Abstract
1. The ratio of ebgA-gene product of ebgC-gene product in the functional aggregate of ebg beta-galactosidases was determined to be 1:1 by isolation of the enzyme from bacteria grown on uniformly radiolabelled amino acids and separation of the subunits by gel-permeation chromatography under denaturing conditions. 2. This datum, taken together with a recalculation of the previous ultracentrifuge data [Hall (1976) J. Mol. Biol. 107, 71-84], analytical gel-permeation chromatography and electron microscopy, strongly suggests an alpha 4 beta 4 quaternary structure for the enzyme. 3. The second chemical step in the enzyme turnover sequence, hydrolysis of the galactosyl-enzyme intermediate, is markedly slower for ebgab, having both Asp-97----Asn and Trp-977----Cys changes in the large subunit, than for ebga (having only the first change) and ebgb (having only the second), and is so slow as to be rate-determining even for an S-glycoside, beta-D-galactopyranosyl thiopicrate, as is shown by nucleophilic competition with methanol. 4. The selectivity of galactosyl-ebgab between water and methanol on a molar basis is 57, similar to the value for galactosyl-ebgb. 5. The equilibrium constant for the hydrolysis of lactose at 37 degrees C is 152 +/- 19 M, that for hydrolysis of allolactose is approx. 44 M and that for hydrolysis of lactulose is approx. 40 M. 6. A comparison of the free-energy profiles for the hydrolyses of lactose catalysed by the double mutant with those for the wild-type and the single mutants reveals that free-energy changes from the two mutations are not in general independently additive, but that the changes generally are in the direction predicted by the theory of Burbaum, Raines, Albery & Knowles [(1989) Biochemistry 28, 9283-9305] for an enzyme catalysing a thermodynamically irreversible reaction. 7. Michaelis-Menten parameters for the hydrolysis of six beta-D-galactopyranosylpyridinium ions and ten aryl beta-galactosides by ebgab were measured. 8. The derived beta 1g values are the same as those for ebgb (which has only the Trp-977----Cys change) and significantly different from those for ebgo (the wild-type enzyme) and ebga. 9. The alpha- and beta-deuterium secondary isotope effects on the hydrolysis of the galactosyl-enzyme of 1.08 and 1.00 are difficult to reconcile with the pyranose ring in this intermediate being in the 4C1 conformation.
Full text
PDF



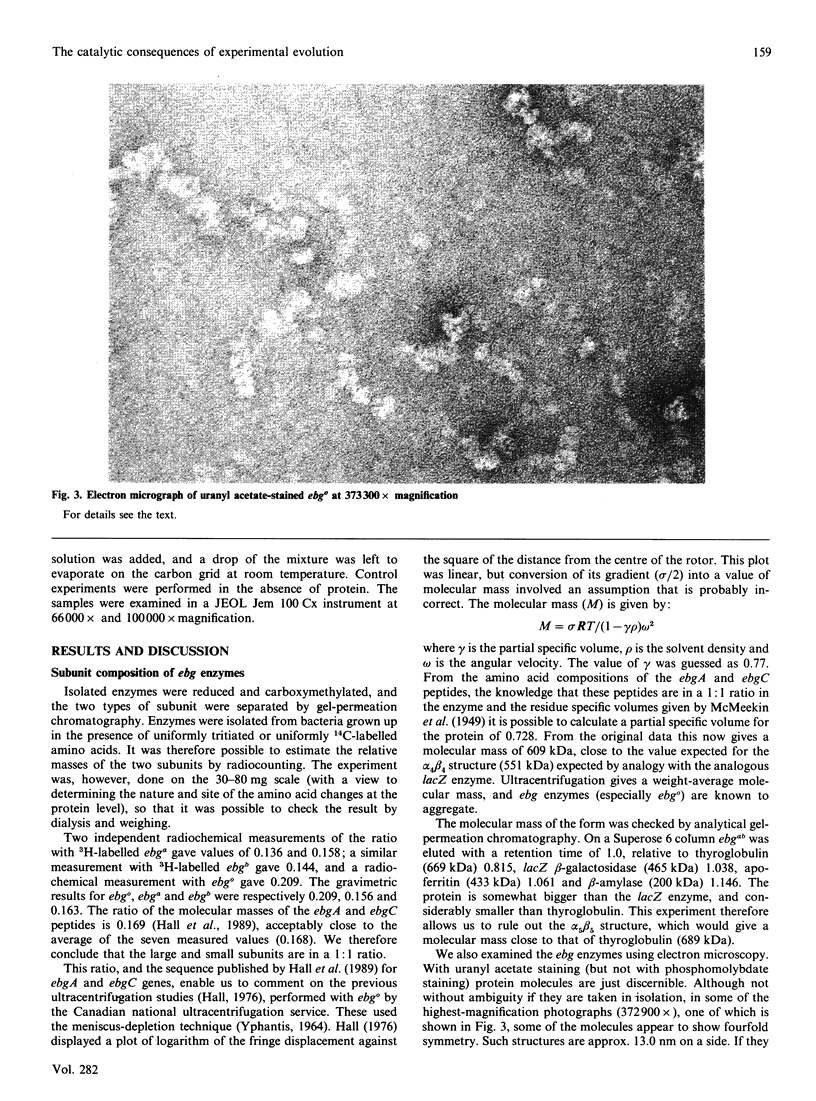

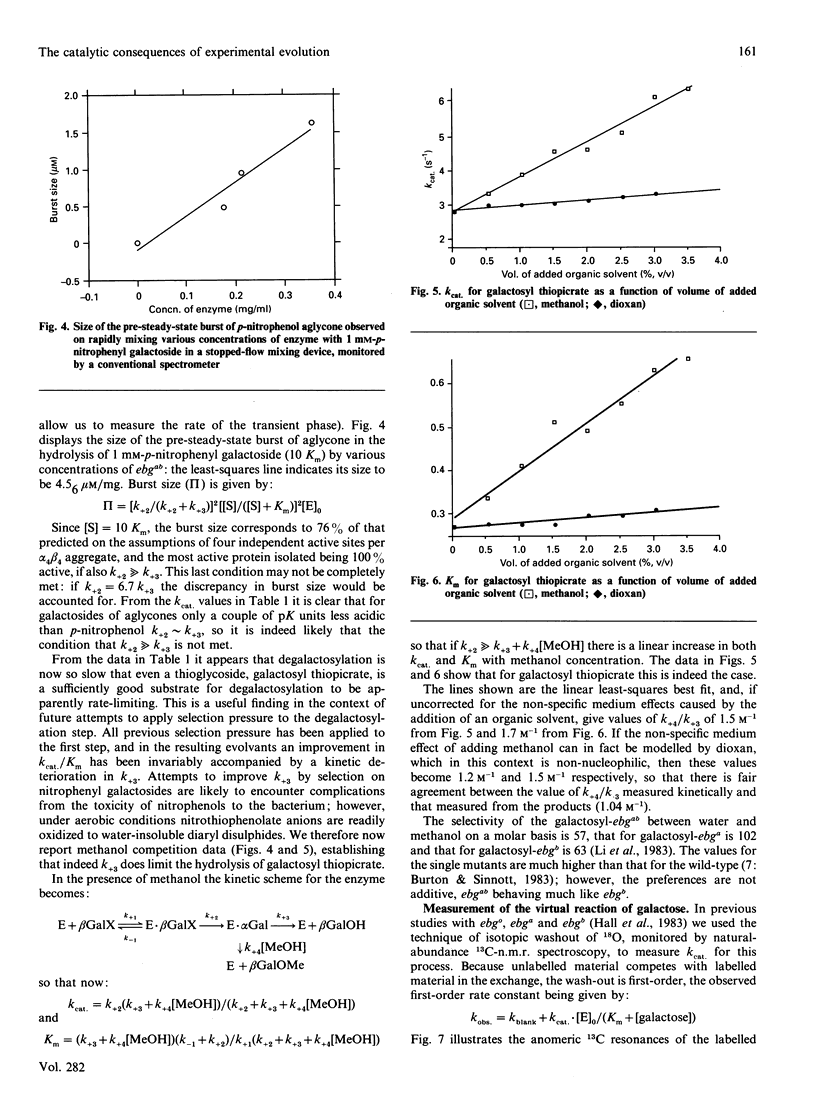
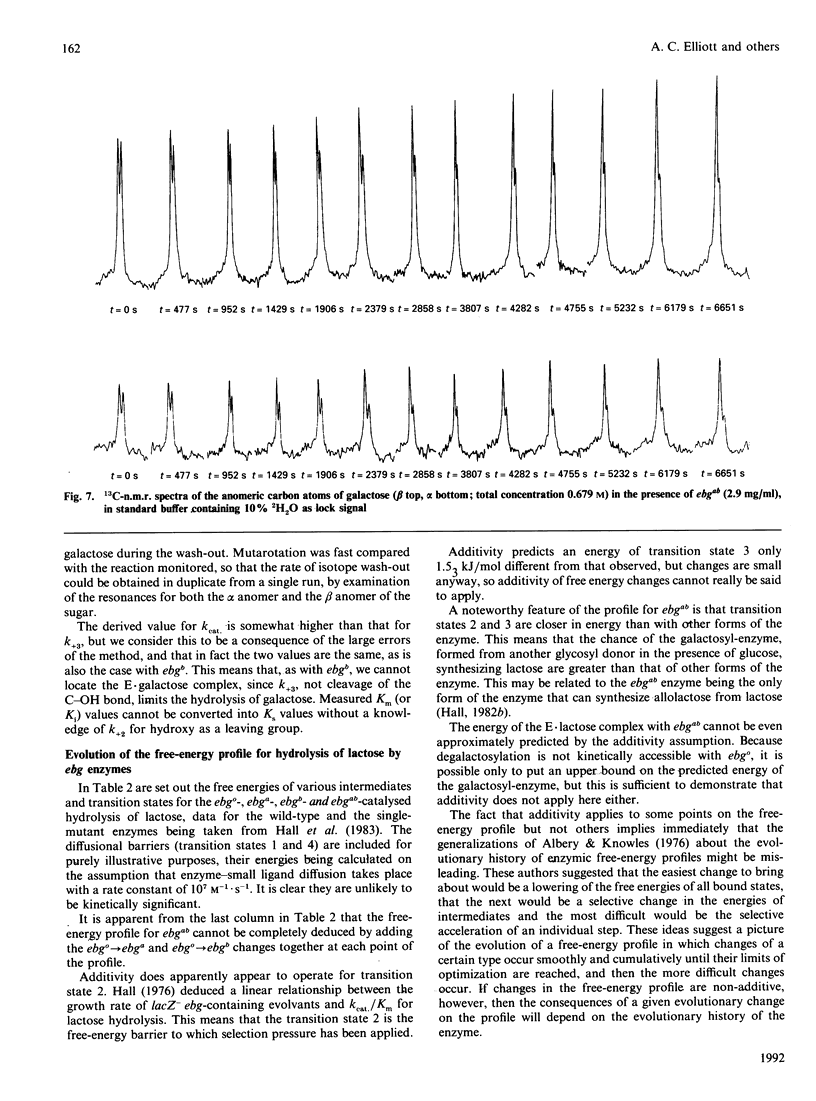

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albery W. J., Knowles J. R. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry. 1976 Dec 14;15(25):5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Raines R. T., Albery W. J., Knowles J. R. Evolutionary optimization of the catalytic effectiveness of an enzyme. Biochemistry. 1989 Nov 28;28(24):9293–9305. doi: 10.1021/bi00450a009. [DOI] [PubMed] [Google Scholar]
- Campbell J. H., Lengyel J. A., Langridge J. Evolution of a second gene for beta-galactosidase in Escherichia coli. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1841–1845. doi: 10.1073/pnas.70.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A. M. Selection and neutrality in lactose operons of Escherichia coli. Genetics. 1989 Nov;123(3):441–454. doi: 10.1093/genetics/123.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A. V., Smith P. J. The active site regions of lacZ and ebg beta-galactosidases are homologous. J Biol Chem. 1983 Sep 10;258(17):10204–10207. [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. The amino acid sequence of beta-galactosidase of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1507–1510. doi: 10.1073/pnas.74.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall B. G., Hartl D. L. Regulation of newly evolved enzymes. II. The ebg repressor. Genetics. 1975 Nov;81(3):427–435. doi: 10.1093/genetics/81.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R. N., Tewari Y. B. A calorimetric and equilibrium investigation of the hydrolysis of lactose. J Biol Chem. 1989 Jun 15;264(17):9897–9900. [PubMed] [Google Scholar]
- Hall B. G., Betts P. W., Wootton J. C. DNA sequence analysis of artificially evolved ebg enzyme and ebg repressor genes. Genetics. 1989 Dec;123(4):635–648. doi: 10.1093/genetics/123.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Changes in the substrate specificities of an enzyme during directed evolution of new functions. Biochemistry. 1981 Jul 7;20(14):4042–4049. doi: 10.1021/bi00517a015. [DOI] [PubMed] [Google Scholar]
- Hall B. G. Directed evolution of a bacterial operon. Bioessays. 1990 Nov;12(11):551–558. doi: 10.1002/bies.950121109. [DOI] [PubMed] [Google Scholar]
- Hall B. G. Experimental evolution of a new enzymatic function. Kinetic analysis of the ancestral (ebg) and evolved (ebg) enzymes. J Mol Biol. 1976 Oct 15;107(1):71–84. doi: 10.1016/s0022-2836(76)80018-6. [DOI] [PubMed] [Google Scholar]
- Hall B. G., Hartl D. L. Regulation of newly evolved enzymes. I. Selection of a novel lactase regulated by lactose in Escherichia coli. Genetics. 1974 Mar;76(3):391–400. doi: 10.1093/genetics/76.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Transgalactosylation activity of ebg beta-galactosidase synthesizes allolactose from lactose. J Bacteriol. 1982 Apr;150(1):132–140. doi: 10.1128/jb.150.1.132-140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Zuzel T. The ebg operon consists of at least two genes. J Bacteriol. 1980 Dec;144(3):1208–1211. doi: 10.1128/jb.144.3.1208-1211.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell E. E., Booth C., Farnum M., Kraut J., Warren M. S. A second-site mutation at phenylalanine-137 that increases catalytic efficiency in the mutant aspartate-27----serine Escherichia coli dihydrofolate reductase. Biochemistry. 1990 Sep 18;29(37):8561–8569. doi: 10.1021/bi00489a009. [DOI] [PubMed] [Google Scholar]
- Huber R. E., Hurlburt K. L. Reversion reactions of beta-galactosidase (Escherichia coli). Arch Biochem Biophys. 1986 Apr;246(1):411–418. doi: 10.1016/0003-9861(86)90487-x. [DOI] [PubMed] [Google Scholar]
- KARLSSON U., KOORAJIAN S., ZABIN I., SJOESTRAND F. S., MILLER A. HIGH RESOLUTION ELECTRON MICROSCOPY ON HIGHLY PURIFIED BETA-GALACTOSIDASE FROM ESCHERICHIA COLI. J Ultrastruct Res. 1964 Jun;10:457–469. doi: 10.1016/s0022-5320(64)80022-8. [DOI] [PubMed] [Google Scholar]
- Li B. F., Holdup D., Morton C. A., Sinnott M. L. The catalytic consequences of experimental evolution. Transition-state structure during catalysis by the evolved beta-galactosidases of Escherichia coli (ebg enzymes) changed by a single mutational event. Biochem J. 1989 May 15;260(1):109–114. doi: 10.1042/bj2600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers F., Messer W. The activity of individual molecules of hybrid -galactosidase reconstituted from the wild-type and an inactive-mutant enzyme. Eur J Biochem. 1973 Apr;34(2):228–231. doi: 10.1111/j.1432-1033.1973.tb02750.x. [DOI] [PubMed] [Google Scholar]
- Pettersson G. Effect of evolution on the kinetic properties of enzymes. Eur J Biochem. 1989 Oct 1;184(3):561–566. doi: 10.1111/j.1432-1033.1989.tb15050.x. [DOI] [PubMed] [Google Scholar]
- SUND H., WEBER K. Studies on the lactose-splitting enzyme. XIII. Quantity and configuration of beta-galactosidase from E. Coli. Biochem Z. 1963;337:24–34. [PubMed] [Google Scholar]
- Sinnott M. L., Withers S. G. The necessity of magnesium cation for acid assistance aglycone departure in catalysis by Escherichia coli (lacZ) beta-galactosidase. Biochem J. 1978 Nov 1;175(2):539–546. doi: 10.1042/bj1750539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes H. W., Betts P. W., Hall B. G. Sequence of the ebgA gene of Escherichia coli: comparison with the lacZ gene. Mol Biol Evol. 1985 Nov;2(6):469–477. doi: 10.1093/oxfordjournals.molbev.a040372. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall B. G. Sequence of the ebgR gene of Escherichia coli: evidence that the EBG and LAC operons are descended from a common ancestor. Mol Biol Evol. 1985 Nov;2(6):478–483. doi: 10.1093/oxfordjournals.molbev.a040373. [DOI] [PubMed] [Google Scholar]
- Ullmann A., Jacob F., Monod J. On the subunit structure of wild-type versus complemented beta-galactosidase of Escherichia coli. J Mol Biol. 1968 Feb 28;32(1):1–13. doi: 10.1016/0022-2836(68)90140-x. [DOI] [PubMed] [Google Scholar]
- Wells J. A. Additivity of mutational effects in proteins. Biochemistry. 1990 Sep 18;29(37):8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- YPHANTIS D. A. EQUILIBRIUM ULTRACENTRIFUGATION OF DILUTE SOLUTIONS. Biochemistry. 1964 Mar;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]



