Summary
IL-23 and IL-17 are well-validated therapeutic targets in autoinflammatory diseases. Antibodies targeting IL-23 and IL-17 have shown clinical efficacy but are limited by high costs, safety risks, lack of sustained efficacy, and poor patient convenience as they require parenteral administration. Here we present designed miniproteins inhibiting IL-23R and IL-17 with antibody-like low picomolar affinities at a fraction of the molecular size. The minibinders potently block cell signaling in vitro, and are extremely stable, enabling oral administration and low-cost manufacturing. The orally administered IL-23R minibinder shows efficacy better than a clinical anti-IL-23 antibody in mouse colitis and has a favorable PK and biodistribution profile in rats. This work demonstrates that orally administered de novo designed minibinders can reach a therapeutic target past the gut epithelial barrier. With high potency, gut stability, and straightforward manufacturability, de novo designed minibinders are a promising modality for oral biologics.
In brief
De novo proteins can be computationally designed with sub-picomolar affinity and extreme stability to enable oral administration, and were effective in a model of colitis.
Graphical Abstract

Introduction
IL-23 cytokine is produced by antigen presenting cells and promotes differentiation and phenotype maintenance of T-helper type 17 (TH17) cells. IL-23 stimulates production of the pro-inflammatory cytokine IL-17 in circulating TH17 as well as tissue-resident innate lymphoid cells (ILC) and ɣδT-cells. IL-23 and IL-17 are genetically and clinically validated therapeutic targets for the treatment of several TH17-mediated autoinflammatory diseases including inflammatory bowel disease (IBD; IL-23 only) and psoriasis (both IL-23 and IL-17). However, existing antibody therapies have several limitations. Only about 30% of IBD patients receiving the anti-IL-23 mAb Stelara achieve remission, and approximately 20% of initial responders lose response over time due to generation of anti-drug antibodies.1–4 Systemic immune suppression puts patients at increased risk for malignancies and serious infections.5 Due to their large molecular size and poor permeability, antibodies are not administered orally but by intravenous infusion or subcutaneous injection, which can be inconvenient and stressful for patients. Systemically administered antibodies generally show poor tissue penetrance, only reaching 5–10% of serum concentration in target tissues after systemic administration.6 Manufacturing and distribution of antibodies is expensive as they are typically produced in mammalian expression systems, need complex purification processes to achieve purity suitable for parenteral administration, and require refrigeration for storage and transport.
A number of oral and topical proteins, peptides, and small molecules are in development as convenient, less immunogenic, inexpensive alternatives to systemically administered antibodies. Oral versions of approved anti-TNFα antibodies (adalimumab, Biora Therapeutics; infliximab, Celltrion and Intract Pharma) promise greater convenience with the same cellular potency but require proprietary formulation to reach the site of action intact, adding to the already high cost of the antibody alone. Oral JAK inhibitors are approved for a number of chronic inflammatory conditions including IBD, but severe side effects have limited their use.7 Oral peptides are in development for psoriasis and IBD, targeting IL-23R (PN-235/JNJ-77242113, Protagonist Therapeutics and Janssen) and a4b7 integrin (PN-943, Protagonist Therapeutics). However, the peptides require noncanonical amino acids and crosslinks to confer resistance to gastrointestinal (GI) proteases, necessitating expensive manufacturing via chemical synthesis. Orally delivered small molecules targeting IL-17A are in development for psoriasis (DC-806 and DC-853, Dice Therapeutics and Eli Lilly); while the affinity of next-generation variant DC-853 is unknown, DC-806 binds IL-17A with only low nanomolar affinity and requires two relatively high daily doses to achieve modest clinical effect.8,9 While the above therapies are more convenient than parenterally administered mAbs, their safety risks, high cost of goods, and limited efficacy are significant downsides.
Computational design methods now enable the design of small (~60 residue) binding proteins with low picomolar affinity, extreme thermostability and resistance to proteolysis, and low immunogenicity.10–16 We reasoned that designed miniprotein inhibitors of IL-23R and IL-17 could address the unmet need for effective, convenient, safe, and low cost therapies for autoinflammatory diseases, and set out to develop such compounds.
Results and Discussion
Computational design yields proteins with low nanomolar affinity for IL-23R and IL-17
IL-23 consists of the p19 subunit unique to IL-23 and the p40 subunit shared with IL-12. The IL-23 receptor is also heterodimeric, with a unique subunit, IL-23R, which binds p19, and a shared subunit, IL-12RB1, which binds p40.17,18 Anti-p40 antibody Stelara, which blocks both IL-23 and IL-12, has seen enormous clinical success. However, preclinical studies demonstrated that IL-23 and not IL-12 drives pathogenic autoinflammation, and therefore subsequent drug discovery efforts have largely focused on targeting the IL-23-specific subunit, p19.19,20 Thus, we aimed to design proteins that disrupt the IL-23R:p19 interaction in order to selectively inhibit IL-23 and not IL-12.
IL-17A and IL-17F monomers pair to form homodimeric (A/A, F/F) and heterodimeric (A/F) cytokines that signal via a ternary complex with receptors IL-17RA and IL-17RC. We selected IL-17A as our initial design target as it is best established among the IL-17 homologs as a mediator of autoinflammatory disease. We aimed to design proteins that bind IL-17A at the surface mediating its interaction with IL-17RA or IL-17RC.
Computational design of binding proteins generally starts from a crystal or cryoEM structure of the target. If a ligand-bound structure is available, critical binding residues (or hotspots) of the ligand may be incorporated into design.21,22 If only the apo structure of the target is available, hotspots may be computationally generated.23 We took a combined approach, using one native hotspot from IL-23p19 cytokine (W156) and additional computationally determined de novo hotspots generated at the p19 interface to seed design. For IL-17A, we exclusively used de novo hotspots generated at the receptor surface. Thousands of computationally designed miniproteins with diverse topologies and experimentally validated stability (scaffolds)11,24,25 were docked at the IL-23R or IL-17A surface such that hotspots were incorporated into the scaffold backbone. Then, with each docked configuration as input, we used the Rosetta molecular modeling suite to optimize scaffold residue identities and conformations at the IL-23R or IL-17A interface for high-affinity binding. Native and de novo hotspots, residues in the scaffold hydrophobic core, and scaffold residues not at the target interface were kept fixed. The resulting designed inhibitor candidates were filtered on computational metrics correlating with binding affinity and monomer stability, and genes encoding the best 15,000 per target were obtained and transformed into yeast for surface display. Yeast were selected for binding to labeled recombinant human IL-23R or IL-17A by multiple successive rounds of fluorescence-activated cell sorting (FACS). Naive and sorted pools were analyzed by next-generation sequencing (NGS) and designs were ranked by their relative enrichment or depletion.
Two IL-23R designs, 23R-1 and 23R-2, and three IL-17A designs, 17–1, 17–2 and 17–3, were highly enriched in the final sorts and were selected for further biochemical characterization and sequence optimization. The IL-23R binding designs are 55- (23R-1) and 54-residue (23R-2) 3-helix bundles comprising a central binding helix that incorporates the native Trp hotspot, and two additional helices that stabilize the central binding helix and make additional contacts with IL-23R (Figure 1A). IL-17A binding designs, 43-residue 3-helix bundle 17–2 and 61-residue ferredoxins 17–1 and 17–3, incorporate de novo generated hotspots at the IL-17A surface that mimic IL-17RA (Figure 1B).
Figure 1. Computational design of IL-23R and IL-17A minibinders.
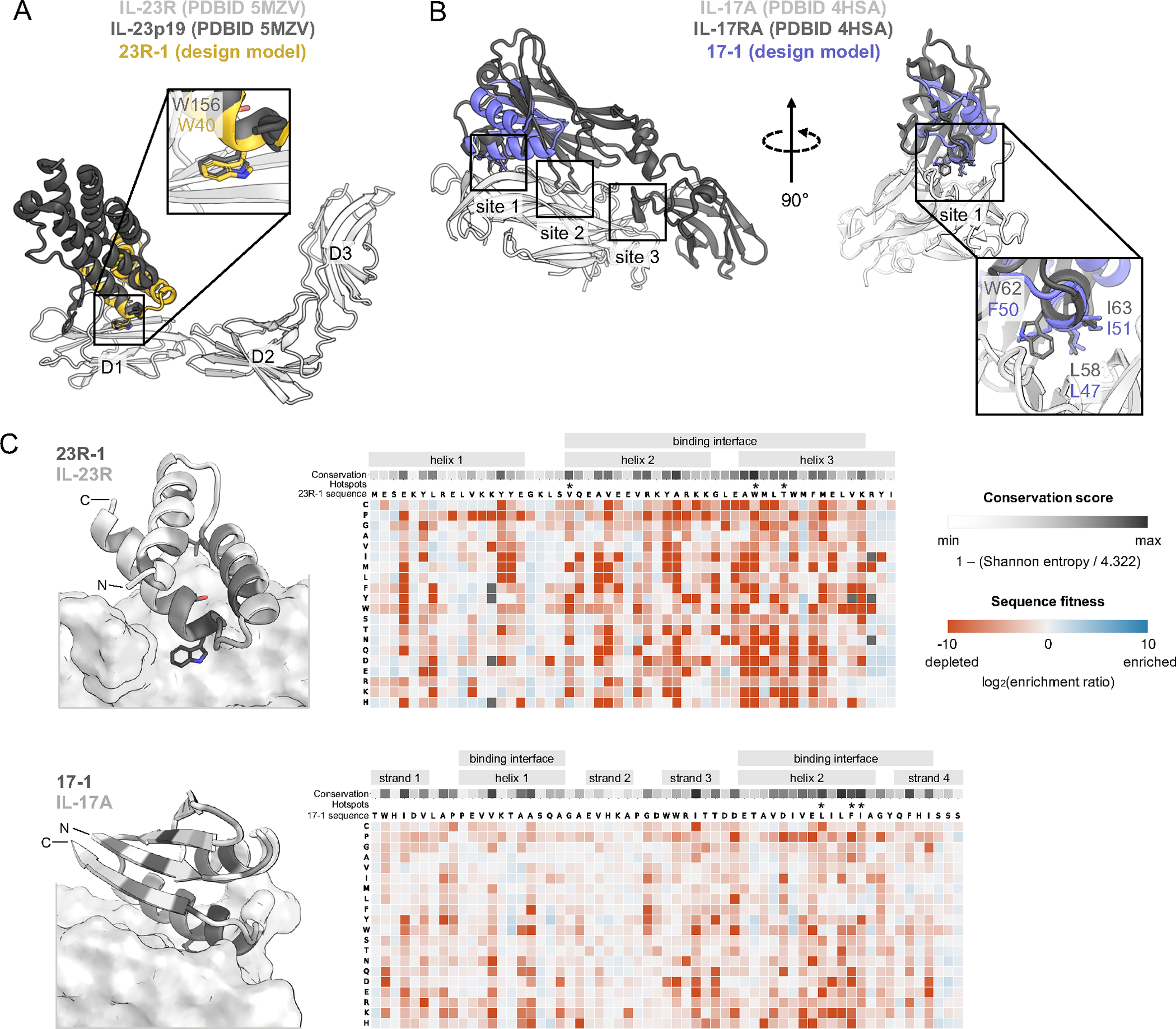
(A) Minibinder 23R-1 was designed to bind IL-23R domain D1 at the IL-23p19 interaction surface, and all designs incorporate native hotspot W156. (B) Minibinder 17–1 was designed to bind IL-17A at site 1 of the IL-17RA interaction surface and incorporates de novo hotspots (purple) that mimic native hotspots from IL-17RA (dark gray). (C) The relative affinity of each mutation was determined using deep mutational scanning. The enrichment (blue) or depletion (red) of each mutation, depicted in 2D heatmaps, represents its impact on affinity relative to the original minibinder sequence (set to 0, white). Positional conservation scores are depicted in a 1D heatmap from minimum (light gray) to maximum (dark gray) per design. Asterisks indicate native and de novo hotspots. See Figure S1 for binding data and Figures S3 and S6 for deep mutational scanning heatmaps of additional IL-23R and IL-17A computationally designed minibinders and peptides.
The binding affinity and potency of the minibinders was determined with biolayer interferometry (BLI) and cell-based signaling assays. Designs were expressed in E. coli and purified. Binding affinities were quantitatively determined with BLI; all designs bound their target with low nanomolar affinity (Figure S1). The minibinders were very stable to heat and chemical denaturant (guanidinium, abbrev. Gdn) in circular dichroism (CD) experiments. IL-23R minibinders 23R-1 and 23R-2 had melting temperatures (Tm) >95°C and very high chemical denaturation midpoints (5 M Gdn for 23R-1, >6 M for 23R-2; Figures 2C and 2D). IL-17A minibinder 17–1 had a Tm of approximately 90°C and Gdn denaturation midpoint of 4 M (Figure S2C). IL-17A minbinder 17–2 had the weakest stability, with a Tm of approximately 70°C and Gdn denaturation midpoint of 2 M (Figure 3C). The minibinders blocked IL-23- or IL-17A-mediated cell signaling in a dose-dependent manner (Figures 2A, 3A, Figure S2A). Figures 2E and 3D provide an overview of the total design and optimization strategy for IL-23R and IL-17A minibinders, respectively, described in detail below.
Figure 2. In vitro potency and stability of the IL-23R minibinders.

(A) Minibinders block IL-23-mediated cell signaling in an engineered IL-23 reporter cell line. Representative curves are shown above (n≥2), and IC50 values reported as mean ± SD of at least two independent experiments (N≥2). (B) The binding affinities of 23R-72 and 23R-91 were determined using SPR. (C) 23R-1 and derivatives, and (D) 23R-2 and derivatives were denatured with heat and/or chemical denaturant guanidinium hydrochloride (Gdn), and helicity (signal at 222 nm) was monitored using circular dichroism. Signal is plotted as a fraction of reference sample (0 M Gdn at 25ºC). (E) Molecular design and optimization workflow. See Figures S1 and S3 for binding data and Figure S4 for biophysical characterization of IL-23R minibinder variants.
Figure 3. In vitro potency and stability of the IL-17 minibinders.

(A) Minibinders block cell signaling mediated by hIL-17A (left), hIL-17F (middle), or hIL-17A/F (right) in an engineered IL-17 reporter cell line. Representative curves are shown above (n≥2), and IC50 values are reported as mean ± SD of at least two independent experiments (N≥2; ND = no data). (B) The binding affinity of lead IL-17A minibinder 17–53 was determined using BLI. We note that the instrument (ForteBio Octet RED96) is not sensitive enough to accurately determine picomolar KDs, but the data nonetheless indicate the very high affinity and slow dissociation rate of 17–53. Data plotted are representative of three independent experiments (N=3). (C) Each minibinder was denatured with heat or chemical denaturant guanidinium hydrochloride (Gdn), and helicity (signal at 222 nm) was monitored using circular dichroism. Signal is plotted as a fraction of reference sample (0 M Gdn at 25°C). (D) Molecular design and optimization workflow. See Figure S1 and S3 for binding analysis and Figure S2 for potency and stability data for other IL-17A minibinder variants.
Saturation mutagenesis data corroborate predicted monomer structure and binding mode
Probing the sequence fitness landscape of the designed proteins provides insight into their 3D structure and binding mode. Site-directed saturation mutagenesis (SSM) libraries were synthesized comprising all possible single-position mutants of 23R-1, 23R-2, 17–1, 17–2 and 17–3, transformed into yeast, and screened for binding to labeled target protein using FACS. Naive and sorted pools were deep sequenced, and the enrichment or depletion of each mutant in the sorted pools was calculated as an estimate for binding fitness. Enrichment per position per amino acid was visualized in a 2D heatmap (Figure 1C and Figure S3A), with blue boxes indicating highly enriched and red boxes highly depleted mutations. An overall sequence conservation score was calculated per amino acid position of each minibinder, visualized in a 1D heatmap located above the enrichment heatmap, colors ranging from light gray (low conservation) to dark gray (high conservation). Positions contributing to the hydrophobic core or binding interface in the design model were conserved (dark gray), while surface positions distal to the interface, which can more readily be mutated without disrupting the minibinder’s 3D structure or binding, were not conserved (light gray). These data suggest that the minibinders are folded and bind the targets as in the computational design models.
In vitro evolution and computational design dramatically improve potency and stability
Orally administered protein antagonist therapeutics must be sufficiently potent and stable in GI conditions to reach the site of action at concentrations that saturate the target and in this case, compete with the native ligand. We therefore sought to further improve the minibinders’ potency and resistance to intestinal proteases. Combinatorial libraries were designed including the mutations most enriched for high-affinity binding in the SSMs, and high affinity variants were selected via multiple successive rounds of FACS. The most enriched variants in the final sort were selected for expression in E. coli and biophysical characterization. Affinity-optimized combinatorial variants were ranked by binding affinity using a single-concentration BLI screen (Figure S3B–C), and for the best variants, kinetic and equilibrium binding constants were determined. The highest affinity IL-23R minibinder variants had 100- to 1,000-fold higher binding affinities compared to the parent computational designs (Figure S1), and 30- to 300-fold higher cellular potencies (Figure 2A). The highest affinity IL-17A minibinder variants had approximately 60-fold improvement in affinity (Figure S1) and cellular potency (Figure 3A). Combinatorial variant sequences were different from parent computational designs by 15–19% (8–10 mutations; Table S1).
Minibinder stability was assessed using circular dichroism as well as timed degradation in simulated gastric and intestinal fluids (SGF, SIF) containing physiological proteases. The affinity-optimized combinatorial variants had similar resistance to heat and chemical denaturant as their precursor computational designs (compare computational design 17–1 to affinity optimized variant 17–16 in Figure S2C, computational design 17–2 to affinity-optimized variant 17–35 in Figure 3C, computational design 23R-1 to affinity optimized variants 23R-3 through 23R-15 in Figure S4A, and computational design 23R-2 to affinity-optimized variants 23R-17 through 23R-24 in Figure S4B). 23R-2-derived combinatorial variant 23R-20 showed similar SIF stability and modestly decreased SGF stability compared to 23R-2 (Figure 4A). To improve minibinder stability while retaining high affinity, intramolecular disulfide(s) were computationally designed in affinity-optimized combinatorial variants. Adding one disulfide to IL-17A combinatorial variant 17–35 (yielding 17–51) significantly improved stability, increasing the Tm from approximately 70°C to 95°C and similarly increasing resistance to chemical denaturant (Figure 3C). 23R-64, an affinity-optimized variant of 23R-1 with one added disulfide, showed significantly improved SIF and SGF stability, with half-lives (t1/2) of approximately 30 minutes and 4 hours, respectively, compared to 23R-1 with t1/2 <5 minutes in both SIF and SGF (Figure 4A). Adding two disulfides to IL-23R combinatorial variant 23R-20 (yielding 23R-49) significantly improved SGF stability, increasing the t1/2 of full-length minbinder from approximately 30 minutes to >24 hours, but decreased SIF stability to a t1/2 of about 5 minutes. To further optimize 23R-64 and 23R-49, SSM libraries were generated and transformed into yeast for surface display. Naive libraries were incubated in SIF, then washed and incubated with labeled IL-23R. Variants that retained binding to IL-23R after SIF treatment were selected via FACS. Mutations most enriched for SIF stability and affinity were included in combinatorial libraries which were sorted under similar conditions as the SSMs, each successive selection increasing the SIF incubation time or concentration of proteases, or decreasing the concentration of labeled IL-23R. Combinatorial variants most enriched in the final sort per library, as well as several variants hand-selected based on SSM data incorporating one to three of the most enriching mutations, were expressed and characterized in vitro.
Figure 4. In vitro and in vivo GI stability of IL-23R minibinders.

(A) Each minibinder or control protein V565, an oral nanobody in development for IBD, were digested in SIF or SGF at 37°C for up to 24 hours (N≥2). In some instances, indicated by white spaces, lanes were isolated in order to maintain consistent order of ladder and samples. (B) SIF and SGF digests were sampled at the indicated timepoints, diluted to 10 nM minibinder (assuming no degradation), and BLI was used to measure residual binding to hIL-23R (N≥2). See Figure S5 for binding analysis after SGF and SIF digest for other IL-23R minibinder variants. (C) A single oral 20 mg/kg dose of 23R-72 or 23R-91 was administered in healthy rats, formulated in either PBS or GI-protective vehicle (GPV), and minibinder concentration measured in serum and target tissues 6 hours after dosing. Mean ± SD, n=2 technical replicates in analysis, N=5 animals per group. Samples falling below the limit of detection (BLOD) were assigned a value of 0. (D) A single oral 140 mg/kg dose of 23R-91 was administered in healthy rats, and serum concentration of minibinder measured at the indicated time points. Mean ± SD, n=2 technical replicates in analysis, N=6 animals per group. All samples at the 6 hour time point (*) were BLOD. See Table S5 for values and Figure S7 for ELISA standard curves.
The best stability-optimized variants, 23R-72 and 23R-91, showed significant improvement in SIF resistance (Figure 4A) and maintained or improved potency (Figure 2A–B). 23R-72 includes three hand-picked mutations (M1P, R8Q, K35W) that were highly enriched in the SIF-treated SSM of 23R-64, and improved SIF t1/2 from approximately 30 minutes (23R-64) to 4–24 hours (23R-72), on par with that of V565, an oral anti-TNFα nanobody in clinical development for IBD (Sorriso Pharmaceuticals) used as a positive control in this assay (Figure 4A). The mutations also modestly improved SGF stability. SDS PAGE shows a small decrease in the molecular weight of 23R-72 as well as precursor 23R-64 early in the SIF digest, which likely indicates cleavage of the inert C-terminal poly-histidine tag as binding to hIL-23R is retained (Figure 4B, Figure S5). 23R-72 has 75 pM affinity for IL-23R (Figure 2B) and similar cellular potency as precursor 23R-64 (Figure 2A). Stability-optimized variant 23R-91, 8 mutations (15%) from precursor 23R-49, shows improved SIF stability, with t1/2 ~60 minutes, and excellent SGF stability (t1/2 4–24 hours; Figure 4A). 23R-91 has a binding affinity lower than the detection limit of the instrument (<1 pM; Biacore 8K) due to an immeasurably slow dissociation rate (Figure 2B), and cellular potency was modestly improved compared to the precursor 23R-49 (Figure 2A). Both 23R-72 and 23R-91 are extremely resistant to heat and chemical denaturant, showing minimal loss of helicity even at 95°C in 6 M guanidinium (Figure 2C and D, Figure S4).
The crystal structure of 23R-91 is very close to the design model
The crystal structure of the most potent IL-23R minibinder, 23R-91, was solved to 1.9 Å resolution and has two copies of 23R-91 in the asymmetric unit (Figure 5; Table S4). The two chains have 0.7 Å (chain A) and 0.4 Å (chain B) Cɑ RMSD to the design model. In the design model, side chain rotamers in the hydrophobic core match those of one or both chains of the crystal structure, while rotamers of surface residues show greater deviation from the crystal structure. The geometry of the disulfide bonds in the design model matches one (C31-C40) or both (C12-C21) chains of the crystal structure. SE-HPLC and LC-MS analyses confirmed that 23R-91 is monomeric in solution and has the expected molecular weight (Figure S4F and H).
Figure 5. The crystal structure of 23R-91 is very close to the design model.
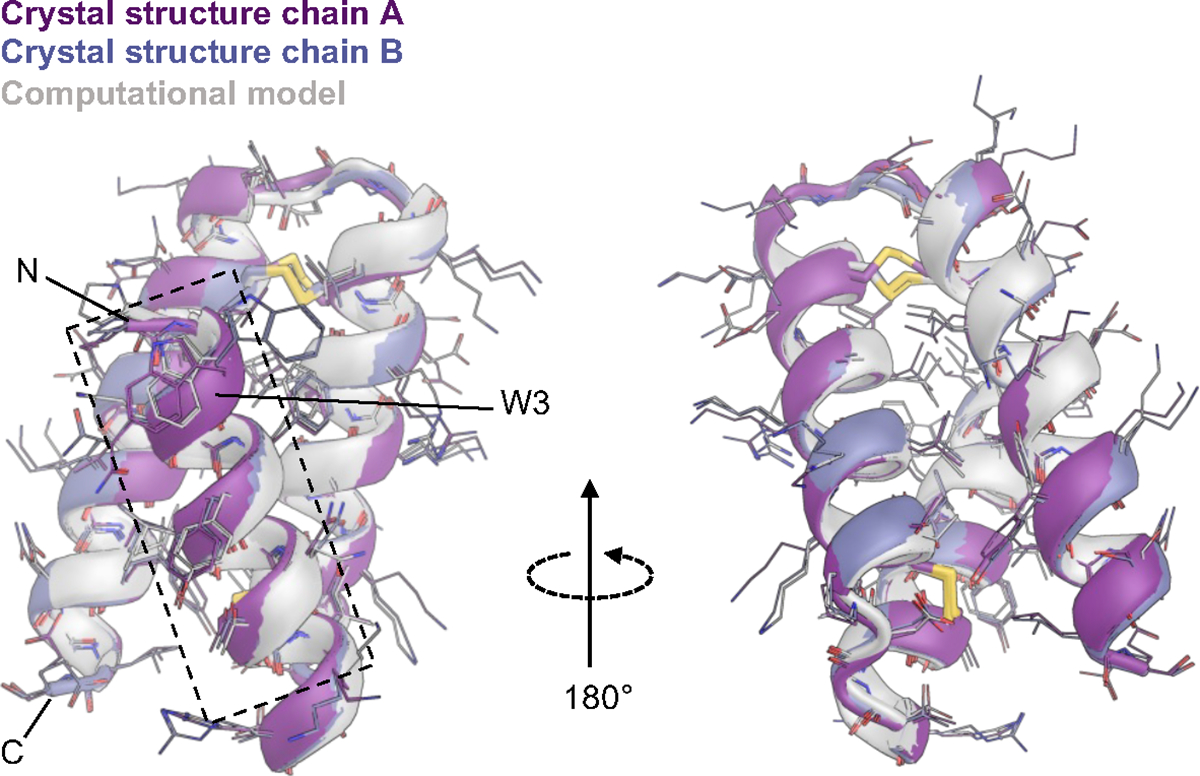
Both chains in the crystal structure asymmetric unit were aligned via Cɑ atoms to the computational model of 23R-91, with 0.7 Å (chain A) and 0.4 Å (chain B) RMSD. The core binding interface, including native hotspot W3, is indicated with a dashed box. See Table S4 for data collection and refinement stats.
Connecting two hIL-17A binding domains with a flexible peptide linker increases potency 2,800-fold through avidity
We sought to further improve the potency of the hIL-17A minibinder by connecting two copies of 17–51 with a flexible peptide linker to avidly bind the IL-17A cytokine homodimer at both symmetric binding sites. The best single-chain minibinder dimer (17–53) has low picomolar affinity with an extremely slow dissociation rate (Figure 3B) compared to the low nanomolar affinity of monomer 17–51 (Figure S1). 17–53 shows a 2,800-fold increase in potency compared to the minibinder monomer (17–51) and 60,000-fold increase compared to the parent computational design (17–2; Figure 3A). 17–53 is 200-fold and 4-fold more potent in blocking hIL-17A-mediated signaling than clinical mAbs secukinumab and bimekizumab, respectively. The linked construct with the shortest linker, 17–52 [linker (GS)10], showed weaker potency than constructs with longer linkers 17–53 [(PAS)8], 17–54 [(PAS)12], and 17–55 [(PAS)20], which may indicate there is a minimum linker length for sterically unhindered engagement of both hIL-17A homodimer binding sites (Figure S2B). 17–51 and 17–53 show significantly weaker inhibition of mouse IL-17A than the human homolog (Figure S2E).
17–53 is highly specific to homodimeric hIL-17A, showing negligible inhibition of hIL-17F- or hIL-17A/F-mediated cell signaling (Figure 3A). The monomer (17–51) and dimer fusion (17–53) minibinders block the hIL-17A/F heterodimeric cytokine with similar relatively weak potency, indicating that the 17–51 binding domain likely only binds weakly to one of the two asymmetric receptor binding sites of hIL-17A/F. Neither 17–51 nor 17–53 bind to homodimeric hIL-17F. As the IL-17F homodimeric cytokine is also a clinically relevant target, we screened the IL-17A minibinder combinatorial libraries for cross-reactivity with hIL-17F, and hits were further optimized for affinity and specificity to hIL-17F by in vitro evolution. The most potent hIL-17F inhibitor, 17–40, blocks hIL-17F-mediated signaling with potency 300-fold greater than hIL-17A-specific minibinder 17–51, 3-fold greater than secukinumab and 1,000-fold worse than bimekizumab (Figure 3A).
3–4 kDa peptide inhibitors of IL-23R are structured and block IL-23-mediated cell signaling
Drug molecular weight influences intestinal permeability and tissue diffusivity, so we sought to reduce the size of the 7–8 kDa IL-23R minibinders in order to increase the concentration of drug at the site of action after oral administration. Design models of the highest affinity 7–8 kDa minibinders were used to seed computational design of 3–4 kDa variants. A small fragment of the 7–8 kDa minibinder central binding helix, including the native Trp hotspot and de novo hotspots, was isolated and then grafted onto 26–32 residue structurally validated peptide scaffolds.12 Designs were filtered using the same computational metrics as in minibinder design described above, and genes encoding the top 3,883 were synthesized and transformed into yeast for surface display and selected for binding to labeled IL-23R, with or without pre-incubation in SIF.
SSM analysis of the three most enriched designs, 26-residue EEH folds with two stabilizing disulfides, demonstrate they are likely folded and bind via the designed interface (Figure S6A). Residues in the hydrophobic core, at the binding interface, and cysteines designed to form disulfide bonds are conserved, while surface positions distal to the binding interface are not conserved. We generated combinatorial libraries including SSM mutants favoring high affinity binding and stability, and screened them as described above for binding to IL-23R after SIF treatment. The most enriched variants were chemically synthesized and characterized. The best 26-residue IL-23R minibinder, 23R-101 (3.2 kDa) is 40 times more potent than a competing IL-23R antagonist peptide PTG-200 (Protagonist Therapeutics/Janssen; Figure S6B). However, 23R-101 is 30 times less potent than the best 7–8 kDa minibinder, 23R-91, and would therefore need to reach concentrations at least 30 times that of 23R-91 in target tissues to achieve similar efficacy. We therefore prioritized the 7–8 kDa minibinders for further in vitro and in vivo characterization.
These results suggest a general strategy for peptide therapeutic discovery: design of larger, high-affinity minibinders followed by grafting of critical binding residues or motifs onto smaller, structured peptide scaffolds.
Minibinders block IL-23- or IL-17-mediated inflammation in primary cell culture and human organoids
Next we determined whether the minibinders could block IL-23- or IL-17A-mediated cell signaling in in vitro systems that mimic the target in vivo environments. IL-23 and IL-17A antagonists are used to treat a variety of autoimmune indications, including IBD (IL-23 only) and psoriasis (IL-17A and IL-23). IBD is characterized by intestinal injury driven by local inflammatory processes in the intestinal lamina propria (LP). 23R-91 efficiently blocked IL-23-mediated cell signaling in cell suspensions from the colon LP and nearby mesenteric lymph nodes (mLN) that were isolated from healthy rats and stimulated ex vivo with anti-CD3 and recombinant rat IL-23 (Figure 6A). The minibinder also blocked signaling in rat splenocytes (Figure 6A). Similarly, IL-23R minibinders block IL-23 mediated signaling in primary human CD4+ T cells (Figure 6B).
Figure 6. IL-23R and IL-17 minibinders block cell signaling in primary cells and organoids derived from human skin cells.
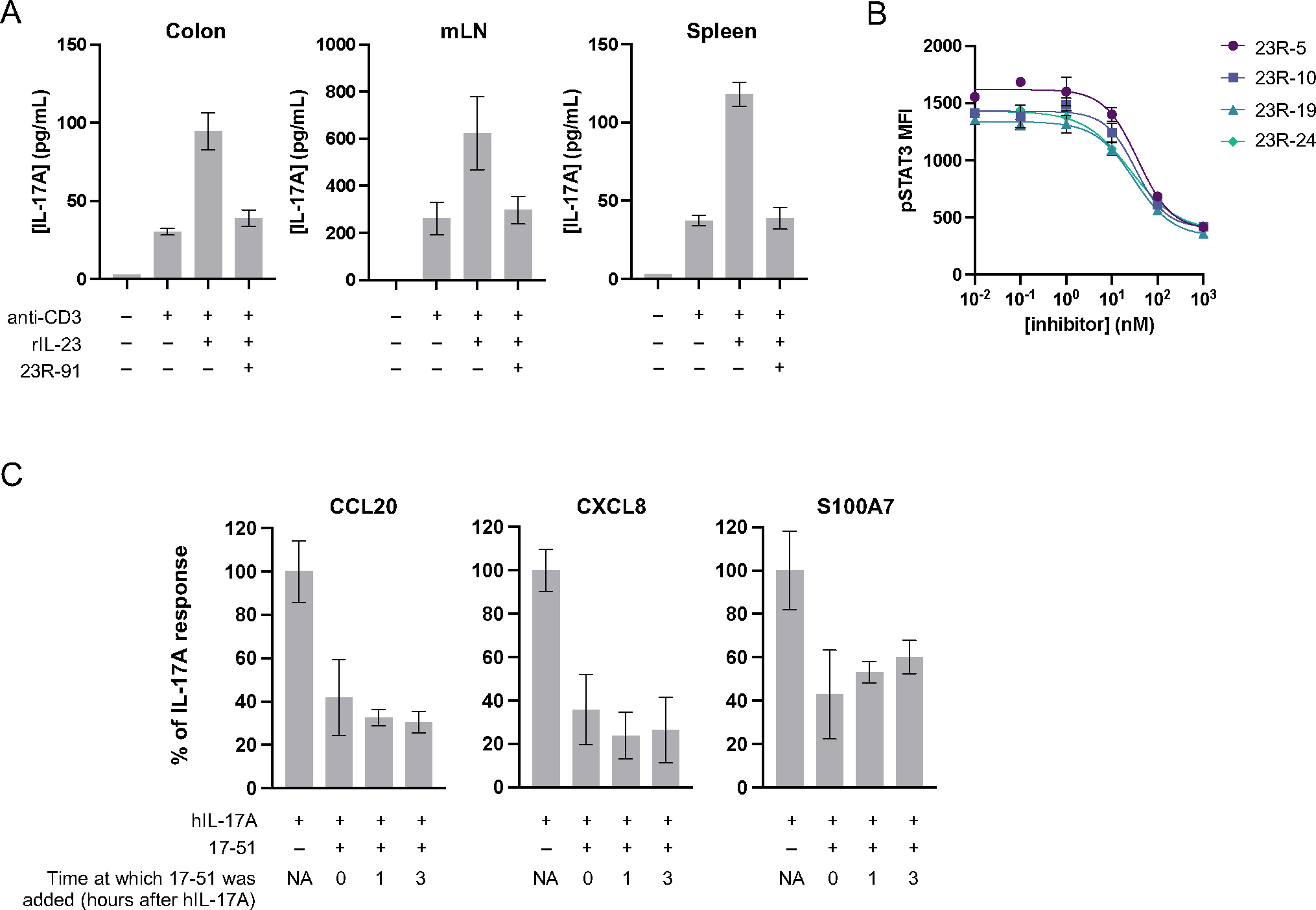
(A) Cell suspensions were prepared from the colon, mLN, and spleen of healthy rats, then stimulated with anti-CD3 and rIL-23 (1 μg/mL for colon, 10 ng/mL for mLN and spleen) with or without minibinder 23R-91 (100 nM). After 24 hours incubation, IL-17A was measured in culture supernatants with ELISA. Anti-CD3 only treatment served as a control showing the extent of IL-23-independent IL-17A production, which is not expected to be inhibited by 23R-91 (n=3 replicates per stimulation condition per two independent experiments). (B) IL-23R minibinders block IL-23 signaling in primary human CD4+ T cells with low nanomolar IC50s. Cells were stimulated with recombinant IL-23 with or without a titration of each minibinder for 20 minutes, and stained for CD4 and phosphorylated STAT3. Mean fluorescence intensity (MFI) of pSTAT3 in CD4+ cells was measured by flow cytometry. Mean values ± SD are shown, n=3. (C) Human epithelial organoids were treated with IL-17A (15 nM) with or without minibinder 17–51 (75 nM) and analyzed by qPCR for downstream markers CCL20, CXCL8 (IL-8), and S100A7. Gene expression data were normalized to housekeeping gene HPRT1. Fold change was calculated relative to an untreated control group and is presented as a percent of the response seen with IL-17A-only treatment. Three organoids per stimulation condition were pooled for qPCR analysis in triplicate (n=3) in each of two independent experiments with unique donors (N=2).
Psoriasis is characterized by skin inflammation, and therefore we used organoids generated from human skin epithelium to study the effect of IL-17A minibinder 17–51. Organoids were cultured and stimulated with recombinant human IL-17A (15 nM). Minibinder 17–51 (75 nM) was added to culture media simultaneously with IL-17A, or 1 or 3 hours after addition of IL-17A. After overnight incubation, organoids were analyzed by qPCR for downstream markers CCL20, CXCL8 (IL-8), and S100A7. Minibinder 17–51 significantly inhibited production of downstream markers in all conditions (Figure 6C).
IL-23R minibinders reach therapeutically relevant concentrations in the GI and serum after oral administration in rats
The integrity of the intestinal barrier is likely to impact PK of oral protein therapies. In IBD patients with active disease, the barrier is disrupted and more permeable, while patients in remission have a more intact, less permeable barrier. To support the use of oral IL-23R minibinders as induction therapy for IBD patients with active disease as well as maintenance therapy for patients in remission, we studied the behavior of 23R-72 and 23R-91 in TNBS rats with a disrupted intestinal barrier and in healthy rats with an intact intestinal barrier. Rats were used in order to capture target-mediated drug deposition, as the IL-23R minibinders cross-react with rat but not mouse IL-23R (Figure S7C–D).
In healthy rats, six hours after a single 20 mg/kg oral dose, minibinder concentration was measured in the intestinal tissues and contents, mLN, or serum using a custom ELISA method. Minibinders were detected at 50–100 nM in the small intestinal contents and not in colon contents, and detected at higher concentrations in the small intestinal tissue (40–200 nM) than colon tissue (2–20 nM), consistent with known transit times in rats (Figure 4C, Table S5).26 Formulation in GI-protective vehicle (GPV; 0.1 M sodium bicarbonate, 200 mg/mL nonfat dry milk) did not significantly impact minibinder concentration in contents or tissues; both minibinders appear to be equally resistant to GI proteases in vivo. Minibinders were not detected in serum at this dose in healthy rats. After a higher single oral dose (140 mg/kg) in healthy rats, 23R-91 was present at a concentration of 73 nM in serum 15 minutes after dose, after which serum concentration decreased rapidly with a half-life of approximately 15 minutes, falling near or below the limit of detection from 3 hours to 24 hours (Figure 4D). Minibinder was not measured in tissues in this study.
In TNBS rats, 23R-72 was administered by oral gavage, and 23R-91 was injected via catheter directly into the cecum, mimicking colonic release formulation. After 9 days of 20 mg/kg TID dosing, rats were sacrificed six hours after the last dose and tissues and serum analyzed for minibinder with ELISA. Generally, both minibinders reached low nanomolar concentrations in GI tissues and demonstrated low systemic bioavailability after oral or intracecal administration, with concentrations near the limit of detection of the assay (1–5 nM) in mLN or serum (Table S5). GI tissue concentrations observed in TNBS rats are generally lower than observed in healthy rats; in IBD and preclinical models of colitis, GI transit time is accelerated relative to a healthy GI, which decreases residence time and could therefore decrease uptake.
The observed absorption of minibinders compares favorably to that of other oral biologic modalities. Antibodies are generally too large (~20x the size of minibinders) and susceptible to degradation in GI conditions to achieve therapeutic concentrations at a reasonable oral dose without sophisticated formulation27,28 or delivery technologies.29–31 Oral nanobodies and peptides engineered for GI stability have shown similar tissue and serum concentrations as our oral minibinders in preclinical studies. Low to mid-nanomolar V565 nanobody (13 kDa) was detected seven hours after a liquid oral dose of 5–10 mg/kg in colon contents of both healthy and TNBS mice and in the serum of TNBS (but not healthy) mice, and in the serum of two out of three healthy monkeys dosed by V565 tablet at 40 mg/kg.32,33 Control nanobodies not engineered for GI stability were quickly degraded in GI fluids in vitro and were not studied in vivo. Low- to mid-nanomolar anti-IL-23R peptides (1.5–3 kDa) have also been detected six hours after a single oral dose of 10 mg/kg in the colon and intestinal tissue, and occasionally in the serum, of healthy rats, and a Phase 1 trial with IL-23R peptide JNJ-77242113 demonstrated peak serum concentrations of up to 10 nM in healthy volunteers.34,35 Engineered nanobodies, peptides, and minibinders are all capable of reaching low- to mid-nanomolar concentrations in target tissues and serum at similar oral doses; whether these concentrations are adequate for therapy, however, depends on the potency of the molecule.
Continuous inhibition of IL-23R over time requires that drug reaches an initial concentration that saturates the target, and subsequent maintenance of saturation over time. Ligands theoretically reach 99% saturation of the target at concentrations 100x the KD of the ligand:target interaction; with KD < 1 pM, 23R-91 would reach 99% saturation at < 100 pM.36 In our preclinical studies, 23R-91 reaches concentrations orders of magnitude higher than 100 pM in target tissues and serum. When drug is cleared over time and free drug concentration goes below the target-saturating concentration, maintenance of IL-23R inhibition depends on the minibinder:target complex half-life and receptor recycling. The extremely slow dissociation rate of 23R-91 measured by SPR (<1 × 10−5 s−1) corresponds to a 23R-91:IL-23R complex half-life of >19 hours37, meaning IL-23R remains inhibited by 23R-91 for hours after free drug is cleared. We expect 23R-91 to block IL-23R recycling, which is induced by interaction with IL-23.38 As free 23R-91 is quickly cleared from the blood (Figure 4D), measuring target engagement over time will further inform development of a clinical formulation and dose regimen that results in continuous IL-23R blockade.
23R-91 has low predicted immunogenicity
Immunogenicity is an important consideration for any protein therapeutic. We hypothesize that the general high stability and solubility of engineered proteins reduces uptake, digestion to fragments, and subsequent presentation of fragments by MHCII in antigen presenting cells, thereby reducing immunogenicity. Several designed proteins have generated very low or undetectable levels of anti-minibinder IgG after repeated systemic dosing in mice14–16 and repeated oral dosing in humans.39 A systemically administered de novo designed IL-2 mimic did not elicit a strong anti-drug response in human clinical trials.40 23R-91 is highly soluble (Figure S4G), a characteristic associated with low immunogenicity.41 As demonstrated above, 23R-91 is stable at low pH and resistant to proteases with diverse recognition sequences, and is therefore unlikely to be efficiently digested to fragments upon endocytosis. If some degree of uptake and digest do occur, 23R-91 fragments have low predicted binding affinity to a variety of MHCII molecules which is associated with low immunogenicity (Table S7).42
Oral IL-23R minibinder is as effective as clinical mAb in a humanized mouse model of colitis
We compared the efficacy of oral 23R-91 to systemically administered guselkumab, a monoclonal antibody in Phase 3 clinical trials for UC and CD, in a humanized mouse model of colitis (Figure 7A). In this model, NOD/SCID/IL2rγnull (NSG) mice deficient in T, B, and NK cells were reconstituted with peripheral blood mononuclear cells (PBMCs) from patients with IBD.43 In NSG-IBD mice, cells expressing IL-23 and IL-23R are primarily human-derived, allowing us to study 23R-91, which binds human IL-23R but not the mouse homolog (Figure S7C–D), and a competing clinical (human-targeted) mAb. After engraftment of human PBMCs (day 1), mice were challenged with intrarectal ethanol on days 7 and 14 to induce colitis. 23R-91 (8 or 80 mg/kg) was administered once daily by oral gavage on days 6, 7, and 13–17. Guselkumab (4 mg/kg) was administered by intraperitoneal injection on days 6 and 13. On day 18, animals were sacrificed and study endpoints assessed, including a clinical score of overall health, colon macroscopic score of inflammation, and colon histopathology (scoring matrices Table S6; study schematic Figure 7A).
Figure 7. Evaluation of 23R-91 in the NSG-IBD humanized mouse model of colitis.

(A) NSG-IBD study design and schematic. N≥5 animals per group in at least 2 independent studies per condition. (B) On the last day of the study, each animal was assigned a clinical score of overall health. At sacrifice, the colon was dissected and assigned a macroscopic score of inflammation, then prepared for histology to assess microscopic features of inflammation and fibrosis. Scores are plotted as mean ± SD. Treatment groups were compared to challenged control using non-parametric Wilcoxon matched-pair tests (ns = no significance, *P <0.033, **P<0.002, 95% confidence interval). See Table S6 for scoring matrices.
Oral 23R-91 at both 8 and 80 mg/kg showed statistically significant improvement in disease scores compared to untreated control and a numerically greater improvement than i.p. guselkumab (Figure 7B). As expected, colon fibrosis was not significantly reduced by any treatment. These data demonstrate that oral 23R-91, a 7 kDa protein, reaches sufficient concentrations in target tissues beyond the gut epithelial barrier to impact disease. This is the first demonstration that a once daily oral protein drug can achieve efficacy in a model of colitis; however, we note that a competing oral anti-IL-23R peptide, JNJ-2113, recently demonstrated efficacy in a rat model of colitis with an unknown dose regimen and clinical efficacy in psoriasis with once or twice daily dosing.35,44
An oral dose of 8 mg/kg is clinically feasible, corresponding to a human of average weight (65 kg) taking a pill with 520 mg active ingredient. While 20, 80, and 140 mg/kg doses used here in PK and efficacy studies are near or beyond the limit of clinical feasibility, similar uptake can likely be achieved at lower mg/kg doses by using a clinical solid oral dosage form (tablet or capsule) that increases local lumenal drug concentration compared to the relatively dilute, high-volume liquid doses formulated simply in PBS used herein. Common polymer coatings for tablets or capsules that enable site-specific release in the desired GI compartment could further concentrate drug at the site of optimal uptake.
Conclusion
Here we demonstrate the potential of de novo designed proteins as oral therapeutics for blocking TH17-mediated inflammation. Our designed 7 kDa IL-23R minibinder has antibody-like potency and extreme resistance to heat, acid, and proteolysis, and our IL-17A minbinder has 200-fold greater potency than the clinical mAb secukinumab and prevents IL-17A-mediated inflammatory signaling in human epithelial organoids. The orally administered IL-23R minibinder is effective in a mouse model of colitis with a clinically relevant dosing scheme (8 mg/kg once daily), and reaches concentrations likely to saturate IL-23R in the serum and intestinal tissues of healthy and TNBS rats after oral administration. While 23R-91 is quickly cleared from the blood, its extremely high affinity and slow dissociation rate may enable continuous target saturation with convenient, once daily oral dosing without engineering for serum half-life extension. The large size of antibodies compared to minibinders limits efficient penetration of the gut epithelial barrier and diffusion into target tissues, and peptides generally must incorporate chemistries that are not genetically encodable in order to confer GI resistance, resulting in expensive manufacturing. Minibinders such as our IL-23R binder combine the advantages of small size for diffusion into target tissues with high stability, affinity and genetic encodability and are thus attractive candidates for development as oral biologics.
Limitations of the study
While minibinder 23R-91 showed efficacy in the NSG-IBD model of colitis with once daily oral dosing, free 23R-91 is quickly cleared from the blood after oral administration. Further investigation and development will be necessary to explore the use of oral minibinders for GI and non-GI indications in which sustained extraintestinal inhibition of IL-23R is desired. High doses of minibinders were used in both PK and efficacy studies. Solid oral dosage forms that enrich lumenal minibinder concentrations at the site of optimal uptake and can thereby decrease the required dose should be evaluated in preclinical species in order to support the clinical use of oral minibinders. We have not demonstrated in vivo efficacy of the IL-17 minibinders; these additional studies will bolster the generality of minibinders as an oral biologic modality.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stephanie Berger (berger389@gmail.com).
Materials availability
DNA sequences of E. coli, yeast and mammalian expression plasmids are available upon request.
Data and code availability
This study did not generate new code. Source data for main and supplemental figures, raw and processed data from deep mutational scanning experiments, and crystal structure data are publicly accessible. Source data for main and supplemental figures and original SDS PAGE images for Figure 4A have been deposited in the Mendeley Data repository with DOI 10.17632/2n9gstvrsy.1. Raw and processed data from deep mutational scanning experiments have been deposited in the NCBI GEO repository with accession ID GSE263250. Data deposition, atomic coordinates, and structure factors reported for the crystal structure of 23R-91 have been deposited in the Protein Data Bank (PDB), http://www.rcsb.org/ with accession ID 8UTK. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell culture
E. coli strains BL21 Star (DE3) (Invitrogen), SHuffle T7 Express (New England Biolabs), and CVB-T7-POL (Avidity) were transformed with plasmid for minibinder expression using the manufacturer’s procedure. Successful transformants were selected by culture on 2% agar containing 50–100 μg/mL kanamycin (for selection of pET29b plasmid) and optionally 10 μg/mL chloramphenicol (CVB-T7-POL only, for selection of pBirAcm plasmid) at 37°C (BL21, CVB-T7-POL) or 30°C (SHuffle T7 Express). A single colony was used to inoculate Terrific Broth II (TBII) media (MP Biomedicals) containing selection antibiotic and grown to confluence overnight at at 37°C (BL21, CVB-T7-POL) or 30°C (SHuffle T7 Express). For expression cultures, TBII media was inoculated with overnight culture at a ratio of 1:50 to 1:100 starter culture:expression media and grown to OD600 0.6–0.8 at 37 °C (BL21, CVB-T7-POL) or 30 °C (SHuffle T7 Express), then expression was induced with IPTG added to 0.5–1 mM overnight at growth temperature of 18–37 °C. 10 mM biotin prepared in TBII and sterile-filtered was added to CVB-T7-POL media at induction.
EBY-100 S. cerevisiae were initially cultured in dropout base medium (MP Biomedicals 114025012-CF) with complete supplement mixture lacking ura and trp (MP Biomedicals 114520512-CF) selective for the yeast strain (-ura) and the transforming plasmid (-trp). Yeast were passaged and subsequently cultured in SDCAA medium (20 g/L dextrose, 6.7 g/L Difco yeast nitrogen base, 5 g/L Bacto casamino acids, 5.4 g/L Na2HPO4, 8.56 g/L NaH2PO4) and protein expression was induced with 2% galactose in SGCAA medium (SDCAA with 20 g/L galactose rather than dextrose).
Expi293F cells (Life Technologies) were grown in Expi293 Expression Medium (Life Technologies), cultured at 37°C with 8% CO2 and shaking at 150 rpm.
IL-23 (IL-23 Bioassay, Promega) reporter cell line was cultured according to the manufacturer’s protocol for single-use assay format. Briefly, cells were thawed and transferred to the culture medium provided with the assay kit and incubated at 37°C with 5% CO2. Cells were not propagated.
IL-17 (HEK-Blue IL-17, Invivogen) reporter cell line was cultured according to the manufacturer’s protocol. Briefly, cells were incubated (37°C, 5% CO2) in growth medium [DMEM, 4.5 g/l glucose, 2 mM L-glutamine, 10% (v/v) heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml Normocin (Invivogen ant-nr-05)] including 1x HEK-Blue selection reagent (Invivogen hb-sel), and passaged at approximately 70% confluency.
Peripheral mononuclear cells (PBMCs) from healthy human donors were obtained from the Stanford Blood Bank and cultured in complete RPMI: RPMI 1640-glutaMAX (Gibco) supplemented with 10% FBS (Gibco), 50 μM 2-mercaptoethanol (βME, Sigma), MEM non-essential amino acids (Gibco), sodium pyruvate (Gibco), 15mM HEPES (Gibco), and penicillin-streptomycin (Gibco) at 37°C with 5% CO2.
Primary rat cells were cultured in RPMI-1640 (Corning) supplemented with 10% FBS (VWR), 1x GlutaMAX (Corning), and penicillin-streptomycin (Corning) at 37°C with 5% CO2.
Human keratinocytes were isolated from neonatal foreskin as described below and cultured in KGM medium (Lonza). Cells were passaged at 65–70% confluency and frozen after two passages using a serum-free cell freezing medium. Epithelial organoids were generated from human keratinocytes as described below and cultured in organoid culture medium [Advanced DMEM/F12 supplemented with 10 mM HEPES, GlutaMAX, 1% pen/strep, 10% R-spondin1 containing conditioning media (in house), 0.2% Primocin, 100 ng/mL rh-Noggin,1mM N-Acetyl-L-cysteine, 1 μM Y27632, 100 ng/ml rh-FGF, 100 ng/mL Forskolin, 2% B-27 supplement and 2 μg/ml heparin solution] at 37°C with 5% CO2.
Rodent models
Male Sprague-Dawley rats (Charles River Laboratories) were acclimated to study conditions for eight to sixteen days prior to dose administration. At dosing, animals were eight weeks of age. Animals were group housed in polycarbonate cages with hardwood chip bedding. Certified Rodent Diet #2016C and 2016CM (Envigo) were provided ad libitum. Water was provided fresh daily, ad libitum. Environmental controls for the animal room were set to maintain a temperature of 20 to 26°C, a relative humidity of 50 ± 20%, and a 12-hour light/12-hour dark cycle. As necessary, the 12-hour dark cycle was interrupted to accommodate study procedures. Animal care including room, cage, and equipment sanitation conformed to the guidelines cited in institutional SOPs.
Female Lewis rats (Envigo) were acclimated to study conditions for at least seven days prior to study start. Animals were 6–8 weeks old at arrival and were housed 4 to 5 per cage in polycarbonate cages with wire tops, wood chip bedding, and suspended food and water bottles. The rats were housed either in large or small rectangular cages (static airflow, approximately 0.10 or 0.15 m2 floor space) or in pie-shaped cages (passive airflow, approximately 0.16 m2 floor space w/ mezzanine level included). During the acclimation and study periods, the animals were housed in a laboratory environment with temperatures ranging 19°C to 25°C and relative humidity of 50 ± 20% and a 12-hour light/12-hour dark cycle. The animals were allowed access ad libitum to Envigo Teklad 8640 diet and fresh municipal tap water. Animal care including room, cage, and equipment sanitation conformed to the guidelines cited in institutional SOPs.
Six- to eight-week-old NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ mice (NSG; Charles River Laboratories) were kept under specific pathogen-free conditions in individually ventilated cages in a facility controlled according to the Federation of Laboratory Animal Science Association (FELASA) guidelines.
METHOD DETAILS
Computational design
We used the crystal structure of human IL-23R in complex with IL-23p19 and IL-23p40 (PDB 5MZV) as a starting point for design. Because specific inhibition of IL-23 and not IL-12 is desired, we aimed to bind IL-23R, the IL-23-specific receptor subunit, and inhibit its interaction with IL-23p19, the IL-23-specific cytokine subunit. From the crystal structure, we first isolated IL-23R and p19 native hotspots L56, W156, L160, and L161. To supplement the native hotspots, a rotamer interaction field (RIF) of de novo hotspots was generated around selected IL-23R residues near the surface of interest: G24, I25, T26, N27, I28, N29, C30, S31, G32, H33, I34, V36, T40, I50, A54, A55, I56, K57, N58, C59, Q60, P61, K63, L64, H65, F66, Y67, K68, N69, G70, I71, K72, P95, H96, A97, S98, M99, Y100, C101, T102, A103, E104, C105, P106, K107, H108, F109, Q110, E111, T112, L113, I114, C115, G116, K117, D118, I119, S120. The RIF residues (disembodied amino acid side chains) were generated such that the side chain atoms form favorable polar and apolar interactions with the given IL-23R surface residues.
Similarly, we used the crystal structure of human IL-17A in complex with IL-17RA (PDB 4HSA) as a starting point for design, with the surface of interest including residues from both chains of the IL-17A homodimer. From chain 1: N40, R44, V46, Q117, E118, I119, L120, R134, L135, K137, I138, L139. From chain 2: L49, N50, I51, H52, N53, N55, T56, T58, R78, E80, P82, E83, R84, Y85, P86, S87, V88, I89, W90, Q117, I119, L120, L122, R123, R124, E125, P126, P127, P130, N131, S132, F133, R134, L135, V140.
In parallel, 12,345 scaffold proteins (inert de novo designed proteins with experimentally validated stability) were roughly placed at the desired IL-23R or IL-17A interaction surface using PatchDock. After RIF generation and initial scaffold placement, scaffolds were docked with higher resolution at the interaction surface such that the backbone atoms of the native hotspot and/or de novo hotspots were matched with appropriate backbone atoms of each scaffold protein, replacing the amino acid previously at that scaffold position. All other scaffold residues, previously computationally optimized for the lowest monomer free energy, were retained. This step generated 130,343 (IL-23R) and 409,045 (IL-17A) docked configurations.
Each docked configuration was input into a Rosetta design protocol to optimize additional scaffold residues at the target interface for high-affinity binding. Only scaffold side chains within 8 Å of the target surface were allowed to mutate. Scaffold sidechains at surface positions further than 8 Å were not allowed to mutate, but were allowed to optimize rotamer conformation. Target residues within 8 Å of the scaffold were allowed to optimize rotamer conformation. All target and scaffold backbone atoms, all scaffold monomer core side chains, and target side chains further than 8 Å from the scaffold were not allowed to move.
Designed target:inhibitor complexes were filtered on metrics thought to predict high-affinity binding, including but not limited to inhibitor monomer free energy, binding energy, shape complementary of the inhibitor to the target surface, buried apolar surface area at the interface, and buried unsatisfied polar atoms. Designs with the best metrics were selected for experimental testing.
Yeast library preparation
DNA encoding the initial design library was commercially synthesized (Agilent). For site saturation mutagenesis (SSM) libraries, in some instances full-length genes were commercially synthesized (Agilent), and in other instances libraries were prepared using overlap PCR with custom primers (Integrated DNA Technologies) as described previously.45 Combinatorial libraries were prepared by gene assembly from custom oligos; oligos were designed such that all included mutations (Table S2) were represented either individually or as degenerate codons encoding two or more desired mutations. Oligo overlap regions had a minimum length of 12 bp and minimum melt temperature of 40 °C, enabling efficient gene assembly.
All yeast libraries, including the initial design library, SSM libraries, and combinatorial libraries, were prepared with 5’ and 3’ overhangs >20 bp to enable homologous recombination with the plasmid backbone (pETCON) for yeast expression and surface display via fusion to Aga2p.46 For initial SSM and combinatorial libraries for affinity-maturation, the reported pETCON3 vector was used. For SSM and combinatorial libraries built with the objective of enhancing stability in simulated intestinal fluid (SIF), a pETCON variant optimized for enhanced proteolytic stability of Aga2p and Myc-tag was used.47
IL-23R and IL-17 minibinder and IL-23R peptide sequences selected from the initial design library and combinatorial libraries for expression in E. coli and further characterization can be found in Table S1.
Fluorescence-activated cell sorting (FACS)
Yeast strain EBY100 was transformed with each library and vector by electroporation and grown in minimal media selective for the yeast strain (-ura) and the transforming plasmid (-trp).48 Expression was induced with 2% galactose. Surface expression was detected with anti-Myc-FITC (Immunology Consultants Laboratory) conjugate, and binding to biotinylated target was detected with streptavidin-PE (Invitrogen).
Selection scheme and sort conditions for each library are detailed in Table S3. The initial design library, and SSM and combinatorial libraries meant for affinity selection were prepared for selection as follows: after 16–24 hours induction, yeast were spun down, washed with PBS with 1% fetal bovine albumin (PBSF), and incubated for 30–60 minutes with biotinylated target at the given concentration. Yeast were then washed with PBSF and incubated for 2–5 minutes with stain solution (1:100 each anti-Myc-FITC and streptavidin-PE), washed, and resuspended for analysis and selection by FACS. FACS consecutive gates were set as follows: (1) cell granularity and size, selecting for yeast cells (BSC vs. FSC); (2) cell morphology, selecting singlets (FSC-height vs. FSC-width); (3) expression, selecting expressors by proxy of the Myc-tag (FITC fluorescence histogram); and (4) binding signal, selecting the top 1–5% relative to total population (PE vs. FITC).
SIF SSM and combinatorial libraries were prepared as follows: after 16–24 hours induction, yeast were spun down, washed with PBSF, resuspended in SIF (recipe described below) at an OD of 2.0, and incubated at 30 °C shaking for 30–90 minutes as noted. After SIF digest, cells were spun down and washed 4 times with 800 uL PBSF, manually aspirating the supernatant each time to ensure complete washing to remove proteases. SIF-treated cells were then treated with biotinylated IL-23R as described above. FACS gates were set similarly, but gate 3 (expressors) was excluded, as the vast majority of pools showed populations of Myc-negative, binding(PE)-positive cells, indicating that the Myc-tag was cleaved leaving binding-competent design variants displayed on the cell surface.
Generally, design and combinatorial libraries were sorted to convergence in 4–7 consecutive rounds, and SSM libraries were sorted in two consecutive rounds and deep sequenced. The concentration of target protein was generally decreased as sorting rounds progressed in order to efficiently separate the highest-affinity variants. In the case of SIF SSM and combinatorial libraries, protease concentrations in SIF as well as the digest duration were increased with consecutive rounds, in addition to decreasing concentration of target.
Deep mutational scanning
From SSM naive and sorted pools, DNA was prepared and sequenced as follows: yeast were lysed with 125 U/ml Zymolase at 37 °C for 5 hr, and DNA was harvested (Zymoprep kit from Zymo Research). Genomic DNA was digested with 2 U/μl Exonuclease I and 0.25 U/μl Lambda exonuclease (New England Biolabs) for 90 min at 30 °C, and plasmid DNA purified with a QIAquick kit (Qiagen). Minibinder genes were PCR amplified using primers that annealed to external regions within the plasmid, followed by a second round of PCR to add flanking sequences for annealing to the Illumina flow cell oligonucleotides and a 6 bp sample identification sequence, or barcode. PCR rounds were 12 cycles each with high-fidelity Phusion polymerase. Barcodes were read on a MiSeq or NextSeq sequencer using either a 300-cycle or 600-cycle reagent kit (Illumina), and sequences were analyzed with adapted scripts from Enrich.49
Protein expression and purification
All minibinders were cloned into the pET29b plasmid for expression from the T7 promoter, between NdeI and XhoI cut sites, incorporating a C-terminal 6-histidine tag for purification by affinity chromatography. E. coli were transformed with the resulting plasmids: strain BL21 Star (DE3) (Invitrogen) for minibinders without disulfides and strain Shuffle T7 Express (New England Biolabs) for minibinders containing disulfide(s). E. coli were grown to OD600 in Terrific Broth II media (MP Biomedicals) at 37 °C (BL21 Star DE3) or 30 °C (Shuffle T7 Express), then expression was induced with IPTG added to 0.5–1 mM overnight at growth temperature of 18–37 °C. Cells were harvested, lysed by sonication, and lysate cleared by centrifugation. Cleared lysate was incubated with NiNTA resin for 30 minutes rocking to allow binding of recombinant protein via the 6-histidine tag, then applied to a gravity column (Biorad), washed and eluted, concentrated and either exchanged into PBS by dialysis or further purified by gel filtration chromatography into PBS (AKTA Pure, Cytiva; Superdex 75 Increase and Superdex S200 Increase columns, Cytiva).
Recombinant human, mouse, or rat IL-23R with an N-terminal secretion tag (BM40) and a C-terminal polyhistidine tag followed by an avi-tag was cloned into the CMVR plasmid for secreted expression in Expi293F cells. hIL-23R was purified from culture supernatants by IMAC, dialyzed into PBS containing 5% glycerol, and concentrated. To support BLI experiments described below, purified hIL-23R was then site-specifically biotinylated via the avi-tag in a reaction catalyzed by recombinant BirA, a biotin ligase (Avidity). Non-biotinylated hIL-23R was used in the custom ELISA method described below.
To support the custom ELISA method described below, 23R-10 was expressed with a C-terminal avi-tag followed by a 6-histidine tag (23R-10-AH) in E. coli strain CVB-T7 POL (Avidity), harboring another plasmid from which the biotin ligase BirA is co-expressed in order to enzymatically biotinylate target protein in the E. coli cytosol. 23R-10-AH was purified from cell lysate with IMAC, dialyzed into PBS, snap-frozen and stored at −80C.
Size exclusion high performance liquid chromatography (SE-HPLC)
To determine minibinder purity, high performance liquid chromatography was used with a size exclusion column and running buffer. Due to the high pI of minibinder 23R-91, it was necessary to use a running buffer consisting of phosphate-buffered saline (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, pH 7.4) with 500 mM arginine to reduce non-specific interactions with the column (Superdex 75 10/300 GL column, GE). 20 μL of sample at 2 mg/mL in PBS at pH 7.4 was injected and analyzed with an isocratic elution at 0.75 mL/min with a run time of 32 minutes. The instrument used was an Agilent 1260 system and absorbance at 280 nm and 260 nm was monitored during the run.
Liquid chromatography-mass spectrometry (LC-MS)
To verify the molecular mass of minibinders, intact LC-MS was used. 5 uL of sample at 0.1 mg/mL was injected onto a Waters Acquity CSH C18 UPLC column and analyzed with an AB Sciex 5600 QTOF. Mobile phase A was H2O with 0.1% formic acid, mobile phase B was acetonitrile with 0.1% formic acid. A gradient elution was performed from 10% B to 100% B over 4 minutes.
Circular dichroism
CD spectra were recorded with a J-1500 Circular Dichroism Spectrometer (JASCO). Proteins were assayed at 40 μM in DPBS free of MgCl2 and CaCl2 (Life Technologies) with guanidinium hydrochloride from 0 to 6 M, and wavelength scans from 260 to 190 nm were measured at 25 °C. For temperature melts, proteins at 40 μM were heated from 25 °C to 95 °C over approximately 1.5 hours, with CD signal at 222 nm measured every 2 degrees, and wavelength scans from 260 to 190 nm measured every 10 degrees.
Biolayer interferometry and surface plasmon resonance
Qualitative and quantitative assessment of binding affinity was performed using biolayer interferometry (BLI; ForteBio Octet RED96 and associated analysis software) and surface plasmon resonance (SPR; Biacore 8K and associated analysis software). Enzymatically biotinlyated target protein (30 nM) was immobilized on streptavidin-coated sensor tips (BLI) or chip (SPR), or target protein fused to Fc domain was immobilized on anti-human IgG tips. BLI sensor tips were then sequentially dipped in wells with: buffer only (baseline), minibinder in solution (association), and buffer only (dissociation). Similar solutions were flowed over the SPR sensor chip. Kinetic constants were determined from the mathematical fit of a 1:1 binding model.
Simulated gastrointestinal fluids digest
Simulated intestinal fluid (SIF) was prepared as recommended by Jantratid et al. (termed FaSSIFv2) with the addition of proteases trypsin and chymotrypsin each at 30 μg/mL.50 This composition is denoted as “1x SIF” in the text and in Table S3 describing SIF SSM and combinatorial library selection conditions. In some instances, protease concentrations were increased to improve differentiation between minibinders and controls; these solutions are denoted as “#x SIF”, where for example “2x SIF” contains 60 μg/mL each trypsin and chymotrypsin. Simulated gastric fluid (SGF) was prepared per the US Pharmacopeia standard: 600 ug/mL pepsin and 34.2 mM NaCl in water, with HCl added to adjust pH to 2.
For qualitative assessment of proteolytic stability, pure recombinant proteins were digested at 37 °C for 24 hours and proteolytic cleavage assessed by SDS PAGE. From concentrated stock solutions, recombinant proteins were added to stock SGF and SIF solutions to a final concentration of 1 mg/mL. At each indicated time point, samples were removed, immediately mixed with load dye and boiled for 5 minutes at 95 °C to quench protease activity. 5 μg protein per sample (assuming no loss to degradation) was run on 16% Tris-tricine or 4–20% Tris-glycine (TGX) polyacrylamide gels (Biorad) alongside 5 μL Precision Plus Protein™ Dual Xtra Prestained Protein Standards (Biorad).
X-ray crystallography
Crystallization experiments for 23R-91 were conducted using the sitting drop vapor diffusion method. Crystallization trials were set up in 200 nL drops using the 96-well plate format at 20°C. Crystallization plates were set up using a Mosquito LCP from SPT Labtech, then imaged using UVEX microscopes from JAN Scientific. Diffraction quality crystals formed in 0.2 M Lithium sulfate 0.1 M Sodium acetate pH 4.5 and 50% (v/v) PEG 400.
Diffraction data were collected at the Advanced Photon Source at beamline 24ID-C. Crystal diffracted to 1.9 Å resolution. X-ray intensities and data reduction were evaluated and integrated using XDS51 and merged/scaled using Pointless/Aimless in the CCP4 program suite.52 Structure determination and refinement starting phases were obtained by molecular replacement using Phaser53 using the designed model structure. Following molecular replacement, the models were improved using phenix.autobuild.54 Structures were refined in Phenix.54 Model building was performed using COOT.55 The final model was evaluated using MolProbity.56 Data collection and refinement statistics are recorded in the Table S4. Data deposition, atomic coordinates, and structure factors reported for in this paper have been deposited in the Protein Data Bank (PDB), http://www.rcsb.org/ with accession code 8UTK.
IL-23 reporter in vitro signaling assay
Commercial IL-23 reporter cells (Promega IL-23 Bioassay) engineered to express luciferase downstream of IL-23R were used to assess inhibition of IL-23-mediated cell signaling. Cells were plated in the inner wells of 96-well tissue culture treated white plates suitable for reading luminescence. Cells were pre-incubated for 30 minutes with a dilution series of each inhibitor, then treated with the EC80 stimulatory concentration (8 ng/mL) of recombinant human IL-23 cytokine (R&D 1290-IL) determined in preceding experiments. After 6 hours incubation with human IL-23, luciferase substrate (Bio-Glo) was added and luminescence read. Inhibitor response was plotted as percent maximum IL-23 or IL-17 stimulation vs. inhibitor concentration, and IC50 values determined using a four-parameter nonlinear curve fit analysis in Prism.
IL-17 reporter in vitro signaling assay
Commercial IL-17 reporter cells (HEK-Blue IL-17, Invivogen) engineered to express secreted embryonic alkaline phosphatase (SEAP) downstream of the IL-17 receptor were used to assess inhibition of IL-17-mediated cell signaling. Cells were cultured per manufacturer’s protocol. Cells were treated with the EC80 stimulatory concentration of recombinant hIL-17A (0.56 ng/mL; R&D 7955-IL), hIL-17F (1.29 ng/mL; R&D 1335-INS), or hIL-17A/F (6.9 ng/mL; R&D BT5837–025) and a titration of each inhibitor or control (media): cytokine and inhibitor were pre-incubated at 37°C for 30 minutes at 10x assay concentration, after which 20 uL of each condition was added to 180 uL cell suspension in media (300,000 cells/mL) in a 96-well tissue culture treated microplate. After overnight incubation at 37°C (5% CO2), 180 uL of supernatant from each well was transferred to a clear 96-well microplate with 20 uL of chromogenic SEAP substrate (QUANTI-Blue, Invivogen) and incubated at 37°C for 2 hours. SEAP activity, as a proxy for IL-17 signaling, was determined by measuring absorbance at 620 nM. Inhibitor response was plotted as percent maximum IL-23 or IL-17 stimulation vs. inhibitor concentration, and IC50 values determined using a four-parameter nonlinear curve fit analysis in Prism.
Human PBMC in vitro IL-23 signaling assay
Human PBMCs were activated for three days with plate-bound anti-CD3 (2.5 μg/mL Biolegend 317326)/ anti-CD28 (5 μg/mL Biolegend 302943) and 100 IU/mL IL-2, rested overnight in complete RPMI and serum-starved for 2 hours. Cells were stimulated with 20 nM recombinant hIL-23 with or without a titration of minibinder for 20 min at 37°C. After incubation, cells were stained with anti-CD4 Pacific Blue (Biolegend 300521), then fixed with 4% PFA (Electron Microscopy Sciences 15710) in PBS for 10 min at room temperature, permeabilized with ice cold methanol, and stained for phosphorylated STAT3 AF647 (BD 557815) as a downstream marker of IL-23 signaling. Cells were analyzed by flow cytometry and pSTAT3 mean fluorescence intensity in CD4+ T cells plotted vs. inhibitor concentration. IC50 values were determined using a three-parameter nonlinear curve fit analysis in Prism.
ELISA method for detection of IL-23R minibinders
A competitive ELISA method was developed in which the concentration of IL-23R minibinder in a sample is quantified by measuring the inhibition of binding human IL-23R (hIL-23R) to a labeled, high-affinity IL-23R minibinder, 23R-10-AH. Preparation of the custom reagents used in this assay, hIL-23R and 23R-10-AH, are described above in Protein expression and purification.
96-well plates (Nunc Maxisorp) were coated with hIL-23R: 100 μL of coating solution (7.2 μg/mL hIL-23R, 40 mM carbonate-bicarbonate pH 9.4, 0.025% BSA) was added to each well, and the plate was sealed and stored overnight at 4°C. Coating solution was decanted, and the plate washed 3x with 300 μL wash buffer (25 mM Tris pH 7.4, 150 mM NaCl, 0.05% Tween 20) per well. The plate was blocked by adding 300 μL blocking buffer (25 mM Tris pH 7.4, 150 mM NaCl, 0.05% Tween 20, 0.5% nonfat dry milk) per well, incubating for 1 hour at room temperature (RT), then solution decanted and the plate blotted on clean paper towels. Blocked plates were then immediately used for the assay. 100 μL of each standard (diluted in appropriate matrix) or sample was added to each well of the assay plate and incubated at RT for 1 hour. Samples were decanted, and the plate washed 3x. 100 μL of solution containing the peroxidase-linked hIL-23R ligand (750 pM 23R-10-AH, 1:1000 dilution of ExtrAvidin-Peroxidase [Millipore Sigma], prepared in blocking buffer) was added to each well and incubated at RT for 15 minutes, then decanted, and the plate washed 3x. 100 uL of peroxidase substrate solution (1-step Ultra TMB, ThermoFisher), pre-equilibrated to RT, was added to each well and incubated at RT in the dark for 30 minutes. 100 μL stop solution (0.16 M sulfuric acid) was added per well, and absorbance at 450 nm measured using a SpectraMax M5 plate reader.
A standard curve was included per assay plate, prepared by spiking minibinder in undiluted serum or tissue homogenate supernatants at the indicated concentrations spanning the dynamic range of the assay (see Figure S12 for standard curves per minibinder, per biological matrix). Quality control samples – minibinder spiked in the relevant matrix at known concentrations – were included in each assay plate, and the assay was deemed valid if each QC value fell within 25% of the expected value. Per assay, the lower limit of detection (LLOD) was defined as the concentration corresponding to 95% of the top best fit A450 value. The upper limit of detection was defined as the concentration corresponding to the bottom best fit A450 value plus 5% of the top best fit value. Standard curves, QC samples, and study samples were all assayed in duplicate technical replicates.
Specimen preparation for ELISA analysis: immediately after collection, blood samples were allowed to clot at ambient temperature prior to centrifugation to obtain serum. When applicable, at sacrifice small intestine and colon contents were separately collected. Small intestine and colon tissues were then rinsed gently with sterile PBS to remove any residual contents before they were snap-frozen and stored at −70–80C. mLN were dissected, trimmed of fat, snap-frozen and stored at −70–80C. Tissues were homogenized: tissue samples were thawed at ambient temperature until just thawed, then submerged in 4 mL homogenization buffer (ice cold sterile PBS containing one Pierce EDTA-Free Protease Inhibitor Tablet [ThermoFisher cat #A32965] per 50 mL) per 1 g tissue. Tissues were homogenized using a handheld homogenizer (OMNI International, with 7 mm tip) at max speed for 30 seconds. Homogenate was centrifuged for 20 minutes at 15,000 × g, 4C, and supernatant transferred to clean microtubes, snap-frozen and stored at −70–80C for later analysis.
Ex vivo rat tissue signaling assay
From healthy rats, cell suspensions were prepared from the colon, mLN, and spleen. All tissues were homogenized in C tubes using a Miltenyi gentleMACS instrument. Lamina propria cells were further isolated from colon homogenates with Liberase and DNAse digest followed by density gradient centrifugation. Spleen homogenates were enriched for CD3+ cells using the EasySep Total Rat CD3 kit. Cells were washed and counted, then 106 cells were transferred to each well of a 24-well treated tissue culture plate. Cells were pre-incubated for 30 minutes with 23R-91 or vehicle (PBS), then transferred to a new 24-well treated tissue culture plate for stimulation with anti-CD3 (plate pre-coated with 2.5 μg/mL for >2 hours) with or without recombinant rat IL-23 (1 μg/mL for colon cells, 10 ng/mL for mLN cells and splenocytes), or vehicle (no anti-CD3 coating, PBS), as noted (n=3 wells per treatment). Cells were incubated for 24 hours at 37°C (5% CO2). Supernatant was analyzed for rat IL-17A and rat IFNɣ with ELISA (Thermo cat. # BMS635 and ERIFNG). Cells were stained (BD fixable viability stain, FVS450) and viability measured using a CytoFlex flow cytometer (Beckman).
Pharmacokinetics and biodistribution of IL-23R minibinders in rats
All rats were handled according to established guidelines and approved protocols. Male Sprague Dawley rats (Charles River Laboratories) were acclimated to study conditions for at least three days prior to dosing. Animals were 6–8 weeks old at administration and were not fasted prior. A single 20 mg/kg dose of 23R-72 or 23R-91, formulated in PBS or in GI-protective vehicle (GPV; 0.1 M sodium bicarbonate, 200 mg/mL nonfat dry milk) was administered by oral gavage, and blood was drawn at 15, 30, 60, 180, and 360 minutes. A single 4 mg/kg dose of 23R-91 was administered intravenously, and blood was drawn at 5, 15, 30, 60, and 120 minutes. Six (oral) or four (IV) hours after dosing, animals were sacrificed by exsanguination (cardiac puncture) under isoflurane anesthesia.
Female Lewis rats (Envigo) were acclimated to study conditions for at least seven days prior to study start. Animals were 6–8 weeks at arrival. On study day −3, rats were distributed into treatment groups based on bodyweight. On study day −2 through 6, rats were given 20 mg/kg 23R-72 or 23R-91, formulated in PBS, administered by oral gavage three times per day (TID) approximately 6 hours apart. On study day −1, rats were fasted for 12–16 hours prior to intrarectal TNBS challenge (20 mg/rat in 50% ethanol/PBS solution) on study day 0. On study day 7, animals were given a final timed dose of test articles as above, then sacrificed under isoflurane anesthesia by bleeding to exsanguination followed by bilateral pneumothorax. In another study, female Lewis rats as described above were administered a single 140 mg/kg dose of 23R-91 by oral gavage, and serum was collected from 15 minutes to 24 hours post dose for minibinder analysis with ELISA.
Minibinder concentration in serum and tissue homogenate supernatants was quantified using the custom ELISA method described above.
Human skin-derived epithelial organoid culture
Epidermal keratinocytes were isolated from foreskin (neonatal circumcision) cut into thin strips and incubated overnight at 4°C with Dispase (STEMCELL Technologies) containing Penicillin-Streptomycin (Gibco). Epidermis was separated from the dermis with fine forceps, cut into small pieces with scissors and transferred into a 15 mL falcon tube containing 2 mL of warm 0,25% Trypsin/EDTA (Gibco). The epidermis was incubated at 37°C for 1–2 min with gentle shaking to allow detachment of the basal layer of keratinocytes. The supernatant was transferred into 5 mL of DMEM supplemented with pen/strep and 10% FBS to inactivate trypsin. This step was repeated 4–5 times. Suspension was filtered through 70 μm to remove undigested pieces of tissues, the cells were collected by centrifugation 5 min at 1200 rpm. Cells were resuspended into DMEM supplemented with 10% FBS and pen/strep and transferred to a 25 cm2 cell culture flask. The next day, cells were washed with warm PBS, and KGM medium (Lonza, serum-free medium for keratinocytes) was added. Cells were passaged when reaching 65–70% confluency and frozen after 2 passages using a serum-free cell freezing medium (Sigma).
Epithelial organoids were generated from human keratinocytes isolated from foreskin (neonatal circumcision) as described for mouse epithelial organoids with few modifications.57 Briefly, four days after seeding in Cultrex Basement membrane extract (BME type 2, R&D), rhIL-17A (15 nM, Peprotech) or PBS was added to the organoid culture medium [Advanced DMEM/F12 supplemented with 10 mM HEPES, GlutaMAX, 1% Penicillin/Streptomycin, 10% R-spondin1 containing conditioning media (in house), 0.2% Primocin, 100 ng/mL rh-Noggin,1mM N-Acetyl-L-cysteine, 1 μM Y27632, 100 ng/ml rh-FGF, 100 ng/mL Forskolin, 2% B-27 supplement and 2 μg/ml heparin solution] and cultured at 37°C with 5% CO2. For inhibition experiments, 0.75 nM of IL-17A minibinder 17–51 was added either simultaneously, 1 hour or 3 hours after addition of rhIL-17A. After overnight incubation (16 hours), organoids were harvested using a nonenzymatic Cultrex organoid harvesting solution (R&D Systems). Total RNA was extracted from organoids using RNeasy Plus Mini Kit (Qiagen), and equal amounts of RNA were reverse-transcribed using the superscript VILO cDNA synthesis kit (Invitrogen).
Expression of human CCL20, IL-8, S100A7, and HPRT1 was quantified using PowerUp SYBR Green PCR Master Mix (Applied Biosystems) and gene-specific primers (see Key Resources Table). Amplification was performed from a 5 ng cDNA template in a final volume of 20 μL in a 96-well PCR plate. Expression of markers downstream of IL-17A (CCL20, IL-8, and S100A7) was normalized to housekeeping gene HPRT1. Fold change was calculated relative to an untreated control group and was plotted as a percent of the response seen with hIL-17A-only treatment using the following formula:
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD28 | Biolegend | 302943; RRID:AB_2616667 |
| Anti-CD3 | Biolegend | 317326; RRID:AB_2749889 |
| Anti-CD4 | Biolegend | 300521; RRID:AB_493099 |
| Anti-Myc-FITC | Immunology Consultants Laboratory | CMYC-45F |
| Anti-pSTAT3 AF647 | BD | 557815; RRID:AB_647144 |
| Anti-rat CD3 | BD | 556970; RRID:AB_396542 |
| Bimekizumab | MedChemExpress | HY-P99280 |
| Guselkumab | Creative Biolabs | TAB-752 |
| Secukinumab | MedChemExpress | HY-P9927 |
| Bacterial and virus strains | ||
| CVB-T7 POL chemically competent E. coli | Avidity | CVB-T7 POL |
| OneShot BL21 Star (DE3) chemically competent E. coli | Invitrogen | C601003 |
| SHuffle T7 Express competent E.coli | New England Biolabs | C3029J |
| Biological samples | ||
| Human foreskin (surgical discards) | The NYULH Center for Biospecimen Research and Development, and Histology and Immunohistochemistry Laboratory | RRID:SCR_018304 RRID:SCR_017930 |
| Human PBMCs, IBD patients | LMU University Hospital | N/A |
| Human peripheral blood mononuclear cells (PBMCs), healthy volunteers | Stanford Blood Bank | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 1-step Ultra TMB-ELISA substrate solution | Thermo | 34028 |
| 2-mercaptoethanol | Sigma | M6250 |
| Advanced DMEM/F-12 | Gibco | 12634010 |
| B-27 Supplement (50X), serum free | Gibco | 17504044 |
| Biotin | Sigma | B4501–1G |
| Blood collection tube, trisodium citrate solution | S-Monovette | 102332 |
| Blotting grade blocker non-fat dry milk | Bio-rad | 1706404XTU |
| Carbonate-bicarbonate buffer, powder | Thermo | 28382 |
| Casamino acids | Gibco Bacto | DF0231–17-2 |
| Cell freezing medium-DMSO serum free | Sigma | C6295–50mL |
| Chloramphenicol | Sigma | C1919 |
| Chymotrypsin | Sigma | C4129–250MG |
| Complete supplement mixture -ura -trp | MP Biomedicals | 114520512-CF |
| Cultrex basement membrane extract, PathClear | R&D | 3432–010-01 |
| Cultrex HA-R-Spondin1-Fc supernatant from 293T cells, organ harvesting solution | R&D | 3710–001-01 |
| Dispase (5U/ml) | STEMCELL Technologies | 7913 |
| DMEM | Gibco | 10938025 |
| DMEM | Fisher | 11–965-092 |
| Dropout base medium | MP Biomedicals | 114025012-CF |
| ExtrAvidin-Peroxidase | Sigma | E2886 |
| Forskolin | Tocris | 1099 |
| Galactose | Thermo | AAA1281318 |
| GlutaMAX | Gibco/Corning | 35050061 |
| HBSS | Sigma | H9394 |
| HEK-Blue selection reagent | Invivogen | hb-sel |
| Heparin Solution | Stem Cells Technologies | 7980 |
| HEPES (1 M) | Gibco | 15630106 |
| HisPur NiNTA Superflow Agarose | Thermo | 25214 |
| IL-23 Bioassay (reporter cell line, single-use vial) | Promega | JA2511 |
| In vitro biotinylation kit | Avidity | BirA500 |
| IPTG | Sigma | 5800-OP |
| Kanamycin sulfate | Sigma | 60615 |
| KGM gold keratinocytes growth medium bulletKit | Lonza | 192060 |
| MEM non-essential amino acids | Gibco | 11140050 |
| N-Acetyl-Lcysteine | Sigma-Aldrich | A9165–5G |
| Normicin | Invivogen | ant-nr-05 |
| One Shot heat inactivated FBS | Thermo | A3160401 |
| Penicillin-Streptomycin (5,000 U/mL) | Gibco | 15070063 |
| Penicillin-Streptomycin (5,000 U/mL) | Thermo | 15070063 |
| Pepsin, procine stomach | Thermo | J6167906 |
| Pierce EDTA-free protease inhibitor tablets | Thermo | A32965 |
| Primocin | Invivogen | ant-pm-05 |
| PTG-200 | WuXi Apptec | N/A |
| QUANTI-Blue, SEAP substrate for HEK-Blue | Invivogen | rep-qbs |
| Recombinant cynomolgus IL-23R | R&D | 10306-IR-050 |
| Recombinant human FGF-10 | R&D | 345-FG-025/CF |
| Recombinant human IL-17A (HEK-Blue IL-17 assay) | R&D | 7955-IL |
| Recombinant human IL-17A (human epithelial organoid) | PeproTech | 200–17 |
| Recombinant human IL-17A/F | R&D | BT5837–025 |
| Recombinant human IL-17F | R&D | 1335-INS |
| Recombinant human IL-23 | R&D | 1290-IL |
| Recombinant human IL-23R | ECD expressed and purified in-house | N/A |
| Recombinant human Noggin | R&D | 6057-NG-025 |
| Recombinant mouse IL-17A | WuXi Biologics | N/A |
| Recombinant mouse IL-23R | ECD expressed and purified in-house | N/A |
| Recombinant rat IL-23 | R&D | 3136-RL |
| Recombinant rat IL-23R | WuXi Biologics | N/A |
| RPMI 1640 | Corning | 15–040-CV |
| RPMI 1640 with GlutaMAX | Gibco | 61870036 |
| Streptavidin-PE | Invitrogen | S866 |
| Terrific Broth II medium | MP Biomedicals | 113046022-CF |
| Trypsin (0.25%), phenol red | Gibco | 15050065 |
| Tween 20 | Fisher | MP1TWEEN201 |
| V565, anti-TNF nanobody | Expressed and purified in-house, using sequence from Crowe 2018 Sci Rep | N/A |
| Xylocain gel | AstraZenica | 1138060 |
| Y-27632 dihydrochloride | Sigma-Aldrich | Y0503 |
| Yeast nitrogen base without amino acids | BD Difco | DF0335–15-9 |
| Critical commercial assays | ||
| Anti-Rat IFNg ELISA | Thermo | ERIFNG |
| Anti-Rat IL-17 ELISA | Thermo | BMS635 |
| EasySep rat total CD3+ T-cell isolation kit | StemCell | 19641 |
| PowerUp SYBR Green Master Mix for qPCR | Applied Biosystems | A25742 |
| RNeasy Plus Mini Kit | Qiagen | 74134 |
| SuperScript VILO cDNA synthesis kit | Invitrogen | 11754050 |
| Deposited data | ||
| 23R-91 crystal structure | RCSB PDB | Accession ID 8UTK |
| Raw (fastq) and processed data from NGS experimnts | NCBI GEO | Accession ID GSE263250 |
| Source data for main and supplemental figures | Mendeley Data | DOI:10.17632/2n9gstvrsy.1 |
| Experimental models: Cell lines | ||
| EBY-100 | ATCC | MYA-4941 |
| Expi239F | Thermo | A14527 |
| HEK-Blue IL-17 cells | Invivogen | hkb-il17 |
| IL-23 reporter cell line, see commercial assays | See above | See above |
| Experimental models: Organisms/strains | ||
| Lewis rats | Envigo | RRID:RGD_737922 |
| NOD.Cg-Prkdcscid II2rgtm1Wjl/SzJ mice | Charles River Laboratories | RRID:IMSR_JAX:005557 |
| Sprague Dawley rats | Charles River Laboratories | RRID:RGD_737891 |
| Oligonucleotides | ||
| CCL20 fwd: AAGTTGTCTGTGTGCGCAAATCC | Integrated DNA Technologies | Custom |
| CCL20 rev: CCATTCCAGAAAAGCCACAGTTTT | Integrated DNA Technologies | Custom |
| CXCL8 (IL-8) fwd: GAGAGTGATTGAGAGTGGACCAC | Integrated DNA Technologies | Custom |
| CXCL8 (IL-8) rev: CACAACCCTCTGCACCCAGTTT | Integrated DNA Technologies | Custom |
| HPRT1 fwd: CATTATGCT GAGGATTTGGAAAGG | Integrated DNA Technologies | Custom |
| HPRT1 rev: CTTGAGCACACAGAGGGCTACA | Integrated DNA Technologies | Custom |
| S100A7 fwd: AGAAGCCAAGCCTGCTGACGAT | Integrated DNA Technologies | Custom |
| S100A7 rev: GTCCTTTTTCTCAAAGACATCGGC | Integrated DNA Technologies | Custom |
| Recombinant DNA | ||
| Design and SSM libraries, oligo pools of full-length genes | Agilent | Custom, sequences available upon request |
| Human or mouse IL-23R cloned in CMVR plasmid | GenScript | Custom, sequences available upon request |
| Overlapping primers for construction of combinatorial libraries | Integrated DNA Technologies | Custom, sequences available upon request |
| pET29b(+) plasmid backbone | Novagen | 69872 |
| pETCON plasmid backbone | Addgene (unpublished) | RRID: Addgene_41522 |
| pETCON4 plasmid backbone (derivative of pETCON selected for increased resistance to trypsin and chymotrypsin) | Maguire 2020 Proteins | Custom |
| Software and algorithms | ||
| CCP4 | Kabsch, 2010 | https://www.ccp4.ac.uk/ |
| Coot | Emsley and Cowtan 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| GraphPad Prism version 10.1.1 | GraphPad | N/A |
| MolProbity | Williams et al. 2018 | http://molprobity.biochem.duke.edu/ |
| NetMHCII version 2.3 | Technical University of Denmark | https://services.healthtech.dtu.dk/services/NetMHCN-2.3 |
| Phaser | McCoy et al. 2007 | https://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| Phoenix WinNonlin®, Version 6.3 | Pharsight Corp | https://www.certara.com/software/phoenix-winnonlin/ |
| Rosetta molecular modeling suite and scripts for protein design | Cao et al. 2022 | https://www.rosettacommons.org/software/license-and-download |
| XDS | Winn et al., 2001 | https://xds.mr.mpg.de/ |
NSG-IBD humanized mouse model of colitis
From ulcerative colitis patients, 60 mL of peripheral blood was collected into trisodium citrate solution (S-Monovette, Sarstedt, Nürnberg, Germany; cat. no.102332.0). Each human donor can provide PBMCs to reconstitute a maximum of 26 mice. Thus, the data presented herein are compiled among multiple donors. As it’s not feasible to coordinate PBMC donations from multiple donors on the same day, mice were reconstituted and treated on staggered timelines per donor. Untreated and anti-IL-23 controls were tested per donor, thus the N for these groups is higher than for 23R-91 treatment groups. N=11, as in the 80 mg/kg oral 23R-91 group, was determined to be sufficient to reach significance.
The blood was diluted with Hankś balanced salt solution (HBSS, Sigma-Aldrich, Deisenhofen, Germany) at a 1:1 ratio. The suspension was loaded onto LeucoSep tubes (Greiner Bio One, Frickenhausen, Germany; cat. no. Z642843). PBMCs were isolated through iterative centrifugation and resuspended in PBS to attain a concentration of 4 × 106 cells per 100 μL.
Six- to eight-week-old NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ mice (NSG; Charles River Laboratories, Sulzfeld, Germany) were kept under specific pathogen-free conditions in individually ventilated cages in a facility controlled according to the Federation of Laboratory Animal Science Association (FELASA) guidelines. Mice were engrafted with 100 μL cell solution into the tail vein on Day 1 of the study.58 Mice were presensitized by rectal application of 150 μL of 10% ethanol on Day 7 using a 1 mm cat catheter (Henry Schein, Hamburg, Germany; cat. no. 2734651). The catheter was lubricated with Xylocain Gel 2% (AstraZeneca, Wedel, Germany; cat. no. 01138060). Rectal application was performed under general anesthesia using 4% isoflurane (Zoetis, Berlin, Germany). After application, mice were kept at an angle of 30° to avoid ethanol dripping. On Day 14, mice were challenged by rectal application of 50% ethanol, following the same protocol. On Day 18, mice were sacrificed. Guselkumab (4 mg/kg, 100 μL; Creative Biolabs) was administered by intraperitoneal injection on Days 6 and 13. 23R-91 (8 or 80 mg/kg, 150 μL in 1% methylcellulose gel) was administered once daily by oral gavage on Days 6, 7, and 13–17.58
The clinical assessment of colitis severity was performed daily according to the scoring system described in Weß et al.58, detailed in Table S6. Daily scores were incorporated into a running total. Animals who suffered from weight loss >20%, rectal bleeding, rectal prolapse, self-isolation or a severity total score >7 were euthanized immediately and not considered in the calculation. All scores were added for statistical analysis.
After euthanizing the mice, their colons were excised, photographed, and a macroscopic score of inflammation was determined based on the criteria described in Table S6.58 The distal sections of the colon were initially preserved in 4% formaldehyde for 24 hours, followed by storage in 70% ethanol, and subsequently underwent standard paraffin embedding. Samples were then sectioned into 3 μm slices, stained with hematoxylin and eosin (H&E) and Sirius Red (SR), and histological disease score assessed based on the criteria described in Table S6.
Group averages ± standard deviations were plotted and each treatment group compared to the challenged control group using the non-parametric Wilcoxon matched-pairs test with GraphPad Prism 10.
Immunogenicity prediction
NetMHCII version 2.3 was used in this study.42 This software was trained to predict MHCII binding based on the Immune Epitope DataBase (IEDB) which contains binding data for over 100,000 peptides to MHCII molecules. The software predicts binding of peptides derived from the input sequence (e.g., 23R-91) to the most common MHCII variants, including 25 alleles for HLA-DR, 20 alleles for HLA-DQ, and 9 alleles for HLA-DP, that have the largest amount of binding data in the IEDB. A percent rank was calculated for each potential binding peptide by comparing its predicted binding affinity to that of one million random peptides. This allowed for the classification of predicted peptide binding as strong (top 2% of predicted binders), weak (top 2–10%), and non-binding (lower 90%). As a benchmark, the 23R-91 sequence (62 amino acids) was compared to that of Neo-2/15 (100 amino acids) and Kuma062 (553 amino acids), two engineered proteins with previously demonstrated low immunogenicity in animals or humans.15,39 Software output values can be found in Table S7.
QUANTIFICATION AND STATISTICAL ANALYSIS
Numbers of technical and biological replicates can be found in the figure legends.
GraphPad Prism version 10.1.1 was used to compute non-parametric Wilcoxon matched-pair tests among groups of the NSG-IBD mouse efficacy study. P values (P* <0.033, P** <0.002, P*** P<0.0001) define significance.
Supplementary Material
Figure S1. Related to Figures 1–3. Binding affinities (KD) of IL-23R and IL-17A minibinders. BLI was used to quantitatively determine the binding constants of initial computational designs (IL23R: 23R-1, 23R-2; IL-17A: 17–1, 17–2, 17–3), combinatorial variants derived from 23R-1 (23R-10, 23R-13, 23R-15), 23R-2 (23R-19, 23R-22, 23R-24), and 17–2 (17–35), a disulfide-stabilized variant of 17–35 (17–51), and a single-chain linked dimer of 17–51 (17–53). Sample data were fit to a single 1:1 binding equation for all IL-23R binders and IL-17A binder 17–53. For all other IL-17A binders, data were fit to a 2:1 binding equation, which reflects that two minibinder molecules in solution may bind to each of two binding sites on immobilized hIL-17A, which are known to have unique affinities that may be influenced by the first binding event. Reported KDs are an average of the KDs determined for the two unique interactions. Plots are representative of at least two independent experiments.
Figure S2. Related to Figure 3. In vitro potency, stability, and species cross-reactivity of IL-17A minibinders. The cellular potencies of (A) 17–1, related affinity-optimized variant 17–16, and related disulfide-stabilized variant 17–45, and (B) single-chain linked dimers of 17–53 having the indicated linker compositions were measured using an engineered IL-17 reporter cell line. Representative curves are shown above (n≥2), and IC50 values are reported as mean ± SD of at least two independent experiments (N≥2). (C) 17–1 and derivatives described above, and (D) additional 17–1 derivative optimized variants were denatured with heat or chemical denaturant guanidinium hydrochloride (Gdn), and helicity (signal at 222 nm) was monitored using circular dichroism. Signal is plotted as a fraction of the reference sample (0 M Gdn at 25ºC). (E) 17–51 and 17–53 inhibit mouse IL-17A with much weaker potency than human IL-17A in the IL-17 reporter cell assay.
Figure S3. Related to Figure 1–3. Saturation mutagenesis of IL-23R and IL-17A minibinders and BLI screening of affinity-optimized combinatorial variants. (A) The relative affinity of each mutation to computational designs targeting IL-23R (23R-B) or IL-17A (17–02, 17–03) was determined using deep mutational scanning. The enrichment (blue) or depletion (red) of each mutation, depicted in 2D heatmaps, represents its impact on affinity relative to the original minibinder sequence (set to 0, white). Positional conservation scores are depicted in a 1D heatmap from minimum (light gray) to maximum (dark gray) per design. Asterisks indicate native and de novo hotspots. (B) Binding of computational designs (23R-1 and 23R-2, black dashed line) and affinity-optimized variants (23R-1 derivatives 23R-3 to 23R-15, 23R-2 derivatives 23R-17 to 23R-22, 23R-24 to 23R-28, colored lines) to hIL-23R immobilized on the BLI sensor tip was measured at 200 nM with BLI. (C) Binding of computational design (17–1, black dashed line) and combinatorial variants (17–1 derivatives 17–4 to 17–20, 17–2 derivatives 17–21 to 17–36, colored lines) to hIL-17A immobilized on the BLI sensor tip was measured at 500 nM.
Figure S4. Related to Figure 2. Additional biophysical characterization of IL-23R minibinders. CD data, collected and represented as described in Figure 2, for affinity-optimized combinatorial IL-23R minibinders derived from (A) 23R-1 and (B) 23R-2, and stability optimized minibinders derived from (C) 23R-1 and (D) 23R-2. (E) Raw CD data for lead IL-23R minibinders 23R-72 and 23R-91 after denaturation with heat. CD signal at 222 nm (helicity) was measured every 2 degrees (left panel), and full wavelength scans from 250 to 200 nm (right panel) were performed every 10 degrees as well as after the sample was cooled to 25°C after heating. Dotted vertical lines mark 222 nm. (F) SE-HPLC: 20 μL of 23R-91 at 2 mg/mL was injected onto a Superdex 75 10/300 GL column. The major peak eluted at 20.078 minutes, consistent with the monomer molecular weight of 7,206 Da. A small second peak eluted at 23.845 minutes; LC-MS analysis identified this peak as 23R-91 with one of the two intramolecular disulfides reduced (−2 Da; data not shown). (G) 23R-91 is soluble at >100 mg/mL; potency of 23R-91 in the IL-23 reporter cell assay is unchanged after concentration to >100 mg/mL. (H) LC-MS analysis of 23R-91 yielded the expected molecule weight (7206.8 Da), as well as a species likely to be 23R-91 with a formyl methionine adduct likely caused by exposure to formic acid during the analytical method (+28 Da). SE-HPLC and LC-MS analyses were performed in at least three independent experiments with various lots of 23R-91.
Figure S5. Related to Figure 4. IL-23R minibinders retain binding capacity after SGF or SIF digest. Stability-optimized IL-23R minibinders (23R-64 derivatives 23R-70, −72, −80, −81, and −82; 23R-49 derivatives 23R-83, −84, −85, −86, −87, −90, −91, and −92) were tested for binding to hIL-23R after digest in SIF or SGF at the indicated time points, using BLI.
Figure S6. Related to Figure 1. Peptides derived from minibinders are folded and block IL-23R-mediated cell signaling. (A) Saturation mutagenesis suggests that 26-residue IL-23R binding peptides (23R-95, 23R-96, 23R-97) have the predicted conformation and binding mode. SSM libraries based on each peptide design sequence were sorted for improved binding to IL-23R, and relative enrichment or depletion of each mutation was determined by deep sequencing the naïve and sorted pools. The enrichment (blue) or depletion (red) of each mutation, depicted in 2D heatmaps, represents its impact on affinity relative to the original peptide sequence (set to 0, white). Positional conservation scores are depicted in a 1D heatmap from minimum (light gray) to maximum (dark gray) per design. Asterisks indicate native and de novo hotspots. (B) Combinatorial libraries including enriching mutations were generated (Table S2) and sorted for improved IL-23R affinity (Table S3). The potency of the highest affinity peptide, 23R-101, in the IL-23 reporter cell assay is shown compared to that of the best 7–8 kDa minibinder (23R-91) and a competing oral anti-IL-23R peptide PTG-200. Representative curves are shown above, and IC50 values are reported as mean ± SD of at least three independent experiments.
Figure S7. Related to Figure 4. ELISA method standard curves for 23R-72 and 23R-91 in biological matrices, and species cross-reactivity of 23R-91. Each 96-well ELISA assay plate included a standard curve of 23R-72 (A) or 23R-91 (B), prepared by spiking the minibinder in undiluted serum or tissue homogenate supernatant at the indicated concentrations spanning the dynamic range of the assay (n=2). The ELISA method was performed as described in Methods. Fit curves were calculated using four parameter logistic regression with Prism. (C) Binding of 23R-91 to human IL-23R or orthologs from species commonly used in preclinical development was compared using BLI. 23R-91 shows high affinity binding to human, macaque, and rat IL-23R, but negligible binding to mouse IL-23R. Recombinant human or mouse IL-23 cytokines were used as positive controls. (D) The model of 23R-91 bound to human IL-23R is shown with positions differing from mouse IL-23R highlighted in pink. Corroborating the BLI data, mouse IL-23R has a single small-to-large mutation (S98Y) at the binding interface, likely responsible for abrogating binding to 23R-91. Rat and macaque IL-23R have no mutations at the core interface (models not shown).
Table S2. Combinatorial library design summary, related to STAR methods.
Table S3. Yeast library FACS conditions, related to STAR methods.
Table S4. Crystallographic data collection and refinement statistics for 23R-91, related to Figure 5.
Table S5. Summary of Rat PK and biodistribution data, related to Figures 4C and 4D.
Table S6. NSG-IBD scoring matrices, related to Figure 7 and STAR methods.
Table S7. Results of immunogenicity prediction of 23R-91 using NetMHCII software, related to STAR methods.
Highlights.
Computational design yielded low- and sub-pM minibinders of IL-17A and IL-23R
IL-23R minibinders are extremely resistant to heat, acid, and proteolysis
Oral IL-23R minibinder is as effective as a clinical mAb in mouse colitis
Acknowledgements
Funding for this research was provided by the Washington Research Foundation Translational Research Grants (S.B., F.S., L.A., C.S., A.B., and D.B.), the Center for Washington Entrepreneurial Research Evaluation & Commercialization Hub (WE-REACH; S.B. and L.A.), The Audacious Project at the Institute for Protein Design (L.C., R.R., M.M., and D.B.), and the Howard Hughes Medical Institute (D.B.). This research was also supported by Mopac Biologics, Inc. Crystallographic diffraction data were collected at the Northeastern Collaborative Access Team beamlines at the Advanced Photon Source, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The NYULH Center for Biospecimen Research and Development and the Histology and Immunohistochemistry Laboratory are supported in part by the Laura and Isaac Perlmutter Cancer Center Support Grant: NIH/NCI P30 CA016087 (D.R., L.G.-M., and S.N.).
Footnotes
Declaration of Interests
S.B., T.Y., I.S.P., L.S., and D.B. are co-founders and shareholders of Mopac Biologics, Inc. S.B., F.S., T.Y., and D.B. are co-inventors on a patent describing the IL-23R minibinders (PCT/US2021/039122), licensed to Mopac Biologics. S.B. is a board member and paid consultant of Mopac Biologics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feagan BG, Sandborn WJ, Gasink C, Jacobstein D, Lang Y, Friedman JR, Blank MA, Johanns J, Gao L-L, Miao Y, et al. (2016). Ustekinumab as Induction and Maintenance Therapy for Crohn’s Disease. N. Engl. J. Med. 375, 1946–1960. [DOI] [PubMed] [Google Scholar]
- 2.Sands BE, Sandborn WJ, Panaccione R, O’Brien CD, Zhang H, Johanns J, Adedokun OJ, Li K, Peyrin-Biroulet L, Van Assche G, et al. (2019). Ustekinumab as Induction and Maintenance Therapy for Ulcerative Colitis. N. Engl. J. Med. 381, 1201–1214. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, Li B, Guo Q, Tang J, Peng B, Ding N, Li M, Yang Q, Huang Z, Diao N, et al. (2022). Systematic review with meta-analysis: loss of response and requirement of ustekinumab dose escalation in inflammatory bowel diseases. Aliment. Pharmacol. Ther. 55, 764–777. [DOI] [PubMed] [Google Scholar]
- 4.Roblin X, Duru G, Papamichael K, Cheifetz AS, Kwiatek S, Berger A-E, Barrau M, Waeckel L, Nancey S, and Paul S (2023). Development of Antibodies to Ustekinumab Is Associated with Loss of Response in Patients with Inflammatory Bowel Disease. J. Clin. Med. Res 12. 10.3390/jcm12103395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janssen Biotech, Inc. (2016). Stelara (ustekinumab) [package insert]. www.accessdata.fda.gov/drugsatfda_docs/label/2016/761044lbl.pdf. Accessed May 26, 2024.
- 6.Shah DK, and Betts AM (2013). Antibody biodistribution coefficients: inferring tissue concentrations of monoclonal antibodies based on the plasma concentrations in several preclinical species and human. MAbs 5, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.U.S. Food and Drug Administration (2019). Xeljanz, Xeljanz XR (tofacitinib): Drug Safety Communication. https://www.fda.gov/safety/medical-product-safety-information/xeljanz-xeljanz-xr-tofacitinib-drug-safety-communication-due-increased-risk-blood-clots-and-death. Accessed May 26, 2024. [Google Scholar]
- 8.Drakos A, Torres T, and Vender R (2024). Emerging Oral Therapies for the Treatment of Psoriasis: A Review of Pipeline Agents. Pharmaceutics 16. 10.3390/pharmaceutics16010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren RB (2023). DC-806, an oral IL-17A inhibitor: Proof-of-concept in adults with mild to moderate psoriasis. Presented at: AAD Annual Meeting; March 17–21, 2023; New Orleans. [Google Scholar]
- 10.Linsky TW, Noble K, Tobin AR, Crow R, Carter L, Urbauer JL, Baker D, and Strauch E-M (2022). Sampling of structure and sequence space of small protein folds. Nat. Commun. 13, 7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocklin GJ, Chidyausiku TM, Goreshnik I, Ford A, Houliston S, Lemak A, Carter L, Ravichandran R, Mulligan VK, Chevalier A, et al. (2017). Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhardwaj G, Mulligan VK, Bahl CD, Gilmore JM, Harvey PJ, Cheneval O, Buchko GW, Pulavarti SVSRK, Kaas Q, Eletsky A, et al. (2016). Accurate de novo design of hyperstable constrained peptides. Nature 538, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park Y-J, et al. (2020). De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chevalier A, Silva D-A, Rocklin GJ, Hicks DR, Vergara R, Murapa P, Bernard SM, Zhang L, Lam K-H, Yao G, et al. (2017). Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva D-A, Yu S, Ulge UY, Spangler JB, Jude KM, Labão-Almeida C, Ali LR, Quijano-Rubio A, Ruterbusch M, Leung I, et al. (2019). De novo design of potent and selective mimics of IL-2 and IL-15. Nature 565, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Case JB, Chen RE, Cao L, Ying B, Winkler ES, Johnson M, Goreshnik I, Pham MN, Shrihari S, Kafai NM, et al. (2021). Ultrapotent miniproteins targeting the SARS-CoV-2 receptor-binding domain protect against infection and disease. Cell Host Microbe 29, 1151–1161.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vignali DAA, and Kuchroo VK (2012). IL-12 family cytokines: immunological playmakers. Nat. Immunol. 13, 722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glassman CR, Mathiharan YK, Jude KM, Su L, Panova O, Lupardus PJ, Spangler JB, Ely LK, Thomas C, Skiniotis G, et al. (2021). Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells. Cell 184, 983–999.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. (2003). Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748. [DOI] [PubMed] [Google Scholar]
- 20.Kikly K, Liu L, Na S, and Sedgwick JD (2006). The IL-23/Th(17) axis: therapeutic targets for autoimmune inflammation. Curr. Opin. Immunol. 18, 670–675. [DOI] [PubMed] [Google Scholar]
- 21.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch E-M, Wilson IA, and Baker D (2011). Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 332, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger S, Procko E, Margineantu D, Lee EF, Shen BW, Zelter A, Silva D-A, Chawla K, Herold MJ, Garnier J-M, et al. (2016). Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer. Elife 5. 10.7554/eLife.20352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao L, Coventry B, Goreshnik I, Huang B, Sheffler W, Park JS, Jude KM, Marković I, Kadam RU, Verschueren KHG, et al. (2022). Design of protein-binding proteins from the target structure alone. Nature 605, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linsky T, Noble K, Tobin A, Crow R, Carter L, Urbauer J, Baker D, and Strauch EM (2021). Sampling of Structure and Sequence Space of Small Protein Folds. bioRxiv. 10.1101/2021.03.10.434454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobin AR, Crow R, Urusova DV, Klima JC, Tolia NH, and Strauch E-M (2023). Inhibition of a malaria host-pathogen interaction by a computationally designed inhibitor. Protein Sci. 32, e4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalziel JE, Young W, Bercik P, Spencer NJ, Ryan LJ, Dunstan KE, Lloyd-West CM, Gopal PK, Haggarty NW, and Roy NC (2016). Tracking gastrointestinal transit of solids in aged rats as pharmacological models of chronic dysmotility. Neurogastroenterol. Motil. 28, 1241–1251. [DOI] [PubMed] [Google Scholar]
- 27.Dodoo CC, Wang J, Basit AW, Stapleton P, Gaisford S (2017). Targeted delivery of probiotics to enhance gastrointestinal stability and intestinal colonisation. Int. J. Pharm. 530, 224–229. [DOI] [PubMed] [Google Scholar]
- 28.Maurer JM, Hofman S, Schellekens RCA, Tonnis WF, Dubois AOT, Woerdenbag HJ, Hinrichs WLJ, Kosterink JGW, and Frijlink HW (2016). Development and potential application of an oral ColoPulse infliximab tablet with colon specific release: A feasibility study. Int. J. Pharm. 505, 175–186. [DOI] [PubMed] [Google Scholar]
- 29.Abramson A, Frederiksen MR, Vegge A, Jensen B, Poulsen M, Mouridsen B, Jespersen MO, Kirk RK, Windum J, Hubálek F, et al. (2022). Oral delivery of systemic monoclonal antibodies, peptides and small molecules using gastric auto-injectors. Nat. Biotechnol. 40, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhalla AK, Al-Shamsie Z, Beraki S, Dasari A, Fung LC, Fusaro L, Garapaty A, Gutierrez B, Gratta D, Hashim M, et al. (2022). A robotic pill for oral delivery of biotherapeutics: safety, tolerability, and performance in healthy subjects. Drug Deliv. Transl. Res. 12, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SN, Stork C, Valenzuela R, Walker M, Smith B, Smith J, Quintana N, Wahl C, Singh S (2023). Evaluation of the pharmacokinetics of glucagon-like-peptide-1 (GLP-1) receptor agonist delivered through the BioJet™ oral biotherapeutic delivery platform in a porcine model. Poster presented at: 59th Annual Meeting of the European Association for the Study of Diabetes, October 2–6, 2023, Hamburg, Germany. [Google Scholar]
- 32.Crowe JS, Roberts KJ, Carlton TM, Maggiore L, Cubitt MF, Clare S, Harcourt K, Reckless J, MacDonald TT, Ray KP, et al. (2018). Preclinical Development of a Novel, Orally-Administered Anti-Tumour Necrosis Factor Domain Antibody for the Treatment of Inflammatory Bowel Disease. Sci. Rep. 8, 4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nurbhai S, Roberts KJ, Carlton TM, Maggiore L, Cubitt MF, Ray KP, Reckless J, Mohammed H, Irving P, MacDonald TT, et al. (2019). Oral Anti-Tumour Necrosis Factor Domain Antibody V565 Provides High Intestinal Concentrations, and Reduces Markers of Inflammation in Ulcerative Colitis Patients. Sci. Rep. 9, 14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhandari A, Bourne G, Cheng X, Frederick BT, Zhang J, Patel DV, and Liu D (2020). Peptide inhibitors of interleukin-23 receptor and their use to treat inflammatory diseases. US Patent 10787490B2.
- 35.Fourie A, Cheng X, Change L, Greving C, Patrick A, Knight B, Polidori D, Patch R, Bhandari A, Liu D, et al. (2023). First-in-class oral peptide systemically targeting the IL-23 pathway. Poster presented at: International Societies for Investigative Dermatology Meeting, May 10–13, 2023, Tokyo, Japan. [Google Scholar]
- 36.Salahudeen MS, and Nishtala PS (2017). An overview of pharmacodynamic modelling, ligand-binding approach and its application in clinical practice. Saudi Pharm J 25, 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corzo J (2006). Time, the forgotten dimension of ligand binding teaching. Biochem. Mol. Biol. Educ. 34, 413–416. [DOI] [PubMed] [Google Scholar]
- 38.Sun R, Hedl M, and Abraham C (2020). IL23 induces IL23R recycling and amplifies innate receptor-induced signalling and cytokines in human macrophages, and the IBD-protective IL23R R381Q variant modulates these outcomes. Gut 69, 264–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pultz IS, Hill M, Vitanza JM, Wolf C, Saaby L, Liu T, Winkle P, and Leffler DA (2021). Gluten Degradation, Pharmacokinetics, Safety, and Tolerability of TAK-062, an Engineered Enzyme to Treat Celiac Disease. Gastroenterology 161, 81–93.e3. [DOI] [PubMed] [Google Scholar]
- 40.Neoleukin Therapeutics Announces Third Quarter 2022 Financial Results and Corporate Update (2022). GlobeNewswire. https://www.globenewswire.com/en/news-release/2022/11/14/2555436/0/en/Neoleukin-Therapeutics-Announces-Third-Quarter-2022-Financial-Results-and-Corporate-Update.html. Accessed May 26, 2024. [Google Scholar]
- 41.Snapper CM (2018). Distinct Immunologic Properties of Soluble Versus Particulate Antigens. Front. Immunol. 9, 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, and Nielsen M (2018). Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unterweger A-L, Rüscher A, Seuß M, Winkelmann P, Beigel F, Koletzko L, Breiteneicher S, Siebeck M, Gropp R, and Aszodi A (2021). NOD/scid IL-2Rγnull mice reconstituted with peripheral blood mononuclear cells from patients with Crohn’s disease reflect the human pathological phenotype. Immun. Inflamm. Dis. 9, 1631–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bissonnette R, Pinter A, Ferris LK, Gerdes S, Rich P, Vender R, Miller M, Shen Y-K, Kannan A, Li S, et al. (2024). An Oral Interleukin-23-Receptor Antagonist Peptide for Plaque Psoriasis. N. Engl. J. Med. 390, 510–521. [DOI] [PubMed] [Google Scholar]
- 45.Procko E, Hedman R, Hamilton K, Seetharaman J, Fleishman SJ, Su M, Aramini J, Kornhaber G, Hunt JF, Tong L, et al. (2013). Computational design of a protein-based enzyme inhibitor. J. Mol. Biol. 425, 3563–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, and Wittrup KD (2006). Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768. [DOI] [PubMed] [Google Scholar]
- 47.Maguire JB, Haddox HK, Strickland D, Halabiya SF, Coventry B, Griffin JR, Pulavarti SVSRK, Cummins M, Thieker DF, Klavins E, et al. (2021). Perturbing the energy landscape for improved packing during computational protein design. Proteins 89, 436–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benatuil L, Perez JM, Belk J, and Hsieh C-M (2010). An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 23, 155–159. [DOI] [PubMed] [Google Scholar]
- 49.Fowler DM, Araya CL, Gerard W, and Fields S (2011). Enrich: software for analysis of protein function by enrichment and depletion of variants. Bioinformatics 27, 3430–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jantratid E, Janssen N, Reppas C, and Dressman JB (2008). Dissolution media simulating conditions in the proximal human gastrointestinal tract: an update. Pharm. Res. 25, 1663–1676. [DOI] [PubMed] [Google Scholar]
- 51.Kabsch W (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 56.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boonekamp KE, Kretzschmar K, Wiener DJ, Asra P, Derakhshan S, Puschhof J, López-Iglesias C, Peters PJ, Basak O, and Clevers H (2019). Long-term expansion and differentiation of adult murine epidermal stem cells in 3D organoid cultures. Proc. Natl. Acad. Sci. U. S. A. 116, 14630–14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weß V, Schuster-Winkelmann P, Karatekin YH, Malik S, Beigel F, Kühn F, and Gropp R (2023). Humanized NSG Mouse Models as a Preclinical Tool for Translational Research in Inflammatory Bowel Diseases. Int. J. Mol. Sci. 24. 10.3390/ijms241512348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figures 1–3. Binding affinities (KD) of IL-23R and IL-17A minibinders. BLI was used to quantitatively determine the binding constants of initial computational designs (IL23R: 23R-1, 23R-2; IL-17A: 17–1, 17–2, 17–3), combinatorial variants derived from 23R-1 (23R-10, 23R-13, 23R-15), 23R-2 (23R-19, 23R-22, 23R-24), and 17–2 (17–35), a disulfide-stabilized variant of 17–35 (17–51), and a single-chain linked dimer of 17–51 (17–53). Sample data were fit to a single 1:1 binding equation for all IL-23R binders and IL-17A binder 17–53. For all other IL-17A binders, data were fit to a 2:1 binding equation, which reflects that two minibinder molecules in solution may bind to each of two binding sites on immobilized hIL-17A, which are known to have unique affinities that may be influenced by the first binding event. Reported KDs are an average of the KDs determined for the two unique interactions. Plots are representative of at least two independent experiments.
Figure S2. Related to Figure 3. In vitro potency, stability, and species cross-reactivity of IL-17A minibinders. The cellular potencies of (A) 17–1, related affinity-optimized variant 17–16, and related disulfide-stabilized variant 17–45, and (B) single-chain linked dimers of 17–53 having the indicated linker compositions were measured using an engineered IL-17 reporter cell line. Representative curves are shown above (n≥2), and IC50 values are reported as mean ± SD of at least two independent experiments (N≥2). (C) 17–1 and derivatives described above, and (D) additional 17–1 derivative optimized variants were denatured with heat or chemical denaturant guanidinium hydrochloride (Gdn), and helicity (signal at 222 nm) was monitored using circular dichroism. Signal is plotted as a fraction of the reference sample (0 M Gdn at 25ºC). (E) 17–51 and 17–53 inhibit mouse IL-17A with much weaker potency than human IL-17A in the IL-17 reporter cell assay.
Figure S3. Related to Figure 1–3. Saturation mutagenesis of IL-23R and IL-17A minibinders and BLI screening of affinity-optimized combinatorial variants. (A) The relative affinity of each mutation to computational designs targeting IL-23R (23R-B) or IL-17A (17–02, 17–03) was determined using deep mutational scanning. The enrichment (blue) or depletion (red) of each mutation, depicted in 2D heatmaps, represents its impact on affinity relative to the original minibinder sequence (set to 0, white). Positional conservation scores are depicted in a 1D heatmap from minimum (light gray) to maximum (dark gray) per design. Asterisks indicate native and de novo hotspots. (B) Binding of computational designs (23R-1 and 23R-2, black dashed line) and affinity-optimized variants (23R-1 derivatives 23R-3 to 23R-15, 23R-2 derivatives 23R-17 to 23R-22, 23R-24 to 23R-28, colored lines) to hIL-23R immobilized on the BLI sensor tip was measured at 200 nM with BLI. (C) Binding of computational design (17–1, black dashed line) and combinatorial variants (17–1 derivatives 17–4 to 17–20, 17–2 derivatives 17–21 to 17–36, colored lines) to hIL-17A immobilized on the BLI sensor tip was measured at 500 nM.
Figure S4. Related to Figure 2. Additional biophysical characterization of IL-23R minibinders. CD data, collected and represented as described in Figure 2, for affinity-optimized combinatorial IL-23R minibinders derived from (A) 23R-1 and (B) 23R-2, and stability optimized minibinders derived from (C) 23R-1 and (D) 23R-2. (E) Raw CD data for lead IL-23R minibinders 23R-72 and 23R-91 after denaturation with heat. CD signal at 222 nm (helicity) was measured every 2 degrees (left panel), and full wavelength scans from 250 to 200 nm (right panel) were performed every 10 degrees as well as after the sample was cooled to 25°C after heating. Dotted vertical lines mark 222 nm. (F) SE-HPLC: 20 μL of 23R-91 at 2 mg/mL was injected onto a Superdex 75 10/300 GL column. The major peak eluted at 20.078 minutes, consistent with the monomer molecular weight of 7,206 Da. A small second peak eluted at 23.845 minutes; LC-MS analysis identified this peak as 23R-91 with one of the two intramolecular disulfides reduced (−2 Da; data not shown). (G) 23R-91 is soluble at >100 mg/mL; potency of 23R-91 in the IL-23 reporter cell assay is unchanged after concentration to >100 mg/mL. (H) LC-MS analysis of 23R-91 yielded the expected molecule weight (7206.8 Da), as well as a species likely to be 23R-91 with a formyl methionine adduct likely caused by exposure to formic acid during the analytical method (+28 Da). SE-HPLC and LC-MS analyses were performed in at least three independent experiments with various lots of 23R-91.
Figure S5. Related to Figure 4. IL-23R minibinders retain binding capacity after SGF or SIF digest. Stability-optimized IL-23R minibinders (23R-64 derivatives 23R-70, −72, −80, −81, and −82; 23R-49 derivatives 23R-83, −84, −85, −86, −87, −90, −91, and −92) were tested for binding to hIL-23R after digest in SIF or SGF at the indicated time points, using BLI.
Figure S6. Related to Figure 1. Peptides derived from minibinders are folded and block IL-23R-mediated cell signaling. (A) Saturation mutagenesis suggests that 26-residue IL-23R binding peptides (23R-95, 23R-96, 23R-97) have the predicted conformation and binding mode. SSM libraries based on each peptide design sequence were sorted for improved binding to IL-23R, and relative enrichment or depletion of each mutation was determined by deep sequencing the naïve and sorted pools. The enrichment (blue) or depletion (red) of each mutation, depicted in 2D heatmaps, represents its impact on affinity relative to the original peptide sequence (set to 0, white). Positional conservation scores are depicted in a 1D heatmap from minimum (light gray) to maximum (dark gray) per design. Asterisks indicate native and de novo hotspots. (B) Combinatorial libraries including enriching mutations were generated (Table S2) and sorted for improved IL-23R affinity (Table S3). The potency of the highest affinity peptide, 23R-101, in the IL-23 reporter cell assay is shown compared to that of the best 7–8 kDa minibinder (23R-91) and a competing oral anti-IL-23R peptide PTG-200. Representative curves are shown above, and IC50 values are reported as mean ± SD of at least three independent experiments.
Figure S7. Related to Figure 4. ELISA method standard curves for 23R-72 and 23R-91 in biological matrices, and species cross-reactivity of 23R-91. Each 96-well ELISA assay plate included a standard curve of 23R-72 (A) or 23R-91 (B), prepared by spiking the minibinder in undiluted serum or tissue homogenate supernatant at the indicated concentrations spanning the dynamic range of the assay (n=2). The ELISA method was performed as described in Methods. Fit curves were calculated using four parameter logistic regression with Prism. (C) Binding of 23R-91 to human IL-23R or orthologs from species commonly used in preclinical development was compared using BLI. 23R-91 shows high affinity binding to human, macaque, and rat IL-23R, but negligible binding to mouse IL-23R. Recombinant human or mouse IL-23 cytokines were used as positive controls. (D) The model of 23R-91 bound to human IL-23R is shown with positions differing from mouse IL-23R highlighted in pink. Corroborating the BLI data, mouse IL-23R has a single small-to-large mutation (S98Y) at the binding interface, likely responsible for abrogating binding to 23R-91. Rat and macaque IL-23R have no mutations at the core interface (models not shown).
Table S2. Combinatorial library design summary, related to STAR methods.
Table S3. Yeast library FACS conditions, related to STAR methods.
Table S4. Crystallographic data collection and refinement statistics for 23R-91, related to Figure 5.
Table S5. Summary of Rat PK and biodistribution data, related to Figures 4C and 4D.
Table S6. NSG-IBD scoring matrices, related to Figure 7 and STAR methods.
Table S7. Results of immunogenicity prediction of 23R-91 using NetMHCII software, related to STAR methods.
Data Availability Statement
This study did not generate new code. Source data for main and supplemental figures, raw and processed data from deep mutational scanning experiments, and crystal structure data are publicly accessible. Source data for main and supplemental figures and original SDS PAGE images for Figure 4A have been deposited in the Mendeley Data repository with DOI 10.17632/2n9gstvrsy.1. Raw and processed data from deep mutational scanning experiments have been deposited in the NCBI GEO repository with accession ID GSE263250. Data deposition, atomic coordinates, and structure factors reported for the crystal structure of 23R-91 have been deposited in the Protein Data Bank (PDB), http://www.rcsb.org/ with accession ID 8UTK. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


