Abstract
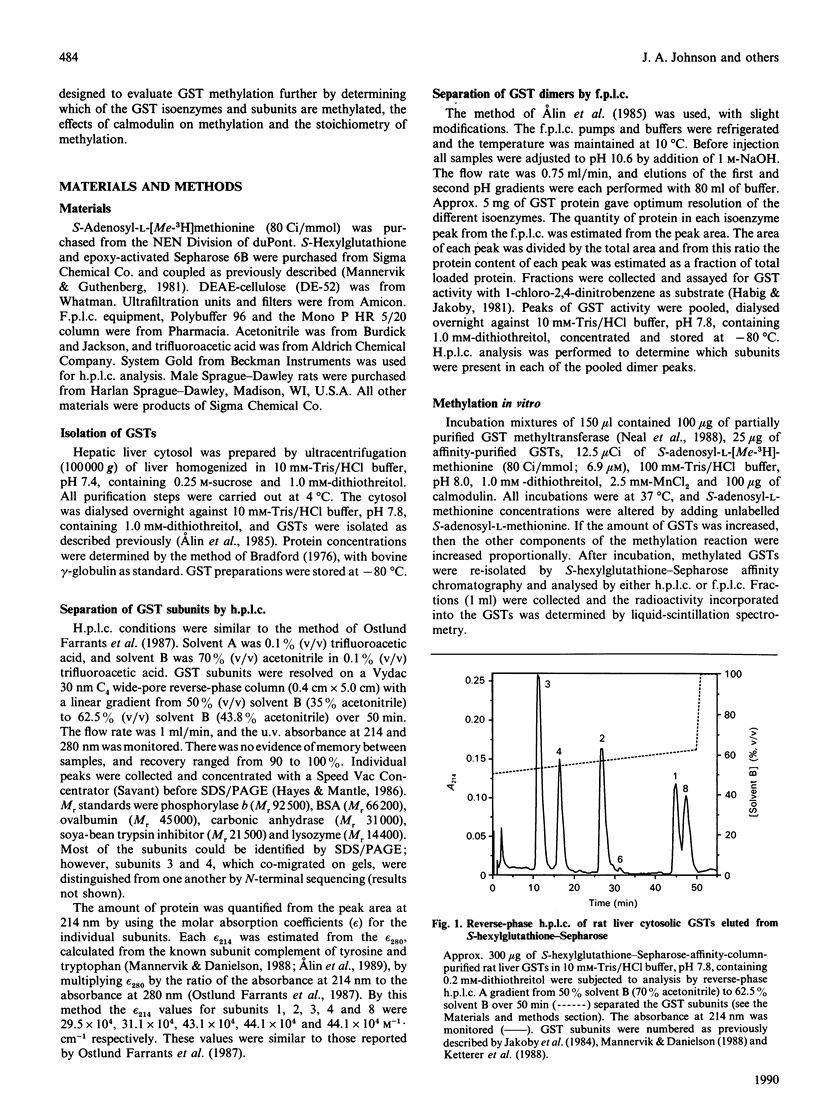
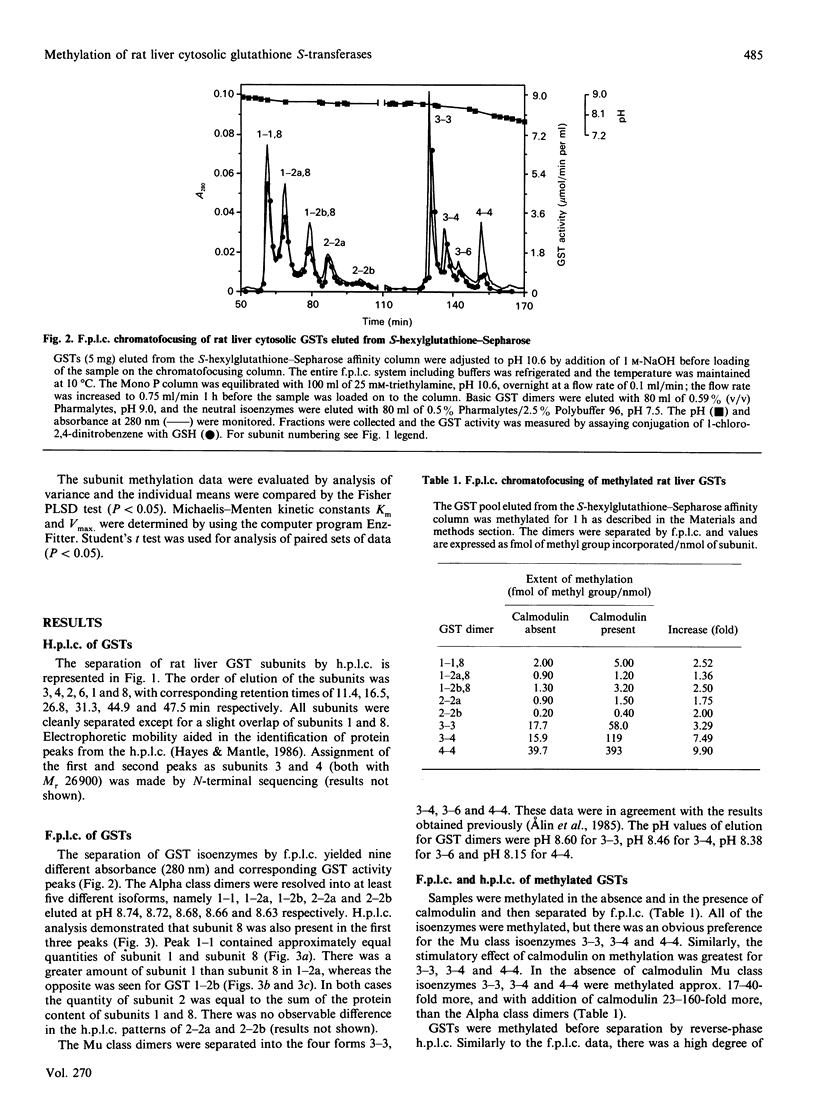
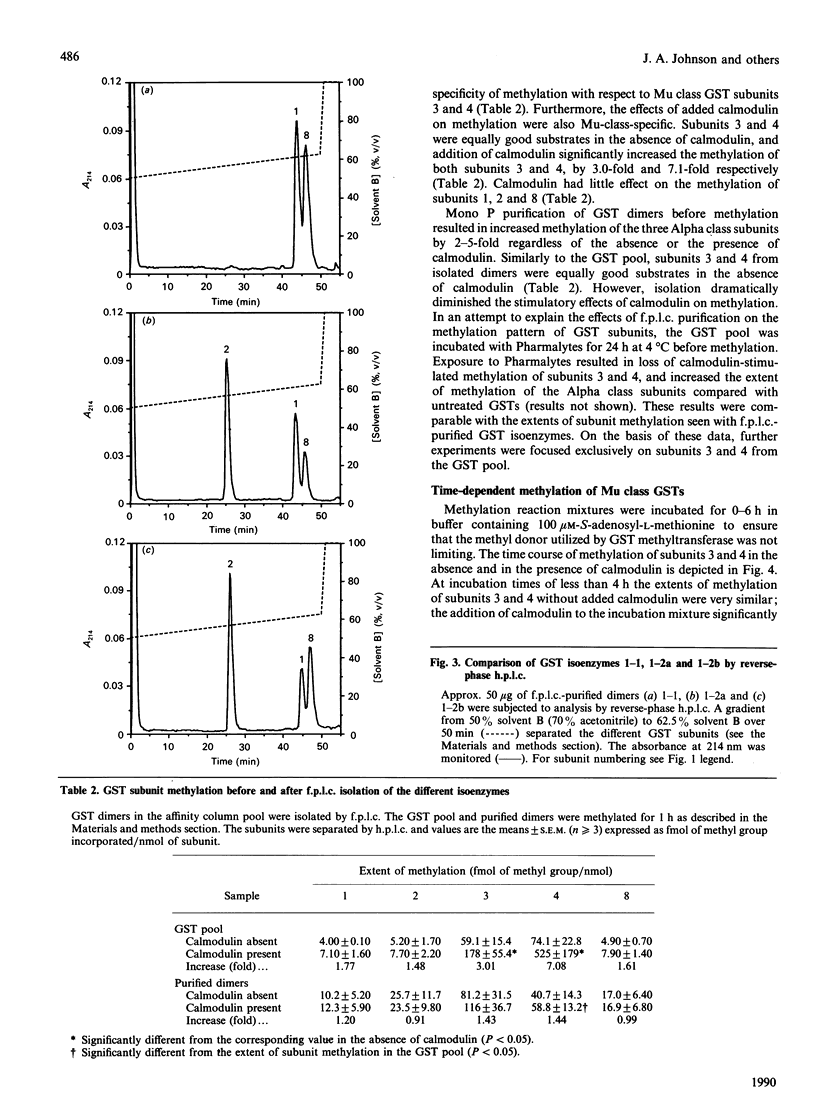
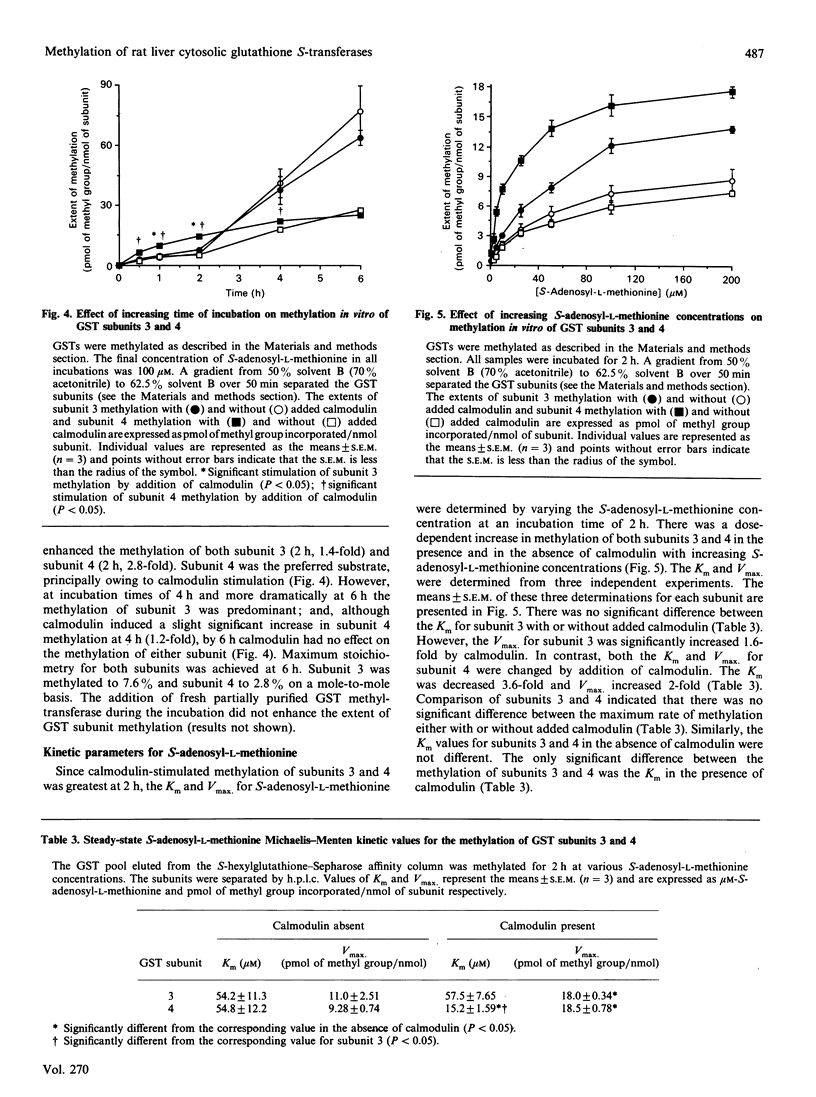
Glutathione S-transferase (GST) subunits in rat liver cytosol were separated by reverse-phase h.p.l.c.; five major proteins were isolated and identified as subunits 1, 2, 3, 4 and 8. F.p.l.c. chromatofocusing resolved the affinity-purified GST pool into nine different isoenzymes. The five basic (Alpha class) dimeric peaks of GST activity were 1-1, 1-2a, 1-2b, 2-2a and 2-2b. Reverse-phase h.p.l.c. analysis revealed that subunit 8 was also present in the protein peaks designated 1-1, 1-2a and 1-2b. The four neutral (Mu class) isoenzymes were 3-3, 3-4, 3-6 and 4-4. The GST pool was methylated in vitro before reverse-phase h.p.l.c. or f.p.l.c. chromatofocusing. Chromatofocusing indicated that the Mu class isoforms (3-3, 3-4 and 4-4) were the primary GSTs methylated, and h.p.l.c. analysis confirmed that subunits 3 and 4 were the major methyl-accepting GST subunits. The addition of calmodulin stimulated the methylation in vitro of GST isoenzymes 3-3, 3-4 and 4-4 by 3.0-, 7.5- and 9.9-fold respectively. Reverse-phase h.p.l.c. also indicated that only the methylation of GST subunits 3 and 4 was stimulated by calmodulin. Basic GST isoenzymes were minimally methylated and the methylation was not enhanced by calmodulin. Investigation of the time course of methylation of GST subunits 3 and 4 indicated that at incubation times less than 4 h the methylation of both Mu class subunits was stimulated by calmodulin, and that under such conditions subunit 4 was the preferred substrate. In contrast, there was essentially no calmodulin-stimulated methylation at incubation times of 4 or 6 h, and the methylation of subunit 3 was predominant. Kinetic parameters at 2 h of incubation were determined in the presence and in the absence of calmodulin. The addition of calmodulin doubled the Vmax. for methylation of both subunits 3 and 4 and decreased the Km of subunit 4 for S-adenosyl-L-methionine 3.6-fold. Finally, methylation was substoichiometric and after 6 h of incubation ranged from 2.8 to 7.6% on a mole-to-mole basis for subunits 4 and 3 respectively.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alin P., Jensson H., Cederlund E., Jörnvall H., Mannervik B. Cytosolic glutathione transferases from rat liver. Primary structure of class alpha glutathione transferase 8-8 and characterization of low-abundance class Mu glutathione transferases. Biochem J. 1989 Jul 15;261(2):531–539. doi: 10.1042/bj2610531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alin P., Jensson H., Guthenberg C., Danielson U. H., Tahir M. K., Mannervik B. Purification of major basic glutathione transferase isoenzymes from rat liver by use of affinity chromatography and fast protein liquid chromatofocusing. Anal Biochem. 1985 May 1;146(2):313–320. doi: 10.1016/0003-2697(85)90545-7. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chang M., Hong Y., Burgess J. R., Tu C. P., Reddy C. C. Isozyme specificity of rat liver glutathione S-transferases in the formation of PGF2 alpha and PGE2 from PGH2. Arch Biochem Biophys. 1987 Dec;259(2):548–557. doi: 10.1016/0003-9861(87)90521-2. [DOI] [PubMed] [Google Scholar]
- Chang M., Rao M. K., Reddanna P., Li C. H., Tu C. P., Corey E. J., Reddy C. C. Specificity of the glutathione S-transferases in the conversion of leukotriene A4 to leukotriene C4. Arch Biochem Biophys. 1987 Dec;259(2):536–547. doi: 10.1016/0003-9861(87)90520-0. [DOI] [PubMed] [Google Scholar]
- Chelsky D., Sobotka C., O'Neill C. L. Lamin B methylation and assembly into the nuclear envelope. J Biol Chem. 1989 May 5;264(13):7637–7643. [PubMed] [Google Scholar]
- Clarke S. Protein carboxyl methyltransferases: two distinct classes of enzymes. Annu Rev Biochem. 1985;54:479–506. doi: 10.1146/annurev.bi.54.070185.002403. [DOI] [PubMed] [Google Scholar]
- Clarke S., Vogel J. P., Deschenes R. J., Stock J. Posttranslational modification of the Ha-ras oncogene protein: evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschenes R. J., Broach J. R. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol Cell Biol. 1987 Jul;7(7):2344–2351. doi: 10.1128/mcb.7.7.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschenes R. J., Stimmel J. B., Clarke S., Stock J., Broach J. R. RAS2 protein of Saccharomyces cerevisiae is methyl-esterified at its carboxyl terminus. J Biol Chem. 1989 Jul 15;264(20):11865–11873. [PubMed] [Google Scholar]
- Gregori L., Marriott D., West C. M., Chau V. Specific recognition of calmodulin from Dictyostelium discoideum by the ATP, ubiquitin-dependent degradative pathway. J Biol Chem. 1985 May 10;260(9):5232–5235. [PubMed] [Google Scholar]
- Habig W. H., Jakoby W. B. Assays for differentiation of glutathione S-transferases. Methods Enzymol. 1981;77:398–405. doi: 10.1016/s0076-6879(81)77053-8. [DOI] [PubMed] [Google Scholar]
- Hayes J. D., Mantle T. J. Anomalous electrophoretic behaviour of the glutathione S-transferase Ya and Yk subunits isolated from man and rodents. A potential pitfall for nomenclature. Biochem J. 1986 Aug 1;237(3):731–740. doi: 10.1042/bj2370731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby W. B., Ketterer B., Mannervik B. Glutathione transferases: nomenclature. Biochem Pharmacol. 1984 Aug 15;33(16):2539–2540. doi: 10.1016/0006-2952(84)90621-x. [DOI] [PubMed] [Google Scholar]
- Kispert A., Meyer D. J., Lalor E., Coles B., Ketterer B. Purification and characterization of a labile rat glutathione transferase of the Mu class. Biochem J. 1989 Jun 15;260(3):789–793. doi: 10.1042/bj2600789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listowsky I., Abramovitz M., Homma H., Niitsu Y. Intracellular binding and transport of hormones and xenobiotics by glutathione-S-transferases. Drug Metab Rev. 1988;19(3-4):305–318. doi: 10.3109/03602538808994138. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Danielson U. H. Glutathione transferases--structure and catalytic activity. CRC Crit Rev Biochem. 1988;23(3):283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Guthenberg C. Glutathione transferase (human placenta). Methods Enzymol. 1981;77:231–235. doi: 10.1016/s0076-6879(81)77030-7. [DOI] [PubMed] [Google Scholar]
- Meister A., Anderson M. E. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- Meister A. Glutathione metabolism and its selective modification. J Biol Chem. 1988 Nov 25;263(33):17205–17208. [PubMed] [Google Scholar]
- Murtaugh T. J., Wright L. S., Siegel F. L. Posttranslational modification of calmodulin in rat brain and pituitary. J Neurochem. 1986 Jul;47(1):164–172. doi: 10.1111/j.1471-4159.1986.tb02845.x. [DOI] [PubMed] [Google Scholar]
- Neal T. L., Wright L. S., Siegel F. L. Identification of glutathione S-transferase as a substrate and glutathione as an inhibitor of in vitro calmodulin-stimulated protein methylation in rat liver cytosol. Biochem Biophys Res Commun. 1988 Oct 14;156(1):368–374. doi: 10.1016/s0006-291x(88)80850-7. [DOI] [PubMed] [Google Scholar]
- Ostlund Farrants A. K., Meyer D. J., Coles B., Southan C., Aitken A., Johnson P. J., Ketterer B. The separation of glutathione transferase subunits by using reverse-phase high-pressure liquid chromatography. Biochem J. 1987 Jul 15;245(2):423–428. doi: 10.1042/bj2450423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik W. K., Kim S. Solubilization and partial purification of protein methylase 3 from calf thymus nuclei. J Biol Chem. 1970 Nov 25;245(22):6010–6015. [PubMed] [Google Scholar]
- Park K. S., Frost B., Tuck M., Ho L. L., Kim S., Paik W. K. Enzymatic methylation of in vitro synthesized apocytochrome c enhances its transport into mitochondria. J Biol Chem. 1987 Oct 25;262(30):14702–14708. [PubMed] [Google Scholar]
- Pyerin W., Taniguchi H., Horn F., Oesch F., Amelizad Z., Friedberg T., Wolf C. R. Isoenzyme-specific phosphorylation of cytochromes P-450 and other drug metabolizing enzymes. Biochem Biophys Res Commun. 1987 Feb 13;142(3):885–892. doi: 10.1016/0006-291x(87)91496-3. [DOI] [PubMed] [Google Scholar]
- Roberts D. M., Rowe P. M., Siegel F. L., Lukas T. J., Watterson D. M. Trimethyllysine and protein function. Effect of methylation and mutagenesis of lysine 115 of calmodulin on NAD kinase activation. J Biol Chem. 1986 Feb 5;261(4):1491–1494. [PubMed] [Google Scholar]
- Siegel F. L., Wright L. S. Calmodulin-stimulated protein methylation in rat liver cytosol. Arch Biochem Biophys. 1985 Mar;237(2):347–353. doi: 10.1016/0003-9861(85)90286-3. [DOI] [PubMed] [Google Scholar]
- Vincent P. L., Siegel F. L. Carboxylmethylation of calmodulin in cultured pituitary cells. J Neurochem. 1987 Nov;49(5):1613–1622. doi: 10.1111/j.1471-4159.1987.tb01035.x. [DOI] [PubMed] [Google Scholar]


