Abstract
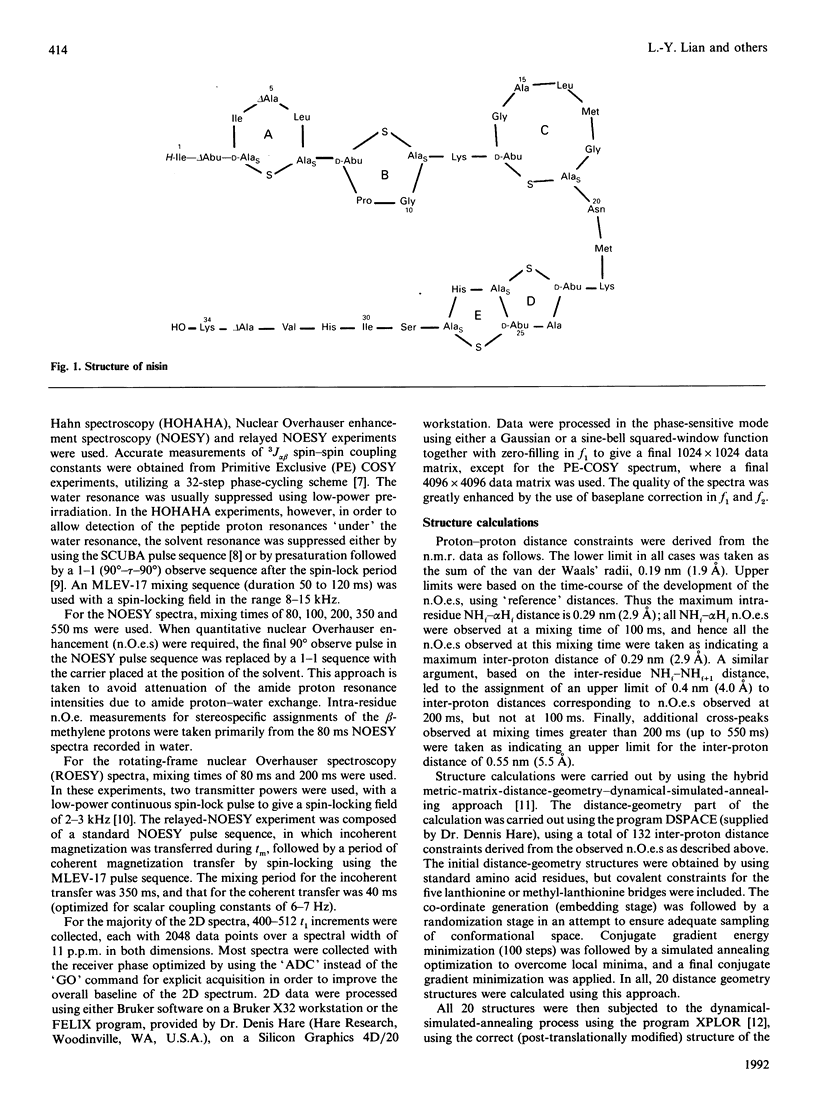
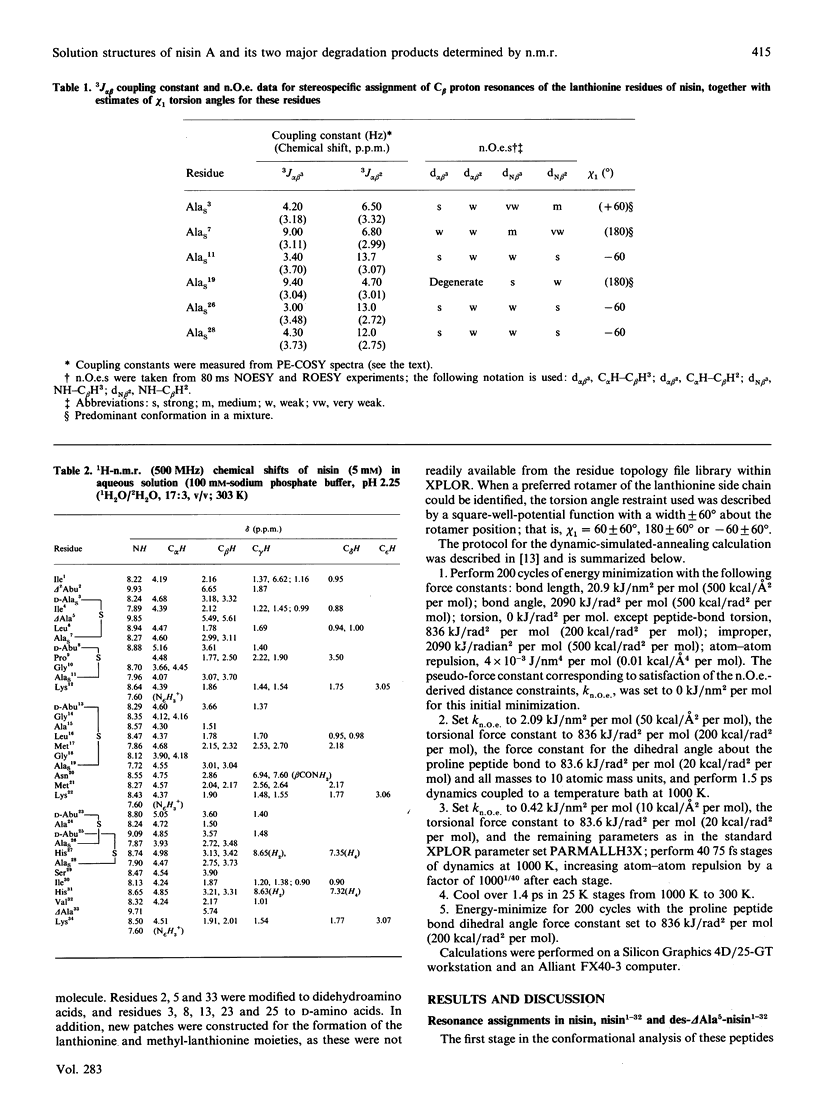
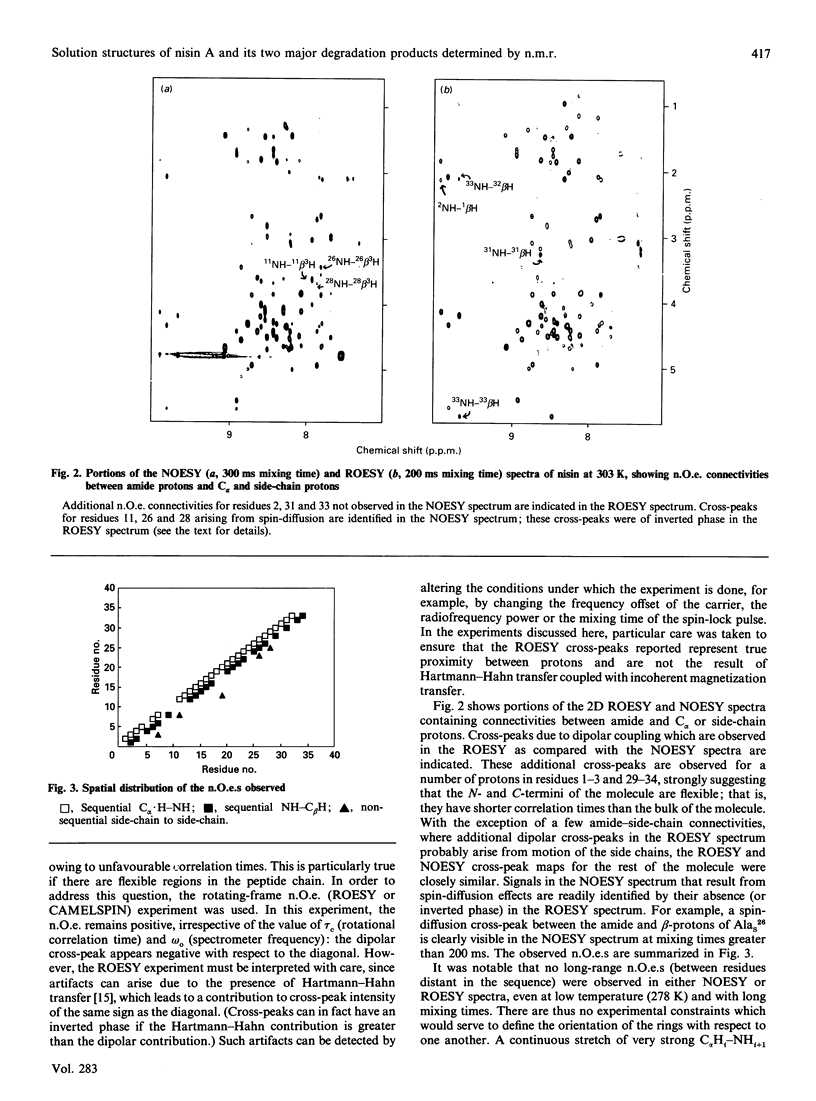
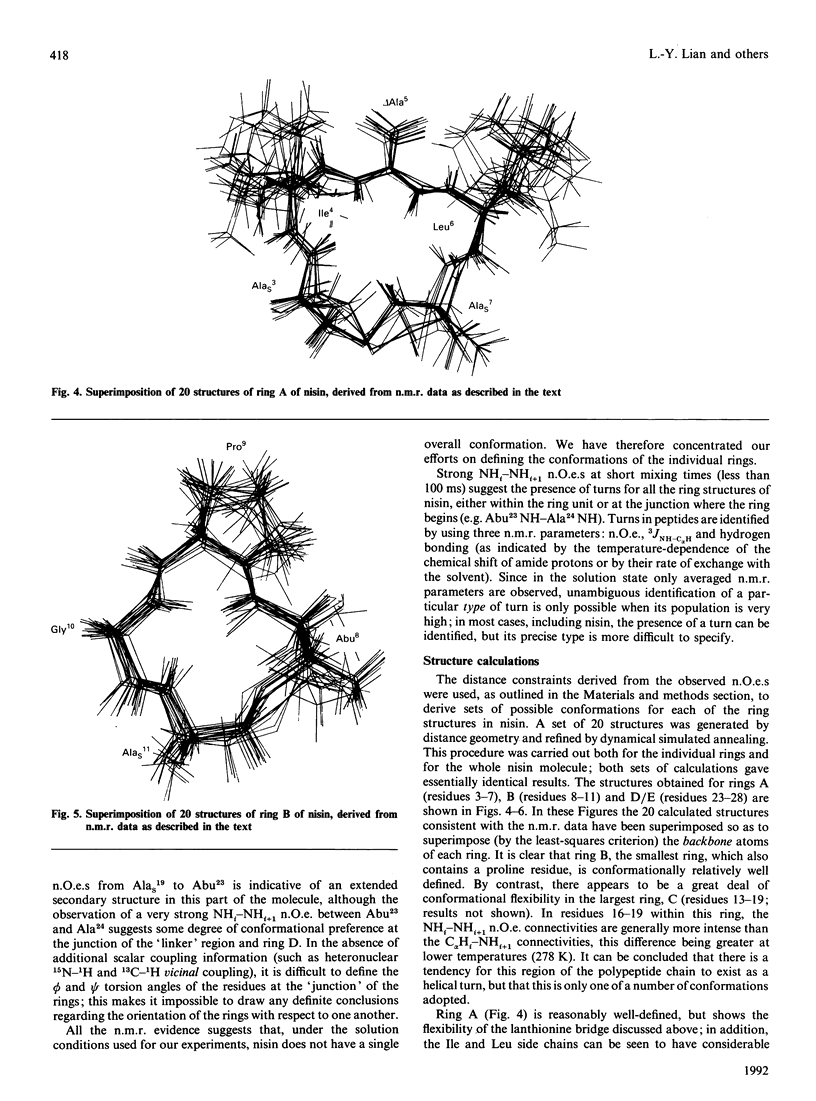
The conformations of nisin and two major degradation products, nisin-(1-32)-peptide (nisin1-32) and des-delta Ala5-nisin1-32 (where delta Ala is alpha beta-didehydroalanine), in aqueous solution have been determined from n.m.r. data. Sequential assignments of the peptides using correlation spectroscopy ('COSY'), homonuclear Hartmann-Hahn spectroscopy ('HOHAHA'), nuclear Overhauser enhancement spectroscopy (NOESY), relayed NOESY and rotating-frame nuclear Overhauser spectroscopy (ROESY) experiments are presented, including stereospecific assignments of beta-methylene protons of the lanthionine residues. ROESY experiments are also used to detect flexible regions in the polypeptide chain. A dynamic-stimulated-annealing approach is used for structural determination. It can be concluded that all these peptides are flexible in aqueous solution, with no experimental evidence of preferred overall conformations; the only defined conformational features are imposed by the presence of the lanthionine residues. Low-temperature studies also reveal that des-delta Ala5-nisin1-32 adopts conformations similar to those when the ring is intact, suggesting that the loss of activity of this degradation product is due to the absence of the delta Ala5 residue rather than to the conformational consequences of ring-opening.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basus V. J. Proton nuclear magnetic resonance assignments. Methods Enzymol. 1989;177:132–149. doi: 10.1016/0076-6879(89)77009-9. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Buchman G. W., Banerjee S., Hansen J. N. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988 Nov 5;263(31):16260–16266. [PubMed] [Google Scholar]
- Dodd H. M., Horn N., Gasson M. J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990 Mar;136(3):555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- Lian L. Y., Yang J. C., Derrick J. P., Sutcliffe M. J., Roberts G. C., Murphy J. P., Goward C. R., Atkinson T. Sequential 1H NMR assignments and secondary structure of an IgG-binding domain from protein G. Biochemistry. 1991 Jun 4;30(22):5335–5340. doi: 10.1021/bi00236a002. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Nilges M., Clore G. M., Gronenborn A. M. Determination of three-dimensional structures of proteins from interproton distance data by hybrid distance geometry-dynamical simulated annealing calculations. FEBS Lett. 1988 Mar 14;229(2):317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]


