Abstract
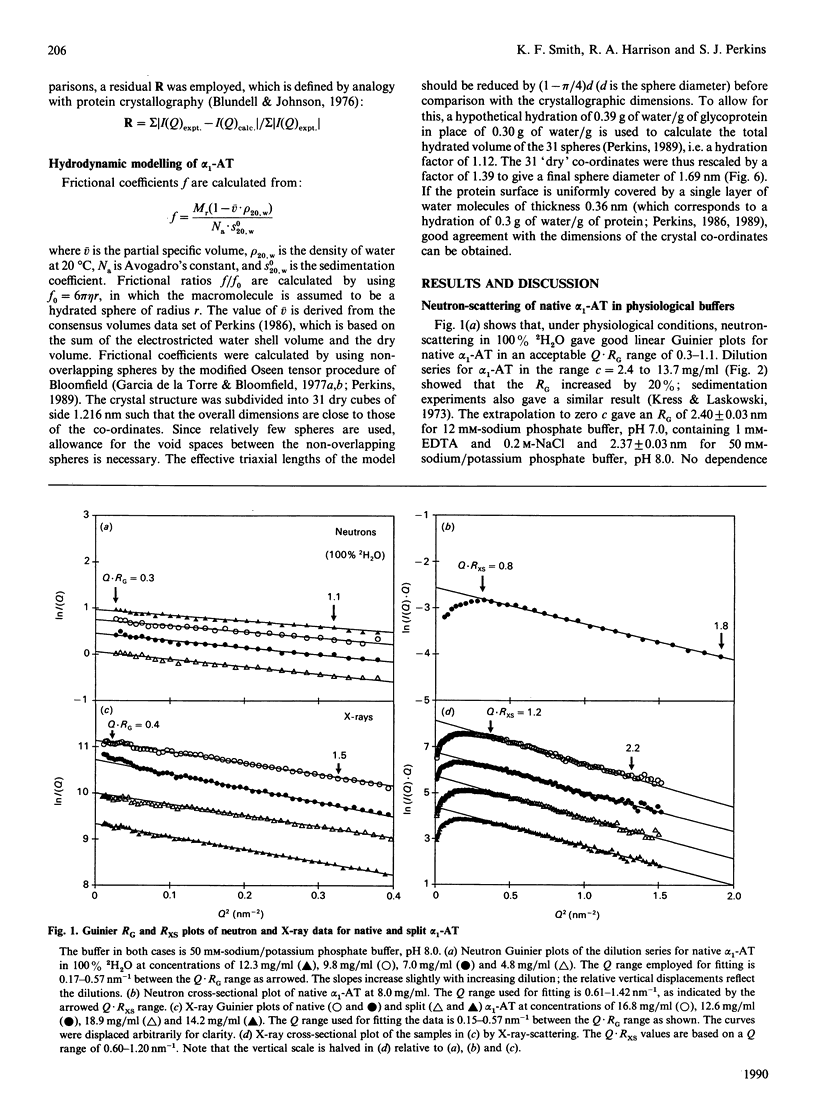
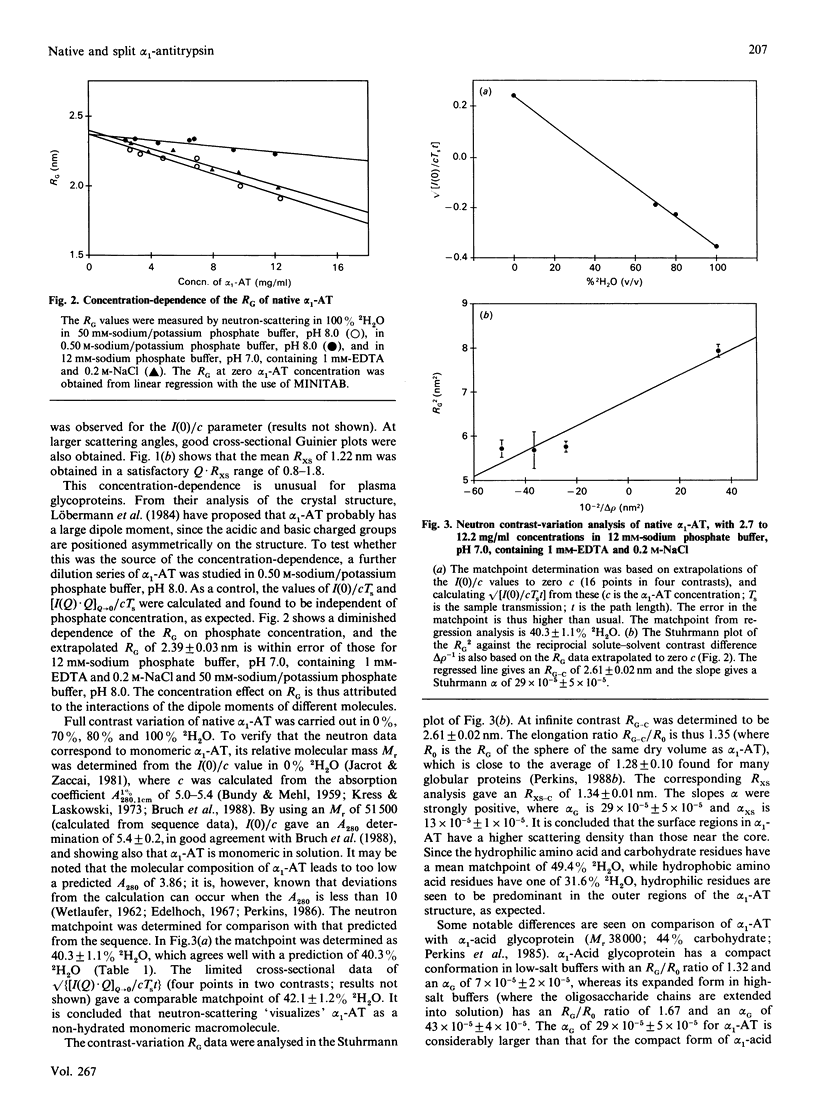
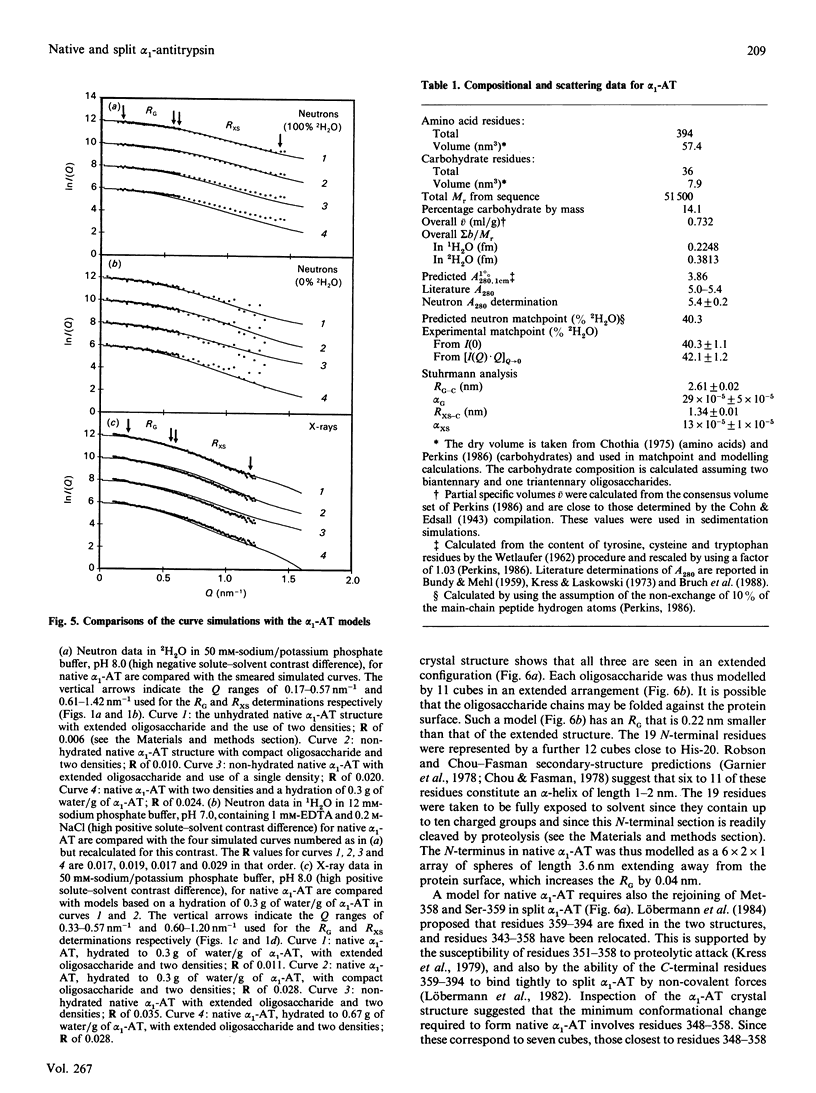
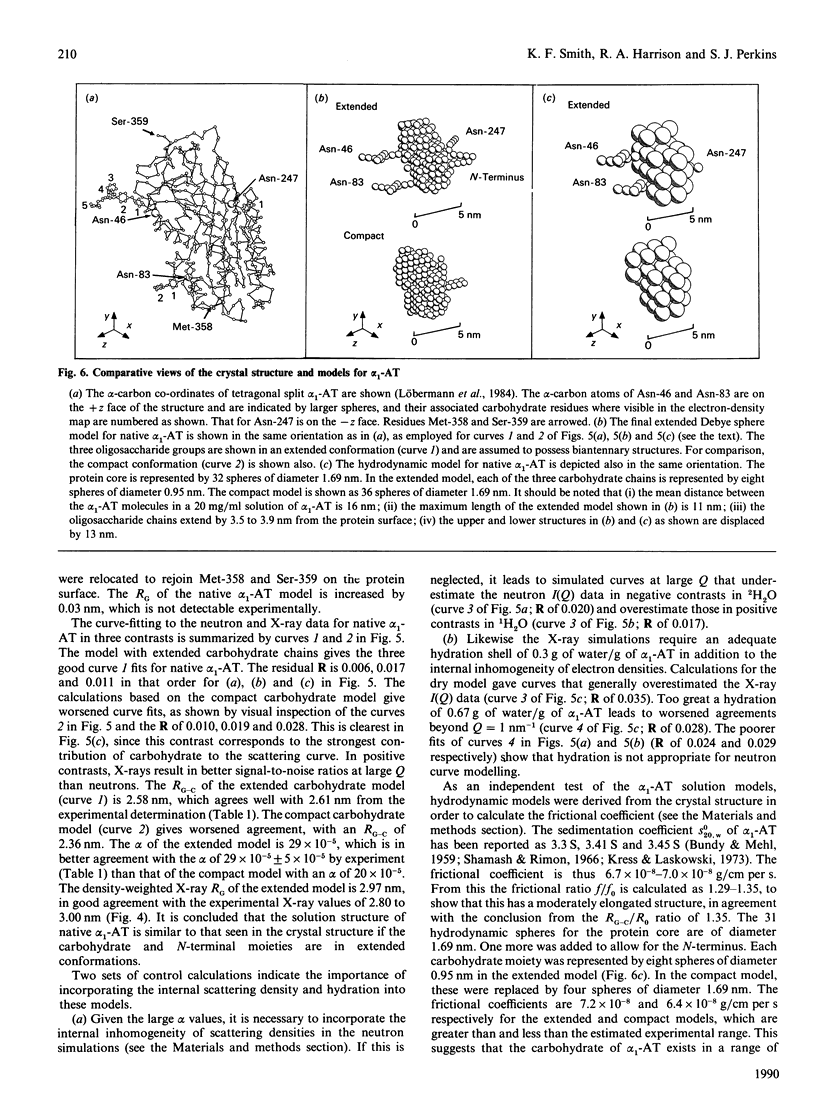
alpha 1-Antitrypsin is the best-characterized member of the serpin (serine-proteinase inhibitor) superfamily. Its solution structure was studied by high-flux neutron-scattering and synchrotron X-ray-scattering. Neutron data show that its absorption coefficient A1% 280,1cm is 5.4. The neutron radius of gyration RG at infinite contrast for native alpha 1-antitrypsin is 2.61 nm, characteristic of a moderately elongated structure, and its cross-sectional RG is 1.34 nm. The internal inhomogeneity of scattering densities within alpha 1-antitrypsin is high at 29 x 10(-5). The X-ray RG is 2.91 nm, in good agreement with the neutron RG of 2.82 nm in 1H2O. This RG is unchanged in reactive-centre-cleaved alpha 1-antitrypsin. These parameters are also unchanged at pH 8 in sodium/potassium phosphate buffers up to 0.6 M. The neutron and X-ray curves for native alpha 1-antitrypsin were compared with Debye simulation based on the crystal structure of reactive-centre-cleaved (papain) alpha 1-antitrypsin. After allowance for residues not visible in the crystallographic electron-density map, and rejoining the proteolysed site between Met-358 and Ser-359 by means of a relatively minor conformational re-arrangement, good agreement to a structural resolution of 4 nm is obtained with the neutron data in two contrasts and with the X-ray data. The structures of the native and cleaved forms of alpha 1-antitrypsin are thus similar within the resolution of solution scattering. This places an upper limit on the magnitude of the presumed conformational changes that occur in alpha 1-antitrypsin on reactive-centre cleavage, as indicated in earlier spectroscopic investigations of the Met-358-Ser-359 peptide-bond cleavage. Methods for scattering-curve simulations from crystal structures are critically assessed. The RG data lead to dimensions of 7.8 nm x 4.9 nm x 2.2 nm for native alpha 1-antitrypsin. The high internal inhomogeneity and the asymmetric shorter semi-axes of 4.9 nm and 2.2 nm suggest that the three oligosaccharide chains of alpha 1-antitrypsin are essentially freely extended into solvent in physiological conditions. This conclusion is also supported by the Debye simulations, and by modelling based on hydrodynamic parameters.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BUNDY H. F., MEHL J. W. Trypsin inhibitors of human serum. II. Isolation of the alpha 1-inhibitor and its partial characterization. J Biol Chem. 1959 May;234(5):1124–1128. [PubMed] [Google Scholar]
- Baldwin J., Chothia C. Haemoglobin: the structural changes related to ligand binding and its allosteric mechanism. J Mol Biol. 1979 Apr 5;129(2):175–220. doi: 10.1016/0022-2836(79)90277-8. [DOI] [PubMed] [Google Scholar]
- Bao J. J., Sifers R. N., Kidd V. J., Ledley F. D., Woo S. L. Molecular evolution of serpins: homologous structure of the human alpha 1-antichymotrypsin and alpha 1-antitrypsin genes. Biochemistry. 1987 Dec 1;26(24):7755–7759. doi: 10.1021/bi00398a033. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bock S. C., Skriver K., Nielsen E., Thøgersen H. C., Wiman B., Donaldson V. H., Eddy R. L., Marrinan J., Radziejewska E., Huber R. Human C1 inhibitor: primary structure, cDNA cloning, and chromosomal localization. Biochemistry. 1986 Jul 29;25(15):4292–4301. doi: 10.1021/bi00363a018. [DOI] [PubMed] [Google Scholar]
- Bruch M., Weiss V., Engel J. Plasma serine proteinase inhibitors (serpins) exhibit major conformational changes and a large increase in conformational stability upon cleavage at their reactive sites. J Biol Chem. 1988 Nov 15;263(32):16626–16630. [PubMed] [Google Scholar]
- Carrell R. W., Boswell D. R., Brennan S. O., Owen M. C. Active site of alpha 1-antitrypsin: homologous site in antithrombin-III. Biochem Biophys Res Commun. 1980 Mar 28;93(2):399–402. doi: 10.1016/0006-291x(80)91090-6. [DOI] [PubMed] [Google Scholar]
- Carrell R. W., Jeppsson J. O., Laurell C. B., Brennan S. O., Owen M. C., Vaughan L., Boswell D. R. Structure and variation of human alpha 1-antitrypsin. Nature. 1982 Jul 22;298(5872):329–334. doi: 10.1038/298329a0. [DOI] [PubMed] [Google Scholar]
- Carrell R. W., Jeppsson J. O., Vaughan L., Brennan S. O., Owen M. C., Boswell D. R. Human alpha 1-antitrypsin: carbohydrate attachment and sequence homology. FEBS Lett. 1981 Dec 7;135(2):301–303. doi: 10.1016/0014-5793(81)80805-8. [DOI] [PubMed] [Google Scholar]
- Carrell R. W., Owen M. C. Plakalbumin, alpha 1-antitrypsin, antithrombin and the mechanism of inflammatory thrombosis. Nature. 1985 Oct 24;317(6039):730–732. doi: 10.1038/317730a0. [DOI] [PubMed] [Google Scholar]
- Chandra T., Stackhouse R., Kidd V. J., Robson K. J., Woo S. L. Sequence homology between human alpha 1-antichymotrypsin, alpha 1-antitrypsin, and antithrombin III. Biochemistry. 1983 Oct 25;22(22):5055–5061. doi: 10.1021/bi00291a001. [DOI] [PubMed] [Google Scholar]
- Chothia C. Structural invariants in protein folding. Nature. 1975 Mar 27;254(5498):304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Conrad H., Mayer A., Thomas H. P., Vogel H. X-ray small-angle scattering from aqueous solutions of oxy-and deoxyhaemoglobin. J Mol Biol. 1969 Apr;41(2):225–229. doi: 10.1016/0022-2836(69)90387-8. [DOI] [PubMed] [Google Scholar]
- Cusack S. Instrumental effects on the scattering curves. J Mol Biol. 1981 Jan 25;145(3):539–541. [PubMed] [Google Scholar]
- De La Torre J. G., Bloomfield V. A. Hydrodynamics of macromolecular complexes. III. Bacterial viruses. Biopolymers. 1977 Aug;16(8):1779–1793. doi: 10.1002/bip.1977.360160813. [DOI] [PubMed] [Google Scholar]
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967 Jul;6(7):1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Hervé G., Moody M. F., Tauc P., Vachette P., Jones P. T. Quaternary structure changes in aspartate transcarbamylase studied by X-ray solution scattering. Signal transmission following effector binding. J Mol Biol. 1985 Sep 5;185(1):189–199. doi: 10.1016/0022-2836(85)90190-1. [DOI] [PubMed] [Google Scholar]
- Ibel K. Comparison of neutron and X-ray scattering of dilute myoglobin solutions. J Mol Biol. 1975 Apr 5;93(2):255–265. doi: 10.1016/0022-2836(75)90131-x. [DOI] [PubMed] [Google Scholar]
- Jeppsson J. O., Laurell C. B., Fagerhol M. Properties of isolated human alpha1-antitrypsins of Pi types M, S and Z. Eur J Biochem. 1978 Feb 1;83(1):143–153. doi: 10.1111/j.1432-1033.1978.tb12078.x. [DOI] [PubMed] [Google Scholar]
- Johnson D., Travis J. Inactivation of human alpha 1-proteinase inhibitor by thiol proteinases. Biochem J. 1977 Jun 1;163(3):639–641. doi: 10.1042/bj1630639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D., Travis J. Structural evidence for methionine at the reactive site of human alpha-1-proteinase inhibitor. J Biol Chem. 1978 Oct 25;253(20):7142–7144. [PubMed] [Google Scholar]
- Ke H. M., Lipscomb W. N., Cho Y. J., Honzatko R. B. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J Mol Biol. 1988 Dec 5;204(3):725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- Krause K. L., Volz K. W., Lipscomb W. N. 2.5 A structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate. J Mol Biol. 1987 Feb 5;193(3):527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- Kress L. F., Kurecki T., Chan S. K., Laskowski M., Sr Characterization of the inactive fragment resulting from limited proteolysis of human alpha1-proteinase inhibitor by Crotalus adamanteus proteinase II. J Biol Chem. 1979 Jun 25;254(12):5317–5320. [PubMed] [Google Scholar]
- Kress L. F., Laskowski M., Sr Large scale purification of alpha-1 trypsin inhibitor from human plasma. Prep Biochem. 1973;3(6):541–552. doi: 10.1080/00327487308061536. [DOI] [PubMed] [Google Scholar]
- Krigbaum W. R., Godwin R. W. Molecular conformation of chymotrypsinogen and chymotrypsin by low-angle x-ray diffraction. Biochemistry. 1968 Sep;7(9):3126–3131. doi: 10.1021/bi00849a015. [DOI] [PubMed] [Google Scholar]
- Kurachi K., Chandra T., Degen S. J., White T. T., Marchioro T. L., Woo S. L., Davie E. W. Cloning and sequence of cDNA coding for alpha 1-antitrypsin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6826–6830. doi: 10.1073/pnas.78.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laurell C. B., Pierce J., Persson U., Thulin E. Purification of alpha1-antitrypsin from plasma through thiol-disulfide interchange. Eur J Biochem. 1975 Sep 1;57(1):107–113. doi: 10.1111/j.1432-1033.1975.tb02281.x. [DOI] [PubMed] [Google Scholar]
- Laurell C. B. Quantitative estimation of proteins by electrophoresis in agarose gel containing antibodies. Anal Biochem. 1966 Apr;15(1):45–52. doi: 10.1016/0003-2697(66)90246-6. [DOI] [PubMed] [Google Scholar]
- Lijnen H. R., Holmes W. E., van Hoef B., Wiman B., Rodriguez H., Collen D. Amino-acid sequence of human alpha 2-antiplasmin. Eur J Biochem. 1987 Aug 3;166(3):565–574. doi: 10.1111/j.1432-1033.1987.tb13551.x. [DOI] [PubMed] [Google Scholar]
- Loebermann H., Tokuoka R., Deisenhofer J., Huber R. Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function. J Mol Biol. 1984 Aug 15;177(3):531–557. [PubMed] [Google Scholar]
- Löbermann H., Lottspeich F., Bode W., Huber R. Interaction of human alpha 1-proteinase inhibitor with chymotrypsinogen A and crystallization of a proteolytically modified alpha 1-proteinase inhibitor. Hoppe Seylers Z Physiol Chem. 1982 Nov;363(11):1377–1388. doi: 10.1515/bchm2.1982.363.2.1377. [DOI] [PubMed] [Google Scholar]
- Mega T., Lujan E., Yoshida A. Studies on the oligosaccharide chains of human alpha 1-protease inhibitor. I. Isolation of glycopeptides. J Biol Chem. 1980 May 10;255(9):4053–4056. [PubMed] [Google Scholar]
- Moody M. F., Vachette P., Foote A. M. Changes in the x-ray solution scattering of aspartate transcarbamylase following the allosteric transition. J Mol Biol. 1979 Oct 9;133(4):517–532. doi: 10.1016/0022-2836(79)90405-4. [DOI] [PubMed] [Google Scholar]
- Ny T., Sawdey M., Lawrence D., Millan J. L., Loskutoff D. J. Cloning and sequence of a cDNA coding for the human beta-migrating endothelial-cell-type plasminogen activator inhibitor. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6776–6780. doi: 10.1073/pnas.83.18.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S. J., Kerckaert J. P., Loucheux-Lefebvre M. H. The shapes of biantennary and tri/tetraantennary alpha 1 acid glycoprotein by small-angle neutron and X-ray scattering. Eur J Biochem. 1985 Mar 15;147(3):525–531. doi: 10.1111/j.0014-2956.1985.00525.x. [DOI] [PubMed] [Google Scholar]
- Perkins S. J. Molecular modelling of human complement subcomponent C1q and its complex with C1r2C1s2 derived from neutron-scattering curves and hydrodynamic properties. Biochem J. 1985 May 15;228(1):13–26. doi: 10.1042/bj2280013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S. J. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur J Biochem. 1986 May 15;157(1):169–180. doi: 10.1111/j.1432-1033.1986.tb09653.x. [DOI] [PubMed] [Google Scholar]
- Perkins S. J. Structural studies of proteins by high-flux X-ray and neutron solution scattering. Biochem J. 1988 Sep 1;254(2):313–327. doi: 10.1042/bj2540313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S. J., Weiss H. Low-resolution structural studies of mitochondrial ubiquinol:cytochrome c reductase in detergent solutions by neutron scattering. J Mol Biol. 1983 Aug 25;168(4):847–866. doi: 10.1016/s0022-2836(83)80078-3. [DOI] [PubMed] [Google Scholar]
- Schmatz W., Kaiser B., Scherm R., Schneider R., Mayer A. Neutron small-angle scattering from aqueous solutions of oxy- and deoxyhaemoglobin. J Mol Biol. 1969 Apr;41(2):231–235. doi: 10.1016/0022-2836(69)90388-x. [DOI] [PubMed] [Google Scholar]
- Shamash Y., Rimon A. The plasmin inhibitors of human plasma. 3. Purification and partial characterization. Biochim Biophys Acta. 1966 May 26;121(1):35–41. doi: 10.1016/0304-4165(66)90346-1. [DOI] [PubMed] [Google Scholar]
- Travis J., Salvesen G. S. Human plasma proteinase inhibitors. Annu Rev Biochem. 1983;52:655–709. doi: 10.1146/annurev.bi.52.070183.003255. [DOI] [PubMed] [Google Scholar]
- Vaughan L., Lorier M. A., Carrell R. W. alpha 1-Antitrypsin microheterogeneity. Isolation and physiological significance of isoforms. Biochim Biophys Acta. 1982 Mar 4;701(3):339–345. doi: 10.1016/0167-4838(82)90237-0. [DOI] [PubMed] [Google Scholar]


