Abstract
The role of the peroxisome proliferator-activated receptor α (PPARα) in regulating hepatitis B virus (HBV) transcription and replication in vivo was investigated in an HBV transgenic mouse model. Treatment of HBV transgenic mice with the peroxisome proliferators Wy-14,643 and clofibric acid resulted in a less than twofold increase in HBV transcription rates and steady-state levels of HBV RNAs in the livers of these mice. In male mice, this increase in transcription was associated with a 2- to 3-fold increase in replication intermediates, whereas in female mice it was associated with a 7- to 14-fold increase in replication intermediates. The observed increases in transcription and replication were dependent on PPARα. HBV transgenic mice lacking this nuclear hormone receptor showed similar levels of HBV transcripts and replication intermediates as untreated HBV transgenic mice expressing PPARα but failed to demonstrate alterations in either RNA or DNA synthesis in response to peroxisome proliferators. Therefore, it appears that very modest alterations in transcription can, under certain circumstances, result in relatively large increases in HBV replication in HBV transgenic mice.
As the host range of hepatitis B virus (HBV) is limited to humans and primates (26), the majority of studies analyzing the mechanisms of HBV transcriptional regulation and viral biosynthesis have been performed in cell culture systems. With cell culture systems, it has been demonstrated that the 3.2-kb viral genome can encode four viral transcripts (3, 33, 35, 38, 41). The levels of the 3.5-, 2.4-, 2.1-, and 0.7-kb transcripts are regulated by the nucleocapsid, large surface antigen, major surface antigen, and X-gene promoters, respectively (29, 43). The 3.5-kb pregenomic transcript is translated into the HBV polymerase and nucleocapsid or core antigen (HBcAg) (24). The HBV polymerase binds to the ɛ stem-loop structure at the 5′ end of the pregenomic RNA, and the RNA plus polymerase complex is encapsidated by dimers of the core polypeptide, generating the immature capsid (17). The HBV polymerase reverse transcribes the pregenomic RNA within the capsid to produce minus-strand HBV DNA and then synthesizes a partial plus-strand HBV DNA, using the minus strand as the template (39). Mature intracellular capsids containing the 3.2-kb partially double-stranded HBV DNA bind to the envelope antigen (HBsAg) in the membrane of the endoplasmic reticulum and subsequently bud into the lumen of the endoplasmic reticulum (10, 18, 42). Mature virus is secreted from the hepatocyte after passing through the endoplasmic reticulum and Golgi apparatus, where the envelope polypeptides are glycosylated (1, 18, 34).
As the 3.5-kb pregenomic HBV RNA is the substrate for the synthesis of virion DNA, it is apparent that regulating the level of transcription of this RNA is likely to have a major influence on viral biogenesis. Consequently, it has been of interest to determine the mechanisms regulating the level of transcription from the nucleocapsid promoter. A variety of studies has identified the cis-acting sequences and trans-acting factors regulating the nucleocapsid promoter activity (4, 5, 14, 22, 40, 44, 47, 48). Several liver-enriched transcription factors, including C/EBP, hepatocyte nuclear factors 3 and 4, retinoid X receptor α (RXRα), and peroxisome proliferator-activated receptor α (PPARα), and ubiquitous transcription factors including Sp1 and RFX1 have been shown to modulate nucleocapsid promoter activity in cell culture (2, 13, 16, 23, 25, 45, 46, 49). The observation that members of the nuclear hormone receptor family of transcription factors can modulate the activity of the nucleocapsid promoter suggested that transcription of the 3.5-kb pregenomic RNA and therefore replication might be influenced by the availability of the ligands for these nuclear hormone receptors. Evidence supporting this contention has been obtained in cell culture where the nuclear hormone receptor ligands 9-cis retinoic acid and clofibric acid, respectively, have been shown to increase the level of transcription from the nucleocapsid promoter in an RXRα- and PPARα-dependent manner. RXRα and PPARα mediate their effects through the peroxisome proliferator response elements (PPREs) located at nucleotide positions −28 to −16 in the nucleocapsid promoter and within the enhancer 1 region of the HBV genome (25).
In this study, these observations have been extended to an in vivo model system of HBV viral replication. By using an HBV transgenic mouse model system of viral replication (12) and the PPARα-null mouse (21), it has been possible to examine the effects of two ligands for PPARα on viral transcription and replication in the presence or absence of the nuclear hormone receptor PPARα. This analysis demonstrated that these ligands activated transcription from responsive cellular promoters in a PPARα-dependent manner, whereas they produced concomitantly limited alterations in the levels of HBV RNAs. Despite these limited effects on HBV transcription, female HBV transgenic mice demonstrated PPARα-dependent 7- to 14-fold increases in viral replication, indicating that modest changes in the level of HBV transcription can be associated with dramatic effects on viral replication intermediates under certain circumstances.
MATERIALS AND METHODS
Transgenic and knockout mice.
The production and characterization of the HBV transgenic mouse lineage 1.3.32 have been described elsewhere (12). These HBV transgenic mice contain a single copy of the terminally redundant, 1.3-genome-length copy of the HBVayw genome integrated into the mouse chromosomal DNA. High levels of HBV replication occur in the livers of these mice. The mice used in the breeding experiments were homozygous for the HBV transgene and were maintained on the C57BL/6 genetic background.
The production and characterization of the PPARα-null mice have been described elsewhere (21). These mice do not express PPARα and are refractory to the pleiotropic effects of peroxisome proliferators. These mice do not display any gross phenotypic defects and are viable and fertile. The mice used in the breeding experiments were maintained on the Sv/129 genetic background.
PPARα-null HBV transgenic mice were generated by mating the HBV transgenic mice with the PPARα-null mice. The resulting F1 mice were subsequently mated with the PPARα-null mice, and the F2 mice were screened for the HBV transgene and PPARα null allele by PCR analysis of tail DNA. Tail DNA was prepared by incubating 1 cm of tail in 500 μl of 100 mM Tris hydrochloride (pH 8.0)–200 mM NaCl–5 mM EDTA–0.2% (wt/vol) sodium dodecyl sulfate containing 100 μg of proteinase K per ml for 16 to 20 h at 55°C. Samples were centrifuged at 14,000 rpm in a microcentrifuge for 5 min, and the supernatant was precipitated with 500 μl of isopropanol. DNA was pelleted by centrifugation at 14,000 rpm in a microcentrifuge for 5 min and subsequently dissolved in 100 μl of 5 mM Tris hydrochloride (pH 8.0)–1 mM EDTA. The HBV transgene was identified by PCR analysis using oligonucleotides XpHNF4-1 (TCGATACCTGAACCTTTACCCCGTTGCCCG; HBV coordinates 1133 to 1159) and CpHNF4-2 (TCGAATTGCTGAGAGTCCAAGAGTCCTCTT; HBV coordinates 1683 to 1658) plus 1 μl of tail DNA. The samples were subjected to 32 amplification cycles involving denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension from the primers at 72°C for 2 min. A PCR product of 551 bp indicated the presence of the HBV transgene. The PPARα null allele was identified by PCR analysis using the oligonucleotides mPPAR1 (CCTGGCCTTCTAAACATAGG; 5′ sequence of exon 8) and mPPAR2 (TCCCTGCTCTCCTGTATGGG; 3′ sequence of exon 8) plus 1 μl of tail DNA. The samples were subjected to 35 amplification cycles involving denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension from the primers at 72°C for 2 min. A PCR product of 319 bp indicated the wild-type PPARα genotype, whereas a PCR product of 1.4 kbp indicated the mutated PPARα genotype. The 20-μl reaction conditions used were as described by the manufacturer (Boehringer Mannheim) and contained 2.5 U of Taq DNA polymerase.
HBV DNA and RNA analysis.
Total DNA and RNA were isolated from livers of HBV transgenic mice as described elsewhere (6, 28). DNA (Southern) and RNA (Northern) filter hybridization analyses were performed with 20 μg of HindIII-digested DNA and 10 μg of total cellular RNA, respectively, as described previously (28). Filters were probed with 32P-labeled HBVayw genomic DNA (9) to detect HBV sequences, the human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA (36) to detect the GAPDH mRNA, and the rat cytochrome P450 4A1 cDNA (21) to detect the CYP4A mRNA.
RNase protection assays were performed with a PharMingen Riboquant kit, and riboprobes were synthesized by using an Ambion Maxiscript kit as described by the manufacturers. Transcription initiation sites for the 3.5-kb HBV transcripts were examined by using 20 μg of total cellular RNA and a 333 (HBV coordinates 1990 to 1658)-nucleotide-long 32P-labeled HBV riboprobe.
Nuclei were prepared from mouse livers by a modification of a previously described procedure (30). Briefly, liver samples were homogenized in a Potter-Elvehjem tissue grinder in 5 ml of 10 mM HEPES (pH 7.9)–25 mM KCl–1.0 mM EGTA–1.0 mM EDTA–0.32 M sucrose–0.15 mM spermine–0.5 mM spermidine–1.0 mM dithiothreitol (DTT)–0.5 mM phenylmethylsulfonyl fluoride (PMSF) containing leupeptin (0.5 μg/ml), aprotinin (1.0 μg/ml), and pepstatin (1.0 μg/ml) (buffer A). Samples were diluted with 10 ml of 10 mM HEPES (pH 7.9)–25 mM KCl–1.0 mM EGTA–1.0 mM EDTA–2.0 M sucrose–0.15 mM spermine–0.5 mM spermidine–1.0 mM DTT–0.5 mM PMSF containing leupeptin (0.5 μg/ml), aprotinin (1.0 μg/ml), and pepstatin (1.0 μg/ml) (buffer B) and centrifuged over two 3.5-ml sucrose cushions of buffer B for 30 min at 24,000 rpm in a Beckman SW41 rotor at 4°C. The two nuclear pellets from each sample were resuspended together in 2.5 ml of buffer A. After the addition of 5 ml of buffer B, the samples were centrifuged over a 3.5-ml sucrose cushion of buffer B for 30 min at 24,000 rpm in a Beckman SW41 rotor at 4°C. The nuclei were resuspended in 1 ml of 20 mM Tris hydrochloride (pH 7.9)–75 mM NaCl–0.5 mM EDTA–50% glycerol–0.15 mM spermine–0.5 mM spermidine–0.85 mM DTT–0.125 mM PMSF containing leupeptin (0.5 μg/ml), aprotinin (1.0 μg/ml), and pepstatin (1.0 μg/ml). Purified nuclei were stored in liquid nitrogen at approximately 107 nuclei per vial until used for nuclear run-on analysis.
Nuclear run-on analysis was performed as previously described (37). The labeled transcripts were hybridized to Hybond N (Amersham) filter strips containing 1 to 2 μg of plasmid DNAs. The plasmids used contained the human GADPH cDNA insert (36), the rat acyl coenzyme A oxidase (ACO) cDNA (21), the rat cytochrome P450 4A1 cDNA (21) and the complete HBVayw genome (9). pBluescript SK(−) plasmid DNA (Stratagene) was used as a negative control.
Results of filter hybridization, RNase protection assays, and nuclear run-on analyses were quantitated by phosphorimaging using a Packard Cyclone storage phosphor system.
HBV antigen analysis.
HBeAg analysis was performed with 20 μl of mouse serum and the HBe enzyme immunoassay as described by the manufacturer (Abbott Laboratories). The level of antigen was determined in the linear range of the assay. Immunohistochemical detection of HBcAg in paraffin-embedded mouse liver sections was performed as previously described (12).
RESULTS
Previous in vitro and cell culture studies have demonstrated that the nuclear hormone receptor PPARα can bind as a heterodimer with RXRα to PPREs in the enhancer 1 region and the nucleocapsid promoter of the HBV genome. As a result of the binding of these factors to their recognition sequences, the level of transcription from the nucleocapsid promoter can be modulated (15, 25). Alteration in the level of transcription from the nucleocapsid promoter would be expected to change the abundance of the pregenomic HBV RNA and consequently the level of viral replication. In an attempt to determine the role of PPARα in modulating viral replication in vivo, we examined the effects of exogenous ligands for PPARα on HBV transcription and replication in the liver of wild-type and PPARα-null HBV transgenic mice.
Effect of PPARα on HBeAg synthesis in HBV transgenic mice.
The nucleocapsid promoter directs the expression of the precore and pregenomic RNAs that are translated to produce HBeAg and the nucleocapsid polypeptide, respectively (26). Consequently, HBeAg expression is an indirect measure of nucleocapsid promoter activity. Therefore, analysis of HBeAg in the sera of HBV transgenic mice was initially used to examine the role of the nuclear hormone receptor PPARα in regulation of HBV transcription from the nucleocapsid promoter.
HBV transgenic mice were bred with PPARα-null mice, and HBV transgenic mice heterozygous (+/−) or homozygous (−/−) for the PPARα null allele were identified in the F2 generation. For these studies, HBV transgenic mice that were heterozygous for the PPARα null allele were used as controls and compared with HBV transgenic mice that were homozygous for the PPARα null allele. Male and female mice of each genotype were assayed for the level of HBeAg in their sera (Table 1). The levels of HBeAg in the sera of male PPARα +/− and −/− mice were very similar. Likewise, the levels of HBeAg in the sera of female PPARα +/− and −/− mice were very similar but lower than those seen in males of both genotypes. These observations indicate that in untreated HBV transgenic mice, the presence or absence of PPARα does not influence the level of HBeAg in the serum. However, the average level of HBeAg in the sera of male HBV transgenic mice is higher, by approximately 50%, than the level of HBeAg in the sera of female HBV transgenic mice, demonstrating that sexually dimorphic expression of this antigen occurs in this model system.
TABLE 1.
Effect of PPARα on HBeAg synthesis in HBV transgenic mice
| Gender | PPARα genotype | No. of mice | Mean (SD) HBeAg A492a |
|---|---|---|---|
| Male | +/− | 9 | 0.56 (0.22) |
| −/− | 10 | 0.62 (0.18) | |
| Female | +/− | 8 | 0.37 (0.19) |
| −/− | 15 | 0.44 (0.19) |
Twenty-microliter volumes of serum were assayed for HBeAg by using the Abbott Laboratories HBe enzyme immunoassay.
To examine the possible role of PPARα in HBV transcription in vivo, HBV transgenic mice were treated with two synthetic PPARα ligands, Wy-14,643 and clofibric acid (8). Following treatment for 7 and 14 days, respectively, age-, gender-, and HBeAg-matched HBV transgenic mice were examined for liver size and serum HBeAg levels (Tables 2 and 3). As previously described, these peroxisome proliferators induce a PPARα-dependent hepatomegaly (21) due to hypertrophy of hepatocytes and hepatocellular hyperplasia (27). Hepatocellular hypertrophy is the primary cause of hepatomegaly and results from a massive increase in peroxisomes within the hepatocytes (27). Treatment of HBV transgenic mice that were heterozygous for a functional PPARα gene with Wy-14,643 for 7 days resulted in a 1.9- to 2.5-fold increase in the size of the liver (Table 2), whereas a 14-day treatment with clofibric acid resulted in a 1.5- to 1.6-fold increase in liver size (Table 3). Mice lacking a functional PPARα gene failed to show this large increase in liver size, as expected from earlier results (21). Treatment of HBV transgenic mice with Wy-14,643 or clofibric acid resulted in a small PPARα-dependent increase in serum HBeAg (Tables 2 and 3). The increase in serum HBeAg with peroxisome proliferators occurred in every PPARα +/− HBV transgenic mouse examined and was somewhat greater in female HBV transgenic mice than in male HBV transgenic mice. HBV transgenic mice lacking PPARα failed to show any increase in serum HBeAg in response to either peroxisome proliferator. These observations are consistent with the possibility that activation of PPARα in HBV transgenic mice results in increased transcription from the nucleocapsid promoter and therefore increased levels of the precore RNA.
TABLE 2.
Effect of Wy-14,643 on HBeAg synthesis in HBV transgenic mice
| Gender | PPARα genotype | Treatmenta | Mean (SD) liver size (% of total body wt) | Mean (SD) HBeAg A492b
|
Mean HBeAg fold induction (day 7 A492/ day 0 A492) | |
|---|---|---|---|---|---|---|
| Day 0 | Day 7 | |||||
| Male | +/− | Control | 5.73 (0.38) | 0.78 (0.07) | 0.71 (0.06) | 0.91 |
| +/− | Wy | 10.90 (0.59) | 0.70 (0.08) | 0.89 (0.07) | 1.27c | |
| −/− | Control | 5.68 (0.45) | 0.76 (0.08) | 0.69 (0.00) | 0.91 | |
| −/− | Wy | 6.17 (0.39) | 0.77 (0.09) | 0.66 (0.06) | 0.86 | |
| Female | +/− | Control | 4.08 (0.44) | 0.48 (0.20) | 0.37 (0.16) | 0.77 |
| +/− | Wy | 10.00 (0.13) | 0.65 (0.05) | 1.05 (0.19) | 1.62c | |
| −/− | Control | 4.40 (0.51) | 0.54 (0.02) | 0.50 (0.14) | 0.93 | |
| −/− | Wy | 5.02 (0.36) | 0.55 (0.16) | 0.43 (0.10) | 0.78 | |
HBV transgenic mice (three mice per group) were fed normal rodent chow (control) or rodent chow containing 0.1% (wt/wt) Wy-14,643 (Wy).
Twenty-microliter volumes of serum were assayed for HBeAg by using the Abbott Laboratories HBe enzyme immunoassay immediately before treatment (day 0) of the mice and on day 7 of treatment.
Statistically significant by paired Student’s t test (P < 0.05).
TABLE 3.
Effect of clofibric acid on HBeAg synthesis in HBV transgenic mice
| Gender | PPARα genotype | Treatmenta | Mean (SD) liver size (% of total body wt) | Mean (SD) HBeAg A492b
|
Mean HBeAg fold induction (day 14 A492/ day 0 A492) | |
|---|---|---|---|---|---|---|
| Day 0 | Day 14 | |||||
| Male | +/− | Control | 4.55 (0.63) | 0.61 (0.04) | 0.54 (0.06) | 0.89 |
| +/− | CA | 6.90 (0.36) | 0.58 (0.04) | 0.69 (0.08) | 1.19c | |
| −/− | Control | 4.99 (0.58) | 0.65 (0.03) | 0.58 (0.04) | 0.89 | |
| −/− | CA | 5.47 (0.07) | 0.66 (0.05) | 0.54 (0.08) | 0.82 | |
| Female | +/− | Control | 4.22 (0.39) | 0.52 (0.05) | 0.41 (0.03) | 0.79 |
| +/− | CA | 6.67 (0.27) | 0.57 (0.14) | 0.70 (0.09) | 1.23 | |
| −/− | Control | 4.16 (0.16) | 0.48 (0.04) | 0.38 (0.07) | 0.79 | |
| −/− | CA | 4.78 (0.36) | 0.49 (0.06) | 0.40 (0.12) | 0.82 | |
HBV transgenic mice (three mice per group) were fed normal rodent chow (control) or rodent chow containing 0.5% (wt/wt) clofibric acid (CA).
Twenty-microliter volumes of serum were assayed for HBeAg by using the Abbott Laboratories HBe enzyme immunoassay immediately before (day 0) treatment of the mice and on day 14 of treatment.
Statistically significant by paired Student’s t test (P < 0.05).
Effect of PPARα on viral transcription and replication in HBV transgenic mice.
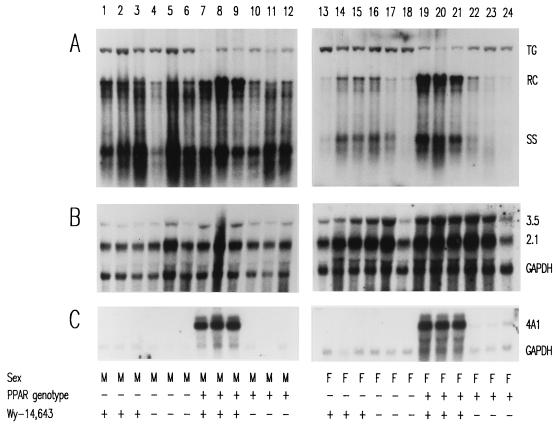
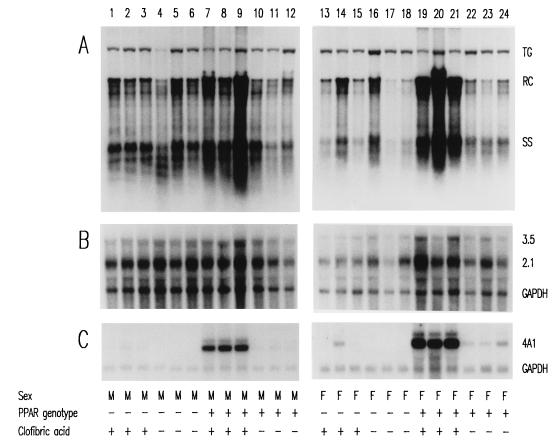
The HBV transgenic mice described above were also examined for the effects of the peroxisome proliferators on the steady-state levels of HBV transcripts and replication intermediates by analysis of the total liver RNA and DNA (Fig. 1 and 2). Induction of the PPARα-responsive cytochrome P450 4A1 (CYP4A) mRNA by peroxisome proliferators confirmed that these compounds mediated PPARα-dependent transcriptional activation of responsive genes in the HBV transgenic mice (Fig. 1C and 2C) (21). HBV transgenic mice lacking PPARα failed to demonstrate increased levels of the CYP4A mRNA in response to peroxisome proliferator treatment, indicating that PPARα is essential for this induction (Fig. 1C and 2C, lanes 1 to 3 and 13 to 15).
FIG. 1.
DNA (Southern) and RNA (Northern) filter hybridization analysis of HBV DNA replication intermediates (A) and transcripts (B) in livers of HBV transgenic mice. Groups of three mice of each gender and genotype were fed rodent chow either with or without Wy-14,643 for 7 days before their livers were analyzed. Induction of the transcript detected by the rat cytochrome P450 4A1 cDNA (4A1) was used as a control to demonstrate the PPARα-dependent activation of transcription by the peroxisome proliferator from a responsive gene. The GAPDH transcript was used as an internal control for quantitation of the HBV 3.5- and 2.1-kb RNAs (3.5 and 2.1). The HBV transgene was used as an internal control for quantitation of the HBV replication intermediates. The probes used were HBVayw genomic DNA (A), HBVayw genomic DNA plus GAPDH cDNA (B) and cytochrome P450 4A1 cDNA plus GAPDH cDNA (C). TG, HBV transgene; RC, HBV relaxed circular replication intermediates; SS, HBV single-stranded replication intermediates; M, male HBV transgenic mouse; F, female HBV transgenic mouse; PPAR genotype −, PPARα knockout (−/−) mouse; PPAR genotype +, PPARα heterozygous (+/−) mouse; Wy-14,643 +, HBV transgenic mouse fed 0.1% (wt/wt) Wy-14,643; Wy-14,643 −, HBV transgenic mouse fed normal rodent chow.
FIG. 2.
DNA (Southern) and RNA (Northern) filter hybridization analysis of HBV DNA replication intermediates (A) and transcripts (B) in livers of HBV transgenic mice. Groups of three mice of each gender and genotype were fed rodent chow either with or without clofibric acid for 14 days before their livers were analyzed. Induction of the transcript detected by the rat cytochrome P450 4A1 cDNA (4A1) was used as a control to demonstrate the PPARα-dependent activation of transcription by the peroxisome proliferator from a responsive gene. The GAPDH transcript was used as an internal control for the quantitation of the HBV 3.5- and 2.1-kb RNAs (3.5 and 2.1). The HBV transgene was used as an internal control for quantitation of the HBV replication intermediates. The probes used were HBVayw genomic DNA (A), HBVayw genomic DNA plus GAPDH cDNA (B), and cytochrome P450 4A1 cDNA plus GAPDH cDNA (C). TG, HBV transgene; RC, HBV relaxed circular replication intermediates; SS, HBV single-stranded replication intermediates; M, male HBV transgenic mouse; F, female HBV transgenic mouse; PPAR genotype −, PPARα knockout (−/−) mouse; PPAR genotype +, PPARα heterozygous (+/−) mouse; clofibric acid +, HBV transgenic mouse fed 0.5% (wt/wt) clofibric acid; clofibric acid −, HBV transgenic mouse fed normal rodent chow.
Analysis of the levels of the HBV 3.5- and 2.1-kb transcripts in the livers of HBV transgenic mice with and without PPARα in the presence or absence of peroxisome proliferators was performed. The steady-state levels of the HBV transcripts vary modestly under these different conditions (Fig. 1B and 2B; Tables 4 and 5). Most notably, the only consistent change in the levels of transcripts was a less than twofold induction of the HBV transcripts in the female PPARα +/− HBV transgenic mice after treatment with peroxisome proliferators (Table 4 and 5). This was somewhat surprising considering the previous cell culture analysis demonstrating that transcription from the nucleocapsid promoter could be increased approximately fourfold by activation of RXRα and PPARα by 9-cis retinoic acid and clofibric acid (25).
TABLE 4.
Effect of Wy-14,643a on HBV RNA and DNA synthesis in HBV transgenic mice
| Gender | PPARα genotype | Mean (SD) 2.1/ GAPDH ratiob
|
Fold induction of HBV 2.1-kb RNA (+Wy/−Wy) | Mean (SD) 3.5/ GAPDH ratioc
|
Fold induction of HBV 3.5-kb RNA (+Wy/−Wy) | Mean (SD) RI/TG ratiod
|
Fold induction of HBV RI (+Wy/−Wy) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| −Wy | +Wy | −Wy | +Wy | −Wy | +Wy | |||||
| Male | +/− | 2.80 (0.50) | 1.54 (0.60) | 0.55 | 0.33 (0.13) | 0.29 (0.20) | 0.88 | 42.58 (21.37) | 99.64 (27.09) | 2.34e |
| −/− | 1.97 (0.07) | 1.60 (0.33) | 0.81 | 0.19 (0.05) | 0.16 (0.00) | 0.84 | 21.35 (9.86) | 24.09 (12.21) | 1.13 | |
| Female | +/− | 1.60 (0.96) | 2.74 (0.27) | 1.71 | 0.50 (0.24) | 0.97 (0.12) | 1.94e | 6.33 (4.78) | 90.97 (62.87) | 14.37e |
| −/− | 2.27 (1.04) | 2.59 (1.12) | 1.14 | 0.55 (0.24) | 0.38 (0.17) | 0.69 | 6.76 (3.28) | 8.14 (5.10) | 1.20 | |
HBV transgenic mice (three mice per group) were fed normal rodent chow (−Wy) or rodent chow containing 0.1% (wt/wt) Wy-14,643 (+Wy) for 7 days before the relative levels of the HBV 3.5- and 2.1-kb RNAs, GAPDH RNA, HBV transgene, and replication intermediate DNAs in the liver (Fig. 1) were measured.
Ratio of level of HBV 2.1-kb RNA to GAPDH RNA, determined from Fig. 1B.
Ratio of level of HBV 3.5-kb RNA to GAPDH RNA, determined from Fig. 1B.
Ratio of level of the HBV replication intermediate DNA (RI) to the HBV transgene DNA (TG), determined from Fig. 1A.
Statistically significant by Student’s t test (P < 0.05).
TABLE 5.
Effect of clofibric acida on HBV RNA and DNA synthesis in HBV transgenic mice
| Gender | PPARα genotype | Mean (SD) 2.1/ GAPDH ratio
|
Fold induction of HBV 2.1-kb RNA (+CA/−CA) | Mean (SD) 3.5/ GAPDH ratio
|
Fold induction of HBV 3.5-kb RNA (+CA/−CA) | Mean (SD) RI/TG ratio
|
Fold induction of HBV RI (+CA/−CA) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| −CA | +CA | −CA | +CA | −CA | +CA | |||||
| Male | +/− | 2.24 (0.46) | 2.29 (0.60) | 1.02 | 0.48 (0.07) | 0.54 (0.22) | 1.12 | 21.30 (12.68) | 56.67 (14.39) | 2.66b |
| −/− | 2.29 (0.38) | 2.41 (0.36) | 1.05 | 0.37 (0.06) | 0.37 (0.05) | 1.00 | 30.80 (17.46) | 22.30 (5.16) | 0.72 | |
| Female | +/− | 2.59 (0.75) | 4.61 (2.94) | 1.78 | 0.30 (0.11) | 0.50 (0.31) | 1.67 | 9.05 (1.08) | 69.35 (16.50) | 7.66b |
| −/− | 2.92 (1.16) | 1.65 (0.40) | 0.57 | 0.37 (0.14) | 0.19 (0.03) | 0.51 | 6.02 (3.30) | 12.14 (8.25) | 2.02 | |
HBV transgenic mice (three mice per group) were fed normal rodent chow (−CA) or rodent chow containing 0.5% (wt/wt) clofibric acid (+CA) for 14 days before the relative levels of the HBV 3.5- and 2.1-kb RNAs, GAPDH RNA, HBV transgene, and replication intermediate DNAs in the liver (Fig. 2) were measured. Other details are as for Table 4, but determined from Fig. 2.
Statistically significant by Student’s t test (P < 0.05).
In addition to examining the HBV transcripts, the levels of the viral replication intermediates in the livers of these HBV transgenic mice were also determined (Fig. 1A and 2A; Tables 4 and 5). In the absence of peroxisome proliferators, PPARα does not influence the level of replication intermediates in the livers of the HBV transgenic mice (Tables 4 and 5). In addition, it is apparent that there is an approximately fourfold-higher level of replication intermediates in male HBV transgenic mice than in female HBV transgenic mice. This difference in replication intermediates is qualitatively similar to the levels of HBeAg in the serum of male and female HBV transgenic mice (Table 1). Treatment of PPARα-null HBV transgenic mice with peroxisome proliferators did not alter the level of replication intermediates observed in livers (Tables 4 and 5). In male HBV transgenic mice expressing PPARα, a modest, two- to threefold increase in HBV replication intermediates was observed in response to treatment with peroxisome proliferators. However, in female PPARα +/− HBV transgenic mice, replication intermediates increased approximately 14-fold with Wy-14,643 treatment and 7-fold with clofibric acid treatment. This effect was not observed in female PPARα-null HBV transgenic mice treated with these PPARα activators. Therefore, despite the modest increase in viral transcription observed in female PPARα +/− HBV transgenic mice in response to peroxisome proliferators, a dramatic PPARα-dependent increase in viral replication intermediates is observed in the livers of these animals. The reason for the sexual dimorphism in the ability to respond to peroxisome proliferators is unclear, but this observation suggests that under certain circumstances minor alterations in HBV transcription can be associated with dramatic alterations in viral replication. Additionally, it should be noted that although female PPARα +/− HBV transgenic mice initially have a lower level of replication intermediates in their livers, treatment with peroxisome proliferators results in similar levels of replication intermediates in male and female PPARα +/− HBV transgenic mice (Fig. 1A and 2A; Tables 4 and 5).
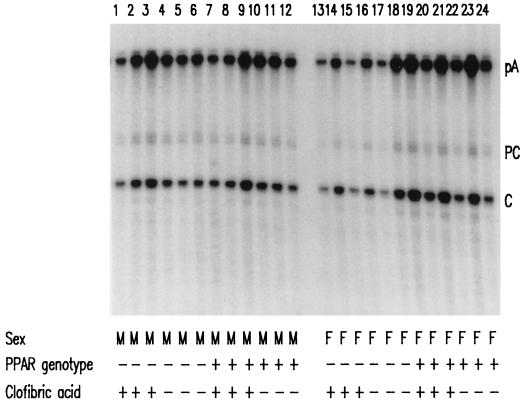
The relative abundance of the precore and pregenomic RNAs in HBV transgenic mice was investigated to determine if activation of PPARα affected the relative utilization of their two start sites (Fig. 3). The presence of PPARα or its activation by clofibric acid did not alter the relative abundance of the precore and pregenomic RNAs as determined by RNase protection analysis. The pregenomic RNA initiation site is utilized approximately five times more than the precore RNA initiation site in HBV transgenic mice irrespective of the presence of PPARα or clofibric acid. As observed for the steady-state levels of the 3.5-kb HBV RNA (Fig. 2B), an approximately 60% increase in the level of the pregenomic RNA is observed in female PPARα +/− HBV transgenic mice treated with clofibric acid (Fig. 3).
FIG. 3.
RNase protection analysis mapping the transcription initiation sites of precore (PC) and pregenomic (C) transcripts from livers of HBV transgenic mice. The 3′ ends of all HBV transcripts corresponding to the polyadenylation sites (pA) of these RNAs also generated a protected fragment in this analysis. Groups of three mice of each gender and genotype were fed rodent chow either with or without clofibric acid for 14 days before their livers were analyzed. The riboprobe used included the HBVayw sequence spanning nucleotide coordinates 1990 to 1658. M, male HBV transgenic mouse; F, female HBV transgenic mouse; PPAR genotype −, PPARα knockout (−/−) mouse; PPAR genotype +, PPARα heterozygous (+/−) mouse; clofibric acid +, HBV transgenic mouse fed 0.5% (wt/wt) clofibric acid; clofibric acid −, HBV transgenic mouse fed normal rodent chow.
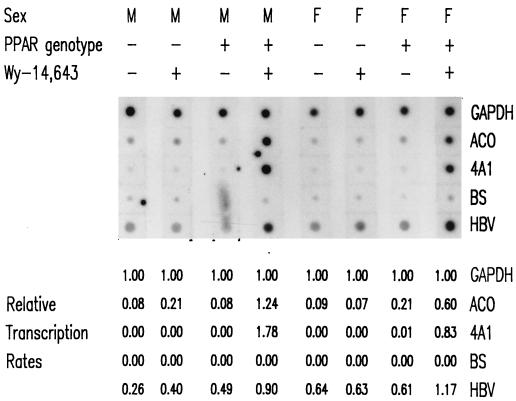
The transcription rate of the HBV RNAs was measured directly by nuclear run-on analysis (Fig. 4) and compared with the steady-state levels of these transcripts observed in the livers of the HBV transgenic mice (Fig. 1B; Table 4). The transcription rate of GAPDH was used as an internal control and designated a relative transcription rate equal to 1.00. The rates of transcription of the PPARα-inducible genes, ACO and CYP4A were low or undetectable in the absence of Wy-14,643 but were similar to the rate for GAPDH in the presence of PPARα and the peroxisome proliferator (Fig. 4). This result is consistent with the observed steady-state levels of these transcripts under these conditions (Fig. 1C) (21). The relative transcription rates of the HBV RNAs were increased approximately twofold in the presence of PPARα and Wy-14,643 (Fig. 4). In the absence of either PPARα or Wy-14,643, the rate of HBV transcription was unaffected. These observations are consistent with the effects of Wy-14,643 on the steady-state levels of the HBV transcripts (Fig. 1B; Table 4) and support the suggestion that a small change in HBV transcription can, under certain circumstances, result in a relatively large increase in viral replication.
FIG. 4.
Nuclear run-on analysis of HBV transcripts in livers of HBV transgenic mice. A mouse of each gender and genotype was fed rodent chow either with or without Wy-14,643 for 7 days before their livers were analyzed. Induction of the transcription rates of the RNAs detected by the rat ACO and cytochrome P450 4A1 (4A1) cDNAs were used as controls to demonstrate the PPARα-dependent increase in transcription rates induced by the peroxisome proliferator from these responsive genes. The rate of transcription from the GAPDH gene was used as an internal control for quantitation of the relative transcription rates of the other genes analyzed. The rate of transcription of the GAPDH gene was designated 1.00. The nonspecific background rate of transcription detected with the pBluescript SK(−) plasmid (BS) control was designated 0.00. The rates of transcription for the HBVayw (HBV), ACO, and 4A1 genes were estimated relative to the GAPDH gene after subtracting the nonspecific background rate of transcription. The probes hybridized to each strip of filter were 32P-labeled transcripts isolated from in vitro-labeled mouse liver nuclei. The DNA on each strip of filter is indicated at the right. M, male HBV transgenic mouse; F, female HBV transgenic mouse; PPAR genotype −, PPARα knockout (−/−) mouse; PPAR genotype +, PPARα heterozygous (+/−) mouse; Wy-14,643 +, HBV transgenic mouse fed 0.1% (wt/wt) Wy-14,643; Wy-14,643 −, HBV transgenic mouse fed normal rodent chow.
Effect of PPARα on viral HBcAg distribution within the livers of HBV transgenic mice.
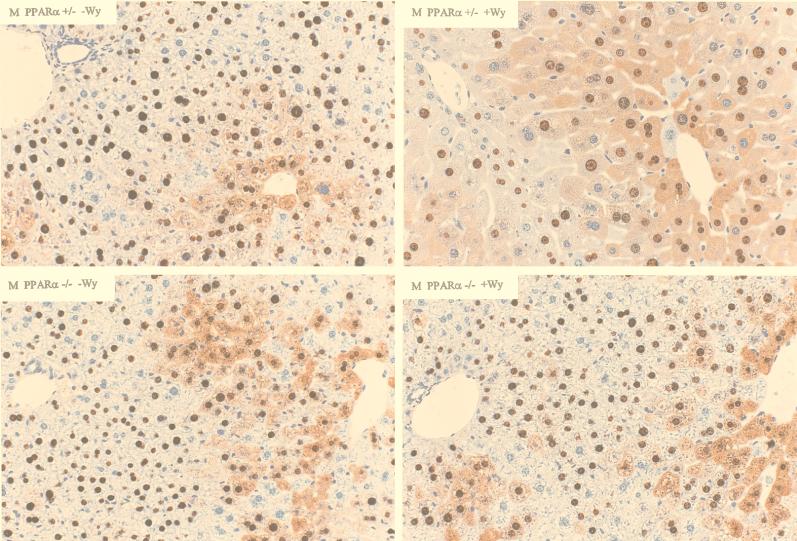
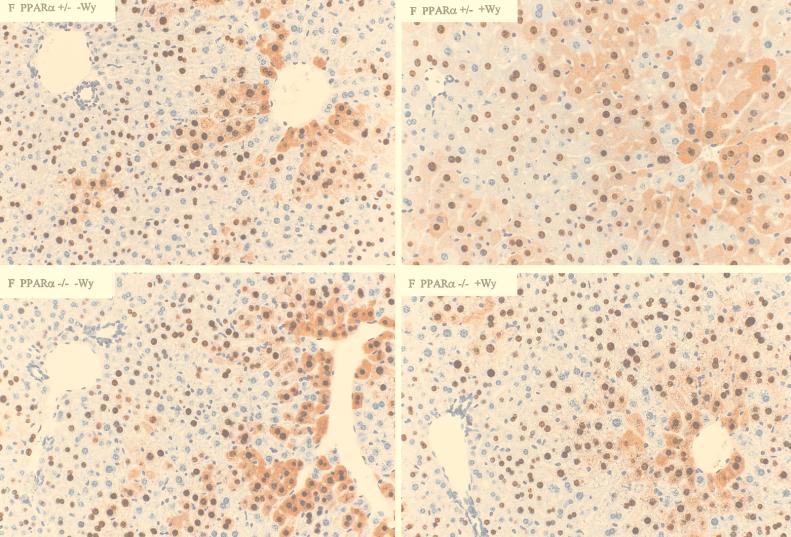
Immunohistochemical analysis of the livers of HBV transgenic mice also demonstrated that peroxisome proliferators influence the distribution of HBcAg within the liver lobule (Fig. 5 and 6). In both male and female PPARα +/− HBV transgenic mice, treatment with Wy-14,643 results in a higher number of hepatocytes that are positive for cytoplasmic HBcAg staining and consequently cytoplasmic staining extends from the centrolobular region much closer to the periportal region of the liver lobule (Fig. 5 and 6). As cytoplasmic HBcAg staining correlates with viral replication (12), this observation is consistent with the increased level of viral replication intermediates found in the livers of these mice. The hepatocytes of the PPARα +/− HBV transgenic mice treated with Wy-14,643 display the expected hypertrophy, whereas this effect and the extended distribution of HBcAg staining is not observed in untreated PPARα +/− HBV transgenic mice and PPARα −/− HBV transgenic mice (Fig. 5 and 6).
FIG. 5.
Immunohistochemical staining of HBcAg in the livers of male HBV transgenic mice (original magnification, ×200). Nuclear staining of HBcAg is observed throughout the liver, whereas cytoplasmic staining is located primarily in the centrolobular hepatocytes in the livers of untreated PPARα +/− HBV transgenic mice (upper left). Treatment of PPARα +/− HBV transgenic mice with Wy-14,643 (upper right) results in hepatocyte hypertrophy and HBcAg staining that extends to the periportal hepatocytes. Livers from Wy-14,643-treated (lower right) or untreated (lower left) PPARα −/− HBV transgenic mice display the same distribution of HBcAg staining as untreated PPARα +/− HBV transgenic mice.
FIG. 6.
Immunohistochemical staining of HBcAg in the livers of female HBV transgenic mice (original magnification, ×200). Nuclear staining of HBcAg is observed throughout the liver, whereas cytoplasmic staining is located primarily in the centrolobular hepatocytes in the livers of untreated PPARα +/− HBV transgenic mice (upper left). Treatment of PPARα +/− HBV transgenic mice with Wy-14,643 (upper right) results in hepatocyte hypertrophy and HBcAg staining that extends to the periportal hepatocytes. Livers from Wy-14,643 treated (lower right) or untreated (lower left) PPARα −/− HBV transgenic mice display the same distribution of HBcAg staining as untreated PPARα +/− HBV transgenic mice.
DISCUSSION
Previously the role of the nuclear hormone receptor PPARα in modulating transcription from the HBV nucleocapsid promoter had been examined in cell culture (25). In transient transfection analysis, RXRα and PPARα in the presence of the activating ligands, 9-cis retinoic acid and clofibric acid, increased transcription from the nucleocapsid promoter approximately fourfold. RXRα and PPARα mediated this effect through the PPREs located in the HBV enhancer 1 region and the nucleocapsid promoter (25). In this study, the effect of activating PPARα with synthetic ligands was investigated in an in vivo model of HBV replication. The in vivo model system used was the HBV transgenic mouse that replicates virus at high levels in the liver (12). If the abundance of the HBV pregenomic RNA were regulated by PPARα activation of transcription from the nucleocapsid promoter, one would anticipate that an increase in this transcript would be associated with an increase in HBV replication. This possibility was examined by treating HBV transgenic mice with the peroxisome proliferators Wy-14,643 and clofibric acid. To determine if these ligands were mediating their effects through PPARα, HBV transgenic mice lacking PPARα were also characterized.
Analysis of the HBV transgenic mice without peroxisome proliferators treatment indicated that under normal physiological conditions PPARα did not influence secretion of HBeAg, HBV transcription, or HBV replication. However, treatment of HBV transgenic mice with the peroxisome proliferators Wy-14,643 and clofibric acid demonstrated that these PPARα ligands increased the levels of secreted HBeAg and HBV transcripts less than twofold (Fig. 1 and 2; Tables 2 to 5). In contrast, the level of viral replication intermediates in male HBV transgenic mice was increased approximately 2.5-fold, whereas the level of replication intermediates in female HBV transgenic mice was increased 7- to 14-fold, by peroxisome proliferator treatment (Fig. 1 and 2; Tables 4 and 5). The increase in viral replication was dependent on PPARα, as PPARα-null HBV transgenic mice failed to show this increase in replication intermediates in response to peroxisome proliferator treatment. Additional evidence that peroxisome proliferators enhanced HBV replication was apparent from immunohistochemical analysis of the livers from HBV transgenic mice (Fig. 5 and 6). HBV transgenic mice displayed cytoplasmic HBcAg in a greater number of hepatocytes after treatment with peroxisome proliferators, suggesting that higher levels of viral replication were occurring in more cells as a result of the activation of PPARα (12).
This analysis demonstrates that female HBV transgenic mice display a dramatic increase in viral replication that is much larger than the observed increase in viral transcription in response to two different peroxisome proliferators (Fig. 1 and 2; Tables 4 and 5). The explanation for this observation is unclear. However, a model in which small alterations in the level of the HBV pregenomic RNA can result in large alterations in replication intermediates offers a possible explanation for these results. In this model, the level of HBV pregenomic RNA in female mice would be slightly lower than the level in male mice. The observation that the ratio of precore to pregenomic RNA is essentially constant in these mice and that the level of secreted HBeAg is higher in male mice than in female mice is consistent with this idea (Fig. 3; Table 1). In female mice, the translation of the core polypeptide from the pregenomic RNA is proposed to be insufficient to permit the efficient formation of capsids from core polypeptide dimers (32, 50, 51). Consequently, many of the core polypeptide dimers are transported into the nucleus of the cell, where they accumulate but do not contribute to viral replication. This pattern of HBcAg immunohistochemical staining is observed in many of the cells surrounding the portal vein where nuclear HBcAg is associated with limited viral replication (Fig. 5 and 6) (12). In contrast, male HBV transgenic mice expressing slightly higher levels of pregenomic RNA would synthesize enough core polypeptide to reach a concentration of core polypeptide dimers sufficient to permit more efficient formation of capsids and consequently more efficient conversion of pregenomic RNA into viral DNA replication intermediates. This possibility is consistent with the observation that male and female HBV transgenic mice express very similar levels of pregenomic RNA but male HBV transgenic mice have approximately fourfold-higher levels of replication intermediates in their livers compared with female HBV transgenic mice (Fig. 1 to 3; Tables 4 and 5). After treatment with peroxisome proliferators, the level of pregenomic RNA in both male and female mice is proposed to be increased to a limited degree but sufficiently to permit the efficient conversion of core polypeptide dimers into nucleocapsids. In this situation, both male and female HBV transgenic mice would be expected to synthesize similar levels of viral replication intermediates, as was observed (Fig. 1 and 2; Tables 4 and 5). This explanation for the PPARα-dependent effects of peroxisome proliferators on viral replication appears the most reasonable. It is consistent with the previous mapping of PPREs to the HBV enhancer 1 region and the nucleocapsid promoter and the observation that transcription from the nucleocapsid promoter can be increased by PPARα and its ligand in cell culture (25).
Alternative explanations for the effects of peroxisome proliferators on viral replication are possible. There may exist in the HBV biosynthetic process a PPARα-sensitive step, other than pregenomic RNA synthesis, that is more critical to viral replication in female mice than in male mice. Stimulation of this hypothetical step could be responsible for the dramatic increase in viral replication observed in female HBV transgenic mice treated with peroxisome proliferators. The putative nature of this step is unknown. However, it is reasonable to assume that the initial event in this process would involve the PPARα-dependent activation of an unknown gene or genes. This gene product would then presumably increase viral replication by activating a posttranscriptional event in the viral life cycle such as pregenomic RNA encapsidation or HBV DNA synthesis.
Two potential alternative explanations for the observed results can probably be excluded. The finding that the level of replication intermediates increases less in male than in female HBV transgenic mice in response to peroxisome proliferators suggests that the simple increase in liver size cannot explain the observed results (Tables 2 and 3). The observation that HBeAg secretion increases in HBV transgenic mice treated with peroxisome proliferators indicates that the increase in intracellular viral replication intermediates is unlikely to be due to retention of virus resulting from a decrease in the ability to secrete the virus from the hepatocytes (Tables 2 and 3).
Regardless of the nature of the PPARα-mediated increase in viral replication, the in vivo demonstration that peroxisome proliferators can increase HBV viral replication intermediates may have important clinical implications. A variety of hypolipidemic drugs mediate their action by activating PPARα (7, 8, 11, 20, 31). Patients receiving these drugs who are also infected with HBV may activate viral replication, and this could have potentially detrimental effects on the outcome of the viral infection. In addition, a variety of dietary and environmental compounds are ligands for PPARα, and exposure to these chemicals might also affect HBV replication in infected individuals (7, 8, 19, 31). As the results in the HBV transgenic mouse model suggest that small alterations in the level of pregenomic RNA levels may have a dramatic effect on viral replication, antagonists directed against PPARα might have therapeutic application in the treatment of HBV infections.
ACKNOWLEDGMENTS
We thank Frank Chisari for providing the HBV transgenic mice and for support and encouragement throughout this study and Stefan Wieland for many helpful discussions. We are grateful to Heike McClary for excellent technical assistance and Margie Chadwell for preparation and staining of tissue sections.
This work was supported by Public Health Service grants AI30070 and AI40696 from the National Institutes of Health.
Footnotes
Publication no. 12404-CB from The Scripps Research Institute.
REFERENCES
- 1.Block T M, Lu X Y, Mehta A, Park J, Blumberg B S, Dwek R. Role of glycan processing in hepatitis B virus envelope protein trafficking. Adv Exp Med Biol. 1998;435:207–216. doi: 10.1007/978-1-4615-5383-0_20. [DOI] [PubMed] [Google Scholar]
- 2.Buckwold V E, Chen M, Ou J H. Interaction of transcription factors RFX1 and MIBP1 with the gamma motif of the negative regulatory element of the hepatitis B virus core promoter. Virology. 1997;227:515–518. doi: 10.1006/viro.1996.8360. [DOI] [PubMed] [Google Scholar]
- 3.Chang C, Jeng K, Hu C, Lo S J, Su T, Ting L-P, Chou C-K, Han S, Pfaff E, Salfeld J, Schaller H. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen I-H, Huang C-J, Ting L-P. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol. 1995;69:3647–3657. doi: 10.1128/jvi.69.6.3647-3657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M, Ou J H. Cell type-dependent regulation of the activity of the negative regulatory element of the hepatitis B virus core promoter. Virology. 1995;214:198–206. doi: 10.1006/viro.1995.9940. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Desvergne B, Wahli W. PPAR: a key nuclear factor in nutrient/gene interactions? In: Baeuerle P A, editor. Inducible gene expression. Boston, Mass: Birkhauser; 1995. pp. 142–176. [Google Scholar]
- 8.Forman B M, Chen J, Evans R M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 10.Gerber M A, Sells M A, Chen M-L, Thung S N, Tabibzadeh S S, Hood A, Acs G. Morphologic, immunohistochemical, and ultrastructural studies of the production of hepatitis B virus in vitro. Lab Investig. 1988;59:173–180. [PubMed] [Google Scholar]
- 11.Gonzalez F J. The role of peroxisome proliferator activated receptor α in peroxisome proliferation, physiological homeostasis, and chemical carcinogenesis. Adv Exp Med Biol. 1997;422:109–125. doi: 10.1007/978-1-4757-2670-1_9. [DOI] [PubMed] [Google Scholar]
- 12.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo W, Chen M, Yen T S B, Ou J-H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honigwachs J, Faktor O, Dikstein R, Shaul Y, Laub O. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J Virol. 1989;63:919–924. doi: 10.1128/jvi.63.2.919-924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huan B, Kosovsky M J, Siddiqui A. Retinoid X receptor α transactivates the hepatitis B virus enhancer 1 element by forming a heterodimeric complex with the peroxisome proliferator-activated receptor. J Virol. 1995;69:547–551. doi: 10.1128/jvi.69.1.547-551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson J L, Raney A K, McLachlan A. Characterization of a functional hepatocyte nuclear factor 3 binding site in the hepatitis B virus nucleocapsid promoter. Virology. 1995;208:147–158. doi: 10.1006/viro.1995.1138. [DOI] [PubMed] [Google Scholar]
- 17.Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamimura T, Yoshikawa A, Ichida F, Sasaki H. Electron microscopic studies of Dane particles in hepatocytes with special reference to intracellular development of Dane particles and their relation with HBeAg in serum. Hepatology. 1981;1:392–397. doi: 10.1002/hep.1840010504. [DOI] [PubMed] [Google Scholar]
- 19.Kliewer S A, Sundseth S S, Jones S A, Brown P J, Wisely G B, Koble C S, Devchand P, Wahli W, Wilson T M, Lenhard J M, Lehmann J M. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and gamma. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroetz D L, Yook P, Costet P, Bianchi P, Pineau T. Peroxisome proliferator-activated receptor α controls the hepatic CYP4A induction adaptive response to starvation and diabetes. J Biol Chem. 1998;273:31581–31589. doi: 10.1074/jbc.273.47.31581. [DOI] [PubMed] [Google Scholar]
- 21.Lee S S T, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 24.Ou J-H, Bao H, Shih C, Tahara S M. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J Virol. 1990;64:4578–4581. doi: 10.1128/jvi.64.9.4578-4581.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raney A K, Johnson J L, Palmer C N A, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raney A K, McLachlan A. The biology of hepatitis B virus. In: McLachlan A, editor. Molecular biology of the hepatitis B virus. Boca Raton, Fla: CRC Press; 1991. pp. 1–37. [Google Scholar]
- 27.Reddy J K, Chu R Y. Peroxisome proliferator-induced pleiotropic responses: Pursuit of a phenomenon. Ann N Y Acad Sci. 1996;804:176–201. doi: 10.1111/j.1749-6632.1996.tb18616.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schaller H, Fischer M. Transcriptional control of hepadnavirus gene expression. Curr Top Microbiol Immunol. 1991;168:21–39. doi: 10.1007/978-3-642-76015-0_2. [DOI] [PubMed] [Google Scholar]
- 30.Schibler U, Hagenbüchle O, Wellauer P K, Pittet A C. Two promoters of different strengths control the transcription of the mouse alpha-amylase gene Amy-1a in the parotid gland and the liver. Cell. 1983;33:501–508. doi: 10.1016/0092-8674(83)90431-2. [DOI] [PubMed] [Google Scholar]
- 31.Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 32.Seifer M, Zhou S, Standring D N. A micromolar pool of antigenically distinct precursors is required to initiate cooperative assembly of hepatitis B virus capsids in Xenopus oocytes. J Virol. 1993;67:249–257. doi: 10.1128/jvi.67.1.249-257.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sells M A, Zelent A Z, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J Virol. 1988;62:2836–2844. doi: 10.1128/jvi.62.8.2836-2844.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stibbe W, Gerlich W H. Variable protein composition of hepatitis B surface antigen from different donors. Virology. 1982;123:436–442. doi: 10.1016/0042-6822(82)90275-6. [DOI] [PubMed] [Google Scholar]
- 35.Sureau C, Romet-Lemonne J-L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 36.Tso J Y, Sun X-H, Kao T, Reece K S, Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985;13:2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsui L V, Guidotti L G, Ishikawa T, Chisari F V. Posttranscriptional clearance of hepatitis B virus RNA by cytotoxic T lymphocyte-activated hepatocytes. Proc Natl Acad Sci USA. 1995;92:12398–12402. doi: 10.1073/pnas.92.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsurimoto T, Fujiyama A, Matsubara K. Stable expression and replication of hepatitis B virus genome in an integrated state in a human hepatoma cell line transfected with the cloned viral DNA. Proc Natl Acad Sci USA. 1987;84:444–448. doi: 10.1073/pnas.84.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Will H, Reiser W, Weimer T, Pfaff E, Buscher M, Sprengle R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaginuma K, Koike K. Identification of a promoter region for 3.6-kilobase mRNA of hepatitis B virus and specific cellular binding protein. J Virol. 1989;63:2914–2920. doi: 10.1128/jvi.63.7.2914-2920.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada G, Sakamoto Y, Mizuno M, Nishihara T, Kobayashi T, Takahashi T, Nagashima H. Electron and immunoelectron microscopic study of Dane particle formation in chronic hepatitis B virus infection. Gastroenterology. 1982;83:348–356. [PubMed] [Google Scholar]
- 43.Yen T S B. Regulation of hepatitis B virus gene expression. Semin Virol. 1993;4:33–42. [Google Scholar]
- 44.Yuh C-H, Chang Y-L, Ting L-P. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuh C-H, Ting L-P. C/EBP-like proteins binding to the functional box-α and box-β of the second enhancer of hepatitis B virus. Mol Cell Biol. 1991;11:5044–5052. doi: 10.1128/mcb.11.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuh C-H, Ting L-P. Differentiated liver cell specificity of the second enhancer of hepatitis B virus. J Virol. 1993;67:142–149. doi: 10.1128/jvi.67.1.142-149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang P, McLachlan A. Differentiation-specific transcriptional regulation of the hepatitis B virus nucleocapsid gene in human hepatoma cell lines. Virology. 1994;202:430–440. doi: 10.1006/viro.1994.1359. [DOI] [PubMed] [Google Scholar]
- 48.Zhang P, Raney A K, McLachlan A. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology. 1992;191:31–41. doi: 10.1016/0042-6822(92)90163-j. [DOI] [PubMed] [Google Scholar]
- 49.Zhang P, Raney A K, McLachlan A. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J Virol. 1993;67:1472–1481. doi: 10.1128/jvi.67.3.1472-1481.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou S, Standring D N. Hepatitis B virus capsid particles are assembled from core- protein dimer precursors. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou S, Yang S Q, Standring D N. Characterization of hepatitis B virus capsid particle assembly in Xenopus oocytes. J Virol. 1992;66:3086–3092. doi: 10.1128/jvi.66.5.3086-3092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]