Abstract
The universal membrane association of positive-strand RNA virus RNA replication complexes is implicated in their function, but the intracellular membranes used vary among viruses. Brome mosaic virus (BMV) encodes two mutually interacting RNA replication proteins: 1a, which contains RNA capping and helicase-like domains, and the polymerase-like 2a protein. In cells from the natural plant hosts of BMV, 1a and 2a colocalize on the endoplasmic reticulum (ER). 1a and 2a also direct BMV RNA replication and subgenomic mRNA synthesis in the yeast Saccharomyces cerevisiae, but whether the distribution of 1a, 2a, and active replication complexes in yeast duplicates that in plant cells has not been determined. For yeast expressing 1a and 2a and replicating BMV genomic RNA3, we used double-label confocal immunofluorescence to define the localization of 1a, 2a, and viral RNA and to explore the determinants of replication complex targeting. As in plant cells, 1a and 2a colocalized on and were retained on the yeast ER, with no detectable accumulation in the Golgi apparatus. 1a and 2a were distributed over most of the ER surface, with strongest accumulation on the perinuclear ER. In vivo labeling with bromo-UTP showed that the sites of 1a and 2a accumulation were the sites of nascent viral RNA synthesis. In situ hybridization showed that completed viral RNA products accumulated predominantly in the immediate vicinity of replication complexes but that some, possibly more mature cells also accumulated substantial viral RNA in the surrounding cytoplasm distal to replication complexes. Additionally, we find that 1a localizes to the ER when expressed in the absence of other viral factors. These results show that BMV RNA replication in yeast duplicates the normal localization of replication complexes, reveal the intracellular distribution of RNA replication products, and show that 1a is at least partly responsible for the ER localization and retention of the RNA replication complex.
The RNA replication complexes of all eukaryotic, positive-strand RNA viruses studied to date are associated with intracellular membranes (36, 39, 46). A variety of studies indicate that interaction of viral RNA replication factors with membranes is important for at least some steps of RNA replication (29, 48). However, the nature and function of this membrane association presently remain uncertain. Moreover, while membrane association is general, the replication complexes of different positive-strand RNA viruses are associated with different intracellular membranes. For example, alphavirus RNA replication occurs exclusively on modified endosomes and lysosomes (12, 30), while poliovirus RNA replication occurs on infection-specific vesicles derived from the membranes of the endoplasmic reticulum (ER), Golgi apparatus, and lysosomes (39), and RNA replication by some plant viruses occurs on chloroplast membranes (13).
A further example of the diversity in membrane targeting of such replication complexes is provided by brome mosaic virus (BMV). BMV encodes RNA replication factors sharing extensive conservation with the endosome-targeted alphavirus replication factors (2, 14) but, in cells of its natural plant hosts, directs assembly of its RNA replication complexes on ER membranes (36). BMV encodes two RNA replication proteins: 1a (109 kDa) contains a C-terminal helicase-like domain and an N-terminal domain required for m7G methyltransferase and m7GMP covalent binding (putative m7G transferase) activities implicated in RNA capping (3), while 2a contains a central polymerase-like domain and an N-terminal region that interacts with 1a (21, 22). 1a and 2a colocalize in infected plant protoplasts at sites of BMV RNA synthesis, which are localized to the ER and do not pass on into the Golgi or later parts of the secretory apparatus (36). Moreover, 1a and 2a copurify from infected cells in an initially membrane-bound state, in combination with a BMV-specific RNA-dependent RNA polymerase activity (27, 33, 34). 1a and 2a are encoded by BMV genomic RNA1 and RNA2, respectively. The third BMV genomic RNA, RNA3, encodes two genes: a 5′-proximal movement gene required for cell-to-cell movement of infection and a 3′-proximal coat protein gene, which is translated from a subgenomic mRNA, RNA4 (4, 28).
1a and 2a can direct RNA replication not only in cells of BMV’s natural plant hosts but also in the yeast Saccharomyces cerevisiae. In particular, when 1a and 2a are expressed in yeast from DNA plasmids and RNA3 is introduced by transfection or in vivo transcription, RNA3 is replicated and the RNA3-encoded subgenomic mRNA, RNA4, is synthesized (17, 20). Accordingly, the use of yeast as an alternate host can assist some studies of viral (19, 44) and cellular (16) functions in BMV replication. BMV RNA3 replication and subgenomic mRNA synthesis in yeast parallel those in plant cells in all aspects tested to date, including dependence on 1a, 2a, and defined cis-acting RNA3 replication and subgenomic mRNA synthesis signals, production of a similar excess of positive-strand over negative-strand RNA, and other respects (17, 20, 32, 44). As the S. cerevisiae genome is more closely related to animal than to plant genomes (6), these results suggest that the essential host features required for BMV replication are fairly widely conserved. Similar to membrane fractions from BMV-infected plant cells, the membrane fractions of yeast cells expressing 1a, 2a, and RNA3 contain 1a, 2a, and BMV RNA-dependent RNA polymerase activity (32). However, the nature of the yeast membrane(s) involved and the intracellular distribution of 1a, 2a, and BMV RNA synthesis sites in yeast have not been examined.
To determine the extent to which the normal membrane localization of BMV RNA replication may or may not be preserved in yeast and to provide a cell biology foundation for studies of viral and host contributions to BMV RNA replication in yeast, we have now examined the distribution in yeast of 1a, 2a, nascent BMV RNA, and BMV RNA replication products. We report here that the localization of BMV replication complexes in yeast closely parallels that in plant cells, that completed viral RNAs remain preferentially localized near replication complexes, and that, like the full BMV RNA replication complex, 1a localizes to the ER in the absence of other viral factors.
MATERIALS AND METHODS
Yeast strain, cell growth, and plasmids.
All experiments were performed with S. cerevisiae YPH500 (MATa ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1). Yeast cells were transformed with plasmids expressing BMV 1a, 2a, or RNA3 by the LiCl-polyethylene glycol method (18) and were grown at 30°C on synthetic liquid or solid medium containing 2% galactose and lacking relevant amino acids or uracil to select for the DNA plasmids present (5). Cells were harvested for analysis at a culture optical density at 600 nm of 0.4 to 0.7. BMV protein 1a was expressed from pB1CT19, a yeast 2μm plasmid containing the BMV 1a open reading frame between the yeast ADH1 promoter and polyadenylation site, plus the HIS2 selectable marker gene (20). Similarly, BMV protein 2a was expressed from pB2CT15, a yeast 2μm plasmid containing the BMV 2a open reading frame between the yeast ADH1 promoter and polyadenylation site, plus the LEU3 selectable marker gene (20). BMV RNA3 was expressed from a yeast CEN4 centromeric plasmid, pB3RQ39, that contains a full-length BMV RNA3 cDNA linked at its 5′ end to the galactose-inducible yeast GAL1 promoter and at its 3′ end to a self-cleaving ribozyme, plus the TRP1 selectable marker gene (17). A URA3-selectable yeast CEN4 plasmid expressing a c-myc-tagged version of EMP47 was kindly provided by Sean Munro (41).
Antibodies.
Anti-2a mouse monoclonal antibodies 6G12 and 10B3 and anti-1a rabbit polyclonal antiserum were used throughout (36). Rabbit polyclonal antiserum against Kar2p was kindly provided by Mark Rose (37). Mouse monoclonal antibodies against c-Myc (9E10) and digoxigenin were from Boehringer Mannheim, while those against Dpm1p and bromodeoxyuridine were from Molecular Probes and Sigma, respectively.
Immunofluorescence.
Fixation of yeast cells with formaldehyde and double-label immunofluorescence staining were performed as described previously (35). Primary antibodies were diluted 1:100 in 1% bovine serum albumin (BSA)–0.05% Nonidet P-40 in phosphate-buffered saline (37.5 mM K2HPO4, 10 mM KH2PO4, 150 mM NaCl) and incubated with the fixed cells overnight at 4°C. After three washes with 1% (BSA)–0.05% Nonidet P-40 in phosphate-buffered saline, donkey anti-rabbit or anti-mouse secondary antibodies conjugated to fluorescein, Texas red, or Alexa 488 (Molecular Probes) were added and incubated for 2 h at room temperature. For nuclear staining, a 10-min incubation with 1 μM To-Pro-3 iodide (Molecular Probes) was added after secondary antibody incubation. Immunofluorescence images were obtained with a Bio-Rad 1024 confocal microscope at the Keck Neural Imaging Laboratory, University of Wisconsin—Madison. To ensure the reproducibility of the results, each experiment was performed three to six times.
Labeling and detection of nascent RNA.
Semi-intact yeast cells were prepared by the spheroplast freeze-thaw procedure of Schlenstedt et al., which permeabilizes the plasma membrane while preserving intracellular membrane structure and functional pathways for such processes as nuclear protein import, protein secretion, and vacuole division (40). After permeabilization, bromo-UTP (BrUTP), MgCl2, and dithiothreitol were added to 10 mM each, and the yeast cells were incubated at 30°C for 5 to 15 min as noted in the text. After two washes in 1 M sorbitol–0.1 M KPO4 (pH 7.5), the cells were fixed in formaldehyde and processed for immunofluorescence as described above.
In situ hybridization.
In situ hybridizations to detect positive-strand BMV RNA3 and RNA4 were performed as described elsewhere (11, 26), with minor modifications. After 45 min of fixation in 5% formaldehyde, yeast cells were washed twice with SP (1.2 M sorbitol, 0.1 M KPO4 [pH 7.5]) and spheroplasted for 30 min at 30°C in SP containing 10 μg of lyticase per ml, 30 mM β-mercaptoethanol, and 20 mM vanadyl ribonucleoside complex (Gibco Life Sciences). The cells were washed with SP, and an aliquot was transferred to polyethyleneimine-coated glass microscope slides and allowed to settle for 15 min. The SP was removed by aspiration, and the cells were covered with 25 μl of 2× SSC (300 mM NaCl, 30 mM sodium citrate [pH 7.0]) containing 40% formamide, 2.5 mg of BSA per ml, 20 mM vanadyl ribonucleoside complex, 120 Units of placental RNase inhibitor (Promega) per ml, 1 mg of salmon sperm DNA per ml, and 2.5 pmol each of four digoxigenin-labeled oligodeoxynucleotides. The oligonucleotides used hybridized to four segments of the coat protein open reading frame common to positive-strand BMV RNA3 and RNA4, i.e., to nucleotides 1480 to 1496, 1541 to 1558, 1757 to 1774, and 1377 to 1394 of RNA3 (1). These oligonucleotides were end labeled with terminal deoxynucleotidyltransferase (Promega) and digoxigenin-dUTP (Boehringer Mannheim) according to the manufacturers’ recommendations. The cells were incubated with the probes for 3 h at 37°C and then washed twice for 5 min with 40% formamide–2× SSC at 37°C and twice for 5 min with 1× SSC at room temperature. The slides then were processed as indicated above for immunofluorescence with anti-BMV 1a and anti-digoxigenin antibodies.
RESULTS
BMV 1a and 2a colocalize in yeast cytoplasm.
As noted in the introduction, yeast cells expressing BMV 1a, 2a, and RNA3 from expression plasmids form active BMV RNA synthesis complexes that replicate RNA3 and synthesize the subgenomic coat protein mRNA encoded by RNA3 (17, 20). Such cells (hereafter referred to as 1a+2a+RNA3 yeast cells) were used for all experiments in this study except as discussed below for Fig. 4B and 6. To visualize and compare the sites of 1a and 2a protein accumulation in 1a+2a+RNA3 yeast cells, we used double-label confocal immunofluorescence microscopy with rabbit polyclonal anti-1a serum and mouse monoclonal anti-2a serum. The antisera used were previously shown to support immunofluorescence localization of 1a and 2a in BMV-infected cells of a natural plant host, barley (36). Figure 1A shows representative immunofluorescence images of 1a+2a+RNA3 yeast cells. In contrast to animal or plant cells, such yeast cells average only around 5 μm in length, requiring high magnification to discern details of intracellular structure. Each row in Fig. 1A presents three images of one focal plane in a single cell, showing 1a (red), 2a (green), and their superposition. The source and independence of 1a and 2a immunofluorescence signals were confirmed in control experiments in which individual primary or secondary antisera for 1a or 2a were omitted, fluors were switched between 1a and 2a secondary antibodies, and yeast cells expressing 1a alone, 2a alone, or neither were treated with both anti-1a and anti-2a sera (Fig. 1B and results not shown).
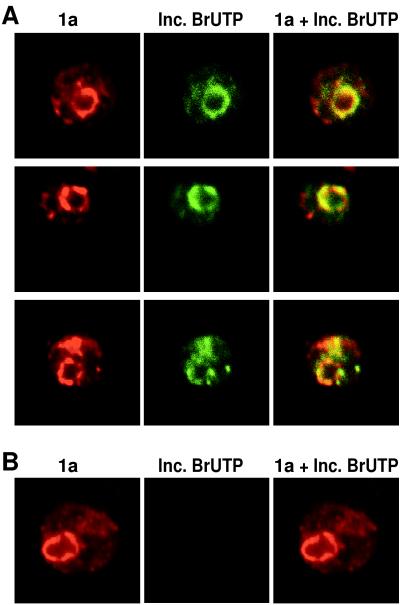
FIG. 4.
(A) Colocalization of newly synthesized BMV RNA with 1a. 1a+2a+RNA3 yeast cells were permeabilized, incubated 10 min with BrUTP, and then fixed and processed for indirect, double-label immunofluorescence with primary antisera that recognize 1a and BrU incorporated into nucleic acid. Rows show 1a (left), incorporated BrUTP (middle), and their superposition (right) for a 0.5 μm optical section of a representative, independent cell. (B) Strong, cytoplasmic BrUTP incorporation is dependent on the presence of a functional BMV RNA replication template. 1a+2a yeast cells were permeabilized, incubated, fixed, and processed as for panel A for indirect immunofluorescence with primary antisera that recognize 1a and BrU incorporated into nucleic acid. Each image is 9 μm per side.
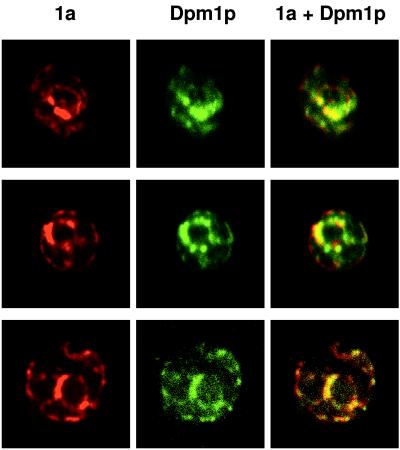
FIG. 6.
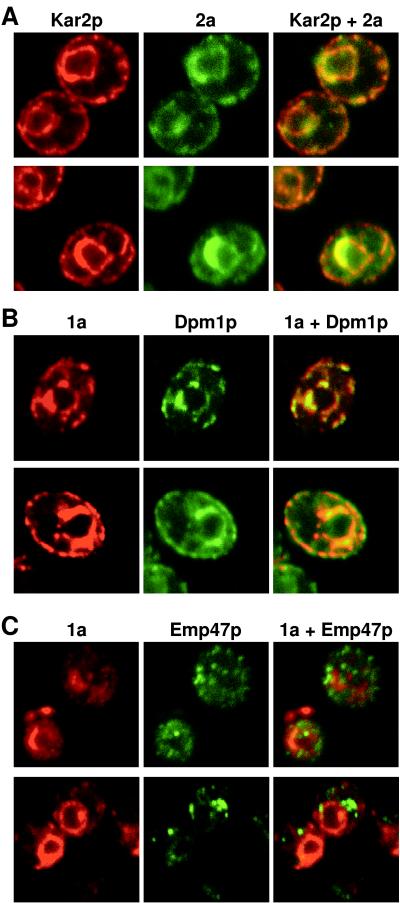
1a colocalizes with ER markers in the absence of 2a or RNA3. Yeast cells expressing 1a but not 2a or RNA3 were processed for indirect, double-label immunofluorescence using primary antisera against 1a and yeast ER protein Dpm1p. Rows show 1a (left), Dpm1p (middle), and their superposition (right) for a 0.5-μm optical section of a representative, independent cell. Each image is 9 μm per side.
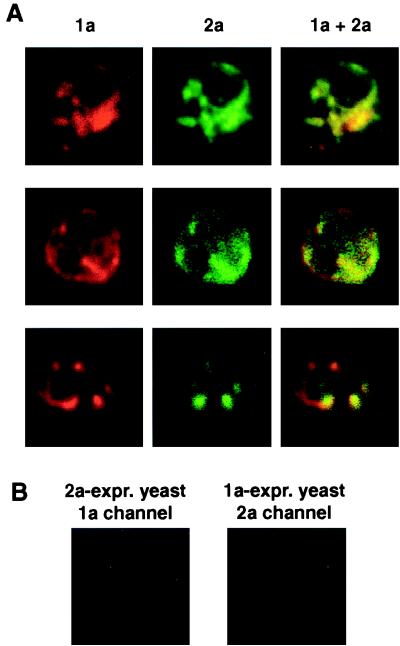
FIG. 1.
BMV 1a and 2a colocalize in 1a+2a+RNA3 yeast cells. (A) 1a+2a+RNA3 yeast cells were processed for indirect, double-label immunofluorescence using rabbit anti-1a and mouse anti-2a antisera followed by anti-rabbit antibodies conjugated to Texas red and anti-mouse antibodies conjugated to Alexa 488. Rows show 1a (left), 2a (middle), and their superposition (right) for a 0.5-μm optical section of a representative, independent cell. Each image is 9 μm per side. (B) Sample negative controls for antibody specificity. Yeast expressing (expr.) 1a only or 2a only were processed for double-label immunofluorescence using both anti-1a and anti-2a primary and secondary antibodies as described above. The images shown illustrate the absence of 1a immunofluorescence signal from yeast expressing 2a only and the absence of 2a immunofluorescence signal from yeast expressing 1a only. See text for additional controls.
In general, the localization of 1a and 2a closely paralleled each other through highly variable, complex patterns in different cells (Fig. 1A). This predominant colocalization of 1a and 2a immunofluorescence was observed across hundreds of cells in numerous independent experiments. However, as in BMV-infected plant cells (36), occasional variations between 1a and 2a immunofluorescence patterns were seen in some cells. The degree to which such variations reflect independent localization of some 1a or 2a, failure to detect all colocalizing 1a or 2a, or artifactual background is not yet clear. Examples of either 1a or 2a immunofluorescence signals without the other were seen. However, sites of 1a immunofluorescence without any detectable 2a immunofluorescence signal (e.g., Fig. 1A, bottom row) were more frequently observed, perhaps because 2a fluorescence was weaker than 1a fluorescence in 1a+2a+RNA3 yeast cells.
1a and 2a colocalize with ER proteins.
To determine the site or sites at which 1a and 2a colocalized, we first compared the distributions of 1a and nuclear DNA in 1a+2a+RNA3 yeast cells. As found previously for BMV-infected barley cells (36), 1a accumulated primarily in partial haloes surrounding the nucleus and in cytoplasmic extensions from these perinuclear haloes (Fig. 2). Since this perinuclear/cytoplasmic distribution of 1a and 2a was similar to their ER-associated distribution in BMV-infected plant cells, we used double-label immunofluorescence to assess whether 1a and 2a also were associated with the ER in yeast cells. As well-characterized yeast ER markers with available antibodies compatible with simultaneous labeling of 2a and 1a, respectively, we used the Kar2 and Dpm1 (dolichol-phosphate mannose synthase) proteins. Kar2p is the yeast homolog of the mammalian chaperone BiP/GRP78 (37). Like BiP, Kar2p is localized to the ER lumen and is commonly used as an ER marker (23, 47). Dpm1p is a transmembrane ER protein (31, 38).
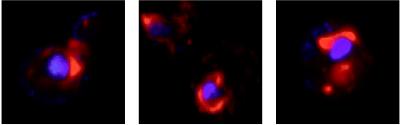
FIG. 2.
BMV 1a accumulates in perinuclear and cytoplasmic regions of 1a+2a+RNA3 yeast cells. 1a+2a+RNA3 yeast cells were processed for 1a immunofluorescence (red) and stained for DNA with To-Pro-3 (blue). Each image (9 μm per side) shows the superimposed 1a and DNA fluorescence patterns from independent, representative cells.
As shown in Fig. 3A, 2a and Kar2p showed intricate but closely corresponding patterns. All sites of significant 2a immunofluorescence were also sites of Kar2p accumulation. Conversely, nearly all sites of Kar2p accumulation showed 2a immunofluorescence. This suggested that under the conditions of 2a expression and cell growth used in these experiments, 2a was distributed over a significant fraction of the ER. Typical features of the 2a/Kar2p distribution included prominent fluorescence in a partial or complete perinuclear ring (verified as perinuclear by double labeling of Kar2p and nuclear DNA stained with To-Pro-3 iodide [Molecular Probes] [results not shown]), variable amounts of cytoplasmic processes, and in some optical sections, labeling at the periphery of the cell. Such appression of a portion of the ER against the plasma membrane is frequently observed for yeast (23, 47), and both 2a and 1a signals were also seen on this peripheral ER (Fig. 3A and B). In general, the strongest 1a and 2a signals were on the perinuclear membrane, which is typical of many yeast ER proteins (31).
FIG. 3.
1a and 2a accumulate on the ER but not the Golgi apparatus in 1a+2a+RNA3 yeast cells. Each row shows immunofluorescence images for a 0.5-μm optical section of representative, independent BMV 1a+2a+RNA3 yeast cells. The cells were processed for indirect, double-label immunofluorescence using primary antisera against BMV 2a and yeast ER protein Kar2p (A), BMV 1a and yeast ER protein Dpm1p (B), and BMV 1a and yeast Golgi protein Emp47p (C). Images of each individual label and their superposition are shown. Each image is 9 μm per side.
Representative optical sections from cells stained for 1a and Dpm1p are shown in Fig. 3B. As expected from the results shown in Fig. 1A and 3A and the established ER localization of Dpm1p, the Dpm1p and 1a distributions correlated closely and were similar to those of Kar2p and 2a. As with Kar2p and 2a, virtually all sites of Dpm1p immunofluorescence also showed 1a immunofluorescence, suggesting that 1a also was distributed over most of the ER.
The close correlation of 1a and 2a localization with the ER markers Kar2p and Dpm1p suggested that 1a and 2a were absent from later compartments in the secretory pathway. To examine this further, we performed simultaneous immunofluorescence labeling of 2a and Emp47p, a Golgi-localized type I transmembrane protein. We used a c-Myc-tagged version of Emp47p that was previously shown to remain localized in a Golgi-associated, punctate pattern (41). In keeping with these prior reports, Emp47p localized in a punctate cytoplasmic pattern distinct from the distribution of 1a (Fig. 3C). Thus, as in plant cells (36), 1a did not detectably accumulate in the Golgi apparatus.
Nascent viral RNA colocalizes with 1a.
Previously we showed that in vitro, BMV RNA-dependent RNA polymerase efficiently incorporates BrUTP into full-length BMV RNAs (36). We further showed that in BMV-infected plant protoplasts, a cytoplasmic, BMV-specific, and actinomycin D-insensitive process incorporates BrUTP into RNase-sensitive material recognized by an antibody that binds bromouridine-containing RNA and that these sites of BMV-specific BrUTP incorporation corresponded to the joint sites of 1a and 2a accumulation (36). To apply similar BrUTP labeling to BMV RNA synthesis in yeast, we first spheroplasted 1a+2a+RNA3 yeast cells. Since directly incubating such spheroplasts in BrUTP did not yield any BrUTP incorporation, we then sought to facilitate BrUTP uptake by permeabilizing the plasma membrane by using established procedures shown to preserve cytoplasmic structure, secretory transport, nuclear protein import, etc. (40). The permeabilized yeast cells then were incubated with BrUTP for 5 to 15 min, fixed, and processed for double immunofluorescence labeling with anti-1a antiserum and an antiserum that recognizes bromouridine-containing RNA but not free BrUTP. Figure 4A shows representative optical sections from 1a+2a+RNA3 yeast cells incubated for 10 min with BrUTP; incubation with BrUTP for 5 or 15 min gave similar results. As in other experiments shown above, 1a (red) localized to a perinuclear ring with projections into the cytoplasm. Like 2a (Fig. 1A and 3), incorporated BrUTP (green) colocalized with the strongest 1a signals, predominantly but not exclusively in the perinuclear region. Moreover, close inspection showed that even fainter sites of 1a accumulation, including those peripheral to the perinuclear region, were also sites of weak but detectable BrUTP incorporation. Conversely, the sites of BrUTP incorporation were also sites of 1a accumulation.
Parallel processing of yeast lacking 1a and 2a did not reveal BrUTP incorporation detectable at this level of sensitivity. To further test whether the BrUTP labeling sites corresponded to sites of BMV RNA synthesis rather than to, e.g., BrUTP binding by 1a or 2a, the same experiment was performed in parallel with yeast cells expressing 1a and 2a but lacking the BMV RNA3 replication template (i.e., 1a+2a yeast cells). As shown in Fig. 4B, 1a immunostaining was unaffected but no BrUTP signal was detected, showing that the signal detected in Fig. 4A was dependent on the presence of a functional BMV RNA replication template. In Fig. 4A and B, the absence of nuclear BrUTP incorporation signals comparable to those of BMV-specific RNA synthesis was expected because of the short labeling periods and because in BMV-infected plant cells, BrUTP incorporation into BMV RNA replication complexes also was much stronger than nuclear transcription (36).
Intracellular distribution of RNA3 and RNA4 replication products.
When yeast cells coexpress 1a, 2a, and wild-type RNA3 from the plasmids used in this study, BMV RNA replication amplifies RNA3 almost 50-fold over the basal level of DNA-derived RNA3 transcripts in yeast cells carrying the RNA3 expression plasmid alone (17, 19). Thus, approximately 98% of the positive-strand RNA3 in 1a+2a+RNA3 yeast cells is derived from 1a- and 2a-directed, RNA-dependent RNA replication rather than from DNA plasmid-directed transcription. In addition, 1a and 2a use negative-strand RNA3 as a template for synthesis of the subgenomic coat protein mRNA, RNA4, which accumulates to approximately 60% of the level of replicated RNA3 (17). No RNA4 can be detected in RNA3-expressing yeast in the absence of 1a and 2a (17, 19).
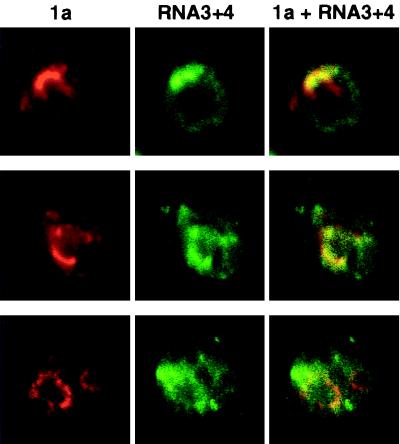
To visualize these RNA products in 1a+2a+RNA3 yeast cells and compare their distribution with that of BMV replication factors, we used in situ hybridization according to approaches previously optimized for yeast (11, 26). To increase signal strength, we probed the yeast with a mixture of four digoxigenin-labeled oligonucleotides complementary to nonoverlapping sequences of BMV RNA3. To further maximize signal strength, all four hybridization probes were complementary to sequences in the coat protein open reading frame, which is present in both RNA3 and RNA4. After yeast fixation and hybridization, the oligonucleotide probes were visualized together with 1a, using antidigoxigenin and anti-1a antisera (Fig. 5). As expected, 1a distributions (red) were similar to those seen previously. The strongest sites of RNA3 and RNA4 accumulation (green) usually coincided with or were in the immediate vicinity of strong sites 1a immunofluorescence, but weaker RNA3 and 4 hybridization signals were also generally diffused through the cytoplasm (Fig. 5, cells in top and middle rows). In a smaller fraction of cells, regions of strong RNA3 and RNA4 hybridization signals were seen adjacent to but separated from sites of 1a accumulation (Fig. 5, cells in bottom row). Possible reasons for such distributions are considered in Discussion.
FIG. 5.
BMV RNA3 and RNA4 accumulate predominantly but not exclusively near 1a. 1a+2a+RNA3 yeast cells were fixed and subjected to in situ hybridization with four digoxigenin-labeled oligodeoxynucleotide probes (see Materials and Methods) complementary to the positive strand of the coat protein gene in RNA3 and its subgenomic mRNA, RNA4. After in situ hybridization, the cells were processed for indirect, double-label immunofluorescence using primary antisera against 1a and digoxigenin. Rows show 1a (left), RNA3 and RNA4 (middle), and their superposition (right) for a 0.5-μm optical section of a representative, independent cell. Each image is 9 μm per side.
1a localizes to the ER in the absence of other viral components.
To explore the independent localization properties of the 1a and 2a proteins, we expressed each separately in yeast. In keeping with Western blotting results showing that 2a accumulation was enhanced by 1a coexpression (16), 2a immunofluorescence in the absence of 1a was significantly less than that in 1a+2a yeast cells. As noted above, 2a immunofluorescence even in 1a+2a cells was notably weaker than 1a immunofluorescence, and the further weakening of 2a immunofluorescence upon omitting 1a resulted in a signal that was too weak and variable to allow us to unambiguously define the intracellular distribution of 2a (see also Discussion).
In contrast, prior Western blotting showed that 1a protein accumulation in vivo is not affected by the presence or absence of 2a (16). Similarly, we found that the strength of 1a immunofluorescence was unchanged when 2a was omitted. Furthermore, the 1a distribution pattern in the absence of 2a (Fig. 6) was not distinguishable from that in 1a+2a yeast cells (Fig. 3B). In both cases, the strongest 1a accumulation usually comprised a partial ring around the nucleus (identified by DNA staining [results not shown]). Narrower strands of 1a immunofluorescence extended from the perinuclear region into the cytoplasm and, in some optical sections, to the periphery of the cell. Double-label immunofluorescence showed that 1a expressed in the absence of 2a colocalized with the ER marker protein Dpm1p (Fig. 6). Thus, in the absence of 2a, RNA3, or any other viral factors, 1a localized to the ER. Moreover, as in 1a+2a+RNA3 cells, 1a expressed alone localized to nearly all sites of Dpm1p fluorescence (Fig. 6), showing that 1a was distributed over most of the ER.
DISCUSSION
Localization of BMV RNA replication and replication factors to the yeast ER.
The assembly of a multiprotein replication complex on intracellular membranes appears to be a general aspect of RNA replication by all positive-strand RNA viruses in their natural hosts. However, unknown variations in virus-host interaction lead even related viruses to direct replication complex assembly to different membrane sites (12, 30, 36). In the present study we have shown that proper targeting of RNA replication factors and the close connection of viral RNA synthesis to specific intracellular membranes is preserved in the BMV-yeast system. In yeast expressing BMV RNA replication factors 1a and 2a and actively replicating BMV RNA3, we found that 1a, 2a, and nascent viral RNA all colocalized in combination with the well-characterized yeast ER markers Kar2p and Dpm1p. These results parallel the ER localization of BMV RNA replication in infected cells of a natural plant host (36), further supporting the suitability of yeast as a model host for studies of BMV RNA replication. Moreover, as discussed below, we have further extended these localization results by showing that 1a independently targets the ER membrane in the absence of other BMV components and by using in situ hybridization to examine the intracellular distribution of previously synthesized RNA replication products.
On close inspection, essentially all sites of 1a accumulation in 1a+2a+RNA3 yeast cells were also visualized as sites of nascent viral RNA synthesis, as detected by BMV-specific BrUTP incorporation (Fig. 4A). Thus, the observed 1a and 2a distributions represented the distribution of active replication complexes. Furthermore, in most yeast cells, 1a and 2a covered a considerable fraction of the ER (Fig. 3A and B). As the yeast cells in this study were harvested while still in exponential-phase growth, the results in Fig. 3 imply that under the conditions used, plasmid-directed expression of 1a plus 2a and BMV RNA replication reached and maintained significant coverage of the ER in pace with the active growth and cytoplasmic expansion of the dividing yeast population. For comparison, in synchronous BMV infections of nondividing plant protoplasts, 1a and 2a are first observed around 3 to 4 h postinoculation at a few punctate sites on the ER and then slowly spread to encompass most of the ER by late in infection (16 to 24 h postinoculation [36]). The steady-state association of BMV replication complexes with most of the yeast ER helps to explain why, after 1a+2a yeast cells are transfected with in vitro transcripts of BMV RNA3 derivatives, these RNA3 derivatives successfully reinitiate replication in most daughter cells and can be maintained indefinitely in dividing yeast populations as free RNA replicons in the absence of an RNA3 plasmid (15, 20). As in plant cells, 1a and 2a in yeast cells remained strictly limited to the ER and did not accumulate detectably in the Golgi apparatus (Fig. 3C). The basis for this ER retention is not yet known.
Distribution of RNA3 and RNA4 replication products.
Since incubation with BrUTP was limited to periods of 5 to 10 min prior to cell harvest, BrUTP incorporation was expected to label primarily nascent RNA or RNA products just completed. Accordingly, BrUTP incorporation was detected only at sites of 1a and 2a accumulation (Fig. 4A). In situ hybridization, by contrast, detects any RNA containing the target sequence, whether synthesized recently or long prior to cell harvest. In keeping with this, the distribution of RNA3 and RNA4 hybridization signals in 1a+2a+RNA3 yeast cells was more variable than that of incorporated BrUTP. While the strongest in situ hybridization signals in most cells were associated with sites of 1a and 2a accumulation, weaker hybridization signals were also diffused throughout the cytoplasm (Fig. 5, top two rows). Moreover, in some cells, substantial RNA3 and RNA4 accumulation was found in areas separated from sites of 1a accumulation (Fig. 5, bottom row). Since free RNA3 turns over rapidly in yeast (19, 44), the accumulation of RNA at these distal sites may involve its interaction with coat protein: in 1a+2a+RNA3 yeast cells, RNA4 production leads to translation of coat protein, which selectively stabilizes BMV RNAs in yeast (24, 44). In addition, some differences in RNA3 and RNA4 distribution may reflect age differences between yeast cells. Yeast cells are not immortal but survive for approximately 25 cycles of asymmetric budding to produce smaller daughter cells (42). Cells with larger accumulations of RNA3 and RNA4 at sites distal to 1a and 2a may represent mature mother cells that have accumulated BMV RNA for many cell cycles. Conversely, in newly budded daughter cells, which constitute 50% of the dividing cell population, BMV RNA may be primarily localized to the vicinity of replication complexes.
Independent ER localization of 1a.
In the yeast system described here, the use of separate plasmids to express functional levels of 1a and 2a provided the opportunity to explore the independent localization of these factors. When 1a was expressed in the absence of 2a or RNA3, the level of its accumulation (16) and immunofluorescence were unchanged from the level in 1a+2a+RNA3 yeast cells. More significantly, 1a expressed alone (Fig. 6) localized to the ER in patterns not distinguishable from those in 1a+2a+RNA3 yeast cells (Fig. 3B). While the basis for the ER localization of 1a is not yet known, host-specific differences in the level of RNA synthesis segregate with the 1a gene in reassortants between BMV strains, implying that 1a interacts directly or indirectly with one or more host components in ways important for RNA replication (8). The nsP1 protein of the alphavirus Semliki Forest virus, which is homologous to the N-terminal half of 1a, also localizes to membranes specifically in the absence of other viral factors: nsP1 appears first on the plasma membrane and then on endosomal and lysosomal vesicles, the normal sites of Semliki Forest virus RNA synthesis (30). nsP1 is palmitoylated, but this modification is not required for its membrane association (25).
While 2a interacts with 1a in vitro (21, 22) and in vivo (9, 43), further work is necessary to determine whether the ER localization of 2a in 1a+2a+RNA3 yeast cells is dependent on 1a. Due to the weaker immunofluorescence of 2a and its reduced accumulation in the absence of 1a (this work and reference 16), it was not possible in this study to unambiguously determine the localization of 2a when expressed alone. To address this, experiments are in progress to increase the sensitivity of 2a localization by alternate detection methods (7).
ACKNOWLEDGMENTS
We thank Mark Rose and Sean Munro for generously providing anti-Kar2p antiserum and a plasmid expressing c-Myc-tagged EMP47p, respectively. Confocal microscopy was performed in the Keck Neural Imaging Laboratory of the University of Wisconsin—Madison.
This research was supported by the National Institutes of Health under grant GM35072. P.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahlquist P, Luckow V, Kaesberg P. Complete nucleotide sequence of brome mosaic virus RNA3. J Mol Biol. 1981;153:23–38. doi: 10.1016/0022-2836(81)90524-6. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist P, Strauss E G, Rice C M, Strauss J H, Haseloff J, Zimmern D. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J Virol. 1985;53:536–542. doi: 10.1128/jvi.53.2.536-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein. J Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison R, Thompson C, Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci USA. 1990;87:1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 6.Baldauf S L, Palmer J D. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., and P. Ahlquist. Unpublished results.
- 8.De Jong W, Ahlquist P. Host-specific alterations in viral RNA accumulation and infection spread in a brome mosaic virus isolate with an expanded host range. J Virol. 1995;69:1485–1492. doi: 10.1128/jvi.69.3.1485-1492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinant S, Janda M, Kroner P A, Ahlquist P. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J Virol. 1993;67:7181–7189. doi: 10.1128/jvi.67.12.7181-7189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doedens J R, Giddings T H, Jr, Kirkegaard K. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol. 1997;71:9054–9064. doi: 10.1128/jvi.71.12.9054-9064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrester W, Stutz F, Roshbash M, Wickens M. Defects in mRNA 3′ end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 12.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier M, Candresse T, Bove J M. Immunocytochemical localization of TYMV-coded structural and nonstructural proteins by the protein A-gold technique. Virology. 1986;151:100–109. doi: 10.1016/0042-6822(86)90107-8. [DOI] [PubMed] [Google Scholar]
- 14.Haseloff J, Goelet P, Zimmern D, Ahlquist P, Dasgupta R, Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci USA. 1984;81:4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa, M., and P. Ahlquist. Unpublished results.
- 16.Ishikawa M, Diez J, Restrepo-Hartwig M, Ahlquist P. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa M, Janda M, Krol M A, Ahlquist P. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janda M, Ahlquist P. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc Natl Acad Sci USA. 1998;95:2227–2232. doi: 10.1073/pnas.95.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 21.Kao C C, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao C C, Quadt R, Hershberger R P, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a from a complex in vitro. J Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koning A J, Roberts C J, Wright R L. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase ioszymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell. 1996;7:769–789. doi: 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krol, M., and P. Ahlquist. Unpublished results.
- 25.Laakkonen P, Ahola T, Kaariainen L. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J Biol Chem. 1996;271:28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- 26.Long R M, Elliot D J, Stutz F, Rosbash M, Singer R H. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- 27.Miller W A, Hall T C. Use of micrococcal nuclease in the purification of highly template dependent RNA-dependent RNA polymerase from brome mosaic virus-infected barley. Virology. 1983;125:236–241. doi: 10.1016/0042-6822(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 28.Mise K, Ahlquist P. Host-specificity restriction by bromovirus cell-to-cell movement protein occurs after initial cell-to-cell spread of infection in nonhost plants. Virology. 1995;206:276–286. doi: 10.1016/s0042-6822(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 29.Molla A, Paul A V, Wimmer E. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J Virol. 1993;67:5932–5938. doi: 10.1128/jvi.67.10.5932-5938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peränen J, Laakkonen P, Hyvönen M, Kääriänen L. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology. 1995;208:610–620. doi: 10.1006/viro.1995.1192. [DOI] [PubMed] [Google Scholar]
- 31.Preuss D, Mulholland J, Kaiser C A, Orlean P, Albright C, Rose M D, Robbins P W, Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- 32.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quadt R, Jaspars E M J. Purification and characterization of brome mosaic virus RNA-dependent RNA polymerase. Virology. 1990;178:189–194. doi: 10.1016/0042-6822(90)90393-6. [DOI] [PubMed] [Google Scholar]
- 34.Quadt R, Kao C C, Browning K S, Hershberger R P, Ahlquist P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redding K, Holcomb C, Fuller R S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose M D, Misra L M, Vogel J P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 38.Rothblatt J, Novick P, Stevens T. Guidebook to the secretory pathway. Oxford, England: Sambrook and Tooze Publications at Oxford University Press; 1994. [Google Scholar]
- 39.Schlegel A, Giddings J, Ladinsky T M, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during polivirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlenstedt G, Hurt E, Doyle V, Silver P A. Reconstitution of nuclear protein transport in semi-intact yeast cells. J Cell Biol. 1993;123:785–798. doi: 10.1083/jcb.123.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schröeder S, Schimmöller F, Singer-Krüger B, Riezman H. The Golgi localization of yeast EMP47p depends on its dilysine motif but is not altered by the ret1-1 mutation in a-COP. J Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair D, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem Sci. 1998;23:131–134. doi: 10.1016/s0968-0004(98)01188-8. [DOI] [PubMed] [Google Scholar]
- 43.Smirnyagina E, Lin N S, Ahlquist P. The polymerase-like core of brome mosaic virus 2a protein, lacking a region interacting with viral 1a protein in vitro, maintains activity and 1a selectivity in RNA replication. J Virol. 1996;70:4729–4736. doi: 10.1128/jvi.70.7.4729-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan M, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teterina N L, Gorbalenya A E, Egger D, Bienz K, Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–8972. doi: 10.1128/jvi.71.12.8962-8972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Meer Y, van Tol H, K. L J, Snijder E J. ORF-1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wooding S, Pelham H. The dynamics of golgi protein traffic visualized in living yeast cells. Mol Biol Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S X, Ahlquist P, Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc Natl Acad Sci USA. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]