Abstract
Hexosaminidases are key enzymes in glycoconjugate metabolism and occur in all kingdoms of life. Here, we have investigated the phylogeny of the GH20 glycosyl hydrolase family in nematodes and identified a β-hexosaminidase subclade present only in the Dorylaimia. We have expressed one of these, HEX-2 from Trichuris suis, a porcine parasite, and shown that it prefers an aryl β-N-acetylgalactosaminide in vitro. HEX-2 has an almost neutral pH optimum and is best inhibited by GalNAc-isofagomine. Toward N-glycan substrates, it displays a preference for the removal of GalNAc residues from LacdiNAc motifs as well as the GlcNAc attached to the α1,3-linked core mannose. Therefore, it has a broader specificity than insect fused lobe (FDL) hexosaminidases but one narrower than distant homologues from plants. Its X-ray crystal structure, the first of any subfamily 1 GH20 hexosaminidase to be determined, is closest to Streptococcus pneumoniae GH20C and the active site is predicted to be compatible with accommodating both GalNAc and GlcNAc. The new structure extends our knowledge about this large enzyme family, particularly as T. suis HEX-2 also possesses the key glutamate residue found in human hexosaminidases of either GH20 subfamily, including HEXD whose biological function remains elusive.
Introduction
Hexosaminidases are enzymes ubiquitous across all domains of life and play multiple roles in glycoconjugate metabolism as they remove nonreducing terminal N-acetylgalactosamine and N-acetylglucosamine residues from glycans, glycolipids, glycoproteins, and glycosaminoglycans. In the case of β-hexosaminidases acting on nonreducing termini, most sequences are found in glycoside hydrolase families GH3 and GH20, which have distinct chemical mechanisms.1 Whereas β-hexosaminidases are primarily catabolic in mammals, in invertebrate species, they are often mediating purposeful processing steps, analogous to Golgi mannosidases, during the maturation of N-linked oligosaccharides.2 However, the biological significance of, and the structural basis for, hexosaminidase-mediated glycan-processing in nonvertebrates are poorly understood.
Four β-hexosaminidase genes are known from mammals: perhaps the most familiar are HEXA and HEXB encoding the α- and β-subunits of the hetero- and homodimeric lysosomal enzymes, which have been shown to be deficient in two storage diseases (Tay-Sachs and Sandhoff diseases, respectively),3 OGA encoding a nucleocytoplasmic O-GlcNAc-specific cleaving activity of family GH84 with roles in signaling4 and HEXDC encoding the nucleocytoplasmic hexosaminidase D, with an uncertain biological role. The latter enzyme5,6 is a member of GH20 subfamily 1 and is only distantly related to HEXA and HEXB, which are in the subfamily 2 (see Figure 1). Hexosaminidase D has a neutral pH optimum and preference for aryl N-acetyl-d-galactosaminides;5−7 this enzyme is apparently significantly responsible for elevated hexosaminidase activity in synovia in rheumatoid arthritis patients.8
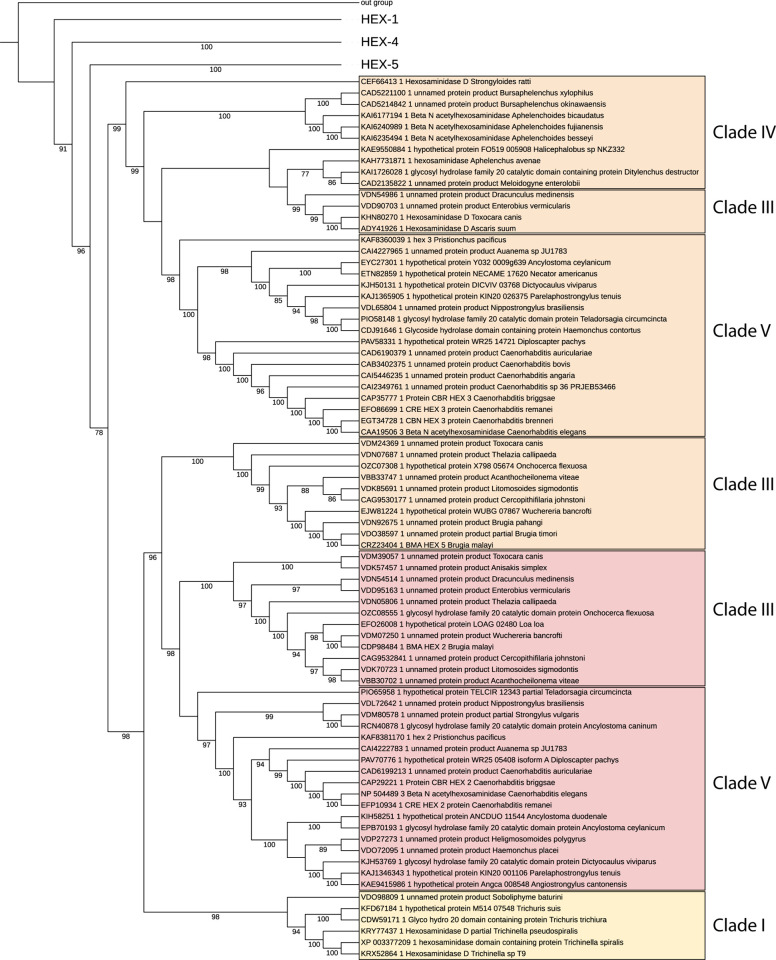
Figure 1.
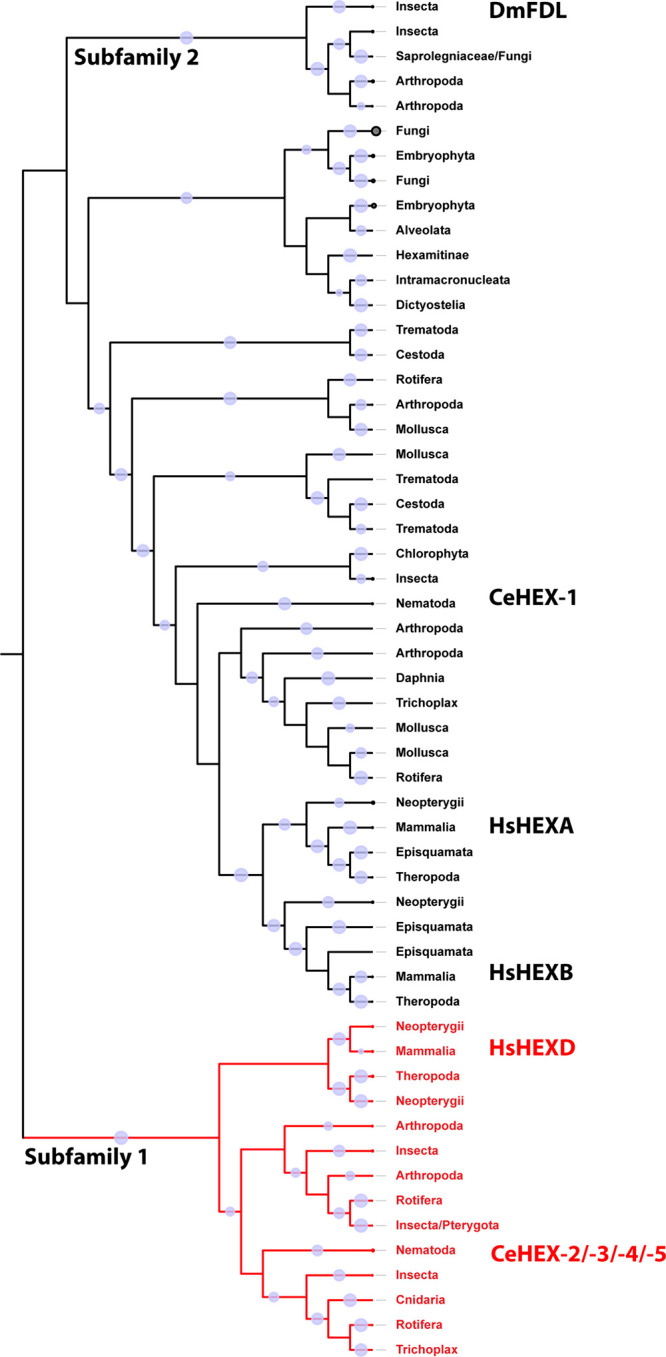
Phylogenic reconstruction of eukaryotic hexosaminidases. Subfamily 2 represents homologues of human HEXA and HEXB (HsHEXA and HsHEXB), C. elegans HEX-1 (CeHEX-1), and insect FDL (DmFDL); subfamily 1 (in red) contains homologues of human HEXD and C. elegans HEX-2/-3/-4/-5. Annotation was done based on known sequences from the literature. FastTree was used to generate an approximate maximum-likelihood phylogenetic tree, which was rooted at the midpoint. Blue circles represent bootstrap support between 70 and 100. Subfamilies 1 and 2 as categorized by Gutternigg et al.2 correspond to clades B and A defined by Intra et al.26
When considering nonvertebrate species, among the best-described hexosaminidases are those from insects. For example, Drosophila melanogaster possesses a number of β-hexosaminidase genes: (i) the cytoplasmic OGA encoding the O-GlcNAc-specific enzyme similar to that in mammals,9 (ii) one GH20 subfamily member 1 (CG7985) with no characterized enzymatic function,10 but phylogenetically relatively “close” to hexosaminidase D and (iii) three members of GH20 subfamily 2 including two chitinolytic and/or broad spectrum enzymes and one N-glycan-specific hexosaminidase.11 The latter is encoded by the fused lobes (fdl) gene named due to the brain morphology defect in the corresponding fruitfly mutant; as enzymes, insect FDL hexosaminidases have a particular specificity for the nonreducing terminal β1,2-GlcNAc linked to the “lower arm” α-1,3-mannose of N-glycans,11−16 thereby removing the GlcNAc transferred by MGAT1 (N-acetylglucosaminyltransferase I). The molecular identification of FDL explained earlier work indicating that a special N-glycan-processing enzyme was present in insect cell microsomes.17
The other major invertebrate model organism, the nematode Caenorhabditis elegans, possesses six β-hexosaminidase genes, but with a different subfamily bias as compared to insects: four of the encoded hexosaminidases belong to GH20 subfamily 1 (HEX-2, -3, -4, and -5), one to subfamily 2 (HEX-1), and one is a proven OGA from GH84 family.2,18 Similar to insect FDL, HEX-2 and -3 have proven activity toward the β-1,2-linked GlcNAc on the lower arm of N-glycans,2 corresponding to a hexosaminidase activity found in C. elegans microsomes;19 additionally, HEX-2 can also cleave nonreducing terminal GalNAc, a property also demonstrated for HEX-4 and -5.2,12 HEX-1, on the other hand, is apparently chitinolytic and is close phylogenetically to human HEXA and HEXB.2 Our use of GFP-promoter constructs suggested different tissue expression patterns for the C. elegans hex genes,2 whereas HPLC/MS-based analyses of hex-2, hex-2;hex-3, and hex-4 mutants showed an impact of their ablation on the N-glycome.2,20,21 Regarding other nematodes, there is little biochemical information regarding homologous enzymes, but a secreted N-glycan-digesting hexosaminidase from Trichinella spiralis, with no defined sequence, has been biochemically characterized22 and may be closest to C. elegans HEX-1 in terms of its properties.
Considering that C. elegans HEX-2 and HEX-3 are FDL-like in terms of their impact on the N-glycome, whereas HEX-4 is GalNAc-specific, we wished to explore the properties of further hexosaminidases from other nematode species. Preliminary database searching suggested that some nematodes have, like C. elegans, a number of GH20 subfamily 1 genes; for instance, Oesophagostomum dentatum, a clade V nematode like C. elegans, has at least HEX-2, -3, and -5 orthologues, whereas Trichinella spiralis and Trichuris suis (both clade I nematodes) appear to have only one subfamily 1 enzyme. On the other hand, O. dentatum lacks N-glycans with terminal GalNAc residues23 and wild-type C. elegans has very few;21 both species rather have chito-oligomer-based antennae for their most complex N-glycans. In contrast, T. suis(24) and T. spiralis(25) are rich in N-glycans containing terminal GalNAc motifs, which may indicate a difference in the hexosaminidase-dependent processing between clade I and V species. Therefore, a thorough phylogenetic analysis was performed and the GH20 subfamily 1 candidate enzyme from T. suis was expressed recombinantly, characterized, and successfully crystallized, yielding the first experimental structure of a eukaryotic subfamily 1 GH20 hexosaminidase.
Results
Phylogeny of GH20 Hexosaminidases
Initially, phylogenetic analyses were performed to recreate a comprehensive evolutionary pathway for GH20 hexosaminidases, based on using almost 4,000 sequences from eukaryotes. The results verify that there are two distinct groups of these hexosaminidases: subfamily 1 and subfamily 2 (Figure 1). Subfamily 2 encompasses the more familiar mammalian HEXA and HEXB as well as the insect FDL and nematode HEX-1 homologues. In subfamily 1, which includes mammalian HEXD, a clearly separated clade of nematode hexosaminidases was observed, which is represented in C. elegans by the previously characterized HEX-2, HEX-3, HEX-4, and HEX-5 enzymes.2,12,21 Each C. elegans hexosaminidase is within its own distinct group of related sequences from other nematodes.
In C. elegans, the earliest separation occurs between HEX-4 and HEX-5 (Figure 2 and Supplementary Figure S1), indicating their specific ability to only remove GalNAc. Later, HEX-3 and HEX-2 evolved and are capable of removing both GalNAc and GlcNAc from glycan structures in vitro.2 The presence of numerous enzymes with similar functions indicates a significant amount of evolutionary pressure or possibly diverse applications for these enzymes. In general, the degree of relatedness within the GH20 clades correlates well with the proposed phylogeny of nematode species (e.g., filarial sequences are grouped together). Additionally, a subclade consisting of Trichuris spp. and Trichinella spp. representatives was identified in which only one hexosaminidase homologue per species, annotated as HEXD, was detected in the database. This finding is unusual compared to other nematodes that possess up to four subfamily 1 GH20 hexosaminidases but reflects that Trichuris spp. and Trichinella spp. are phylogenetically distinct from the majority of nematodes, falling within nematode clade I as defined by Blaxter.27,28 Based on phylogenetic analysis, it can be inferred that Trichuris and Trichinella enzymes are likely to have a similar activity to that of C. elegans HEX-2; thus, we designated the theoretical KFD87184 sequence as T. suis HEX-2. Overall, T. suis HEX-2 has around 40% identity over ca. 500 residues with C. elegans HEX-2 and HEX-3 (Supplementary Figure S2) and shares the His/Asn-Xaa-Gly-Yaa-Asp-Glu motif with many other GH20 hexosaminidases (Supplementary Figure S3), whereby this sequence is shifted toward the N-terminus of subfamily 1 sequences as compared to subfamily 2.
Figure 2.
Nematode hexosaminidase phylogeny. Maximum likelihood (IQ-TREE) phylogeny of the nematode GH20 hexosaminidases. The HEX-2 and HEX-3 branches are highlighted and annotated with the groups of related species in terms of the Nematoda clades as defined by Blaxter;27,28Supplementary Figure S1A,B shows all the different nematode hexosaminidase branches. The D. melanogaster FDL sequence was used as an out group. Bootstrap values of >70 are shown. In the HEX-2 branch, the sequences highlighted in yellow represent a subclade of sequences in clade I species, which only have one subfamily 1 member each.
Characterization of HEX-2
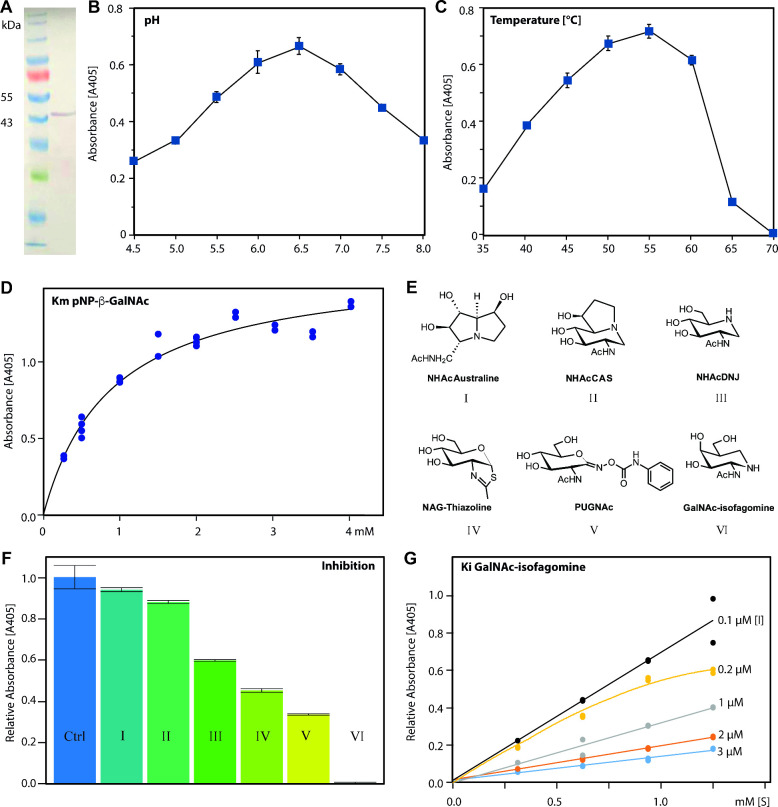
To determine whether T. suis HEX-2 had an activity similar to that of C. elegans HEX-2, we cloned the predicted open reading frame, excluding the sequence encoding the N-terminal cytoplasmic, transmembrane and stem domains (i.e., residues 85–620 or 137–620 of the predicted sequence, Supplementary Figure S2), into a Pichia vector for secreted expression. Only constructs with a C-terminal His-tag could be purified by immobilized metal affinity chromatography. Resulting ‘short’ or ‘long’ forms of the protein with apparent molecular weights of 50 or 75 kDa (Figure 3A and Supplementary Figure S4) were also verified by tryptic peptide mapping. In terms of enzymatic activity, we first examined the properties of T. suis HEX-2 using artificial aryl glycoside substrates. While pNP-β-GlcNAc was a poor substrate, there was excellent activity toward pNP-β-GalNAc (Supplementary Figure S4). It was observed that within the linear range of product formation with respect to time and in the presence of McIlvaine buffers, optimal activity was at pH 6–7 (Figure 3B), similar to the values for the C. elegans homologues HEX-2 and HEX-4.26 The optimal temperature was 50–60 °C (Figure 3C), similar to C. elegans HEX-2 and HEX-3.12 Incubation with a range of pNP-β-GalNAc substrate concentrations and 5 ng of T. suis HEX-2 allowed for the determination of an apparent Km value of 0.9 mM (Figure 3D), which is also in the range for other characterized nematode hexosaminidases.
Figure 3.
Activity of recombinant T. suis HEX-2 with a simple substrate. (A) Anti-His western blot of the purified recombinant C-terminally His6-tagged “short” form of T. suis HEX-2 expressed in Pichia. (B) pH dependency of activity toward pNP-β-GalNAc of recombinant T. suis HEX-2 assayed at 37 °C for 1 h using a range of McIlvaine buffers. (C) Temperature dependency of recombinant T. suis HEX-2. (D) Michaelis–Menten curve for T. suis HEX-2 with pNP-β-GalNAc as substrate. (E, F) Inhibition of T. suis HEX-2 protein using pNP-GalNAc as a substrate (5 mM) and six different competitive inhibitors (0.5 mM). (G) Graphical representation of the data obtained with GalNAc-isofagomine to calculate Ki as fitted by Prism (GraphPad). Each assay was performed in duplicate or triplicate and error bars indicate standard deviations; in panels (F) and (G), relative absorbance is in comparison to the activity of the uninhibited enzyme.
Six different known hexosaminidase inhibitors were tested (PUGNAc, NHAcDNJ, NHAcCAS, NHAc-Australine, NAG-Thiazoline, GalNAc-isofagomine,29−34Figure 3E) with T. suis HEX-2. After preincubation of the competitive inhibitors with the enzyme, pNP-β-GalNAc was again used as a substrate and the activity was determined. The highest degree of inhibition as compared to the control was observed with GalNAc-isofagomine and the least with NHAc-Australine (Figure 3F). The Ki for GalNAc-isofagomine was determined to be 0.6 μM (Figure 3G), a value lower than that determined for Streptomyces plicatus β-N-acetylhexosaminidase.34
Specificity of T. suis HEX-2
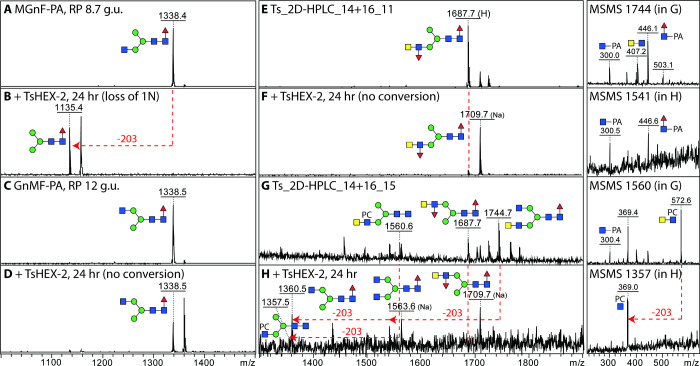
T. suis HEX-2 was tested with typical N-glycan substrates and shown to remove only one GlcNAc from a GnGn-dabsyl-N-glycopeptide (m/z 2060), but both GalNAc residues as well as just one terminal GlcNAc from a βGNβGN-dabsyl-N-glycopeptide (m/z 2467, with two LacdiNAc units) (Figure 4A–E). In order to test the arm specificity, RP-HPLC analysis of a pyridylamino-N-glycan (GnGn) before and after incubation with T. suis HEX-2 was performed; the shift to later retention time was indicative of removing solely the “lower” arm GlcNAc (Figure 4F) and dependent on the pH of the reaction mixture (Supplementary Figure S4D). Another test of the specificity was to take RP-HPLC-purified core fucosylated N-glycans from Dirofilaria immitis with either a lower or an upper arm GlcNAc;35 in the case of the former, the nonreducing terminal GlcNAc was removed (Figure 5A,B). In contrast, T. suis HEX-2 did not remove the upper arm of GlcNAc (Figure 5C,D). Regarding more complicated structures with LacdiNAc-based antennae, the activity of T. suis HEX-2 toward two selected N-glycan fractions derived from T. suis itself were selected. While the glycan with a fucosylated LacdiNAc was resistant to HEX-2 (Figure 5E,F), the structure with a phosphorylcholine-substituted LacdiNAc did lose the terminal GalNAc, as also reflected by the MS/MS data (Figure 5G,H)
Figure 4.
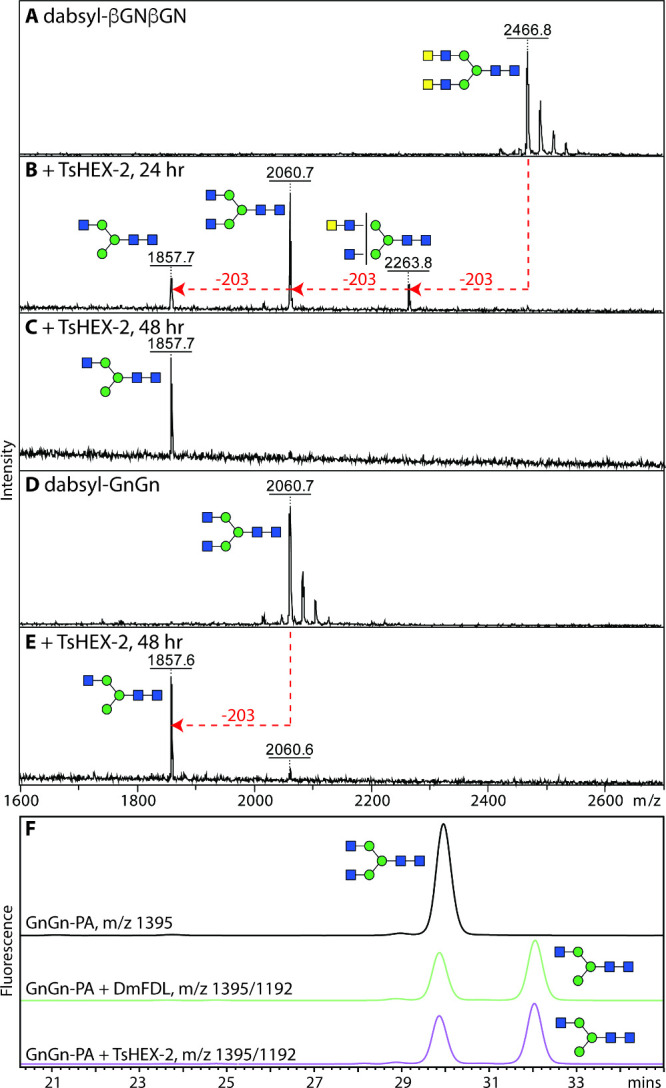
Activity of recombinant T. suis HEX-2 with biantennary glycan substrates. (A–E) MALDI-TOF MS analysis of incubations of dabsyl glycopeptides carrying either GnGn (m/z 2060) and βGNβGN (m/z 2467, with two LacdiNAc units) before (A/C) or after treatment with T. suis HEX-2 for 24 h (B), 48h (C/E). (F) RP-HPLC chromatogram of GnGn-PA (m/z 1395) before (black) or after treatment with either insect FDL (green) or T. suis HEX-2 (blue); a shift to later elution time is indicative of removal of the “lower” nonreducing terminal GlcNAc residue.36 Red lines with arrows indicate losses of HexNAc residues from the substrates. Glycans are depicted according to the Symbol Nomenclature for Glycans (SNFG).
Figure 5.
Activity of recombinant T. suis HEX-2 with complex nematode glycan substrates. (A–D) MALDI-TOF MS of Dirofilaria immitis glycans before (A/C) or after incubation with T. suis HEX-2 (B/D); while one Hex3HexNAc3Fuc isomer (MGnF, m/z 1338, eluting at 8.7 g.u. on RP-HPLC35) was sensitive (B), the second isomer (GnMF, eluting at 12 g.u.) was resistant. (E–H) MALDI-TOF MS of 2D-HPLC purified T. suis glycans before (E/G) and after incubation (F/H) with T. suis HEX-2; while two glycans with fucosylated LacdiNAc motifs (m/z 1687 as [M + H]+ or 1709 as [M + Na]+) were not digested, structures with either nonsubstituted or phosphorylcholine-substituted LacdiNAc (m/z 1744 and 1560 as [M + H]+) were sensitive to T. suis HEX-2 (respective products of m/z 1563 and 1360 as [M + Na]+ and 1357 as [M + H]+), resulting in alterations in the MS/MS spectra (loss of B-ion HexNAc2PC0–1 fragments of m/z 407 and 572). Note that the addition of HEX-2 results in a shift to sodiated ions for neutral glycans. Glycans are depicted according to the Symbol Nomenclature for Glycans (SNFG).
X-ray Crystallography
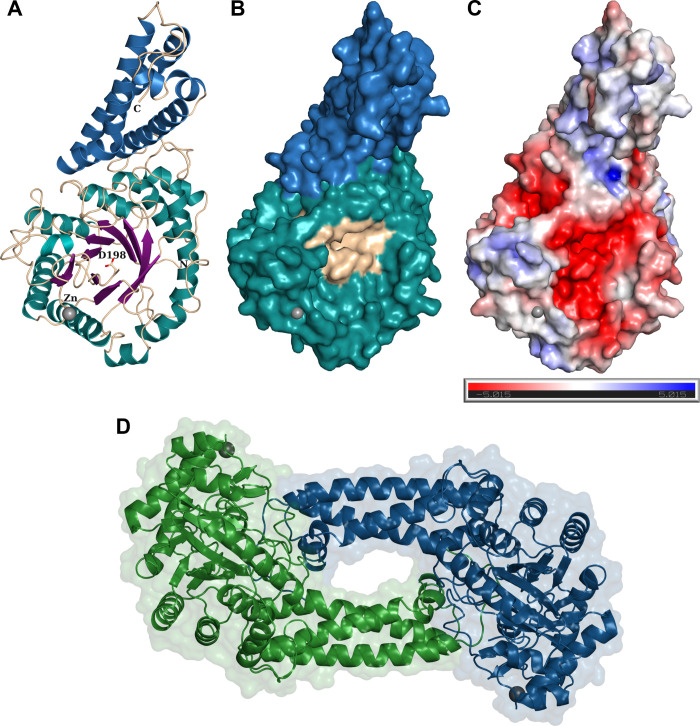
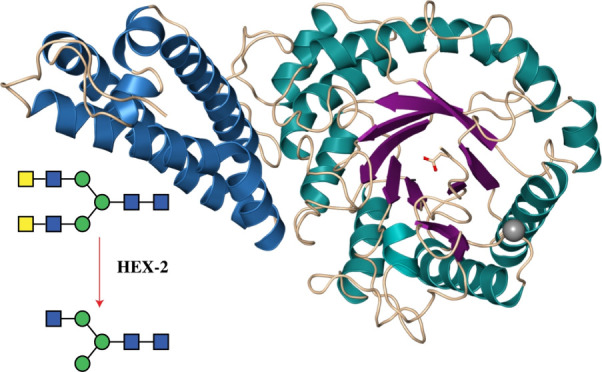
In order to gain insight into the specificity of T. suis HEX-2, we solved its X-ray crystal structure at a resolution of 2.55 Å in a C2 space group (Table 1). Overall, HEX-2 displays a modular structure with a N-terminal catalytic domain taking the shape of an (β/α)8 or TIM barrel followed by a C-terminal three helix bundle, whose function remains unknown. There is one protomer in the asymmetric unit (Figure 6A). Analysis of the interfaces and assemblies with PDBePISA37 revealed one interface leading to the formation of a crystallographic dimer around the 2-fold axis. The interface represents only 13% of the solvent accessible area (2610 Å) and includes 15.8% of the total residues located in surface loops of the C-terminal part of the three helices bundle interacting with the surface loops connecting strands and helices 2, 3, 7, and 8 of the TIM (Figure 6B). The complex formation significance score (CSS) of 0.3 indicates an auxiliary role of the interface in the dimer formation implying an unstable or weak dimer or a crystallographic artifact; other GH20 enzymes are indeed known to exist as dimers in solution, including murine HexD and OfHex16,38 and HEX-2 migrated on native gel electrophoresis in multimeric forms (Supplementary Figure S4B). Although the enzyme is predicted to be N-glycosylated, no electron density for even a core GlcNAc could be detected. Disordered regions at the N- and C-terminus correspond to the probable stem and the hexahistidine tag, respectively. The structure also revealed the presence of a Zn2+ ion (Figure 6A) not far from the binding pocket. There is also a disordered surface loop between the Zn2+ site and the binding site which resulted in the lack of electron density between residues Glu199 and Arg220 so they could not be modeled (Supplementary Figure S5). Analysis of the closest related structure, the GH20C β-hexosaminidase from Streptococcus pneumoniae,39 indicates that this region should contain an α-helix. Additionally, the positioning of the Zn2+ ion implies a noncatalytic/structural role, potentially in the stabilization of this disordered loop upon substrate binding. Experiments indeed showed that up to 10 mM Zn(II) or EDTA had only minor effects on the enzyme activity (Supplementary Figure 4E,F). The N-terminal region of the crystallized protein with no observed electron density (residues 85–137 of the full theoretical KFD87184 sequence, corresponding to residues 1–53 of the construct) does not align well with C. elegans HEX-2 (Supplementary Figure S2) and was susceptible to proteolysis; as the “short” form of the enzyme lacking this region was active, it is assumed that the stem domain extends as far as Phe138 of the full theoretical sequence.
Table 1. Data Collection and Refinement Statisticsa.
| data collection | |
|---|---|
| beamline | SOLEIL Proxima-1 |
| wavelength | 0.97856 |
| space group | C2 |
| unit cell dimensions | 96.12 59.04 105.98 90.00 114.49 90.00 |
| resolution (Å) | 43.74–2.55 (2.66–2.55) |
| Nb reflections | 116,794 (14,395) |
| Nb unique reflections | 17,835 (2,157) |
| Rmerge | 0.067 (0.782) |
| Rmeas | 0.080 (0.931) |
| Rpim | 0.043 (0.500) |
| mean I/σI | 14.1 (2.1) |
| completeness (%) | 99.9 (99.9) |
| redundancy | 6.5 (6.7) |
| CC1/2 | 0.999 (0.843) |
| refinement | |
|---|---|
| resolution (Å) | 43.74–2.55 |
| no. reflections | 17,268 |
| no. free reflections | 836 |
| Rwork/Rfree | 0.189/0.259 |
| rmsd bond lengths (Å) | 0.0120 |
| rmsd bond angles (deg) | 1.99 |
| rmsd chiral (Å3) | 0.0864 |
| clashscore | 11 |
| No. atoms/Bfac (Å2) | |
| protein | 3,688/44.5 |
| water | 45/32.3 |
| Zn | 1/81.3 |
| ramachandran | |
| allowed/favored (%) | 99.3/93.4 |
| outliers | 3 |
| PDB Code | 8QK1 |
Values in parentheses are for the high-resolution shell.
Figure 6.
3D-structural analysis of recombinant T. suis HEX-2. (A) Cartoon representation of the X-ray crystal structure of T. suis HEX-2 at 2.55 Å. The TIM barrel and helix bundle are colored differently; the key catalytic Asp residue is represented as ball and sticks and Zn2+ ion as a gray ball. (B) Surface representation of HEX-2 with the helix bundle colored in blue, the TIM barrel in green and the active site area in wheat. (C) ±5 kT/e electrostatic potential of HEX-2 in PyMOL plotted on the solvent-accessible surface and calculated with APBS plugin.40 (D) Surface and cartoon representations of the HEX-2 potential dimer.
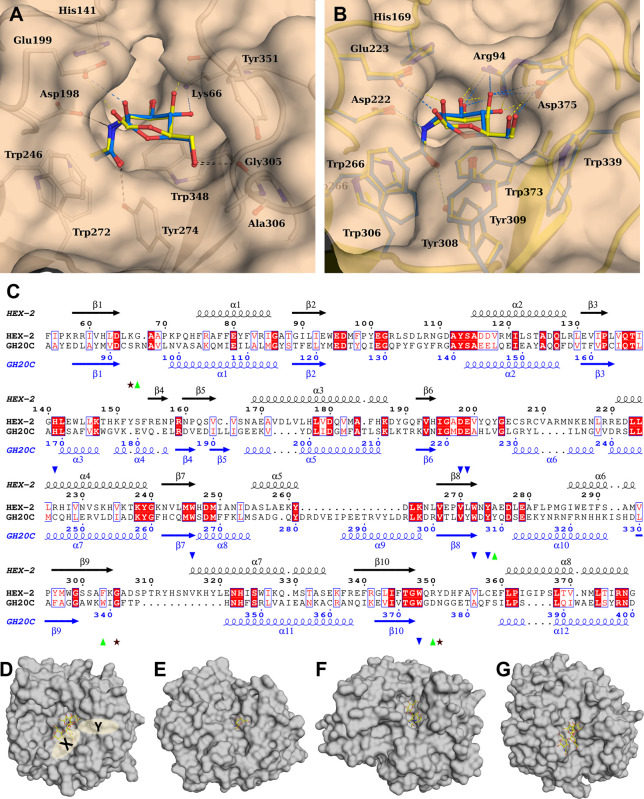
As the enzymatic activity experiments indicated that T. suis HEX-2 removes GalNAc and GlcNAc residues from glycan substrates, its newly resolved catalytic domain was superimposed on that of S. pneumoniae GH20C (Supplementary Figure S6), which has been cocrystallized with the GalNAc and GlcNAc reaction products, which are also surrogates for actual substrates. It is noted that those monosaccharides are distorted in the catalytic site. 265 residues out of 320 aligned with a rmsd of 1.9 Å and major differences are observed at the level of surface loops in particular those surrounding the active site pocket and in particular the −1 subsite as described below. This structural analysis indicated that either monosaccharide is capable of effectively binding to the predicted −1 subsite (Figure 7A), whereby the key contacts will be conserved with the 1-hydroxyl and 2-acetamido groups. Based on the presented structure, a number of residues are predicted to participate in substrate binding within the structure (Table 2, Figure 7). These include the Asp198 and Glu199 of the His/Asn-Xaa-Gly-Xaa-Asp-Glu motif shared with other GH20 hexosaminidases (i.e., residues 282 and 283 of the full theoretical sequence), whereby the Asp and Glu are the polarizing and general acid/base residues as demonstrated by studies on members of this retaining enzyme family, including a photoaffinity labeling study on human hexosaminidase B.41 Four aromatic groups forming the bottom (Trp348) and the walls of the active site pocket around the acetamido group (Trp246, Trp272, and Tyr274) are also conserved (Figure 7). There are, though, differences in terms of contacts with the monosaccharide for hydroxyls at the 3, 4, and 6 positions and in the binding side topology around the hydroxymethyl group resulting from differences in several surface loops between HEX-2 and GH20C (Figure 7). The loop containing Arg94 in GH20C is a little shorter in HEX-2, but the side chain nitrogen of Lys66 occupies the same position as the NH1 atom of Arg94 and therefore could interact with the 3-hydroxyl but also with the 4-hydroxyl in GlcNAc. In GH20C, the 4 and 6-hydroxyls interact with the carboxylate atoms of Asp375 while the hydroxymethyl conformation is blocked by hydrophobic interactions with Trp339 and Tyr309 (Figure 7B). In HEX-2, those residues are replaced by the side chains of Tyr351, Gly305, and Ala275, respectively. The sequence of the surface loops containing Gly305 and Tyr351 are nonconserved leading to a very different conformation, which, along with the replacement of Tyr309 by an alanine, results in altered interactions and a more open active site pocket in HEX-2. In order to avoid steric clashes with Tyr351, the hydroxymethyl has to rotate to stack along the aromatic ring and in that orientation the 6-hydroxyl group would make an H-bond with the main chain oxygen of Gly305 (Figure 7A); thus, it can be expected that conformational changes upon substrate binding would optimize the interactions and result in ordering of the missing amino acids 200–220. The differences in sequence and hence in conformation of six surface loops surrounding the −1 subsite have a strong impact in the overall architecture of the active site and in the formation of additional subsite (in HEX-2:65–74, 195–206, 247–254, 273–285, 305–315, 348–360). In HEX-2, the loop containing Lys66 corresponding to the one containing Arg94 in GH20C, will prohibit the formation of a −2 subsite as found in the Bifidobacterium bifidum lacto-N-biosidase LNBase (Figure 7D–F).42 The other five loops create two grooves that could accommodate at least +1 and +2 subsites and a branched glycan (Figure 7D). The X groove is also found in other hexosaminidases such as in the endoglycosidase E GH20 from Enterococcus faecalis, but is shallower in HEX-2 (Figures 7D,F).43
Figure 7.
Modeling and alignments of the binding site pocket of the T. suis HEX-2 catalytic domain. (A) Interactions of HEX-2 with manually docked GalNAc (yellow) and GlcNAc (cyan). Position of Glu199 has been modified to the one expected upon binding. (B) Interactions of S. pneumoniae GH20C β-hexosaminidase with GalNAc (yellow carbons; PDB-ID: 5AC4) and GlcNAc (cyan carbons; PDB-ID: 5AC5). Amino acids are represented by balls and sticks; H-bonds are displayed in dash lines of corresponding color. (C) Alignment of HEX-2 with GH20-C highlighting the amino acids involved in the active site with the one conserved (blue triangles), those only found in HEX-2 (green triangles) and those only in GH20-C (brown stars). Alignment made by Clustal Omega44 and figure drawn with ESPript 3.0.45 Surface representation of the TIM barrel of T. suis HEX-2 (D), S. pneumoniae GH20C in complex with GlcNAc (E, PDB-ID: 5AC4), Bifidobacterium bifidum Lacto-N-biosidase LNBase in complex with LNB-NHAcAUS (F, PDB-ID: 5BXT) and Enterococcus faecalis endoglycosidase E GH20 domain (G, PDB-ID: 7PUL). Figures are drawn in the same orientation after overlay on the HEX-2 catalytic domain to illustrate active site architecture with ligand represented in balls and sticks. The ligand of PDB-ID: 2YLA was manually docked in HEX-2 and EndoE.
Table 2. List of Residues Potentially Involved in the Binding of Substrates as Compared to S. pneumoniae GH20C β-Hexosaminidasea.
| substrate binding | T. suis HEX-2 | S. pneumoniaeβ-hexosaminidase |
|---|---|---|
| GalNAc/GlcNAc H-bond O3 | K66 | R94 |
| GalNAc/GlcNAc H-bond N2 | D198 | D222 |
| GalNAc/GlcNAc H-bond O1 | E199 | E223 |
| GalNAc/GlcNAc hydrophobic N-Acetyl | W246 | W266 |
| GalNAc/GlcNAc hydrophobic N-acetyl | W272 | W306 |
| GalNAc/GlcNAc O7 | Y274 | Y308 |
| GalNAc/GlcNAc hydrophobic ring | W348 | W373 |
| GalNAc O6/GlcNAc O4 & O6 | G305 | D375 |
| GalNAc/GlcNAc hydrophobic O6 | Y351 |
Residue numbering is for the crystallized forms.
Discussion
The phylogenetic analysis of eukaryotic GH20 hexosaminidases presented here has provided new insights into the origin of these enzymes in nematodes (Figures 1 and 2). As indicated by previous studies, it was confirmed that the aforementioned HEXD is the most similar human homologue to the subfamily 1 hexosaminidases present in nematodes.26 Furthermore, the phylogenetic reconstruction revealed that the four major branches of subfamily 1 hexosaminidases found in nematodes derive from a single ancestor, indicating that the genes evolved through numerous duplications and later speciation. While most nematodes analyzed possess multiple predicted subfamily 1 GH20 hexosaminidases, the examined clade I species (Dorylaimia; Soboliphyme baturini, Trichuris spp., and Trichinella spp.) have apparently only one such sequence forming a separate GH20 subbranch (Figure 2).
Previously published studies have suggested that subfamily 1 GH20 enzymes prefer aryl β-N-acetylgalactosaminides over β-N-acetylglucosaminides and are generally more efficiently inhibited by galacto-epimers of hexosaminidase inhibitors as compared to those in the gluco-configuration2,7,39 with our unpublished data on C. elegans HEX-2 also indicating a reduction in Ki for Gal-PUGNAc as opposed to PUGNAc (500-fold). In the case of T. suis HEX-2, low activity toward pNP-β-GlcNAc was detected, but it had a high activity with pNP-β-GalNAc and GalNAc-isofagomine was the most effective inhibitor of those tested (Figure 3), demonstrating its close relationship with other subfamily 1 enzymes.
The situation with N-glycan substrates is more complex than for the simple aryl glycosides: while only the “lower” GlcNAc was removed from biantennary glycans with nonreducing terminal GlcNAc residues, all terminal GalNAc residues and one subterminal GlcNAc was lost from biantennary glycans with LacdiNAc motifs (Figures 4 and 5). This is akin to the activity of C. elegans HEX-2; thus, the phylogenetic designation of the T. suis enzyme as an HEX-2 matches its enzymatic activity. On the other hand, C. elegans HEX-3 only removes the “lower” GlcNAc, but C. elegans HEX-4 is completely GalNAc-specific. Thus, despite the galacto-epimer bias for the simple substrates and inhibitors as for other subfamily 1 enzymes, T. suis HEX-2 can also digest a specific GlcNAc-containing linkage at slightly acidic pH in the same manner as insect FDL enzymes, which are members of GH20 subfamily 2. In contrast, plant hexosaminidases, which remove nonreducing terminal GlcNAc, can remove both such residues from N-glycans.2,12
Overall, we assume that T. suis HEX-2 has a role in the biosynthesis of the major paucimannosidic N-glycans known to occur in this parasite.24 The subtlety of its specificity toward LacdiNAc-type substrates are of interest: while C. elegans HEX-4 can remove GalNAc from fucosylated and phosphorylcholine-modified LacdiNAc-type motifs,46 the presence of an antennal fucose appears to block the action of T. suis HEX-2. Unlike C. elegans, the T. suis N-glycome is rich in antennal fucose motifs but lacks the phosphorylcholine-modified chito-oligomer modifications found in a variety of other nematodes; also C. elegans HEX-4 appears to have a role in N-glycan biosynthesis in the Golgi apparatus and ablation of its gene leads to an increase in GalNAc-containing N-glycans in the model nematode.21 Thus, the fine biosynthetic control mechanism involving LacdiNAc-containing glycans may be lacking in T. suis and so may be a partial explanation for its species-specific glycome.47 On the other hand, the catabolism of GalNAc- and GlcNAc-containing N-glycans at acidic pH may be performed in T. suis by HEX-1, which belongs to GH20 subfamily 2; at least, the potentially related T. spiralis enzyme can degrade such structures.22
The GH20C enzyme from Streptococcus pneumoniae,39 which is the closest related protein (29% identity) with a crystal structure, has some bias toward GalNAc, but far less than T. suis HEX-2. Nevertheless, as GH20C was also cocrystallized with GalNAc, GlcNAc, and some inhibitors, the superimposition with its structure is the most meaningful possible approximation; these structures were the basis for predicting potential interactions of T. suis HEX-2 with GalNAc and GlcNAc in subsite −1 (Figure 6B). The two enzymes only superposed well in the catalytic domain (Figure 7A). Although the modeled conformations of the two monosaccharides are subtly different, H-bond interactions of the anomeric hydroxyl group with Glu199 (corresponding to Glu223 of GH20C) and the 2-acetamido group with Asp198 and Tyr274 (corresponding to Asp222 and Tyr308 of GH20C) would be conserved as well as the hydrophobic or stacking interactions with aromatic rings of Trp246, Trp 272, and Trp348 (corresponding to Trp residues 266, 306, and 373 of GH20C). The cocrystallization with GH20C indicates a different H-bonding pattern of the 4-hydroxyl groups of GalNAc and GlcNAc to either guanidino amino group of Arg 94 (Figure 7B), enabling binding of both monosaccharides; the corresponding Lys66 in HEX-2 only has a single side chain amino group, which may affect relative specificities for the two pNP-substrates as well as the inhibitors. Although the in silico prediction using AlphaFold was close to the model from the crystal structure (Figure 7C and Supplementary Figure S5), some deviations were found, thereby highlighting the value of an experimentally based approach. The presence of a Zn2+ ion near the proposed active site was not expected, its role is unknown but this is found in some other glycosidases, including Golgi mannosidase II.48 Another example monosaccharide-releasing hexosaminidase with a high GalNAc-bias is the recently crystallized Paenobacillus TS12 NgaP2 (PDB 8K2L);49 despite overall structural superposition of their catalytic regions being possible, a meaningful comparison regarding side chains determining substrate specificity is difficult as NgaP2, assigned to the GH123 family, has only 11% identity with HEX-2.
Few comparisons can be made to eukaryotic GH20 hexosaminidases as the four proteins for which there are crystallographic data are all of subfamily 2. Nevertheless, the key role of an Asp-Glu pair and a Tyr residue up to 100 amino acids toward the C-terminus in binding GalNAc, GlcNAc, or inhibitors is conserved.50−52 However, the general architecture of subfamily 2 enzymes contrasts with that of T. suis HEX-2, whereby the active site is closer to the N-terminus in subfamily 1. As most GH20 crystallographic studies are of bacterial enzymes, the crystal structure described here is particularly valuable, as it is the first eukaryotic one from this subfamily. Thus, this structure and data presented here coupled with further studies will allow for a better understanding of the substrate specificities of invertebrate hexosaminidases and how they have evolved.
Experimental Procedures
Phylogenetic Analyses
Eukaryotic Tree
To find all eukaryotic sequences, which belong to the GH20 hexosaminidase family, the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) was used.53 Two protein families IPR015883 (subfamily 1) and IPR025705 (subfamily 2) were found, and all eukaryotic data was downloaded and stored on a local disk. The data has been processed to decrease the number of input sequences; any sequences below 300 or over 750 amino acids were removed. Next, sequences were used to build an alignment with MAFFT54 and then the TrimAl tool was used for alignment trimming.55 Thereafter, the data was used as input to calculate a new phylogeny tree using the FastTree tool56 on the local computer and visualized with iTOL.57
Nematode Tree
Characterized GH20 sequences from C. elegans were taken from the Wormbase database (gene names, HEX-1 – CE07499; HEX-2 – CE36785; HEX-3 – CE41720; HEX-4 – CE46668; HEX-5 – CE53609) and used as a query the whole Nematode proteome (NCBI 11.01.2023) using the hidden Markov Models algorithm from phmmer. All found sequences were used to build an alignment with MAFFT54 and subsequently the final approximately maximum-likelihood phylogenetic tree was built with IQ-tree.58 The resulting phylogeny tree was limited to one homologue per species and visualized with iTOL.57
Cloning and Purification
The T. suis HEX-2 open reading frame sequence, excluding the region encoding the cytosolic, transmembrane, and stem domains (i.e., residues 85–620 of the predicted protein), was synthesized by GenScript, based on the sequence with NCBI database ID KFD67184. The hexosaminidase sequence was cloned into the pPICZαA plasmid (Invitrogen) without the native stop codon using the Gibson Assembly Cloning Kit (primers: Forward/Long: 5′-AGAGAGGCTGAAGCTGAATTCACGATGAAAGTGTATCGATGGCGA-3′, Forward/Short: 5′-AGAGAGGCTGAAGCTGAATTCACGGTGTTTATTCCGAAACGT-3′, Reverse: 5′-GACGGCACGCGTCGTATCGATAG-3′). Ligation products were transformed into 5-alpha competent Escherichia coli (New England Biolabs, C2987) prior to selection on zeocin. The sequenced expression vectors were linearized and transformed into P. pastoris (GS115 strain), and colonies were selected on Zeocin and expression performed with methanol induction at 30 °C as previously described.12
Purification of the recombinant proteins from the culture media was performed with an ÄKTA go protein purification system with HisTrap High Performance 1 mL column (Cytiva); samples were applied in binding buffer (25 mM sodium phosphate, 150 mM NaCl, pH 7.4), and the column was washed before using a gradient of elution buffer (25 mM sodium phosphate, 150 mM NaCl, 500 mM imidazole, pH 7.4). After purification, eluted fractions were tested for purity by SDS-PAGE; fractions of interest were pooled, concentrated using an Amicon Ultra-0.5, Ultracel-30 Membrane with a 30 kDa cutoff (Merck Millipore), and exchanged into storage buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.5). His-tagged forms of the hexosaminidases were detectable after western blotting using the anti-His monoclonal antibody (1:10000; Sigma-Aldrich) and alkaline-phosphatase conjugated antimouse IgG (1:10000; Sigma-Aldrich). The resulting secreted long (536 residues corresponding to residues 85–620 of the predicted protein) and short (residues 135–620 of the predicted protein, i.e., 51–536 encoded by the construct) forms both had a C-terminal His-tag. An alternative long form with an N-terminal His/FLAG-tag was also expressed (Supplementary Figure S4A) and was active, but lost the N-terminal tag due to proteolysis prior to purification. To examine the monomeric or multimeric status of HEX-2 (Supplementary Figure S4B), native gel electrophoresis was performed as for SDS-PAGE, except for exclusion of SDS and reducing agents from the sample buffer and gel; the resulting gel was fixed in a solution of 40% (v/v) ethanol and 10% (v/v) acetic acid, incubated with 0.125% (w/v) glutaraldehyde, 0.2% (w/v) sodium thiosulfate, 6.8% (w/v) sodium acetate in 30% (v/v) ethanol, and then with 0.25% (w/v) silver nitrate and 0.015% (v/v) formaldehyde prior to development overnight in 2.5% (w/v) sodium carbonate and 0.0075% (v/v) formalehyde.
Hexosaminidase Assays
The standard enzyme activity test was performed in 96-well plates. Typically, a mixture of 2.5 μL of pNP-β-GalNAc (100 mM in dimethyl sulfoxide), 46.5 μL of McIlvaine buffer pH 6.5,59 and 1 μL of enzyme was incubated for 1 h at 37 °C; 200 μL of stop solution (0.4 M glycine/NaOH, pH 10.4) was added and the absorbance measured with an Infinite 200 PRO instrument (Tecan). Inhibitors were prepared as previously reported.30,34,60−63 For tests with remodelled glycopeptides12 or 2D-HPLC fractions,35,47 a 1 μL aliquot was mixed with 0.2 μL enzyme and 0.8 μL of 50 mM ammonium acetate solution, pH 6.5. After overnight incubation at 37 °C, 0.5 μL of the mixture was analyzed by MALDI-TOF-MS (Autoflex Speed, Bruker, Bremen) with 6-aza-2-thiothymine (ATT) as the matrix; data were analyzed with the Flexanalysis (Bruker) program.
X-ray Crystallography
Initial protein crystallization screening was performed using the robotized HTXlab platform (EMBL, Grenoble, France) in a sitting drop vapor diffusion setup by mixing 100 nL of protein solution (5.1 mg/mL) and 100 nL of crystallization solution prior storage at 20 °C in a visible and UV Imaging Robot. A second screening was performed using different commercially available crystallization screens at CERMAV in a hanging drop vapor diffusion setup by mixing 1 μL of protein solution (6 mg/mL) and 1 μL of crystallization solution. The screening plate was kept in a vibration-free incubator (Molecular Dimensions, Calibre Scientific, Rotherham, UK) at 19 °C. Crystal clusters were obtained from condition 18 of the Clear Strategy Screen II (Molecular dimensions) consisting of 20% PEG 1500, 0.15 M potassium thiocyanate, and 0.1 M Tris pH 7.5. 15% PEG 1000 were added to the mother liquor as a cryoprotectant prior to mounting a single crystal in a cryoloop (Molecular Dimensions) and flash freezing in liquid nitrogen. The diffraction data were collected at Synchrotron SOLEIL, Beamlines, Proxima-1 (Saint-Aubin, France) using an Eiger 16 M detector (Table 1). The XDS64 and XDSme65 were used to process the data and further steps were performed with CCP4, version 8.25–27.66,67 As the crystal diffracted anisotropically, data was processed using the STARANISO server and the aimless CCP4 program. The structure of HEX-2 was solved by molecular replacement where AlphaFold68 was used to generate a search model for PHASER.69 Iterated maximum likelihood refinement and manual building of the resulting electron density maps were respectively performed using REFMAC 5.870 and Coot.71 Five percent of the reflections were used for cross-validation analysis, and the behavior of Rfree was employed to monitor the refinement strategy. Water molecules were added by using Coot and subsequently manually inspected.
Acknowledgments
This work was funded by the FWF-funded BioTOP doctoral programme [Austrian Science Fund W 1224] and by an FWF grant [P29466] to I.B.H.W.; I.A. is funded by the “Top Vet Science” Program of the University of Veterinary Medicine Vienna. This work benefited from access to the EMBL HTX lab, supported by iNEXT Discovery, project number 871037, funded by the Horizon 2020 program of the European Commission. We acknowledge the synchrotron SOLEIL (Saint Aubin, France) for access to beamlines Proxima 1 (Proposal Number 20210859) and for the technical support of Pierre Legrand. We would like to thank Valérie Chazalet and Emilie Gillon for their technical help during Z.D.’s stay at CERMAV.
Glossary
ABBREVIATIONS
- HEX
hexosaminidase
- PC
phosphorylcholine
- pNP
p-nitrophenyl
Data Availability Statement
Data described in the manuscript are shown in the figures; the coordinates of the T. suis HEX-2 crystal structure were in the Protein Data Bank (PDB) under code 8QK1.72
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.4c00187.
full phylogeny of nematode hexosaminidases; predicted sequence of T. suis HEX-2 as well as recombinant forms and alignments of T. suis HEX-2 with C. elegans HEX-2 and other hexosaminidases; further data on the recombinant T. suis HEX-2; further views of the X-ray crystal data (PDF)
Accession Codes
HEX-2 from Trichuris suis has the NCBI protein accession number KFD67184.
Author Present Address
# Institut für Mikrobiologie, Universität Innsbruck, A-6020 Innsbruck, Austria
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. Z.D. performed bioinformatic analyses and kinetics experiments; L.N., I.A., Z.D prepared constructs and purified protein, Z.D. and A.V. crystallized, collected data and solved the HEX-2 crystal structure, K.P., L.N. and Z.D. performed the glycomics experiments and interpreted data, K.J.B and K.A.S. synthesized inhibitors, Z.D. prepared draft text and figures, A.V. and I.B.H.W., funding acquisition and supervision, I.B.H.W. prepared final versions of the text and most figures. All authors have reviewed and agreed to the published version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Vocadlo D. J.; Withers S. G. Detailed comparative analysis of the catalytic mechanisms of β-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry 2005, 44, 12809–12818. 10.1021/bi051121k. [DOI] [PubMed] [Google Scholar]
- Gutternigg M.; Kretschmer-Lubich D.; Paschinger K.; Rendić D.; Hader J.; Geier P.; Ranftl R.; Jantsch V.; Lochnit G.; Wilson I. B. H. Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants and insects. J. Biol. Chem. 2007, 282, 27825–27840. 10.1074/jbc.M704235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahuran D. J. Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim. Biophys. Acta 1999, 1455, 105–138. 10.1016/S0925-4439(99)00074-5. [DOI] [PubMed] [Google Scholar]
- Alonso J.; Schimpl M.; van Aalten D. M. O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling?. J. Biol. Chem. 2014, 289, 34433–34439. 10.1074/jbc.R114.609198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T.; Suzuki K. Neutral hydrolases of rat brain. Preliminary characterization and developmental changes of neutral β-N-acetylhexosamindases. Biochim. Biophys. Acta 1980, 615, 402–413. 10.1016/0005-2744(80)90507-0. [DOI] [PubMed] [Google Scholar]
- Gutternigg M.; Rendić D.; Voglauer R.; Iskratsch T.; Wilson I. B. H. Mammalian cells contain a second nucleocytoplasmic hexosaminidase. Biochem. J. 2009, 419, 83–90. 10.1042/BJ20081630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteen M. G.; Oehler V.; Nemčovičová I.; Wilson I. B. H.; Vocadlo D. J.; Gloster T. M. Mechanism of Human Nucleocytoplasmic Hexosaminidase D. Biochemistry 2016, 55, 2735–2747. 10.1021/acs.biochem.5b01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pásztói M.; Sódar B.; Misják P.; Pálóczi K.; Kittel A.; Tóth K.; Wellinger K.; Géher P.; Nagy G.; Lakatos T.; Falus A.; Buzás E. I. The recently identified hexosaminidase D enzyme substantially contributes to the elevated hexosaminidase activity in rheumatoid arthritis. Immunol. Lett. 2013, 149, 71–76. 10.1016/j.imlet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Sinclair D. A.; Syrzycka M.; Macauley M. S.; Rastgardani T.; Komljenovic I.; Vocadlo D. J.; Brock H. W.; Honda B. M. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 13427–13432. 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzioğlu Kara E.; Kiral F. R.; Öztürk Çolak A.; Çelik A. Generation and characterization of inner photoreceptor-specific enhancer-trap lines using a novel piggyBac-Gal4 element in Drosophila. Arch. Insect Biochem. Physiol. 2020, 104, e21675 10.1002/arch.21675. [DOI] [PubMed] [Google Scholar]
- Léonard R.; Rendić D.; Rabouille C.; Wilson I. B. H.; Préat T.; Altmann F. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J. Biol. Chem. 2006, 281, 4867–4875. 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- Dragosits M.; Yan S.; Razzazi-Fazeli E.; Wilson I. B. H.; Rendić D. Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology 2015, 25, 448–464. 10.1093/glycob/cwu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler C.; Aumiller J. J.; Jarvis D. L. A fused lobes gene encodes the processing β-N-acetylglucosaminidase in Sf9 cells. J. Biol. Chem. 2008, 283, 11330–11339. 10.1074/jbc.M710279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y.; Chen L.; Qu M.; Chen Q.; Yang Q. Biochemical characterization of a novel β-N-acetylhexosaminidase from the insect Ostrinia furnacalis. Arch Insect Biochem Physiol 2013, 83, 115–126. 10.1002/arch.21099. [DOI] [PubMed] [Google Scholar]
- Nomura T.; Ikeda M.; Ishiyama S.; Mita K.; Tamura T.; Okada T.; Fujiyama K.; Usami A. Cloning and characterization of a β-N-acetylglucosaminidase (BmFDL) from silkworm Bombyx mori. J. Biosci Bioeng 2010, 110, 386–391. 10.1016/j.jbiosc.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Geisler C.; Jarvis D. L. Identification of genes encoding N-glycan processing β-N-acetylglucosaminidases in Trichoplusia ni and Bombyx mori: Implications for glycoengineering of baculovirus expression systems. Biotechnol. Prog. 2010, 26, 34–44. 10.1002/btpr.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann F.; Schwihla H.; Staudacher E.; Glössl J.; März L. Insect cells contain an unusual, membrane-bound β-N- acetylglucosaminidase probably involved in the processing of protein N-glycans. J. Biol. Chem. 1995, 270, 17344–17349. 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- Hanover J. A.; Forsythe M. E.; Hennessey P. T.; Brodigan T. M.; Love D. C.; Ashwell G.; Krause M. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 11266–11271. 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Cao P.; Chen S.; Spence A. M.; Zhu S.; Staudacher E.; Schachter H. Synthesis of paucimannose N-glycans by Caenorhabditis elegans requires prior actions of UDP-GlcNAc:α3-D-mannoside β1,2-N-acetylglucosaminyltransferase, α3,6-mannosidase II and a specific membrane-bound β-N-acetylglucosaminidase. Biochem. J. 2003, 372, 53–64. 10.1042/bj20021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S.; Bleuler-Martinez S.; Plaza D. F.; Künzler M.; Aebi M.; Joachim A.; Razzazi-Fazeli E.; Jantsch V.; Geyer R.; Wilson I. B. H.; Paschinger K. Galactosylated fucose epitopes in nematodes: increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. J. Biol. Chem. 2012, 287, 28276–28290. 10.1074/jbc.M112.353128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K.; Wöls F.; Yan S.; Jin C.; Vanbeselaere J.; Dutkiewicz Z.; Arcalis E.; Malzl D.; Wilson I. B. H. N-glycan antennal modifications are altered in Caenorhabditis elegans lacking the HEX-4 N-acetylgalactosamine-specific hexosaminidase. J. Biol. Chem. 2023, 299, 103053 10.1016/j.jbc.2023.103053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. F.; Gounaris K. Characterisation of a secreted N-acetyl-β-hexosaminidase from Trichinella spiralis. Mol. Biochem. Parasitol. 2006, 145, 84–93. 10.1016/j.molbiopara.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Jiménez-Castells C.; Vanbeselaere J.; Kohlhuber S.; Ruttkowski B.; Joachim A.; Paschinger K. Gender and developmental specific N-glycomes of the porcine parasite Oesophagostomum dentatum. Biochim. Biophys. Acta 2017, 1861, 418–430. 10.1016/j.bbagen.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. B. H.; Paschinger K. Sweet secrets of a therapeutic worm: Mass spectrometric N-glycomic analysis of Trichuris suis. Anal Bioanal Chem. 2016, 408, 461–471. 10.1007/s00216-015-9154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle W.; Haslam S. M.; Olivier V.; Appleton J. A.; Morris H. R.; Dell A. Phosphorylcholine-containing N-glycans of Trichinella spiralis: identification of multiantennary lacdiNAc structures. Glycobiology 2000, 10, 941–950. 10.1093/glycob/10.9.941. [DOI] [PubMed] [Google Scholar]
- Intra J.; Pavesi G.; Horner D. S. Phylogenetic analyses suggest multiple changes of substrate specificity within the glycosyl hydrolase 20 family. BMC Evol. Biol. 2008, 8, 214. 10.1186/1471-2148-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. Nematodes: the worm and its relatives. PLoS Biol. 2011, 9, e1001050 10.1371/journal.pbio.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparative genomics of the major parasitic worms. Nat. Genet. 2019, 51, 163–174. 10.1038/s41588-018-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch M.; Hoesch L.; Vasella A.; Rast D. M. N-acetylglucosaminono-1,5-lactone oxime and the corresponding (phenylcarbamoyl)oxime. Novel and potent inhibitors of β-N-acetylglucosaminidase. Eur. J. Biochem. 1991, 197, 815–818. 10.1111/j.1432-1033.1991.tb15976.x. [DOI] [PubMed] [Google Scholar]
- Stubbs K. A.; Bacik J. P.; Perley-Robertson G. E.; Whitworth G. E.; Gloster T. M.; Vocadlo D. J.; Mark B. L. The development of selective inhibitors of NagZ: increased susceptibility of Gram-negative bacteria to β-lactams. Chembiochem 2013, 14, 1973–1981. 10.1002/cbic.201300395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropak M. B.; Reid S. P.; Guiral M.; Withers S. G.; Mahuran D. Pharmacological enhancement of β-hexosaminidase activity in fibroblasts from adult Tay-Sachs and Sandhoff Patients. J. Biol. Chem. 2004, 279, 13478–13487. 10.1074/jbc.M308523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluvinage B.; Ghinet M. G.; Brzezinski R.; Boraston A. B.; Stubbs K. A. Inhibition of the exo-β-D-glucosaminidase CsxA by a glucosamine-configured castanospermine and an amino-australine analogue. Org. Biomol Chem. 2009, 7, 4169–4172. 10.1039/b913235j. [DOI] [PubMed] [Google Scholar]
- Mark B. L.; Vocadlo D. J.; Knapp S.; Triggs-Raine B. L.; Withers S. G.; James M. N. Crystallographic evidence for substrate-assisted catalysis in a bacterial β-hexosaminidase. J. Biol. Chem. 2001, 276, 10330–10337. 10.1074/jbc.M011067200. [DOI] [PubMed] [Google Scholar]
- Mark B. L.; Vocadlo D. J.; Zhao D.; Knapp S.; Withers S. G.; James M. N. Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 β-N-acetylhexosaminidase inhibitor. J. Biol. Chem. 2001, 276, 42131–42137. 10.1074/jbc.M107154200. [DOI] [PubMed] [Google Scholar]
- Martini F.; Eckmair B.; Neupert C.; Štefanić S.; Jin C.; Garg M.; Jiménez-Castells C.; Hykollari A.; Yan S.; Venco L.; Varón Silva D.; Wilson I. B. H.; Paschinger K. Highly modified and immunoactive N-glycans of the canine heartworm. Nat. Commun. 2019, 10, 75. 10.1038/s41467-018-07948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiya N.; Kurono M.; Ishihara H.; Tejima S.; Endo S.; Arata Y.; Takahashi N. Structural analysis of N-linked oligosaccharides by a combination of glycopeptidase, exoglycosidases, and high-performance liquid chromatography. Anal. Biochem. 1987, 163, 489–499. 10.1016/0003-2697(87)90253-3. [DOI] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Liu T.; Liu F.; Qu M.; Qian X. A novel β-N-acetyl-D-hexosaminidase from the insect Ostrinia furnacalis (Guenee). FEBS J. 2008, 275, 5690–5702. 10.1111/j.1742-4658.2008.06695.x. [DOI] [PubMed] [Google Scholar]
- Robb M.; Robb C. S.; Higgins M. A.; Hobbs J. K.; Paton J. C.; Boraston A. B. A second β-hexosaminidase encoded in the Streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans. J. Biol. Chem. 2015, 290, 30888–30900. 10.1074/jbc.M115.688630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurrus E.; Engel D.; Star K.; Monson K.; Brandi J.; Felberg L. E.; Brookes D. H.; Wilson L.; Chen J.; Liles K.; Chun M.; Li P.; Gohara D. W.; Dolinsky T.; Konecny R.; Koes D. R.; Nielsen J. E.; Head-Gordon T.; Geng W.; Krasny R.; Wei G. W.; Holst M. J.; McCammon J. A.; Baker N. A. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liessem B.; Glombitza G. J.; Knoll F.; Lehmann J.; Kellermann J.; Lottspeich F.; Sandhoff K. Photoaffinity labeling of human lysosomal β-hexosaminidase B. Identification of Glu-355 at the substrate binding site. J. Biol. Chem. 1995, 270, 23693–23699. 10.1074/jbc.270.40.23693. [DOI] [PubMed] [Google Scholar]
- Hattie M.; Ito T.; Debowski A. W.; Arakawa T.; Katayama T.; Yamamoto K.; Fushinobu S.; Stubbs K. A. Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis. Chem. Commun. 2015, 51, 15008–15011. 10.1039/C5CC05494J. [DOI] [PubMed] [Google Scholar]
- García-Alija M.; Du J. J.; Ordóñez I.; Diz-Vallenilla A.; Moraleda-Montoya A.; Sultana N.; Huynh C. G.; Li C.; Donahue T. C.; Wang L.-X.; Trastoy B.; Sundberg E. J.; Guerin M. E. Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE. Nat. Commun. 2022, 13, 1137. 10.1038/s41467-022-28722-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Higgins D. G. The Clustal Omega Multiple Alignment Package. Methods Mol. Biol. 2021, 2231, 3–16. 10.1007/978-1-0716-1036-7_1. [DOI] [PubMed] [Google Scholar]
- Robert X.; Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–324. 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R.; Hykollari A.; Eckmair B.; Malzl D.; Dragosits M.; Palmberger D.; Wang P.; Wilson I. B. H.; Paschinger K. The underestimated N-glycomes of lepidopteran species. Biochim. Biophys. Acta 2017, 1861, 699–714. 10.1016/j.bbagen.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmair B.; Gao C.; Mehta A. Y.; Dutkiewicz Z.; Vanbeselaere J.; Cummings R. D.; Paschinger K.; Wilson I. B. H. Recognition of highly branched N-glycans of the porcine whipworm by the immune system. Mol. Cell Proteomics 2024, 23, 100711 10.1016/j.mcpro.2024.100711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N.; Kuntz D. A.; Rose D. R. Golgi α-mannosidase II cleaves two sugars sequentially in the same catalytic site. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 9570–9575. 10.1073/pnas.0802206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida T.; Hiraoka S.; Usui K.; Ishiwata A.; Sengoku T.; Stubbs K. A.; Tanaka K.; Deguchi S.; Fushinobu S.; Nunoura T. Genetic and functional diversity of β-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis. Nat. Commun. 2024, 15, 3543. 10.1038/s41467-024-47653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T.; Strater N.; Schuette C. G.; Klingenstein R.; Sandhoff K.; Saenger W. The X-ray crystal structure of human β-hexosaminidase B provides new insights into Sandhoff disease. J. Mol. Biol. 2003, 328, 669–681. 10.1016/S0022-2836(03)00311-5. [DOI] [PubMed] [Google Scholar]
- Lemieux M. J.; Mark B. L.; Cherney M. M.; Withers S. G.; Mahuran D. J.; James M. N. Crystallographic structure of human β-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J. Mol. Biol. 2006, 359, 913–929. 10.1016/j.jmb.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark B. L.; Mahuran D. J.; Cherney M. M.; Zhao D.; Knapp S.; James M. N. Crystal structure of human β-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J. Mol. Biol. 2003, 327, 1093–1109. 10.1016/S0022-2836(03)00216-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A.; Bouvier J. T.; Davidson D. B.; Imker H. J.; Sadkhin B.; Slater D. R.; Whalen K. L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854, 1019–1037. 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K.; Kuma K.; Toh H.; Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S.; Silla-Martínez J. M.; Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N.; Dehal P. S.; Arkin A. P. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 2010, 5, e9490 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I.; Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T.; Schmidt H. A.; von Haeseler A.; Minh B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlvaine T. C. A buffer solution for colorimetric comparison. J. Biol. Chem. 1921, 49, 183–186. 10.1016/S0021-9258(18)86000-8. [DOI] [Google Scholar]
- Knapp S.; Vocadlo D.; Gao Z.; Kirk B.; Lou J.; Withers S. G. NAG-thiazoline, An N-Acetyl-β-hexosaminidase Inhibitor That Implicates Acetamido Participation. J. Am. Chem. Soc. 1996, 118, 6804–6805. 10.1021/ja960826u. [DOI] [Google Scholar]
- Furneaux R. H.; Gainsford G. J.; Manson J. M.; Tyler P. C. The chemistry of castanospermine, part I: synthetic modifications at C-6. Tetrahedron 1994, 50, 2131–2160. 10.1016/S0040-4020(01)85075-4. [DOI] [Google Scholar]
- Beer D.; Maloisel J.-L.; Rast D. M.; Vasella A. Synthesis of 2-Acetamido-2-deoxy-D-gluconhydroximolactone- and Chitobionhydroximolactone-Derived N-Phenylcarbamates, Potential Inhibitors of β-N-Acetylglucosaminidase. Helv. Chim. Acta 1990, 73, 1918–1922. 10.1002/hlca.19900730714. [DOI] [Google Scholar]
- Knapp S.; Zhao D. Synthesis of the sialidase inhibitor siastatin B. Org. Lett. 2000, 2, 4037–4040. 10.1021/ol0066680. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand P.XDSME: XDS Made Easier, In GitHub Repos; 2017https://github.com/legrandp/xdsme.
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre J.; Atanasova M.; Bagdonas H.; Ballard C. B.; Basle A.; Beilsten-Edmands J.; Borges R. J.; Brown D. G.; Burgos-Marmol J. J.; Berrisford J. M.; Bond P. S.; Caballero I.; Catapano L.; Chojnowski G.; Cook A. G.; Cowtan K. D.; Croll T. I.; Debreczeni J. E.; Devenish N. E.; Dodson E. J.; Drevon T. R.; Emsley P.; Evans G.; Evans P. R.; Fando M.; Foadi J.; Fuentes-Montero L.; Garman E. F.; Gerstel M.; Gildea R. J.; Hatti K.; Hekkelman M. L.; Heuser P.; Hoh S. W.; Hough M. A.; Jenkins H. T.; Jimenez E.; Joosten R. P.; Keegan R. M.; Keep N.; Krissinel E. B.; Kolenko P.; Kovalevskiy O.; Lamzin V. S.; Lawson D. M.; Lebedev A. A.; Leslie A. G. W.; Lohkamp B.; Long F.; Maly M.; McCoy A. J.; McNicholas S. J.; Medina A.; Millan C.; Murray J. W.; Murshudov G. N.; Nicholls R. A.; Noble M. E. M.; Oeffner R.; Pannu N. S.; Parkhurst J. M.; Pearce N.; Pereira J.; Perrakis A.; Powell H. R.; Read R. J.; Rigden D. J.; Rochira W.; Sammito M.; Sanchez Rodriguez F.; Sheldrick G. M.; Shelley K. L.; Simkovic F.; Simpkin A. J.; Skubak P.; Sobolev E.; Steiner R. A.; Stevenson K.; Tews I.; Thomas J. M. H.; Thorn A.; Valls J. T.; Uski V.; Uson I.; Vagin A.; Velankar S.; Vollmar M.; Walden H.; Waterman D.; Wilson K. S.; Winn M. D.; Winter G.; Wojdyr M.; Yamashita K. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr. D Struct Biol. 2023, 79, 449–461. 10.1107/S2059798323003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; Bridgland A.; Meyer C.; Kohl S. A. A.; Ballard A. J.; Cowie A.; Romera-Paredes B.; Nikolov S.; Jain R.; Adler J.; Back T.; Petersen S.; Reiman D.; Clancy E.; Zielinski M.; Steinegger M.; Pacholska M.; Berghammer T.; Bodenstein S.; Silver D.; Vinyals O.; Senior A. W.; Kavukcuoglu K.; Kohli P.; Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 586, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 32–41. 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Skubák P.; Lebedev A. A.; Pannu N. S.; Steiner R. A.; Nicholls R. A.; Winn M. D.; Long F.; Vagin A. A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz Z.; Varrot A.. Crystal structure of Trichuris suis β-N-acetyl-D-hexosaminidase - HEX-2 in apo form; RCSB PDB; 2023 10.2210/pdb8QK1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the manuscript are shown in the figures; the coordinates of the T. suis HEX-2 crystal structure were in the Protein Data Bank (PDB) under code 8QK1.72