Abstract
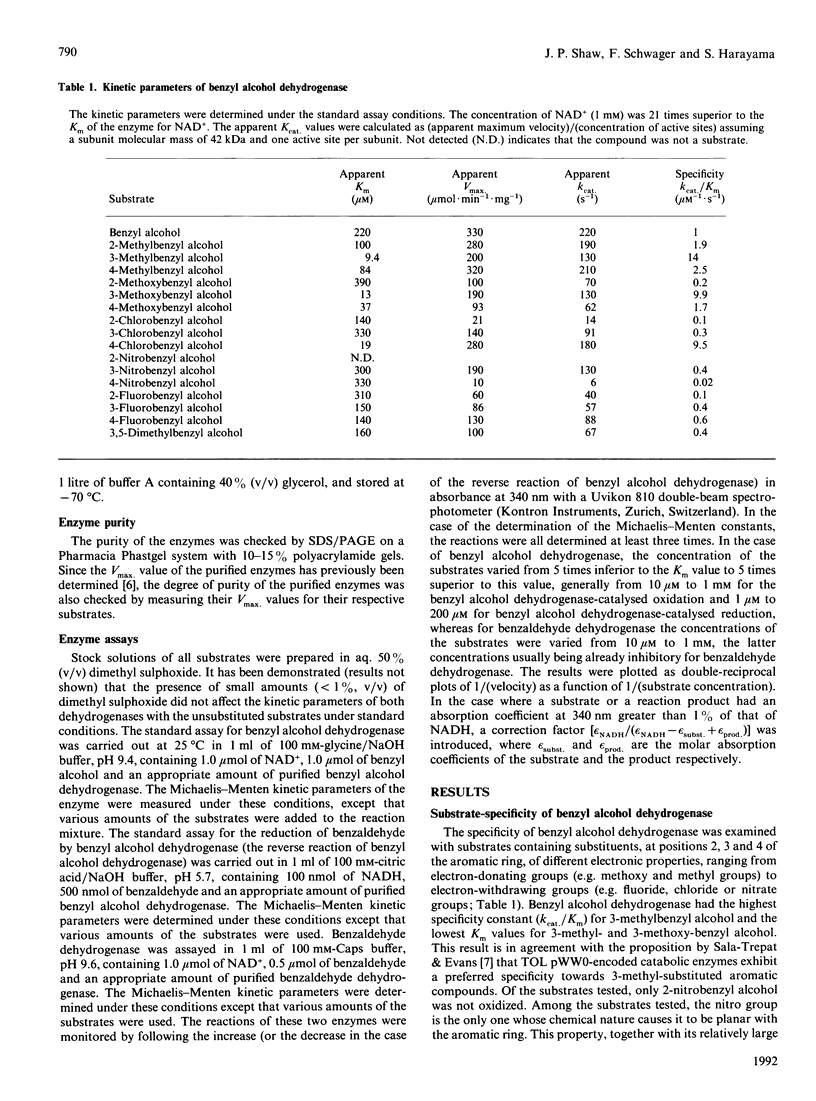
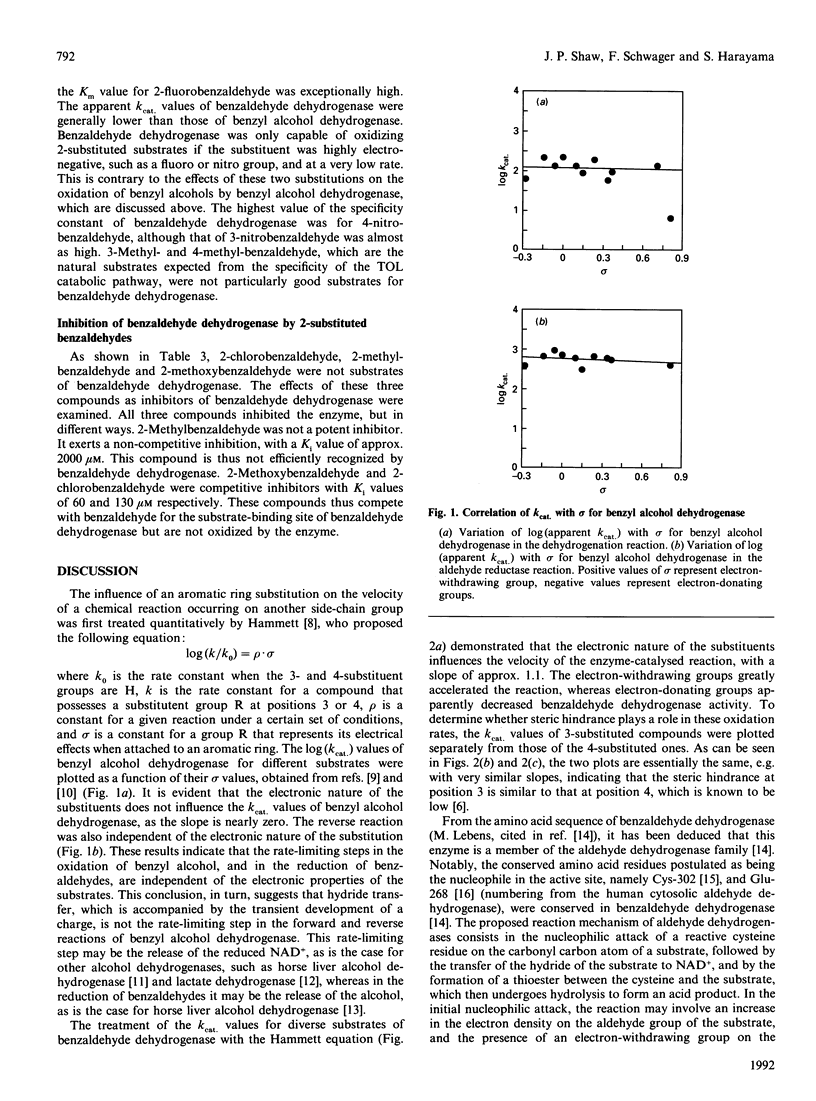
The substrate-specificities of benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase, encoded by TOL plasmid pWW0 of Pseudomonas putida mt-2, were determined. The rates of benzyl alcohol dehydrogenase-catalysed oxidation of substituted benzyl alcohols and reduction of substituted benzaldehydes were independent of the electronic nature of the substituents at positions 3 and 4. Substitutions at position 2 of benzyl alcohol affected the reactivity of benzyl alcohol dehydrogenase: the velocity of the benzyl alcohol dehydrogenase-catalysed oxidation was lower for compounds possessing electron-withdrawing substitutions. In the reverse reaction of benzyl alcohol dehydrogenase, none of the substitutions tested influenced the apparent kcat. values. The rates of benzaldehyde dehydrogenase-catalysed oxidation of substituted benzaldehydes were influenced by the electronic nature of the substitutions: electron-withdrawing groups at positions 3 and 4 favoured the oxidation of benzaldehydes. Substitution at position 2 of benzaldehyde greatly diminished the benzaldehyde dehydrogenase-catalysed oxidation. Substitution at position 2 with electron-donating groups essentially abolished reactivity, and only substitutions that were strongly electron-withdrawing, such as nitro and fluoro groups, permitted enzyme-catalysed oxidation. The influence of the electronic nature and the position of substitutions on the aromatic ring of the substrate on the velocity of the catalysed reactions provided some indications concerning the transition state during the oxidation of the substrates, and on the rate-limiting steps of the enzymes. Pseudomonas putida mt-2 containing TOL plasmid pWW0 cannot grow on toluene derivatives substituted at position 2, nor can it grow on 2-substituted benzyl alcohols or aldehydes. One of the reasons for this may be the substrate-specificities of the benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abriola D. P., Fields R., Stein S., MacKerell A. D., Jr, Pietruszko R. Active site of human liver aldehyde dehydrogenase. Biochemistry. 1987 Sep 8;26(18):5679–5684. doi: 10.1021/bi00392a015. [DOI] [PubMed] [Google Scholar]
- Bernhard S. A., Dunn M. F., Luisi P. L., Schack P. Mechanistic studies on equine liver alcohol dehydrogenase. I. The stoichiometry relationship of the coenzyme binding sites to the catalytic sites active in the reduction of aromatic aldehydes in the transient state. Biochemistry. 1970 Jan 6;9(1):185–192. doi: 10.1021/bi00803a024. [DOI] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Wubbolts M., Rose K., Leppik R. A., Timmis K. N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989 Sep;171(9):5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel J., Kaiser R., Jörnvall H. Mitochondrial aldehyde dehydrogenase from human liver. Primary structure, differences in relation to the cytosolic enzyme, and functional correlations. Eur J Biochem. 1985 Nov 15;153(1):13–28. doi: 10.1111/j.1432-1033.1985.tb09260.x. [DOI] [PubMed] [Google Scholar]
- Holbrook J. J., Lodola A., Illsley N. P. Histidine residues and the enzyme activity of pig heart supernatant malate dehydrogenase. Biochem J. 1974 Jun;139(3):797–800. doi: 10.1042/bj1390797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn J. M., Harayama S., Timmis K. N. DNA sequence determination of the TOL plasmid (pWWO) xylGFJ genes of Pseudomonas putida: implications for the evolution of aromatic catabolism. Mol Microbiol. 1991 Oct;5(10):2459–2474. doi: 10.1111/j.1365-2958.1991.tb02091.x. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5182–5186. doi: 10.1073/pnas.84.15.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitson T. M., Hill J. P., Midwinter G. G. Identification of a catalytically essential nucleophilic residue in sheep liver cytoplasmic aldehyde dehydrogenase. Biochem J. 1991 Apr 1;275(Pt 1):207–210. doi: 10.1042/bj2750207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh R. W., Fewson C. A. Benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II from Acinetobacter calcoaceticus. Substrate specificities and inhibition studies. Biochem J. 1988 Oct 15;255(2):653–661. [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Mermod N., Timmis K. N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987 Nov;1(3):293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Shaw J. P., Harayama S. Purification and characterisation of TOL plasmid-encoded benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase of Pseudomonas putida. Eur J Biochem. 1990 Aug 17;191(3):705–714. doi: 10.1111/j.1432-1033.1990.tb19179.x. [DOI] [PubMed] [Google Scholar]
- Tsai C. S., Senior D. J. Chemical studies of high-Km aldehyde dehydrogenase from rat liver mitochondria. Biochem Cell Biol. 1991 Feb-Mar;69(2-3):193–197. doi: 10.1139/o91-028. [DOI] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]


