Abstract
The function of the X protein in the life cycle of mammalian hepadnaviruses is unclear. Based on tissue culture experiments it has been suggested that this protein represents a transcriptional transactivator which might be essential for the expression of the viral core gene. Here we have examined whether the activity of the human hepatitis B virus (HBV) core gene in vivo depends on X coexpression. To this end we compared core gene expression between four lineages of transgenic mice carrying the HBV core gene in cis arrangement with the X gene (cex lineage) and six lineages containing a modified construct in which the start codon of the X gene had been deleted (ce lineage). Whereas all cex lineages consistently exhibited a high-level hepatic core gene expression, the liver-specific core gene expression pattern of the ce lineages was heterogenous with four lineages virtually not expressing the core gene. This defect was due to a strongly reduced transcription since no core mRNA could be detected by Northern blotting. To test whether core gene expression could be restored by providing an intact X gene in trans, we crossbred mice of two lines which expressed no core mRNA or core protein with transgenic mice expressing the X-gene product under the transcriptional regulation of the liver-specific major-urinary-protein promoter/enhancer (MUP-X mice). The introduction of the MUP-X transgene induced core mRNA expression and core protein biosynthesis in the livers of the double-transgenic mice. This demonstrates that the X-gene product has the capacity to transactivate HBV core gene expression in vivo.
The hepadnaviruses comprise a small group of enveloped DNA viruses which are characterized by extreme species and organ specificity (6). The most prominent members of this virus group are the human hepatitis B virus (HBV), the woodchuck hepatitis virus (WHV), the ground squirrel hepatitis virus, and the duck hepatitis virus. Whereas the genomes of all hepadnaviruses comprise three open reading frames (ORFs) encoding the core, envelope, and polymerase proteins, mammalian hepadnaviruses possess an additional, fourth, ORF. This ORF was designated X since its function remained unclear. As was shown in the WHV (5) and HBV (16, 19, 25) systems, the X-gene product (pX) is expressed at least during certain stages of hepadnaviral infection.
Since the position of the X ORF within the hepadnavirus genome resembled the locations of the Tat and Tax ORFs in the human immunodeficiency virus and human T-cell leukemia virus genomes, it was suggested that pX might represent a transcriptional transactivator (17). Later it could be shown in a variety of tissue culture systems that pX does in fact have transactivating properties (4, 24, 29; for a review, see reference 22). In contrast to retroviral transactivators which specifically exert their function on the viral long terminal repeat element, however, pX was found to transactivate a wide variety of cellular and viral genes.
In spite of these findings, the precise function of pX during the viral life cycle remained unknown. X-defective hepadnaviral genomes are replication competent after transfection into differentiated hepatoma cell lines (2, 14, 26). On the other hand, studies performed in the WHV system unequivocally demonstrated that an intact X gene is required for the successful establishment of hepadnavirus infection in vivo (3, 31). Although these studies proved that pX exerts an essential function during the hepadnavirus life cycle, they did not identify what this function was. Because of the known transactivating potential of pX, it was suggested that the lack of infectivity of the X-gene mutants was due to a defect in viral gene expression. In this respect, regulation of the core gene promoter appeared to be a particularly interesting potential target for pX since this promoter is preferentially active in hepatocytes and is believed to contribute to the liver tropism of the hepadnaviruses (1, 9, 12, 28, 30).
The aim of the present study was to examine the transactivating potential of the X-gene product on the HBV core gene in an in vivo situation. As has been pointed out, expression of hepadnavirus genes after transfection of cloned viral DNA into tissue culture cells is mostly pX independent. For this reason, the suitability of such in vitro systems for the analysis of pX function is questionable. We therefore decided to analyze the transactivating properties of this protein in transgenic mice, which more closely approximate the situation encountered in a normal infection.
Recently, we have generated four lineages of transgenic mice carrying the HBV core and X genes in cis arrangement (cex lineages [20]). These mice expressed the X mRNA in all organs investigated and exhibited a high-level HBV core protein expression in the liver. To investigate the role of pX for HBV core gene expression, we generated six additional mouse lineages which carried the same transgene as the cex mice except that the start codon of the X gene was deleted (ce lineages). In four of these lineages virtually no hepatocellular core gene expression could be detected, suggesting an impairment of HBV core gene activity due to the lack of X coexpression. Here we tested whether core gene expression could be restored in these mice by introducing a functional X gene in trans. To this end, we crossbred mice from two core-negative ce lines with mice of transgenic lineages expressing the X gene under the transcriptional regulation of the major-urinary-protein (MUP) promoter (8). Our data indicate that pX has the capacity to transactivate HBV core gene expression in vivo and support the hypothesis that the biological function of pX may be the transcriptional transactivation of the core gene.
MATERIALS AND METHODS
Generation of transgenic mice.
Animals were bred at the Animal Research Unit of the University of Ulm under strict specific-pathogen-free conditions. All analyses presented in this report were performed with hemizygous animals.
Production and characteristics of cex-transgenic mice coexpressing the HBV core and X genes (ayw subtype) under authentic promoter control and MUP-X-757- and MUP-X-760-transgenic mice (expressing the X gene under control of the MUP promoter/enhancer) have been described (8, 20). In previous works (20, 21) lineages cex-1, cex-2, cex-4, and cex-5 had been designated cexL, cexC, cexIV, and cexV, respectively. For production of ce-transgenic lineages, plasmid p24.6/29, which had been used previously for generation of the cex mice, was cut with NcoI, and the single-stranded protruding ends containing the start codon of the X gene were removed by treatment with mung bean nuclease. This mutation has already been used in previous studies (4) to prevent the expression of a functional X-gene product. After religation and removal of vector sequences, the HBV fragment was microinjected into C57BL/6J (B6) or F2(B6 × CBA/Ca) (F2) embryos according to standard procedures. Two of the six ce-transgenic lineages, designated ce-1 and ce-2, were directly obtained on a B6 background, whereas the other lineages were derived from F2 founders and backcrossed to the B6 strain for at least three generations before expression analysis. Cex- and ce-transgenic animals were identified by PCR amplification of transgenic sequences using primers GAG ATG GGG TTA CTC TCT and CCT TGT AAG TTG GCG AGA or in few experiments by detection of hepatitis B e antigen (HBeAg) in the serum using a commercially available diagnostic test kit (HBe or HBe II; Abbott, Wiesbaden, Germany). MUP-X-transgenic mice were identified by PCR amplification of transgene-specific sequences using the MUP-promoter-specific plus primer TGT AGC CAC GAT CAC AAG AA and the X-specific minus primer GGT GAA GCG AAG TGC ACA.
Transgene copy number.
Genomic DNA was prepared from representatives of all cex and ce lineages and was dot blotted onto nylon membranes. DNA was equilibrated using a glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific 32P-labelled probe. For determination of transgene copy number the membranes were hybridized with an HBV-specific probe. Single-copy integrations were verified by selective PCR analysis as described previously (20).
Northern blotting.
For Northern blotting, snap-frozen organs were pulverized in liquid nitrogen in a microdismembrator (Braun Biotech International GmbH, Melsungen, Germany), and total RNA was prepared using the RNEasy Midi Kit (Qiagen). Twenty-five micrograms of total RNA was separated on a formaldehyde gel, blotted onto nylon membranes, and hybridized either with an HBV core (BglII fragment of the core ORF)- or a GAPDH-specific 32P-labelled probe.
Quantitation of HBcAg.
For quantitation of hepatitis B core antigen (HBcAg) in murine organs, the Abbott IMX microparticle HBe assay system, which detects both HBeAg and HBcAg (20), was used. The test was calibrated with recombinant HBcAg prepared from Escherichia coli. For analysis, 10 mg of snap-frozen pulverized tissue was suspended in TNE containing 1% Triton X-100 which was then diluted such that the HBcAg concentration fell within the range of the calibration curve.
Immunohistology.
Tissue specimens were fixed in 4% formaldehyde solution in phosphate-buffered saline (pH 7.2). HBcAg-specific immunostaining was performed with paraffin sections using the avidin-biotin complex method (11). Paraffin sections were treated with a commercial target unmasking fluid (Dianova, Hamburg, Germany) in a microwave oven before antibody incubation. The sections were incubated overnight at 4°C with a 1:4,000-diluted polyclonal rabbit antiserum specific for both HBcAg and HBeAg (23). Specifically bound antibodies were detected with a biotinylated secondary antibody and subsequent incubation with phosphatase-conjugated streptavidin (Biogenex, San Ramon, Calif.) and staining with naphthol AS-BI phosphate in combination with hexazotized new fuchsine (E. Merck AG, Darmstadt, Germany). Alternatively, phosphatase activity was revealed using a commercial 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium substrate solution (DAKO Diagnostika, Hamburg, Germany). Endogenous avidin-binding activity was reduced by pretreatment of the sections with avidin and biotin solutions (Zymed Laboratories, San Francisco, Calif.). Negative controls comprised naive rabbit serum and tissues from nontransgenic mice.
Sequence analysis.
To investigate for the presence of an intact translational start codon of the X gene in cex mice and for the absence of this ATG codon in ce mice, total liver RNA was obtained from representatives of all lineages as mentioned above. The RNA was treated with DNase I (amplification grade; Life Technologies, Paisley, Scotland) and was reverse transcribed with primer AAG GAT CCG TCG ACA TCG ATA ATA CGA CTC ACT ATA GGG ATT TTT TTT TTT TTT TTT using the SuperScript II reverse transcription kit (Life Technologies). The resulting liver-specific cDNAs as well as samples of leukocytic genomic DNA were amplified using the plus primer GAT CCA TAC TGC GGA ACT, which anneals downstream from the initiation site of X-gene transcription (27), and the minus primer CCC GCG CAG GAT CCA GTT, which anneals shortly downstream from the translational start signal of the X ORF. In order to investigate the integrity of the core gene promoter in transgenic mice, leukocytic DNA was amplified using primer GGG AAG CTT GGG TAT ACA TTT AAA CCC, which anneals to the 5′ terminus of the cex and ce constructs, and primer GGG AAG CTT GAG TAA CTC CAC AGT AGC, which anneals shortly downstream from the translational start signal of the c-ORF. Sequence analyses of cloned amplification products were performed by MWG Biotech (Ebersberg, Germany).
RESULTS
Impairment of core gene expression in transgenic livers by X-gene inactivation.
Recently, we have generated four lineages (designated cex lines) of transgenic mice carrying the HBV core ORF with all upstream regulatory elements known to be relevant for core gene expression together with an intact X gene (Fig. 1, construct cex) (20). These mice expressed high levels of both HBc and HBe protein. To investigate whether pX was required for core gene expression, the start codon of the X gene was deleted as described in the methods section (Fig. 1, construct ce). Since the X ATG does not overlap with any regulatory element of the HBV core gene, its deletion should not influence core gene expression directly. On the other hand, this mutation was previously shown to prevent the expression of a functional X-gene product (4).
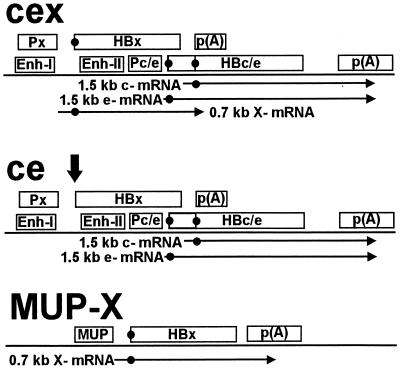
FIG. 1.
DNA constructs used for the generation of transgenic mice. The construct used for generation of cex-transgenic mice (20) comprised a fragment of the HBV genome containing the core ORF (HBc/e), the core promoter (Pc/e), and the enhancer elements (Enh-I and Enh-II). Since the X gene colocalizes with these sequences, the cex construct carried a functional X gene. Px, X promoter; HBx, X-ORF; p(A), polyadenylation signal. To provide a functional p(A) site for the core gene, a short HBV sequence including the authentic HBV p(A) signal was added downstream of this fragment. The construct, used for the generation of ce-transgenic mice, was obtained by deleting the translational start codon of the X gene (indicated by the vertical arrow) in the cex construct. In MUP-X-transgenic mice (8) the X gene was regulated by the liver-specific promoter/enhancer complex of the MUP gene. ORFs and transcriptional regulatory elements are depicted as boxes. The mRNA species expected to be expressed by the various DNA constructs are presented as horizontal arrows. The relevant translational start codons are indicated by black dots. Core gene transcription of cex- and ce-transgenic mice should result in the expression of a 1.5-kb transcript.
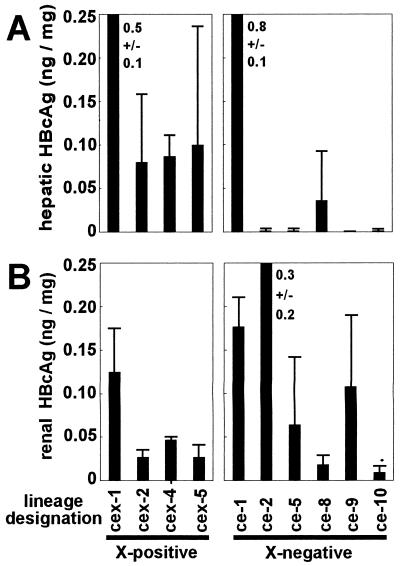
Upon microinjection of the X-defective construct into murine zygotes, six transgenic lineages were established and designated as ce lineages. Using dot blot analysis of genomic DNA, the transgene copy numbers of all cex and ce lineages were determined (cex-1, 1 copy; cex-2, 1 copy; cex-4, approximately 5 copies; cex-5, approximately 10 copies; ce-1, 1 copy; ce-2, approximately 10 copies; ce-5, approximately 100 copies; ce-8, 1 copy; ce-9, approximately 10 copies; ce-10, approximately 10 copies). The presence of the translational start codon of the X gene in cex lineages and the absence of this ATG in ce lineages, respectively, was confirmed in representatives of all lines by sequence analyses of PCR products amplified from genomic DNA and from liver-specific cDNA, respectively (data not shown). To investigate whether X-gene inactivation affected core gene expression, we first compared the HBcAg concentrations in the livers of X-positive cex mice and X-negative ce mice. As is depicted in the left diagram of Fig. 2A, a high-level HBcAg expression could be observed in the livers of all cex mice which carried the intact X gene. In contrast, the ce lineages exhibited a heterogenous hepatic core gene expression pattern. Whereas the livers of ce-1 and ce-8 transgenic mice exhibited respectively high and low levels of HBcAg expression, virtually no HBcAg was detectable in livers from four of the six X-defective lineages (Fig. 2A, right, ce-2, ce-5, ce-9, and ce-10). The hepatic HBcAg expressions of X-positive cex lineages and X-negative ce lineages were found to differ significantly (as determined by analysis of variance, P < 0.05).
FIG. 2.
HBc/eAg expression in organs of X-positive cex- and X-negative ce-transgenic mice. Mean HBcAg concentrations were quantitated in livers (A) and kidneys (B) of transgenic mice originating from four cex lineages (cex-1, cex-2, cex-4, and cex-5) and six ce lineages (ce-1, ce-2, ce-5, ce-8, ce-9, and ce-10). The data shown represent means of three independent determinations performed with samples of different 50- to 65-day-old males plus standard deviations. Note that four of six X-deficient lineages expressed virtually no hepatic HBcAg whereas their renal HBcAg expression was the same as that of the other lineages.
The low level of core gene expression in ce mice was not due to a defect or a general silencing of the transgene.
One trivial explanation for the virtual lack of core gene expression observed in four of the six ce lineages was that the transgene might have been rendered inactive due to a rearrangement which could have occurred during the integration process. Alternatively, integration could have taken place at unfavorable sites, resulting in the silencing of the regulatory elements important for core gene expression.
In order to address these points, first the integrity of the transgenes was examined in cex lineages cex-1 and cex-2 and ce lineages ce-1, ce-2, and ce-9 by amplifying the core promoter region from genomic DNA as described in Materials and Methods. All PCR products showed the expected sizes. The specificity of the reaction products was controlled by sequence analyses. These experiments revealed that all transgenic lineages investigated lacked HBV nucleotide 2431 (nucleotide positions are relative to the HBc ATG with the A being nucleotide 1) upstream of the X ORF. Further analyses demonstrated that this point deletion was also present in the plasmids used for generation of the transgenic mouse lines. However, since both the ce and the cex mice carried the same mutation, it could not explain the low level of core gene expression observed in the ce animals.
To test whether there was a general inactivation of the transgene, quantitative core protein analyses were performed with the kidneys. It is well documented that the core gene enhancer is not strictly liver specific in transgenic mice but rather is active in a variety of organs, in particular in the kidneys. As is shown in Fig. 2B, a significant level of core gene expression could be found in the kidneys of all cex and ce lineages. This shows that the observed lack of hepatic core protein expression found in four ce lineages was not due to a general silencing of the transgene.
Low levels of core protein expression in ce mice correlates with strongly reduced core mRNA levels.
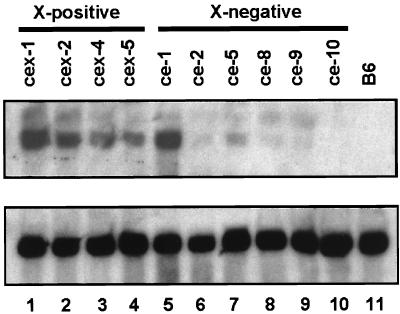
Since the pX protein can transactivate the core gene in vitro, it appeared likely that the strongly reduced core protein levels observed in four of the six ce lineages was due to an impaired core gene transcription. To test this, we performed Northern blot analyses with total liver RNA prepared from all cex- or ce-line mice. As is shown in Fig. 3, lanes 1 to 5, a good expression of the core gene transcript could be detected in livers of those cex and ce mice which exhibited high-level HBcAg expression (mice from all four X-intact cex lineages and from the X-defective lineage ce-1). In livers of mice from the other five ce lines, only minute amounts of this transcript could be detected (Fig. 3, lanes 6 to 10). The good correlation between HBcAg and core mRNA expression shows that pX probably does not exert a posttranscriptional effect on the core gene products but rather might be required for efficient core gene transcription.
FIG. 3.
Hepatic core gene transcription of X-positive cex lineages and X-negative ce lineages. Northern blots prepared with 25 μg of total liver RNA originating from mice of the four X-intact cex lineages and the six X-defective ce-transgenic lineages were hybridized with a HBV core-gene-specific (upper part) or GAPDH-specific (lower part) probe. The 1.5-kb core-specific mRNA was well expressed in livers from lineages with high-level HBcAg expression (all X-positive cex lines and the X-defective lineage ce-1, lanes 1 to 5) but was virtually not expressed in livers from lineages lacking or with low-level core protein expression. X-negative ce lineages ce-2, ce-5, ce-8, ce-9, and ce-10, lanes 6 to 10, respectively. B6, nontransgenic control.
The X-gene product can transactivate core gene expression in transgenic livers.
As has been described above, four of the six transgenic mouse lineages which lacked a functional X gene expressed virtually no core mRNA in the liver. This allowed us to directly address the question whether the X protein can exert a transactivating effect on the core gene in vivo. To date, this important issue has only been analyzed in permanent cell lines which differ significantly from fully differentiated hepatocytes. Transgenic mice of lineages ce-2 and ce-9 were chosen for these experiments since these animals express minimal amounts of core protein in the liver but exhibit high-level HBcAg expression in the kidneys.
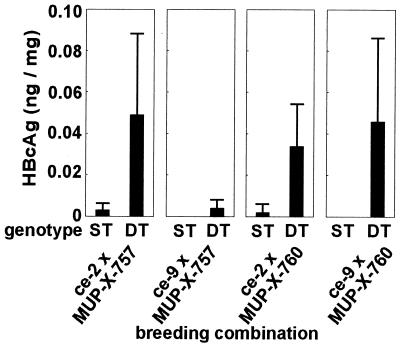
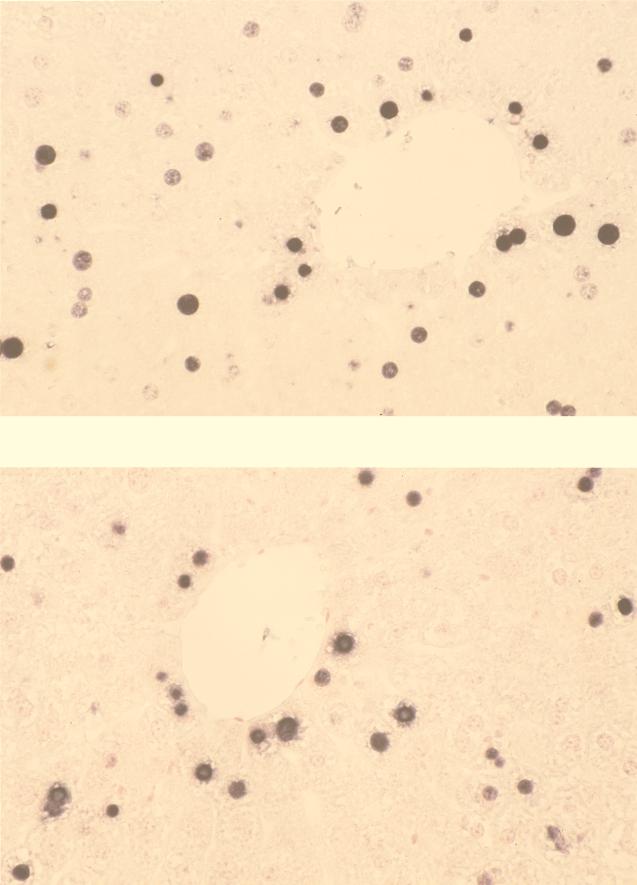
Hemizygous transgenic mice of lineages ce-2 and ce-9 were crossbred with hemizygous animals from two mouse lineages expressing a functional HBV X gene under the transcriptional control of the strong, liver-specific MUP promoter (MUP-X-757 and MUP-X-760 mice [8]). As is depicted in Fig. 4, in all four breeding combinations tested the double-transgenic (DT) mice exhibited significantly (as determined by the Wilcoxon test) increased HBcAg concentrations in the liver in comparison to their single-transgenic (ST) littermates. The enhancement factors of core protein expression were 18, 56, 16, and 331 for breeding combinations ce-2 × MUP-X-757, ce-9 × MUP-X-757, ce-2 × MUP-X-760, and ce-9 × MUP-X-760, respectively. To test whether there was any difference with respect to hepatolobular or subhepatocellular HBcAg expression between the F1(ce × MUP-X) mice which express the X gene in trans and the cex mice which express this gene in cis, liver sections were examined by immunohistochemistry. The results are depicted in Fig. 5. In the livers of both F1(ce × MUP-X) transgenic variants tested, HBcAg-positive staining was restricted to the nuclei of hepatocytes located in the centrolobular region of the hepatic lobule. No HBcAg could be detected in the periportal regions. An identical expression pattern had previously been detected in cex-transgenic livers (20). Hence, the HBcAg expression pattern was independent from the constructs employed for transgene generation. As expected, no staining could be observed in sections derived from the X-negative ce mice (data not shown).
FIG. 4.
Induction of HBcAg expression in livers of X-negative ce mice by the MUP-X transgene. X-gene-defective ce-2 or ce-9 mice lacking hepatic HBcAg expression were crossbred with MUP-X-757 or MUP-X-760 animals expressing high amounts of X mRNA in their livers (8). HBcAg was quantitated in livers of ceST and F1(ce × MUP-X) DT offspring. The data shown represent mean values of at least five determinations performed with samples of different 50- to 65-day-old mice plus standard deviations. In all four breeding combinations tested, a significant difference of mean hepatic HBcAg concentrations could be detected between ST and DT mice (as determined by the Wilcoxon test).
FIG. 5.
Hepatolobular and subcellular distribution pattern of HBcAg expression. Liver sections of F1(ce-2 × MUP-X-757) (upper panel) and F1(ce-9 × MUP-X-760) (lower panel) mice were analyzed immunohistochemically with a polyclonal antibody recognizing all epitopes of the core protein (final magnification, ×490). HBcAg-specific staining was restricted to the nuclei located in the centrolobular liver regions. A similar HBcAg staining pattern had been previously observed in cex-transgenic mice (20). No HBcAg staining was detectable in liver sections of ce-2 or ce-9 ST controls (data not shown).
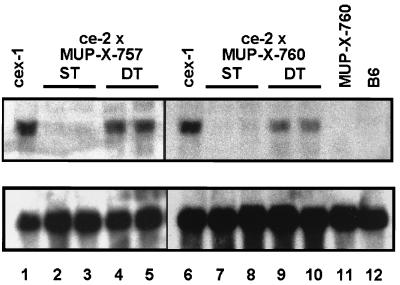
As is clear from these data, liver-specific core protein biosynthesis could be restored in ce mice by the introduction of an intact X gene. To investigate whether this effect was due to enhanced mRNA levels, Northern blotting was performed with total liver RNA prepared from ce ST and F1(ce × MUP-X) DT mice. As is shown in Fig. 6, the expected core gene transcript could be detected in the DT (lanes 4, 5, 9, and 10) but not in the ST (lanes 2, 3, 7, and 8) mice. The enhancement factors of core mRNA expression were 28 and 6 for breeding combinations ce-2 × MUP-X-757 and ce-2 × MUP-X-760, respectively. In fact, trans expression of the X gene resulted in mRNA levels which were comparable to those found in the cex mice which contain this gene in a cis arrangement. These data clearly demonstrate that pX has the potential to transactivate core gene expression in vivo.
FIG. 6.
Induction of core gene transcription in livers of X-negative ce mice by the MUP-X transgene. X-gene-defective ce-2 mice lacking hepatic HBcAg expression were crossbred with X-mRNA-expressing MUP-X-757 and MUP-X-760 animals, respectively. Core gene transcription in livers of ce ST (lanes 2, 3, 7, and 8) and F1(ce × MUP-X) DT (lanes 4, 5, 9, and 11) offspring mice of various breeding combinations was analyzed by Northern blotting using an HBV core-gene-specific probe (upper part). Total liver RNA of cex-1-transgenic mice (lanes 1 and 6) and nontransgenic B6 mice (lane 12) were analyzed in parallel as positive and negative controls, respectively. A GAPDH-specific probe was used to equilibrate the RNA (lower part). Note that with introduction of the MUP-X transgene, expression of the core-gene-specific 1.5-kb transcript was induced in livers of ce-transgenic mice.
DISCUSSION
The function of the HBV X-gene product during the viral life cycle is unknown. The discoveries of Spandau and Lee (24), Zahm et al. (29), Colgrove et al. (4), and many others (22) have shown that pX can transactivate the transcription of HBV-specific as well as many unrelated genes. However, since all these studies were performed in vitro, the relevance to HBV transcription in vivo has been unclear. There have been reports suggesting that HBV mutants with defective X genes replicate after transfection of cloned viral DNA into hepatoma cells (2, 14, 26), raising the question whether the X gene encodes an essential viral protein at all. However, in all three studies it was observed that gene expression and/or replication was reduced in hepatoma cells transfected with HBV X mutants. More importantly, recently two groups presented unequivocal evidence showing that a hepadnavirus which lacks a functional X gene is noninfectious in vivo (3, 31). Since HBV infects only humans and chimpanzees, these experiments were carried out with WHV. In several well-controlled studies both groups failed to detect any sign of infection after the injection of WHV X-mutant DNA into the liver, a method which otherwise reliably results in viremia. It appears possible that the low number of genomes which enter a hepatocyte during a normal infection, which in most cases is a single copy, is not sufficiently well transcribed to allow for the initial amplification of the covalently closed circular DNA which is a prerequisite for long-term virus production. In transfection experiments this limitation might be compensated by the large amount of DNA which is usually taken up by the cells. Taken together, the available data support two conclusions. Firstly, pX is essential for the establishment of a hepadnavirus infection in vivo, and secondly, in vitro systems may be of limited value for the analysis of pX function.
One major drawback of all in vivo systems is that it is usually not possible to determine why a certain virus mutant is no longer infectious. Consequently, while it could be shown in the aforementioned studies that pX is definitely important for infectivity, its role during the viral life cycle remained unresolved. To approach this problem, we decided to analyze the transactivating properties of the HBV X-gene product in an in vivo situation. These experiments were performed with transgenic mice, which not only can be easily manipulated but also represent a good equivalent of a natural infection. In a previous study, we generated several transgenic mouse lines using a construct containing the HBV X and core genes together with all upstream sequences which are known to be important for their expression. These lines consistently expressed high levels of HBc and HBe protein as well as core specific mRNA in the liver, showing that this construct was suitable to analyze the significance of pX for hepatic core gene expression. To this end, we deleted the start codon of the X gene and used this modified construct to generate six additional transgenic lines.
As is shown here, the hepatic core protein expression pattern of the X-deficient lineages was heterogenous. Two lineages exhibited low-level and a high-level hepatic HBcAg expression, respectively, whereas four of the lines expressed virtually no core protein in the liver. Why the core gene was expressed in the livers of two of the lineages while it was silent in the four other lines is unclear. The most probable explanation is that in these lines integration of the transgene took place at a favorable chromosomic site with a high basic expression level, which is a general problem when gene expression is studied in transgenic mice (10). However, statistical analysis of hepatic core gene expression between all cex and ce lineages showed a significant effect of X coexpression on the magnitude of core gene expression in the liver, suggesting that pX has the potential to increase core gene expression. These findings are reminiscent of data published previously by Nagashima et al. (18). However, this study suffered from the fact that only few X-deficient lineages and no X-intact controls were examined, making the results difficult to interpret.
To directly test the hypothesis that HBx can enhance core gene expression, we crossbred mice from two core-negative ce lines with mice which expressed the X protein under foreign promoter control. Our data convincingly demonstrate that core gene expression can be restored by providing an active X gene in trans, and we believe that the most plausible explanation for this result is that pX transactivated the regulatory elements important for core gene transcription. In this context it is important to mention that due to their structure the HBV sequences which had been introduced into the transgenic lines cannot give rise to replication-competent core particles. Thus, it can be ruled out that the observed effect was due to a stimulated replication.
Our interpretation that the HBx is important for efficient hepatic HBV gene expression in vivo is supported by observations which were made during attempts to generate transgenic mice expressing a replication-competent over-length HBV construct. When 1.1- and 1.2-genome-length constructs lacking the 5′ X sequences were used to produce transgenic mice, only low levels of HBV expression and replication were observed (7). High-level HBV replication was only observed with a 1.3-genome-length construct that contained two copies of the X transcription unit, one at each end. Whether this finding was due to the position of the X gene or due to the amount of X protein expressed is unclear. It is not possible to reliably detect the X protein in organs by existing methods, e.g., Western blotting or immunohistology. There are reports claiming successful detection of HBx in vivo (13, 15, 25), but others had difficulty reproducing these data (8, 19, 20). Therefore, it is not possible to compare the amounts of X protein expressed in the 1.1-, 1.2-, or 1.3-genome-length or the MUP-X mice with the amount of X protein expressed during a natural infection.
Taken together, the available data clearly demonstrates that pX has the capacity to enhance the expression of the HBV core gene in vitro and in vivo, supporting the hypothesis that the natural function of pX is to enhance the transcription of the core gene during the viral life cycle.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Re 1030/2-2 and Schl 270/1-3). The Heinrich Pette Institut is financially supported by the Freie and Hansestadt Hamburg and the Bundesministerium für Gesundheit.
We appreciate the excellent assistance of Johann Derksen, Nikolay Derksen, Martina Fransewitz, Iris Gastrock-Balitsch, Susanne Knehr, Timur Muratow, and Gabriele Spindler.
REFERENCES
- 1.Billet O, Grimber G, Levrero M, Seye K, Briand P, Joulin V. In vivo activity of the hepatitis B virus core promoter: tissue specificity and temporal regulation. J Virol. 1995;69:5912–5916. doi: 10.1128/jvi.69.9.5912-5916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum H E, Zhang Z S, Galun E, von-Weizsacker F, Garner B, Liang T J, Wands J R. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H, Kaneko S, Girones R, Anderson R, Hornbuckle W, Tennant B, Cote P, Gerin J, Purcell R, Miller R. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colgrove R, Simon G, Ganem D. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J Virol. 1989;63:4019–4026. doi: 10.1128/jvi.63.9.4019-4026.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dandri M, Schirmacher P, Rogler C E. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganem D, Varmus H. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 7.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guidotti L G, Matzke B, Pasquinelli C, Shoenberger J M, Rogler C E, Chisari F V. The hepatitis B virus precore protein inhibits HBV replication in transgenic mice. J Virol. 1996;70:7056–7061. doi: 10.1128/jvi.70.10.7056-7061.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Chen M, Yen T S, Ou J H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer R E, Krumlauf R, Camper S A, Brinster R L, Tilghman S M. Diversity of alpha-fetoprotein gene expression in mice is generated by a combination of separate enhancer elements. Science. 1987;235:53–58. doi: 10.1126/science.2432657. [DOI] [PubMed] [Google Scholar]
- 11.Hsu S, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabelled antibody (PAP) procedures. J Histochem Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 12.Hu K Q, Siddiqui A. Regulation of the hepatitis B virus gene expression by the enhancer element I. Virology. 1991;181:721–726. doi: 10.1016/0042-6822(91)90906-r. [DOI] [PubMed] [Google Scholar]
- 13.Kim C M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 14.Koike K, Shirakata Y, Yaginuma K, Arii M, Takada S, Nakamura I, Hayashi Y, Kawada M, Kobayashi M. Oncogenic potential of hepatitis B virus. Mol Biol Med. 1989;6:151–160. [PubMed] [Google Scholar]
- 15.Lee T H, Finegold M J, Shen R F, DeMayo J L, Woo S L, Butel J S. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J Virol. 1990;64:5939–5947. doi: 10.1128/jvi.64.12.5939-5947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levrero M, Stemler M, Pasquinelli C, Alberti A, Jean-Jean O, Franco A, Balsano C, Diop D, Brechot C, Melegari M, et al. Significance of anti-HBx antibodies in hepatitis B virus infection. Hepatology. 1991;13:143–149. [PubMed] [Google Scholar]
- 17.Miller R H, Robinson W S. Common evolutionary origin of hepatitis B virus and retroviruses. Proc Natl Acad Sci USA. 1986;83:2531–2535. doi: 10.1073/pnas.83.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagashima H, Imai M, Iwakura H. Aberrant tissue specific expression of the transgene in transgenic mice that carry the hepatitis B virus genome defective in the X gene. Arch Virol. 1993;132:381–397. doi: 10.1007/BF01309547. [DOI] [PubMed] [Google Scholar]
- 19.Pfaff E, Salfeld J, Gmelin K, Schaller H, Theilmann L. Synthesis of the X-protein of hepatitis B virus in vitro and detection of anti-X antibodies in human sera. Virology. 1987;158:456–460. doi: 10.1016/0042-6822(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 20.Reifenberg K, Löhler J, Pudollek H P, Schmitteckert E, Spindler G, Köck J, Schlicht H J. Long term expression of the hepatitis B virus core e- and X-proteins does not cause organ pathology in transgenic mice. J Hepatol. 1997;26:119–130. doi: 10.1016/s0168-8278(97)80018-9. [DOI] [PubMed] [Google Scholar]
- 21.Reifenberg K, Deutschle T, Wild J, Hanano R, Gastrock-Balitsch I, Schirmbeck R, Schlicht H J. The hepatitis B virus e antigen cannot pass the murine placenta efficiently and does not induce CTL immune tolerance in H-2b mice in utero. Virology. 1998;243:45–53. doi: 10.1006/viro.1998.9033. [DOI] [PubMed] [Google Scholar]
- 22.Rossner M. The hepatitis B virus x-gene product: a promiscuous transcriptional activator. J Med Virol. 1992;36:101–117. doi: 10.1002/jmv.1890360207. [DOI] [PubMed] [Google Scholar]
- 23.Schlicht H J, Schaller H. The secretory core protein of human hepatitis B virus is expressed on the cell surface. J Virol. 1989;63:5399–5404. doi: 10.1128/jvi.63.12.5399-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spandau D F, Lee C H. trans-Activation of viral enhancers by the hepatitis B virus X protein. J Virol. 1988;62:427–434. doi: 10.1128/jvi.62.2.427-434.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Q, Schroder C H, Hofmann W J, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109–1120. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- 26.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaginuma K, Nakamura I, Takada S, Koike K. A transcription initiation site for the hepatitis B virus X gene is directed by the promoter-binding protein. J Virol. 1993;67:2559–2565. doi: 10.1128/jvi.67.5.2559-2565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuh C, Ting L. The genome of hepatitis B virus contains a second enhancer: cooperation of two elements within this enhancer is required for its function. J Virol. 1990;64:4281–4287. doi: 10.1128/jvi.64.9.4281-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahm P, Hofschneider P H, Koshy R. The HBV X-ORF encodes a transactivator: a potential factor in viral hepatocarcinogenesis. Oncogene. 1988;3:169–177. [PubMed] [Google Scholar]
- 30.Zhang P, Raney A, McLachlan A. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology. 1992;191:31–41. doi: 10.1016/0042-6822(92)90163-j. [DOI] [PubMed] [Google Scholar]
- 31.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]