Abstract
To replicate, a retrovirus must synthesize a cDNA copy of the viral RNA genome and integrate that cDNA into a chromosome of the host. We have investigated the role of a host cell cofactor, HMG I(Y) protein, in integration of human immunodeficiency virus type 1 (HIV-1) and Moloney murine leukemia virus (MoMLV) cDNA. Previously we reported that HMG I(Y) cofractionates with HIV-1 preintegration complexes (PICs) isolated from freshly infected cells. PICs depleted of required components by treatment with high concentrations of salt could be reconstituted by addition of purified HMG I(Y) in vitro. Here we report studies using immunoprecipitation that indicate that HMG I(Y) is associated with MoMLV preintegration complexes. In mechanistic studies, we show for both HIV-1 and MoMLV that each HMG I(Y) monomer must contain multiple DNA binding domains to stimulate integration by HMG I(Y)-depleted PICs. We also find that HMG I(Y) can condense model HIV-1 or MoMLV cDNA in vitro as measured by stimulation of intermolecular ligation. This reaction, like reconstitution of integration, depends on the presence of multiple DNA binding domains in each HMG I(Y) monomer. These data suggest that binding of multivalent HMG I(Y) monomers to multiple cDNA sites compacts retroviral cDNA, thereby promoting formation of active integrase-cDNA complexes.
Several well-studied recombination enzymes have been found to require accessory proteins for efficient function. Many of these cofactors are small DNA-binding proteins which help the recombinase-DNA complexes adopt their active conformation. For example, in the case of phage lambda integration, the host protein integration host factor (IHF) binds and bends phage DNA, thereby facilitating formation of active lambda integrase-DNA complexes (21). Architectural DNA-binding proteins can also be important in formation of transcription complexes, as has been shown for HMG I(Y) in vertebrate cells (15, 28, 44, 45). Here we examine the role of HMG I(Y) in another setting, as an architectural cofactor for retroviral cDNA integration.
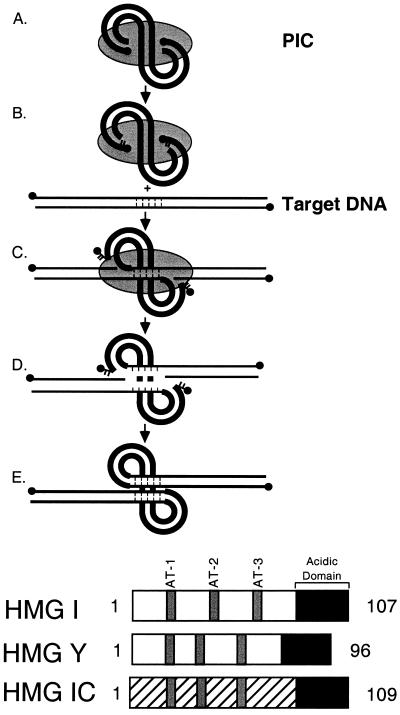
The DNA breaking and joining reactions mediating retroviral integration are well understood (Fig. 1, top panel) (for reviews, see references 12 and 23;1x). Initially, the virus-encoded integrase enzyme binds the ends of the viral cDNA and cleaves to remove two nucleotides from each 3′ end (Fig. 1, top panel, parts A and B) (5, 13, 33, 42). The cleavage reaction may help prepare a homogeneous substrate for subsequent reaction steps. The recessed 3′ ends are then joined by integrase to the target DNA (Fig. 1, top panel, part C) (6, 13, 32). The resulting integration intermediate is then resolved, probably by host DNA repair enzymes, to yield the integrated provirus (Fig. 1, top panel, parts D and E).
FIG. 1.
(Top) Diagram of the integration pathway. The HIV cDNA is shown by the thick lines, and proteins of the PIC are shown by the gray shading. Target DNA is shown by the thin lines. DNA 5′ ends are marked with solid circles. Note that product D corresponds to the integration intermediate marked “II” in later figures. (Bottom) HMG I family proteins. The conserved A/T hook DNA binding domains are shown in gray, and the acidic C-terminal domains are shown in black.
Integration-competent replication intermediates can also be isolated from virus-infected cells and studied in vitro. Such preintegration complexes (PICs) can direct the joining of the viral cDNA to an exogenously added target (2, 14, 18). The cDNA ends in PICs are protected by bound proteins from attack by nucleases or recombination complexes (11, 39, 46), and the two cDNA ends are apposed by bound proteins (39). In the human immunodeficiency virus type 1 (HIV-1) system, the virus-encoded integrase, matrix, and reverse transcriptase proteins cofractionated with PICs, and some studies also indicate association of nucleocapsid and Vpr (4, 17, 20, 39). The host protein HMG I(Y) has also been found to cofractionate (discussed below) (16).
In previous studies, it was found that PICs of HIV-1 could be depleted of components by gel filtration in buffers containing high concentrations of salt, resulting in a loss of integration activity in vitro. Activity could be reconstituted by addition of an extract from uninfected cells. Fractionation of such extracts identified HMG I(Y) as the most prominent reconstitution activity (16). HMG I(Y) was subsequently found to cofractionate with PICs and to be removed by the salt-stripping procedure. Importantly, the reconstituting activity could be depleted with an anti-HMG I(Y) antibody, but not other antibodies (16). HMG I(Y) was also found to boost activity of salt-stripped PICs from Moloney murine leukemia virus (MoMLV) (37) and stimulate the activity of purified integrases under some (26), although not all (9), conditions in vitro.
The HMG I(Y) family consists of three members (Fig. 1, bottom panel). HMG I and HMG Y are expressed from the same gene, differing by alternative mRNA splicing. The two are often referred to together as HMG I(Y). HMG I-C is closely related, but transcribed from a different gene (7). Each protein contains three A/T hook DNA binding domains, each of which can bind to A/T-rich DNA sequences.
Several other candidates for integrase cofactors have been proposed based on studies in vitro. PICs of MoMLV, when treated with high salt concentrations, do not carry out normal intermolecular integration, but instead use their own cDNA as an integration target (autointegration). BAF protein was isolated based on its ability to block autointegration by salt-stripped PICs of MoMLV (35). BAF also displayed a high specific activity for reconstitution of salt-stripped HIV-1 PICs (11), though BAF has not been shown to be associated with PICs or to be important in vivo. The virus-encoded nucleocapsid (NC) protein has been found to promote activity of integrase (8, 9, 34). Under some conditions in vitro, NC can greatly stimulate “coupled joining,” integration of pairs of cDNA ends into target DNA with the correct biological spacing between the points of joining (9). Ini-1 protein (31) and HMG-1 protein [not to be confused with HMG I(Y), an unrelated protein] (1) have also been proposed to serve as integration cofactors based on studies in vitro. Clearly further studies are necessary to distinguish factors that are important in vivo from proteins displaying nonbiological in vitro activities.
Here we present data to (i) strengthen the idea that HMG I(Y) family of proteins are important for cDNA integration in vivo and (ii) elucidate aspects of the mechanism with studies in vitro. Many of the experiments were carried out in both the MoMLV and HIV-1 systems to investigate the generality of our conclusions. Here we used immunoprecipitation to demonstrate that HMG I(Y) is bound to MoMLV PICs isolated from virus-infected cells, complementing data on cofractionation from the HIV-1 system (16). In mechanistic studies, we found that HMG I(Y) derivatives can support reconstitution of depleted PICs as long as each HMG I(Y) monomer contains at least two of the three DNA binding domains. Recently others have proposed that multiple DNA binding domains are important for stimulating reactions with purified integrase (26). We also demonstrated that the HIV-1 long terminal repeats (LTRs) contain multiple DNA binding sites for HMG I(Y). Tests in vitro revealed that binding of HMG I(Y) apposed LTR DNAs in a manner that promoted intermolecular ligation of dilute DNA solutions. As with assays of PIC reconstitution, these assays of DNA condensation required multiple DNA binding domains in each HMG I(Y) monomer. Taken together, these studies strengthen the idea that HMG I(Y) acts as an architectural cofactor for retroviral cDNA integration complexes and suggest a model for its role.
MATERIALS AND METHODS
Depletion and reconstitution of MoMLV and HIV-1 PICs.
For MoMLV PICs, the salt-stripping and reconstitution procedure was performed as described previously (37). HIV-1 PICs for this study were generated with HIV-based vectors as described previously (24). HIV-1 PICs were depleted by gel filtration under high-salt conditions. The PICs were first incubated in buffer A (20 mM HEPES [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol [DTT], 10 μg of aprotinin per ml) containing 300 mM KCl for 10 min at room temperature and then centrifuged through a Sepharose CL-4B column in buffer A–300 mM KCl. PICs were then incubated in buffer A–600 mM KCl for 10 min and passed through a Sepharose CL-4B column in buffer A–600 mM KCl. The 600 mM KCl-stripped PICs were then concentrated with a Microcon-100 ultrafiltration unit (Millipore). Twenty-five microliters (∼3 × 107 cDNA copies) was mixed with HMG I(Y), the mixture was incubated at room temperature for 10 min, and then the salt concentration was lowered to 150 mM by the addition of 75 μl of buffer A–40% glycerol (lacking KCl). The reaction mixture was then incubated at room temperature for 10 min. Integration reactions were carried out by addition of 1 μg of target DNA (linearized φX174 RFII), and the mixtures were incubated at 37°C for 1 h. DNA was purified by treatment with proteinase K–sodium dodecyl sulfate (SDS)–EDTA, phenol extraction, and ethanol precipitation. DNAs were analyzed by Southern blots and probed with 32P-labeled LTR sequences.
Purification of recombinant MoMLV integrase.
To prepare MoMLV integrase for affinity purification of antibodies, plasmid pETINH (30) (expressing MoMLV integrase fused to a hexahistidine tag) was transformed into BL21(DE3), and cultures were induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the optical density at 595 nm (OD595) of bacteria reached 0.6. After 2.5 h of induction at 30°C, the bacteria were pelleted, resuspended in 1× binding buffer (Novagen) with 10 mM β-mercaptoethanol and l0 μg of lysozyme per ml, and incubated at 4°C for 30 min. The lysate was then sonicated and pelleted at 15,000 × g for 30 min. The pellet was resuspended in 1× binding buffer with 6 M guanidine-HCl and 10 mM β-mercaptoethanol. The resuspended pellet was pelleted again at 15,000 × g for 30 min. The supernatant, which contained integrase, was harvested and loaded on a nickel column preequilibrated in 1× binding buffer with 6 M guanidine-HCl. The column was washed with 10 bed volumes of 1× binding buffer with 6 M guanidine-HCl followed by 10 bed volumes of 1× binding buffer containing 25 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-l-propanesulfonate}, but lacking 6 M guanidine-HCl. Integrase was eluted by gradual increase of the imidazole concentration in 1× binding buffer with 25 mM CHAPS. Fractions were collected, analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and found to yield a single band of the expected mobility. To raise antibodies against MoMLV integrase, proteins were further purified by SDS-PAGE and electroelution.
Purification of antibodies on antigen affinity columns.
Purified HMG I(Y) and MoMLV integrase (IN) proteins were coupled to immobilized diaminodipropylamine (DADPA) resins according to the manufacturer's instructions (Pierce). Sera were loaded on HMG I(Y) and IN affinity columns, the columns were washed, and the antibodies were eluted by Tris-glycine buffer (pH 2.5). The eluate was immediately neutralized by addition of 1/20 volume of 1 M Tris (pH 8.0) and dialysis against phosphate-buffered saline.
Immunoprecipitation of partially purified PICs.
MoMLV PICs were harvested from infected cells as described previously (2, 37). Partially purified PICs were obtained by gel filtration through a Sepharose CL-4B column. Fifty microliters of partially purified PICs or salt-stripped PICs (about 3 × 107 cDNA copies) was mixed with 5 μl of antigen affinity-purified antibodies or control preimmune serum and incubated on ice for 1 h. Thirty microliters of protein A/G beads (Santa Cruz Biotechnology) was added, and the reaction mixtures were rocked at 4°C for 1 h. The beads were washed three times with buffer A–150 mM KCl and suspended in 100 μl of buffer A–150 mM KCl with 30% glycerol. One microgram of linear φX174 DNA was added to the beads, and integration reactions were carried out at 37°C for 1 h. DNA products were purified from beads by SDS-EDTA-proteinase K treatment followed by phenol-chloroform extraction and ethanol precipitation. The recovered DNA was separated by electrophoresis on 0.75% agarose gel and analyzed by Southern blotting with a 32P-labeled MoMLV LTR probe. For a control reaction, the affinity-purified anti-HMG I(Y) antibody (15 μg) was incubated with 10 μg of HMG I(Y) for 1 h prior to addition of PICs.
Purification of HMG I family proteins.
HMG I family proteins [HMG I(Y), HMG I(Y) 1–90, and HMG I-C] were expressed in BL21(DE3) cells and induced by IPTG. The cell pellet was resuspended in 10 mM Tris (pH 8.0). The cells were lysed by the addition of lysozyme (l0 μg/ml) and sonication. Two volumes of 7.5% perchloric acid was then added, and the suspension was Dounce homogenized 20 times at 4°C. The suspension was spun at 1,000 × g at 4°C for 30 min. Concentrated HCl (0.3 volume) and acetone (6 volumes) were added to the supernatant and incubated at 4°C for 2 h. The mixture was spun at 3,000 rpm at 4°C for 30 min. The pellet was washed with cold acetone and dried. The pellet was resuspended in buffer B (20 mM Tris [pH 8.0], 0.1 mM EDTA, 1 mM DTT) and loaded onto a fast protein liquid chromatography Mono-S column and eluted by a gradient with buffer B containing 0 to 1 M NaCl. The fractions containing HMG I family proteins were combined and dialyzed against buffer B with 100 mM NaCl and 20% glycerol. SDS-PAGE analysis revealed a single band of the expected mobility. It has recently been reported that purification of HMG I family proteins by acid extraction, while convenient, yields HMG I preparations with reduced activity compared to other methods (41). This may account for the large amounts of protein required for in vitro reactions reported here.
Peptides matching A/T hooks were synthesized on an Applied Biosystems Synergy 432A peptide synthesizer and confirmed by mass spectrometry. The peptide sequences were as follows: AT-1 peptide, DGTEKRGRPRKQPPVSPG; AT-2 peptide, VPTPKRPRGRPKGSKNKGAA; and AT-3 peptide, TTPGRKPRGRPKKLEK.
DNase I footprinting of HMG I bound to the HIV-1 LTR.
The LTR DNA used for footprinting was derived from the plasmid pTA-LTR (9), which contains an HIV-1 LTR flanked by artificial NdeI sites. pTA-LTR was cleaved with AvaI, which cuts within the LTR DNA, or NdeI. DNA strands were labeled on the 5′ end by sequential treatment with phosphatase, then kinase and [γ-32P]ATP, or on the 3′ end by filling in with 32P-labeled deoxynucleoside triphosphates (dNTPs) and Klenow fragment. LTR DNA was then further cleaved with NdeI or AvaI to liberate labeled fragments. Labeled DNA fragments were gel isolated and recovered by binding to glass powder. Footprinting reaction mixtures contained 10 mM Tris (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, 100 μg of bovine serum albumin (BSA) per ml, and 0.1 U of DNase I per ml in a final volume of 25 μl. Reaction mixtures were incubated for 2 min at room temperature, and then the reactions were stopped by addition of 75 μl of 92% ethanol, 0.7 M NH4OAc, and 70 μg of tRNA per ml. Reaction mixtures were precipitated, rinsed with 70% ethanol, and resuspended in 5 μl of Tris-EDTA. Reaction products were denatured by heating in 10 μl of sequencing gel loading dye and then separated on 6% denaturing polyacrylamide gels and visualized by autoradiography.
DNA gel retardation analysis of HMG I family proteins.
Oligonucleotide probes were labeled at the 5′ end with 32P. Binding of HMG I, HMG Y, and HMG I-C was assayed in a mixture of 10 mM Tris (pH 7.5), 50 mM KCl, 0.5 mM MgCl2, 0.1 mM EDTA, 5% glycerol, 0.25 μg of BSA per ml, and 2 nM annealed oligonucleotide in a final volume of 30 μl. Proteins were added to a final concentration of 200, 400, or 800 pM and incubated at room temperature for 30 min. Five microliters of the reaction mixture was loaded onto a 6% native gel in 0.4× Tris-borate-EDTA and run at 300 V at 4°C. Gels were dried and analyzed by autoradiography.
Assay of condensation of LTR DNA by HMG I(Y) and mutant derivatives.
The reaction mixture contained 10 ng of 5′ 32P-labeled LTR (bearing Nde-complementary ends), 4 μl of 10× ligation buffer (New England Biolabs), and HMG I(Y) protein or mutant derivatives in a final volume of 40 μl. The reaction was initiated by the addition of 1 μl of T4 DNA ligase (400 U/μl; New England Biolabs) and incubated at 16°C. Five microliters of the reaction mixture was withdrawn at the indicated time points, and the reaction was terminated by addition of 35 μl of stop solution (0.5% SDS, 10 mM EDTA, 1 mg of proteinase K per ml). After overnight incubation at 50°C, the DNA was purified by phenol-chloroform extraction and separated on a 1% agarose gel. The gels were then dried and visualized by autoradiography and PhosphorImager analysis.
RESULTS
Immunoprecipitation of active MoMLV PICs with antibodies against HMG I(Y) or MoMLV integrase.
To complement studies in the HIV-1 system showing that HMG I(Y) cofractionated with PICs (16), we have investigated by immunoprecipitation, whether HMG I(Y) is bound to MoMLV PICs. PICs were prepared by infection of NIH 3T3 cells with MoMLV virus, followed by detergent permeabilization as described previously (2, 37). PICs were partially purified by gel filtration, and then affinity-purified antibodies were added and complexes were captured by incubation with protein A/G beads. Beads were washed to remove the unbound material and resuspended in integration reaction buffer. Target DNA was then added, and the integration reaction was carried out with PICs bound to the beads. Product DNAs were released from the beads by treatment with proteinase K and SDS, purified, and separated by electrophoresis in agarose gels. DNAs were transferred to a nylon support and probed with labeled MoMLV LTR DNA sequences.
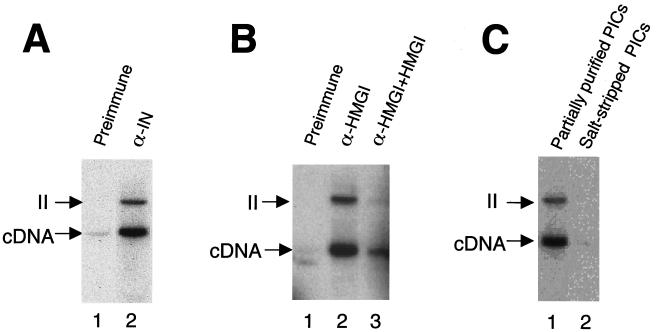
Immunoprecipitation with preimmune serum (Fig. 2A, lane 1), yielded little cDNA. Immunoprecipitation with an anti-MoMLV integrase antibody, in contrast, yielded abundant cDNA products (Fig. 2A, lane 2). Some of these retained integration activity (Fig. 2A, lane 2 [“II” indicates the integration intermediate produced by covalent joining to target DNA; this species corresponds to D in Fig. 1, top panel]). Immunoprecipitation with an anti-HMG I(Y) antibody was then compared (Fig. 2B). Mock immunoprecipitation resulted in no recovery of cDNA (Fig. 2B, lane 1), while addition of the purified anti-HMG I(Y) antibody yielded abundant cDNA (Fig. 2B, lane 2). Addition of HMG I protein blocked recovery (Fig. 2B, lane 3). Immunoprecipitation of salt-stripped PICs was then tested, since previous work indicated that bound HMG I(Y) should be removed by this treatment (16, 37). The anti-HMG I(Y) antibody was unable to immunoprecipitate salt-stripped PICs as expected (compare Fig. 2C, lanes 1 and 2).
FIG. 2.
Immunoprecipitation (IP) of active PICs with antigen affinity-purified antibodies against HMG I(Y). Partially purified or salt-stripped PICs were immunoprecipitated with the indicated antibodies and protein A/G beads. Integration reactions were carried out with PICs bound to beads. The cDNA arrows mark the unreacted cDNA. II, integration intermediate. (A) IP with antibodies against MoMLV integrase. Lane 1, IP with preimmune serum; lane 2, IP with affinity-purified anti-integrase antibody (α- IN). (B) IP with HMG I(Y) antibody. Lane 1, IP with preimmune serum; lane 2, IP with anti-HMG I(Y) antibody; lane 3, IP with anti-HMG I(Y) antibody and free HMG I(Y) added as a competitor. (C) Salt stripping blocks immunoprecipitation with the anti-HMG I(Y) antibody. Lane 1, IP of partially purified PICs with anti-HMG I(Y) antibody; lane 2, IP of salt-stripped PICs with anti-HMG I(Y) antibody. Note that the production of the circular autointegration product seen in subsequent figures is a consequence of salt stripping and so is absent here. Characterization of the structures of integration products can be found in references 3, 19, 35, 37, and 40.
In summary, antibodies against HMG I(Y) or MoMLV integrase were able to immunoprecipitate PICs of MoMLV. Immunoprecipitation with the HMG I(Y) antibody could be blocked by addition of HMG I(Y) protein or prior salt stripping of PICs. This complements data from the HIV-1 system, in which HMG I(Y) was found to copurify with PICs and an anti-HMG I(Y) antibody was able to deplete reconstitution activity released from PICs by high-salt treatment (16). HMG I(Y) also copurifies with MoMLV PICs (unpublished data).
Reconstitution of salt-stripped PICs with deletion derivatives of HMG I.
To investigate the mechanism of reconstitution by HMG I, we tested deletion derivatives of HMG I protein for the ability to restore activity to salt-stripped PICs. We and others have found that HMG I(Y) can stimulate purified integrase from HIV-1 (26), but comparison with reactions with salt-stripped PICs reveals that the degree of stimulation is greater with the in vivo-assembled complexes (unpublished data). Thus, we assayed stimulation by using PICs instead of purified integrase.
PICs of MoMLV and HIV-1 were first partially purified and then subjected to gel filtration in high-salt buffers as described in Materials and Methods. HMG I(Y) and derivatives (Fig. 3A) were added to complexes in the presence of high salt concentrations, preincubated for 10 min at room temperature, and then the salt concentration was reduced to 150 mM by dilution. Target DNA was added and incubated with the reconstituted PICs. Product DNAs were then deproteinized and analyzed on Southern blots probed with labeled LTR DNA.
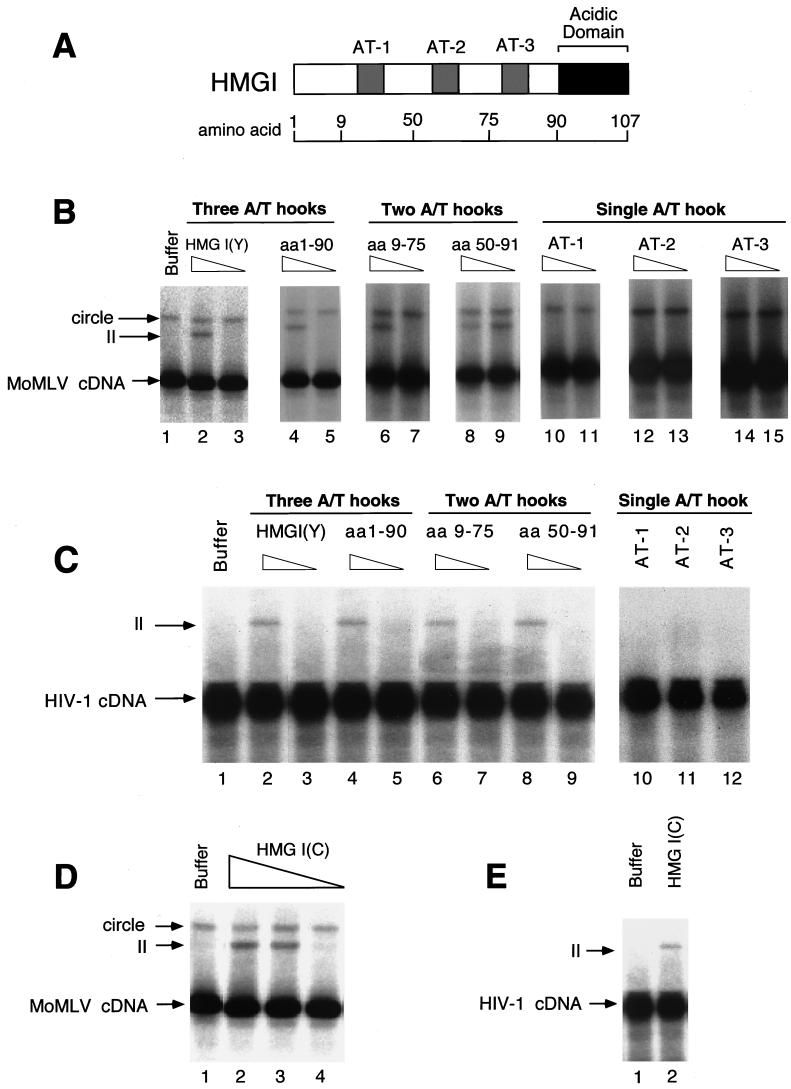
FIG. 3.
Reconstitution of integration activity with HMG I(Y) deletion derivatives and HMG I-C. (A) Diagram of HMG I illustrating the locations of breakpoints studied. (B) Reconstitution of salt-stripped MoMLV PICs with deletion derivatives of HMG I(Y) proteins. Integration was carried out in vitro and analyzed on Southern blots probed with a labeled MoMLV LTR. HMG I(Y) derivatives used for reconstitution are indicated above the autoradiograms. II, integration intermediate produced by integration in vitro; circle, autointegration product; cDNA, unreacted viral DNA; aa, amino acids. Triangles above the lane indicate 1 or 0.2 μg was added. Lanes: 1, no added protein; 2 to 3, reconstitution with full-length HMG I; 4 and 5, reconstitution with an HMG I(Y) derivative lacking the acidic C-terminal domain; 6 to 9, reconstitution with HMG I(Y) derivatives containing two A/T hooks; 10 to l5, reconstitution with single A/T hooks. Molarities for 1 μg in 100-μl reaction mixtures were as follows: full-length HMG I(Y), 0.85 μM; 1–90, 1 μM; 9–75, 1.25 μM; 50–91, 2 μM; AT-1, 4.7 μM; AT-2, 4.7μM; and AT-3, 5 μM. (C) Reconstitution of salt-stripped HIV-1 PICs with deletion derivatives of HMG I(Y). Assay and labeling are as in panel B, except that the triangles indicate 5 or 1 μg was added. The assays of single A/T hooks contained 5 μg. (D and E) Reconstitution of MoMLV PICs (D) and HIV-1 PICs (E) with HMG I-C. The amounts added were 5, 1, or 0.2 μg for MoMLV or 5 μg for HIV-1.
Lanes containing salt-stripped MoMLV PICs revealed two DNA forms: the unreacted cDNA and a slow-migrating species corresponding to the circular DNA product of autointegration (Fig. 3B, lane 1). As was reported previously, depletion by salt stripping promotes autointegration in the MoMLV system (35–37). Reconstitution of PICs with HMG I yielded integration products (Fig. 3B, lane 2 [II, integration intermediate]). HMG Y also supported reconstitution (data not shown). A derivative of HMG I lacking amino acids 91 to 108, the acidic C-terminal domain, was also capable of reconstituting integration activity (Fig. 3B, lane 4). Derivatives containing amino acids 9 to 75 or 50 to 91, either A/T hooks 1 plus 2 or 2 plus 3, also reconstituted activity (Fig. 3B, lanes 6, 8, and 9). The 50–91 protein displayed the highest specific activity for reconstitution, retaining activity at 0.2 μg per reaction. Derivatives containing single A/T hooks were unable to restore activity (Fig. 3B, lanes 10 to 15). Single A/T hook-containing peptides have been reported to have a Kd for AT sites of around 150 nM (27), but no reconstitution was seen here at peptide concentrations as high as 500 μM peptide (data not shown). Thus, the peptide probably bound the cDNA, but failed to support integration.
A similar study of HIV-1 PICs revealed that derivatives with two A/T hooks were sufficient for reconstitution in this system as well (Fig. 3C, lanes 6 and 8). Single A/T hooks were not sufficient for reconstitution of HIV-1 PICs, as with MoMLV PICs (Fig. 3C, lanes 10 to 12). These observations suggest a role for multivalent binding of HMG I(Y) monomers to pairs of DNA sites.
To explore the requirements for reconstitution of salt-stripped PICs further, HMG I-C, the third family member, was also tested. PICs of MoMLV or HIV-1 were salt stripped and then incubated with HMG I-C (Fig. 3D and E). Addition of HMG I-C restored activity to MoMLV PICs and partial activity to HIV-1 PICs. Thus, HMG I-C is also a candidate integration cofactor. HMG I-C is expressed embryonically and so is not likely to be important for HIV infection of adults, but the protein is expressed in many cell lines. In summary, all three members of the HMG I family are competent for stimulating integration of salt-depleted PICs.
HMG I binding to the HIV-1 LTR.
Next the HMG I(Y) binding sites in the cDNA were mapped. Studies with retroviral vectors reveal that most of the cDNA sequences are dispensable for integration, so our analysis focused on the required LTR sequences. HMG I(Y) has been reported previously to bind to any stretch of five or more A/T or T/A base pairs (38, 43). The LTR sequences of HIV-1 (NL4-3 strain) and MoMLV contain nine and seven candidate sites, respectively, which thus constitute expected binding sites.
To investigate whether the sites in the HIV-1 LTR indeed bind HMG I, DNase I protection experiments were carried out. DNA fragments from the HIV-1 LTR were end labeled, incubated with HMG I, and treated with DNase I. DNA fragments were separated on a DNA sequencing-type gel and visualized by autoradiography.
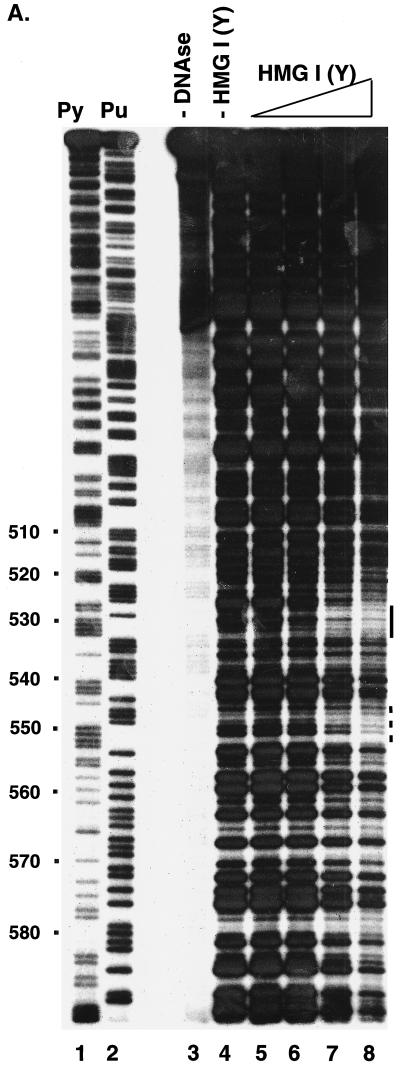
A representative experiment is shown in Fig. 4A, assaying HIV-1 LTR DNA from residue 295 to residue 634 (coordinates as in the NL4-3 genome, left LTR). The addition of DNase I yielded a pattern of partial digestion (Fig. 4A, lane 4). Addition of HMG I displayed several sites of protection, one at residues 527 to 532, and a weaker site around 550 (Fig. 4A, lanes 5 to 8, bar beside the gel). The degree of protection from DNase I digestion by binding of HMG I was somewhat modest, occurring near the concentration at which the DNA became fully coated with protein. This could be due to weakly selective binding by HMG I or easy displacement of bound HMG I by the DNase I enzyme under the conditions tested. Seven sites displayed the clearest reductions in DNase I digestion, located at residues 11 to 15, 22 to 27, 113 to 116, 250 to 254, 426 to 431, 440 to 444, and 527 to 532 (Fig. 4A) (data not shown). All of these contain five or more sequential A or T residues, except site 113, which contains four sequential A/T base pairs. For some of the sites predicted by the primary sequence, DNase I cleavage of naked DNA was so weak in the region of the site that possible protection could not be assessed. In summary, the sequences bound by HMG I were mostly as predicted by the primary DNA sequence.
FIG. 4.
Analysis of binding of HMG I(Y) to the HIV-1 LTR. (A) Analysis by DNase I protection. A restriction fragment containing sequences 295 to 634 from the HIV-1 LTR was labeled with 32P on the 5′ end at an artificial NdeI site at the U5 edge of the LTR. Lanes: 1, cleavage at pyrimidine (Py) residues; 2, cleavage at purine (Pu) residues; 3, uncleaved DNA; 4 to 9, 0.1 U of DNase I per ml; 4, no added HMG I(Y); 5, 0.8 nM HMG I(Y); 6, 2 nM HMG I(Y); 7, 7 nM HMG I(Y); 8, 22 nM HMG I(Y). (B) Oligonucleotide probes used in the gel retardation assay. The wild-type probe corresponds to residues 404 to 467 of the HIV LTR (NL4-3, left LTR). MUT, mutant; sub, substitution. (C) Gel retardation analysis of binding to the wild-type probe by HMG I or HMG Y. The mobilities of the free probe and observed complexes are indicated beside the gel. Lanes 1 to 5 contained the following concentrations of HMG I: 1, 0.13 nM; 2, 0.8 nM; 3, 4 nM; 4, 20 nM; 5, 100 nM. Lanes 6 to 9 contained the following concentrations of HMG Y: 6, 0.8 nM; 7, 4 nM; 8, 20 nM; 9, 100 nM. (D) Rescue of binding to mutant sites by inosine substitution. The DNA substrates studied are marked above the gel. W.T., wild type. Lanes 1 to 7, no added protein; lanes 8 to 14, 1 nM HMG I.
To confirm and extend the findings with DNase I protection, one pair of HMG I(Y) binding sites was studied in more detail by gel retardation assays. The two sites at residues 404 to 467 (NL4-3 strain, left LTR) were chosen for study, since they yielded strong DNase I footprints. A duplex oligonucleotide matching the pair of sites was synthesized (Fig. 4B) and incubated with either HMG I or HMG Y. Protein-DNA complexes were separated by gel electrophoresis and analyzed by autoradiography. Addition of 4 nM HMG I or Y yielded single discrete complexes (Fig. 4C, lanes 3 and 7). Addition of higher quantities yielded multiple slower-migrating species, probably arising either from binding of multiple HMG I monomers to a single probe DNA or binding of single HMG I monomers to multiple probe DNAs.
The contacts between HMG I and the 404 to 467 sites were next investigated. A-to-G substitution mutations in single HMG I binding sites had little effect on binding (Fig. 4D, compare lane 8 with lanes 9 and 11). Given that each of the complex bands showed the same electrophoretic mobility, this suggests that a single HMG I monomer is binding to the probe DNA. Mutation of both sites, however, abolished binding at the concentration tested (1 nM) (Fig. 4D, lane 13).
Substitution of inosine was then used to probe the mechanism of specific binding. Inosine can pair with C or T. Inosine resembles A in the minor groove and G in the major groove of DNA. Thus, I substitutions can be used to probe the side of the helix contacted by a protein. Substitution of an inosine residue for a guanine residue is expected to promote binding if the protein tested favors binding to A in the minor groove (45).
Substitution of I for G in the doubly mutant oligonucleotide restored binding, indicating that HMG I is binding to the DNA minor groove at the LTR 404 to 467 sites (Fig. 4D, compare lanes 13 and 14). Substitutions in single sites improved binding only slightly, since the single site mutations showed only modest decreases in binding to begin with (Fig. 4D, lanes 10 and 12).
Gel retardation assays were also carried out to test binding of HMG I-C to the 404 to 467 sites. Complex bands were seen, generally paralleling results with HMG I(Y) (data not shown).
In summary, the sites predicted to bind HMG I(Y) in the HIV LTR were mostly found to bind as expected. We did not analyze the MoMLV LTR, but based on the HIV data, we expect most of the predicted MoMLV sites will support binding as well.
LTR DNA condensation by HMG I.
We next investigated whether HMG I could promote the association of distant DNA sites, since this is an activity potentially involved in reconstitution of salt-depleted PICs. Linear LTR DNA molecules in dilute solution were incubated with HMG I or deletion derivatives. We reasoned that the proximity of the LTR DNA ends could be assayed by addition of T4 DNA ligase. Thus, association of LTR DNAs mediated by HMG I would be revealed by increased formation of intermolecular ligation product. For this experiment, DNAs were used which contained a single full-length LTR sequence flanked by artificial NdeI restriction sites (9). LTR DNAs were end-labeled with 32P, incubated with HMG I proteins, and then treated with ligase. After ligation, products were separated by gel electrophoresis and quantitated by PhosphorImager. The kinetics of ligation were monitored over 1 h. LTR DNAs of HIV-1 and MoMLV were studied (Fig. 5).
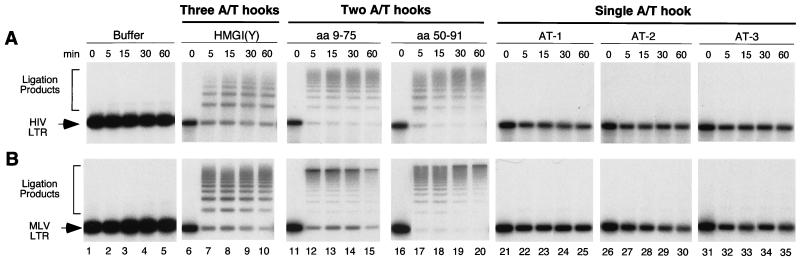
FIG. 5.
DNA ligation promoted by HMG I(Y) and deletion derivatives. The HMG I proteins used in ligation reactions are as indicated above the gels (designations as in Fig. 3). LTR, the end-labeled DNA substrate matching a single LTR DNA. (A) HIV-1 LTR. (B) MoMLV LTR. The time of ligation is indicated in minutes above the gel. Lanes: 1 to 5, no HMG I; 6 to 10, 1 μM HMG I(Y); 11 to 15, 1.5 μM HMG I(Y) 9–75; 16 to 20, 1.5 μM HMG I(Y) 50–91; 21 to 25, 3 μM AT-1; 26 to 30, 3 μM AT-2; 31 to 35, 3 μM AT-3. Concentrations were adjusted to maintain the same molarity of A/T hook DNA binding domains.
In the absence of HMG I (Fig. 5A and B, lanes 1 to 5), little ligation was seen, since the DNA concentration (0.06 nM) was too low for efficient ligation. However, addition of 1 μM HMG I resulted in efficient conversion of labeled LTR DNA to higher-molecular-weight forms (Fig. 5, lanes 6 to 10). After less than 5 min of incubation with ligase, more than 50% of the starting DNA was converted to ligation products.
We next studied the HMG I deletion mutants used in the tests of reconstitution of PICs described above. Concentrations of deletion mutants were adjusted to maintain the same concentrations of A/T hook DNA binding domains in each reaction. Addition of the peptides containing two A/T hooks also resulted in more than half of the input DNA becoming ligated after 5 min. Ligation in the presence of the deletions proceeded even more quickly than with wild-type HMG I (Fig. 5, lanes 11 to 20). The peptides containing single A/T hooks did not promote ligation efficiently (Fig. 5, lanes 21 to 35), failing to convert half the starting DNA to ligation product even after 60 min. Thus the ability of each peptide to promote ligation of LTR DNAs closely paralleled the ability to reconstitute PICs. This supports the model that HMG I promotes integration by bringing together distant DNA domains.
DISCUSSION
In this study and in previous work (16), we have presented four lines of evidence supporting the idea that HMG I(Y) is a likely cofactor for integration in vivo. (i) We found that HMG I(Y) is associated with MoMLV PICs from infected cells by using immunoprecipitation with an affinity-purified anti-HMG I(Y) antibody. Salt stripping of PICs or preincubation of the antibody with HMG I(Y) protein blocked immunoprecipitation. (ii) In previous work, we demonstrated cofractionation of HMG I(Y) with HIV-1 PICs through several purification steps (16). Salt stripping of HIV-1 PICs depleted HMG I(Y). (iii) Reconstitution activity derived from HIV-1 PICs could be depleted with an anti-HMG I(Y) antibody (16). (iv) Northern blot studies of HMG I(Y) revealed that it is expressed widely, including in tissues known to support infection by HIV-1 and MoMLV, placing the cofactor protein in the tissue sites of viral replication (7, 29, 47; unpublished data). Taken together, the above data support the idea that HMG I(Y) is a cofactor for replication in vivo.
Are the other proteins proposed as integration cofactors (BAF, NC, HMG-1, and Ini-1) active as such in vivo? It will be crucial to obtain further data to strengthen or disprove these proposals. Some of the proteins that can reconstitute integration in vitro (BAF and NC), like HMG I(Y) can also promote DNA aggregation, suggesting a common mechanism for in vitro action. NC mutations have been reported that act in vivo at a step after reverse transcription is mostly completed, indicating a potential role in integration (22). NC proteins containing these mutant changes are defective for stimulating integrase in vitro (9). Ongoing studies are investigating the role of NC and these mutants during infection. BAF, Ini-1, and HMG-1 have not yet been shown to be associated with PICs or to stimulate integration in vivo.
How might binding of HMG I(Y) promote activity of PICs? HMG I(Y) has not been found to bind to integrase directly (16, 26), consistent with models in which HMG I(Y) acts by binding to the cDNA. Data presented here and previously support a model in which HMG I(Y) may act by bridging distant DNA segments (16, 26, 37). HMG I(Y) contains multiple DNA binding domains (A/T hooks), so that binding of an HMG I(Y) monomer to widely spaced DNA sites could appose distant DNA domains (7). In the nuclear magnetic resonance structure of HMG I amino acids 51 to 90 bound to DNA, each of the two A/T hooks binds a separate DNA oligonucleotide (27). The HMG I(Y) derivatives that supported reconstitution of PICs, which contained two or more A/T hooks, matched exactly with those that could promote intermolecular ligation of linear LTR DNAs in dilute solution. This parallel supports the idea that apposition of distant DNA sites is the mechanism operating in both assays.
One possible role for multivalent DNA binding by HMG I(Y) could have been target DNA capture. An HMG I(Y) monomer bound to LTR DNA might use a free A/T hook to bind target DNA and promote integration at that site. However, a study of 61 HIV-1 integration sites in human DNA showed no enrichment for HMG I(Y) sites, arguing against this model (10).
HMG I(Y) may act by promoting conversion of integrase from an inactive to an active conformation. For HIV-1, integrase is apparently inactivated by salt stripping of PICs, but reactivated by reconstitution with HMG I(Y). The inactive HIV-1 integrase protein must remain bound to the cDNA in high-salt buffer to survive gel filtration, providing evidence for a bound but inactive conformation. Studies with inhibitors have also suggested that integrase may exist in functional and nonfunctional conformations (25). Perhaps high-salt treatment disrupts integrase-DNA interactions at the active site, so that the viral DNA end is no longer correctly positioned, but integrase is not fully released from the cDNA. The situation differs for MoMLV integrase, in which some of the salt-stripped complexes are able to carry out autointegration. However, upon reconstitution with HMG I(Y), autointegration is not reduced, but new complexes become active for intermolecular integration, again indicating that HMG I(Y) causes previously inactive complexes to become active. These results parallel studies of the role of the architectural DNA-binding protein IHF in phage lambda integration. Lambda integrase can bind to lambda DNA in the absence of IHF, but does not carry out efficient integrative recombination. Bending of DNA allows lambda integrase to make the multivalent contacts with DNA that are required for full activity. Data presented here support a related model in which condensation of retroviral cDNA by HMG I(Y) stabilizes integrase in an active conformation.
ACKNOWLEDGMENTS
We thank members of the Bushman laboratory and the Salk Institute Infectious Disease Laboratory for helpful discussions. We thank D. Thanos for the HMG I expression vector, M. Roth for pETINH, C. Bewley for the 9–75 and 50–91 HMG I derivatives, and J. Maher for the HMG I-C expression vector. We also thank Lynn Artale for artwork and for helping to prepare the manuscript.
L.L. was supported in part by the Rau Foundation. This work was supported by NIH grants GM56553 and AI34786 to F.D.B., the James B. Pendleton Charitable Trust, and the Berger Foundation. F.D.B. is a Scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.Aiyar A, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 3.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent complex and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley G W, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 7.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 8.Carteau S, Batson S C, Poljak L, Mouscadet J-F, de Rocquigny H, Darlix J-L, Roques B P, Käs E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carteau S, Gorelick R J, Bushman F D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carteau S, Hoffmann C, Bushman F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 13.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 14.Ellison V, Abrams H, Roe T, Lifson J, Brown P. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFN Beta gene enhancer by transcriptional factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 16.Farnet C, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 17.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 20.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;17:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 21.Goodman S D, Nash H A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989;341:251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- 22.Gorelick R J, Gagliardi T D, Bosche W J, Wiltrout T A, Coren L V, Chabot D J, Lifson J D, Henderson L E, Arthur L O. Strict conservation of the retroviral nucleocapsid (NC) protein zinc-finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant NC zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 23.Hansen M S T, Carteau S, Hoffmann C, Li L, Bushman F. Retroviral cDNA integration: mechanism, applications and inhibition. In: Setlow J K, editor. Genetic engineering. Principles and methods. Vol. 20. New York, N.Y: Plenum Press; 1998. pp. 41–62. [DOI] [PubMed] [Google Scholar]
- 24.Hansen M S T, Smith G J I, Kafri T, Molteni V, Siegel J S, Bushman F D. Integration complexes derived from HIV vectors for rapid assays in vitro. Nat Biotechnol. 1999;17:578–582. doi: 10.1038/9886. [DOI] [PubMed] [Google Scholar]
- 25.Hazuda D J, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler J A, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller M D. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 26.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huth J R, Bewley C A, Nissen M S, Evans J N S, Reeves R, Gronenborn A M, Clore G M. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 28.John S, Reeves R B, Lin J-X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor α-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF -κB family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson K R, Lehn D A, Elton T S, Barr P J, Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y) J Biol Chem. 1988;263:18338–18342. [PubMed] [Google Scholar]
- 30.Jonsson C B, Donzella G A, Roth M J. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993;268:1–8. [PubMed] [Google Scholar]
- 31.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 32.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 33.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J-L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee M S, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Farnet C M, Anderson W F, Bushman F D. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J Virol. 1998;72:2125–2131. doi: 10.1128/jvi.72.3.2125-2131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maher J F, Nathans D. Multivalent DNA-binding properties of the HMG-I proteins. Proc Natl Acad Sci USA. 1996;93:6716–6720. doi: 10.1073/pnas.93.13.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller M D, Wang B, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J Virol. 1995;69:3938–3944. doi: 10.1128/jvi.69.6.3938-3944.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves R, Nissen M S. Purification and assays for high mobility group HMG-I(Y) protein function. Methods Enzymol. 1999;304:155–188. doi: 10.1016/s0076-6879(99)04011-2. [DOI] [PubMed] [Google Scholar]
- 42.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solomon M J, Strauss F, Varshavsky A. A mammalian high mobility group protein recognizes any stretch of six A.T base pairs in duplex DNA. Proc Natl Acad Sci USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-KB-dependent virus induction of the human IFN-B gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 45.Thanos D, Maniatis T. Virus induction of human IFNB gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 46.Wei S-Q, Mizuuchi K, Craigie R. Footprints of the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X, Benson K F, Ashar H R, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]