Abstract
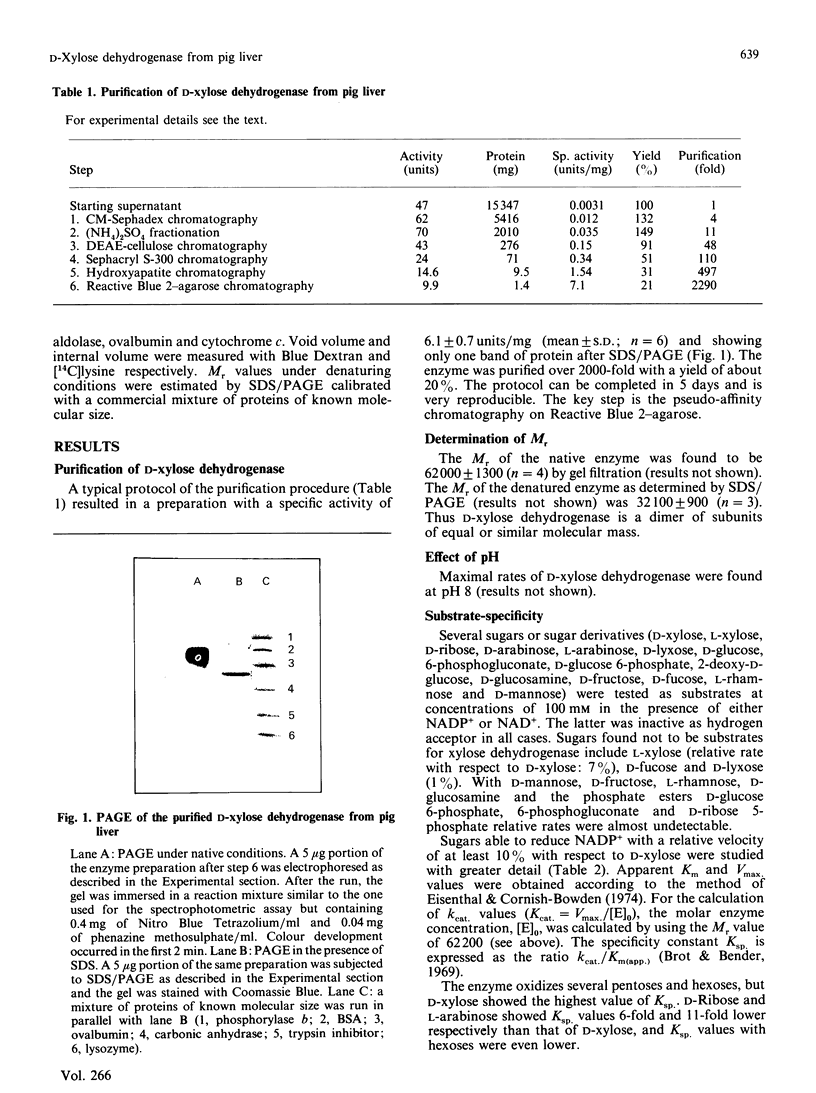
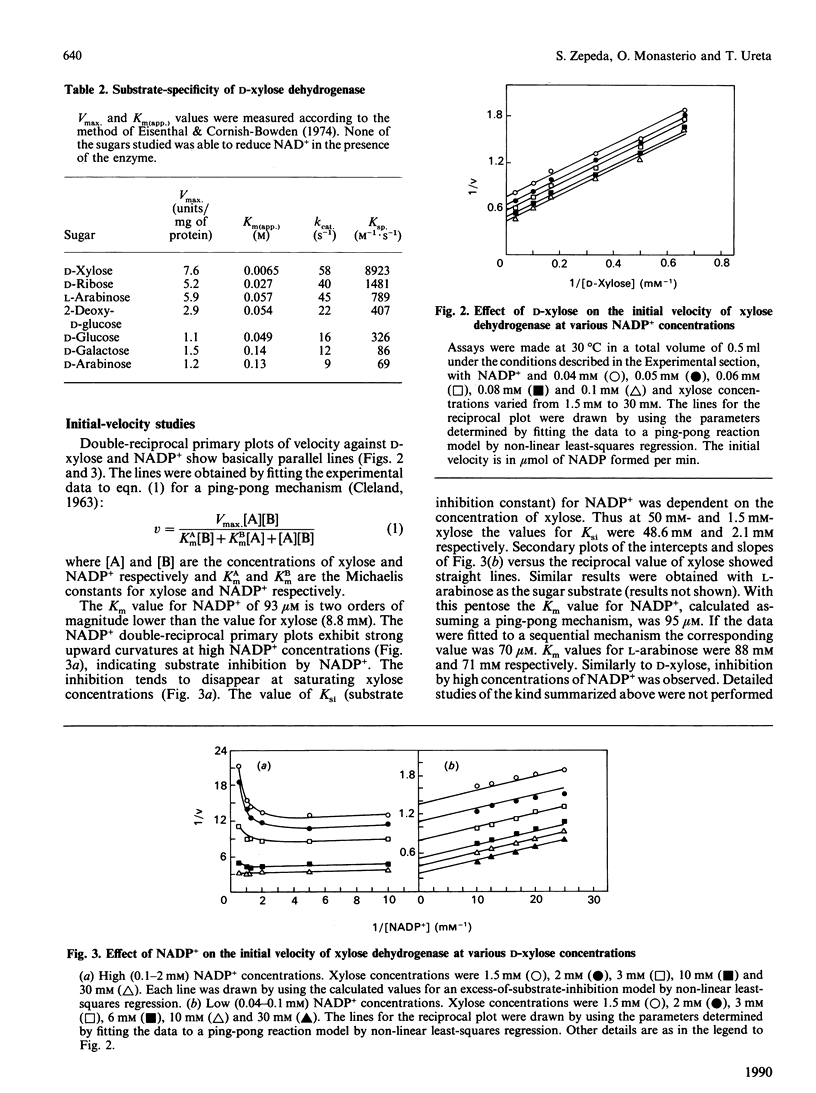
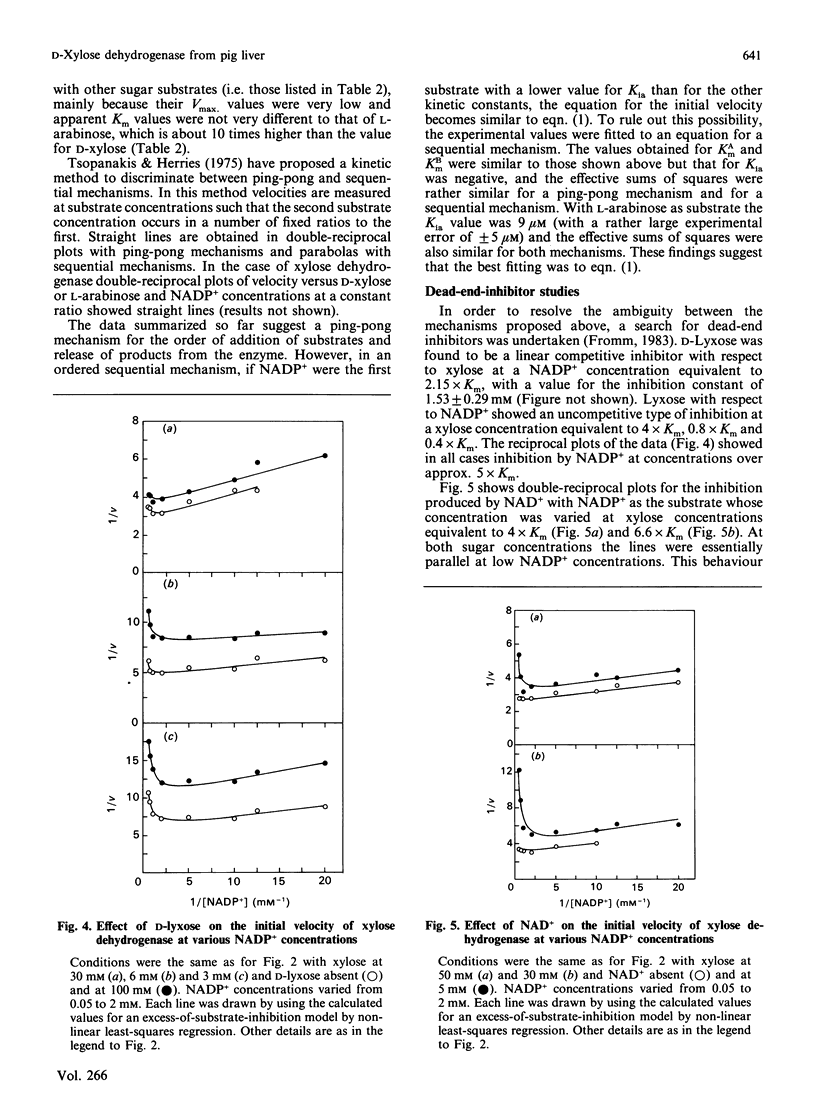
An NADP(+)-dependent D-xylose dehydrogenase from pig liver cytosol was purified about 2000-fold to apparent homogeneity with a yield of 15% and specific activity of 6 units/mg of protein. An Mr value of 62,000 was obtained by gel filtration. PAGE in the presence of SDS gave an Mr value of 32,000, suggesting that the native enzyme is a dimer of similar or identical subunits. D-Xylose, D-ribose, L-arabinose, 2-deoxy-D-glucose, D-glucose and D-mannose were substrates in the presence of NADP+ but the specificity constant (ratio kcat./Km(app.)) is, by far, much higher for D-xylose than for the other sugars. The enzyme is specific for NADP+; NAD+ is not reduced in the presence of D-xylose or other sugars. Initial-velocity studies for the forward direction with xylose or NADP+ concentrations varied at fixed concentrations of the nucleotide or the sugar respectively revealed a pattern of parallel lines in double-reciprocal plots. Km values for D-xylose and NADP+ were 8.8 mM and 0.99 mM respectively. Dead-end inhibition studies to confirm a ping-pong mechanism showed that NAD+ acted as an uncompetitive inhibitor versus NADP+ (Ki 5.8 mM) and as a competitive inhibitor versus xylose. D-Lyxose was a competitive inhibitor versus xylose and uncompetitive versus NADP+. These results fit better to a sequential compulsory ordered mechanism with NADP+ as the first substrate, but a ping-pong mechanism with xylose as the first substrate has not been ruled out. The presence of D-xylose dehydrogenase suggests that in mammalian liver D-xylose is utilized by a pathway other than the pentose phosphate pathway.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aird B. A., Heinrikson R. L., Westley J. Isolation and characterization of a prokaryotic sulfurtransferase. J Biol Chem. 1987 Dec 25;262(36):17327–17335. [PubMed] [Google Scholar]
- Allen S. H., Patil J. R. Studies on the structure and mechanism of action of the malate-lactate transhydrogenase. J Biol Chem. 1972 Feb 10;247(3):909–916. [PubMed] [Google Scholar]
- Baumann P., Wright B. E. The phosphofructokinase of Dictyostelium discoideum. Biochemistry. 1968 Oct;7(10):3653–3661. doi: 10.1021/bi00850a044. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- CLELAND W. W. The kinetics of enzyme-catalyzed reactions with two or more substrates or products. I. Nomenclature and rate equations. Biochim Biophys Acta. 1963 Jan 8;67:104–137. doi: 10.1016/0006-3002(63)91800-6. [DOI] [PubMed] [Google Scholar]
- Carper W. R., Chang K. W., Thorpe W. G., Carper M. A., Buess C. M. Arabinose (fucose) dehydrogenase from pig liver. II. Steady-state kinetics. Biochim Biophys Acta. 1974 Jul 17;358(1):49–56. doi: 10.1016/0005-2744(74)90257-5. [DOI] [PubMed] [Google Scholar]
- Carper W. R., Toews M. L., Thompson R. E., Buess C. M. A kinetic study of pig liver glucose dehydrogenase. Arch Biochem Biophys. 1976 Jul;175(1):312–320. doi: 10.1016/0003-9861(76)90513-0. [DOI] [PubMed] [Google Scholar]
- Carrasco A., Pincheira G., Ureta T. Genetic and biochemical characterization of D-arabinose dehydrogenase from Neurospora crassa. J Bacteriol. 1981 Jan;145(1):164–170. doi: 10.1128/jb.145.1.164-170.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline A. L., Hu A. S. Enzymatic characterization and comparison of three sugar dehydrogenases from a pseudomonad. J Biol Chem. 1965 Nov;240(11):4493–4497. [PubMed] [Google Scholar]
- Cline A. L., Hu A. S. Some physical properties of three sugar dehydrogenases from a pseudomonad. J Biol Chem. 1965 Nov;240(11):4498–4502. [PubMed] [Google Scholar]
- Cline A. L., Hu A. S. The isolation of three sugar dehydrogenases from a psuedomonad. J Biol Chem. 1965 Nov;240(11):4488–4492. [PubMed] [Google Scholar]
- Cornish-Bowden A., Endrenyi L. Robust regression of enzyme kinetic data. Biochem J. 1986 Feb 15;234(1):21–29. doi: 10.1042/bj2340021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- DOUDOROFF M., PALLERONI N. J. Characterization and properties of 2-keto-3-deoxy-D-arabonic acid. J Biol Chem. 1956 Nov;223(1):499–508. [PubMed] [Google Scholar]
- Eisenthal R., Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974 Jun;139(3):715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Hiyama N. Isolation and characterization of L-fucose dehydrogenase from rabbit liver. J Biochem. 1979 Nov;86(5):1559–1565. doi: 10.1093/oxfordjournals.jbchem.a132673. [DOI] [PubMed] [Google Scholar]
- Gershman H., Abeles R. H. Deuterium isotope effects in the oxidation of alcohols in vitro and in vivo. Arch Biochem Biophys. 1973 Feb;154(2):659–674. doi: 10.1016/0003-9861(73)90021-0. [DOI] [PubMed] [Google Scholar]
- Gold M. H., Segel I. H. Neurospora crassa protein kinase. Purification, properties, and kinetic mechanism. J Biol Chem. 1974 Apr 25;249(8):2417–2423. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maijub A. G., Pecht M. A., Miller G. R., Carper W. R. Arabinose (fucose) dehydrogenase from pig liver. I. Isolation and characterization. Biochim Biophys Acta. 1973 Jul 5;315(1):37–42. doi: 10.1016/0005-2744(73)90126-5. [DOI] [PubMed] [Google Scholar]
- Metzger R. P., Copp E. F., Metzger S. A., Wick A. N. Dehydrogenation of glucose and xylose catalyzed by rat and sheep liver microsomal and soluble fractions. Metabolism. 1970 Aug;19(8):587–597. doi: 10.1016/0026-0495(70)90015-6. [DOI] [PubMed] [Google Scholar]
- Metzger R. P., Wick A. N. Partial purification of rat liver D-arabinose dehydrogenase. Biochem Biophys Res Commun. 1967 Mar 21;26(6):742–747. doi: 10.1016/s0006-291x(67)80136-0. [DOI] [PubMed] [Google Scholar]
- Mobley P. W., Metzger R. P. The physical properties NAD-dependent L-fucose dehydrogenase from sheep liver. Arch Biochem Biophys. 1978 Feb;186(1):184–188. doi: 10.1016/0003-9861(78)90478-2. [DOI] [PubMed] [Google Scholar]
- Mobley P. W., Metzger R. P., Wick A. N. NAD-dependent L-fucose dehydrogenase from sheep liver. Arch Biochem Biophys. 1970 Jul;139(1):83–86. doi: 10.1016/0003-9861(70)90047-0. [DOI] [PubMed] [Google Scholar]
- Newton M. F., Nash H. R., Peters J., Andrews S. J. Xylose dehydrogenase-1, a new gene on mouse chromosome 7. Biochem Genet. 1982 Aug;20(7-8):733–745. doi: 10.1007/BF00483970. [DOI] [PubMed] [Google Scholar]
- Pincheira G., Leon G., Ureta T. Aldosugar dehydrogenases from Neurospora crassa Partial purification and characterization of D-arabinose: NAD dehydrogenase. FEBS Lett. 1973 Feb 15;30(1):111–114. doi: 10.1016/0014-5793(73)80630-1. [DOI] [PubMed] [Google Scholar]
- Purich D. L., Fromm H. J. The kinetics and regulation of rat brain hexokinase. J Biol Chem. 1971 Jun 10;246(11):3456–3463. [PubMed] [Google Scholar]
- Ropson I. J., Powers D. A. A novel dehydrogenase reaction mechanism for hexose-6-phosphate dehydrogenase isolated from the teleost Fundulus heteroclitus. J Biol Chem. 1988 Aug 25;263(24):11697–11703. [PubMed] [Google Scholar]
- SEGAL S., FOLEY J. B. The metabolic fate of C14 labeled pentoses in man. J Clin Invest. 1959 Feb;38(2):407–413. doi: 10.1172/JCI103815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H., Sarney J., McGuire E. J., Roseman S. Isolation of diphosphopyridine nucleotide-dependent L-fucose dehydrogenase from pork liver. J Biol Chem. 1969 Sep 10;244(17):4785–4792. [PubMed] [Google Scholar]
- Schiwara H. W., Domschke W., Domagk G. F. Uber die Zucker-Dehydrogenasen in der Säugetierleber. I. Differenzierung verschiedener Zucker-Dehydrogenasen in der Schweineleber durch Disk-Electrophorese und Ionenaustausch-cnromatographie. Hoppe Seylers Z Physiol Chem. 1968 Nov;349(11):1575–1581. [PubMed] [Google Scholar]
- Ureta T., Radojković J. Chromatographic and electrophoretic evidence for several sugar dehydrogenases in mammalian liver. FEBS Lett. 1970 Jul 29;9(2):57–60. doi: 10.1016/0014-5793(70)80311-8. [DOI] [PubMed] [Google Scholar]
- VAN HEYNINGEN R. Metabolism of xylose by the lens; calf lens in vitro. Biochem J. 1958 Aug;69(4):481–491. doi: 10.1042/bj0690481a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen R., Schachter H. L-Fucose metabolism in mammals. I. Port liver L-fuconate hydro-lyase. Can J Biochem. 1972 Jul;50(7):798–806. doi: 10.1139/o72-111. [DOI] [PubMed] [Google Scholar]



