Abstract
Scanning x-ray microdiffraction of complex tissues and materials is an emerging method for the study of macromolecular structures in situ, providing information on the way molecular constituents are arranged and interact with their microenvironment. Acting as a bridge between high-resolution images of individual constituents and lower resolution microscopies that generate global views of material, scanning microdiffraction provides an approach to study the functioning of complex tissues across multiple length scales. Here, we discuss the methodology, summarize results from recent studies, and discuss the potential of the technique for future studies coordinated with other biophysical techniques.
Introduction
The molecular processes underlying biological development or disease progression involve interactions across length scales that span from molecules to whole tissue. High-resolution approaches such as cryoEM, X-ray crystallography, and solid-state NMR (ssNMR) are capable of determining the structure of macromolecules and molecular assemblies at atomic resolution. But the relevance of these structures to biological processes often requires placing them in their biological context. Optical microscopy can map the presence of molecules within tissue.
But there is a huge gap between the resolution of these two classes of techniques and many biologically critical processes take place at these intermediate length scales. X-ray scattering is capable of generating structural information at a higher resolution than optical techniques but scattering from complex tissues or materials has rarely been attempted because the heterogeneous mixture of constituents makes data interpretation difficult. The availability of micro- and nano-beams of increased brilliance combined with recent advances in detector technology enables measurement of scattering from small volumes which, in favorable cases, are populated largely by a single constituent. This makes possible the collection of high-resolution data in situ and the generation of information on the organization and interactions of molecular constituents within the microenvironment in which they are embedded. Studies of the structure of complex soft materials [1], biomaterials, such as plant cell walls [2], bones [3], biological cells [4,5], or pathological protein aggregates [6·], have demonstrated the flexibility and power of the technique.
Scanning X-ray microdiffraction (XMD) can be thought of as a multiple resolution technique. The cellular-level resolution is defined by the beam diameter and step size as the sample is scanned across the beam. For instance, a 5 μ beam scanned over a 5 μ raster results in information about the distribution of molecular structures with 5 μ resolution. But at each raster point, a full diffraction pattern is collected, and the nature of those diffraction patterns defines the molecular-level resolution. For instance, scattering from amyloid fibrils that include the prototypical 4.7 Å cross-β reflection may provide structural data to a resolution of 4.7 Å or better while simultaneously mapping the distribution of these structures over a 5 μ grid.
The application of this technique to biomolecular and biomaterial systems is producing insights into molecular processes that are largely inaccessible by conventional techniques. To take advantage of these opportunities, many synchrotron facilities have established beamlines customized for scanning XMD, including BioCAT and GM/CA CAT at the Advanced Photon Source (APS), LiX at NSLS–II, ID13 at ESRF, P03, P10 at Petra III, and I22 at Diamond. Figure 1 is a diagram of a typical XMD experiment and the form of data it generates. Here we review the technological advances enabling this work and describe examples of its use to characterize the molecular organization of complex materials and tissues.
Figure 1.
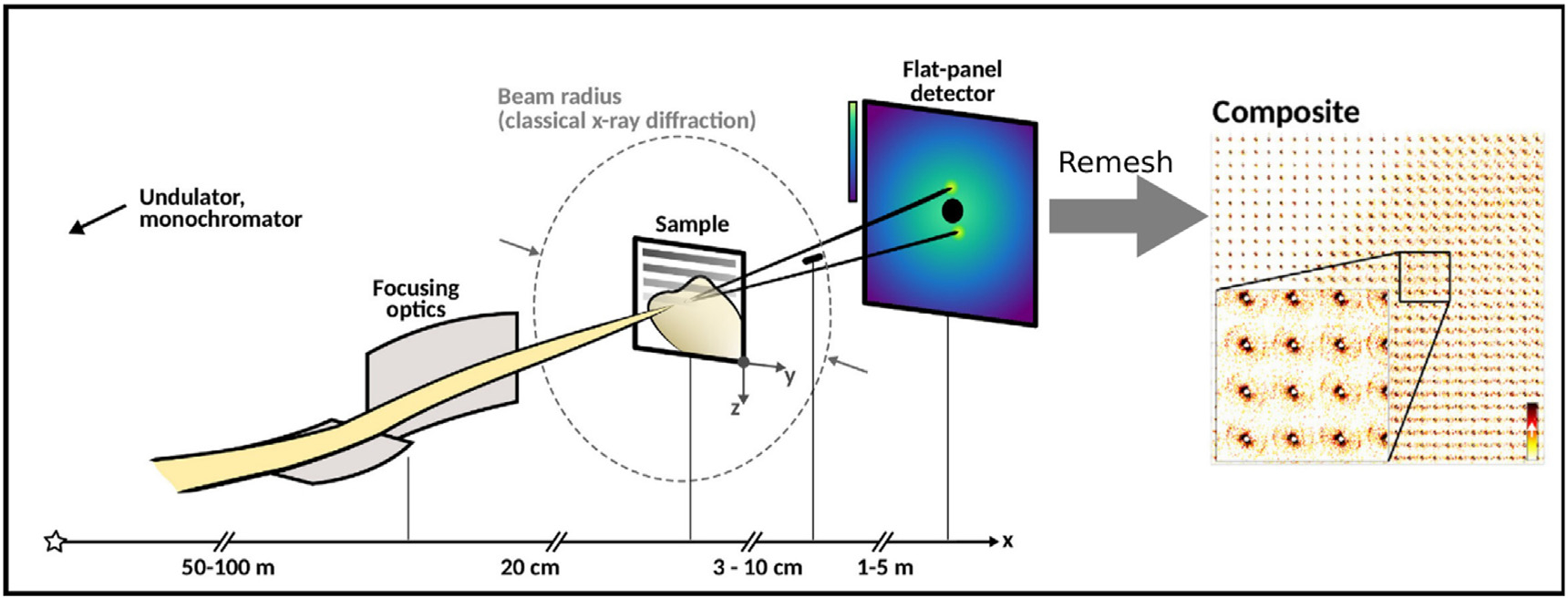
Schematic of scanning X-ray microdiffraction. Monochromatic X-rays are focused at sample by X-ray optics. Diffraction pattern is collected by one or more 2D detectors. The diffraction patterns are regrouped to their corresponding scan positions to compose scanning X-ray microdiffraction image. The images can be computed to show the spatial distribution of any attribute of the scattering patterns, making possible the mapping of multiple material properties simultaneously [Nicolas J et al., 2019, Progress in Biophysics and Molecular Biology].
To make possible production of beam sizes from 10 μ down to 100 nm [7], a number of breakthrough x-ray optical technologies were developed. Compound refractive lenses can generate μ-sized x-ray beams with simple alignment [8]. Diffractive lenses, based on Fresnel zone plate concepts, can focus x-rays to 20 nm [9]. Recently, 10 nm x-ray beams have been achieved using ultra–smooth Kirkpatrick–Baez mirrors [10]. To supplement these developments, highly efficient detectors with very high data collection rates [11] have been developed to enable fast scanning using a range of small-angle and wide-angle (S/WAXS) data collection arrangements, customized to the needs of particular projects [7], greatly expanding the kinds of structural questions amenable to study by XMD.
Application to macromolecular crystallography
The use of microbeams for studies of biomaterials emerged originally for use in macromolecular crystallography [12], driven by the potential for studying macromolecular structures using microcrystals or even highly ordered crystalline domains of very limited size embedded in larger, less-well-ordered crystals [12]. These advances made possible, for instance, the groundbreaking studies of G-protein coupled receptors that could not have been achieved at a conventional crystallographic beam line [13].
Hierarchical structure of biomaterials
Scanning XMD is particularly well suited to the study of hierarchical organization in biological tissues and materials: detailed mapping of the orientation and density of cellulose fibrils within the cell walls of wood from Norwegian spruce trees revealed a helical arrangement responsible for its remarkable mechanical properties [2], providing a basis for advanced considerations of wood engineering [14]. Heterogeneity in the skin-core structure of spider silk revealed by XMD [15] identified two amorphous, fine silk fibers with distinct mesoscale features co-existing in the center of an orb-web. Motivated by an interest in developing advanced adhesives, XMD was used to study the nano-scale structure of spider feet with a focus on the attachment hairs (setae) responsible for the ability of spiders to walk up walls and across ceilings [16· ·]. Figure 2a shows the variation of intensity and orientation of scattering as observed using a 250 nm x-ray beam scanned across a hair when it was attached or removed from a surface. These measurements were used to calculate gradients of the mechanical properties responsible for supporting attachment, stabilizing adhesion, and withstanding high stress at detachment.
Figure 2.
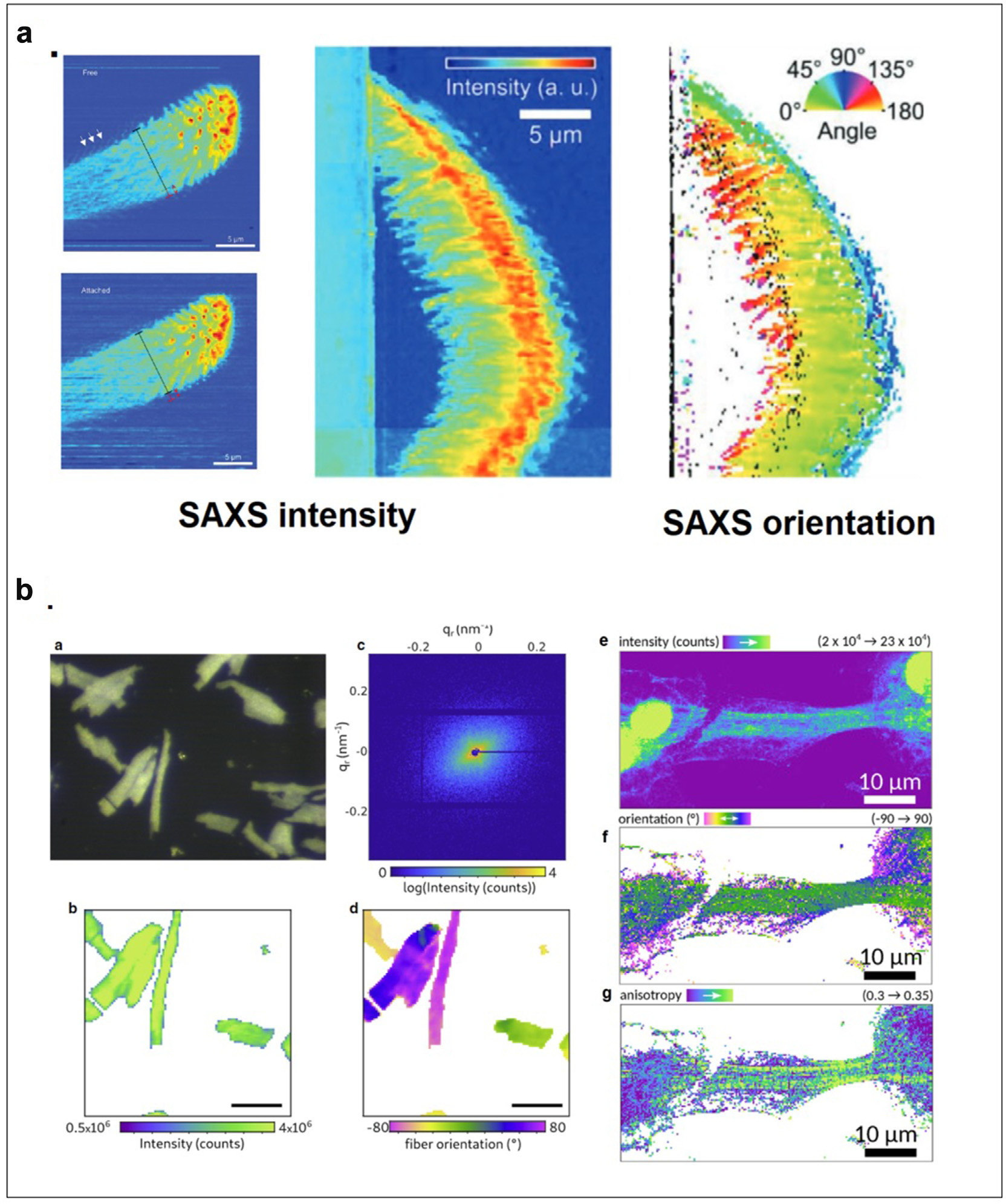
(a) Physical basis of adhesion by spider attachment hairs. Top view of attachment hair freestanding and attached (right) as visualized by WAXS intensities. Side view of attached hair (middle) and the distribution of orientations in attachment fibrils as determined from SAXS intensity and orientation (Flenner et al., 2020). (b) Structural organization of cardiomyocytes (a) Optical micrograph of a freeze-dried cardiomyocyte recorded with the beamline on-axis microscope (brightfield). (b) X-ray darkfield image from the integrated signal. (c) Diffraction pattern from one scan point. (d) Orientation of constituent actomyosin fibrils is determined by the orientation of diffraction. Scale bars for (a), (b), and (d) 50 mm. (a-d from Reichardt et al., 2020). (e) X-ray darkfield map of an iPS-derived cardiomyocyte. (f) Orientation of actomyosin fibers obtained by PCA analysis. The anisotropy (g) contrasts the highly oriented actomyosin filaments with modulation originating from the striated actomyosin filaments (e-g from Nicolas et al., 2019).
The cuticle of shrimp has been studied extensively due to its remarkable mechanical toughness. The impact of mechanical compression on the intensity of the (002) reflection from chitin was used [17·] as a strain sensor to measure the distribution of sub-micron deformation of lamella in the exo- and endo-cuticle and demonstrated that the material toughness arises from a combination of the α-chitin fiber networks deforming elastically while the surrounding matrix deforms plastically prior to systematic failure.
Cardiac cells and tissues
The intricate, hierarchical organization of cardiomyocytes — the fundamental structural unit of cardiac muscle — had proven particularly reticent to structural analysis by conventional techniques but scanning microdiffraction has proven particularly well suited to its study [18, 19••, 20••]. For instance, the diffraction signal of the actomyosin contractile unit was recorded from living cardiomyocytes bringing muscle diffraction to the scale of single cells as shown in Figure 2b. In coordination with the scanning diffraction studies, coherent optics were used to perform holographic imaging and tomography on a single cardiomyocyte [19· ·]. This allowed the extension of the length scales covered by scanning XMD and made possible the reconstruction of the electron density of an entire freeze-dried cardiomyocyte, visualizing the three-dimensional arrangement of myofibrils, sarcomeres, and mitochondria with a voxel size less than 50 nm.
Pathological structures in Alzheimer’s disease
Aggregation of proteins or peptides into pathological deposits in brain tissue is a hallmark of many neurodegenerative diseases, such as Alzheimer’s disease or Parkinson’s disease (PD). The formation of amyloid fibrils occurs by the stacking of β-strands that run perpendicular to the fibril axis and are stabilized by axial hydrogen bonds, providing amyloid cores with great stability. The fibrils are highly polymorphic and may be affected by variations in constituents and environmental factors such as pH, temperature, and chemical modification [5,21]. High-resolution images of many structural polymorphs of these fibrils have been determined by cryo-electron microscopy and solid-state NMR [22· ·]. But, to probe the molecular mechanisms underlying the neurotoxicity associated with these fibrils, investigators have turned to their study in the context of the tissue where their relevance to disease progression can be examined. The locations of structural polymorphs of amyloid have been mapped in situ using scanning XMD of histological thin sections of human brain tissue. Changes in scattering intensity in the prototypical 4.7 Å cross-β reflection have been used to map variation in fibril structure from core to margin of plaques [23,24] as shown in Figure 3. The shape of the 4.7 Å reflection exhibited significant differences between that observed in scattering from plaques in a typical AD case and in resilient cases that exhibit high plaque burdens in the absence of overt dementia [24], suggesting that variation in fibril structure might modulate the progression of disease.
Figure 3.

Amyloid fibril structure and spatial organization. (a) A diagram of the core cross-β structure common to protofilaments of all amyloid fibrils. Fibrils are formed by the longitudinal stacking of β sheets with a 4.7 Å repeat distance. Fibrils may contain different numbers of protofilaments [22· ·]. (b) Cross section of in vitro assembled Aβ42 peptides as calculated from SAXS pattern superimposed on a corresponding electron microscope reconstruction (left); as reconstructed from SAXS data (middle); with the corresponding SAXS data (right) [23]. (c) Structure of a dense amyloid plaque as visualized by optical microscopy after staining with congo red dye (left); as visualized by the intensity of the 4.7 A reflection (middle); and a map of fibril orientation as determined from the orientation of the 4.7 A reflection (right) [24].
Pathological structures in PD
Amyloid fibrils composed of α-synuclein are a major component of Lewy bodies (LBs), the principal pathological hallmark of PD. The structure of LBs has been studied in situ using synchrotron Fourier tansform infra red spectroscopy (FTIR) and XMD [6·,25,26]. This work was able to distinguish two distinct morphological regions of the LB, with fibril structures differentially concentrated in the halo, distinct from a dense protein and lipid core. The involvement of metal ions in the in vitro aggregation of α-synuclein fibrils [27] motivated the use of X-ray fluorescence microscopy to determine their distribution in situ [28, 29••]. The combination of X-ray diffraction and X-ray fluorescence revealed the correlation of the presence of heavy metal ions with the accumulation of α-synuclein fibrils within LB.
Three-dimensional studies and multi-modal imaging
Although most studies involve the mapping of structure or structural parameters in two dimensions, scanning microdiffraction tensor tomography (TT) has made possible mapping of fibrillar-like structures in three dimensions. The three-dimensional arrangement of collagen matrix within bone [30,31] and myelin sheath of neuronal cells within brain tissue [32· ·] have been characterized in this way. Results on the organization of collagen in bone are shown in Figure 4. This work demonstrated that a SAXS computed tomogram can be constructed using multiple micro-focused X-ray raster scans taken with the sample oriented at different angles relative to the x-ray beam. Using a sample with oriented fiber structure, they demonstrated that SAXS TT could be used to reconstruct the distribution of fibrillar structures within the sample in three dimensions. Assuming the sample could be separated into a series of voxels (each of which has an associated tensor), each scan could be thought of as a projection of scattering from voxels in the beam path multiplied by appropriate weights. Thus, the voxel size could be smaller than the beam size. Using this approach, the organization of collagen fibrils within bone tissue was mapped in three dimensions. These studies are continuing, with a demonstration of the structural variation of collagen within bone tissue as correlated to the involvement of biomineral and artificial implants [33, 34•, 35].
Figure 4.
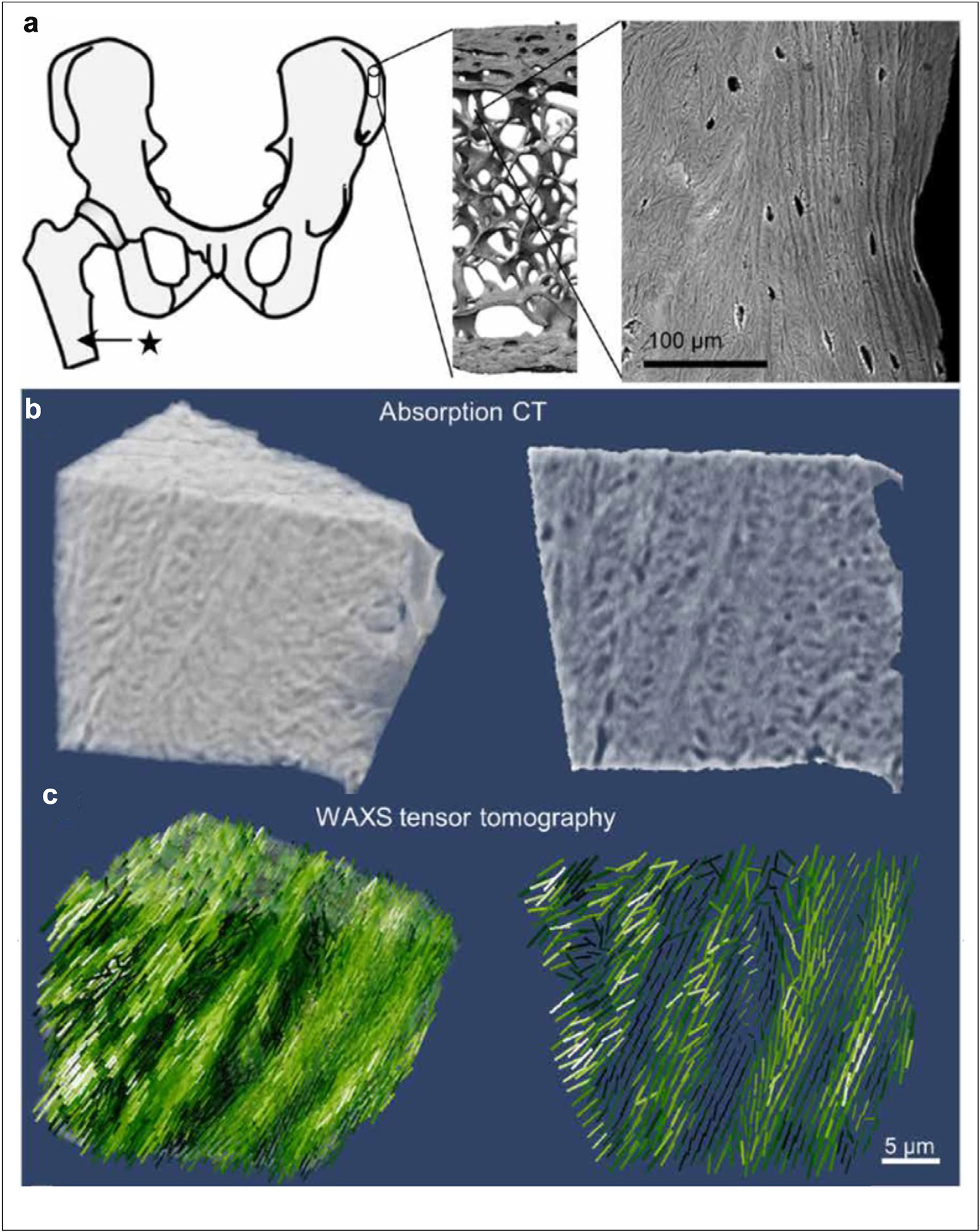
Mapping the 3D orientation of nanostructures in human bone (a) anatomical location of the sampling site of the bone cube, its hierarchical structure from laboratory microcomputed tomography (microCT) and scanning electron microscopy (SEM). (b) High-resolution absorption tomogram (left) with the two-dimensional image (right) of a central section. (c) Three-dimensional distribution of orientations reconstructed from the WAXS tensor.
SAXS–TT has also been used for the study of the structure of myelin sheath within nerve tissue [32••,36] and to quantify myelin structural alterations in demyelinated mouse brain. These results demonstrated that SAXS–TT could be a reliable, non–destructive stain-free imaging method for the study of the molecular basis of myelin–related diseases [32,36].
Furthermore, scanning XMD can be combined with x-ray spectroscopies such as X-ray fluorescence [28, 29••] or phase contrast imaging (holography, etc.) [37], which are feasible with specialized hardware, making possible direct correlation of multiscale structure with mapping of element distribution and cell morphology.
Sample preparation
In examination of heterogeneous materials, smaller scattering volumes are desired since they have a higher chance of being relatively homogeneous in composition and thereby more likely to give rise to interpretable scattering patterns. Nevertheless, obtaining data with an adequate signal-to-noise ratio from small scattering volumes may require X-ray exposures that result in radiation damage. Some materials are far more resistant to radiation damage than others, and preliminary studies are generally required to determine the maximum dose tolerable by a sample. Given the experience of macromolecular crystallography, data collection at cryogenic temperatures is likely to lower the radiation damage incurred, but no systematic evaluation of the benefit of this for XMD has yet been made. de la Mora et al. [38] demonstrated data collection at both room temperature and 100 K by sample cooling with a gaseous-nitrogen stream at ID13 ESRF. To prevent tissue damage due to water crystal formation during freezing, it may be necessary to add cryoprotectants as is now routine for macromolecular cryocrystallography [39]. If this can be done effectively, many of the methods developed for crystallography should be applicable to the handling of thin, frozen sections.
An alternative to freezing of tissue is the use of fixed tissue which usually involves cross-linking of constituents with formaldehyde and dehydration with ethanol to lock the relative positions of materials in place. This treatment preserves most protein secondary structures [40] and epitopes [41] but may disrupt the tertiary structure of proteins. Fibrillar structures such as neuropathological amyloid fibrils appear to be nearly impervious to these treatments.
For two-dimensional studies of thin-sections, the section thickness should be roughly the same as the x-ray beam diameter because the increase in sample thickness leads to the mixing of scattering from features at different depths in the section. For studies of fixed, histological sections of human brain tissue using a 5 μ X-ray beam, Liu et al. [24] chose to use 20 μ thick sections to provide a larger scattering volume resulting in an increase in signal-to-noise ratio without increasing the X-ray dose.
For three-dimensional studies, such as those described above on bone [30,31], samples ~ 40 × 40 × 40 μ3 were used and the cellular-scale resolution was determined by a combination of the beam size and the step size of the angular rotation of the sample.
Data processing for scanning XMD
Processing the significant volume of data produced by scanning XMD, often coordinated with data from other microscopies, provides continuing challenges. The number of diffraction patterns in one scan of a sample has increased from thousands of images (diffraction patterns) in step scans with a 10 μ beam to millions of images in fly scans utilizing a ~ 10 nm beam. It is impossible to manually examine this number of diffraction patterns and automatic data reduction and processing are essential. Current 2D detectors widely used for scanning S/WAXS microdiffraction, collect all relevant intensity, symmetry and orientation information at high data rates. Most software packages used for solution or fiber diffraction are ill-suited to this data volume. For instance, Fit2D is a commonly used, C-based computation GUI for processing 1D plots (azimuthal integration) or 2D patterns (a polar transformation) but exhibits severe limits for processing large volumes of data and suffers from a lack of compatibility with other analysis programs. ESRF has developed a software package called pyFAI, a python–based high-performance software [42,43]. PyFAI allows CPU and GPU parallel computation with a single pattern processing time of about 20 ms. Based on pyFAI, groups at Bessy and Petra synchrotrons developed DPDAK for processing and visualizing scanning XMD patterns [44]. py4xs, developed at LiX at NSLS-II, is another software package designed for scanning XMD, capable of processing data from multiple detectors simultaneously [45]. Other processing tools, such as Xi-CAM and blusky, are well-designed software packages for diffraction pattern analysis [46,47]. Customized programs have been developed for extracting specific scattering attributes from scanning microdiffraction data, such as reciprocal space position, or orientation of fibrillar constituents within the tissue [20••,24,48], the recent demonstration of the use of software for the automatic extraction of orientation and symmetry information from diffraction data [49] and the demonstration of the use of a Gaussian optimization method for extraction of structural features from sparsely sampled data sets [50]. Machine learning has been applied to disambiguate data from complex mixtures of constituents [51]. These advanced image processing and optimization methods have shown high efficiency and reliability in some applications and have great potential for automatic processing, analysis, and interpretation of scanning XMD data in the future.
Conclusions
There are few available techniques for generating moderate resolution structural information about macromolecules in the context of complex materials and tissues. The widespread availability of micro- and nanobeams at synchrotron x-ray sources is opening many opportunities for studies of this kind. When the scattering volume is reduced, the number of constituents contributing to the data is lowered, and in scattering volumes where a single constituent dominates, detailed structural information can often be extracted. Detailed analysis of this data often relies on the use of high-resolution images of isolated molecules or molecular assemblies, which in the absence of in situ information may be of uncertain biological relevance. Scanning XMD provides a bridge from these high-resolution structures of molecular constituents to lower-resolution optical micrographs that may locate constituents but cannot provide information on molecular organization or configuration. As such it represents an important link in comprehensive biophysical characterizations of complex tissues and materials.
Acknowledgements
This work was supported in part by the National Institutes of Health, R21-AG068972.
Footnotes
Conflict of interest statement
Nothing declared.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Lutz-Bueno V, Zhao J, Mezzenga R, Pfohl T, Fischer P, Liebi M: Scanning-SAXS of microfluidic flows: nanostructural mapping of soft matter. Lab Chip 2016, 16:4028–4035. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenegger H, Müller M, Paris O, Riekel C, Fratzl P: Imaging of the helical arrangement of cellulose fibrils in wood by synchrotron X-ray microdiffraction. J Appl Crystallogr 1999, 32:1127–1133. [Google Scholar]
- 3.Fratzl P, Weinkamer R: Nature’s hierarchical materials. Prog Mater Sci 2007, 52:1263–1334. [Google Scholar]
- 4.Weinhausen B, Saldanha O, Wilke RN, Dammann C, Priebe M, Burghammer M, Sprung M, Köster S: Scanning X-ray nanodiffraction on living eukaryotic cells in microfluidic environments. Phys Rev Lett 2014, 112, 088102. [Google Scholar]
- 5.Bernhardt M, Priebe M, Osterhoff M, Wollnik C, Diaz A, Salditt T, Rehfeldt F: X-ray micro-and nanodiffraction imaging on human mesenchymal stem cells and differentiated cells. Biophys J 2016, 110:680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.•. Araki K, Yagi N, Aoyama K, Choong CJ, Hayakawa H, Fujimura H, Nagai Y, Goto Y, Mochizuki H: Parkinson’s disease is a type of amyloidosis featuring accumulation of amyloid fibrils of α-synuclein. Proc Natl Acad Sci USA 2019, 116:17963–17969. Aggregates of a-synuclein within LB were found to form a cross-β structure of amyloid fibrils by scanning XMD
- 7.Riekel C, Burghammer M, Davies R: Progress in micro-and nano-diffraction at the ESRF ID13 beamline. In IOP Conference Series. Mater Sci Eng 2010, 14. p. 012013. IOP Publishing. [Google Scholar]
- 8.Snigirev A, Kohn V, Snigireva I, Lengeler B: A compound refractive lens for focusing high-energy X-rays. Nature 1996, 384:49–51. [DOI] [PubMed] [Google Scholar]
- 9.Vila-Comamala J, Pan Y, Lombardo J, Harris WM, Chiu WK, David C, Wang Y: Zone-doubled Fresnel zone plates for high-resolution hard X-ray full-field transmission microscopy. J Synchrotron Radiat 2012, 19:705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimura H, Handa S, Kimura T, Yumoto H, Yamakawa D, Yokoyama H, Matsuyama S, Inagaki K, Yamamura K, Sano Y, Tamasaku K: Breaking the 10 nm barrier in hard-X-ray focusing. Nat Phys 2010, 6:122–125. [Google Scholar]
- 11.Broennimann C, Eikenberry EF, Henrich B, Horisberger R, Huelsen G, Pohl E, Schmitt B, Schulze-Briese C, Suzuki M, Tomizaki T, Toyokawa H: The PILATUS 1M detector. J Synchrotron Radiat 2006, 13:120–130. [DOI] [PubMed] [Google Scholar]
- 12.Cusack S, Belrhali H, Bram A, Burghammer M, Perrakis A, Riekel C: Small is beautiful: protein micro-crystallography. Nat Struct Biol 1998, 5:634–637. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF: Crystal structure of the human β 2 adrenergic G-protein-coupled receptor. Nature 2007, 450:383–387. [DOI] [PubMed] [Google Scholar]
- 14.Farid T, Rafiq MI, Ali A, Tang W: Transforming wood as next-generation structural and functional materials for a sustainable future. EcoMat 2022, 4, e12154. [Google Scholar]
- 15.Riekel C, Burghammer M, Rosenthal M: Skin-core morphology in spider flagelliform silk. Appl Phys Lett 2019, 115:123702. [Google Scholar]
- 16.••. Flenner S, Schaber CF, Krasnov I, Stieglitz H, Rosenthal M, Burghammer M, Gorb SN, Müller M: Multiple mechanical gradients are responsible for the strong adhesion of spider attachment hair. Adv Mater 2020, 32:2002758. Scanning XMD was utilized to demonstrate the nanostructured attachment system of spider feet, which allow spiders to move vertically on a smooth surface.
- 17.•. Zhang Y, Garrevoet J, Wang Y, Roeh JT, Terrill NJ, Falkenberg G, Dong Y, Gupta HS: Molecular to macroscale energy absorption mechanisms in biological body armour illuminated by scanning X-ray diffraction with in situ compression. ACS Nano 2020, 14:16535–16546. Scanning XMD determine how concurrent strain varied multi-scale structure.
- 18.Singh JP, Young JL: The cardiac nanoenvironment: form and function at the nanoscale. Biophys Rev 2021, 13:625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.••. Reichardt M, Neuhaus C, Nicolas JD, Bernhardt M, Toischer K, Salditt T: X-ray structural analysis of single adult cardiomyocytes: tomographic imaging and microdiffraction. Biophys J 2020, 119:1309–1323. Combining scanning XMD and xray holography and xray tomography to visualize 3D structure of myosin, sarcomeres and mitochondria within muscle.
- 20.••. Nicolas JD, Bernhardt M, Schlick SF, Tiburcy M, Zimmermann WH, Khan A, Markus A, Alves F, Toischer K, Salditt T: X-ray diffraction imaging of cardiac cells and tissue. Prog Biophys Mol Biol 2019, 144:151–165. Scanning XMD was used to determine the structural variation during tissue development in cardiac muscle cell.
- 21.Lattanzi V, André I, Gasser U, Dubackic M, Olsson U, Linse S: Amyloid β 42 fibril structure based on small-angle scattering. Proc Natl Acad Sci USA 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.••. Gallardo R, Ranson NA, Radford SE: Amyloid structures: much more than just a cross-β fold. Curr Opin Struct Biol 2020, 60:7–16. High resolution structure of amyloid fibrils resolved by diffraction and cryo-EM shows that amyloid fold is unexpected diverse and complex, this diversity may play an essential role of fibrils in diseases.
- 23.Roig-Solvas B, Makowski L: Calculation of the cross-sectional shape of a fibril from equatorial scattering. J Structural Biol 2017, 200(3):248–257, 10.1016/j.jsb.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Costantino I, Venugopalan N, Fischetti RF, Hyman BT, Frosch MP, Gomez-Isla T, Makowski L: Amyloid structure exhibits polymorphism on multiple length scales in human brain tissue. Sci Rep 2016, 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M: α-Synuclein in Lewy bodies. Nature 1997, 388: 839–840. [DOI] [PubMed] [Google Scholar]
- 26.Araki K, Yagi N, Ikemoto Y, Yagi H, Choong CJ, Hayakawa H, Beck G, Sumi H, Fujimura H, Moriwaki T, Nagai Y: Synchrotron FTIR micro-spectroscopy for structural analysis of Lewy bodies in the brain of Parkinson’s disease patients. Sci Rep 2015, 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky VN, Li J, Fink AL: Evidence for a partially folded intermediate in α-synuclein fibril formation. J Biol Chem 2001, 276:10737–10744. [DOI] [PubMed] [Google Scholar]
- 28.Carboni E, Nicolas JD, Töpperwien M, Stadelmann-Nessler C, Lingor P, Salditt T: Imaging of neuronal tissues by x-ray diffraction and x-ray fluorescence microscopy: evaluation of contrast and biomarkers for neurodegenerative diseases. Biomed Opt Express 2017, 8(10):4331–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.••. Joppe K, Nicolas JD, Grünewald TA, Eckermann M, Salditt T, Lingor P: Elemental quantification and analysis of structural abnormalities in neurons from Parkinson’s-diseased brains by X-ray fluorescence microscopy and diffraction. Biomed Opt Express 2020, 11(7):3423–3443. Scanning XMD combined with XRF was used to study the structure and elemental composition of neurons in substantia nigra of Parkinson diseases.
- 30.Schaff F, Bech M, Zaslansky P, Jud C, Liebi M, Guizar-Sicairos M, Pfeiffer F: Six-dimensional real and reciprocal space small-angle X-ray scattering tomography. Nature 2015, 527:353–356. [DOI] [PubMed] [Google Scholar]
- 31.••. Grünewald TA, Liebi M, Wittig NK, Johannes A, Sikjaer T, Rejnmark L, Gao Z, Rosenthal M, Guizar-Sicairos M, Birkedal H, Burghammer M: Mapping the 3D orientation of nanocrystals and nanostructures in human bone: indications of novel structural features. Sci Adv 2020, 6:eaba4171. Combining both SAXS-TT and WAXS-TT to reconstruct 3D orientation of both nanostructure and biomineral.
- 32.••. Georgiadis M, Schroeter A, Gao Z, Guizar-Sicairos M, Liebi M, Leuze C, McNab JA, Balolia A, Veraart J, Ades-Aron B, Kim S: Nanostructure-specific X-ray tomography reveals myelin levels, integrity and axon orientations in mouse and human nervous tissue. Nat Commun 2021, 12:1–13. SAXS–TT could be used as a non-destructive imaging technique to quantify myelin structural alterations in nervous tissues.
- 33.Grünewald TA, Rennhofer H, Hesse B, Burghammer M, StanzlTschegg SE, Cotte M, Löffler JF, Weinberg AM, Lichtenegger HC: Magnesium from bioresorbable implants: distribution and impact on the nano-and mineral structure of bone. Biomaterials 2016, 76:250–260. [DOI] [PubMed] [Google Scholar]
- 34.•. Liebi M, Lutz-Bueno V, Guizar-Sicairos M, Schönbauer BM, Eichler J, Martinelli E, Löffler JF, Weinberg A, Lichtenegger H, Grünewald TA: 3D nanoscale analysis of bone healing around degrading Mg implants evaluated by X-ray scattering tensor tomography. Acta Biomater 2021, 134:804–817. Applying SAXS-TT and elemental analysis to determine the effects of implant on 3D nanostructure of biomineral in bone.
- 35.Palle J, Wittig NK, Kubec A, Niese S, Rosenthal M, Burghammer M, Grünewald TA, Birkedal H: Nanobeam X-ray fluorescence and diffraction computed tomography on human bone with a resolution better than 120 nm. J Struct Biol 2020, 212, 107631. [DOI] [PubMed] [Google Scholar]
- 36.Georgiadis M, Schroeter A, Gao Z, Guizar-Sicairos M, Novikov DS, Fieremans E, Rudin M: Retrieving neuronal orientations using 3D scanning SAXS and comparison with diffusion MRI. Neuroimage 2020, 204, 116214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernhardt M, Nicolas JD, Osterhoff M, Mittelstädt H, Reuss M, Harke B, Wittmeier A, Sprung M, Köster S, Salditt T: Correlative microscopy approach for biology using X-ray holography, Xray scanning diffraction and STED microscopy. Nat Commun 2018, 9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de la Mora E, Coquelle N, Bury CS, Rosenthal M, Holton JM, Carmichael I, Garman EF, Burghammer M, Colletier JP, Weik M: Radiation damage and dose limits in serial synchrotron crystallography at cryo-and room temperatures. Proc Natl Acad Sci USA 2020, 117:4142–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garman EF, Schneider TR: Macromolecular cryocrystallography. J Appl Crystallogr 1997, 30:211–237. [Google Scholar]
- 40.Zohdi V, Whelan DR, Wood BR, Pearson JT, Bambery KR, Black MJ: Importance of tissue preparation methods in FTIR micro-spectroscopical analysis of biological tissues: ‘traps for new users. PLoS One 2015, 10, e0116491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werner M, Chott A, Fabiano A, et al. : Effect of formalin fixation and processing on immunohistochemistry. Am J Surg Pathol 2000, 24:1016–1019. [DOI] [PubMed] [Google Scholar]
- 42.Kieffer J, Karkoulis D: PyFAI, a versatile library for azimuthal regrouping. J Phys Conf 2013, 425:202012. IOP Publishing. [Google Scholar]
- 43.Kieffer J, Wright JP: PyFAI: a Python library for high performance azimuthal integration on GPU. Powder Diffr 2013, 28:S339–S350. [Google Scholar]
- 44.Benecke G, Wagermaier W, Li C, Schwartzkopf M, Flucke G, Hoerth R, Zizak I, Burghammer M, Metwalli E, Müller-Buschbaum P, Trebbin M: A customizable software for fast reduction and analysis of large X-ray scattering data sets: applications of the new DPDAK package to small-angle X-ray scattering and grazing-incidence small-angle X-ray scattering. J Appl Crystallogr 2014, 47:1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Liu J, Chodankar S, Antonelli S, DiFabio J: Scanning structural mapping at the life science X-ray scattering beamline. J Synchrotron Radiat 2022, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pandolfi RJ, Allan DB, Arenholz E, Barroso-Luque L, Campbell SI, Caswell TA, Blair A, De Carlo F, Fackler S, Fournier AP, Freychet G: Xi-cam: a versatile interface for data visualization and analysis. J Synchrotron Radiat 2018, 25:1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arkilic A, Allan DB, Caswell TA, Li L, Lauer K, Abeykoon S: Towards integrated facility-wide data acquisition and analysis at NSLS-II. Synchrotron Radiat News 2017, 30:44–45. [Google Scholar]
- 48.Nicolas JD, Bernhardt M, Markus A, Alves F, Burghammer M, Salditt T: Scanning X-ray diffraction on cardiac tissue: automatized data analysis and processing. J Synchrotron Radiat 2017, 24:1163–1172. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Lhermitte J, Tian Y, Zhang Z, Yu D, Yager KG: Healing Xray scattering images. IUCrJ 2017, 4:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noack MM, Doerk GS, Li R, Streit JK, Vaia RA, Yager KG, Fukuto M: Autonomous materials discovery driven by Gaussian process regression with inhomogeneous measurement noise and anisotropic kernels. Sci Rep 2020, 10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park WB, Chung J, Jung J, Sohn K, Singh SP, Pyo M, Shin N, Sohn KS: Classification of crystal structure using a convolutional neural network. IUCrJ 2017, 4:486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]


