Abstract
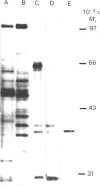
Streptomyces glaucescens has a DNAase whose synthesis is under nutritional control. We have purified this enzyme to apparent homogeneity by phosphocellulose chromatography followed by heparin-agarose, Cibacron Blue F3-GA-Sepharose and Sephadex G-75 chromatography and MonoQ f.p.l.c. The enzyme had an apparent Mr of 39,600 and a pI of approx. 8.15. The Mr of the native enzyme estimated by gel chromatography was 49,000. The DNAase had a pH optimum of 7.5 and an absolute requirement for bivalent cations in the reaction buffer. It was inhibited by high salt concentrations, chelating agents or phosphate-containing compounds and was stimulated by dimethyl sulphoxide. The activity was greatly diminished unless dithiothreitol or 2-mercaptoethanol was included in the reaction mixture. Reagents such as Hg2+ or iodoacetate strongly inhibited the enzyme. The nuclease hydrolysed both double-stranded and single-stranded DNA, showing greater affinity for double-stranded DNA, and no detectable hydrolysis of RNA. The enzyme produced nicks in double-stranded DNA, generating 3'-hydroxy and 5'-phosphate termini, and degraded circular DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aparicio J. F., De los Reyes-Gavilán C. G., Barbés C., Hardisson C., Sánchez J. A non-specific deoxyribonuclease with restriction function in Streptomyces glaucescens. J Gen Microbiol. 1988 Aug;134(8):2345–2351. doi: 10.1099/00221287-134-8-2345. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange as an assay for class II restriction endonucleases. Methods Enzymol. 1980;65(1):28–36. doi: 10.1016/s0076-6879(80)65007-1. [DOI] [PubMed] [Google Scholar]
- Blakesley R. W., Dodgson J. B., Nes I. F., Wells R. D. Duplex regions in "single-stranded" phiX174 DNA are cleaved by a restriction endonuclease from Haemophilus aegyptius. J Biol Chem. 1977 Oct 25;252(20):7300–7306. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- Chaconas G., van de Sande J. H. 5'-32P labeling of RNA and DNA restriction fragments. Methods Enzymol. 1980;65(1):75–85. doi: 10.1016/s0076-6879(80)65012-5. [DOI] [PubMed] [Google Scholar]
- Engel P., Ullah A. H. Purification and characterization of an endonuclease (E.C. 3.1.30.1) from Streptomyces tendae. Prep Biochem. 1988;18(2):137–152. doi: 10.1080/00327488808062517. [DOI] [PubMed] [Google Scholar]
- Fuchs R., Blakesley R. Guide to the use of type II restriction endonucleases. Methods Enzymol. 1983;100:3–38. doi: 10.1016/0076-6879(83)00043-9. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisson C., Manzanal M. B., Salas J. A., Suárez J. E. Fine structure, physiology and biochemistry of arthrospore germination in Streptomyces antibioticus. J Gen Microbiol. 1978 Apr;105(2):203–214. doi: 10.1099/00221287-105-2-203. [DOI] [PubMed] [Google Scholar]
- Hnilica L. S., Grimes S. R., Chiu J. F. Electrophoretic fractionation of histones utilizing starch gels and sodium dodecyl sulfate--urea gels. Methods Cell Biol. 1978;17:211–222. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R., Wu R. Terminal transferase-catalyzed addition of nucleotides to the 3' termini of DNA. Methods Enzymol. 1980;65(1):43–62. doi: 10.1016/s0076-6879(80)65009-5. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Marley G. M. Purification and properties of HindII and HindIII endonucleases from Haemophilus influenzae Rd. Methods Enzymol. 1980;65(1):104–108. doi: 10.1016/s0076-6879(80)65015-0. [DOI] [PubMed] [Google Scholar]
- Spector T. Refinement of the coomassie blue method of protein quantitation. A simple and linear spectrophotometric assay for less than or equal to 0.5 to 50 microgram of protein. Anal Biochem. 1978 May;86(1):142–146. doi: 10.1016/0003-2697(78)90327-5. [DOI] [PubMed] [Google Scholar]
- Sànchez J., Barbès C., Hernandez A., de los Reyes Gavilàn C. R., Hardisson C. Restriction-modification systems in Streptomyces antibioticus. Can J Microbiol. 1985 Oct;31(10):942–946. doi: 10.1139/m85-177. [DOI] [PubMed] [Google Scholar]
- Yanagida T., Ogawara H. Deoxyribonucleases in Streptomyces. J Antibiot (Tokyo) 1980 Oct;33(10):1206–1207. doi: 10.7164/antibiotics.33.1206. [DOI] [PubMed] [Google Scholar]