Abstract
Smads perform pivotal functions in the intracellular signaling of transforming growth factor-β (TGF-β). TGF-β-mediated activation of TGF-β type I receptor stimulates the phosphorylation of Smad2 and Smad3 and subsequent heteromeric complex formation with Smad4. The heteromeric Smad complexes translocate into the nucleus where they, in co-operation with co-activators and co-repressors, regulate transcriptional responses. Here we investigated the possible co-activator function of P/CAF in TGF-β/Smad signaling. P/CAF was found to interact directly with Smad3 in vitro. Moreover, Smad2 and Smad3 interacted with P/CAF upon TGF-β type I receptor activation in cultured mammalian cells. The interaction involves the MH2 domain of Smad3 and the N-terminal region of P/CAF. P/CAF potentiated the transcriptional activity of heterologous Gal4–Smad2 and Gal4–Smad3 fusion proteins. In addition, P/CAF potentiated the TGF-β/Smad3-induced transcriptional responses, which could be further enhanced by co-activators p300 and Smad4. P/CAF may, therefore, activate Smad-mediated transcriptional responses independently or in co-operation with p300/CBP. Our results indicate a direct physical and functional interplay between two negative regulators of cell proliferation, Smad3 and P/CAF.
INTRODUCTION
Transforming growth factor-β (TGF-β) regulates a variety of cellular responses, including proliferation, differentiation, apoptosis and migration (1,2). TGF-β elicits its cellular effects by inducing a heteromeric complex of two serine/threonine kinase receptors, the TGF-β type II (TβR-II) and TGF-β type I (TβR-I) receptors (2,3). In the complex, TβR-I is phosphorylated by the constitutively active TβR-II kinase. The activated TβR-I kinase initiates signaling through phosphorylation of specific receptor-regulated Smads (R-Smads), i.e. Smad2 and Smad3 (2,3). Type I receptor phosphorylation occurs at two serine residues in the SXS motifs in the absolute C-termini of R-Smads (4,5). Subsequently, activated R-Smads form heteromeric complexes with Smad4, which acts as a common-partner Smad (Co-Smad) (3,6–8). R- and Co-Smads share two conserved regions at their N- and C-termini, which are called Mad homology region 1 (MH1) and Mad homology region 2 (MH2), respectively. The N-terminal regions in Smad3 and Smad4 bind directly, albeit with low affinity, to DNA. The target for this DNA-binding activity is the Smad binding element (SBE) (6–8). SBE contains a 5′-GACA-3′ core sequence, which is of critical importance in promoter regions of certain TGF-β responsive genes (8). The MH2 domain mediates homo- and heteromeric complex formation and contributes to the Smad transcriptional activity (see below) (6–8). MH1 and MH2 domains interact with and repress the activity of each other (9). In R-Smads this intramolecular repression can be relieved by type I receptor phosphorylation, which allows R-Smads to form complexes with Smad4 (2,3,7,8). Both domains have been shown to associate with a large number of transcription factors (6,8,10). Mice deficient in a particular Smad have developmental defects; Smad2- and Smad4-deficient mice die as early as day E6.5, whereas Smad3 null mice are viable (11). Smad2 and Smad4 genes are frequently mutated in particular types of cancer and have been classified as putative tumor suppressor genes (12).
The transcriptional coactivators p300/CBP have been shown to interact in a ligand-dependent manner with R-Smads (13–20). p300/CBP have intrinsic histone acetyltransferase (HAT) activity, which facilitates transcription by decreasing chromosome condensation through histone acetylation and by increasing the accessibility of transcription factors with the basal transcription machinery (21–23). Thus, p300/CBP positively regulate Smad-mediated transcriptional activation. R-Smads interact with p300/CBP mainly through their MH2 domains. In p300/CBP, the Smad interaction has been mapped at the C-terminus although the N-terminal region of p300/CBP may also contribute (13–20).
The co-activator P/CAF was originally discovered through its association with p300/CBP (24). Both P/CAF and p300/CBP have intrinsic HAT activity, but with different substrate specificity and differential effects. P/CAF and p300 acetylate p53 at distinct lysine residues (25,26). The acetylase domain of P/CAF is required for MyoD-dependent activation, whereas p300 functions as an adaptor protein in MyoD activation for which the acetylase activity is dispensable (27). P/CAF is also known to be a part of a large complex of more than 20 proteins in HeLa cells (28,29). However, p300/CBP is not present as a stoichiometric component in this complex (28). P/CAF has also been shown to interact with transcription factors, including liganded nuclear hormone receptors (30–32), NF-Y (33) and cyclin D (34). Recent data support the notion of a critical role of P/CAF in the control of growth inhibition, differentiation and apoptosis. P/CAF and proteins that associate with it, the so-called P/CAF acetylase complex, may act as a tumor suppressors (28).
We now present evidence for a hitherto unknown direct link between two proteins with putative tumor suppressor function, P/CAF and Smad3; P/CAF interacts with Smad3 upon TGF-β type I receptor activation and co-operates with Smad3 in stimulating TGF-β/Smad-mediated transcriptional responses.
MATERIALS AND METHODS
Cell culture
HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) containing 10% fetal calf serum (FCS; Sigma) and 1× MEM non-essential amino acids (Sigma). MDA-MB468, COS7 and 293T cells were maintained in DMEM containing 10% FCS. All media were supplemented with 100 IU/ml penicillin, 100 µg/ml streptomycin and 2 mM glutamine.
Luciferase assay
One day prior to transfection, HepG2 and MDA-MB468 cells were seeded at 2.5 × 105 cells/well in 6-well plates. The cells were transfected using the calcium phosphate co-precipitation method, as previously described (35). In all experiments, sea-pansy luciferase (pRL-TK; Promega) activity was measured to normalize for transfection efficiency. Each transfection was carried out in triplicate and repeated at least twice.
Plasmid constructions
Expression constructs for constitutively active TGF-β type I receptor, also termed activin receptor-like kinase (ALK)5, Myc–Smad3 and Flag–Smad4 have been described previously (36). Expression constructs for Gal4 DNA-binding domain (pGAD424) and Gal4–Smad2 were obtained from Dr J. Massagué (Memorial Sloan-Kettering Cancer Center, New York, NY; 37). HA–p300 and Flag–P/CAF were provided by Dr R. Derynck (University of California, San Francisco, CA; 13) and Dr T. Kouzarides (Wellcome/CRC Institute, Cambridge University, Cambridge, UK; 38), respectively. Flag–p300, Flag–Smad3, 6× Myc–Smad2 and 6× Myc–Smad3 were generously provided by Dr K. Miyazono (The Cancer Institute of Japanese Foundation for Cancer Research, Tokyo, Japan; 15,39). A Gal4 DNA-binding domain fusion with Smad3 was created by subcloning of Smad3 into pGAD424. P/CAF, P/CAF(1–348) and P/CAF(349–832) were cloned by PCR and inserted into the Flag–pCDEF3 or 6× Myc–pCDNA3 vectors (15). Mammalian expression constructs for the Smad3 mutants in Figure 1a were made by PCR technique and subcloning in 6× Myc–pCDNA3 vector. Luciferase transcriptional reporter constructs, pGL3ti–(SBE)4 and Gal4–M1-Luc, were described previously (35,40).
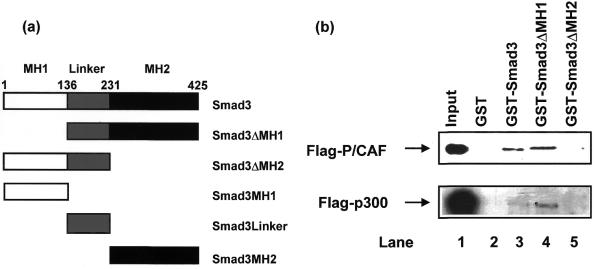
Figure 1.
P/CAF interacts with Smad3 as analyzed by a GST-pull-down assay. (a) A schematic presentation of the Smad3 mutants is shown. (b) GST-pull-down assays were performed in the presence of recombinant Flag–p300 or Flag–P/CAF produced in insect cells. Interaction between Smad3 and P/CAF or p300 was detected by western blotting with Flag antibody. Flag–P/CAF or Flag–p300 (1 µg) was applied in lane 1.
Immunoprecipitation and western blotting
Combinations of Smads, P/CAF and p300, in the presence or absence of ALK5 constitutively active (ALK5ca), were transfected in COS7 cells at 1.2 × 106 cells/10-cm dish using Fugene 6 (Boehringer Mannheim). Forty hours after transfection, cells were lysed in 1 ml of TNE buffer [10 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM phenylmethylsulphonyl fluoride (PMSF), 5 µg/ml leupeptin and 100 U/ml Trasylol]. The cell lysates were precleared with protein A–Sepharose beads (Pharmacia) and incubated with Flag M5 antibody (Sigma) for 2 h at 4°C. Subsequently, protein G–Sepharose beads (Pharmacia) were added to the reaction mixture and incubated for 30 min at 4°C. After washing the immunoprecipitates with high salt buffer (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 1% Triton X-100, 1 mM PMSF, 5 µg/ml leupeptin and 100 U/ml Trasylol) three times and with TNE buffer once, immunoprecipitates, as well as aliquots of total cell lysates, were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to a Hybond-C Extra membrane (Amersham). The membrane was subsequently probed with Flag M5 antibody or Myc 9E10 monoclonal antibody (Santa Cruz). Primary antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse antibody (Amersham) and a chemiluminescent substrate.
Preparation of nuclear extract
293T cells were seeded at 3.5 × 106/10-cm dish 1 day before transfection. The cells were transfected with Flag–Smad3 (2.5 µg), 6× Myc–P/CAF (2.5 µg) in the presence or absence of ALK5ca (2.5 µg) using Fugene 6. Forty hours later, nuclear extracts were prepared by the method of Schreiber et al. (41). In brief, the cells were suspended in 400 µl of 10 mM HEPES, pH 7.9, containing 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT and 0.5 mM PMSF for 15 min on ice. Then, 25 µl of 10% NP-40 were added to the suspension. After centrifugation, the pellet was incubated with 20 mM HEPES, pH 7.9, containing 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 mM PMSF for 15 min at 4°C. Subsequently, the mixture was centrifuged to get a clear supernatant which was defined as the nuclear extract. The protein concentration of all nuclear extracts was ∼6 µg/µl.
Preparation of purified proteins from baculovirus
Flag–P/CAF and Flag–p300 were expressed in Sf9 cells by infecting recombinant baculovirus and purified to homogeneity.
Electromobility shift assay (EMSA)
EMSAs were performed as previously described (42) with minor modifications. The 4× wild-type (WT) oligonucleotide (35), which consists of four repeats of SBE, was end-labeled with [γ-32P]ATP (Amersham) using T4 DNA polynucleotide kinase. Nuclear extract (6 µg) and 32P-labeled 4× WT (10 fmol) were incubated for 30 min at 25°C in a solution of 20 mM HEPES, pH 7.9, 30 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 0.8 mM NaPi, 20% glycerol, 4 mM spermidine, 0.3 µg/µl of poly(dI·dC) and 0.25 µg/µl of salmon sperm DNA at a final volume of 20 µl. Where indicated, a 200-fold molar excess of cold competitors [4× WT or 4× mutant (Mu) oligonucleotide (35)] was included in the reaction mixture. Protein–DNA complexes were analyzed in 5% non-denaturing polyacrylamide gels containing 0.5× TBE (0.045 M Tris–borate, 0.001 M EDTA, pH 8.0).
GST-pull-down assay
Smad3 and its mutants were subcloned in pGEX4T-1 (Pharmacia). GST proteins from Escherichia coli were purified according to the manufacturer’s instructions (Pharmacia). Flag–P/CAF or Flag–p300 (1 µg) was precleared with GST immobilized to GSH–Sepharose 4B (Pharmacia) for 30 min at 4°C. Subsequently, recombinant Flag–P/CAF or Flag–p300 was incubated with GST–Smad3 or its mutants immobilized to GSH–Sepharose 4B for 2 h at 4°C and washed three times with 20 mM Tris–HCl, pH 7.4, containing 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM PMSF and 100 U/ml Trasylol. After loading the samples on the SDS–PAGE gel, proteins were blotted on a Hybond-C extra membrane and detected with anti-FlagM5 antibody using the chemiluminescence method.
RESULTS
P/CAF binds directly to the Smad3 MH2 region
Smads need to recruit transcriptional co-activators to activate gene transcription (3,6–8). The Smad transcriptional factors and the transcriptional co-activator P/CAF mediate growth inhibition, differentiation and apoptotic signals (11,28); both are putative tumor suppressor gene products and may therefore functionally cooperate. In addition, the co-activator P/CAF has been shown to bind directly to sequence-specific transcriptional activators independently of p300/CBP (30–34). We therefore examined the possibility of a direct interaction between P/CAF and Smad3 using a GST-pull-down assay; GST alone, GST–Smad3, GST–Smad3ΔMH1 and GST–Smad3ΔMH2 were incubated with Flag–P/CAF or Flag–p300 purified from Sf9 cells infected with baculovirus. As shown in Figure 1b, GST–Smad3ΔMH1 as well as GST–Smad3 could interact with Flag–P/CAF, but not GST or GST–Smad3ΔMH2. p300 associated with the MH2 domain of Smad3, as previously reported (13–20). Thus, P/CAF can bind directly to the Smad3 MH2 domain.
Activation of TGF-β type I receptor induces the association of TGF-β/activin R-Smads with P/CAF
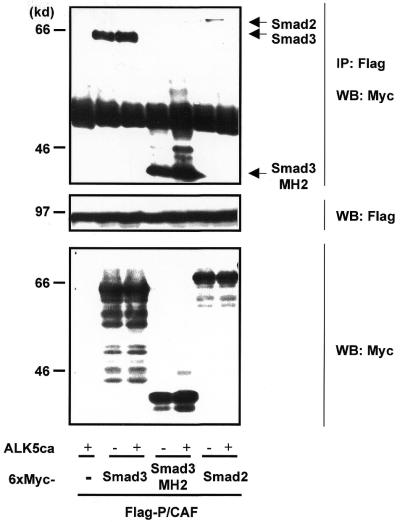
Smad3 is highly similar to Smad2 in its amino acid sequences and both of them belong to the TGF-β/activin-Smad subfamily (2,3,7,8). We examined whether P/CAF can associate with Smad2 and Smad3 in cultured mammalian cells. COS7 cells were transfected with combinations of expression plasmids for P/CAF, Smads and an ALK5ca. ALK5ca stimulates the TGF-β pathway in a ligand-independent manner (43). As shown in Figure 2, P/CAF could interact with Smad3 or Smad3MH2 in the absence of ALK5ca. This interaction was increased slightly by the co-transfection of ALK5ca; the degree to which the binding is increased varied between experiments. The ligand-independent interaction may be caused by the predominant nuclear localization of the transfected Smad3, in particular when highly overexpressed (44,45); endogenous Smad3 was found to be confined to the cytoplasm in the absence of ligand stimulus (36) and P/CAF is a nuclear protein (24). Smad2 did not associate with P/CAF in the absence of ALK5ca, but this association was detected in the presence of ALK5ca. Smad3 bound with higher affinity to P/CAF than did Smad2. Of note, Smad3 is a much stronger transcriptional activator than Smad2 (data not shown).
Figure 2.
P/CAF interacts with Smad2 and Smad3 upon TGF-β type I receptor activation. Upper panel, the interaction of 6× Myc–Smad3 and its mutants with Flag–P/CAF. 6× Myc–Smad3, 6× Myc–Smad3MH2 or 6× Myc–Smad2 was co-transfected with Flag–P/CAF with or without ALK5ca in COS7 cells. Immunoprecipitations were performed with Flag antibody, and co-immunoprecipitated Smads were detected by western blotting with Myc antibody. The expression of Flag–P/CAF and 6× Myc–Smads was measured by applying one-fiftieth of total cell lysate on SDS–PAGE followed by western blotting with Flag antibody (middle panel) or Myc antibody (lower panel).
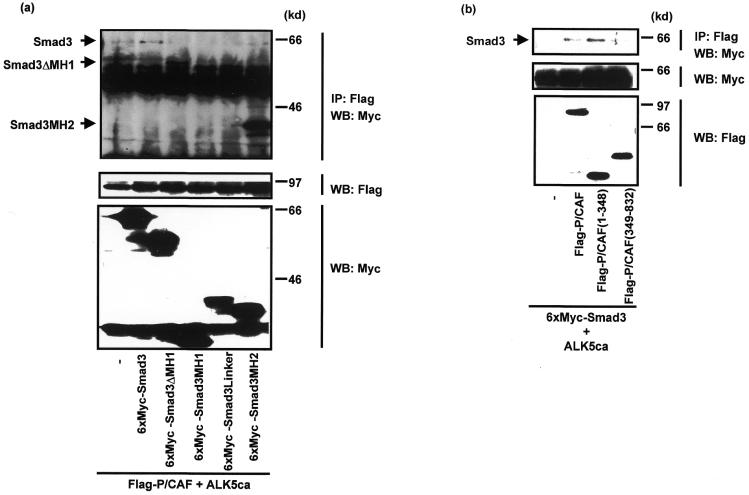
Considering the differential binding of Smad2 versus Smad3 to P/CAF, we have focused on the interaction of P/CAF with Smad3 in subsequent experiments. To determine which domain(s) of Smad3 interact(s) with P/CAF in COS7 cells, we made constructs directing the expression of several 6× Myc-tagged deletion mutants of Smad3 (see Fig. 1a). Each mutant was co-transfected with Flag-tagged P/CAF and ALK5ca in COS7 cells and cell lysates were immunoprecipitated with Flag antibody followed by western blotting with Myc antibody. As shown in Figure 3a, P/CAF bound to Smad3, Smad3ΔMH1 and Smad3MH2, but not to Smad3MH1 or Smad3Linker. These results are consistent with the GST-pull-down experiment.
Figure 3.
Determination of interacting domain in COS7 cells. (a) P/CAF can bind to the MH2 domain of Smad3. 6× Myc–Smad3 or its mutants (Fig. 1a) was co-transfected with Flag–P/CAF and ALK5ca in COS7 cells. The cell lysates of COS7 cells were subjected to immunoprecipitation with Flag antibody followed by western blotting with Myc antibody (upper panel). The expression of Flag–P/CAF and 6× Myc–Smad3 or its mutants was measured by applying one-fiftieth of total cell lysate on SDS–PAGE followed by western blotting with Flag antibody (middle panel) or Myc antibody (lower panel). (b) Smad3 associates with the N-terminal region of P/CAF. 6× Myc–Smad3 was co-transfected with Flag–P/CAF, Flag–P/CAF(1–348) or Flag–P/CAF(349–832) in the presence of ALK5ca in COS7 cells. The cell lysates were subjected to immunoprecipitation with Flag antibody followed by western blotting with Myc antibody (upper). The expression of Smad3 and P/CAF or its mutants in total lysates is also shown in the middle and lower panels, respectively.
Determination of the Smad3 interaction domain in P/CAF
P/CAF, which consists of 832 amino acid residues, contains a HAT domain (amino acid residues 349–658) with similarity to yeast GCN5 and a bromodomain (amino acid residues 742–832) which binds to acetylated lysine residues (24,28,46). To map the Smad interaction domain in P/CAF we expressed two mutants of P/CAF, P/CAF(1–348) and P/CAF(349–832) with Smad3 in COS7 cells. An interaction of P/CAF(1–348) with Smad3 was detected, but P/CAF(349–832) marginally interacted with Smad3 (Fig. 3b), suggesting that P/CAF mainly interacted with Smad3 through its N-terminal domain.
P/CAF is a co-activator for Smad3
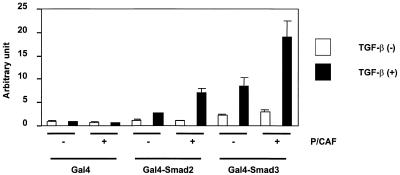
Smads have been shown to have intrinsic transcriptional activity when fused to the Gal4 DNA-binding domain (37,47). In order to determine if P/CAF could affect the intrinsic transcriptional activity, cells were transfected with a luciferase reporter gene containing multiple Gal4-binding sites upstream of a minimal promoter, various Gal4–Smad constructs and P/CAF (Fig. 4). Following transfection, cells were grown in the absence or presence of TGF-β1. Expression of P/CAF enhanced the transcriptional activity of Gal4–Smad2 and Gal4–Smad3 upon stimulation with TGF-β1. P/CAF had no effect on the luciferase activity when cells were transfected with only the Gal4 DNA-binding domain (Fig. 4). These results demonstrate the co-activator function of P/CAF for Smad2 and Smad3.
Figure 4.
P/CAF potentiates the transcriptional activity of Gal4–Smad2 and Gal4–Smad3. HepG2 cells were co-transfected with Gal4–M1-Luc and Flag–P/CAF and plasmids encoding the indicated Gal4–Smad in the absence (open bars) or presence (solid bars) of TGF-β1. All data are presented as relative values of Gal4 alone without TGF-β1.
To study the effect of P/CAF on a TGF-β/Smad-mediated transcriptional response, HepG2 cells were transfected with a P/CAF expression vector and an SBE-driven reporter plasmid, termed pGL3ti–(SBE)4, containing Smad3 and Smad4 binding elements (35). P/CAF is able to induce TGF-β1-dependent luciferase activity in a dose-dependent manner (data not shown). The potentiating effect of P/CAF was similar to that obtained previously with p300/CBP (13–20), i.e. weak but significant; this suggests that P/CAF and p300/CBP are present at near to optimal levels.
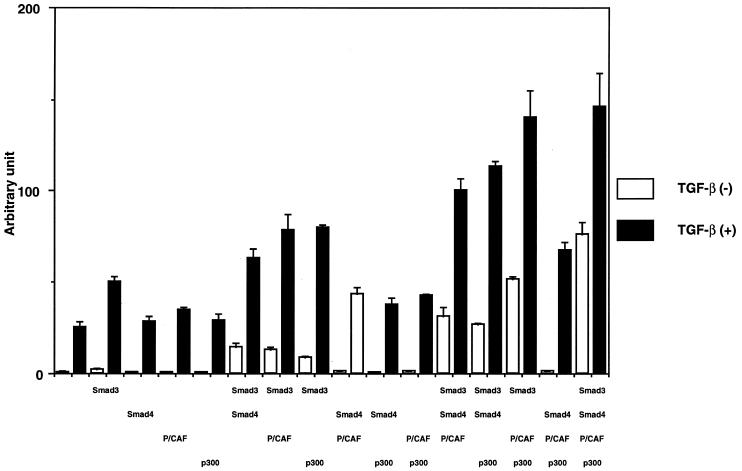
Smad4 and p300 are co-activators of Smad3-mediated transcriptional responses and P/CAF is a CBP-interacting protein. HepG2 cells were therefore transfected with different combinations of P/CAF, Smad3, Smad4 and p300, together with pGL3ti–(SBE)4; after stimulation with TGF-β1, transcriptional responses were analyzed. The combination of P/CAF and Smad3 enhanced the reporter activity to a level similar to that obtained in cells transfected with Smad3 and Smad4 (Fig. 5). P/CAF was also found to cooperate with p300 individually and in combination with other components (Fig. 5). We also used the p800neoLuc reporter, which contains 800 nt of the PAI-1 promoter (48). Similar to the results with the synthetic SBE containing reporter in Figure 5, the basal activity was induced by co-transfection of Smad3, Smad4, P/CAF and p300 (data not shown).
Figure 5.
P/CAF stimulates Smad3-induced transcription together with p300 and Smad4. HepG2 cells were transfected with pGL3ti–(SBE)4 and the indicated combinations of expression plasmids for Myc–Smad3, Flag–Smad4, Flag–P/CAF and HA–p300 in the absence (open bars) or presence (solid bars) of TGF-β1. All data are presented as relative values of mock transfected cells without TGF-β1 treatment.
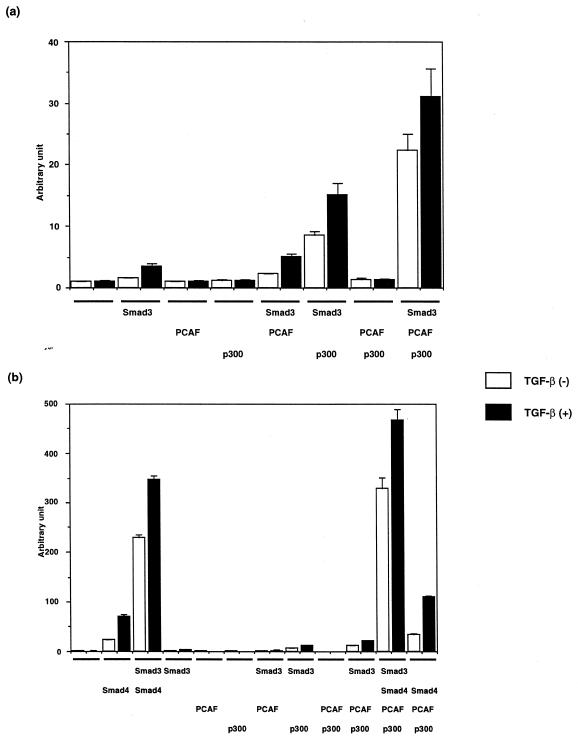
Smad4 is known to recruit R-Smads after ligand stimulation, whereafter the complex is translocated to the nucleus (2,3,7,8). To examine whether P/CAF can cooperate with Smad3 in the absence of Smad4, the pGL3ti–(SBE)4 reporter was co-transfected with various combinations of P/CAF, Smad3 and p300 in MDA-MB468 cells, which genetically lack endogenous Smad4 (Fig. 6). As previously reported, TGF-β1 weakly stimulated transcription upon transfection of Smad3, and P/CAF or p300 alone had no effect on the luciferase activity even in the presence of TGF-β1. Co-transfection of either P/CAF or p300 with Smad3 enhanced the basal activity of luciferase, but not the TGF-β1-dependent activity of the promoter-reporter gene. P/CAF and p300 were less efficient than Smad4 in increasing the basal luciferase activity of pGL3ti–(SBE)4. Triple transfection of P/CAF, p300 and Smad3 in MDA-MB468 cells dramatically increased the basal luciferase activity, compared with single or double transfection of P/CAF, p300 and Smad3. Introduction of Smad4 with various combinations of the other components stimulated the basal transcriptional levels in particular. Together, these results suggest that P/CAF supports the transcriptional function of Smad3 together with p300 and/or Smad4.
Figure 6.
P/CAF activates the basal transcription of pGL3ti–(SBE)4 in MDA-MB468 cells in the absence (a) or presence (b) of Smad4. MDA-MB468 cells were transfected with pGL3ti–(SBE)4 and the indicated combinations of Myc–Smad3, Flag–Smad4, Flag–P/CAF and HA–p300 without (open bars) or with (solid bars) TGF-β1. All data are presented as relative values of mock transfected cells without TGF-β1 treatment.
Contribution of P/CAF to DNA binding of Smad3
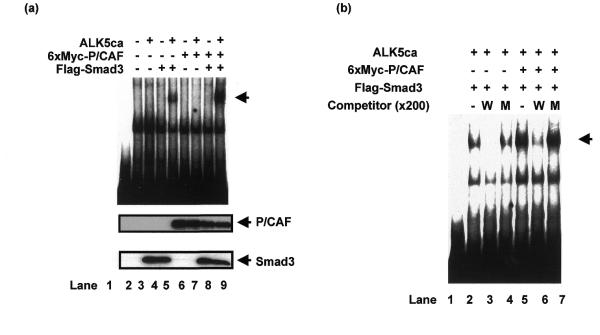
To test the possibility that P/CAF affects the ability of Smad3 to bind SBE, we performed EMSAs using the nuclear extracts from 293T cells transfected with combination(s) of Smad3, P/CAF and ALK5ca (Fig. 7a). Nuclear extracts from 293T cells transfected with Smad3 alone showed no specific binding to the 4× WT SBE probe. However, a specific shifted band appeared when a nuclear extract from 293T cells transfected with both Smad3 and ALK5ca was used. Interestingly, a nuclear extract from cells transfected with P/CAF, Smad3 and ALK5ca enhanced the intensity of the shifted band (Fig. 7a, lane 9). We could not see any specific bands when nuclear extract from 293T cells transfected with P/CAF alone was incubated with the probe (Fig. 7a, lane 6). The specificity of the shifted complex was shown by the disappearance of the band when 4× WT with a 200-fold excess of the cold competitor was included in the assay (Fig. 7b, lane 5 versus lane 6), whereas 4× Mu, which has three mutations in SBE, did not abolish the complex (Fig. 7b, lane 5 versus lane 7). Similar results were obtained when nuclear extracts from 293T cells transfected with Smad3 and ALK5ca was used (Fig. 7b, lanes 2–4). We confirmed that Smad3 and P/CAF in each sample were expressed at similar levels (Fig. 7a). We also found that recombinant P/CAF induced a stronger binding of recombinant phosphorylated Smad3 to DNA (data not shown), which is consistent with the nuclear extract from 293T cells co-transfected with P/CAF, Smad3 and ALK5ca. These experiments suggest that P/CAF increases the affinity of Smad3 for its cognate SBE binding motif.
Figure 7.
P/CAF enhances the binding of Smad3 to SBE. (a) Gel shift analysis using nuclear extracts from 293T cells. Gel shift assays were performed using nuclear extract from 293T cells transfected with the indicated combination of Flag–Smad3, 6× Myc–P/CAF and ALK5ca. 4× WT was used as a probe. Lane 1, no protein added. The specific DNA complex is indicated with an arrow. The expression of Flag–Smad3 and 6× Myc–P/CAF was measured by applying one-fiftieth of total lysate on SDS–PAGE followed by western blotting with Flag and Myc antibodies, respectively. (b) Specificity of the Smad3–DNA interaction. Cold competitor (200-fold excess) of 4× WT (W) or 4× Mu (M) was incubated with nuclear extracts from 293T cells. Lane 1, no protein added. The specific DNA complex is indicated with an arrow.
DISCUSSION
In the present study we have shown that the co-activator P/CAF physically associates and functionally co-operates with Smad3 in TGF-β-induced transcriptional responses. P/CAF is thus required for most optimal TGF-β/Smad3-dependent promoter activation. Furthermore, the P/CAF-mediated potentiation of TGF-β/Smad3-driven synthetic reporter constructs containing multimerized SBEs (Fig. 5), as well as endogenous PAI-1 promoter reporter constructs (data not shown), could be further enhanced by co-activators p300 and Smad4. The MH2 domain of Smad3 interacted directly with P/CAF; the N-terminal region in P/CAF was found to be critical for Smad3 binding. Interestingly, p300 has also been shown to interact with R-Smad MH2 domains (13–20) and the N-terminal domain of P/CAF was found to be necessary for the interaction with CBP (32). Taken together, Smad3/Smad4 complex may thus form in vivo transcription factor complexes either with P/CAF or p300 alone or involving both of these components.
Our studies extend the repertoire of P/CAF-interacting transcription factors with Smad3. p300/CBP also interact with multiple transcription factors and this has been established as a possible means for integrating the effects of multiple signaling pathways. For example, Stat3 and Smad1 are brought together within one functionally synergizing complex upon stimulation by bone morphogenetic protein and leukemia inhibitory factor through their association with p300 (49). Whether cross-talk of Smad3 with other signaling pathways is mediated through P/CAF is an interesting topic for future research.
We observed that the TGF-β-dependent sequence-specific DNA binding of Smad3 was enhanced by P/CAF (Fig. 7). Interestingly, we did not observe a slower migrating protein–DNA complex upon the addition of P/CAF to Smad3. It is possible that the stoichiometry of Smad3 bound to DNA changes upon P/CAF binding without changing the mobility of the retarded protein complex. However, whereas the Flag antibody, which recognizes Flag–Smad3, produced a supershifted band in a gel shift assay, we could not detect a supershifted band upon addition of a Myc antibody, which recognizes 6× Myc–P/CAF (data not shown). Another possibility is that P/CAF, which has an intrinsic HAT activity, may induce acetylation of Smad3 and acetylated Smad3 may bind DNA with higher affinity. A similar phenomenon has been reported for p300 following acetylation of p53 and GATA1 (25,26,50,51). However, although we found that Smad3 is acetylated, we were unable to observe a difference in the acetylation with or without TGF-β receptor activation (data not shown). Yet another option is that a putative P/CAF–Smad3 complex bound to DNA cannot withstand the EMSA conditions. However, in preliminary experiments using a DNA affinity precipitation, using biotinylated multimerized SBE on cell lysates from cells transfected with P/CAF, activated type I receptor and Smad3, followed by western blotting, we were unable to demonstrate a P/CAF interaction with phosphorylated Smad3 bound to DNA (data not shown). The DNA precipitation method should detect the interaction of P/CAF and Smad3 independent of their possible dissociation upon gel electrophoresis. Alternatively, P/CAF may increase the affinity of Smad3 to DNA by inducing a conformational change in Smad3, and possibly the binding of P/CAF to Smad3 may be mutually exclusive with the binding of Smad3 to DNA. Taking all our data together, it is this possibility that we favor most to explain why the addition of P/CAF enhances Smad3 binding without changing the mobility of the Smad3–SBE complex in EMSA. However, further studies are needed to distinguish between these possibilities. It is of interest to note that Suzuki et al. (52) also found that the interaction of co-activator p300 to transcription factor Sp1 increased Sp1–DNA binding, without resulting in a shift of the DNA–protein complex in EMSA; it was proposed that p300 may induce a conformational change in Sp1 leading to an Sp1 with increased affinity for DNA, but with reduced affinity for p300. Furthermore, Pardali et al. (53) recently reported that DNA binding of Sp1 was enhanced without changing the mobility of DNA–protein complexes in EMSA.
In conclusion, after phosphorylation by type I receptor, Smad3 forms a heteromeric complex with Smad4, which translocates from the cytosol to the nucleus, whereafter Smad3 can bind to P/CAF, which promotes its DNA binding. P/CAF may activate Smad-mediated transcriptional responses independently or in co-operation with p300.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Fumiko Itoh and Susanne Grimsby for excellent technical assistance and Drs T. Kouzarides and A. Moustakas for valuable discussions. We are grateful to Drs Y. Nakatani (Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA) for recombinant P/CAF and N. Ferrara (Genentech, South San Francisco, CA) for recombinant TGF-β1. In addition, we thank Drs J. Massagué, R. Derynck, T. Kouzarides, K. Miyazono and J. A. Langer (Robert Wood Johnson Medical School, Piscataway, NJ) for expression plasmids. This work was supported by the Dutch Cancer Society (NKI 2000-2217).
REFERENCES
- 1.Roberts A.B. and Sporn,M.B. (1990). In Sporn,M.B. and Roberts,A.B. (eds), Peptide Growth Factors and Their Receptors, Part 1. Springer-Verlag, Berlin, Germany, pp. 419–472.
- 2.Massagué J. (1998) Annu. Rev. Biochem., 67, 753–791. [DOI] [PubMed] [Google Scholar]
- 3.Massagué J. and Chen,Y.-G. (2000) Genes Dev., 14, 627–644. [PubMed] [Google Scholar]
- 4.Abdollah S., Macias-Silva,M., Tsukazaki,T., Hayashi,H., Attisano,L. and Wrana,J.L. (1997) J. Biol. Chem., 272, 27678–27685. [DOI] [PubMed] [Google Scholar]
- 5.Souchelnytskyi S., Tamaki,K., Engstrom,U., Wernstedt,C., ten Dijke,P. and Heldin,C.-H. (1997) J. Biol. Chem., 272, 28107–28115. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R., Zhang,Y. and Feng,X.H. (1998) Cell, 95, 737–740. [DOI] [PubMed] [Google Scholar]
- 7.Attisano L. and Wrana,J.L. (2000) Curr. Opin. Cell Biol., 12, 235–243. [DOI] [PubMed] [Google Scholar]
- 8.Massagué J. and Wotton,D. (2000) EMBO J., 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hata A., Lo,R.S., Wotton,D., Lagna,G. and Massagué,J. (1997) Nature, 388, 82–87. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y. and Derynck,R. (1999) Trends Cell Biol., 9, 274–279. [DOI] [PubMed] [Google Scholar]
- 11.Goumans M.-J. and Mummery,C. (2000) Int. J. Dev. Biol. 44, 253–265. [PubMed] [Google Scholar]
- 12.de Caestecker M.P., Piek,E. and Roberts,A.B. (2000) J. Natl Cancer Inst., 92, 1388–1402. [DOI] [PubMed] [Google Scholar]
- 13.Feng X.H., Zhang,Y., Wu,R.-Y. and Derynck,R. (1998) Genes Dev., 12, 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janknecht R., Wells,J.N. and Hunter,T. (1998) Genes Dev., 12, 2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishihara A., Hanai,J., Okamoto,N., Yanagisawa,J., Kato,S., Miyazono,K. and Kawabata,M. (1998) Genes Cells, 3, 613–623. [DOI] [PubMed] [Google Scholar]
- 16.Pouponnot C., Jayaraman,L. and Massagué,J. (1998) J. Biol. Chem., 273, 22865–22868. [DOI] [PubMed] [Google Scholar]
- 17.Shen X., Hu,P.P., Liberati,N.T., Datto,M.B., Frederick,J.P. and Wang,X.-F. (1998) Mol. Biol. Cell, 9, 3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topper J.N., DiChiara,M.R., Brown,J.D., Williams,A.J., Falb,D., Collins,T. and Gimbrone,M.A.,Jr (1998) Proc. Natl Acad. Sci. USA, 95, 9506–9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima K., Yanagisawa,M., Arakawa,H., Kimura,N., Hisatsune,T., Kawabata,M., Miyazono,K. and Taga,T. (1999) Science, 284, 479–482. [DOI] [PubMed] [Google Scholar]
- 20.Pearson K.L., Hunter,T. and Janknecht,R. (1999) Biochim. Biophys. Acta, 1489, 354–364. [DOI] [PubMed] [Google Scholar]
- 21.Kuo M.-H. and Allis,C.D. (1998) Bioessays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- 22.Shikama N., Lyon,J. and La Thangue,N.B. (1997) Trends Cell Biol., 7, 230–236. [DOI] [PubMed] [Google Scholar]
- 23.Xu L., Glass,C.K. and Rosenfeld,M.G. (1999) Curr. Opin. Genet. Dev., 9, 140–147. [DOI] [PubMed] [Google Scholar]
- 24.Yang X.-J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- 25.Liu L., Scolnick,D.M., Trievel,R.C., Zhang,H.B., Marmorstein,R., Halazonetis,T.D. and Berger,S.L. (1999) Mol. Cell. Biol., 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakaguchi K., Herrera,J.E., Saito,S., Miki,T., Bustin,M., Vassilev,A., Anderson,C.W.and Appella,E. (1998) Genes Dev., 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puri P.L., Sartorelli,V., Yang,X.-J., Hamamori,Y., Ogryzko,V.V., Howard,B.H., Kedes,L., Wang,J.Y.J., Graessmann,A., Nakatani,Y. and Levrero,M. (1997) Mol. Cell, 1, 35–45. [DOI] [PubMed] [Google Scholar]
- 28.Schiltz R.L. and Nakatani,Y. (2000) Biochim. Biophys. Acta, 1470, M37–M53. [DOI] [PubMed] [Google Scholar]
- 29.Ogryzko V.V., Kotani,T., Zhang,X., Schiltz,R.L., Howard,T., Yang,X.-J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- 30.Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- 31.Blanco J.C., Minucci,S., Lu,J., Yang,X.-J., Walker,K.K., Chen,H., Evans,R.M., Nakatani,Y. and Ozato,K. (1998) Genes Dev., 12, 1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korzus E., Torchia,J., Rose,D.W., Xu,L., Kurokawa,R., Mclnerney,E.M., Mullen,T.-M., Glass,C.K. and Rosenfeld,M.G. (1998) Science, 279, 703–707. [DOI] [PubMed] [Google Scholar]
- 33.Currie R.A. (1998) J. Biol. Chem., 273, 1430–1434. [DOI] [PubMed] [Google Scholar]
- 34.McMahon C., Suthiphongchai,T., DiRenzo,J. and Ewen,M.E. (1999) Proc. Natl Acad. Sci. USA, 96, 5382–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonk L.J., Itoh,S., Heldin,C.-H., ten Dijke,P. and Kruijer,W. (1998) J. Biol. Chem., 273, 21145–21152. [DOI] [PubMed] [Google Scholar]
- 36.Nakao A., Imamura,T., Souchelnytskyi,S., Kawabata,M., Ishisaki,A., Oeda,E., Tamaki,K., Hanai,J., Heldin,C.-H., Miyazono,K. and ten Dijke,P. (1997) EMBO J., 16, 5353–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F., Pouponnot,C. and Massagué,J. (1997) Genes Dev., 11, 3157–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid J.L., Bannister,A.J., Zegerman,P., Martinez-Balbas,M.A. and Kouzarides,T. (1998) EMBO J., 17, 4469–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawabata M., Inoue,H., Hanyu,A., Imamura,T. and Miyazono,K. (1998) EMBO J., 17, 4056–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryant G.O., Martel,L.S., Burley,S.K. and Berk,A.J. (1996) Genes Dev., 10, 2491–2504. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber E., Matthias,P., Muller,M.M. and Schaffner,W. (1989) Nucleic Acids Res., 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dennler S., Itoh,S., Vivien,D., ten Dijke,P., Huet,S. and Gauthier,J.-M. (1998) EMBO J., 17, 3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieser R., Wrana,J.L. and Massagué,J. (1995) EMBO J., 14, 2199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y., Lebrun,J.-J. and Vale,W. (1996) Proc. Natl Acad. Sci. USA, 93, 12992–12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Musci,T. and Derynck,R. (1997) Curr. Biol., 7, 270–276. [DOI] [PubMed] [Google Scholar]
- 46.Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K., Zhou,M.-M. and Zhou,M.-M. (1999) Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- 47.Liu F., Hata,A., Baker,J.C., Doody,J., Carcamo,J., Harland,R.M. and Massagué,J. (1996) Nature, 381, 620–623. [DOI] [PubMed] [Google Scholar]
- 48.Abe M., Harpel,J.G., Metz,C.N., Nunes,I., Loskutoff,D.J. and Rifkin,D.B. (1994) Anal. Biochem., 216, 276–284. [DOI] [PubMed] [Google Scholar]
- 49.Nakashima K., Yanagisawa,M., Arakawa,H., Kimura,N., Hisatsune,T., Kawabata,M., Miyazono,K. and Taga,T. (1999) Science, 284, 479–482. [DOI] [PubMed] [Google Scholar]
- 50.Boyes J., Byfield,Y., Nakatani,Y. and Ogryzko,V. (1998) Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W. and Bieker,J.J. (1998) Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki T., Kimura,A., Nagai,R. and Horikoshi,M. (2000) Genes Cells, 5, 29–41. [DOI] [PubMed] [Google Scholar]
- 53.Pardali K., Kurisaki,A., Morén,A., ten Dijke,P., Kardassis,D. and Moustakas,A. (2000) J. Biol. Chem., 275, 29244–29256. [DOI] [PubMed] [Google Scholar]