profile GBM proteomic dynamics in vivo and can be more widely applied to other diseases and microenvironmental niches. For example, NSCs play a role in other disorders such as neurodegenerative disease and stroke (69–71), while GBM makes use of other malignancy-promoting niches such as the perivascular niche (72–74). Proteomic characterization in these cell populations is extremely important because of posttranscriptional regulation of stem cell fate and low transcript-protein correlation (28, 29, 75), meaning that protein-based signaling analysis will give a more accurate picture of the biological pathways at play. Using the MetRS L274G system or other cell-specific proteomic isolation methods to further illuminate the proteomic dynamics of GBM-niche cross-talk and SVZ neurogenesis will lead to discoveries in the field and, potentially, the development of new therapeutic strategies.
This study is based on the concept of interaction of NSC/NPCs with GBM in the human brain. However, it is well known that neurogenesis is markedly decreased in the adult human brain, with only populations of quiescent NSCs located in the SVZ and remarkably little new neuron generation (76). Despite a strong decline in cell proliferation and differentiation with aging, functional NSCs are still able to be isolated from the SVZ of adult and aged humans, both from intraoperative specimens and postmortem tissue (10, 19, 77–81), indicating a persistence of NSCs throughout life. That being said, the decreased neurogenic nature of these cells makes exploring other cell types of the SVZ, such as the ependymal layer, important in fully defining the bidirectional interaction between the SVZ and GBM. In the human brain, LV-proximal GBM would likely be interacting with a mix of quiescent and senescent NSC/NPCs, based on the median age of diagnosis of 65 years (1) and the increase of senescence with aging. Pushing this NSC population more toward senescence over quiescence may have severe consequences for patient outcome. For one, senescent NPCs would be highly unlikely to successfully re-enter the cell cycle (82), making brain repair following an injury such as tumor resection more challenging. In addition, many senescence-associated secretory phenotype factors contribute to glioma growth and recurrence, including interleukin-6 (IL-6), IL-8, and hepatocyte growth factor (HGF) (83–85). An accumulation of senescent cells in the nearby SVZ may therefore further promote GBM growth and prevent brain healing.
In our study, we use primary cultured GBM-derived BTICs and expose them to hfNPCs. Ideally, this interaction should be assessed between BTICs and NPCs derived from adult humans in an experimental setup. However, obtaining viable brain samples from the human SVZ or dentate gyrus, sourced from noncancer patients, is not a routine procedure in surgery. Therefore, using primary cultured cells for this study is practically unattainable. An alternative approach could entail using induced pluripotent stem cells (iPSC)–derived NPCs, which offers a viable solution. Nevertheless, before deeming them a suitable substitute, it is imperative to thoroughly examine the added complexity and artificial nature inherent in iPSC cultures. This includes evaluating their ability to accurately replicate the biology of adult NPCs, which warrants comprehensive investigation.
On the basis of our results, we propose that NSC/NPCs contribute to GBM progression. This has some distressing implications for current clinical trials using human NSCs for therapeutic delivery in patients with GBM. Because of the homing nature of NSCs to tumor masses in vivo, several preclinical and clinical studies have used NSCs in stem cell therapies as delivery systems for oncolytic virus or as mediators of enzyme/prodrug therapies (86–89). The clinical trials derived from these studies are still in early phases, though phase 1 results suggest no safety concerns with NSC-based treatment. If these treatments are successful, then careful screening of individual NSC lines will need to be performed to ensure that each line used is not secreting large amounts of stemness-promoting growth factors and that these lines do not result in CTSB up-regulation in the patient glioma, which would then lead to downstream resistance to standard-of-care therapy.
Ultimately, our results indicate that there is malignancy-promoting cross-talk between GBM and the SVZ that disrupts normal neurogenesis through activation of senescence while promoting tumor progression through CTSB up-regulation. Targeting this pathway using CTSB inhibitors may be a useful therapeutic approach to treat patients with these tumors. The future use of nascent proteomic labeling methods to perform cell-specific research in the tumor microenvironment will give substantial insight into the biology of GBM.
MATERIALS AND METHODS
Experimental animals
Animal experiments were fully approved by the Mayo Clinic Institutional Animal Care and Use Committee (protocols A00002260-16 and A00004969-19). Mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited facility abiding by all federal and local regulations. For NSC proteomic labeling experiments, C57BL6/J Nestin-Cre/ERT2 transgenic mice (Nes-cre/ERT2, the Jackson Laboratory, no. 016261) (90, 91) were crossed with STOPflox R26-MetRS L274G transgenic mice (STOPflox R26-MetRS*, the Jackson Laboratory, no. 028071) (92). Animals were backcrossed in a C57BL6 background. Genotypes used for all NSC labeling experiments were NestinCreERT2/+; MetRSL274G/L274G.
Cre activity for NSC proteomic labeling was induced via TAM (Sigma-Aldrich no. T5648). TAM dissolved in sterile corn oil was administered intraperitoneally at a dosage of 140-mg TAM/kg body weight per day over a period of 5 days. Animals were monitored daily until the end of study for changes in weight and physical signs of toxicity or distress.
Cell culture
All cell culture was performed in sterile conditions using aseptic technique. Cells were monitored for mycoplasma contamination once per month and were discarded if positive. Syngeneic murine glioma line 261 (GL261) was cultured in adherent conditions in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 200 μM l-glutamine (GlutaMAX, Gibco), antibiotic-antimycotic solution, and 10% fetal bovine serum (FBS). Patient-derived BTICs were isolated and cultured as previously described by our group (93). Briefly, BTICs were obtained from freshly dissociated patient GBM tumors, and cells were cultured in suspension with complete media consisting of DMEM/F12 (Gibco) supplemented with antibiotic-antimycotic solution, B-27, epidermal growth factor (EGF; 20 ng/ml; PeproTech), and fibroblast growth factor (FGF; 20 ng/ml; PeproTech). BTIC line GBM1A was originally established as line 0913 by Vescovi and co-workers (94). Cells were passaged at least three times and characterized for self-renewal, differentiation capacity, and tumor formation capacity before being characterized as BTICs. This method of culture has been shown to successfully recapitulate several aspects of the original patient tumor (93). Table 1 shows patient characteristics and references for each BTIC line. Primary lines of hfNPCs were previously obtained at the Johns Hopkins University from freshly dissociated elective abortion fetal brain tissue following informed consent as described (95). hfNPCs were cultured in complete media of DMEM/F12 supplemented with antibiotic-antimycotic solution, B-27, EGF, FGF, leukemia inhibitory factor (10 ng/ml; Millipore), and heparin (5 μg/ml). BTICs and hfNPCs were cultured as neurospheres in suspension until needed for experimentation. All cells were used between 3 and 14 passages from patient isolation for experiments. For adherent experiments, tissue culture flasks were coated with laminin (10 μg/ml in PBS) for 2 hours at 37°C before plating cells.
Table 1. BTIC cell line patient characteristics.
Table of BTIC lines used in study with accompanying patient age at collection, biological sex, molecular subtype, methylation subtype, copy number variation (CNV) gains, CNV losses, MGMT methylation, and references for previous publication.
| BTIC line | Patient age at surgery | Biological sex | Molecular subtype | Methylation subtype | CNV gains | CNV losses | MGMT methylated | Reference |
|---|---|---|---|---|---|---|---|---|
| GBM1A | – | M | Classical/Proneural | Indeterminate | MDM4, MYCN, TERT, EGFR, CDK6, MET, KIAA1549/BRAF | RB1, TP53 | Y | (32, 94) |
| QNS120 | 59 | M | Classical | RTKII | FGFR3/TACC3, PDGFRA, TERT, MET, MYBL1, CCND2 | MDMA, MYB, CDKN2A/B, PTCH1, PTEN, MGMT, CCND1, NF1, C19MC | Y | (93) |
| GBM965 | 61 | F | Classical | RTKII | EGFR, CDK6, MET, KIAA1549/BRAF | CDKN2A/B, NF1 | Y | (32, 114) |
Intracranial tumor implantation
For NSC proteomic labeling experiments in mice of BL6 background, GL261 cells were transduced via lentivirus to stably express CMV-mCherry and CMV-luciferase PGK-puromycin (96) (mCh-luc). pLV-mCherry was a gift from Pantelis Tsoulfas (Addgene plasmid no. 36084; http://n2t.net/addgene:36084; RRID: Addgene_36084). pLenti CMV Puro LUC (w168-1) was a gift from E. Campeau and P. Kaufman (Addgene plasmid no. 17477; http://n2t.net/addgene:17477; RRID: Addgene_17477). For human BTIC injection into athymic nude mice, cells were transduced to express PGK-GFP-IRES-LUC-W (GFP-luc) (97). pHAGE PGK-GFP-IRES-LUC-W was a gift from D. Kotton (Addgene plasmid no. 46793; http://n2t.net/addgene:46793; RRID: Addgene_46793). Following transduction, intracranial implantation of cells was performed. Briefly, mice were placed under anesthesia and fitted into a stereotactic frame. For murine glioma experiments, 5.0 × 104 GL261 mCh-luc+ cells in a volume of 1 μl of PBS were injected into the right brain hemisphere. For human BTIC experiments, 3.5 × 105 GFP-luc+ BTICs were implanted in 2 μl of PBS. Injection in LV-proximal and LV-distal locations was performed as previously reported (16) but with a slightly more lateral LV-proximal injection. In millimeters relative to bregma, LV-proximal injections were anterior-posterior (AP): 1.0, lateral (L):1.35, and dorsal (D): 2.3, while LV-distal locations were AP: 1.0, L: 2.1, and D: 2.3. Vehicle-injected mice (PBS only) at the LV-proximal location were included to account for NSC response to the intracranial injection. Tumor engraftment was monitored with bioluminescence following intraperitoneal injection of luciferin. For lysate preparation, mice were intracardially perfused with 50 ml of ice-cold 0.9% saline with heparin (0.5 mg/ml). For IHC, animals were perfused with saline with heparin and then 4% paraformaldehyde (PFA) in PBS at study end point. For survival experiments, mice were maintained following GBM xenograft until reaching humane end point criteria defined as weight loss greater than or equal to 20% of body weight, inability to ambulate, inability to reach food or water, ataxia, paraplegia, inability to right oneself, or a body condition score of 1 or less using the Institutional Animal Care and Use Committee–approved scoring system.
ANL administration
For animal experiments, ANL (Iris Biotech) was fully dissolved in sterile 0.9% saline solution. NaOH was added to pH = 7.4 and the ANL solution was filtered through a 0.22-μm-pore syringe filter. ANL was administered intraperitoneally to mice at a dosage of 200 mg/kg per day over 5 days. Nonlabeled controls for NSC labeling experiments consisted of littermates that received only corn oil vehicle and ANL. For cell culture experiments, ANL was prepared in sterile DMEM/F12 to a 4 mM concentration of pH = 7.0.
Immunohistochemistry
For IHC analysis, paraffin-embedded brain sections (10 μm) were deparaffinized and rehydrated followed by antigen retrieval with sodium citrate buffer (10 mM, pH = 6.0). For fluorescent IHC, rehydrated sections or thawed frozen sections were washed, permeabilized, blocked with 10% normal goat serum, and incubated with appropriate primary antibodies at the dilutions described in Table 2 overnight at 4°C. The next day, sections were washed and incubated in secondary antibodies diluted in blocking solution (1:500) for 1 hour at room temperature. Sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) before coverslipping. For immunocytochemistry (ICC), cells were washed twice, fixed with 4% PFA for 15 min, and then went through the same staining protocol beginning at the permeabilization step. Preparations were imaged using an LSM 880 confocal microscope (Zeiss).
Table 2. Primary antibodies used in study.
Table of primary antibodies used in the study with vendor, catalog number, host species, experimental use, and dilutions.
| Target | Manufacturer and catalog number | Host species | Use | Dilution |
|---|---|---|---|---|
| CTSB | Cell Signaling Technology no. 31718S | Rabbit | IHC and Western | 1:100–1:1000 |
| Dcx | Cell Signaling Technology no. 4604S | Rabbit | IHC | 1:500 |
| GAPDH | Santa Cruz Biotechnology no. sc-47724 | Mouse | Western | 1:5000 |
| GFP | Aves Labs no. GFP1010 | Chicken | IHC | 1:500 |
| Thermo Fisher Scientific no. A-11122 | Rabbit | IHC | 1:500–1:1000 | |
| Ki67 | Thermo Fisher Scientific no. MA5-14520 | Rabbit | IHC | 1:150 |
| Mash1 | BD Pharminigen no. 556604 | Mouse | IHC | 1:100 |
| Nestin | BD Pharminigen no. 556309 | Mouse | IHC | 1:100 |
| Olig2 | MilliporeSigma no. AB9610 | Rabbit | IHC | 1:500 |
| p16 | Abcam no. ab241543 | Rat | IHC | 1:250 |
| p21 | Abcam no. ab188224 | Rabbit | IHC | 1:100 |
| 53BP1 | Novus Biologicals no. NB100-304 | Rabbit | IHC | 1:1000 |
| pHH3 | Cell Signaling Technology no. 9701S | Rabbit | IHC | 1:250 |
| S100β | Abcam no. ab52642 | Rabbit | IHC | 1:100 |
| Slfn5 | Atlas Antibodies no. HPA017760 | Rabbit | IHC | 1:100 |
Cell viability assay
Cells were plated in a black 96-well plate with a clear bottom and allowed to attach in complete media overnight. The following day, media was exchanged to base medium without growth factors. AlamarBlue viability reagent (Invitrogen) was added to total 10% of well volume. Fluorescence was measured with excitation/emission spectra 540/600. Viability was measured at 0, 4, 24, 48, and 72 hours following addition of reagent.
Transwell chamber coculture
Transwell chamber inserts with 0.4-μm pores (Corning) in either a 6-well plate or a 24-well plate format were used to coculture BTICs with hfNPCs. Cells were first allowed to adhere to the bottom of cell culture plates overnight in complete media. The next day, media was replaced with base media lacking growth factors, and the Transwell inserts were placed on top. Cells to be cocultured were placed into the top chamber in base media. Cells were incubated together for 48 hours before appropriate analysis.
Transwell migration
Transwell chamber inserts with 8.0-μm pores (Corning) in a 24-well plate format were used to analyze migration. On the basis of optimization experiments, 4.0 × 104 cells were placed into the top chamber. To ensure migration, an FBS gradient of 2.5% in the bottom chamber (500 μl) and 0.5% in the top chamber (250 μl) was established in media in the absence of added growth factors. Plates were incubated for 24 hours before processing. Cells remaining in the top chamber were removed with a cotton swab, while cells that successfully migrated to the bottom of the membrane were fixed and labeled with DAPI before being imaged at nine fields per membrane at a ×10 magnification using a fluorescence microscope. Cells per field were quantified using ImageJ software.
FUNCAT of newly synthesized proteins in vivo
FUNCAT labeling in tissue sections was performed according to previously published protocols (98). Briefly, slides were washed and blocked as described for IHC. ANL-labeled proteins were clicked by mixing 200 μM triazole ligand (TBTA), 500 μM TCEP, 5 μM fluorescent alkyne tag, and 200 μM CuSO4 in PBS and adding to sections for 3.5 hours at room temperature under gentle rotation. Slides were washed twice with PBS containing 1% Tween 20 and 500 μM EDTA for 15 min, followed by two washes with PBS containing 0.1% Triton X-100. Slides were then counterstained using DAPI or underwent IHC staining beginning from the blocking step.
Lysate preparation
For NSC MetRS* experiments, the ipsilateral SVZ to intracranial injection site was dissected out of the brain, and wet tissue weight was measured. Tubes containing tissue were snap frozen in dry ice and stored at −80°C until lysate extraction. Tissue was manually homogenized in 12× volume of lysis buffer consisting of PBS with 1% SDS, 1% Triton X-100, 1× protease inhibitors, and Benzonase (≥250 U/ml). Lysates were sonicated and incubated at room temperature for 30 min to allow for complete lysis before centrifuging at 16,000g and retaining supernatant for downstream analysis.
For cell culture experiments, lysis buffer was added to flasks containing cells, which were then scraped and placed into tubes on ice. Lysates were sonicated and incubated on ice for 15 min with intermittent vortexing before centrifuging at 16,000g and retaining supernatant as whole-cell lysate.
BONCAT and purification of cell-specific nascent proteome
For gel imaging, 100 μg of tissue lysate was first reduced with 25 mM dithiothreitol (DTT) at 80°C for 15 min. Following reduction, lysates were alkylated with 50 mM iodoacetamide (Thermo Fisher Scientific) at room temperature for 1 hour and 30 min rotating at 1000 rpm and protected from light. This alkylation step was repeated for a total of two times. Proteins were then clicked to 10 μM tetramethylrhodamine-diarylcyclooctyne moiety (TAMRA-DBCO) using strain-promoted click chemistry for 1 hour at room temperature protected from light. Excess DBCO reagent was removed using Amicon Ultra Centrifugal Columns with a 3-kDa cutoff (Millipore). A volume of 30 μl of resulting solution was run on an SDS–polyacrylamide gel electrophoresis gel and imaged using the ChemiDoc MP System (Bio-Rad). Imaging was followed by Coomassie Blue staining for total protein quantification.
For purification of ANL-labeled proteins, 1 mg of proteins were first reduced and alkylated as described for gel imaging. Proteins were then precipitated in ice-cold acetone and redissolved in fresh lysate buffer. Following protein concentration quantification by bicinchoninic acid assay (BCA), lysates were clicked to 10 μM DBCO-S-S-PEG3-biotin (BroadPharm) for 6 hours at room temperature. Excess DBCO reagent was removed with PD G-25 SpinTrap columns (Cytiva), and proteins were incubated with Streptavidin Sepharose High Performance affinity resin (Cytiva) for 16 hours at 4°C in rotation. Resin was washed three times with 1% Triton X-100 and 0.15% SDS in 1× PBS, three times with 1% Triton X-100 and 0.2% SDS in 1× PBS, and three times with 1× PBS. Proteins were eluted from beads using 50 mM DTT and 0.1% SDS in 1× PBS over 4 hours at room temperature.
LFQ mass spectrometry sample preparation
LFQ sample preparation was carried out similar to that previously described (99) but with slight updated modification. The submitted samples were brought up to 100 μl with water. The proteins were precipitated using an acetone precipitation procedure. Protein pellet was air dried, dissolved in 10 μl of 8 M urea/0.4 M ammonium bicarbonate, reduced with 1 μl of 45 mM DTT at 37°C for 30 min, and subsequently alkylated with 1 μl of 100 mM iodoacetamide at room temperature for 30 min in the dark. The solution was then diluted with 27 μl of water and digested with 1 μl of trypsin (0.5 μg/μl) at 37°C overnight. The tryptic digestion was quenched by acidifying with 2 μl of 20% trifluoroacetic acid (TFA). The peptide solution was then desalted using a mini Reverse Phase (RP) C18 desalting column (The Nest Group, Ipwich, MA). The eluted peptides were then dried in a SpeedVac and stored at −80°C. The dried samples were then redissolved in 5 μl of 70% formic acid (FA) and 35 μl of 0.1% TFA. An aliquot was taken to obtain total digested protein amount. A 1:10 dilution of Pierce Retention Time Calibration Mixture (catalog no. 88321, Thermo Fisher Scientific, Waltham, MA) was added to each sample before injecting onto the ultraperformance liquid chromatography (UPLC)–coupled Q-Exactive Plus mass spectrometer system for normalization of the LFQ data.
LFQ data collection
Data-dependent acquisition liquid chromatography with tandem mass spectrometry (LC-MS/MS) data collection for LFQ was performed on a Thermo Fisher Scientific Q-Exactive Plus mass spectrometer connected to a Waters nanoACQUITY UPLC system equipped with a Waters Symmetry C18 180 μm by 20 mm trap column and a 1.7-μm, 75 μm by 250 mm nanoACQUITY UPLC column (35°C). The digests were diluted to 0.05 μg/μl with 0.1% TFA before injecting 5 μl of each triplicate analysis in block-randomized order. To ensure a high level of identification and quantitation integrity, a resolution of 120,000 and 30,000 was used for MS and MS/MS data collection, respectively. MS and MS/MS (from higher-energy C-trap dissociation) spectra were acquired using a 3-s cycle time with dynamic exclusion on. All MS (profile) and MS/MS (centroid) peaks were detected in the Orbitrap. Trapping was carried out for 3 min at 5 μl/min in 99% buffer A (0.1% FA in water) and 1% buffer B [0.075% FA in acetonitrile (ACN)] before eluting with linear gradients that reach 25% B at 150 min, 50% B at 170 min, and 85% B at 175 min; then back down to 3% at 182 min. Two blanks (first 100% ACN, second buffer A) followed each injection to ensure there was no sample carry over.
Proteomic data analysis
The collected LC-MS/MS LFQ data were processed with Progenesis QI software (Nonlinear Dynamics, version.4.2) with protein identification carried out using the Mascot search algorithm (Matrix Science, v. 2.7). The Progenesis QI software performs feature/peptide extraction, chromatographic/spectral alignment (one run was chosen as a reference for alignment), data filtering, and quantitation of peptides and proteins. A normalization factor for each run was calculated to account for differences in sample load between injections as well as differences in ionization. The normalization factor was determined by comparing the abundance of the spike in Pierce Retention Time Calibration mixture among all the samples. The experimental design was set up to group multiple injections from each run. The algorithm then tabulated raw and normalized abundances and maximum fold change for each feature in the dataset. The combined MS/MS spectra were exported as .mgf files (Mascot generic files) for database searching. The Mascot search results were exported as .xml files using a significance peptide cutoff of P < 0.05 [must have at least peptide count of “2,” unique peptides count of “1,” and confidence score of greater than or equal to 35 (~95% confidence in ID)] and protein false discovery rate (FDR) of 1%. The .xml search result and then imported into the Progenesis QI software, where search hits were assigned to corresponding aligned spectral features. Relative protein-level fold changes were calculated from the sum of all unique and nonconflicting, normalized peptide ion abundances for each protein on each run. Three to five biological replicates were processed for each condition. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (100) via the PRIDE (101) partner repository with the dataset identifier PXD044773.
Proteins that had a fold change ANOVA FDR < 0.05 were considered differentially expressed and were analyzed for enrichment of GO terms and KEGG pathways using the g:Profiler web-based tool. For significance thresholds, the statistical domain scope was set to “Only annotated genes” and the Benjamini-Hochberg–corrected FDR < 0.05. Protein-protein interaction networks were obtained using the STRING database (102) with an interaction combined cutoff score of 0.7 and imported into the Cytoscape software platform (103) for visualization.
TUNEL assay
Fixed brain sections were rehydrated and underwent antigen retrieval before being processed for TUNEL assay using the Click-iT Plus TUNEL Alexa Fluor 647 Assay Kit for In Situ Detection (Invitrogen) according to the manufacturer’s instructions. Unstained sections were included as a control.
Immunoblotting
A total of 10 to 20 μg of proteins were subjected to electrophoresis on polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Blots were blocked in 5% bovine serum albumin in Tris-buffered saline with 0.1% Tween-20 (TBST) for 1 hour at room temperature before being incubated with the appropriate primary antibody in blocking buffer overnight at 4°C (see antibody table above). The following day, membranes were washed with TBST before being incubated with horseradish peroxidase–conjugated secondary antibody (1:5000 dilution) in blocking buffer for 1 hour at room temperature. Blots were then washed again with TBST, incubated with enhanced chemiluminesence substrate, and imaged using the MyECL Imager system (Thermo Fisher Scientific) with a charge-coupled device camera. Densitometry analysis was performed using ImageJ Fiji (104).
Lentiviral transduction
BTICs were adhered and transduced with lentiviral particles in the presence of polybrene (8 μg/ml) using a multiplicity of infection of 100. Following infection, cells were selected for lentiviral incorporation using puromycin (0.5 μg/ml) over a period of at least 6 days. KDs were confirmed before performing experimentation. All KD experiments were performed within four passages of lentiviral transduction.
Real-time qPCR
RNA was extracted using TRIzol (Thermo Fisher Scientific), and RNA cleanup was performed using the RNeasy Mini Kit (Qiagen). RNA concentration was measured with a NanoDrop (Thermo Fisher Scientific) and reverse transcribed using iScript reverse transcriptase (Bio-Rad). Quantitative real-time PCR was performed using Power SYBR Green Master Mix (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene. Primers used were GAPDH forward 5′-ACCTGCCAAGTATGA-TGACATCA-3′, GAPDH reverse 5′-CCCTCAGATGCCTGCTT-CAC-3′, CTSB forward 5′-ACAACGTGGACATGAGCTACT-3′, and CTSB reverse 5′-TCGGTAAACATAACTCTCTGGGG-3′.
EdU proliferation assay
Proliferation of cells was measured using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen). Cells were treated with 10 μM EdU for 1 hour before they were harvested, fixed, clicked, and analyzed for EdU incorporation. Unstained controls were included. Flow cytometry was performed using the CytoFLEX (Beckman Coulter). Cells positive for Alexa Fluor 647 were considered to be in S phase and actively proliferating.
Time-lapse migration
Glass-bottom 96-well plates (Cellvis) were treated with l-poly-ornithine (Sigma-Aldrich) for 1 hour, followed by laminin coating. Cells were seeded on the plate at a density of 1.0 × 104 to 1.5 × 104 cells per well and monitored for attachment before replacing media and placing in the LiveCyte (Phasefocus) cell imager. Cells were imaged every 20 min over a period of 24 to 48 hours. Quantification was performed by the LiveCyte software, with further graphical analysis being performed in GraphPad and R.
Limiting dilution assay
hfNPCs or BTICs were plated in ultra low-attachment 96-well plates with 400, 200, 100, 50, 25, 12, 6, and 3 cells per well (12 wells per dilution). After 2 weeks, the fraction of wells containing neurospheres (tight, nonadherent masses >50 μm in diameter) were recorded. The log of the fraction of nonresponding wells was calculated and used for graphing results. Significance was determined through chi-square test for stem cell frequency and pairwise comparisons for differences in stem cell frequencies using the Extreme Limiting Dilution Assay web tool (105).
Recombinant CTSB experiments in GBM and NPC cell lines
CTSB (Sino Biological) was dissolved in base media and diluted to a final concentration of 100 nM, in the same range as previously published (106). The dilution of CTSB was determined using a dilution curve in a cell viability assay compared to NT media control. For experiments, cells were treated for 48 hours in base media lacking growth factors unless otherwise noted.
Senescence Green and LysoTracker assay
hfNPCs were plated on a glass-bottom 96-well plate and maintained in the presence or absence of CTSB for 48 hours. For an additional 1 hour, LysoTracker Red DND-99 (Invitrogen) was added to the cells at a final concentration of 50 nM. Cells were then washed twice with PBS and fixed with 2% PFA before being stained for β-galactosidase activity with the CellEvent Senescence Green Detection Kit (Invitrogen) per manufacturer’s instructions.
Intraoperative LV-proximal and LV-distal sample collection
All deidentified samples and patient data were collected following Mayo Clinic Institutional Review Board approved protocols and were collected only after obtaining informed consent from the patient. Brain tumor tissue was obtained during standard-of-care craniotomy for GBM resection by specialized neurosurgeons. Location of obtained biopsy samples was determined with intraoperative surgical navigation on the StealthStation S7 Surgical Navigation System (Medtronic PLC, Minneapolis, MN). Intraoperative screenshots of biopsy location were collected using the surgical navigation system, and sample distance from the LV was determined by a physician using medical imaging software (Qreads Clinical Image Viewer, integrated to the Mayo Clinic EMR system). Tumors were considered LV-proximal when sample distance to LV was <10 mm.
RNA sequencing and analysis
Cells were grown in 25-cm2 flasks, transduced with lentivirus, and selected with puromycin until 80% confluent. Total RNA was isolated using TRIzol separation (Thermo Fisher Scientific) combined with RNeasy column cleanup and deoxyribonuclease I treatment (Qiagen). Samples were diluted to a concentration of 60 ng/μl in nuclease-free water before shipping to the Mayo Clinic Genome Analysis Core. Samples were sequenced on the NovaSeq 6000 (Illumina) with paired-end index read and the TruSeq v2 library. The data discussed in this publication have been deposited in National Center for Biotechnology Information’s (NCBI’s) Gene Expression Omnibus (GEO) (107) and are accessible through GEO Series accession number GSE243836.
The raw RNA sequencing paired-end reads for the samples were processed through the Mayo RNA-Seq bioinformatics pipeline, MAP-RSeq version 3.1.4 (108). Briefly, MAP-RSeq uses the very fast, accurate and splice-aware aligner, STAR (109), to align reads to the reference human genome build hg38. The aligned reads are then processed through a variety of modules in a parallel fashion. Gene and exon expression quantification was performed using the Subread package (110) to obtain both raw and normalized [(Fragments Per Kilobase of transcript per Million (FPKM)] reads. Known and novel gene isoforms were assembled and quantified using StringTie (111) to enable detection of alternative spliced isoforms. Last, comprehensive analyses were run on the aligned reads to assess quality of the sequenced libraries.
Using the raw gene counts report from MAP-RSeq, genes differentially expressed between the groups were assessed using the bioinformatics package edgeR 2.6.2 (112). Genes found different between the groups are reported along with their magnitude of change (log2 scale) and their level of significance (q value <5%). Further analysis was performed using GSEA for hallmark signature gene sets to identify relevant biological pathways (113), which were considered significant if the FDR q value was below 0.250.
Statistical analysis
All box-and-whisker plots have boxes extending from the 25th to the 75th percentiles with the line in the box indicating the median. The whiskers extend to the smallest and largest values. Bar chart data are represented as means ± SD unless otherwise noted. Statistical analysis and graphical rendering were performed using GraphPad Prism software and R Studio. Normal distribution of the data was evaluated using the Shapiro-Wilk normality test. For comparisons across two normally distributed groups, Student’s t test was performed. For matched patient sample IHC, the paired sample t test was performed. For comparisons among three normally distributed groups, ANOVA with Tukey’s post hoc correction was performed. For survival analysis, the Mantel-Cox log-rank test was used and was corrected for multiple tests when more than two groups were analyzed. The level of significance was determined as P < 0.05. For all figures, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Acknowledgments
This publication was made possible by support of the Mayo Clinic Center for Biomedical Discovery (CBD), Mayo Clinic, Rochester, MN. We thank B. Edenfield for assistance in IHC and scientific illustrator S. Graepel for creating Fig. 8. In addition, we thank F. Collin and J. Kanyo at the Keck MS & Proteomics Resource for support with proteomics sample preparation and data collection. We thank the MS & Proteomics Resource at Yale School of Medicine for providing the necessary mass spectrometers and the accompanying biotechnology tools funded, in part, by the Yale School of Medicine and by the Office of The Director, National Institutes of Health (S10OD02365101A1, S10OD019967, and S10OD018034). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. E.S.N. and M.N.R. would like to thank the Mayo Clinic Graduate School of Biomedical Sciences for support.
Funding: E.S.N. was supported by NINDS F31NS120605, the Mayo Clinic Regenerative Sciences Training Program, and the Uihlein family scholarship award. E.S.N., L.A.W., V.K.J., M.N.R., N.Z., A.C., and H.G.-C. were supported by NINDS K01NS110930 and the Uncle Kory Foundation. A.Q.-H. was supported by the Mayo Clinic Clinician Investigator award, the William J. and Charles H. Mayo Named Professorship, the Monica Flynn Jacoby Endowed Chair, and the Uihlein Neuro-Oncology Research Fund.
Author contributions: Conceptualization: E.S.N. and H.G.-C. Methodology: E.S.N., M.M.B., A.R.-F., N.Z., W.R., T.T.L., A.Q.-H., and H.G.-C. Resources: E.S.N., M.M.B., D.M., A.R.-F., W.R., T.T.L., K.L.C., P.Z.A., A.Q.-H., and H.G.-C. Investigation: E.S.N., L.A.W., V.K.J., M.M.B., M.N.R., E.J., A.R.-F., N.Z., A.C., and T.T.L. Validation: E.S.N., L.A.W., M.M.B., M.N.R., N.Z., A.C., T.T.L., A.Q.-H., and H.G.-C. Formal analysis: E.S.N., L.A.W., V.K.J., M.N.R., D.M., E.J., Y.W.A., T.T.L., and H.G.-C. Visualization: E.S.N., V.K.J., D.M., E.J., P.S., and H.G.-C. Project administration: E.S.N., T.T.L., P.Z.A., A.Q.-H., and H.G.-C. Supervision: E.S.N. and P.S., W.R., T.T.L., P.Z.A., A.Q.-H., and H.G.-C. Software: E.J. Funding acquisition: E.S.N., P.S., P.Z.A., A.Q.-H., and H.G.-C. Writing—original draft: E.S.N. and H.G.-C. Writing—review and editing: all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in this paper are present in the paper, the Supplementary Materials, and/or online data repository. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (100) via the PRIDE (101) partner repository with the dataset identifier PXD044773. The RNA sequencing data discussed in this publication have been deposited in NCBI’s GEO (107) and are accessible through GEO Series accession number GSE243836.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Legends for tables S1 and S2
Table S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Ostrom Q. T., Price M., Neff C., Cioffi G., Waite K. A., Kruchko C., Barnholtz-Sloan J. S., CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2016-2020. Neuro Oncol. 25, iv1–iv99 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartkowiak T., Lima S. M., Hayes M. J., Mistry A. M., Brockman A. A., Sinnaeve J., Leelatian N., Roe C. E., Mobley B. C., Chotai S., Weaver K. D., Thompson R. C., Chambless L. B., Ihrie R. A., Irish J. M., An immunosuppressed microenvironment distinguishes lateral ventricle-contacting glioblastomas. JCI Insight 8, e160652 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaichana K. L., McGirt M. J., Frazier J., Attenello F., Guerrero-Cazares H., Quinones-Hinojosa A., Relationship of glioblastoma multiforme to the lateral ventricles predicts survival following tumor resection. J. Neurooncol. 89, 219–224 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Lim D. A., Cha S., Mayo M. C., Chen M. H., Keles E., VandenBerg S., Berger M. S., Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol. 9, 424–429 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mistry A. M., Hale A. T., Chambless L. B., Weaver K. D., Thompson R. C., Ihrie R. A., Influence of glioblastoma contact with the lateral ventricle on survival: A meta-analysis. J. Neurooncol 131, 125–133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nestler U., Lutz K., Pichlmeier U., Stummer W., Franz K., Reulen H. J., Bink A.; 5-ALA Glioma Study Group , Anatomic features of glioblastoma and their potential impact on survival. Acta Neurochir. 157, 179–186 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Steed T. C., Treiber J. M., Taha B., Engin H. B., Carter H., Patel K. S., Dale A. M., Carter B. S., Chen C. C., Glioblastomas located in proximity to the subventricular zone (SVZ) exhibited enrichment of gene expression profiles associated with the cancer stem cell state. J. Neurooncol 148, 455–462 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Luskin M. B., Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron 11, 173–189 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Quiñones-Hinojosa A., Sanai N., Soriano-Navarro M., Gonzalez-Perez O., Mirzadeh Z., Gil-Perotin S., Romero-Rodriguez R., Berger M. S., Garcia-Verdugo J. M., Alvarez-Buylla A., Cellular composition and cytoarchitecture of the adult human subventricular zone: A niche of neural stem cells. J. Comp. Neurol. 494, 415–434 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Sanai N., Tramontin A. D., Quiñones-Hinojosa A., Barbaro N. M., Gupta N., Kunwar S., Lawton M. T., McDermott M. W., Parsa A. T., Manuel-García Verdugo J., Berger M. S., Alvarez-Buylla A., Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature 427, 740–744 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Reynolds B. A., Weiss S., Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Zarco N., Norton E., Quiñones-Hinojosa A., Guerrero-Cázares H., Overlapping migratory mechanisms between neural progenitor cells and brain tumor stem cells. Cell. Mol. Life Sci. 76, 3553–3570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lathia J. D., Mack S. C., Mulkearns-Hubert E. E., Valentim C. L., Rich J. N., Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bexell D., Gunnarsson S., Nordquist J., Bengzon J., Characterization of the subventricular zone neurogenic response to rat malignant brain tumors. Neuroscience 147, 824–832 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Norton E. S., Whaley L. A., Ulloa-Navas M. J., García-Tárraga P., Meneses K. M., Lara-Velazquez M., Zarco N., Carrano A., Quiñones-Hinojosa A., García-Verdugo J. M., Guerrero-Cázares H., Glioblastoma disrupts the ependymal wall and extracellular matrix structures of the subventricular zone. Fluids Barriers CNS 19, 58 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripari L. B., Norton E. S., Bodoque-Villar R., Jeanneret S., Lara-Velazquez M., Carrano A., Zarco N., Vazquez-Ramos C. A., Quiñones-Hinojosa A., de la Rosa-Prieto C., Guerrero-Cázares H., Glioblastoma proximity to the lateral ventricle alters neurogenic cell populations of the subventricular zone. Front. Oncol. 11, 650316 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cebrian-Silla A., Nascimento M. A., Redmond S. A., Mansky B., Wu D., Obernier K., Romero Rodriguez R., Gonzalez-Granero S., García-Verdugo J. M., Lim D. A., Álvarez-Buylla A., Single-cell analysis of the ventricular-subventricular zone reveals signatures of dorsal and ventral adult neurogenesis. eLife 10, e67436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couturier C. P., Nadaf J., Li Z., Baig S., Riva G., Le P., Kloosterman D. J., Monlong J., Nkili Meyong A., Allache R., Degenhard T., Al-Rashid M., Guiot M. C., Bourque G., Ragoussis J., Akkari L., Quintana F. J., Petrecca K., Glioblastoma scRNA-seq shows treatment-induced, immune-dependent increase in mesenchymal cancer cells and structural variants in distal neural stem cells. Neuro Oncol. 24, 1494–1508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donega V., van der Geest A. T., Sluijs J. A., van Dijk R. E., Wang C. C., Basak O., Pasterkamp R. J., Hol E. M., Single-cell profiling of human subventricular zone progenitors identifies SFRP1 as a target to re-activate progenitors. Nat. Commun. 13, 1036 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dulken B. W., Buckley M. T., Navarro Negredo P., Saligrama N., Cayrol R., Leeman D. S., George B. M., Boutet S. C., Hebestreit K., Pluvinage J. V., Wyss-Coray T., Weissman I. L., Vogel H., Davis M. M., Brunet A., Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizrak D., Levitin H. M., Delgado A. C., Crotet V., Yuan J., Chaker Z., Silva-Vargas V., Sims P. A., Doetsch F., Single-cell analysis of regional differences in adult V-SVZ neural stem cell lineages. Cell Rep. 26, 394–406.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neftel C., Laffy J., Filbin M. G., Hara T., Shore M. E., Rahme G. J., Richman A. R., Silverbush D., Shaw M. L., Hebert C. M., Dewitt J., Gritsch S., Perez E. M., Gonzalez Castro L. N., Lan X., Druck N., Rodman C., Dionne D., Kaplan A., Bertalan M. S., Small J., Pelton K., Becker S., Bonal D., Nguyen Q. D., Servis R. L., Fung J. M., Mylvaganam R., Mayr L., Gojo J., Haberler C., Geyeregger R., Czech T., Slavc I., Nahed B. V., Curry W. T., Carter B. S., Wakimoto H., Brastianos P. K., Batchelor T. T., Stemmer-Rachamimov A., Martinez-Lage M., Frosch M. P., Stamenkovic I., Riggi N., Rheinbay E., Monje M., Rozenblatt-Rosen O., Cahill D. P., Patel A. P., Hunter T., Verma I. M., Ligon K. L., Louis D. N., Regev A., Bernstein B. E., Tirosh I., Suvà M. L., An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel A. P., Tirosh I., Trombetta J. J., Shalek A. K., Gillespie S. M., Wakimoto H., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L., Louis D. N., Rozenblatt-Rosen O., Suvà M. L., Regev A., Bernstein B. E., Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varn F. S., Johnson K. C., Martinek J., Huse J. T., Nasrallah M. P., Wesseling P., Cooper L. A. D., Malta T. M., Wade T. E., Sabedot T. S., Brat D., Gould P. V., Wöehrer A., Aldape K., Ismail A., Sivajothi S. K., Barthel F. P., Kim H., Kocakavuk E., Ahmed N., White K., Datta I., Moon H. E., Pollock S., Goldfarb C., Lee G. H., Garofano L., Anderson K. J., Nehar-Belaid D., Barnholtz-Sloan J. S., Bakas S., Byrne A. T., D’Angelo F., Gan H. K., Khasraw M., Migliozzi S., Ormond D. R., Paek S. H., van Meir E. G., Walenkamp A. M. E., Watts C., Weiss T., Weller M., Palucka K., Stead L. F., Poisson L. M., Noushmehr H., Iavarone A., Verhaak R. G. W., Glioma progression is shaped by genetic evolution and microenvironment interactions. Cell 185, 2184–2199.e16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan C. W., Verhaak R. G., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S. A., Ciriello G., Yung W. K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D. D., van Meir E. G., Prados M., Sloan A., Black K. L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D. W., Guha A., Iacocca M., O’Neill B. P., Foltz G., Myers J., Weisenberger D. J., Penny R., Kucherlapati R., Perou C. M., Hayes D. N., Gibbs R., Marra M., Mills G. B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P. W., Haussler D., Getz G., Chin L.; TCGA Research Network , The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kjell J., Fischer-Sternjak J., Thompson A. J., Friess C., Sticco M. J., Salinas F., Cox J., Martinelli D. C., Ninkovic J., Franze K., Schiller H. B., Götz M., Defining the adult neural stem cell niche proteome identifies key regulators of adult neurogenesis. Cell Stem Cell 26, 277–293.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravi V. M., Will P., Kueckelhaus J., Sun N., Joseph K., Salié H., Vollmer L., Kuliesiute U., von Ehr J., Benotmane J. K., Neidert N., Follo M., Scherer F., Goeldner J. M., Behringer S. P., Franco P., Khiat M., Zhang J., Hofmann U. G., Fung C., Ricklefs F. L., Lamszus K., Boerries M., Ku M., Beck J., Sankowski R., Schwabenland M., Prinz M., Schüller U., Killmer S., Bengsch B., Walch A. K., Delev D., Schnell O., Heiland D. H., Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639–655.e13 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Wang L. B., Karpova A., Gritsenko M. A., Kyle J. E., Cao S., Li Y., Rykunov D., Colaprico A., Rothstein J. H., Hong R., Stathias V., Cornwell M., Petralia F., Wu Y., Reva B., Krug K., Pugliese P., Kawaler E., Olsen L. K., Liang W. W., Song X., Dou Y., Wendl M. C., Caravan W., Liu W., Cui Zhou D., Ji J., Tsai C. F., Petyuk V. A., Moon J., Ma W., Chu R. K., Weitz K. K., Moore R. J., Monroe M. E., Zhao R., Yang X., Yoo S., Krek A., Demopoulos A., Zhu H., Wyczalkowski M. A., McMichael J. F., Henderson B. L., Lindgren C. M., Boekweg H., Lu S., Baral J., Yao L., Stratton K. G., Bramer L. M., Zink E., Couvillion S. P., Bloodsworth K. J., Satpathy S., Sieh W., Boca S. M., Schürer S., Chen F., Wiznerowicz M., Ketchum K. A., Boja E. S., Kinsinger C. R., Robles A. I., Hiltke T., Thiagarajan M., Nesvizhskii A. I., Zhang B., Mani D. R., Ceccarelli M., Chen X. S., Cottingham S. L., Li Q. K., Kim A. H., Fenyö D., Ruggles K. V., Rodriguez H., Mesri M., Payne S. H., Resnick A. C., Wang P., Smith R. D., Iavarone A., Chheda M. G., Barnholtz-Sloan J. S., Rodland K. D., Liu T., Ding L.; Clinical Proteomic Tumor Analysis Consortium , Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 39, 509–528.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baser A., Skabkin M., Kleber S., Dang Y., Gülcüler Balta G. S., Kalamakis G., Göpferich M., Ibañez D. C., Schefzik R., Lopez A. S., Bobadilla E. L., Schultz C., Fischer B., Martin-Villalba A., Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Ramos-Fresnedo A., Pullen M. W., Perez-Vega C., Domingo R. A., Akinduro O. O., Almeida J. P., Suarez-Meade P., Marenco-Hillembrand L., Jentoft M. E., Bendok B. R., Trifiletti D. M., Chaichana K. L., Porter A. B., Quiñones-Hinojosa A., Burns T. C., Kizilbash S. H., Middlebrooks E. H., Sherman W. J., The survival outcomes of molecular glioblastoma IDH-wildtype: A multicenter study. J. Neurooncol. 157, 177–185 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Chen L., Guerrero-Cazares H., Ye X., Ford E., McNutt T., Kleinberg L., Lim M., Chaichana K., Quinones-Hinojosa A., Redmond K., Increased subventricular zone radiation dose correlates with survival in glioblastoma patients after gross total resection. Int. J. Radiat. Oncol. Biol. Phys. 86, 616–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lara-Velazquez M., Zarco N., Carrano A., Phillipps J., Norton E. S., Schiapparelli P., Al-Kharboosh R., Rincon-Torroella J., Jeanneret S., Corona T., Segovia J., Jentoft M. E., Chaichana K. L., Asmann Y. W., Quiñones-Hinojosa A., Guerrero-Cazares H., Alpha 1-antichymotrypsin contributes to stem cell characteristics and enhances tumorigenicity of glioblastoma. Neuro Oncol. 23, 599–610 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrano A., Zarco N., Phillipps J., Lara-Velazquez M., Suarez-Meade P., Norton E. S., Chaichana K. L., Quiñones-Hinojosa A., Asmann Y. W., Guerrero-Cázares H., Human cerebrospinal fluid modulates pathways promoting glioblastoma malignancy. Front. Oncol. 11, 624145 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin E. Y., Cooper D. D., Abbott K. L., Lennon J., Nagaraja S., Mackay A., Jones C., Vogel H., Jackson P. K., Monje M., Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell 170, 845–859.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutecki P. A., Neuronal excitability: Voltage-dependent currents and synaptic transmission. J. Clin. Neurophysiol. 9, 195–211 (1992). [PubMed] [Google Scholar]
- 36.Huang J., Wu C., Kloeber J. A., Gao H., Gao M., Zhu Q., Chang Y., Zhao F., Guo G., Luo K., Dai H., Liu S., Huang Q., Kim W., Zhou Q., Zhu S., Wu Z., Tu X., Yin P., Deng M., Wang L., Yuan J., Lou Z., SLFN5-mediated chromatin dynamics sculpt higher-order DNA repair topology. Mol. Cell 83, 1043–1060.e10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panier S., Boulton S. J., Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15, 7–18 (2014). [DOI] [PubMed] [Google Scholar]
- 38.d’Adda di Fagagna F., Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Rayess H., Wang M. B., Srivatsan E. S., Cellular senescence and tumor suppressor gene p16. Int. J. Cancer 130, 1715–1725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yosef R., Pilpel N., Papismadov N., Gal H., Ovadya Y., Vadai E., Miller S., Porat Z., Ben-Dor S., Krizhanovsky V., p21 maintains senescent cell viability under persistent DNA damage response by restraining JNK and caspase signaling. EMBO J. 36, 2280–2295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leeman D. S., Hebestreit K., Ruetz T., Webb A. E., McKay A., Pollina E. A., Dulken B. W., Zhao X., Yeo R. W., Ho T. T., Mahmoudi S., Devarajan K., Passegué E., Rando T. A., Frydman J., Brunet A., Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowman R. L., Wang Q., Carro A., Verhaak R. G., Squatrito M., GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 19, 139–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mistry A. M., Wooten D. J., Davis L. T., Mobley B. C., Quaranta V., Ihrie R. A., Ventricular-subventricular zone contact by glioblastoma is not associated with molecular signatures in bulk tumor data. Sci. Rep. 9, 1842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuizumi N., Harada Y., Kuniya T., Sunabori T., Koike M., Wakabayashi M., Ishihama Y., Suzuki Y., Kawaguchi D., Gotoh Y., Maintenance of neural stem-progenitor cells by the lysosomal biosynthesis regulators TFEB and TFE3 in the embryonic mouse telencephalon. Stem Cells 39, 929–944 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Venkataramani V., Tanev D. I., Strahle C., Studier-Fischer A., Fankhauser L., Kessler T., Körber C., Kardorff M., Ratliff M., Xie R., Horstmann H., Messer M., Paik S. P., Knabbe J., Sahm F., Kurz F. T., Acikgöz A. A., Herrmannsdörfer F., Agarwal A., Bergles D. E., Chalmers A., Miletic H., Turcan S., Mawrin C., Hänggi D., Liu H. K., Wick W., Winkler F., Kuner T., Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573, 532–538 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Venkatesh H. S., Morishita W., Geraghty A. C., Silverbush D., Gillespie S. M., Arzt M., Tam L. T., Espenel C., Ponnuswami A., Ni L., Woo P. J., Taylor K. R., Agarwal A., Regev A., Brang D., Vogel H., Hervey-Jumper S., Bergles D. E., Suvà M. L., Malenka R. C., Monje M., Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alapati K., Gopinath S., Malla R. R., Dasari V. R., Rao J. S., uPAR and cathepsin B knockdown inhibits radiation-induced PKC integrated integrin signaling to the cytoskeleton of glioma-initiating cells. Int. J. Oncol. 41, 599–610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gondi C. S., Lakka S. S., Yanamandra N., Olivero W. C., Dinh D. H., Gujrati M., Tung C. H., Weissleder R., Rao J. S., Adenovirus-mediated expression of antisense urokinase plasminogen activator receptor and antisense cathepsin B inhibits tumor growth, invasion, and angiogenesis in gliomas. Cancer Res. 64, 4069–4077 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Kenig S., Alonso M. B., Mueller M. M., Lah T. T., Glioblastoma and endothelial cells cross-talk, mediated by SDF-1, enhances tumour invasion and endothelial proliferation by increasing expression of cathepsins B, S, and MMP-9. Cancer Lett. 289, 53–61 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Lakka S. S., Gondi C. S., Yanamandra N., Olivero W. C., Dinh D. H., Gujrati M., Rao J. S., Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene 23, 4681–4689 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Malla R. R., Gopinath S., Alapati K., Gorantla B., Gondi C. S., Rao J. S., uPAR and cathepsin B inhibition enhanced radiation-induced apoptosis in gliomainitiating cells. Neuro Oncol. 14, 745–760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aggarwal N., Sloane B. F., Cathepsin B: Multiple roles in cancer. Proteomics Clin. Appl. 8, 427–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruan H., Hao S., Young P., Zhang H., Targeting cathepsin B for cancer therapies. Horiz. Cancer Res. 56, 23–40 (2015). [PMC free article] [PubMed] [Google Scholar]
- 54.Haddad A. F., Young J. S., Amara D., Berger M. S., Raleigh D. R., Aghi M. K., Butowski N. A., Mouse models of glioblastoma for the evaluation of novel therapeutic strategies. Neurooncol. Adv. 3, vdab100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Kharboosh R., ReFaey K., Lara-Velazquez M., Grewal S. S., Imitola J., Quiñones-Hinojosa A., Inflammatory mediators in glioma microenvironment play a dual role in gliomagenesis and mesenchymal stem cell homing: Implication for cellular therapy. Mayo Clin. Proc. Innov. Qual. Outcomes 4, 443–459 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szadvari I., Krizanova O., Babula P., Athymic nude mice as an experimental model for cancer treatment. Physiol. Res. 65, S441–S453 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., de Carvalho A. C., Lyu S., Li P., Li Y., Barthel F., Cho H. J., Lin Y. H., Satani N., Martinez-Ledesma E., Zheng S., Chang E., Sauvé C. G., Olar A., Lan Z. D., Finocchiaro G., Phillips J. J., Berger M. S., Gabrusiewicz K. R., Wang G., Eskilsson E., Hu J., Mikkelsen T., DePinho R. A., Muller F., Heimberger A. B., Sulman E. P., Nam D. H., Verhaak R. G. W., Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogawa J., Pao G. M., Shokhirev M. N., Verma I. M., Glioblastoma model using human cerebral organoids. Cell Rep. 23, 1220–1229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rybin M. J., Ivan M. E., Ayad N. G., Zeier Z., Organoid models of glioblastoma and their role in drug discovery. Front. Cell. Neurosci. 15, 605255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakod P. S., Kim Y., Rao S. S., Biomimetic models to examine microenvironmental regulation of glioblastoma stem cells. Cancer Lett. 429, 41–53 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Tang M., Xie Q., Gimple R. C., Zhong Z., Tam T., Tian J., Kidwell R. L., Wu Q., Prager B. C., Qiu Z., Yu A., Zhu Z., Mesci P., Jing H., Schimelman J., Wang P., Lee D., Lorenzini M. H., Dixit D., Zhao L., Bhargava S., Miller T. E., Wan X., Tang J., Sun B., Cravatt B. F., Muotri A. R., Chen S., Rich J. N., Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 30, 833–853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans H. T., Bodea L. G., Götz J., Cell-specific non-canonical amino acid labelling identifies changes in the de novo proteome during memory formation. eLife 9, e52990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prabhakar P., Pielot R., Landgraf P., Wissing J., Bayrhammer A., van Ham M., Gundelfinger E. D., Jänsch L., Dieterich D. C., Müller A., Monitoring regional astrocyte diversity by cell type-specific proteomic labeling in vivo. Glia 71, 682–703 (2023). [DOI] [PubMed] [Google Scholar]
- 64.Schiapparelli L. M., Xie Y., Sharma P., McClatchy D. B., Ma Y., Yates J. R. III, Maximov A., Cline H. T., Activity-induced cortical glutamatergic neuron nascent proteins. J. Neurosci. 42, 7900–7920 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azizian N. G., Sullivan D. K., Nie L., Pardo S., Molleur D., Chen J., Weintraub S. T., Li Y., Selective labeling and identification of the tumor cell proteome of pancreatic cancer in vivo. J. Proteome Res. 20, 858–866 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du Y., Li X., Yan W., Zeng Z., Han D., Ouyang H., Pan X., Luo B., Zhou B., Fu Q., Lu D., Huang Z., Li Z., Deciphering the in vivo dynamic proteomics of mesenchymal stem cells in critical limb ischemia. Front. Cell Dev. Biol. 9, 682476 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han D., Yang J., Zhang E., Liu Y., Boriboun C., Qiao A., Yu Y., Sun J., Xu S., Yang L., Yan W., Luo B., Lu D., Zhang C., Jie C., Mobley J., Zhang J., Qin G., Analysis of mesenchymal stem cell proteomes in situ in the ischemic heart. Theranostics 10, 11324–11338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin S., Lee S., Choi S., Park N., Kwon Y., Jeong J., Ju S., Chang Y., Park K., Ha C., Lee C., Characterization of the secretome of a specific cell expressing mutant methionyl-tRNA synthetase in co-culture using click chemistry. Int. J. Mol. Sci. 23, 6527 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Curtis M. A., Faull R. L., Eriksson P. S., The effect of neurodegenerative diseases on the subventricular zone. Nat. Rev. Neurosci. 8, 712–723 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Dillen Y., Kemps H., Gervois P., Wolfs E., Bronckaers A., Adult neurogenesis in the subventricular zone and its regulation after ischemic stroke: Implications for therapeutic approaches. Transl. Stroke Res. 11, 60–79 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Zhang R. L., Zhang Z. G., Chopp M., Ischemic stroke and neurogenesis in the subventricular zone. Neuropharmacology 55, 345–352 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calabrese C., Poppleton H., Kocak M., Hogg T. L., Fuller C., Hamner B., Oh E. Y., Gaber M. W., Finklestein D., Allen M., Frank A., Bayazitov I. T., Zakharenko S. S., Gajjar A., Davidoff A., Gilbertson R. J., A perivascular niche for brain tumor stem cells. Cancer Cell 11, 69–82 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Pietras A., Katz A. M., Ekström E. J., Wee B., Halliday J. J., Pitter K. L., Werbeck J. L., Amankulor N. M., Huse J. T., Holland E. C., Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 14, 357–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schiffer D., Annovazzi L., Casalone C., Corona C., Mellai M., Glioblastoma: Microenvironment and niche concept. Cancers 11, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Domingo-Muelas A., Duart-Abadia P., Morante-Redolat J. M., Jordán-Pla A., Belenguer G., Fabra-Beser J., Paniagua-Herranz L., Pérez-Villalba A., Álvarez-Varela A., Barriga F. M., Gil-Sanz C., Ortega F., Batlle E., Fariñas I., Post-transcriptional control of a stemness signature by RNA-binding protein MEX3A regulates murine adult neurogenesis. Nat. Commun. 14, 373 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanai N., Nguyen T., Ihrie R. A., Mirzadeh Z., Tsai H. H., Wong M., Gupta N., Berger M. S., Huang E., Garcia-Verdugo J. M., Rowitch D. H., Alvarez-Buylla A., Corridors of migrating neurons in the human brain and their decline during infancy. Nature 478, 382–386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kirschenbaum B., Nedergaard M., Preuss A., Barami K., Fraser R. A., Goldman S. A., In vitro neuronal production and differentiation by precursor cells derived from the adult human forebrain. Cereb. Cortex 4, 576–589 (1994). [DOI] [PubMed] [Google Scholar]
- 78.Leonard B. W., Mastroeni D., Grover A., Liu Q., Yang K., Gao M., Wu J., Pootrakul D., van den Berge S. A., Hol E. M., Rogers J., Subventricular zone neural progenitors from rapid brain autopsies of elderly subjects with and without neurodegenerative disease. J. Comp. Neurol. 515, 269–294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moe M. C., Westerlund U., Varghese M., Berg-Johnsen J., Svensson M., Langmoen I. A., Development of neuronal networks from single stem cells harvested from the adult human brain. Neurosurgery 56, 1182–1190 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Palmer T. D., Schwartz P. H., Taupin P., Kaspar B., Stein S. A., Gage F. H., Progenitor cells from human brain after death. Nature 411, 42–43 (2001). [DOI] [PubMed] [Google Scholar]
- 81.van Strien M. E., Sluijs J. A., Reynolds B. A., Steindler D. A., Aronica E., Hol E. M., Isolation of neural progenitor cells from the human adult subventricular zone based on expression of the cell surface marker CD271. Stem Cells Transl. Med. 3, 470–480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ashraf H. M., Fernandez B., Spencer S. L., The intensities of canonical senescence biomarkers integrate the duration of cell-cycle withdrawal. Nat. Commun. 14, 4527 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fletcher-Sananikone E., Kanji S., Tomimatsu N., Di Cristofaro L. F. M., Kollipara R. K., Saha D., Floyd J. R., Sung P., Hromas R., Burns T. C., Kittler R., Habib A. A., Mukherjee B., Burma S., Elimination of radiation-induced senescence in the brain tumor microenvironment attenuates glioblastoma recurrence. Cancer Res. 81, 5935–5947 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Riviere-Cazaux C., Carlstrom L. P., Neth B. J., Olson I. E., Rajani K., Rahman M., Ikram S., Mansour M. A., Mukherjee B., Warrington A. E., Short S. C., von Zglinicki T., Brown D. A., Burma S., Tchkonia T., Schafer M. J., Baker D. J., Kizilbash S. H., Kirkland J. L., Burns T. C., An untapped window of opportunity for glioma: Targeting therapy-induced senescence prior to recurrence. NPJ Precis. Oncol. 7, 126 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salam R., Saliou A., Bielle F., Bertrand M., Antoniewski C., Carpentier C., Alentorn A., Capelle L., Sanson M., Huillard E., Bellenger L., Guégan J., Le Roux I., Cellular senescence in malignant cells promotes tumor progression in mouse and patient glioblastoma. Nat. Commun. 14, 441 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aboody K. S., Najbauer J., Metz M. Z., D’Apuzzo M., Gutova M., Annala A. J., Synold T. W., Couture L. A., Blanchard S., Moats R. A., Garcia E., Aramburo S., Valenzuela V. V., Frank R. T., Barish M. E., Brown C. E., Kim S. U., Badie B., Portnow J., Neural stem cell-mediated enzyme/prodrug therapy for glioma: Preclinical studies. Sci. Transl. Med. 5, 184ra159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahmed A. U., Thaci B., Tobias A. L., Auffinger B., Zhang L., Cheng Y., Kim C. K., Yunis C., Han Y., Alexiades N. G., Fan X., Aboody K. S., Lesniak M. S., A preclinical evaluation of neural stem cell-based cell carrier for targeted antiglioma oncolytic virotherapy. J. Natl. Cancer Inst. 105, 968–977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fares J., Ahmed A. U., Ulasov I. V., Sonabend A. M., Miska J., Lee-Chang C., Balyasnikova I. V., Chandler J. P., Portnow J., Tate M. C., Kumthekar P., Lukas R. V., Grimm S. A., Adams A. K., Hébert C. D., Strong T. V., Amidei C., Arrieta V. A., Zannikou M., Horbinski C., Zhang H., Burdett K. B., Curiel D. T., Sachdev S., Aboody K. S., Stupp R., Lesniak M. S., Neural stem cell delivery of an oncolytic adenovirus in newly diagnosed malignant glioma: A first-in-human, phase 1, dose-escalation trial. Lancet Oncol. 22, 1103–1114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Portnow J., Synold T. W., Badie B., Tirughana R., Lacey S. F., D’Apuzzo M., Metz M. Z., Najbauer J., Bedell V., Vo T., Gutova M., Frankel P., Chen M., Aboody K. S., Neural stem cell-based anticancer gene therapy: A first-in-human study in recurrent high-grade glioma patients. Clin. Cancer Res. 23, 2951–2960 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lagace D. C., Whitman M. C., Noonan M. A., Ables J. L., DeCarolis N. A., Arguello A. A., Donovan M. H., Fischer S. J., Farnbauch L. A., Beech R. D., DiLeone R. J., Greer C. A., Mandyam C. D., Eisch A. J., Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 27, 12623–12629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun M. Y., Yetman M. J., Lee T. C., Chen Y., Jankowsky J. L., Specificity and efficiency of reporter expression in adult neural progenitors vary substantially among nestin-CreERT2 lines. J. Comp. Neurol. 522, 1191–1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alvarez-Castelao B., Schanzenbächer C. T., Hanus C., Glock C., Tom Dieck S., Dörrbaum A. R., Bartnik I., Nassim-Assir B., Ciirdaeva E., Mueller A., Dieterich D. C., Tirrell D. A., Langer J. D., Schuman E. M., Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol. 35, 1196–1201 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Garcia C. A., Bhargav A. G., Brooks M., Suárez-Meade P., Mondal S. K., Zarco N., ReFaey K., Jentoft M., Middlebrooks E. H., Snuderl M., Carrano A., Guerrero-Cazares H., Schiapparelli P., Sarabia-Estrada R., Quiñones-Hinojosa A., Functional characterization of brain tumor-initiating cells and establishment of GBM preclinical models that incorporate heterogeneity, therapy, and sex differences. Mol. Cancer Ther. 20, 2585–2597 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., Vescovi A., Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64, 7011–7021 (2004). [DOI] [PubMed] [Google Scholar]
- 95.Guerrero-Cazares H., Lavell E., Chen L., Schiapparelli P., Lara-Velazquez M., Capilla-Gonzalez V., Clements A. C., Drummond G., Noiman L., Thaler K., Burke A., Quiñones-Hinojosa A., Brief report: Robo1 regulates the migration of human subventricular zone neural progenitor cells during development. Stem Cells 35, 1860–1865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., Kaufman P. D., A versatile viral system for expression and depletion of proteins in mammalian cells. PLOS ONE 4, e6529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson A. A., Murphy G. J., Hamakawa H., Kwok L. W., Srinivasan S., Hovav A. H., Mulligan R. C., Amar S., Suki B., Kotton D. N., Amelioration of emphysema in mice through lentiviral transduction of long-lived pulmonary alveolar macrophages. J. Clin. Invest. 120, 379–389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tom Dieck S., Müller A., Nehring A., Hinz F. I., Bartnik I., Schuman E. M., Dieterich D. C., Metabolic labeling with noncanonical amino acids and visualization by chemoselective fluorescent tagging. Curr. Protoc. Cell. Biol. 7, 7.11.1–7.11.29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Henderson M. X., Wirak G. S., Zhang Y. Q., Dai F., Ginsberg S. D., Dolzhanskaya N., Staropoli J. F., Nijssen P. C., Lam T. T., Roth A. F., Davis N. G., Dawson G., Velinov M., Chandra S. S., Neuronal ceroid lipofuscinosis with DNAJC5/CSPα mutation has PPT1 pathology and exhibit aberrant protein palmitoylation. Acta Neuropathol. 131, 621–637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deutsch E. W., Bandeira N., Perez-Riverol Y., Sharma V., Carver J. J., Mendoza L., Kundu D. J., Wang S., Bandla C., Kamatchinathan S., Hewapathirana S., Pullman B. S., Wertz J., Sun Z., Kawano S., Okuda S., Watanabe Y., MacLean B., MacCoss M. J., Zhu Y., Ishihama Y., Vizcaíno J. A., The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 51, D1539–D1548 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D. J., Prakash A., Frericks-Zipper A., Eisenacher M., Walzer M., Wang S., Brazma A., Vizcaíno J. A., The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., von Mering C., STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu Y., Smyth G. K., ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 106.Peng Y., Zhang M., Zheng L., Liang Q., Li H., Chen J. T., Guo H., Yoshina S., Chen Y. Z., Zhao X., Wu X., Liu B., Mitani S., Yu J. S., Xue D., Cysteine protease cathepsin B mediates radiation-induced bystander effects. Nature 547, 458–462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kalari K. R., Nair A. A., Bhavsar J. D., O’Brien D. R., Davila J. I., Bockol M. A., Nie J., Tang X., Baheti S., Doughty J. B., Middha S., Sicotte H., Thompson A. E., Asmann Y. W., Kocher J. P., MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics 15, 224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liao Y., Smyth G. K., Shi W., The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schiapparelli P., Guerrero-Cazares H., Magaña-Maldonado R., Hamilla S. M., Ganaha S., Fernandes E. G. L., Huang C. H., Aranda-Espinoza H., Devreotes P., Quinones-Hinojosa A., NKCC1 regulates migration ability of glioblastoma cells by modulation of actin dynamics and interacting with cofilin. EBioMedicine 21, 94–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Legends for tables S1 and S2
Table S3
Tables S1 and S2