Abstract
Escherichia coli DNA polymerase IV encoded by the dinB gene is involved in untargeted mutagenesis. Its human homologue is DNA polymerase κ (Polκ) encoded by the DINB1 gene. Our recent studies have indicated that human Polκ is capable of both error-free and error-prone translesion DNA synthesis in vitro. However, it is not known whether human Polκ also plays a role in untargeted mutagenesis. To examine this possibility, we have measured the fidelity of human Polκ during DNA synthesis from undamaged templates. Using kinetic measurements of nucleotide incorporations and a fidelity assay with gapped M13mp2 DNA, we show that human Polκ synthesizes DNA with extraordinarily low fidelity. At the lacZα target gene, human Polκ made on average one error for every 200 nucleotides synthesized, with a predominant T→G transversion mutation at a rate of 1/147. The overall error rate of human Polκ is 1.7-fold lower than human Polη, but 33-fold higher than human Polβ, a DNA polymerase with very low fidelity. Thus, human Polκ is one of the most inaccurate DNA polymerases known. These results support a role for human Polκ in untargeted mutagenesis surrounding a DNA lesion and in DNA regions without damage.
INTRODUCTION
Mutagenesis is a key factor to evolution, human hereditary diseases and cancer. It is now clear that mutagenesis is an active biological response to genotoxic stress conditions, such as cellular exposure to radiations and many chemical carcinogens. Following DNA damage, the resulting mutagenesis is divided into two categories depending on sites of occurrence: targeted mutagenesis and untargeted mutagenesis. The former induces mutations opposite a DNA lesion; while the latter induces mutations in undamaged regions of DNA. Untargeted mutagenesis normally refers to the mutagenic consequence of exogenously induced DNA damage, but at genomic sites without the actual lesion, including a physically separated chromosome, phage or virus that had never been damaged. Untargeted mutagenesis differs from spontaneous mutagenesis in that the latter is the combined end point of errors of the cellular replication machinery and mutagenesis opposite background DNA damage. Spontaneous mutagenesis occurs in the absence of exogenous DNA damaging agents.
In Escherichia coli, both targeted mutagenesis and untargeted mutagenesis are largely under the control of the SOS system that can be switched on by DNA damaging agents. Targeted mutagenesis is mainly mediated by the UmuCD pathway, also known as the damage-induced mutagenesis pathway (1,2). DNA polymerase (Pol) V composed of the UmuCD′2 protein complex is responsible for the translesion DNA synthesis step (3–5). Untargeted mutagenesis requires DNA polymerase IV, encoded by the dinB gene (6). Artificially overexpressing the dinB gene in E.coli is sufficient to trigger untargeted mutagenesis without exogenous DNA damage (7). Both targeted mutagenesis and untargeted mutagenesis empower E.coli cells the genomic flexibility, i.e. accelerated genetic changes in the presence of genotoxic stress conditions such as DNA damage.
In eukaryotes, the REV3 mutagenesis pathway is believed to play an important role in targeted mutagenesis. Originally discovered in the yeast Saccharomyces cerevisiae, this mutagenesis pathway has been recently recognized to also function in humans (8–12). The REV3 mutagenesis pathway involves multiple proteins, including REV1. REV1 possesses a dCMP transferase activity capable of inserting a dCMP opposite a template apurinic/apyrimidinic (AP) site (10,13). The role of REV1 in targeted mutagenesis opposite other DNA lesions remains to be elucidated. Yeast Rev1, yeast Rad30, E.coli UmuC and E.coli DinB form the prototypic members of the UmuC superfamily (14,15). RAD30 codes for DNA Polη in eukaryotes (16). In humans, Polη is encoded by the xeroderma pigmentosum variant (XPV) gene, whose inactivation leads to the hereditary XPV disease (17,18). A role for Polη in both error-free and error-prone lesion bypasses has been shown recently by in vitro studies (16,17,19–22). The in vivo function of human Polη in error-free lesion bypass following UV radiation has been unequivocally established by the phenotypes of XPV patients (23). A human homologue of the E.coli dinB has been identified, DINB1 (24,25). Human DINB1 protein is a DNA polymerase (26,27) and is designated Polκ by several laboratories (15,26,28). Our recent in vitro studies show that human Polκ is capable of both error-free and error-prone translesion DNA synthesis (28). Therefore, it appears that proteins of the UmuC superfamily are involved in various mechanisms of lesion bypass in response to unrepaired DNA damage during replication.
Most recently, the eighth S.cerevisiae DNA polymerase was identified as the TRF4 gene product and was named Polκ (29), which is required for sister chromatid cohesion. Our analysis by protein sequence alignment indicates that this polymerase has no relation to the human Polκ encoded by the DINB1 gene. In fact, this yeast does not contain a sequence homologue of the BINB1 gene.
Much less is known about untargeted mutagenesis in eukaryotes. However, there are indications that untargeted mutagenesis does occur in eukaryotes including mammals. For example, after replicating shuttle vectors containing a site-specific AAF in COS cells, untargeted mutagenesis was observed surrounding the lesion (30,31). Since human Polκ is capable of translesion DNA synthesis in vitro (26,28), the possibility is raised that Polκ may be involved in untargeted mutagenesis surrounding a lesion during lesion bypass by this polymerase. Furthermore, overexpression of mouse DINB1 gene in mouse cells results in elevated mutagenesis (25), implicating Polκ in a more general mechanism of untargeted mutagenesis.
To gain insights into the possible role of Polκ in untargeted mutagenesis surrounding a DNA lesion and in DNA regions without damage, we have measured the fidelity of human Polκ during DNA synthesis from undamaged templates. In this report, we show that human Polκ synthesizes DNA with extraordinarily low fidelity, thus supporting a role for Polκ in untargeted mutagenesis in humans.
MATERIALS AND METHODS
Materials
A mouse monoclonal antibody against the His6 tag was purchased from Qiagen (Valencia, CA). Alkaline phosphatase conjugated anti-mouse IgG was from Sigma (St Louis, MO). Pfu DNA polymerase was from Stratagene (La Jolla, CA). The Klenow fragment of E.coli DNA polymerase I was from BRL (Bethesda, MD). The yeast rad30 deletion mutant strain BY4741rad30Δ (MATa his3 leu2 met15 ura3 rad30Δ) was purchased from Research Genetics (Huntsville, AL). Human testis cDNA and human placenta cDNA were purchased from Clontech Laboratories (Palo Alto, CA). Gapped M13mp2 DNA (32) for initial studies was provided by Thomas Kunkel (National Institute of Environmental Health Sciences, Research Triangle Park, NC). Escherichia coli strains MC1061 and CSH50 were provided by Dale Mosbaugh (Oregon State University, Corvallis, OR).
Plasmid constructions
The human DINB1 cDNA was obtained by polymerase chain reaction (PCR) amplification from human testis cDNAs using Pfu DNA polymerase and two primers: 5′-CGGGATCCATGGATAGCACAAAGGAGAAG-3′ and 5′-CCCAAGCTTCACCTTCTTATCCCATTCCTAATTTC-3′. The resulting 2.9 kb PCR product was then cloned into the BamHI and HindIII sites of the yeast expression vector pECUh6 (28), yielding pECUh6–hDINB1. The human XPV cDNA was obtained by PCR amplification from human testis cDNAs using Pfu DNA polymerase and two primers: 5′-CGGGATCCATGGCTACTGGACAGGATCGAG-3′ and 5′-ACGCGTCGACCATTGT-ACCCGGCCGAG-3′. The resulting 2.6 kb PCR product was then cloned into the BamHI and SalI sites of the yeast expression vector pECUh6 (28), yielding pECUh6–XPV. The human POLB cDNA was obtained by PCR amplification from human placenta cDNAs using Pfu DNA polymerase and two primers: 5′-GCTCTAGAATGAGCAAACGGAAGGCGC-3′ and 5′-CCCAAGCTTCCCATTGGGTTGTGTCTGCG-3′. The resulting 1.1 kb PCR product was then cloned into the XbaI and HindIII sites of the yeast expression vector pEGUh6 (33), yielding pEGUh6–hPOLB.
Purification of human Polκ, Polη and Polβ
To express human Polκ and Polη, yeast rad30 deletion mutant cells containing pECUh6–hDINB1 or pECUh6–XPV were grown at 30°C for 2 days in minimum medium containing 2% dextrose. After 10-fold dilution in 16 l of YPD medium (2% Bacto-peptone, 1% yeast extract, 2% dextrose), cells were grown for 6 h at 30°C. Expression of the human genes was induced by adding CuSO4 to 0.3 mM and growth for another 3 h. To express human Polβ, yeast SX46A (MATa ade2 his3-532 trp1-289 ura3-52) cells harboring pEGUh6–hPOLB were grown in minimal medium containing 2% sucrose for 2 days. Expression of Polβ was induced by diluting the culture 10-fold in 16 l of YPG medium (2% Bacto-peptone, 1% yeast extract, 2% galactose) supplemented with 0.5% sucrose and incubation for 15 h at 30°C with shaking. Collected cells were homogenized by Zirconium beads in a Bead-beater in an extraction buffer containing 50 mM Tris–HCl pH 7.5, 1 M KCl, 10% sucrose, 20% glycerol, 5 mM β-mercaptoethanol and protease inhibitors (34,35). The clarified extract (∼130 ml) was loaded onto a HiTrap chelating column charged with NiSO4 (2 × 5 ml; Amersham Pharmacia Biotech, Piscataway, NJ), followed by washing the column sequentially with 100 ml of Ni buffer A (20 mM KH2PO4 pH 7.4, 1 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol and protease inhibitors) containing 10 mM imidazole and 100 ml of Ni buffer A containing 35 mM imidazole. Bound proteins were eluted with a linear gradient of 35–108 mM imidazole. The His-tagged human Polκ, Polη or Polβ were identified by western blots using a mouse monoclonal antibody specific to the His6 tag.
To purify Polκ and Polη further, the pooled nickel column samples were concentrated by PEG 10 000 and desalted through 5 × 5 ml Sephadex G-25 columns in FPLC buffer A (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 10% glycerol and 5 mM β-mercaptoethanol) containing 100 mM KCl. The resulting samples (∼40 ml each) were loaded onto a Mono S HR5/5 column (Amersham Pharmacia Biotech) separately and eluted with a 30 ml linear gradient of 100–400 mM KCl in FPLC buffer A. Both Polκ and Polη were eluted at ∼250 mM KCl. Fractions containing Polκ or Polη were pooled and concentrated by PEG 10 000. The samples were then loaded separately onto an FPLC Superdex 200 gel filtration column equilibrated with FPLC buffer A containing 300 mM KCl and eluted in the same buffer.
To purify human Polβ further, the nickel column fractions were concentrated and desalted as described above. Then, the sample was loaded onto an FPLC Resource S column (1 ml) and eluted using the same buffer system and gradient as described in Polκ and Polη purifications. Polβ was eluted at ∼230 mM KCl. The Resource S fractions were concentrated and loaded onto an FPLC Superdex 200 column using the same method as in Polκ and Polη purifications. Polβ was eluted at ∼40 kDa position.
DNA polymerase assays
A standard DNA polymerase reaction (10 µl) contained 25 mM KH2PO4 pH 7.0, 5 mM MgCl2, 5 mM dithiothreitol, 100 µg/ml bovine serum albumin, 50 µM each of dATP, dCTP, dTTP and dGTP, 50 fmol of a primed DNA template, 10% glycerol and a purified DNA polymerase. After incubation at 30°C for 10 min, reactions were terminated with 7 µl of a stop solution (20 mM EDTA, 95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol). Reaction products were separated by electrophoresis on a 20% polyacrylamide gel containing 8 M urea and visualized by autoradiography. Primer extension was quantitated by scanning densitometry of the autoradiogram using the SigmaGel Software (Sigma) for analysis.
Kinetic measurements of DNA polymerases
Kinetic analyses of human DNA polymerases were performed as previously described (36,37). Briefly, DNA polymerase assays were performed using 50 fmol of a primed DNA template, 0.7 ng (7 fmol) of purified human Polκ and increasing concentrations of each dNTP (dATP, dCTP, dTTP or dGTP). The DNA primer, 5′-GGAAGAAGAAGTATGTT-3′, was labeled at its 5′-end with 32P. The four DNA templates used were: template A, 5′-CCTTCTTCATTCAAACATACTTCTTCTTCC-3′; template G, 5′-CCTTCTTCATTCGAACATACTTCTTCTTCC-3′; template C, 5′-CCTTCTTCATTCCAACATACTTCTTCTTCC-3′; and template T, 5′-CCTTCTTCATTCTAACATACTTCTTCTTCC-3′, with the analyzed template base underlined. After incubation for 10 min at 30°C under standard DNA polymerase assay conditions, reaction products were separated by electrophoresis on a 20% denaturing polyacrylamide gel. The percentage of primers extended by the polymerase was calculated following scanning densitometry of the extended DNA band(s) and the remaining primer band on the autoradiogram. Product formed (P) was derived from the calculation: P = % primer extension × 50 fmol. Observed enzyme velocity (v) was obtained from the calculation: v = P/10 min. Then, the observed enzyme velocity was plotted as a function of the dNTP concentrations. The plotted data was fitted by a non-linear regression curve to the Michaelis–Menten equation, v = (Vmax × [dNTP])/(Km + [dNTP]), using the SigmaPlot software. Vmax and Km of a DNA polymerase for the incorporation of the correct and the incorrect nucleotides were obtained from the fitted curve. Relative polymerase selectivity of incorrect versus correct nucleotides (finc) was finally calculated from the equation: finc = (Vmax/Km)incorrect/(Vmax/Km)correct (37).
M13mp2 DNA synthesis fidelity assays
DNA synthesis reaction mixture (10 µl) contained 25 mM KH2PO4 (pH 7.0), 5 mM dithiothreitol, 100 µg/ml bovine serum albumin, 30% glycerol, 100 µM dNTP, 8 fmol of gapped M13mp2 DNA (407 nt gap expanding into the lacZα gene) and 400 fmol of purified human Polκ, 205 fmol of purified human Polη or 1200 fmol of purified human Polβ. To completely fill the gap, excess amounts of DNA polymerases were needed (32), especially for human Polβ, due to the distributive mode of DNA synthesis by these polymerases. After incubation at 30°C for 1 h, the reaction was terminated by mixing 2 µl of the reaction mixture with 2 µl of 40 mM EDTA. The remaining reaction mixture (8 µl) was analyzed by electrophoresis on a 0.8% agarose gel to monitor the gap filling reaction. Then, the terminated reaction mixture was diluted 5-fold in water and 1 µl was transfected by electroporation into competent cells (50 µl) of E.coli strain MC1061. In order to detect mutant M13mp2 phage plaques, the transfected cells were plated on plates containing IPTG, X-gal and a lawn of E.coli strain CSH50 as described by Bebenek and Kunkel (32). M13mp2 phage plaques were developed following incubation of the plates at 37°C overnight. Mutant M13mp2 phage plaques (light blue and colorless) were picked and plaque-purified before DNA sequencing.
RESULTS
Purification of human Polκ, Polη and Polβ
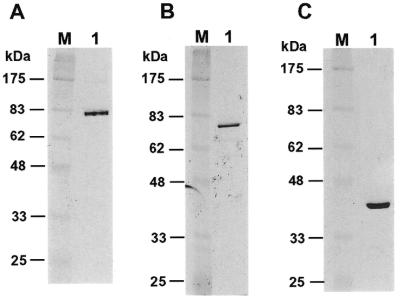
Purified human Polκ was used to study its DNA synthesis fidelity in vitro. For comparison, human Polη and Polβ were also purified. To facilitate the purification of human Polκ, Polη and Polβ, we tagged all three proteins with six histidine residues at their N-termini and expressed them in the yeast S.cerevisiae cells. The yeast rad30 deletion mutant cells were used for Polκ and Polη expression to avoid potential contamination by the yeast Polη. The three human DNA polymerases were purified separately to apparent homogeneity (Fig. 1). The identity of these purified polymerases were confirmed by western blot analyses using a mouse monoclonal antibody specific to the His6 tag (data not shown). The three polymerases were biochemically active as tested by DNA polymerase assays (data not shown) (28). Human Polη and Polβ migrated as 77 and 42 kDa proteins, respectively, consistent with their calculated molecular weights of 78 and 38 kDa, respectively (Fig. 1B and C). Human Polκ migrated as an 81 kDa protein on a 10% SDS–polyacrylamide gel, faster than its calculated molecular weight of 99 kDa (Fig. 1A). However, there are indications suggesting that human Polκ migrated abnormally faster than predicted from its molecular weight (28). Thus, our purified human Polκ, Polη and Polβ are apparently the full-length proteins.
Figure 1.
Analysis of purified human Polκ, Polη and Polβ. (A) Purified human Polκ (200 ng), (B) Polη (80 ng) and (C) Polβ (230 ng) were analyzed by electrophoreses on 10% SDS–polyacrylamide gels and visualized by silver staining of the gels (lanes 1). Protein size markers (lanes M) are indicated on the left.
Human Polκ does not possess a 3′→5′ proofreading nuclease activity
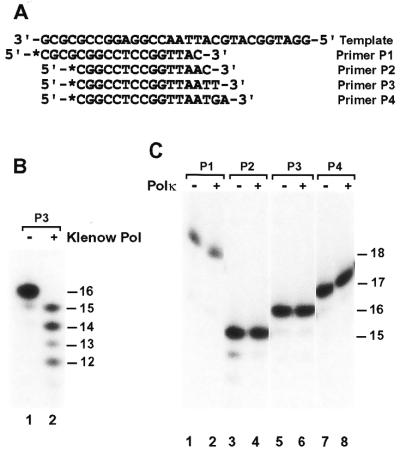
To examine whether Polκ contains a proofreading nuclease activity, we labeled several primers at their 5′-ends with 32P and annealed them to a 30mer DNA template (Fig. 2A). These DNA substrates contained a TC mismatch (Fig. 2A, primer P1), an AC mismatch (Fig. 2A, primer P2), a CT mismatch (Fig. 2A, primer P3) or a GA mismatch (Fig. 2A, primer P4) at the primer 3′-end. In the absence of dNTPs, a 3′→5′ proofreading nuclease would recognize a mismatch at the primer 3′-end and subsequently remove one or a few nucleotides from the 3′-end of the primer. Under the conditions used, the proofreading nuclease activity of the Klenow fragment of E.coli DNA polymerase I was readily detected after 2 min incubation (Fig. 2B, compare lanes 1 and 2). In contrast, human Polκ did not degrade the mismatched primers even after 30 min incubation in the absence of dNTPs (Fig. 2C). These results show that human Polκ does not possess a 3′→5′ proofreading nuclease activity.
Figure 2.
Proofreading nuclease assays of purified human Polκ. (A) The DNA template and four primers used for nuclease activity assays. The primers were labeled with 32P at their 5′-ends as indicated by an asterisk and annealed individually to the template. (B) DNA substrates (50 fmol) containing annealed primer P3 were incubated without (Klenow Pol, –) or with (Klenow Pol, +) the purified Klenow fragment of E.coli DNA polymerase I (1 U) for 2 min at 30°C in the DNA polymerase assay buffer without dNTPs. Reaction products were separated by electrophoresis on a 20% denaturing polyacrylamide gel. (C) Using the indicated DNA substrates, proofreading nuclease assays were similarly performed with (Polκ, +) or without (Polκ, –) purified human Polκ (1.3 ng, 13 fmol) as in (B). However, the incubation time was extended to 30 min. DNA size markers (in nucleotides) are indicated on the right.
Nucleotide incorporations by human Polκ
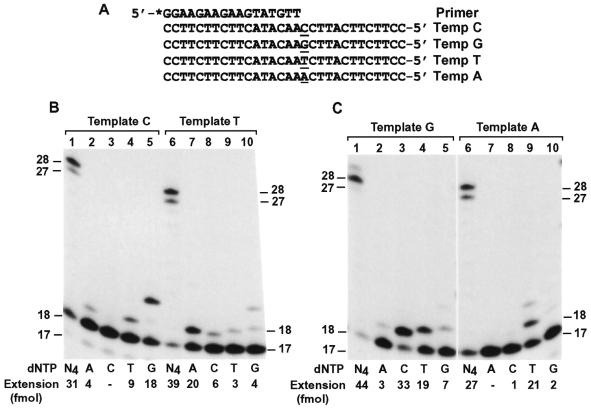
To examine the base pairing property during DNA synthesis catalyzed by human Polκ, we performed DNA polymerase assays in the presence of only one deoxyribonucleoside triphosphate using 7 fmol of Polκ and 50 fmol of primed 30mer DNA templates. A 17mer primer was labeled with 32P at its 5′-end and annealed to templates C, G, T and A (Fig. 3A). Opposite the template CC sequence, two G residues were incorporated by Polκ as the major event (Fig. 3B, lane 5). Less frequently, T and A were also incorporated at a decreasing rate (Fig. 3B, lanes 4 and 2). Opposite the template T, A was predominantly incorporated (Fig. 3B, lane 7). Less frequently, C, G and T were also incorporated (Fig. 3B, lanes 8–10). Opposite the template G, a C residue was predominantly incorporated by Polκ (Fig. 3C, lane 3). Less frequently, T, G and A were also incorporated at a decreasing rate (Fig. 3C, lanes 4, 5 and 2). Opposite the template A, T was predominantly incorporated by Polκ (Fig. 3C, lane 9) and, although to a much lesser extent, G was also incorporated (Fig. 3C, lanes 10). In the presence of all four dNTPs as controls, human Polκ efficiently extended the primers near the end of the templates (Fig. 3B and C, lanes 1 and 6). It was consistently observed that human Polκ stopped DNA synthesis one or two nucleotides before the end of the template (Fig. 3B and C, lanes 1 and 6) (27,28). These results show that human Polκ preferentially incorporates the correct base opposite the template C, T, G or A, but is prone to misincorporations.
Figure 3.
Base pairing specificity of human Polκ. (A) Each DNA template (Temp) was annealed with the 17mer primer that was labeled at its 5′-end with 32P as indicated by an asterisk. The four template bases examined are underlined. (B and C) Polymerase assays were performed with 50 fmol of the indicated DNA templates and 7 fmol (0.7 ng) of purified human Polκ in the presence of a single dNTP or all four dNTPs (N4) as indicated. DNA size markers (in nucleotides) are indicated on the sides. Quantitation of extended primers is shown at the bottom of the gels.
Kinetic measurements of base selection by human Polκ
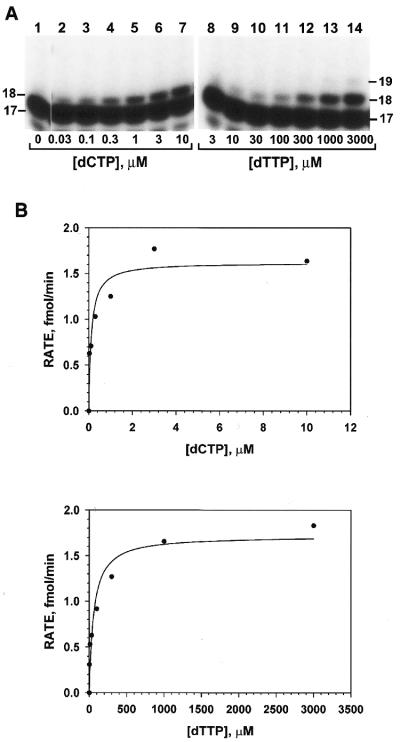
To obtain a quantitative measurement of base selections by human Polκ, we measured its nucleotide incorporation fidelity using a steady state kinetic analysis (36,37). DNA polymerase assays were performed with increasing concentrations of a single dNTP using 50 fmol of a primed 30mer DNA template (Fig. 3A) and 7 fmol of purified human Polκ. Opposite the template G, correct C incorporation was observed at very low dCTP concentrations (Fig. 4A, lanes 2–7). However, for T misincorporation, primer extension occurred at much higher dTTP concentrations (Fig. 4A, lanes 8–14). These base incorporation results were quantitated and the observed incorporation velocities (v) were calculated. Experimental v values were plotted against the dNTP substrate concentrations and fitted into the Michaelis–Menten equation, v = (Vmax × [dNTP])/(Km + [dNTP]), as shown in Figure 4B. From the fitted curve, the kinetic parameters Vmax and Km were obtained and listed in Table 1.
Figure 4.
Kinetic analysis of C and T incorporations opposite a template G. (A) Polymerase assays were performed at 30°C for 10 min using 7 fmol (0.7 ng) of purified human Polκ, 50 fmol of the template G (Fig. 3A) and increasing concentrations of dCTP or dTTP as indicated. Primer extension products were separated from the 32P-labeled 17mer primer by electrophoresis on a 20% denaturing polyacrylamide gel and visualized by autoradiography. DNA size markers (in nucleotides) are indicated on the sides. (B) Results in (A) were quantitated and the rate of nucleotide incorporation (primer extension) was graphed as a function of dCTP or dTTP concentrations. The scattered plots were then fitted into the Michaelis–Menton equation (see Materials and Methods). The Vmax and Km values obtained from the fitted curve were listed in Table 1.
Table 1. Kinetic measurement of nucleotide incorporation by human Polκ.
| dNTP | Vmax (fmol/min) | Km (µM) | Vmax/Km | finca |
|---|---|---|---|---|
| Template A | ||||
| dATP | 0.81 ± 0.11 | 270 ± 134 | 0.003 | 5.2 × 10–4 |
| dGTP | 1.7 ± 0.16 | 118 ± 35 | 0.014 | 2.4 × 10–3 |
| dCTP | 1.8 ± 0.08 | 230 ± 37 | 0.0078 | 1.3 × 10–3 |
| dTTP | 1.1 ± 0.07 | 0.19 ± 0.08 | 5.8 | 1 |
| Template G | ||||
| dATP | 2.6 ± 0.07 | 110 ± 9.9 | 0.024 | 1.8 × 10–3 |
| dGTP | 0.7 ± 0.009 | 41 ± 24 | 0.017 | 1.3 × 10–3 |
| dCTP | 1.6 ± 0.13 | 0.12 ± 0.05 | 13.3 | 1 |
| dTTP | 1.7 ± 0.14 | 59 ± 22 | 0.029 | 2.2 × 10–3 |
| Template C | ||||
| dATP | 2.8 ± 0.06 | 50 ± 4.5 | 0.056 | 4.5 × 10–2 |
| dGTP | 0.67 ± 0.1 | 0.58 ± 0.26 | 1.2 | 1 |
| dCTP | 1.3 ± 0.33 | 989 ± 634 | 0.0013 | 1.1 × 10–3 |
| dTTP | 2.0 ± 0.16 | 29 ± 12 | 0.069 | 5.8 × 10–2 |
| Template T | ||||
| dATP | 1.9 ± 0.23 | 0.29 ± 0.18 | 6.6 | 1 |
| dGTP | 1.4 ± 0.19 | 9.1 ± 6.6 | 0.15 | 2.3 × 10–2 |
| dCTP | 2.1 ± 0.04 | 22 ± 2.2 | 0.095 | 1.4 × 10–2 |
| dTTP | 1.4 ± 0.07 | 49 ± 13 | 0.029 | 4.4 × 10–3 |
afinc = (Vmax/Km)incorrect/(Vmax/Km)correct.
Using this method, we systematically measured the Vmax and Km values of all 16 possible base incorporations opposite the four template bases (Table 1). As indicated by the Vmax/Km values (Table 1), template T was most likely to lead to misincorporations, whereas template A was least likely to lead to misincorporations. Based on the Vmax/Km results (Table 1), G and C misincorporations opposite template T occurred most frequently; while C misincorporation opposite template C and A misincorporation opposite template A occurred least frequently. Discrimination between right and wrong nucleotides as indicated by finc (Table 1) show that human Polκ significantly misincorporated nucleotides opposite template A, G, C and T. These kinetic measurements suggest that human Polκ is a low fidelity DNA polymerase.
Mismatch extension by human Polκ
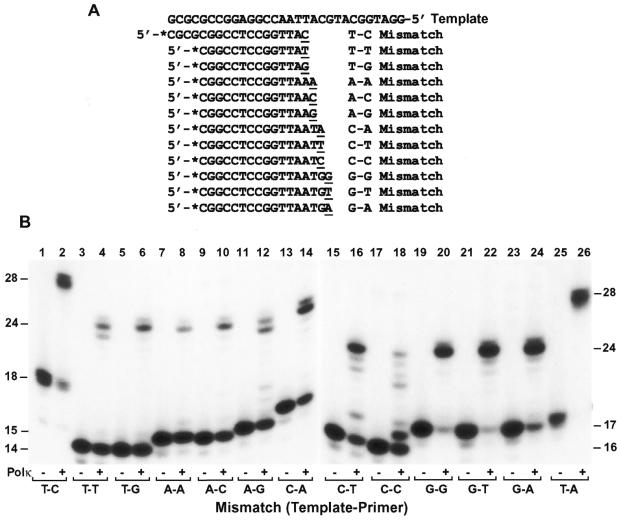
Mutation formation during normal DNA synthesis is determined by two steps: (i) misincorporation followed by (ii) mismatch extension by the DNA polymerase. To determine if human Polκ is able to extend misincorporated bases, we performed mismatch extension assays using 40 fmol of the purified polymerase and 50 fmol of a primed DNA template containing a mismatch at the primer 3′-end. All 12 possible mismatches were examined (Fig. 5A). As shown in Figure 5B, at the polymerase concentration used, all mismatches were extended by human Polκ, but with very different efficiencies. While G–G, G–T and T–C (template–primer) mismatches were extended relatively efficiently, A–A mismatch was extended with least efficiency (Fig. 5B). In comparison, a Watson–Crick base pair such as T–A was extended most efficiently by human Polκ (Fig. 5B, lane 26).
Figure 5.
Mismatch extension by human Polκ. (A) Various primers labeled at their 5′-ends with 32P as indicated by asterisks were annealed to the 30mer template, generating 12 possible mismatches at the primer 3′-ends. (B) Mismatched substrates were incubated with (Polκ, +) or without (Polκ, –) DNA Polκ (4 ng, 40 fmol) under standard polymerase assay conditions. Lanes 25 and 26, the 30mer template was annealed with the 17mer primer 5′-*CGCGCGGCCTCCGGTTA-3′ generating a correct T–A base pair at the primer 3′-end. DNA size markers (in nucleotides) are indicated on the sides. It should be noted that the primers in the T–C (lane 2) and T–A DNA substrates (lane 26) are 4 nt longer at the 5′-end than other primers.
In most cases, primer extension by human Polκ stopped one or two nucleotides before the end of the template, regardless of correct base pair or mismatch at the primer 3′-end prior to extension (Fig. 5B, lanes 2, 6, 8, 10, 14, 20, 22, 24 and 26). With T–T and A–G mismatches, however, significant amounts of the primers were extended one nucleotide shorter than others (23mer DNA band) (Fig. 5B, lanes 4 and 12). The 23mer extension products were probably derived from –1 misalignment prior to extension, as the primer T could pair with the next template A in the T–T mismatch substrate and the primer G could pair with the next template C in the A–G mismatch substrate (Fig. 5A). Consistent with this interpretation, a tendency for E.coli DNA polymerase IV (DinB protein) to make –1 template–primer misalignment has been observed (6,7). With C–T and C–C mismatches, several shorter bands were observed (Fig. 5B, lanes 16 and 18), which might result from multiple and/or various template–primer misalignments. These results show that human Polκ can readily extend some mismatched base pairs, although not as efficiently as extending a correct base pair; and suggest that some mismatch extensions may be mediated by the template–primer misalignments.
DNA synthesis fidelity of human Polκ
To determine if frequent misincorporations and the ability of mismatch extension by human Polκ would indeed lead to a high mutation rate, we measured the fidelity of this polymerase in synthesizing 229 nt of the target lacZα gene (from nucleotide –84 to +145) (32). For comparison, we also measured the fidelity of purified human Polη and Polβ. To perform the fidelity assay, a gapped M13mp2 DNA was prepared containing a 407 nt gap (32). Within the gap lies the lacZα template DNA strand. The gap was then filled in by purified human Polκ in a standard DNA polymerase reaction buffer. Complete gap filling by human Polκ slowed the DNA migration to an identical position as the nicked circular M13mp2 control DNA on a 0.8% agarose gel (compare Fig. 6A and B). Polη and Polβ also completely filled in the gap in M13mp2 DNA (Fig. 6C and D). After gap filling by the purified polymerases, the phage DNA was then transfected into the E.coli MC1061 cells for mutant identification. In the presence of IPTG and X-gal, wild-type M13mp2 phage produced dark blue plaques, whereas mutants were identified as light blue or colorless phage plaques (32). Human Polκ yielded a mutation frequency of 30% at the lacZα target gene, which was similar to human Polη (49%), but considerably higher than human Polβ (2.7%) (Table 2).
Figure 6.
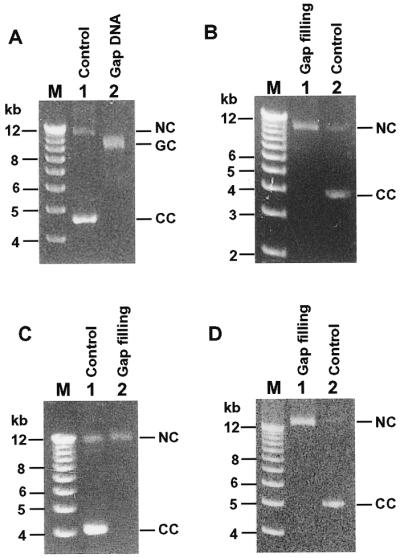
Gap filling DNA synthesis by purified human Polκ, Polη and Polβ. (A) Analysis of gapped M13mp2 DNA (Gap DNA) by a 0.8% agarose gel. Control, double-stranded M13mp2 DNA without gap. (B–D) Gapped M13mp2 DNA (8 fmol) was incubated with purified human Polκ (400 fmol), Polη (205 fmol) and Polβ (1200 fmol), respectively, for gap filling DNA syntheses (Gap filling) (see Materials and Methods). Gap filling products (6.4 fmol of M13mp2 DNA) were analyzed by electrophoreses on 0.8% agarose gels in the presence of 0.5 µg/ml ethidium bromide. NC, nicked circular DNA; GC, gapped circular DNA; CC, closed circular DNA.
Table 2. DNA synthesis fidelity of human Polκ.
| DNA polymerasea | Mutation frequencyb | Point mutation (%) | Deletion (%) | Insertion (%) | Average mutations per targetc | Average mutation rate |
|---|---|---|---|---|---|---|
| Polκ | 3.0 × 10–1 | 87.3 | 9.3 | 3.4 | 3.8 | 5.0 × 10–3 |
| Polη | 4.9 × 10–1 | 91.8 | 7.2 | 1.0 | 3.9 | 8.3 × 10–3 |
| Polβ | 2.7 × 10–2 | 36.6 | 58.5 | 4.9 | 1.3 | 1.5 × 10–4 |
aHuman DNA polymerases κ, η and β were purified to apparent homogeneity and 400, 205 and 1200 fmol, respectively, were used in the M13mp2 DNA synthesis fidelity assays.
bIn the M13mp2 fidelity assay, ∼60% phage plaques were derived from the gap filling strand (minus strand) (32). Thus, mutation frequency = (mutant plaques/total plaques) × 1/0.6. For the Polκ experiment, 2144 phage plaques were scored, of which 82 light blue and 301 colorless mutant phage plaques were observed. For the Polη experiment, 1577 phage plaques were scored, of which 142 light blue and 323 colorless mutant phage plaques were observed. For the Polβ experiment, 2349 phage plaques were scored, of which 3 light blue and 35 colorless mutant phage plaques were observed.
cMutant phage clones sequenced in Polκ, η and β experiments were 53, 53 and 32, respectively. The target sequence is 229 nt of the E.coli lacZα gene (from nucleotide –84 to +145).
To identify the specific mutations during DNA synthesis by human Polκ from the undamaged template, we isolated 53 mutant M13mp2 phage clones and sequenced the target gene. Within the 229 nt target sequence, 50 of the 53 mutant clones contained multiple mutations, up to 8 mutation sites per target sequence and 4 tandem double-base substitutions were observed. On average, 3.8 mutation sites per target sequence were produced by Polκ (Table 2). The overall error rate of Polκ is thus one mutation for every 200 nucleotides synthesized (3.8 × 3.0 × 10–1/229 = 5.0 × 10–3 = 1/200). For comparison, we also sequenced 53 Polη- and 32 Polβ-induced mutants. Polη produced 3.9 mutations per target sequence (Table 2). In contrast, Polβ produced 1.3 mutations per target sequence, as expected (Table 2). The overall error rate of Polκ is 1.7-fold lower than human Polη, but 33-fold higher than human Polβ. Thus, human Polκ and Polη are two of the most inaccurate DNA polymerases known, synthesizing DNA with unprecedented high error rates.
Major mutations produced by Polκ and Polη were base substitutions, followed by deletions (Table 2). Most deletion mutations were single base deletions. One nucleotide deletion mutations of –T, –G, –C and –A (on the DNA +strand) were scored 5, 4, 2 and 2 times, respectively, among the 204 total mutations at the lacZα target sequence. One nucleotide insertion mutations of +A, +G, +C and +T (on the DNA +strand) were scored 2, 2, 1 and 1 times, respectively. Additionally, one mutant lacZα sequence contained a stretch of 73-nt deletion and a stretch of 16-nt insertion. These results show that human Polκ synthesizes DNA with extraordinarily low fidelity.
Specificity of base substitution mutations by human Polκ
Base substitution error rates by human Polκ are summarized in Table 3. Based on the total number of mutations produced opposite template A, G, C and T (base substitutions in Table 3 plus deletions and insertions), template bases were copied by human Polκ with very different fidelity and followed the order from most accurate to most inaccurate: A > C > G > T. All 12 possible base substitutions were observed. As a result of C mispairing with template T, T→G transversion was the most frequent mutation event during DNA synthesis by Polκ with an error rate of 1/147 (Table 3). The second highest base substitution error rate observed was T→C transition as a result of G mispairing with template T (Table 3). The third highest base substitution error rate observed was G→A transition as a result of T mispairing with template G (Table 3). The lowest base substitution error rate (1/10 000) observed was A→C transversion as a result of G mispairing with template A (Table 3).
Table 3. Error rates of human Polκ for base substitution mutationsa.
| Template base (number sequenced)b | Mismatch (template–primer) | Mutation | Detected substitutions | Error rate |
|---|---|---|---|---|
| A (3021) | A–A | A→T | 3 | 3.0 × 10–4 |
| A–G | A→C | 1 | 1.0 × 10–4 | |
| A–C | A→G | 8 | 7.9 × 10–4 | |
| G (2597) | G–A | G→T | 9 | 1.0 × 10–3 |
| G–G | G→C | 8 | 1.0 × 10–3 | |
| G–T | G→A | 13 | 1.5 × 10–3 | |
| C (3286) | C–A | C→T | 11 | 1.0 × 10–3 |
| C–C | C→G | 3 | 2.7 × 10–4 | |
| C–T | C→A | 15 | 1.4 × 10–3 | |
| T (3233) | T–G | T→C | 26 | 2.4 × 10–3 |
| T–C | T→G | 74 | 6.8 × 10–3 | |
| T–T | T→A | 7 | 6.5 × 10–4 |
aThe polymerase fidelity assay was performed (see Materials and Methods) using 8 fmol of gapped M13mp2 DNA and 400 fmol of purified human Polκ.
bTotal number of template bases sequenced. There are 57, 49, 62 and 61 template A, G, C and T bases, respectively, in the 229 nt lacZα target sequence. Fifty-three M13mp2 mutant clones were sequenced.
DISCUSSION
Using three different approaches, we found that purified human Polκ synthesizes DNA with extraordinarily low fidelity. In the first approach, we examined base misincorporations by human Polκ opposite template C, T, G and A under the standard polymerase assay conditions. Significant misincorporations were observed. In the second approach, we measured kinetic parameters of base incorporations by human Polκ. These two methods yielded consistent results, indicating that human Polκ significantly incorporates incorrect bases, especially opposite a template T, C or G. Furthermore, both approaches show that human Polκ is catalytically most efficient opposite a template G. In the third approach, we determined the fidelity of human Polκ during in vitro synthesis of a 229 nt lacZα sequence. This method reflects the ability of Polκ to incorporate incorrect bases and to subsequently extend mismatches. It detects both point and frameshift mutations (32). Low fidelity DNA synthesis of human Polκ was again clearly demonstrated.
Human Polβ has been recognized as a low fidelity DNA polymerase (38). Remarkably, human Polκ is 33-fold more inaccurate than human Polβ. Human Polκ generates on average one mutation for every 200 nucleotides synthesized, while human Polη generates on average one mutation for every 120 nucleotides synthesized. Extraordinarily low fidelity of human Polη has also been reported most recently by Matsuda et al. (39). Hence, human Polκ and Polη are in the same category of unprecedented low fidelity DNA polymerases. Although the overall error rates between human Polκ and Polη are similar, the mutation specificity generated by these two polymerases is quite different. While the most frequent base substitution mutations observed with human Polκ is T→G (C incorporation opposite template T), followed by T→C (G incorporation opposite T), the most frequent base substitution mutations observed with human Polη is T→C, followed by A→T (A incorporation opposite A) (our unpublished data) (39).
Overexpressing the E.coli dinB gene in E.coli cells results in higher spontaneous mutations, mostly –1 deletions (7). Surprisingly, deletions were not the major mutations produced during DNA synthesis by human Polκ, accounting for only 9.3% of the total mutations, which is much lower than 87% of base substitutions. It would be very informative to examine the intrinsic fidelity of E.coli DNA polymerase IV using the M13mp2 DNA synthesis fidelity assay and compare to human Polκ. In the M13mp2 fidelity assay, due to the distributive mode of DNA synthesis by human Polκ, Polη and Polβ, these polymerases were used in excess amounts relative to DNA in order to completely fill in the 407-nt gap. Nevertheless, the error rates of base substitutions observed in this assay (Table 3) are in good agreement with the error rates of base misincorporations (Table 1) obtained by kinetic measurements where much lower polymerase to DNA ratio was used. Thus, it is unlikely that excess amounts of the polymerase used in the M13mp2 fidelity assay could drastically affect the intrinsic fidelity of the polymerase. Supporting this conclusion, Matsuda et al. (39) observed similar mutation frequencies of purified human Polη during synthesis of the M13mp2 DNA gap at several polymerase concentrations.
Most recently, Tang et al. (4) examined base misincorporations by E.coli polymerase IV using steady state kinetic analyses. It was reported that E.coli polymerase IV incorporated nucleotides 5–10-fold more accurately than E.coli polymerase V (4). Since the reported finc for E.coli polymerase IV ranged mostly from 10–4 to 10–5 (4), the result would predict that this polymerase is not remarkably error-prone, which seems to be inconsistent with the in vivo results of untargeted mutagenesis by polymerase IV (7,40). Most recently, using kinetic analysis, Johnson et al. (27) also examined nucleotide miscincorporations by purified human DINB1 protein. Our results significantly differ from those of Johnson et al. (27). For example, it was reported that G incorporation opposite template A was the most frequent misincorporation (27). Our kinetic measurements indicate that this incorporation is among the least frequent misincorporation and our M13mp2 fidelity assays show that mutations resulting from this misincorporation represent the lowest base substitutions. In general, kinetic measurements of Johnson et al. (27) predict human Polκ to be a significantly more accurate DNA polymerase than what we have observed. The precise basis for this discrepancy is presently unknown. While our human Polκ contained a short N-terminal His6 tag, the human DINB1 protein isolated by Johnson et al. (27) contained at its N-terminus a bulky GST protein (26 kDa) and the E.coli polymerase IV used by Tang et al. (4) contained at its N-terminus a bulky maltose binding protein (42.7 kDa). This raises the possibility that the fused bulky proteins may have somewhat altered the properties of the human Polκ and the E.coli DNA polymerase IV.
Both Polη and Polκ are members of the recently identified UmuC superfamily of proteins (14,15). The important role of Polη in response to UV radiation has been well demonstrated by the phenotypes of the human hereditary disease XPV (23). The role of Polκ in human biology, however, is not well understood. Our biochemical studies indicate that human Polκ is a novel lesion bypass enzyme. Purified human Polκ is capable of error-free bypass opposite a template (–)-trans-anti-benzo[a]pyrene-N2-dG bulky lesion and error-prone bypass opposite a template 8-oxoguanine, abasic site and AAF-adducted guanine (28). It is conceivable that lesion bypass DNA polymerases may contain a loose and flexible active site pocket to accommodate damaged DNA templates for translesion DNA synthesis. Consequently, lesion bypass polymerases may have lost stringent constraints at their active sites for high fidelity Watson–Crick base pairings. This hypothesis predicts that all lesion bypass polymerases are associated with a very low DNA synthesis fidelity. Supporting this hypothesis, the lesion bypass enzymes Polη and Polκ of humans are associated with extraordinarily low DNA synthesis fidelity. Further supporting the hypothesis, human Polι is able to incorporate one nucleotide opposite several DNA lesions, notably a template abasic site (41); and this polymerase is found to be highly error-prone on undamaged DNA templates (41,42).
Due to the high error rate of human Polκ, lesion bypass by this polymerase may expose the surrounding DNA sequences to a much higher probability of mutations leading to untargeted mutagenesis. Lesion bypass by Polκ could not only gain survival for the cell, but also may empower the cell to evolve and adapt through mutations at both the targeted and untargeted sequences. Additionally, the highly error-prone nature of human Polκ also supports a role for this polymerase in untargeted mutagenesis in undamaged regions of DNA. Indeed, overexpressing the mouse DINB gene, a condition that may mimic presumptive up-regulation of the gene under genotoxic stress conditions, results in elevated spontaneous mutation rates in mouse cells (25). It is likely that the untargeted mutagenesis function of DinB protein may have been evolutionally conserved from E.coli to humans. Our results presented here and in another report (28) support two functions for human Polκ: (i) DNA lesion bypass and (ii) untargeted mutagenesis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Thomas Kunkel for providing us with the gapped M13mp2 DNA for initial studies. We thank Dale Mosbaugh for the E.coli strains MC1061 and CSH50. We thank Thomas Kunkel and Katarzyna Bebenek for technical advice on the M13mp2 DNA synthesis fidelity assay. This work was supported by a New Investigator Award in Toxicology from Burroughs Wellcome Fund and a THRI grant from the University of Kentucky.
REFERENCES
- 1.Goodman M.F. (2000) Trends Biochem. Sci., 25, 189–195. [DOI] [PubMed] [Google Scholar]
- 2.Goodman M.F. and Tippin,B. (2000) Curr. Opin. Genet. Dev., 10, 162–168. [DOI] [PubMed] [Google Scholar]
- 3.Tang M., Shen,X., Frank,E.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (1999) Proc. Natl Acad. Sci. USA, 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang M., Pham,P., Shen,X., Taylor,J.S., O’Donnell,M., Woodgate,R. and Goodman,M.F. (2000) Nature, 404, 1014–1018. [DOI] [PubMed] [Google Scholar]
- 5.Reuven N.B., Arad,G., Maor-Shoshani,A. and Livneh,Z. (1999) J. Biol. Chem., 274, 31763–31766. [DOI] [PubMed] [Google Scholar]
- 6.Wagner J., Gruz,P., Kim,S.R., Yamada,M., Matsui,K., Fuchs,R.P. and Nohmi,T. (1999) Mol. Cell, 4, 281–286. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.R., Maenhaut-Michel,G., Yamada,M., Yamamoto,Y., Matsui,K., Sofuni,T., Nohmi,T. and Ohmori,H. (1997) Proc. Natl Acad. Sci. USA, 94, 13792–13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin W., Wu,X. and Wang,Z. (1999) Mutat. Res., 433, 89–98. [DOI] [PubMed] [Google Scholar]
- 10.Lin W., Xin,H., Zhang,Y., Wu,X., Yuan,F. and Wang,Z. (1999) Nucleic Acids Res., 27, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs P.E., Wang,X.D., Li,Z., McManus,T.P., McGregor,W.G., Lawrence,C.W. and Maher,V.M. (2000) Proc. Natl Acad. Sci. USA, 97, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakumo Y., Roth,T., Ishii,H., Rasio,D., Numata,S., Croce,C.M. and Fishel,R. (2000) J. Biol. Chem., 275, 4391–4397. [DOI] [PubMed] [Google Scholar]
- 13.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- 14.McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg E.C., Feaver,W.J. and Gerlach,V.L. (2000) Proc. Natl Acad. Sci. USA, 97, 5681–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson R.E., Prakash,S. and Prakash,L. (1999) Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 17.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999) Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- 19.Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999) EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.-S. and Wang,Z. (2000) J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Yuan,F., Wu,X., Rechkoblit,O., Taylor,J.-S., Geacintov,N.E. and Wang,Z. Nucleic Acids Res. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleaver J.E. and Kraemer,K.H. (1989) In Scriver,C.R., Beaudet,A.L., Sly,W.S. and Valle,D (eds), The Metabolic Basis of Inherited Disease, 6th edn. McGraw-Hill Book Co., New York, NY, pp. 2949–2971.
- 24.Gerlach V.L., Aravind,L., Gotway,G., Schultz,R.A., Koonin,E.V. and Friedberg,E.C. (1999) Proc. Natl Acad. Sci. USA, 96, 11922–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogi T., Kato,T.,Jr, Kato,T. and Ohmori,H. (1999) Genes Cells, 4, 607–618. [DOI] [PubMed]
- 26.Ohashi E., Ogi,T., Kusumoto,R., Iwai,S., Masutani,C., Hanaoka,F. and Ohmori,H. (2000) Genes Dev., 14, 1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson R.E., Prakash,S. and Prakash,L. (2000) Proc. Natl Acad. Sci. USA, 97, 3838–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Yuan,F., Wu,X., Wang,M., Rechkoblit,O., Taylor,J.-S., Geacintov,N.E. and Wang,Z. (2000) Nucleic Acids Res., 28, 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Castano,I.B., De Las Penas,A., Adams,C. and Christman,M.F. (2000) Science, 289, 774–779. [DOI] [PubMed] [Google Scholar]
- 30.Shibutani S., Suzuki,N. and Grollman,A.P. (1998) Biochemistry, 37, 12034–12041. [DOI] [PubMed] [Google Scholar]
- 31.Gentil A., Le Page,F., Margot,A., Lawrence,C.W., Borden,A. and Sarasin,A. (1996) Nucleic Acids Res., 24, 1837–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bebenek K. and Kunkel,T.A. (1995) Methods Enzymol., 262, 217–232. [DOI] [PubMed] [Google Scholar]
- 33.Wu X., Braithwaite,E. and Wang,Z. (1999) Biochemistry, 38, 2628–2635. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z., Wu,X. and Friedberg,E.C. (1995) Methods, 7, 177–186. [Google Scholar]
- 35.Xin H., Lin,W., Sumanasekera,W., Zhang,Y., Wu,X. and Wang,Z. (2000) Nucleic Acids Res., 28, 2847–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Washington M.T., Johnson,R.E., Prakash,S. and Prakash,L. (1999) J. Biol. Chem., 274, 36835–36838. [DOI] [PubMed] [Google Scholar]
- 37.Creighton S., Bloom,L.B. and Goodman,M.F. (1995) Methods Enzymol., 262, 232–256. [DOI] [PubMed] [Google Scholar]
- 38.Osheroff W.P., Jung,H.K., Beard,W.A., Wilson,S.H. and Kunkel,T.A. (1999) J. Biol. Chem., 274, 3642–3650. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda T., Bebenek,K., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2000) Nature, 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- 40.Brotcorne-Lannoye A. and Maenhaut-Michel,G. (1986) Proc. Natl Acad. Sci. USA, 83, 3904–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Yuan,F., Wu,X. and Wang,Z. (2000) Mol. Cell. Biol., 20, 7099–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tissier A., McDonald,J.P., Frank,E.G. and Woodgate,R. (2000) Genes Dev., 14, 1642–1650. [PMC free article] [PubMed] [Google Scholar]