Abstract
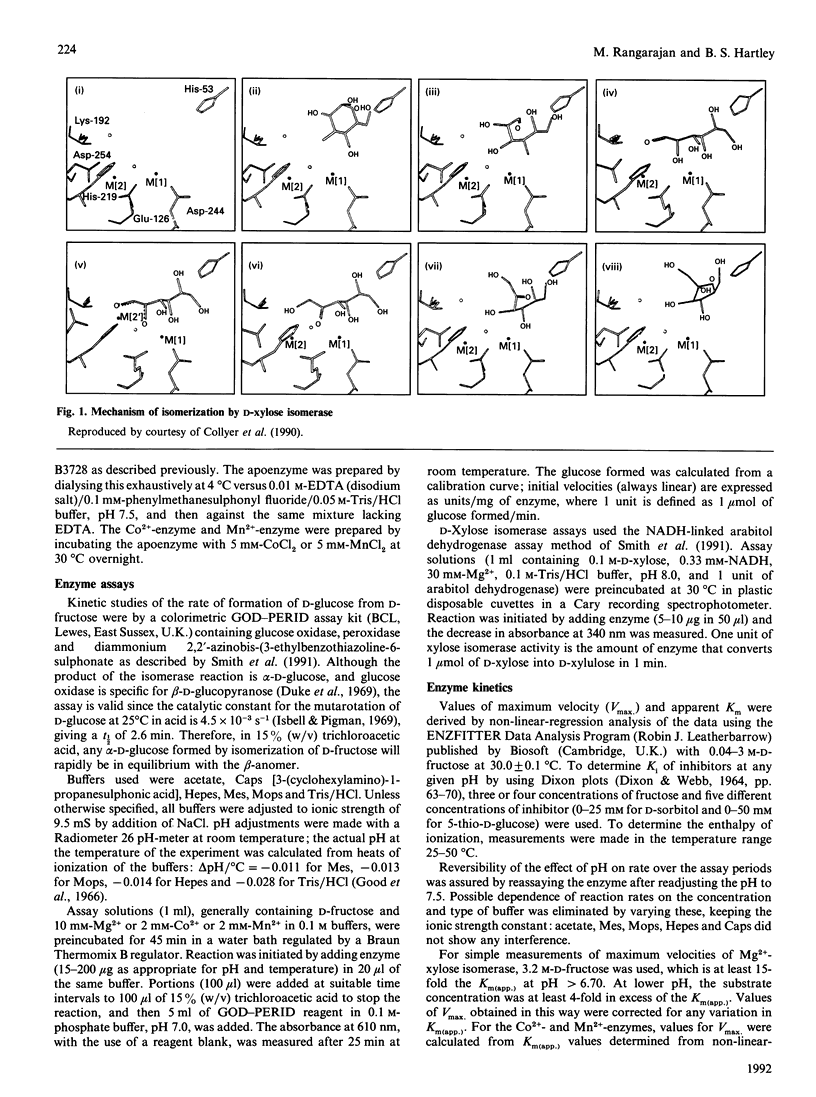
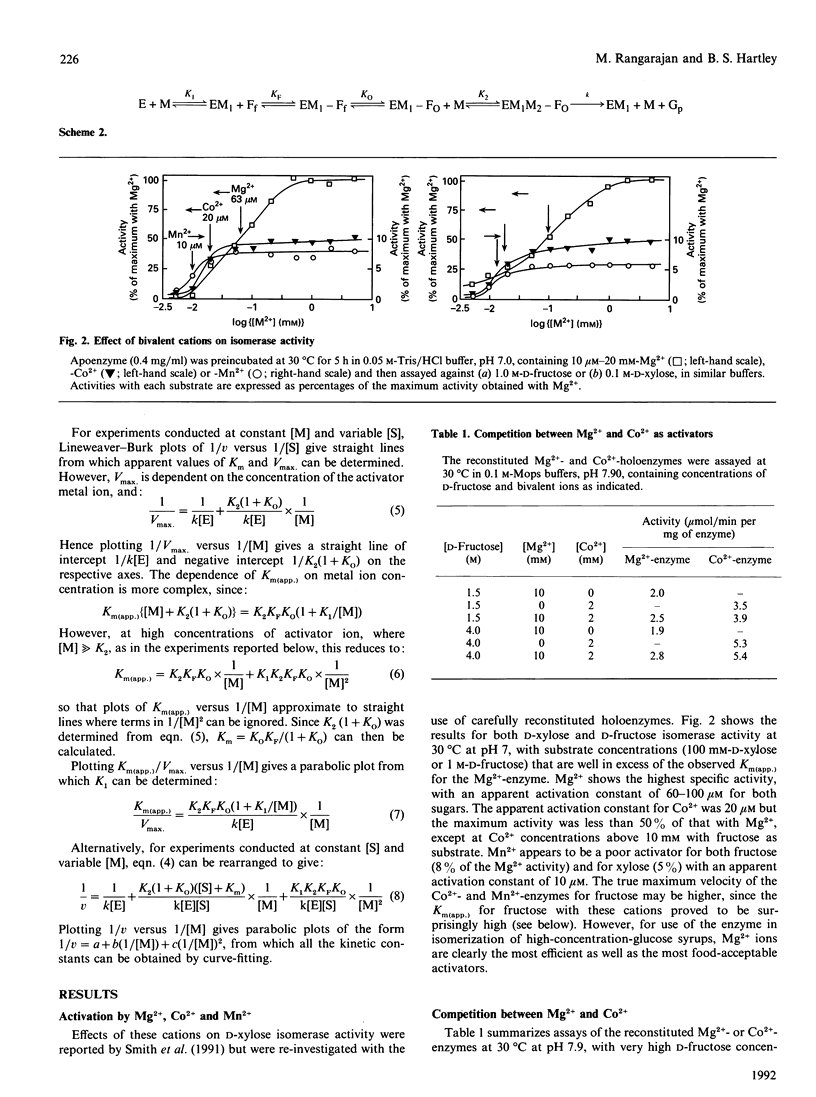
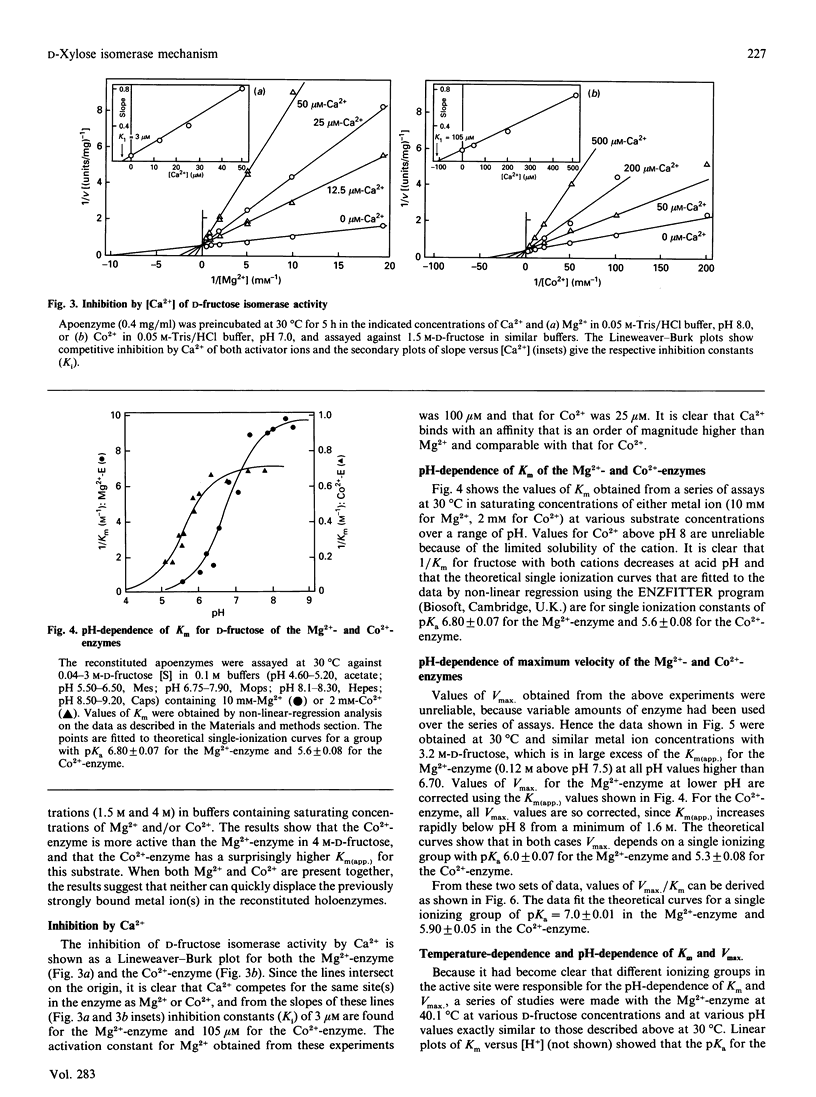
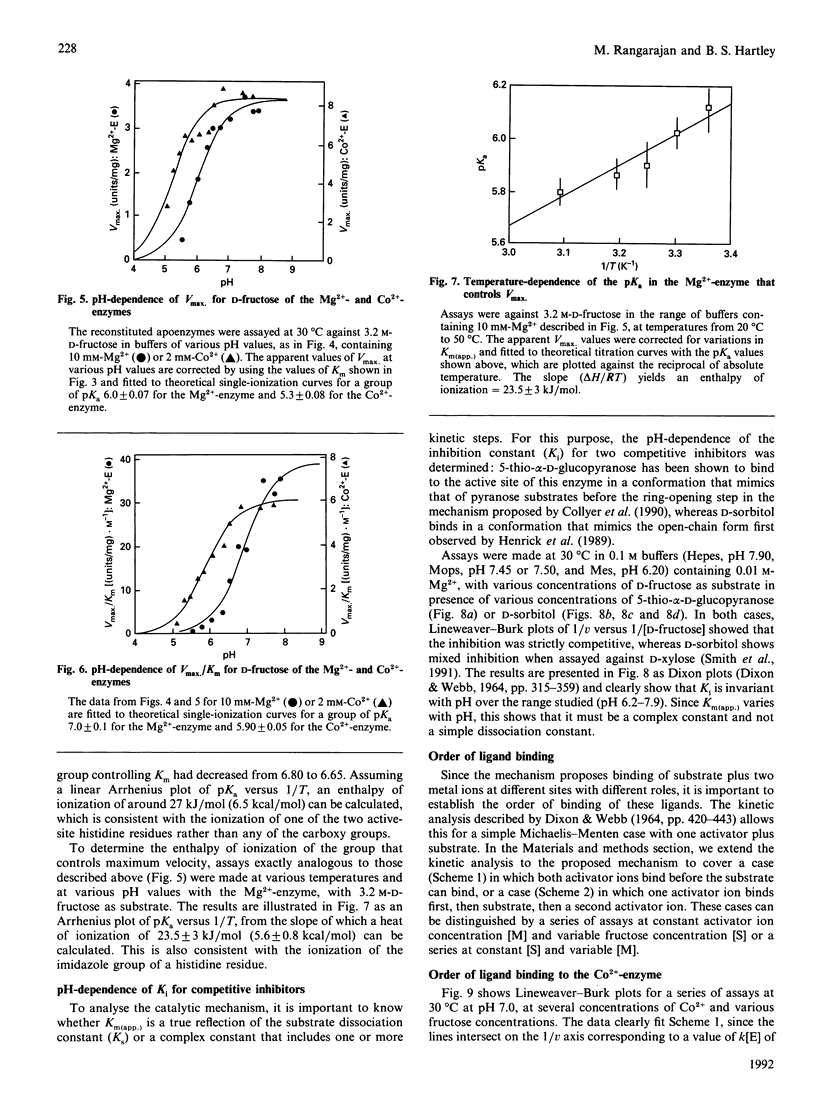
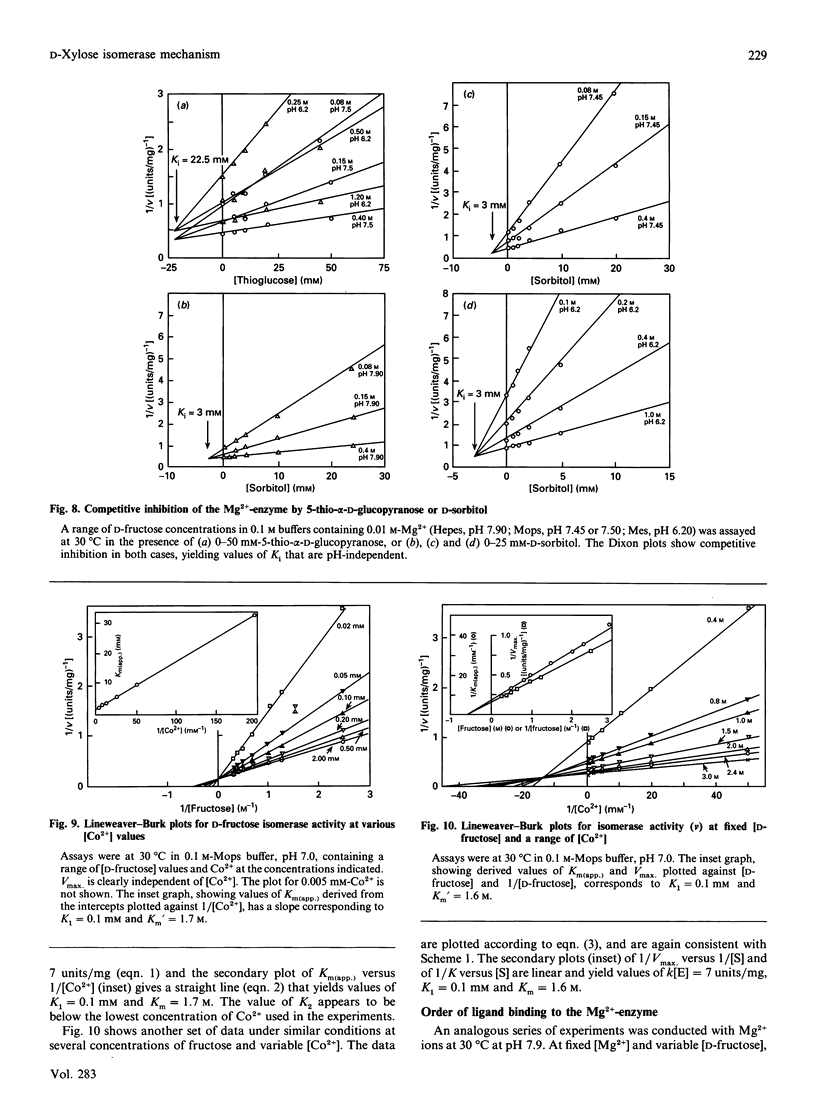
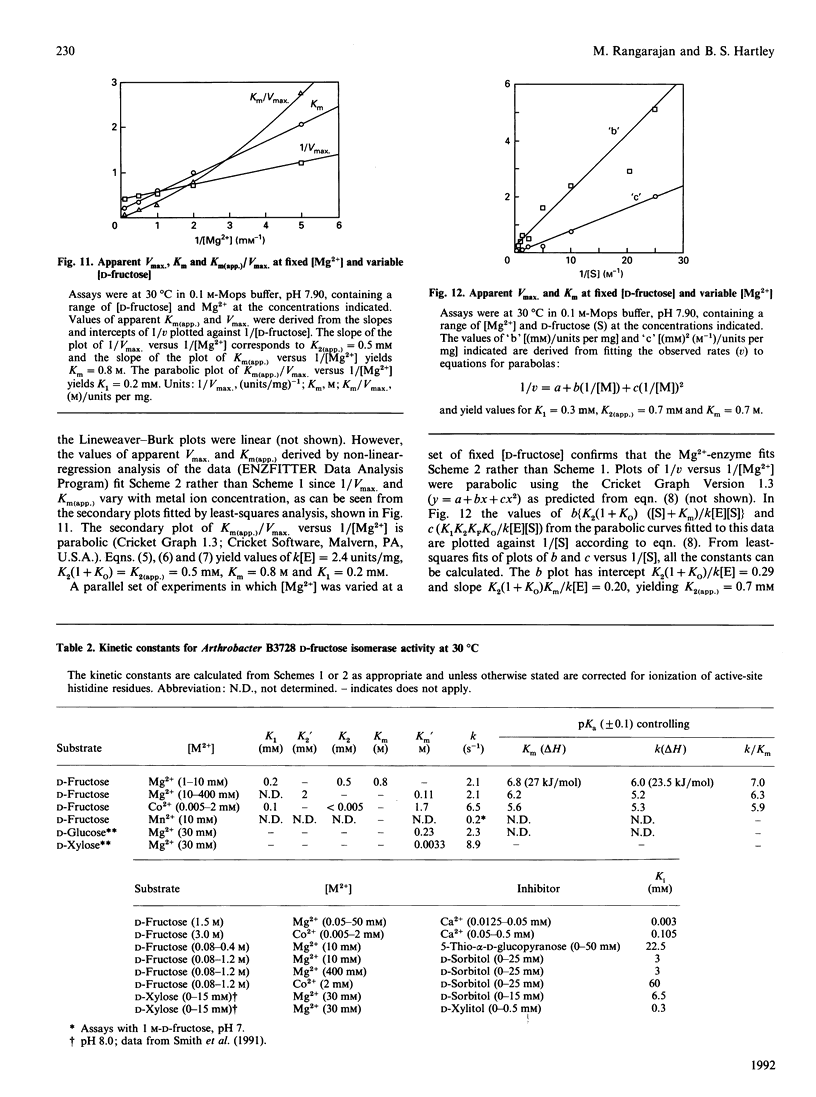
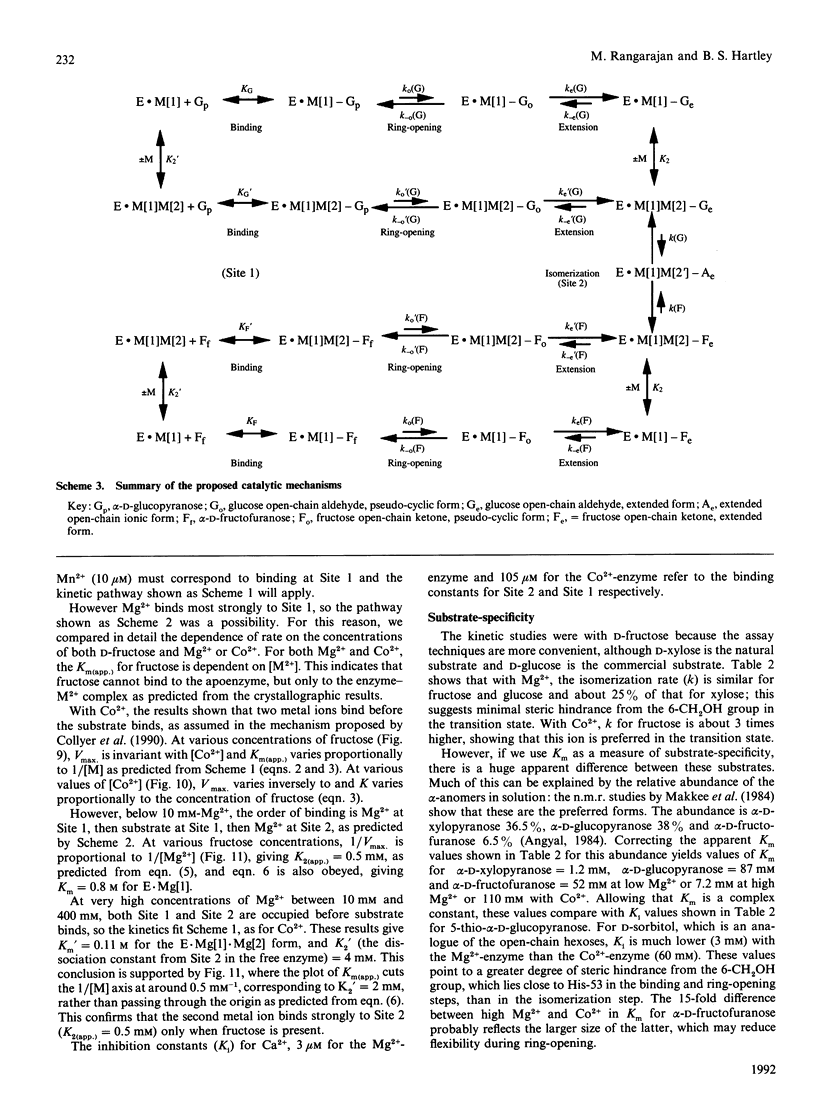
The mechanism of D-fructose isomerization by Arthrobacter D-xylose isomerase suggested from X-ray-crystallographic studies was tested by detailed kinetic analysis of the enzyme with various metal ions at different pH values and temperatures. At D-fructose concentrations used in commercial processes Mg2+ is the best activator with an apparent dissociation constant of 63 microM; Co2+ and Mn2+ bind more strongly (apparent Kd 20 microM and 10 microM respectively) but give less activity (45% and 8% respectively). Ca2+ is a strict competitive inhibitor versus Mg2+ (Ki 3 microM) or Co2+ (Ki 105 microM). The kinetics show a compulsory order of binding; Co2+ binds first to Site 2 and then to Site 1; then D-fructose binds at Site 1. At normal concentrations Mg2+ binds at Site 1, then D-fructose and then Mg2+ at Site 2. At very high Mg2+ concentrations (greater than 10 mM) the order is Mg2+ at Site 1, Mg2+ at Site 2, then D-fructose. The turnover rate (kcat.) is controlled by ionization of a residue with apparent pKa at 30 degrees C of 6.0 +/- 0.07 (Mg2+) or 5.3 +/- 0.08 (Co2+) and delta H = 23.5 kJ/mol. This appears to be His-219, which is co-ordinated to M[2]; protonation destroys isomerization by displacing M[2]; Co2+ binds more strongly at Site 2 than Mg2+, so competes more strongly against H+. The inhibition constant (Ki) for the two competitive inhibitors 5-thio-alpha-D-glucopyranose and D-sorbitol is invariant with pH, but Km(app.) in the Mg[1]-enzyme is controlled by ionization of a group with pKa 6.8 +/- 0.07 and delta H = 27 kJ/mol, which appears to be His-53. This shows that Km(app.) is a complex constant that includes the rate of the ring-opening step catalysed by His-53, which explains the pH-dependence. In the Mg[1]Mg[2]-enzyme or Co[1]Co[2]-enzyme, the pKa is lower (6.2 +/- 0.1 or 5.6 +/- 0.08) because of the extra adjacent cation. Hence the results fit the previously proposed pathway, but show that the mechanisms differ for Mg2+ and Co2+ and that the rate-limiting step is isomerization and not ring-opening as previously postulated.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blow D. M., Birktoft J. J., Hartley B. S. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969 Jan 25;221(5178):337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- Bock K., Meldal M., Meyer B., Wiebe L. Isomerization of D-glucose with glucose-isomerase. A mechanistic study. Acta Chem Scand B. 1983;37(2):101–108. doi: 10.3891/acta.chem.scand.37b-0101. [DOI] [PubMed] [Google Scholar]
- Callens M., Tomme P., Kersters-Hilderson H., Cornelis R., Vangrysperre W., De Bruyne C. K. Metal ion binding to D-xylose isomerase from Streptomyces violaceoruber. Biochem J. 1988 Feb 15;250(1):285–290. doi: 10.1042/bj2500285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collyer C. A., Henrick K., Blow D. M. Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift. J Mol Biol. 1990 Mar 5;212(1):211–235. doi: 10.1016/0022-2836(90)90316-E. [DOI] [PubMed] [Google Scholar]
- Good N. E., Winget G. D., Winter W., Connolly T. N., Izawa S., Singh R. M. Hydrogen ion buffers for biological research. Biochemistry. 1966 Feb;5(2):467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- Henrick K., Collyer C. A., Blow D. M. Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively. J Mol Biol. 1989 Jul 5;208(1):129–157. doi: 10.1016/0022-2836(89)90092-2. [DOI] [PubMed] [Google Scholar]
- Isbell H. S., Pigman W. Mutarotation of sugars in solution. II. Catalytic processes, isotope effects, reaction mechanisms, and biochemical aspects. Adv Carbohydr Chem Biochem. 1969;24:13–65. [PubMed] [Google Scholar]
- Lee C. Y., Bagdasarian M., Meng M. H., Zeikus J. G. Catalytic mechanism of xylose (glucose) isomerase from Clostridium thermosulfurogenes. Characterization of the structural gene and function of active site histidine. J Biol Chem. 1990 Nov 5;265(31):19082–19090. [PubMed] [Google Scholar]
- Loviny-Anderton T., Shaw P. C., Shin M. K., Hartley B. S. D-Xylose (D-glucose) isomerase from Arthrobacter strain N.R.R.L. B3728. Gene cloning, sequence and expression. Biochem J. 1991 Jul 1;277(Pt 1):263–271. doi: 10.1042/bj2770263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigman W., Isbell H. S. Mutarotation of sugars in solution. 1. History, basic kinetics, and composition of sugar solutions. Adv Carbohydr Chem Biochem. 1968;23:11–57. [PubMed] [Google Scholar]
- Smith C. A., Rangarajan M., Hartley B. S. D-Xylose (D-glucose) isomerase from Arthrobacter strain N.R.R.L. B3728. Purification and properties. Biochem J. 1991 Jul 1;277(Pt 1):255–261. doi: 10.1042/bj2770255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangrysperre W., Callens M., Kersters-Hilderson H., De Bruyne C. K. Evidence for an essential histidine residue in D-xylose isomerases. Biochem J. 1988 Feb 15;250(1):153–160. doi: 10.1042/bj2500153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangrysperre W., Van Damme J., Vandekerckhove J., De Bruyne C. K., Cornelis R., Kersters-Hilderson H. Localization of the essential histidine and carboxylate group in D-xylose isomerases. Biochem J. 1990 Feb 1;265(3):699–705. doi: 10.1042/bj2650699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. M., Schray K. J., Mildvan A. S. Proton magnetic relaxation studies of the interaction of D-xylose and xylitol with D-xylose isomerase. Characterization of metal-enzyme-substrate interactions. J Biol Chem. 1975 Dec 10;250(23):9021–9027. [PubMed] [Google Scholar]


