Abstract
Epidermal stem cells (EPSCs) are essential for maintaining skin homeostasis and ensuring a proper wound healing. During in vitro cultivations, EPSCs give rise to transient amplifying progenitors and differentiated cells, finally forming a stratified epithelium that can be grafted onto patients. Epithelial grafts have been used in clinics to cure burned patients or patients affected by genetic diseases. The long-term success of these advanced therapies relies on the presence of a correct amount of EPSCs that guarantees long-term epithelial regeneration. For this reason, a deeper understanding of self-renewal and differentiation is fundamental to fostering their clinical applications.
The coordination between energetic metabolism (e.g., glycolysis, tricarboxylic acid cycle, oxidative phosphorylation, and amino acid synthesis pathways), molecular signalling pathways (e.g., p63, YAP, FOXM1, AMPK/mTOR), and epigenetic modifications controls fundamental biological processes as proliferation, self-renewal, and differentiation. This review explores how these signalling and metabolic pathways are interconnected in the epithelial cells, highlighting the distinct metabolic demands and regulatory mechanisms involved in skin physiology.
Keywords: Epidermal stem cells, Signaling pathways, Metabolism
Introduction
The epidermis serves as a crucial barrier against environmental insults, pathogens, and water loss. The maintenance of its integrity relies on a delicate balance between stem cell proliferation, progenitor differentiation, and cell death [1, 2]. Understanding the metabolic pathways that support these processes is vital for developing therapeutic strategies for skin disorders and enhancing regenerative medicine approaches.
Epidermal stem cells (EPSCs) are located within the basal layer of the epidermis, the lowermost layer of the skin. These stem cells are anchored to the basal membrane, a critical structure that delineates the boundary between the epidermis and the underlying dermis. The basal membrane provides essential support and signals that maintain the stem cells in their undifferentiated state [1].
As EPSCs begin the differentiation process, they gradually migrate upwards through the various strata of the epidermis. This journey takes them from the basal layer, where they are initially situated, through the spinous and granular layers, and ultimately to the outermost layer, the stratum corneum. During this upward movement, stem cells undergo significant morphological and functional changes, transitioning from undifferentiated, proliferative cells into specialized, terminally differentiated keratinocytes that form the skin’s protective outer barrier [3, 4].
In vitro cultures of human primary keratinocytes have been extensively used in clinical settings as they can generate a functional epidermal or corneal graft to treat burned patients or patients affected by limbal stem cells deficiency (LSCD) and epidermolysis bullosa (EB) [5–13]. During in vitro cultivation, keratinocytes isolated from the basal layer of the epidermis give rise to three types of clones: holoclones, meroclones, and paraclones [14, 15]. Holoclone is the clone originated from the stem cell and is endowed with the highest proliferative and self-renewing capacity [7]. Transient amplifying progenitors give rise to meroclones and paraclones with limited proliferative capacity. The long-lasting regeneration and repair processes rely on long-lived holoclone-forming stem cells producing short-lived meroclone- and paraclone-forming cells that eventually give rise to terminally differentiated keratinocytes [7, 14, 16]. The unique property of holoclone-forming stem cells to replenish differentiated cells, thus fostering wound healing, is also the main characteristic to consider for generating potentially perpetual epithelial grafts for regenerative medicine applications. In this framework, an adequate number of holoclone-forming stem cells is essential to sustaining long-term epithelial regeneration [7].
Metabolic requirements in somatic stem and differentiated cells
Metabolic reprogramming of energy production is one of the hallmarks of cell differentiation, which drives morphology reshaping; the metabolic core is the oxidation of glucose, and its tuning and deviation are the driving forces dictating cell fate. Human embryonic stem cells (hESCs) and human pluripotent stem cells (hPSCs) are a fine example of such metabolic re-programming [17]. As soon as they exit pluripotency, the glycolysis-based energy production is switched to a mechanism that is heavily dependent on oxidative phosphorylation (OXPHOS) (Fig. 1) [18–21].
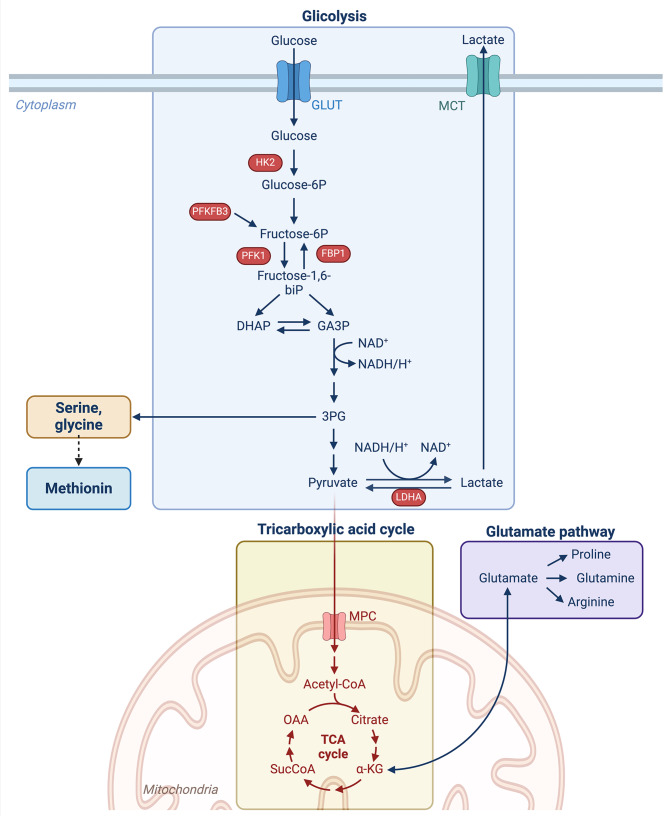
Fig. 1.
Interconnection between glycolysis, TCA cycle, and amino acids metabolism. During glycolysis, glucose is broken down to two molecules of pyruvate. One of the glycolysis intermediate, 3PG, can be converted to serine, glycine and indirectly to methionine (See Fig. 5 for details). Pyruvate is translocated to mitochondria for the oxidative decarboxylation into acetyl-CoA. Acetyl-CoA is oxidated via the TCA cycle and sustains the production of ATP from ADP in the oxidative phosphorylation process. α-KG produced during the TCA cycle, can be converted to glutamate and vice-versa. Glutamate is the starting point for the production of proline, glutamine and arginine. Abbreviations: GLUT = glucose transporter; MCT = monocarboxylate transporter; HK2 = hexokinase 2; PFKFB3 = 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3; PFK1 = 6-phosphofructokinase; FBP1 = fructose-bisphosphatase 1; fructose-1,6-biP = fructose-1,6-biphosphate; DHAP = dihydroxyacetone phosphate; GA3P = glyceraldehyde 3-phosphate; TCA= tricarboxylic acid cycle; 3PG = 3-phosphoglycerate; LDHA = lactate dehydrogenase A; α-KG = α-ketoglutarate; Acetyl-CoA = acetyl coenzyme A
During glycolysis, glucose is broken down into two molecules of pyruvate, each containing three carbons. All the enzymatic steps of the glycolytic pathway (ten steps, of which seven reversible and three irreversible) occur in the cytosol and are conventionally divided into two main phases: the energy investment phase and the energy payoff phase. In the energy investment phase, two molecules of ATP are consumed to phosphorylate glucose, bringing to higher energy levels the corresponding derivative (i.e., glyceraldehyde-3-phosphate, GA3P in Fig. 1) and to ensure the subsequent exergonic, payoff phase yielding pyruvate. The fate of pyruvate depends on the metabolic signature of the cell: only those cells with highly aerobic metabolism direct the large part of cytosolic pyruvate to mitochondria for the oxidative decarboxylation into acetyl-CoA. Its oxidation into CO2, via the tricarboxylic acid (TCA) cycle, sustains the O2-dependent mitochondrial proton motive force used to produce ATP from ADP (namely, oxidative phosphorylation). To avoid redox unbalancing, the mitochondrial processes are fine-tuned to balance electron donors and acceptors availability. Conversely, some cells account mainly for glycolysis to ensure ATP supply and/or take advantage of glycolytic intermediates for anabolic purposes; excess cytosolic pyruvate is generally converted into lactate to refresh the NAD+ pool (Fig. 1).
Adult stem cells (SCs) also prefer glycolysis to OXPHOS as it is a quicker way to produce ATP (although less efficient than OXPHOS) to sustain their high proliferative rate [17]. Indeed, since SCs often reside in niches where oxygen levels can fluctuate, the glycolytic signature allows them to function efficiently even when oxygen is low, and hypoxia could activate them to foster wound healing [22]. In addition, this strategy minimizes the possible production of reactive oxygen species (ROS) in case of the high rate of OXPHOS and shortage of electron acceptor (i.e., O2). As mentioned above, in parallel to fast energy production, glycolysis also provides some intermediates of biosynthetic pathways (i.e., the carbon and reductive equivalents) that are used to generate biomass (e.g., nucleotides, amino acids, and lipids) essential for cell division and growth [23].
The same metabolic reprogramming can be observed during epidermis differentiation. EPSCs primarily rely on cytoplasmic glycolysis as their main pathway for generating energy (ATP). Conversely, differentiating cells prefer mitochondrial OXPHOS for ATP production, as it generates much more ATP per glucose molecule compared to glycolysis [24–26].
The relevance of tuning glycolytic rate (and lactate production) as a strategy of keratinocyte differentiation has been recently demonstrated [24]. The gluconeogenic enzyme FBP1 catalyzes the hydrolysis of fructose-1,6-bisphosphate to fructose 6-phosphate, thus de facto affecting pyruvate accumulation; its activity impairs the glycolytic rate promoting differentiation and inhibiting proliferation in a glycolysis-dependent manner [24].
Once activated, murine hair follicle stem cells (HFSCs) quickly divide to generate a new hair. Metabolites analysis revealed that these cells exhibit higher levels of glycolysis-derived metabolites and produce significantly more lactate compared to their progeny. In particular, the Authors provide evidence that HFSC activation relies on the activity of the enzyme lactate dehydrogenase (LDHA) responsible for lactate production [25].
These findings give a first hint on how the energetic metabolism orchestrates proliferation and differentiation in epithelial cells. The tight connection between the signalling pathways and metabolic signature(s) during proliferation and differentiation is detailed in this review.
p63 controls biochemical networks in epidermal cells
p63 is a member of the p53-p73 family of transcription factors. It is a key transcription factor controlling the squamous epithelia and is required to sustain the proliferative potential of cultured epithelial cells [27, 28]. Its importance has been studied in mice where its genetic deletion causes significant defects in limb and craniofacial development, together with the absence of epidermis and other stratified epithelia [27, 29–32]. In particular, the proliferative and regenerative potential of epithelial stem cells is dependent on the ΔNp63α isoform, which is highly expressed in holoclones (namely, clones originated from EPSCs), and its expression progressively declines during keratinocyte clonal conversion [27, 33, 34].
Moreover, p63 has been identified as an orchestrator of epidermal cells’ metabolic activity. Indeed, it controls the expression of genes linked to glucose metabolism and response to oxidative stress [35, 36] (Fig. 2).
Fig. 2.
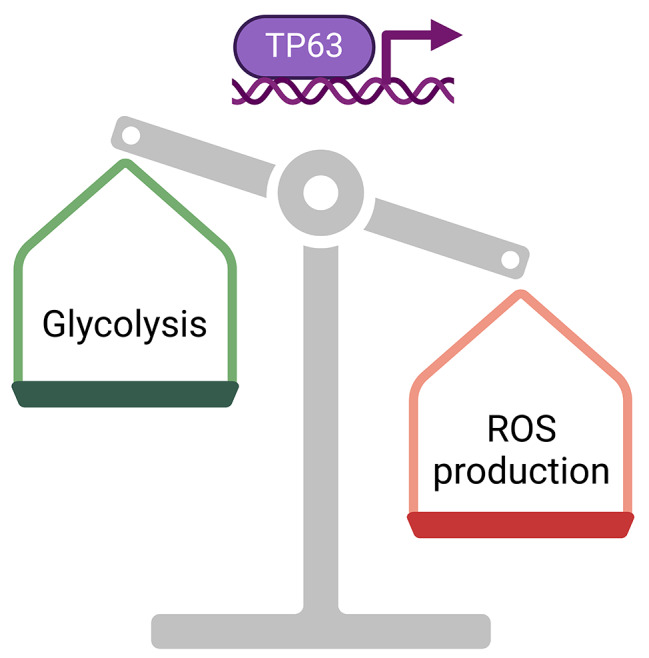
Effects of TP63 transcription factor on cellular metabolism. As a consequence of TP63-target-genes transcription, glycolysis is enhanced, and the production of reactive oxygen species (ROS) decreased
Viticchiè et al. reported that p63 positively regulates the expression of hexokinase 2 (HK2), which is localized in the mitochondrial membrane and catalyzes the first glycolytic step [35]. p63 depletion in epidermal cells caused a reduction in the expression of glutathione peroxidase 2 (GPX2), mitochondrial superoxide dismutase (SOD2), and NADPH quinone oxidoreductase (NQO1), impaired mitochondrial basal respiration, induced mitochondrial membrane hyperpolarization, and increased intracellular ROS. These metabolic alterations are mediated by HK2 as silencing HK2 resulted in similar effects. Overall, the Authors concluded that p63 acts through HK2 to couple the glucose metabolism with the mitochondrial function, limiting oxidative stress [35].
p63 has been linked to another fundamental player of glycolysis, the tightly regulated enzyme 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) [36]. PFKFB3 is an enzyme with a dual role, facilitating the conversion of fructose-6-phosphate to fructose-2,6-bisphosphate and vice versa. Fructose-2,6-bisphosphate serves as an activator for phosphofructokinase-1 (PFK1), a crucial enzyme in glycolysis responsible for converting fructose-6-phosphate into fructose-1,6-bisphosphate (Fig. 1). This conversion, mediated by PFK1, is pivotal in regulating the pace of glycolysis and is susceptible to allosteric inhibition by ATP. It has been reported that overexpression of PFKFB3 inhibited keratinocyte differentiation. Oppositely, its loss inhibited proliferation and promoted differentiation in clonogenic keratinocytes, thus suggesting a tight connection between glucose metabolism and the balance between cell proliferation and differentiation [36].
The above-mentioned role of the gluconeogenic enzyme FBP1 in promoting keratinocyte differentiation has been negatively associated with TP63 [24]. Indeed, following FBP1 deletion, increased glycolysis led to increased production of citrate, which undergoes cytoplasmic translocation, and it is converted into acetyl-CoA via ATP-citrate lyase (ACLY). Acetyl-CoA serves as a crucial signaling molecule, regulating various cellular functions such as energy metabolism and cell division by impacting the acetylation patterns of multiple proteins. Coherently, histone 3 acetylation on lysine 9 (H3K9-Ac) increases upon FBP1 loss and is associated with enhanced expression of growth-related genes, such as CDC6 and TP63 [24].
The YAP-FOXM1 axis and glycolysis tuning
Yes-associate protein 1 (YAP1, also known as YAP) is a transcriptional co-regulator that acts together with its homolog, WW Domain Containing Transcription Regulator 1 (WWTR1 or TAZ). YAP/TAZ are components of the Hippo pathway and mechanotransduction effectors, controlling gene expression by binding to TEAD transcription factors [37, 38].
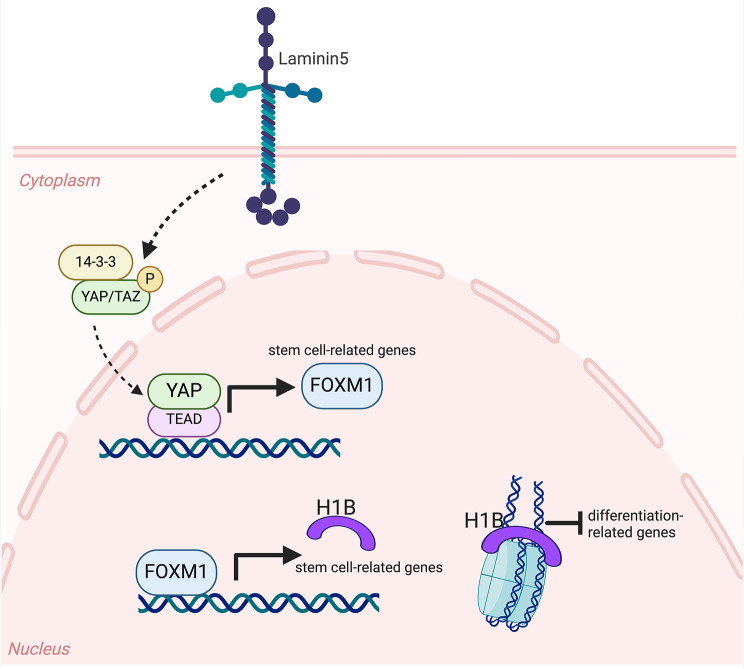
In human EPSCs, the YAP/TAZ activity is mainly controlled by Laminin5 (also known as Laminin-332), a heterotrimeric protein consisting of three chains (encoded by the genes LAMB3, LAMA3, and LAMG2) located at the dermal-epidermal junction. Genetic deletion of the LAMB3 gene causes a devastating skin disease called junctional EB (JEB), characterized by rapid EPSC exhaustion due to the loss of YAP/TAZ activity [39, 40] (Fig. 3). Self-renewal potential can be rescued after enforced expression of YAP, even if adhesion properties are not restored, providing an excellent model for uncoupling self-renewal from adhesion [39].
Fig. 3.
Transcriptional regulation of stemness in EPSCs. Laminin5 positively controls the phosphorylation of YAP/TAZ that translocate into the nucleus and induces FOXM1 transcription. FOXM1 then induces H1B transcription that binds to the nucleosomes and inhibits the transcription of differentiated-related genes
YAP/TAZ activity has been connected to the glycolytic flux in a cellular model of epithelial breast cancer [41]. In this context, YAP/TAZ activity is regulated by glycolysis through the enzyme PFK1, which interacts with YAP/TAZ’s DNA-binding platforms, TEADs, increasing its transcriptional activity [42]. This connection between YAP/TAZ and glycolysis could be relevant also in non-tumorigenic context.
In EPSCs, YAP induces the expression of another transcription factor, the Forkhead Box M1 (FOXM1), whose expression is tightly restricted to self-renewing holoclone-forming cells and essential for maintaining self-renewal [43] (Fig. 3). Its enforced expression increases the number of holoclones, while FOXM1 deletion selectively reduces holoclone-forming cells in culture. Finally, acting downstream of YAP, FOXM1 also rescues the self-renewal potential in JEB-derived epidermal cells [43].
Moreover, FOXM1 activates the expression of Linker Histone H1B, one of the proteins that envelop DNA around the nucleosome core, also involved in gene expression control [44]. H1B binds to the promoter regions of differentiated genes, and its absence causes a coherent upregulation of differentiation genes and downregulation of self-renewal-related genes [45] (Fig. 3).
FOXM1 protects proliferating keratinocytes against oxidative stress by regulating PRDX, SOD2, and GPX2 expression. A similar protective role was found in the A253 cell line (deriving from the human submaxillary salivary gland epidermoid carcinoma), in which FOXM1 downregulation causes an increase in ROS levels coupled with a raised cell death [46].
FOXM1 influences glycolysis and OXPHOS rate also in myeloma cells: FOXM1-deficient myeloma cells exhibit reduced oxygen consumption, lower expression of HK2, glucose transporter GLUT1, and LDHA, which promotes growth and drug resistance through lactate production. Taken together, these results suggest that FOXM1 might have a protective role in cancer cells as it promotes their growth and inhibits apoptosis, hence implying that FOXM1 could be a potential therapeutic target [47].
Mitochondria and oxidative stress in epithelial cells
Mitochondrial metabolism influences cell and tissue physiology by generating ROS. ROS are produced during oxidative metabolism by partially reducing molecular oxygen (O2), leading to the formation of superoxide anions (O2•−). The electron transport chain (ETC) comprises five protein complexes bound to the inner mitochondrial membrane. ETC utilizes a series of electron transfer reactions to generate ATP, and it is the main source of O2•− production in the mitochondria [48]. O2•− is then converted into hydrogen peroxide (H2O2) by cellular superoxide dismutases (SODs). Oxidation of cysteine thiol groups (–SH) by H2O2 results in the formation of disulfide (–SS–) or sulfenyl amide (–SN–) bonds, altering protein function. Cellular systems involving thioredoxin and glutathione reductases restore these oxidatively modified residues to their reduced state. Recently, it was demonstrated that ROS are crucial in regulating skin physiology. Indeed, their production triggers numerous cellular responses, such as activating HIF-1α independently of hypoxia and recruiting immune cells during skin infections [49, 50]. Mitochondrial transcription factor A (TFAM) is responsible for the transcription of mitochondrial genes encoding ETC subunits. In TFAM-deficient mice, ROS levels are reduced, and incomplete epidermal differentiation and hair follicle formation are observed. The Authors suggest that these phenotypes are due to an impairment in the ROS signal that cannot activate Notch and β-catenin [49]. More skin alterations caused by mutated mitochondrial-related genes are widely reviewed in Feichtinger et al. [50].
In addition, altered mitochondrial membrane and activity have been highlighted also in epidermolysis bullosa simplex (EBS) [51]. EBS-causing mutations involve genes encoding for cytoskeletal-forming proteins, such as keratin5 (KRT5) and keratin14 (KRT14). Keratins are a family of intermediate filament proteins that form along with other cytoskeletal elements (e.g., actin filaments and microtubules) a dense network within keratinocytes, providing structural integrity and mechanical stability to the cell. Moreover, they are involved in various cellular functions, such as differentiation, migration, and wound healing. Even if with less frequency, EBS can be caused by mutations in Plectin or Destonin, proteins located in the hemidesmosome that anchor keratin filaments to the plasma membrane [52, 53]. In KRT14-mutated keratinocytes, mitochondria are dispersed in the cytoplasm other than their normal perinuclear localization, and the oxygen consumption rate of EBS primary cells shows a three-fold decrease of basal and maximal values compared to keratinocytes from healthy donors [51]. However, the mechanisms linking alteration in the keratin network and mitochondrial activity need to be further elucidated. Of note, similar mitochondrial dysfunctions have also been observed in muscle cells derived from EBS patients affected by PLEC1 mutation [54].
The role of mitochondrial stress in EPSCs has been studied in mice, where the mitochondrial antioxidant enzyme SOD2 has been knocked out [55]. Deficiency in SOD2 induces cellular senescence and diminishes the proliferative capacity, promoting the differentiation of epidermal stem/progenitor cells. Such differentiation fosters the wound-healing process in young mice. However, in aged mice, a long-term decreased proliferation of SCs causes a delay in wound healing due to premature SC exhaustion. These data are coherent with the previous reported effect of the p63-dependent SOD2 downregulation [35]. Similar effect has been observed in human keratinocytes by the same Authors, but the relevance of this finding in human EPSCs has not been fully addressed [55].
AMPK, a sensor of cellular energy in wound healing
AMP-activated protein kinase (AMPK) is activated in response to an increase in the AMP/ATP ratio, indicating low cellular energy levels. AMPK regulates lipid metabolism by inhibiting fatty acid synthesis and promoting fatty acid oxidation to generate ATP (Fig. 4).
Fig. 4.
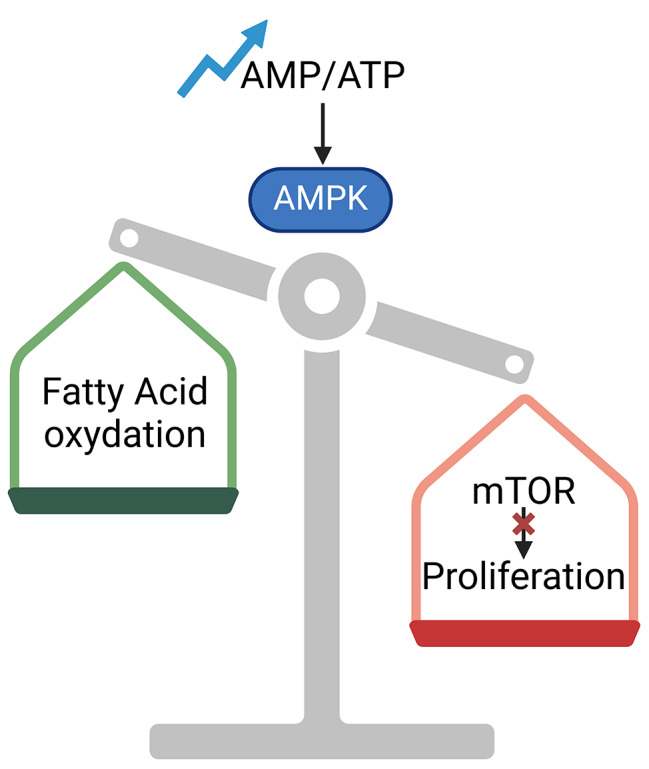
Effects of AMPK/mTOR pathway on cellular behavior. Increased AMP/ATP ratio triggers AMPK activation. AMPK promotes fatty acid oxidation and inhibits mTOR pro-proliferative effect. Abbreviations: AMPK = AMP-activated protein kinase; mTOR = mechanistic Target of Rapamycin
AMPK is also a negative regulator of the mechanistic Target of Rapamycin (mTOR) pathway. mTOR is a serine/threonine kinase, and it is organized into two distinct complexes, namely mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Each complex contains several subunits and represents a central regulator of cell growth and proliferation. On the one hand, keratinocyte hyperproliferation is necessary during wound healing, a very dynamic and complicated process. A robust activation of mTORC1 and mTORC2 was detected in keratinocytes at the wound edge [56].
The treatment with everolimus, a mTOR inhibitor, leads to developing skin ulcerations and chronic wounds, further confirming the clinical relevance of mTOR activation in mediating wound healing [57].
On the other hand, keratinocyte hyperproliferation must be finely controlled to ensure that epidermal differentiation occurs properly. In fact, a dysregulated differentiation leads to hyperproliferative skin conditions, such as psoriasis, which is characterized by scaly plaques and epidermal thickenings. Following rapamycin-mediated inhibition of mTOR, AMPK slows down keratinocyte proliferation, preventing epidermis hyperproliferation and scale plaque formation [58] (Fig. 4). This notion was further confirmed by a recent study showing that rapamycin treatment in healthy skin conditions leads to the formation of a thinner epidermis with lower expression of differentiation markers, such as IVL and SPINK5 [59]. In conclusion, the fine control of the AMPK/mTOR pathway ensures that keratinocytes proliferate at a controlled rate, maintaining proper skin thickness and function [60].
Amino acid synthesis in the differentiation process and epigenetic memory
If, during proliferation, amino acids provide building blocks for protein biosynthesis, during differentiation, specific amino acids contribute to the synthesis of structural proteins, modulate epigenetic regulation, and influence cell signalling pathways. Among the metabolic pathways involving amino acids, glutamine, serine, glycine and methionine are relevant for keratinocyte physiology (see Figs. 1 and 5).
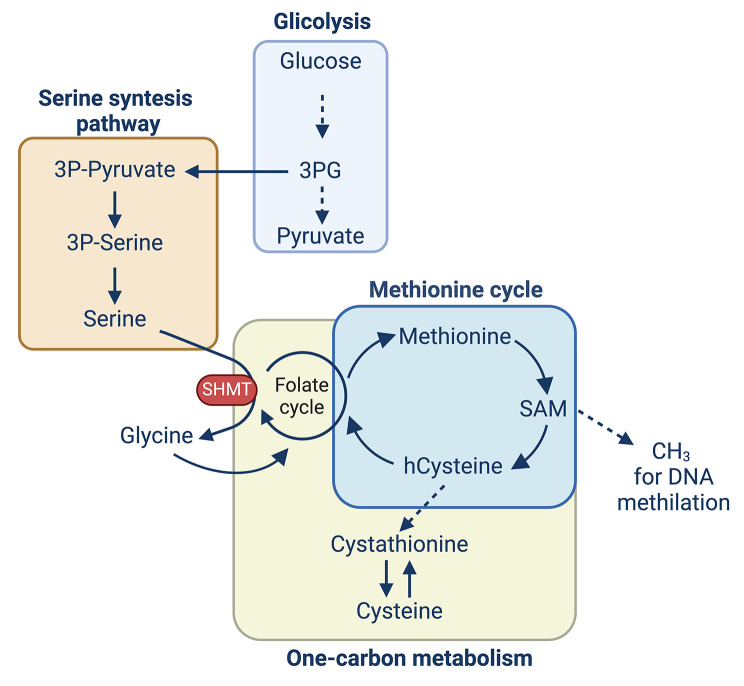
Fig. 5.
Interconnection between glycolysis, serine biosynthesis, one-carbon metabolism, and methionine cycle. 3PG produced during glycolysis enters the serine synthesis pathway, a series of reactions that produce serine. Serine can then be converted to glycine thanks to the SHMT enzyme. Glycine participates in the folate cycle to generate additional one-carbon units. Folate is necessary for the methionine cycle, in which methionine is precursor of SAM, the methyl donor used by DNMTs to methylate DNA and histones. One-carbon metabolism comprised folate cycle and methionine cycle. Abbreviations: 3PG = 3-phosphoglycerate; SHMT = serine hydroxy methyltransferase; hCysteine = homocysteine
Glutamine serves as a key energy source, feeding into the TCA cycle through its conversion to α-ketoglutarate, fostering ATP production (Fig. 1). Moreover, glutamine provides nitrogen for synthesizing of nucleotides and amino acids, respectively, essential for DNA replication and protein synthesis during cell division. Glutamine metabolism contributes to the production of glutathione via glutamate, a crucial antioxidant that protects keratinocytes from oxidative stress, maintaining cellular health and promoting self-renewal [61, 62]. In HFSCs, it has been shown to be essential for lineage progression and fate reversibility, thanks to mTOR2 and AKT activation [63].
This pathway is also important in the most differentiated layer of the epidermis, as Transglutaminase I (TG1; encoded by the gene TGM1) catalyzes the cross-linking of identical or different substrate proteins through the formation of covalent bonds between glutamine and lysine residues. This biochemical reaction is required for the formation of the cornified envelope. TG1 is activated by intracellular calcium levels physiologically rising during differentiation [64]. Indeed, mutations of the TGM1 gene cause a rare genetic skin disease called Lamellar Ichthyosis (LI). LI manifests as widespread skin scaling, extreme dryness, erythema, hyperkeratosis, and significant scaling, severely affecting patients’ quality of life [65, 76].
Serine and glycine enter the one-carbon metabolism, a collection of interconnected biochemical pathways, facilitating the transfer of one-carbon units vital for numerous cellular processes (Fig. 4). This metabolic network plays a crucial role in synthesizing of nucleotides, amino acids, and other essential biomolecules, as well as in regulating gene expression through methylation. In addition to extracellular consumption, serine can be derived from the serine synthesis pathway (SSP), wherein glucose-derived 3-phosphoglycerate (3-PG) is converted to serine through the sequential activity of enzymes phosphoglycerate dehydrogenase (PHGDH), phosphoserine transaminase (PSAT1), and phosphoserine phosphatase (PSPH). Moreover, serine can be converted into glycine through the enzyme serine hydroxymethyltransferase (SHMT). This reaction also produces a one-carbon unit in the form of 5,10-methylenetetrahydrofolate (5,10-methylene-THF), a key intermediate in one-carbon metabolism essential for the synthesis of purines (components of DNA and RNA) and the methylation of DNA, proteins, and lipids. Glycine participates in the folate cycle, where it can be converted back to serine or used to generate additional one-carbon units (Fig. 5). Notably, it is also a precursor of the key antioxidant glutathione.
First insights on the fundamental role of serine metabolism were gained from mice models of squamous cell carcinoma progression [66]. In mice, sustained SOX2 expression induces pretumorigenic lesions marked by the expansion of KRT14-positive progenitors and induction of the tumor SC marker CD44. Premalignant cells were collected from wild-type and SOX2+ mice, and among the non-essential amino acids, only serine was differentially consumed. Although SHMT1/2 enzymes expression levels were comparable, SOX2+ cells revealed reduced glucose-derived serine synthesis. Notably, premalignant SOX2+ cells also inhibit de novo serine synthesis through SSP by increasing mitochondrial pyruvate consumption, which results in endogenous serine auxotrophy. Hence, SOX2+ cells rely solely on extracellular serine for proliferation. Wild-type cells, instead, are still able to derive serine from glucose metabolism [66].
The function of these pathways in human epidermal cells has been studied by Cappello et al. [67]. They showed that keratinocyte proliferation requires mainly extracellular serine as the intracellular de novo serine biosynthesis is unable to compensate for the lack of extracellular one. Both the cytoplasmic (SHMT1) and mitochondrial (SHMT2) enzymes decrease during differentiation. Specific downregulation of SHMT2 caused a decreased and reduced synthesis of bioenergetic metabolites. These findings are relevant to treat psoriatic lesions, as in human psoriatic sections, SHMT2 is highly expressed, and reduced expression of SHMT2 rescues the epidermal thickening and scaling in a psoriatic mouse model [67].
Methionine is a precursor for S-adenosylmethionine (SAM), a universal methyl donor used in DNA and histone methylation that regulates gene expression patterns necessary for differentiation (Fig. 5). This process is catalysed by DNA methyltransferases (DNMTs), which use SAM as a methyl donor. SAM is synthesized from methionine in the methionine cycle, which is influenced by the availability of dietary nutrients and metabolic intermediates such as Folate and Vitamin B12. Folate is converted to tetrahydrofolate (THF), which then participates in the conversion of homocysteine to methionine, facilitating SAM production. Deficiencies in folate or vitamin B12 can alter DNA methylation patterns, affecting gene expression [68].
DNMT1 and ubiquitin-like with PHD and ring finger domains 1 (UHRF1), that are responsible for maintaining DNA methylation patterns during DNA replication, are abundant in epidermal undifferentiated cells, where it is necessary to sustain proliferative capacity and inhibit differentiation [69]. Moreover, EZH1 and 2, the histone methyltransferases that catalyse the repressive marks (trimethylation of histone H3 at lysine 27 -H3K27me3), have been studied during hair follicle morphogenesis and wound healing in mouse models, suggesting their fundamental role in these processes [70]. Conversely, in mouse skin, the histone demethylase JMJD3 depletion blocked differentiation, while active JMJD3 dominantly induced it [71]. These results suggest that methionine metabolism and epigenetic regulation tightly control mammalian epidermal differentiation and could have an important impact on pathological conditions, as reviewed in Moltrasio et al. [72]. These genes illustrate the complex interplay between metabolism and epigenetics in the epidermis, highlighting how metabolic states can influence gene expression and cellular behavior through epigenetic mechanisms. Understanding these connections provides insights into skin biology and potential therapeutic targets for skin disorders.
Conclusion
Understanding the connections between metabolism, self-renewal and differentiation opens new therapeutic avenues. The metabolic pathways in the epidermis are intricately linked to the functions and fate of both stem cells and differentiated cells [17]. Advances in metabolomics and single-cell technologies continue to uncover the complexities of epidermal metabolism, offering insights into skin biology and potential therapeutic targets [73, 74, 76]. Metabolic interventions, such as dietary modifications, supplementation with metabolic cofactors, or drugs targeting metabolic enzymes, can potentially foster cellular reprogramming and restore normal gene expression [61, 75].
The interplay between metabolism and epigenetics also extends to the concept of epigenetic memory, where past metabolic states can leave lasting marks on the epigenome. For instance, metabolic shifts during development, differentiation, or disease states can establish specific epigenetic patterns that persist even when the original metabolic signals are no longer present [26]. Such interplay has profound implications for understanding how environmental factors, such as diet and stress, can have long-term effects on gene expression and phenotype.
By delineating these pathways, we can better understand how to manipulate them for improved skin health and disease management.
Acknowledgements
We thank all lab members for helpful feedback and comments on this manuscript. We apologize to those we were unable to cite within this review. All the Figures were created with BioRender.com.
Author contributions
M.P.P., A.R., S.R., collected the literature and wrote the manuscript; E.E. conceptualized the review, collected the literature, and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by PRIN 2022 (the European Union - NextGenerationEU (PNRR) – No. 2022KRTKXS) to EE.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maria Pia Polito, Alessio Romaldini .
References
- 1.Díaz-García D, Filipová A, Garza-Veloz I, Martinez-Fierro ML. A beginner’s introduction to skin stem cells and Wound Healing. Int J Mol Sci. 2021;22(20):11030. 10.3390/ijms222011030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee HJ, Kim M. Skin barrier function and the Microbiome. Int J Mol Sci. 2022;23(21):13071. 10.3390/ijms232113071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gambardella L, Barrandon Y. The multifaceted adult epidermal stem cell. Curr Opin Cell Biol. 2003;15(6):771–7. 10.1016/j.ceb.2003.10.011 [DOI] [PubMed] [Google Scholar]
- 4.Blanpain C, Fuchs E. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344(6189):1242281. 10.1126/science.1242281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rama P, Matuska S, Paganoni G, Spinelli A, De Luca M, Pellegrini G. Limbal stem-cell therapy and long-term corneal regeneration. N Engl J Med. 2010;363(2):147–55. 10.1056/NEJMoa0905955 [DOI] [PubMed] [Google Scholar]
- 6.Gallico GG, O’Connor NE, Compton CC, Kehinde O, Green H. Permanent coverage of large burn wounds with autologous cultured human epithelium. N Engl J Med. 1984;311(7):448–51. 10.1056/NEJM198408163110706 [DOI] [PubMed] [Google Scholar]
- 7.Hirsch T, Rothoeft T, Teig N, Bauer JW, Pellegrini G, De Rosa L, et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551(7680):327–32. 10.1038/nature24487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellegrini G, Ranno R, Stracuzzi G, Bondanza S, Guerra L, Zambruno G, et al. The control of epidermal stem cells (holoclones) in the treatment of massive full-thickness burns with autologous keratinocytes cultured on fibrin1. Transplantation. 1999;68(6):868–79. 10.1097/00007890-199909270-00021 [DOI] [PubMed] [Google Scholar]
- 9.Pellegrini G, Traverso CE, Franzi AT, Zingirian M, Cancedda R, De Luca M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 1997;349(9057):990–3. 10.1016/S0140-6736(96)11188-0 [DOI] [PubMed] [Google Scholar]
- 10.Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12(12):1397–402. 10.1038/nm1504 [DOI] [PubMed] [Google Scholar]
- 11.Bauer JW, Koller J, Murauer EM, De Rosa L, Enzo E, Carulli S, et al. Closure of a large chronic wound through transplantation of gene-corrected epidermal stem cells. J Invest Dermatology. 2017;137(3):778–81. 10.1016/j.jid.2016.10.038 [DOI] [PubMed] [Google Scholar]
- 12.De Rosa L, Enzo E, Zardi G, Bodemer C, Magnoni C, Schneider H, et al. Hologene 5: a phase II/III clinical trial of combined cell and Gene Therapy of Junctional Epidermolysis Bullosa. Front Genet. 2021;12:705019. 10.3389/fgene.2021.705019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Rosa L, Enzo E, Palamenghi M, Sercia L, De Luca M. Stairways to Advanced therapies for Epidermolysis Bullosa. Cold Spring Harb Perspect Biol. 2022;a041229. [DOI] [PMC free article] [PubMed]
- 14.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84(8):2302–6. 10.1073/pnas.84.8.2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enzo E, Cattaneo C, Consiglio F, Polito MP, Bondanza S, De Luca M. Clonal analysis of human clonogenic keratinocytes. In: Methods in Cell Biology [Internet]. Elsevier; 2022 [cited 2022 Aug 26]. pp. 101–16. https://linkinghub.elsevier.com/retrieve/pii/S0091679X22000267 [DOI] [PubMed]
- 16.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation keratinizin colonies from single cell is. Cell. 1975;6(3):331–43. 10.1016/S0092-8674(75)80001-8 [DOI] [PubMed] [Google Scholar]
- 17.Ito K, Ito K. Metabolism and the control of cell fate decisions and Stem Cell Renewal. Annu Rev Cell Dev Biol. 2016;32(1):399–409. 10.1146/annurev-cellbio-111315-125134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu W, Gaeta X, Sahakyan A, Chan AB, Hong CS, Kim R, et al. Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell. 2016;19(4):476–90. 10.1016/j.stem.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, et al. Glycolysis-mediated changes in Acetyl-CoA and histone Acetylation Control the early differentiation of embryonic stem cells. Cell Metabol. 2015;21(3):392–402. 10.1016/j.cmet.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 20.Varum S, Rodrigues AS, Moura MB, Momcilovic O, Iv CAE, Ramalho-Santos J, et al. Energy Metabolism in Human pluripotent stem cells and their differentiated counterparts. PLoS ONE. 2011;6(6):e20914. 10.1371/journal.pone.0020914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30(24):4860–73. 10.1038/emboj.2011.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong WX, Hu MS, Esquivel M, Liang GY, Rennert RC, McArdle A, et al. The role of Hypoxia-Inducible factor in Wound Healing. Adv Wound Care. 2014;3(5):390–9. 10.1089/wound.2013.0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P, Yang J, Liu X, Huang C, Tao Y, Shen P, et al. FBP1 orchestrates keratinocyte proliferation/differentiation and suppresses psoriasis through metabolic control of histone acetylation. Cell Death Dis. 2024;15(6):392. 10.1038/s41419-024-06706-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores A, Schell J, Krall AS, Jelinek D, Miranda M, Grigorian M, et al. Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat Cell Biol. 2017;19(9):1017–26. 10.1038/ncb3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner RN, Piñón Hofbauer J, Wally V, Kofler B, Schmuth M, De Rosa L, et al. Epigenetic and metabolic regulation of epidermal homeostasis. Exp Dermatol. 2021;30(8):1009–22. 10.1111/exd.14305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci. 2001;98(6):3156–61. 10.1073/pnas.061032098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Giovannini S, Wang T, Fang J, Li P, Shao C, et al. p63: a crucial player in epithelial stemness regulation. Oncogene. 2023;42(46):3371–84. 10.1038/s41388-023-02859-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–13. 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- 30.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–16. 10.1016/S1097-2765(00)80275-0 [DOI] [PubMed] [Google Scholar]
- 31.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in Stratified Epithelia. Cell. 2007;129(3):523–36. 10.1016/j.cell.2007.02.045 [DOI] [PubMed] [Google Scholar]
- 32.Pecorari R, Bernassola F, Melino G, Candi E. Distinct interactors define the p63 transcriptional signature in epithelial development or cancer. Biochem J. 2022;479(12):1375–92. 10.1042/BCJ20210737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Iorio E, Barbaro V, Ferrari S, Ortolani C, De Luca M, Pellegrini G. Q-FIHC: quantification of fluorescence immunohistochemistry to analyse p63 isoforms and cell cycle phases in human limbal stem cells. Microsc Res Tech. 2006;69(12):983–91. 10.1002/jemt.20375 [DOI] [PubMed] [Google Scholar]
- 34.Pellegrini G, Golisano O, Paterna P, Lambiase A, Bonini S, Rama P, et al. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J Cell Biol. 1999;145(4):769–82. 10.1083/jcb.145.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viticchiè G, Agostini M, Lena AM, Mancini M, Zhou H, Zolla L, et al. p63 supports aerobic respiration through hexokinase II. Proc Natl Acad Sci USA. 2015;112(37):11577–82. 10.1073/pnas.1508871112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamanaka RB, Mutlu GM. PFKFB3, a direct target of p63, is required for proliferation and inhibits differentiation in Epidermal Keratinocytes. J Invest Dermatology. 2017;137(6):1267–76. 10.1016/j.jid.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–83. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 38.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Rosa L, Secone Seconetti A, De Santis G, Pellacani G, Hirsch T, Rothoeft T, et al. Laminin 332-Dependent YAP dysregulation depletes epidermal stem cells in Junctional Epidermolysis Bullosa. Cell Rep. 2019;27(7):2036–e20496. 10.1016/j.celrep.2019.04.055 [DOI] [PubMed] [Google Scholar]
- 40.Chakravarti S, Enzo E, de Barros M, Rizzarda Mafezzoni B, Pellegrini G. Genetic disorders of the Extracellular Matrix: from cell and gene therapy to future applications in Regenerative Medicine. 2022;Annual Review of Genomics and Human Genetic, review. [DOI] [PubMed]
- 41.Santinon G, Enzo E, Dupont S. The sweet side of YAP/TAZ. Cell Cycle. 2015;14(16):2543–4. 10.1080/15384101.2015.1062328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enzo E, Santinon G, Pocaterra A, Aragona M, Bresolin S, Forcato M, et al. Aerobic glycolysis tunes YAP / TAZ transcriptional activity. EMBO J. 2015;34(10):1349–70. 10.15252/embj.201490379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enzo E, Secone Seconetti A, Forcato M, Tenedini E, Polito MP, Sala I, et al. Single-keratinocyte transcriptomic analyses identify different clonal types and proliferative potential mediated by FOXM1 in human epidermal stem cells. Nat Commun. 2021;12(1):2505. 10.1038/s41467-021-22779-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan C, Fan Y. Role of H1 linker histones in mammalian development and stem cell differentiation. Biochim et Biophys Acta (BBA) - Gene Regul Mech. 2016;1859(3):496–509. 10.1016/j.bbagrm.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polito MP, Marini G, Fabrizi A, Sercia L, Enzo E, De Luca M. Biochemical role of FOXM1-dependent histone linker H1B in human epidermal stem cells. Cell Death Dis. 2024;15(7):508. 10.1038/s41419-024-06905-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smirnov A, Panatta E, Lena A, Castiglia D, Di Daniele N, Melino G, et al. FOXM1 regulates proliferation, senescence and oxidative stress in keratinocytes and cancer cells. Aging. 2016;8(7):1384–97. 10.18632/aging.100988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng Y, Sun F, Thornton K, Jing X, Dong J, Yun G, et al. FOXM1 regulates glycolysis and energy production in multiple myeloma. Oncogene. 2022;41(32):3899–911. 10.1038/s41388-022-02398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nolfi-Donegan D, Braganza A, Shiva S. Mitochondrial electron transport chain: oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020;37:101674. 10.1016/j.redox.2020.101674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal. 2013;6(261):ra8–8. 10.1126/scisignal.2003638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feichtinger RG, Sperl W, Bauer JW, Kofler B. Mitochondrial dysfunction: a neglected component of skin diseases. Exp Dermatol. 2014;23(9):607–14. 10.1111/exd.12484 [DOI] [PubMed] [Google Scholar]
- 51.Vetter A, Jahn K, Bouameur JE, Kiritsi D, Magin TM. Epidermolysis Bullosa Simplex Keratinocytes Show disturbed mitochondrial positioning and activity. J Invest Dermatol. 2020;140(7):1438–e14425. 10.1016/j.jid.2019.10.023 [DOI] [PubMed] [Google Scholar]
- 52.De Rosa L, Latella MC, Secone Seconetti A, Cattelani C, Bauer JW, Bondanza S, et al. Toward combined cell and gene therapy for Genodermatoses. Cold Spring Harb Perspect Biol. 2020;12(5):a035667. 10.1101/cshperspect.a035667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cattaneo C, Enzo E, De Rosa L, Sercia L, Consiglio F, Forcato M, et al. Allele-specific CRISPR-Cas9 editing of dominant epidermolysis bullosa simplex in human epidermal stem cells. Mol Ther. 2024;32(2):372–83. 10.1016/j.ymthe.2023.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schröder R, Kunz WS, Rouan F, Pfendner E, Tolksdorf K, Kappes-Horn K, et al. Disorganization of the Desmin Cytoskeleton and mitochondrial dysfunction in plectin-related Epidermolysis Bullosa Simplex with muscular dystrophy. J Neuropathol Exp Neurol. 2002;61(6):520–30. 10.1093/jnen/61.6.520 [DOI] [PubMed] [Google Scholar]
- 55.Velarde MC, Demaria M, Melov S, Campisi J. Pleiotropic age-dependent effects of mitochondrial dysfunction on epidermal stem cells. Proc Natl Acad Sci USA. 2015;112(33):10407–12. 10.1073/pnas.1505675112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Squarize CH, Castilho RM, Bugge TH, Gutkind JS. Accelerated Wound Healing by mTOR activation in genetically defined mouse models. PLoS ONE. 2010;5(5):e10643. 10.1371/journal.pone.0010643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown A, Neumayer D, Rafieé-Tari Z, Krieg T, Eming SA. [Delayed wound healing during therapy of cutaneous graft-versus-host disease with everolimus]. Hautarzt. 2014;65(6):553–5. 10.1007/s00105-014-2762-y [DOI] [PubMed] [Google Scholar]
- 58.Gao M, Si X. Rapamycin ameliorates psoriasis by regulating the expression and methylation levels of tropomyosin via ERK1/2 and mTOR pathways in vitro and in vivo. Exp Dermatol. 2018;27(10):1112–9. 10.1111/exd.13745 [DOI] [PubMed] [Google Scholar]
- 59.Nanba D, Sakabe J, Mosig J, Brouard M, Toki F, Shimokawa M, et al. Low temperature and mTOR inhibition favor stem cell maintenance in human keratinocyte cultures. EMBO Rep. 2023;24(6):e55439. 10.15252/embr.202255439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Cui B, Chen Z, Ding X. The regulation of skin homeostasis, repair and the pathogenesis of skin diseases by spatiotemporal activation of epidermal mTOR signaling. Front Cell Dev Biol. 2022;10:950973. 10.3389/fcell.2022.950973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cibrian D, De La Fuente H, Sánchez-Madrid F. Metabolic pathways that control skin homeostasis and inflammation. Trends Mol Med. 2020;26(11):975–86. 10.1016/j.molmed.2020.04.004 [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Zhao F, Xu C, Zhang Q, Ren H, Huang X, et al. Metabolic reprogramming in skin wound healing. Burns Trauma. 2024;12:tkad047. 10.1093/burnst/tkad047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim CS, Ding X, Allmeroth K, Biggs LC, Kolenc OI, L’Hoest N, et al. Glutamine metabolism controls stem cell fate reversibility and long-term maintenance in the hair follicle. Cell Metabol. 2020;32(4):629–e6428. 10.1016/j.cmet.2020.08.011 [DOI] [PubMed] [Google Scholar]
- 64.Eckert RL, Kaartinen MT, Nurminskaya M, Belkin AM, Colak G, Johnson GVW, et al. Transglutaminase regulation of cell function. Physiol Rev. 2014;94(2):383–417. 10.1152/physrev.00019.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fischer J, Bourrat E. Genetics of inherited ichthyoses and related diseases. Acta Derm Venereol. 2020;100(7):adv00096. 10.2340/00015555-3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baksh SC, Todorova PK, Gur-Cohen S, Hurwitz B, Ge Y, Novak JSS, et al. Extracellular serine controls epidermal stem cell fate and tumour initiation. Nat Cell Biol. 2020;22(7):779–90. 10.1038/s41556-020-0525-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cappello A, Mancini M, Madonna S, Rinaldo S, Paone A, Scarponi C, et al. Extracellular serine empowers epidermal proliferation and psoriasis-like symptoms. Sci Adv. 2022;8(50):eabm7902. 10.1126/sciadv.abm7902 [DOI] [PubMed] [Google Scholar]
- 68.Crider KS, Yang TP, Berry RJ, Bailey LB, Folate, Methylation DNA. A review of Molecular mechanisms and the evidence for Folate’s role. Adv Nutr. 2012;3(1):21–38. 10.3945/an.111.000992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463(7280):563–7. 10.1038/nature08683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25(5):485–98. 10.1101/gad.2019811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22(14):1865–70. 10.1101/gad.1673508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moltrasio C, Romagnuolo M, Marzano AV. Epigenetic mechanisms of epidermal differentiation. IJMS. 2022;23(9):4874. 10.3390/ijms23094874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Polito MP, Marini G, Palamenghi M, Enzo E. Decoding the human epidermal complexity at single-cell resolution. Int J Mol Sci. 2023. [DOI] [PMC free article] [PubMed]
- 74.Paganelli A, Righi V, Tarentini E, Magnoni C. Current knowledge in skin metabolomics: updates from Literature Review. Int J Mol Sci. 2022;23(15):8776. 10.3390/ijms23158776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mustfa SA, Maurizi E, McGrath J, Chiappini C. Nanomedicine approaches to negotiate local biobarriers for Topical Drug Delivery. Adv Ther. 2021;4(1):2000160. 10.1002/adtp.202000160 [DOI] [Google Scholar]
- 76.Laura Sercia, O. Romano, G. Marini, E. Enzo, M. Forcato, Laura De Rosa, Michele De Luca, A cellular disease model towards gene therapy of TGM1-dependent Lamellar Ichthyosis, Molecular Therapy - Methods & Clinical Development, 2024, 101311, ISSN 2329-0501, https://doi.org/10.1016/j.omtm.2024.101311.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.