Abstract
A yeast two-hybrid screen for cellular proteins that interact with the murine leukemia virus (MuLV) Gag protein resulted in the identification of nucleolin, a host protein known to function in ribosome assembly. The interacting fusions contained the carboxy-terminal 212 amino acids of nucleolin [Nuc(212)]. The nucleocapsid (NC) portion of Gag was necessary and sufficient to mediate the binding to Nuc(212). The interaction of Gag with Nuc(212) could be demonstrated in vitro and was manifested in vivo by the NC-dependent incorporation of Nuc(212) inside MuLV virions. Overexpression of Nuc(212), but not full-length nucleolin, potently and specifically blocked MuLV virion assembly and/or release. A mutant of MuLV, selected to specifically disrupt the binding to Nuc(212), was found to be severely defective for virion assembly. This mutant harbors a single point mutation in capsid (CA) adjacent to the CA-NC junction, suggesting a role for this region in Moloney MuLV assembly. These experiments demonstrate that selection for proteins that bind assembly domain(s) can yield potent inhibitors of virion assembly. These experiments also raise the possibility that a nucleolin-Gag interaction may be involved in virion assembly.
The retroviral gag gene products play crucial roles at late stages in the viral life cycle, mediating virion assembly and RNA packaging (reviewed in references 65 and 72). The gag gene of the type C retroviruses is expressed as a precursor protein that moves to the plasma membrane, assembles into large structures, and induces the formation and release of virion particles. gag mutations have often been found to block virion assembly (1, 33, 61), and expression of the gag gene alone in mammalian cells is sufficient to direct the formation and release of “bald particles” from the cell (19, 63). Thus, the Gag precursor is both necessary and sufficient for assembly, earning the protein the name “particle making machine” (21). In addition, Gag is important early in infection, during virus entry; many mutations in the gag gene have no effect on assembly but rather block early stages of infection (3, 17, 31, 73). These studies indicate that some of the Gag domains are actively involved in the process of uncoating, reverse transcription, and perhaps nuclear transport and entry.
During and after the assembly process, the Gag protein of Moloney murine leukemia virus (Mo-MuLV) is cleaved by the viral protease into four products found in the mature virion: matrix (MA), p12, capsid (CA), and nucleocapsid (NC). The MA domain is required for membrane targeting of Gag and for virion assembly (57). The p12 protein includes an L domain that is required for late stages of viral assembly and efficient release from the cell (74). In addition, some mutations in p12 block early events (17, 75). The CA domain of Mo-MuLV is important for virion assembly, as most deletions and many point mutations in CA block this process. A few point mutations do not affect assembly but rather block the early stages of infection (3, 37). The NC domain is a highly basic sequence containing a single Cys-His box, a conserved zinc-binding motif (CX2CXnHX4C) found in most Gag proteins. Although NC is not absolutely required for assembly, it contains a sequence, termed the I (interaction) domain (65), that is important for multimerization of Gag, and deletion of NC results in a drastic reduction in particle production. Point mutations in NC often affect the process of encapsidation of the viral RNA (32, 48, 56), and very subtle mutations, in which Cys and His residues in the Cys-His box are changed to His or Cys, result in defects in reverse transcription of the RNA or the stability of the resulting DNA (31).
The complex and diverse activities of the Gag protein raise the possibility that cellular factors are involved in regulating Gag functions. Indeed, several cellular gene products have been suggested to serve as negative or positive factors for Gag activities during the retrovirus life cycle. Cyclophilin A, a prolyl isomerase thought to be involved in protein folding, is an example of a cellular factor that interacts with human immunodeficiency virus type 1 (HIV-1) Gag and serves as a positive factor for virus replication (44). Cyclophilin A is incorporated at substantial levels into the HIV-1 virion particle, and this incorporation enhances virus infectivity (25). A number of other Gag-interacting proteins have recently been identified. These include H03, a putative histidyl-tRNA synthetase that binds to MA of HIV-1 (41); KIF4, a microtubule-associated motor protein in the kinesin superfamily that interacts with different retroviral Gag proteins, including that of Mo-MuLV and HIV-1 (40, 67); actin and other cytoskeletal proteins (ezrin, moesin, and cofilin) found in HIV-1 virions (50); and translation elongation factor 1α, bound to MA and NC domains of HIV-1 Gag (14). Proteins may also interact with Gag protein early in infection; one example is the Fv-1 gene product, which blocks virus infection through effects on the CA domain (7, 20, 30, 51). It is likely that many other cellular proteins interact with Gag and are also involved in Gag functions.
To identify new host proteins that bind to Gag, we have applied the yeast two-hybrid system to screen a mouse cDNA expression library for proteins that interact with an MuLV Gag polyprotein serving as the bait. One protein recovered repeatedly was identified as a fragment of nucleolin, a host protein known to shuttle between the nucleus and the cytoplasm and to function in ribosome biogenesis. Here we describe the interaction of MuLV Gag with the C-terminal portion of nucleolin. We find that this interaction can potently inhibit virus assembly. Furthermore, a single point mutation adjacent to the CA-NC boundary, selected to abolish this interaction, also profoundly diminished assembly. Thus, these experiments indicate that the region of Gag that contains the CA-NC junction is important for Mo-MuLV assembly and that this region can serve as a target for inhibiting assembly. These data also raise the possibility that nucleolin may play a role in this process.
MATERIALS AND METHODS
Yeast plasmids.
The yeast expression vector pGADNOT encodes the C-terminal Gal4 activation domain (Gal4AD) and carries the LEU2 marker (44); plasmids pSH2-1 (35) and pBTM116 (6) encode the N-terminal LexA DNA-binding domain (LexADB) and carry HIS3 and TRP1 markers, respectively; plasmids pMA424 (45) and pAS1 (23), generously provided by S. Elledge, Baylor College of Medicine, encode the N-terminal Gal4 DNA-binding domain (Gal4DB), with HIS3 and TRP1 markers, respectively. Plasmids containing the Gag sequences of HIV-1, simian immunodeficiency virus type 1 (SIV-1), Mason-Pfizer monkey virus (MPMV), Mo-MuLV, N- and B-tropic MuLVs, and Rous sarcoma virus (RSV) were as described previously (4, 42, 44). The NC domain of the Mo-MuLV Gag was removed by partial digestion with MscI plus SalI, and the deleted DNA was transferred into pSH2-1 to generate a LexA-gagΔNC fusion. The CA-NC and NC domains of Mo-MuLV were amplified by PCR (Expand High Fidelity PCR system [Boehringer Mannheim]) according to the manufacturer's protocol, using oligonucleotides 5′TCGCAGGGATCCCCCTCCGCGCAGGA3′ and 5′GCGGGATCCGAGCCACTGTCGTTAGTGG3′, respectively, together with oligonucleotide 5′CGCGCGGTCGACAGGAGGGAGG3′ (nonviral sequences are in boldface) from plasmid pNCA (16). The amplified fragments were digested with BamHI and SalI and cloned into the same sites in pSH2-1. The resulting plasmids, pSH2-1-CANC and pSH2-1-NC, encode an N-terminal LexADB fused in frame to the CA-NC and NC domains, respectively. A construct expressing LexADB fused to lamin (39) was generously provided by R. Sternglanz (Stony Brook, N.Y.). Plasmid pGADNOT-Nuc(212) was isolated from the yeast two-hybrid screen and harbors a cDNA encoding the last 212 amino acids of the cellular protein nucleolin [Nuc(212)]. A construct expressing LexADB fused to Nuc(212) was made by transferring an EcoRI-BglII fragment from pGADNOT-Nuc(212) (EcoRI and BglII sites are located at a short polylinker at the 5′ end of the cDNA and at the plasmid backbone, respectively) to plasmid vector pBTM116, creating pBTM-Nuc(212).
Yeast strains.
Saccharomyces cerevisiae strains GGY::171 and CTY10-5d (39) were generously provided by R. Sternglanz. BY3171 is the MaV103 reverse two-hybrid S. cerevisiae strain (70), generously provided by E. Harlow (Massachusetts General Hospital).
Yeast two-hybrid library construction and screening.
Mouse cDNAs were obtained from a lambda phage library of cDNAs derived from the murine cell line WEHI-3 (a BALB/c macrophage/monocyte-derived cell line) in the directional UNI-ZAP vector (Stratagene). Roughly 106 plaques were pooled, the phage were propagated in liquid culture, and DNA was prepared by standard procedures (59). Inserts were excised by cleavage of NotI and XhoI (resulting in a short polylinker attached to the 5′ end of the cDNAs) and separated by electrophoresis on a 1% agarose gel. DNA inserts in the range of 0.5 to 4 kb were isolated and ligated into the NotI and SalI sites in the yeast vector pGADNOT, and the products were used to transform bacteria. A total of seven pools of at least 105 colonies were generated and used to prepare large amounts of plasmid DNA. More than 98% of the DNAs contained inserts. Since this library is directional, one clone in three should be in frame with the Gal4AD, and all should be the correct orientation.
To screen for Gag-interacting proteins, yeast strain CTY10-5d was cotransformed with a 1:1 mixture of pLexADB-Ngag plasmid and a given Gal4AD library plasmid pool, selecting for transformation by both plasmids. The colonies were transferred onto nitrocellulose filters and assayed for β-galactosidase (β-Gal) activity as described elsewhere (44). Blue transformants were restreaked and retested, and DNA was recovered from positive clones. The DNA was used to transform Escherichia coli strain Leu− BAI, and Leu+ transformants were selected to allow recovery of the cDNA-containing plasmid pGADNOT. These plasmids were then retested in yeast for activation of β-Gal to confirm their ability to activate in concert with plasmid pLexADB-Ngag. The sequence of each insert in these plasmids was determined and compared with entries in the GenBank database.
Reverse two-hybrid system.
A BamHI-SalI fragment, containing the Mo-MuLV CA and NC sequences from plasmid pSH2-1-CANC, was cloned into the same sites in the yeast expression vector pAS-1. The resulting plasmid, pAS1-CANC, encodes a Gal4DB-CA-NC fusion. This plasmid was randomly mutated using an E. coli mutator strain (XL-1 Red; Stratagene) according to the manufacturer's protocol, and the library of mutagenized DNA was used to transform E. coli strain ElectroMAX DH10B (GIBCO-BRL) to yield about 5 × 106 independent colonies. To obtain a rough estimate of the mutagenesis efficiency of the XL-1 Red strain, a parallel random mutagenesis procedure was performed on plasmid pUC18, and the mutated plasmid library was isolated and used to transform E. coli DH5α. Colony staining for β-Gal activity revealed that about 1% of the colonies were white.
Yeast strain BY3171 harboring plasmid pGADNOT-Nuc(212) was transformed with the randomly mutated pAS1-CANC library and plated on medium lacking Leu and Trp and containing 0.1% 5-fluoro-orotic acid (5-FOA). Out of approximately 104 transformants, only three 5-FOA-resistant colonies were obtained. Plasmid DNA was recovered and used to transform Trp− His− Leu− KC8 bacteria (Clontech). The pAS1-CANC plasmids recovered were used to transform yeast strain BY3171 harboring plasmid pGADNOT-Nuc(212) and were confirmed not to activate the reporter. Restriction digestion of these plasmids revealed that one was a rearranged plasmid lacking the insert. DNA sequence analysis of the other two plasmids showed that only one was mutated in the CA-NC sequence. This mutation (L477P) changed a leucine codon (CTA) at position 477 of Gag to a proline codon (CCA).
To confirm that no other mutations in the backbone of the plasmid contributed to the lack of interaction with the Gal4AD-Nuc(212) fusion protein, the CA-NC sequence was excised by BamHI and SalI digestion and then cloned into the same sites in plasmid pGADNOT. The resulting plasmid, pGADNOT-L477P, was transformed together with pBTM-Nuc(212) into yeast strain CTY10-5d, and Leu+ Trp+ colonies were selected. No activation of the lacZ gene was observed in these colonies, indicating that the L477P mutation is sufficient to disrupt the CA-NC–Nuc(212) interaction. In addition, these experiments demonstrated that L477P disrupts this interaction whether the CA-NC sequence is fused to Gal4DB or to Gal4AD.
In vitro binding experiments.
The Nuc(212) cDNA insert was excised from plasmid pGADNOT-Nuc(212) by cleavage at an XbaI site (located at a short polylinker at the 5′ end of the cDNA) and a BglII site (located at the pGADNOT plasmid backbone). This insert was cloned into the same sites in plasmid pGEX2TKPL, a derivative of pGEX-2TK (Pharmacia), to form a glutathione S-transferase (GST)–Nuc(212) fusion. Cultures of E. coli DH5α (200 ml) harboring pGEXT2TKPL or pGEXT2TKPL-Nuc(212) were grown to an optical density at 600 nm of 0.5, induced by the addition of isopropyl-β-d-thiogalactoside (IPTG) to a final concentration of 1 mM, and incubated at 37°C for 2 h. The bacteria were chilled on ice, washed twice with TNEN (50 mM Tris-HCl [pH 7.5], 0.5% Nonidet P-40 [NP-40], 10 mM EDTA, 50 mM NaCl), and 25% of the bacterial pellet was resuspended in 1 ml of TNENI solution (TNEN solution mixed with 2 mM phenylmethylsulfonyl fluoride (PMSF), aprotinin [1 μg/ml], leupeptin [1 μg/ml], and pepstatin A [1 μg/ml]). The bacteria were disrupted by sonication, and the cell debris was removed by centrifugation. Extracts of mammalian cells containing Gag proteins were prepared by lysing Mo-MuLV-infected NIH 3T3 cells with ice-cold TNENI (1 ml per 100-mm-diameter dish); the lysates were passed through a 19-gauge needle and clarified by centrifugation. A 500-μl aliquot of each bacterial lysate was mixed with 500 μl of the mammalian cell extract. After 1 h of rotation at 4°C, a slurry of glutathione-agarose beads (20 μl; Sigma) was added, and incubation was continued for 1 h at 4°C. The beads were then washed four times with TNENI solution and boiled in 60 μl of 2× loading buffer (125 mM Tris-HCl [pH 6.8], 2.5% sodium dodecyl sulfate [SDS], 20% glycerol, 50 μl of β-mercaptoethanol/ml); 20 μl of this mixture was analyzed by Western blotting. In typical experiments, the bound protein obtained from 15 to 20% of the original lysate was loaded per lane; to allow comparison with the input protein, 2% of the original 1-ml lysate was loaded directly in a parallel lane.
Mammalian expression plasmids.
Plasmid pNLENV-1 (46), which lacks the simian virus 40 (SV40) origin of replication, was used to express HIV-1 gag and pol genes (NL4-3 strain). pHIV-HSA, which contains the SV40 origin of replication, carries the HIV-1 genome (HXB2 strain) with a portion of its envelope gene substituted by the mouse heat-stable antigen gene (58) (generously provided by Manfred Schubert, National Institutes of Health, Bethesda, Md.).
Plasmid pXM-MoZipWT expresses a modified Mo-MuLV Gag in which the BZip domain from human CREB (77) was fused to the capsid C terminus, replacing the NC domain (E. Barklis, personal communication). The plasmid was generously provided by E. Barklis (Oregon Health Sciences University).
Mo-MuLV was expressed from plasmid pNCA (16) or from pNCS, a derivative carrying an SV40 origin of replication in the plasmid backbone. To introduce the L477P mutation into the Mo-MuLV provirus, an XhoI-SalI fragment was excised from pNCS and from pGADNOT-L477P and cloned into the same sites in pBluescript KS (Stratagene) to create pBLS-wt and pBLS-L477P, respectively. An internal XhoI-NruI fragment (560 bp) was excised from pBLS-wt and replaced with the corresponding fragment derived from pBLS-L477P, to create pBLS-wt-L477P. The mutated gag sequence was then exchanged for the wild-type gag sequence in pNCS, using the XhoI-SalI sites, to form pNCS-L477P. The L477P change abolished an AluI site, which was used to confirm the presence of this mutation.
To construct a protease-defective Mo-MuLV, the protease sequence in plasmid pBLS-wt was mutated by using the GeneEditor in vitro site-directed mutagenesis system (Promega) as instructed by the manufacturer along with the oligonucleotide 5′CGTCACCTTCCTGGTCGCGACTGGGGCCCAACAC3′ (bold letters represent changes from the wild-type sequence); the mutation results in replacement of the aspartic acid in the protease catalytic site with alanine. The existence of this mutation was verified by the presence of a new NruI site and by additional DNA sequence analysis. The mutated protease sequence was then exchanged for the wild-type sequence in pNCS, using the XhoI-SalI sites, to form plasmid pNCS-Prot−.
To create a Mo-MuLV with a deletion in the RT, a BclI-BlpI fragment was deleted from pNCA; the termini were blunted with the Klenow fragment of DNA polymerase I and ligated. An XhoI-SacII fragment harboring the deletion was then exchanged for the wild-type sequence in pNCS, using the same sites, to create pNCSΔRT.
A construct expressing hemagglutinin (HA)-tagged cyclophilin A was generously provided by J. Luban (Columbia University).
A C-terminally Myc-tagged, full-length nucleolin was constructed as follows. Total RNA extracted from WEHI cells was subjected to reverse transcription using oligo(dT) as a primer and SuperScript RNase H− RT (GIBCO-BRL) according to the manufacturer's protocol. The open reading frame of nucleolin (8) was amplified from the WEHI cDNA by PCR (Expand High Fidelity PCR system) as instructed by the manufacturer, using the oligonucleotides 5′CGGAATTCCGGCGCCGTAATCCGCCACC3′ and 5′TCCGCCCGGGCATTCAAACTTCGTCTTCTTTCC3′ (nucleotides that flank the nucleolin sequence are in boldface). The resulting amplified fragment contains an EcoRI site, 20 nucleotides of the 5′ untranslated region just upstream of the nucleolin start codon, and the nucleolin open reading frame with an SrfI site fused at its 3′ (lacking the stop codon). This fragment was cloned into the SmaI site in plasmid pUC18, using a SureClone ligation kit (Pharmacia) as instructed by the manufacturer, creating pUC18-1b. The EcoRI-SrfI fragment was excised from plasmid pUC18-1b and cloned into the same sites in pMT21myc (22), creating a full-length open reading frame of the mouse nucleolin cDNA, fused at its 3′ end in frame to the Myc epitope. N-terminally HA-tagged Nuc(212) was expressed from plasmid pCGN-Nuc(212), which was constructed by excision of the Nuc(212) cDNA insert from pGADNOT-Nuc(212), using XbaI and BglII sites (located at a short polylinker at the 5′ end of the cDNA and at the pGADNOT plasmid backbone, respectively), and was cloned into the XbaI and BamHI sites of the mammalian expression vector pCGN (66). An N-terminally HA-tagged, full-length nucleolin was constructed as follows. The full-length open reading frame of nucleolin was amplified from plasmid pUC18-1b by PCR, using the oligonucleotides 5′GGACCTTCTAGAATGGTGAAGCTCGCAAAGGCTGGC3′ and 5′CGGCGGAGATCTCTATTCAAACTTCGTCTTCTTTCC3′ (nucleotides that flank the nucleolin sequence are in boldface). The amplified PCR fragment was digested with XbaI and BamHI (an internal BamHI site exists in the C-terminal portion of the nucleolin sequence) and cloned into the same sites in pCGN-Nuc(212), to generate pCGN-Nucleolin. This plasmid encodes the full-length nucleolin cDNA fused at its amino terminus in frame to the HA epitope. Both HA-tagged and Myc-tagged full-length nucleolin migrated with the same apparent size as the endogenous nucleolin when extracts of cells expressing these proteins were analyzed by Western blotting using a monoclonal antibody against the N terminus of wild-type nucleolin (data not shown).
Transformation of 293T and COS-7 cells.
293T cells were transfected using calcium phosphate (52). Subconfluent COS-7 cells were transfected using DEAE-dextran (55). Expression was analyzed 2 days after transfection for both 293T and COS-7 cells.
RT assay.
Exogenous RT assay mixtures (68) were incubated for various times, and the radioactivity of the DNA product was quantitated by PhosphorImager analysis (26). Relative RT activity was calculated from the slope of the graph of radioactivity in DNA (in arbitrary pixel units), plotted against the reaction time (in minutes).
Equilibration centrifugation of virion particles.
Virions were purified on 25%/45% sucrose step gradients (62). Virions were also analyzed on 20 to 60% continuous sucrose gradients by centrifugation for 16 h at 80,000 × g at 4°C. Gradient fractions of 0.5 ml were collected; proteins were precipitated with trichloroacetic acid in the presence of 1 μg of bovine serum albumin, resuspended, and analyzed by SDS-gel electrophoresis and Western blotting.
Subtilisin digestion of virions.
COS-7 cells (3×100-mm plates) were transfected with 5 μg of pNCS expressing Mo-MuLV, together with 5 μg of plasmid expressing HA-Nuc(212). Two days later the virions were purified from culture supernatants by sucrose step gradients, and virion pellets were resuspended in subtilisin buffer (40 mM Tris [pH 8], 2 mM CaCl2). Various amounts of subtilisin (Boehringer Mannheim D-68298) were added; after incubation at room temperature, reactions were stopped by adding PMSF and aprotinin to final concentrations of 2 mM and 1 μg/ml, respectively. Particles were purified through a 25% sucrose cushion, and the pellets were resuspended and boiled in 2× loading buffer. For subtilisin treatment in the presence of detergent, NP-40 was added to a final concentration of 0.2%; after the reactions were stopped, the mixture was supplemented with an equal volume of 2× loading buffer and boiled.
Western blot analysis and antibodies.
Western blot analyses were performed using 10 to 12% polyacrylamide gels and Immobilon-P transfer membrane (Millipore) according to the manufacturer's protocol. Peroxidase-conjugated secondary antibodies were detected by staining the membrane with ECL Western blotting detection reagent (Amersham Pharmacia Biotech) according to the manufacturer's protocol. Goat polyclonal anti-Gag antiserum, raised against Rauscher MuLV, was obtained from the National Cancer Institute (product no. 79S-804). This antiserum cross-reacts with Mo-MuLV Gag and was used at a 1:5,000 dilution. Monoclonal anti-p15E envelope antibody, in culture supernatants of hybridoma line 42-114 (53), was diluted 1:2 before use. Mouse monoclonal antibody against HIV-1 capsid (HIV-018-48171) was purchased from Capricorn, Scarborough, Maine, and used at a 1:5,000 dilution. Mouse monoclonal antibody against the HA epitope (Boehringer Mannheim) was used at a final concentration of 0.4 μg/ml. Peroxidase-conjugated polyclonal antisera raised against goat immunoglobulin G (Boehringer Mannheim) and mouse immunoglobulin G (Amersham Life Sciences) were used at 1:10,000 and 1:3,000 dilutions, respectively.
Transformation of Phoenix cells and assays for transduction.
293T-based, amphotropic or ecotropic Phoenix helper cell lines (34) (generously provided by G. Nolan, Stanford University) were transfected with 2 μg of a Mo-MuLV-based, green fluorescent protein (GFP)-containing retroviral vector (MFG-GFP; generously provided by G. Nolan) and the indicated plasmid DNAs, using the calcium phosphate method (52). The next day the supernatants were replaced with 3 ml of fresh medium. Two days after transfection, the cells were analyzed by fluorescence-activated cell sorting (FACS) for GFP fluorescence. Mock-transfected cells were used to determine the background autofluorescence level of GFP-negative cells. Total fluorescence of the GFP-positive cell population was calculated by multiplying the percentage of GFP-positive cells with their mean fluorescence value. Supernatants of transfected cells were filtered through a 0.45-μm-pore-size filter (GelmanSciences) and diluted 1:3 with culture medium; then Polybrene was added to a final concentration of 5 μg/μl. These virus preparations (2 ml) were used to infect NIH 3T3 cells (2 × 105 cells in 60-mm-diameter dishes) for 2 h. Two days later, fluorescence of the infected, GFP-positive cells was measured by FACS and total fluorescence was calculated. Normalized infectivity was calculated by dividing the total fluorescence of infected cells by the total fluorescence of transfected cells. FACS analysis of GFP-positive cells revealed that between 15 and 45% of the transfected Phoenix cells were transfected and that between 0.1 and 55% of the NIH 3T3 cells were infected.
RESULTS
Yeast two-hybrid screen for MuLV Gag-interacting proteins.
To screen for cellular proteins that interact with the MuLV Gag protein, the LexADB-Ngag fusion protein was used as a bait to screen a mouse cDNA library of sequences expressed as fusions to the Gal4AD. Out of approximately 106 yeast colonies screened, we identified 12 plasmids that reproducibly activated the lacZ reporter gene in the presence of LexADB-Ngag. Three of these clones were independent isolates of the same cDNA, approximately 1.3 kb long, showing complete identity to mouse nucleolin (8). In these clones the Gal4AD sequence was fused in frame to the last 212 amino acids of nucleolin [Nuc(212)]. Nucleolin is a ubiquitous, abundant 100-kDa phosphoprotein with a complex structure and diverse activities (for a recent review, see reference 29). The carboxy two-thirds of nucleolin consists of four RNA-binding domains, also called RNA recognition motifs (RRM) (8–10, 18), followed by a fifth distinct domain, rich in glycine, dimethylarginine, and phenylalanine, called a glycine-arginine-rich (GAR) domain (36, 43). The GAR domain also binds RNA (28, 36, 49) and possesses a nucleic acid helicase activity (27, 69). The region recovered in the yeast plasmid contains a partial copy of the third RRM, a complete copy of the fourth RRM, and the complete GAR helicase region. Three other clones recovered in the screen had no similarity to nucleolin but showed similarity to the DEAD box family of RNA helicases; the corresponding gene was named GIP2.
Gag-Nuc(212) interactions in the yeast two-hybrid system.
To evaluate the range of Gag proteins that interact with Nuc(212), various pairs of plasmids encoding different fusion proteins were tested in the yeast two-hybrid system (Table 1). Gag proteins of Mo-MuLV and of N- and B-tropic viruses fused to LexADB (LexADB-Mgag, -Ngag, and -Bgag, respectively) all interacted strongly with Gal4AD-Nuc(212). Gag proteins from RSV and SIV fused to Gal4DB (Gal4DB-RSVgag and SIVgag, respectively) also interacted strongly with Gal4AD-Nuc(212). However, the Gag protein of HIV-1 fused to Gal4DB (Gal4DB-HIV) showed only weak interaction, and the Gag protein of MPMV fused to Gal4DB (Gal4DB-MPMV) did not detectably interact. All of these Gag constructs did not interact with Gal4AD alone, and Gal4AD-Nuc(212) did not interact with an irrelevant protein (lamin) fused to LexADB, with LexADB alone, or with Gal4DB alone. These results indicate that a specific interaction occurs between Nuc(212) and many, though not all, Gag proteins in the yeast two-hybrid system.
TABLE 1.
β-Gal activity in yeast expressing different Gag and Nuc(212) hybrid proteinsa
| Protein | Partner
|
|
|---|---|---|
| Gal4AD-Nuc(212) | Gal4AD | |
| LexADB-Ngag | + | − |
| LexADB-Bgag | + | − |
| LexADB-Mgag | + | − |
| LexADB-CANC | + | − |
| LexADB-MgagΔNC | − | − |
| LexADB-NC | + | − |
| LexADB-lamin | − | − |
| LexADB | − | − |
| Gal4DB-Mgag | + | − |
| Gal4DB-RSVgag | + | − |
| Gal4DB-MPMVgag | − | − |
| Gal4DB-SIVgag | + | − |
| Gal4DB-HIVgag | +/− | − |
| Gal4DB | − | − |
Yeast were transformed with different combinations of plasmids encoding the indicated fusion proteins. The LexADB or Gal4DB fusion proteins were expressed in yeast strain CTY10-5d or GGY::171, respectively. Yeast colonies were lifted and stained for β-Gal activity as described elsewhere (5). Entries indicate the presence or absence of blue color with transformants. +/− indicates only faint blue color.
The region of the Mo-MuLV Gag protein required for interaction with Nuc(212) was determined by testing LexADB fusions with various domains of Gag for interaction with Gal4AD-Nuc(212) (Table 1). A LexADB fusion containing MA, p12, and CA but lacking NC (LexADB-MgagΔNC), did not interact with Gal4AD-Nuc(212), indicating that the NC domain was required. A fusion containing the CA and NC domains, LexADB-CANC, interacted as strongly as the complete Gag protein. LexADB fused to NC alone (LexADB-NC) did interact, though this interaction was somewhat weaker than the interaction with LexADB-CANC. These data suggest that the NC domain is necessary and sufficient for the interaction of Nuc(212) and that addition of the CA domain to the NC domain may stabilize or enhance this interaction.
Nuc(212) binds Mo-MuLV Gag in vitro.
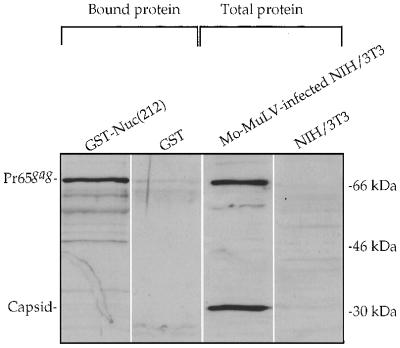
To verify the results from the yeast two-hybrid system, we tested for the ability of Nuc(212) to bind to Mo-MuLV Gag in vitro. A GST fusion protein containing Nuc(212), or GST protein alone, was expressed in bacteria, extracted, and incubated with lysates of Mo-MuLV-infected NIH 3T3 cells. The GST and GST-Nuc(212) proteins were then recovered by binding to glutathione beads. The beads were washed, and the bound proteins were eluted and analyzed by SDS-gel electrophoresis followed by Western blotting using polyclonal anti-Gag antisera (Fig. 1). Analysis of the initial Mo-MuLV-infected NIH 3T3 lysates revealed the presence of the full-length Gag precursor, Pr65gag, and the mature CA domain, p30. The GST-Nuc(212) fusion protein efficiently bound the full-length Gag protein but not CA. In contrast, beads containing the GST protein alone did not bind the full-length Gag from the same lysate. These data demonstrate that Nuc(212) can specifically bind to the full-length Mo-MuLV Gag protein in vitro.
FIG. 1.
In vitro binding of Mo-MuLV Gag to Nuc(212). An extract of Mo-MuLV-infected NIH 3T3 cells was divided and mixed with extracts of bacteria expressing either GST or GST-Nuc(212); the GST proteins were recovered on glutathione-coated beads. Proteins in whole lysates and proteins bound to the beads and eluted were analyzed by Western blotting using a polyclonal antibody against capsid protein. Migration of size marker is shown on the right. Positions of Pr65gag and capsid are indicated on the left.
Incorporation of Nuc(212) into Mo-MuLV virions.
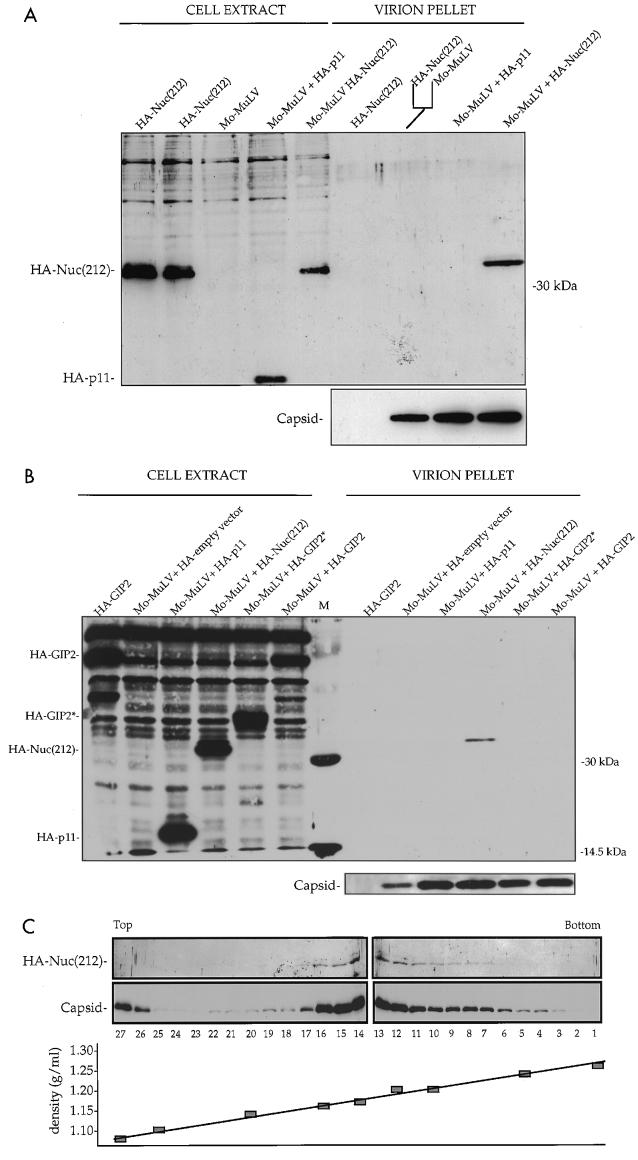
To investigate whether Nuc(212) can interact with Mo-MuLV Gag in vivo, we tested for the incorporation of Nuc(212) into virion particles. COS-7 cells were transfected with DNAs containing the complete Mo-MuLV genome together with DNA encoding the Nuc(212) fragment tagged with an influenza virus HA epitope. Plasmid DNA encoding an irrelevant HA-tagged protein (annexin II light chain; hereafter called HA-p11) was used as a negative control. Virions were purified from the culture supernatants of the transfected cells by sucrose step gradients, and virion proteins recovered in the pellets were analyzed by gel electrophoresis followed by Western blotting (Fig. 2A). Probing the membrane with monoclonal anti-HA antibody revealed that the HA-Nuc(212) protein was present at high levels in the virions isolated from cultures expressing both Mo-MuLV and HA-Nuc(212) protein. No HA-Nuc(212) protein was detected in virion preparations from cultures that separately expressed HA-Nuc(212) or Mo-MuLV alone, nor was it detected if the supernatants from the singly transfected cells were mixed before the virion purification step. Coexpression of Mo-MuLV with HA-p11 did not result in the recovery of HA-p11 in the virion pellet. These data indicate that Nuc(212) interacts with Mo-MuLV in vivo and is associated with the virions.
FIG. 2.
In vivo binding of Mo-MuLV Gag to Nuc(212). Five micrograms of plasmid expressing Mo-MuLV and/or 5 μg of plasmid expressing the indicated HA-tagged protein were transiently expressed in COS-7 cells. All plasmids contained the SV40 origin of replication. Two days later the cells were extracted, and the virions were purified from supernatants, analyzed by Western blotting with a monoclonal antibody against the HA epitope (top panels), and reprobed with a polyclonal antibody against the capsid protein (bottom panels). (A and B) Virions purified through 25 and 45% sucrose step gradients, followed by a second purification step through a 25% sucrose cushion. About 2% of each cell extract and 30% of each virion pellet were analyzed by Western blotting. The fork-like line indicates that the supernatants of the indicated transfections were mixed prior to virion purification. M, protein size marker. Positions of migration of size marker bands (right) and of capsid and HA-tagged proteins (left) are shown. (C) Virions purified through a 25% sucrose cushion, followed by purification through a 20 to 60% continuous sucrose gradient. 1 to 27, gradient fraction numbers. Top and Bottom designate the top and bottom gradient fractions, respectively. The locations of HA-Nuc(212) and capsid are shown on the left. The plot of the gradient density is shown at the bottom.
Not all proteins that interact with Gag in the yeast system are incorporated into virions in mammalian cells. Two segments of GIP2, another protein that also interacted strongly with Mo-MuLV NC in the yeast two-hybrid system, were expressed with HA tags (HA-GIP2, and HA-GIP2*) and tested for incorporation into virions (Fig. 2B). Although these proteins were expressed at high levels in the virion-producing cells, neither HA-GIP2 protein could be detected in the purified virions released by these cells. Thus, the interaction of Nuc(212) with Gag is distinct in that it results in efficient incorporation into virion particles.
To probe the association of Nuc(212) with Mo-MuLV particles further, we transiently expressed in COS-7 cells both Mo-MuLV and HA-Nuc(212), purified the virions from supernatants of transfected cultures by sucrose step gradients, and then separated the particles by equilibrium centrifugation in a 20 to 60% linear sucrose gradient. Fractions were collected from the gradient and analyzed by Western blotting (Fig. 2C). Probing the fractions with antibodies directed against either the HA tag or the capsid protein revealed that the peak of the HA-Nuc(212) coincided precisely with the peak of the capsid protein. Control experiments showed that Mo-MuLV virions could be readily separated from an irrelevant membrane protein, the Tyro3 receptor (data not shown). These results suggest that the Nuc(212) is genuinely associated with the viral particles.
Nuc(212) is incorporated inside Mo-MuLV virions.
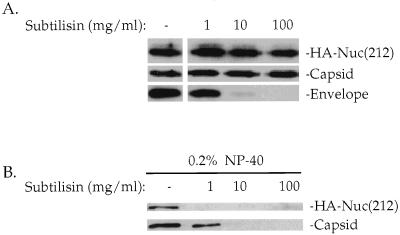
HA-Nuc(212) might in principle copurify with Mo-MuLV particles because of an association with membrane vesicles with a density similar to the density of the virions (50) or because it can bind to the outer surface of the budding virions. Cellular proteins associated with vesicles, however, are sensitive to digestion by subtilisin, while proteins in the virion core are protected from digestion by the virion envelope (50). To test whether Nuc(212) was localized inside the viral envelope, virions were purified by sucrose step gradients from supernatants of COS-7 cells that coexpressed Mo-MuLV and HA-Nuc(212) and digested with various concentrations of subtilisin (50). After digestion, the particles were purified through a 25% sucrose cushion, and proteins in the pellets were separated by SDS-gel electrophoresis and detected by Western blotting. The envelope protein on the virion surface was degraded by treatment of the virions with low levels of protease, but the internal Gag protein and the HA-Nuc(212) protein were protected from digestion even at very high levels of protease (Fig. 3A). The virion-associated Gag and HA-Nuc(212) proteins were digested by low levels of protease, however, if the treatments were performed in the presence of detergent to disrupt the virion membrane (Fig. 3B). These experiments strongly suggest that the HA-Nuc(212) is incorporated inside the virions.
FIG. 3.
Subtilisin treatment of Mo-MuLV virions. COS-7 cells were transiently transfected with plasmids expressing Mo-MuLV and HA-Nuc(212) protein, and virions were purified as for Fig. 2A. Twenty percent of the pellet was digested with the indicated amount of subtilisin, after which PMSF and aprotinin were added to terminate the digestion. (A) Subtilisin digestion in the absence of detergent for 17 h at room temperature, followed by particle purification through 25% sucrose cushion. Thirty percent of the pellet was analyzed by Western blotting. (B) Subtilisin digestion in the presence of 0.2% NP-40 for 1 h at room temperature. Twenty percent of the sample was analyzed by Western blotting. The membranes were probed with a monoclonal anti-HA epitope antibody reprobed with a polyclonal anticapsid serum, and probed again with a monoclonal antienvelope antibody, as indicated on the left. The locations of HA-Nuc(212) and capsid are shown on the right.
Nuc(212) incorporation into virions requires the NC domain.
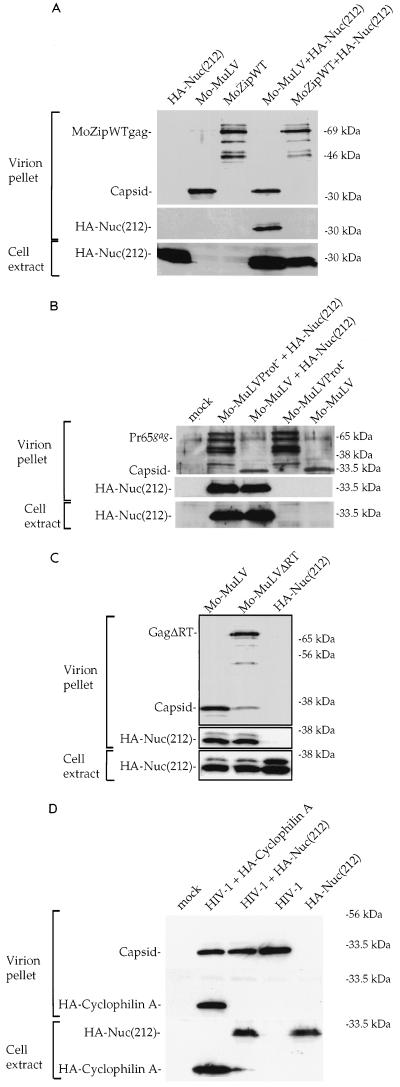
The incorporation of Nuc(212) into virions might be mediated by nonspecific interactions with the membrane, or even by the flow of protein by mass action into assembling particles, or could result from direct binding to Gag. To determine whether Nuc(212) incorporation required a specific interaction with the Gag NC domain, we tested both a modified Mo-MuLV Gag lacking its NC domain and the HIV-1 Gag for the ability to incorporate Nuc(212). Deletion of NC from Mo-MuLV Gag causes a drastic reduction in the yield of virions, precluding a direct test of a mutant Gag simply lacking NC. However, Zhang et al. generated an assembly-competent variant of the Mo-MuLV Gag (MoZipWT) in which the NC domain was replaced by a foreign protein, the BZip domain from human CREB (77). The substituted region of the resulting construct, pXM-MoZipWT, replaces the assembly function of the NC domain to allow the formation of high levels of immature virion particles. COS-7 cells were transfected with DNAs expressing MoZipWT and HA-Nuc(212), and virions were recovered on sucrose step gradients as before. Western blot analysis showed that although substantial levels of particles were produced, no HA-Nuc(212) associated with these particles was detected (Fig. 4A). Thus, the incorporation of Nuc(212) into Mo-MuLV virions required the presence of the NC domain.
FIG. 4.
In vivo incorporation of HA-Nuc(212) by different Gag proteins. The ability to incorporate HA-Nuc(212) by different viruses was compared to that of Mo-MuLV. COS-7 transfection, virion purification, and Western blot analysis were done as described for Fig. 2A. The lower panel shows analysis of the cell extracts for the expression of HA-Nuc(212), and the upper panels show analysis of the virion pellets for the expression of HA-Nuc(212) and Gag proteins. Shown is comparison between Mo-MuLV and Mo-MuLV with a modified Gag protein in which the NC domain was replaced with the BZip domain from human CREB (MoZipWT) (A), Mo-MuLV with an inactivated protease (Mo-MuLVProt−) (B), Mo-MuLV lacking RT (Mo-MuLVΔRT) (C), and HIV-1 (D). Note that the deletion in Mo-MuLVΔRT causes poor Gag-Pol processing although this virus contains a wild-type protease. HIV-1 was expressed from plasmid pHIV-HSA.
The pXM-MoZipWT construct expresses only the altered Gag protein and does not allow incorporation of Gag-Pol, processing of Gag by the viral protease, or virion maturation. To test whether these properties were responsible for the inability of the variants to incorporate Nuc(212), additional mutants of Mo-MuLV were used in the assay. Mo-MuLV carrying a point mutation that inactivated the protease (Mo-MuLVProt−) and a variant with a deletion affecting RT (Mo-MuLVΔRT) were able to induce high levels of virions; when coexpressed with HA-Nuc(212), the resulting virions were found to contain high levels of the tagged protein, similar to the levels in wild-type virions (Fig. 4B and C). Thus, Gag processing and an intact Gag-Pol protein are not required for incorporation of Nuc(212). Thus, the failure of MoZipWT to incorporate Nuc(212) must be attributed to its lack of an NC domain.
The initial tests for interaction of Nuc(212) with retroviral Gags in yeast indicated only a very weak interaction with the HIV-1 Gag (Table 1). To test for the incorporation of Nuc(212) into virions assembled by the HIV-1 Gag, COS-7 cells were transfected with DNAs containing the HIV-1 provirus and DNA encoding Nuc(212), and virions were collected and analyzed as before. Particles assembled by HIV-1 Gag did not incorporate detectable HA-Nuc(212) but did incorporate HA-tagged cyclophilin A, known to be packaged into HIV-1 virions (Fig. 4D). Examination of very high amounts of HIV-1 particles, encoded by a different construct expressing high levels of HIV Gag, similarly failed to detect any incorporated HA-Nuc(212) protein (data not shown). Estimates from these blots suggest that there was at least 30-fold less incorporation of HA-Nuc(212) than HA-cyclophilin A into the HIV virions. As before, HA-Nuc(212) could not be detected in mock virion preparations from supernatants of control cultures that separately expressed either HA-Nuc(212), Mo-MuLV, MoZipWT, or HIV alone. These data strongly suggest that HA-Nuc(212) protein is specifically incorporated into Mo-MuLV particles, and not generally assembled into budding viruses, and that this incorporation is dependent not on the presence of Gag-Pol protein or on Gag processing but rather on the presence of the Mo-MuLV NC domain.
Overexpression of Nuc(212), but not full-length nucleolin, inhibits retroviral infectivity.
The interaction of Nuc(212) with Mo-MuLV Gag in vivo could have various consequences; very high-level expression of Nuc(212) might sequester Gag, block Gag-Gag interactions, or otherwise interfere with normal virion assembly. To test this possibility, we developed a quantitative assay which measures the effect of high-level expression of a candidate inhibitory gene on the ability of a producer cell line to release transducing virus. Phoenix cells, which constitutively express the Mo-MuLV Gag, Pol, and Env proteins, were cotransfected with a retroviral vector DNA transducing the GFP marker and a DNA encoding a potential inhibitor. The transfected cells could be analyzed by flow cytometry to determine the number of cells transfected and the level of expression of the GFP marker. The virus produced by these cells could then be collected, and the yield of infectious virus could be determined by infecting fresh NIH 3T3 cells and analyzing these recipients for the GFP marker by flow cytometry. To achieve high-level expression of the inhibitors, the DNAs encoding these products contained the SV40 origin of replication and thus would be amplified to high copy number by the T antigen present in the Phoenix cells.
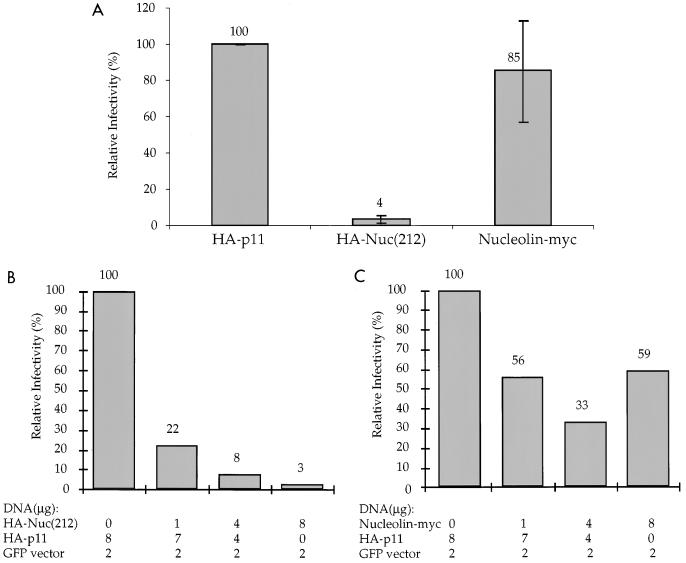
Transfection of the Phoenix cells with the GFP vector and various test DNAs showed efficient expression of the GFP marker and no inhibition of expression in the transfected cells by most of the DNAs (data not shown). Expression of HA-Nuc(212), however, caused a drastic reduction in the production of transducing virus. In three experiments, coexpression of the HA-Nuc(212) caused an average 25-fold reduction in the infectivity of the retroviral vector compared to the control HA-p11 protein (Fig. 5A). Coexpression of a full-length nucleolin carrying a Myc epitope tag at the C terminus (nucleolin-Myc) did not significantly reduce the vector titer. To test whether the inhibition was dose dependent, various ratios of the DNA encoding HA-Nuc(212) to the vector expressing HA-p11 were used; the results showed a dose-dependent decrease of the titer of the retroviral vector by increasing levels of HA-Nuc(212) (Fig. 5B). The nucleolin-Myc construct did not show a similar inhibition (Fig. 5C). These results suggest that the high-level expression of Nuc(212) can virtually abolish release of infectious virus, without significant effect on the level of marker gene expression in virus-producing cells.
FIG. 5.
Overexpression of HA-Nuc(212) in a helper cell line reduces the infectivity of the released virions. (A) Phoenix helper cells were transfected with 8 μg of HA-p11, HA-Nuc(212), or nucleolin-Myc expression plasmid, together with 2 μg of a GFP-containing retroviral vector. The HA- and Myc-tagged proteins, but not the GFP vector, were expressed from plasmids containing the SV40 origin of replication. The graph represents the average of three independent experiments. In each experiment, normalized infectivity was calculated (see Materials and Methods) for each transfection. Infectivity is reported relative to the HA-p11 control. (B and C) Phoenix helper cells were transfected with the indicated plasmid mixtures, and the normalized infectivity was calculated. Infectivity is reported relative to the control, where no HA-Nuc(212) (B) or nucleolin-Myc (C) was expressed.
To test whether Nuc(212) could block virus infection during virus entry, COS-7 cells were transfected with DNAs overexpressing HA-Nuc(212), or HA-p11 as a control, and then were challenged with GFP transducing virus preparations pseudotyped with the vesicular stomatitis virus G envelope protein. Similar numbers of infected cells were observed for both HA-Nuc(212) and HA-p11-expressing cells (data not shown). Thus, overexpression of HA-Nuc(212) did not block susceptibility to virus infection. These results also indicate that overexpression of the protein did not cause a gross block to cellular metabolism.
High-level expression of Nuc(212) inhibits Mo-MuLV virion release.
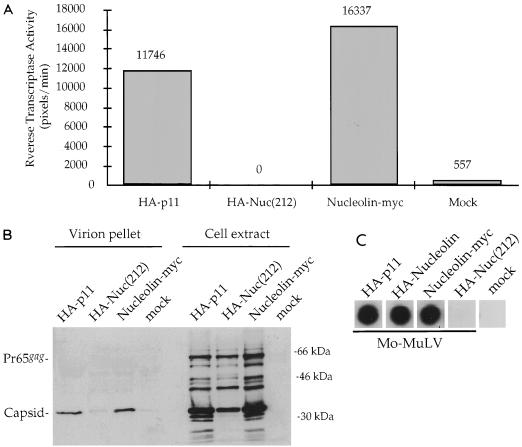
The inhibition of release of infectious GFP virus mediated by Nuc(212) could be attributed either to a reduction in the infectivity of the released virus or to a reduction in the actual number of particles released by the cells. To determine which was the case, we tested whether the expression of HA-Nuc(212) could block the production of virion particles expressed from a wild-type Mo-MuLV provirus introduced along with the expression plasmid. 293T cells were cotransfected with Mo-MuLV DNA and candidate inhibitor constructs, and the culture supernatants were assayed for RT activity as a measure of virion particle release. The coexpression of HA-Nuc(212) almost completely abolished the release of RT, whereas the control protein HA-p11 had no such effect (Fig. 6A). The full-length nucleolin-Myc had no inhibitory effect.
FIG. 6.
Overexpression of HA-Nuc(212) in producer cells reduces Mo-MuLV release. 293T cells were transiently transfected with 8 μg of HA-p11, HA-Nuc(212), or nucleolin-Myc expression plasmid, together with 2 μg of plasmid expressing Mo-MuLV. The HA- and Myc-tagged proteins, but not Mo-MuLV, were expressed from plasmids containing the SV40 origin of replication. Levels of viral protein expression were determined 2 days later. (A) Quantitative RT assay (see Materials and Methods) of unpurified virions in supernatants of transfected cells. The HA- or Myc-tagged proteins that were coexpressed with Mo-MuLV are indicated below the columns. RT activity is represented in arbitrary pixel units, quantitated by a PhosphorImager, divided by the reaction time in minutes. Mock, transfection without adding plasmid DNA. (B) Cell extracts and purified virions from the experiment described for panel A were analyzed by the Western blot procedure as described for Fig. 2A. The membrane was probed with polyclonal antibodies against capsid. Positions of migration of size marker bands (right) and of full-length Pr65gag and capsid (left) are shown. (C) Exogenous RT assay of unpurified virions from supernatants of transfected cells. The HA- or Myc-tagged proteins indicated at the top were coexpressed with Mo-MuLV. MoZipWT virus that lacks RT was used as a mock control.
To test for the inhibition of release of viral proteins directly, culture supernatants were collected, virions were purified by sedimentation through sucrose density step gradients, and the proteins were analyzed by SDS-gel electrophoresis followed by Western blotting with an anticapsid antiserum (Fig. 6B). Overexpression of HA-Nuc(212) caused a drastic reduction in the levels of CA protein (p30) recovered in the virion fractions. Examination of the intracellular viral proteins in the transfected cells showed high levels of the Pr65gag precursor, comparable to the control, and a modest reduction in the yield of mature capsid and other processing intermediates seen in the uninhibited controls. Thus, HA-Nuc(212) did not prevent Gag expression but reduced its processing to mature forms and strongly inhibited its release into the culture medium.
To test whether the different abilities of HA-Nuc(212) and nucleolin-Myc to inhibit RT release could be attributed to the different positions and identities of the epitope tag, a full-length nucleolin with an HA tag at the N terminus (HA-nucleolin) was constructed and tested (Fig. 6C). This full-length version, like nucleolin-Myc, showed no inhibition of RT release.
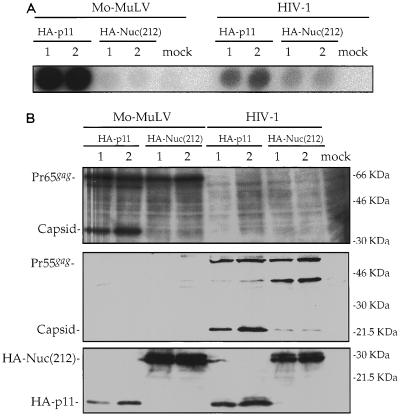
If the inhibition of particle release requires interaction between Nuc(212) and Gag, there should be less inhibition of HIV-1 assembly, since the interaction of Nuc(212) with HIV-1 Gag is weak. Coexpression of HA-Nuc(212) with the HIV-1 genome in 293T cells caused only a very modest reduction in the levels of RT recovered in the culture supernatant (Fig. 7A). Controls with the Mo-MuLV genome showed the expected nearly complete loss of released RT (a 200-fold reduction for Mo-MuLV, versus only a 2.4-fold reduction for HIV-1). Western blot analysis of extracts from the producer cells showed that the Gag proteins of Mo-MuLV and HIV-1 were both produced in high amounts; processing of the Mo-MuLV Gag was strongly reduced by Nuc(212), while processing of the HIV-1 Gag was only moderately affected (Fig. 7B). Cotransformation of the cells with a luciferase expression construct resulted in good expression of the marker, with no significant inhibition in the levels of activity caused by Nuc(212) (data not shown). These results suggest that overexpression of HA-Nuc(212) does not have a general cytotoxic effect on protein production, but rather specifically inhibits virion assembly or release of Mo-MuLV, and that this inhibition is much more effective for Mo-MuLV than for HIV-1.
FIG. 7.
Overexpression of HA-Nuc(212) strongly reduces Mo-MuLV, but not HIV-1, particle release. 293T cells were transiently transfected in duplicates (1 and 2) with 0.1 μg of plasmid expressing luciferase, 2 μg of plasmid expressing either Mo-MuLV or HIV-1, and 8 μg of plasmid expressing either HA-p11 or HA-Nuc(212). All plasmids except those expressing the viruses contained the SV40 origin of replication. HIV-1 was expressed from pNLENV-1. (A) Exogenous RT assay with unpurified virions in supernatants of transfected cells. Mock, transfection without adding plasmid DNA. (B) Cell extracts were analyzed by Western blotting, and the membrane was probed with a polyclonal anti-MuLV capsid serum (top), monoclonal anti-HIV-1 capsid antibody (middle), and monoclonal anti-HA epitope antibody (bottom). Positions of migration of size marker bands (right) and of full-length Mo-MuLV Gag (Pr65gag), Mo-MuLV capsid, full-length HIV-1 (Pr55gag), HIV-1 capsid, HA-Nuc(212), and HA-p11 (left) are shown.
Isolation of a Mo-MuLV Gag mutant with a single point mutation unable to interact with Nuc(212).
One possible explanation for the ability of Nuc(212) to inhibit assembly is that it binds to and masks residues in the Gag protein that are important for assembly. Thus, Gag mutants that are specifically selected for the loss of the ability to bind to Nuc(212) might also show defects in assembly. To identify such noninteracting mutants, we used the reverse two-hybrid system (70, 71). A plasmid expressing a Gal4DB fusion to the Mo-MuLV Gag CA-NC region (pAS1-CANC) was subjected to random mutagenesis by passage through a mutator E. coli strain. The DNA was then introduced into the reverse two-hybrid yeast strain BY3171 already expressing the Gal4AD-Nuc(212) protein, and Ura− transformants were isolated by selection for growth on 5-FOA. Out of approximately 104 transformants placed under selection, a single clone was isolated and retested as a true noninteracting mutant (see Materials and Methods). Nucleotide sequence analysis of the clone revealed a single nucleotide substitution changing a single codon and specifying a change of the penultimate residue of CA, a leucine, to proline (L477P). The location of this change near the CA-NC boundary is consistent with the contribution of both CA and NC to the interaction with Nuc(212).
To further demonstrate that only the L477P mutation, and not alterations outside the CA-NC domain, was responsible for the loss of interaction with Nuc(212), the CA-NC sequences were subcloned from the original plasmid into pGADNOT to create a mutant version of Gal4AD-CANC. Tests showed that the mutant L477P Gal4AD-CANC failed to interact with LexADB-Nuc(212) in the yeast strain CTY10-5d (Table 2). The mutant did, however, continue to form multimers with LexADB-CANC and to interact normally with LexADB-GIP2. These results indicate that the mutant Gal4AD-CANC L477P protein was properly expressed and still able to interact with most of its binding partners. Thus, the mutation caused a specific block in the interaction between Mo-MuLV CA-NC and Nuc(212).
TABLE 2.
β-Gal activity in yeast expressing CA-NC, CA-NC L477P, and Nuc(212) hybrid proteinsa
| Protein | Partner
|
||
|---|---|---|---|
| Gal4AD-CANC L477P | Gal4AD-CANC | Gal4AD | |
| LexADB-Nuc(212) | − | + | − |
| LexADB-GIP2 | + | + | − |
| LexADB-CANC | + | + | − |
| LexADB | − | − | − |
Yeast strain CTY10-5d was transformed with different combinations of plasmids encoding the indicated fusion proteins. Yeast colonies were lifted and stained for β-Gal activity as described in the footnote to Table 1. Entries indicate the presence or absence of blue color with transformants.
The L477P mutant of Mo-MuLV fails to assemble virion particles.
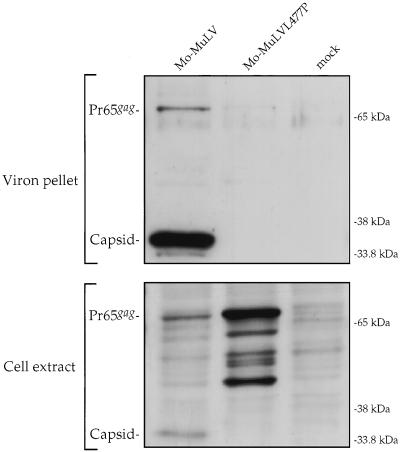
To determine the effects of the L477P mutation on viral replication and virion assembly, the mutation was transferred to the complete proviral genome. Rat fibroblast cells were transfected with the mutant DNA, and the spread of virus in the cultures was tested by assaying for RT activity in the culture medium. Whereas wild-type viral DNAs induced the appearance of virus and associated RT after 4 days, the mutant DNA did not result in the formation of detectable virus even after 1 month of culture (data not shown). These results suggested that some step in the life cycle was blocked by the mutation. To test for the ability of the mutant to mediate virion assembly and release directly, 293T cells were transfected with the L477P mutant or the wild-type DNA, and the yield of virus released into the culture supernatant was measured by RT assays. Whereas the wild-type DNA induced the release of high levels of virion-associated RT, the mutant DNA produced dramatically reduced RT levels, only slightly above background (data not shown). Virion particles were isolated from these culture supernatants, and the levels of CA protein in the virions and in cell lysates were assessed by Western blotting (Fig. 8). The preparations of wild-type virions showed substantial levels of CA protein, while the L477P mutant preparations contained no detectable CA. In contrast, the intracellular lysates showed higher levels of the Gag proteins in cells expressing the L477P mutant than in those expressing the wild-type. Thus, the L477P mutant is defective in assembly or release, and the Gag products accumulate within the producer cell. As for most assembly-defective mutants, processing of the Gag precursor is impaired in the L477P mutant.
FIG. 8.
Mo-MuLV harboring the L477P mutation fails to assemble. 293T cells were transfected with 10 μg of plasmid expressing either Mo-MuLV or L477P mutant. Mock, transfection without adding plasmid DNA. Cell extracts (bottom) and purified virions (top) were analyzed by Western blotting as described for Fig. 2A. The membrane was probed with polyclonal anticapsid serum. Positions of migration of size marker bands (right) and of full-length Gag (Pr65gag) and capsid (left) are shown.
DISCUSSION
The results presented here demonstrate that the C-terminal portion of nucleolin interacts specifically with the MuLV Gag protein in a variety of settings. The interaction could be readily detected in yeast as binding between two fusion proteins: in vitro, through the binding of GST-Nuc(212) to the MuLV Gag precursor; and in vivo, through the NC-dependent incorporation of Nuc(212) into MuLV virion particles. We suggest that the interaction was mediated by a direct protein-protein contact rather than by a nucleic acid bridge between the two. Very small alterations of Gag—a single amino acid substitution—could abolish the interaction without affecting interactions with other proteins and without any obvious change in the RNA binding sequences. Additionally, HIV NC binds RNA but does not bind nucleolin well. Nevertheless, we note that both Gag and nucleolin are nucleic acid-binding proteins, and it is plausible that their interaction could be stimulated or stabilized by bridging nucleic acids.
Nuc(212) interacted with the Gags from many, though not all, retroviruses. The strongest signals were obtained with the MuLVs, while only very weak signals were observed with MPMV and HIV-1. The broad interactions with many Gags presumably reflected a conserved structure of the NC domain, the major site of interaction, among many but not all retroviruses. A surprising consequence of this interaction is the incorporation of Nuc(212) into the virion particle. This incorporation required the presence of the NC domain, and moreover of an NC from a virus that could interact with Nuc(212). Thus, a variant virus in which the NC domain was replaced by a foreign protein did not incorporate Nuc(212), nor could HIV-1 Gag mediate detectable incorporation of the protein. These results correlate completely with the binding seen in yeast and suggest that a direct binding to Gag is required for incorporation. However, it should be noted that the binding of proteins to Gag was not sufficient to mediate the similar incorporation of all such proteins; GIP2, another Gag-interacting protein, bound to the same region of Gag but was not detectably incorporated. The incorporation of Nuc(212) was seen in cells overexpressing an epitope-tagged C-terminal portion of nucleolin, and it remains unclear whether this reflects a role for the endogenous nucleolin in the assembly process (see below). At a minimum, the incorporation of Nuc(212) is strong evidence for its binding to Gag in vivo. We are currently investigating whether Nuc(212) can be used as a targeting domain to direct the efficient encapsidation of fusions to heterologous proteins.
The inhibition of virion release by Nuc(212) was the most striking effect of its high-level overexpression in virus-infected cells. The inhibition was strongly dose dependent; at low levels of expression, there was no effect on virion production, and the only consequence was the incorporation of Nuc(212) into virions. But if the levels of Nuc(212) were very high, virtually all virion release could be blocked. This inhibition was extremely potent; of many Gag-interacting proteins that we have tested, Nuc(212) caused by far the most significant block to virus production. The inhibition was manifested by strong decreases in the release of virus titer, virion-associated RT, and virion proteins, suggesting a block to virion release per se rather than merely an effect on virus infectivity. There are several possible mechanisms by which Nuc(212) could inhibit assembly. Because nucleolin is involved in several steps of ribosome biogenesis, overexpression of a truncated form of nucleolin might be supposed to interfere with general translation, which could lead to reduced amounts of released virus. However, while overexpression of Nuc(212) reduced Gag levels in the supernatants, it did not significantly affect the levels of coexpressed luciferase activity, strongly suggesting that the inhibition of virus release is not the result of a general toxic effect. More importantly, similar levels of Pr65gag were produced in cells overexpressing Nuc(212) and in control cells, arguing that the protein specifically blocks assembly per se. In addition, while Mo-MuLV release was strongly inhibited, HIV-1 release was only slightly reduced, indicating that a tight interaction with Gag is required.
One likely mechanism by which assembly could be inhibited is that Nuc(212) could bind to and directly prevent the multimerization of the Gag precursors required for virion assembly. In this view, Nuc(212) may serve as an “intracellular antibody.” The binding of Nuc(212) to NC could mask either NC-NC or NC-RNA interactions, which are thought to play a role in assembly (11, 13, 15, 65). It might also be that the binding of Nuc(212) disrupts assembly by masking the CA-NC junction. Indeed, the L477P mutation, which is adjacent to this junction, disrupted the binding to Nuc(212) and also dramatically inhibited virion assembly. Thus, the CA-NC boundary is a region required for assembly, and masking it or changing its structure results in a defect in assembly. Supporting this idea is the recent observation that Mo-MuLV Gag proteins can be cross-linked at their C-terminal CA residues to form dimers by a cysteine-specific cross-linking agent (47). In addition, an analogous region in HIV-1 (the C-terminal capsid-p2 domain of Pr55gag) is part of the minimal gag sequences needed for efficient assembly of retrovirus-like particles (2). The reduction in processing of the Gag precursor observed in Nuc(212)-expressing cells is consistent with this model; activation of the viral protease and Gag cleavage depends on normal multimerization of Gag and Gag-Pol proteins associated with assembly (44, 57, 61).
Another possibility is that the Nuc(212) fragment acts as a dominant negative allele that interferes with a positive assembly function of the endogenous wild-type nucleolin during virion assembly. In this case, the wild-type nucleolin would need to bind to Gag to promote its assembly. The NC domain contains the I domain, an important assembly domain for many retroviruses (65), and nucleolin binding may be a part of that domain's function. It has been suggested that a nonspecific interaction of NC with RNA may also be required for assembly (11, 13, 15). Nucleolin could possibly be involved in promoting the binding of RNA to Gag during this process. Indeed, nucleolin has been suggested to serve as an RNA chaperone, capable of delivering RNA to assembling ribosomal proteins and assisting in their folding and binding (for a recent review, see reference 29). The assembly of a retrovirus particle has many similarities to the assembly of a ribosome, and it is thus possible that nucleolin helps deliver viral RNA to the assembling Gag proteins. Interestingly, nucleolin has been shown to associate with the capsid protein of adeno-associated virus type 2 and has been proposed to have a role in virion assembly in this system (54).
If nucleolin is important for Mo-MuLV assembly, then Gag mutants that cannot bind should show defects for virion assembly. The L477P substitution is one such mutant. This single substitution strongly blocked the interaction with Nuc(212), with minimal effects on its ability to interact with other proteins; concomitantly, the mutation caused a nearly complete block to virus production and the accumulation of Gag in the cell. This finding does not prove but is consistent with the possibility that an interaction with nucleolin plays a positive role in virion production. If so, a direct binding should be demonstrated between Mo-MuLV Gag protein and endogenous nucleolin or its cleavage products (24, 64), which has been reported in different contexts and various intracellular locations (12, 38, 60, 76). However, our attempts to identify the HA- or Myc-tagged full-length nucleolin in assembled virions or as proteins bound to Mo-MuLV Gag in coimmunoprecipitation experiments have failed (data not shown). The availability of only poor polyclonal antisera against nucleolin and of monoclonal antibodies specific only for epitope tags, which may not be retained on many of nucleolin's cleavage products, have frustrated our efforts to test the binding of such molecules to Gag. We have not been able to obtain any direct evidence of an involvement of nucleolin in Mo-MuLV assembly, and such a physiological role still remains only a possibility.
In summary, we have identified a specific interaction of Nuc(212) with the Mo-MuLV Gag precursor. The binding of Nuc(212) resulted in a strong inhibition of virion assembly or release in vivo. Nuc(212) could serve as a model for other inhibitors of virion assembly, including potentially therapeutic antivirals that would similarly bind to NC or other domains required for assembly. It also raises the possibility that endogenous nucleolin may play a positive role in the assembly of Mo-MuLV particles. Finally, the experiments presented here indicate a critical involvement of the CA-NC boundary in the process of Mo-MuLV assembly. Further experiments should help determine the exact step in virion formation blocked by CA-NC mutations and the fate of mutant Gags that are unable to form particles.
ACKNOWLEDGMENTS
We thank Paul Jolicoeur, Rolf Sternglanz, Ed Harlow, Erik Barklis, Jeremy Luban, Gary Nolan, Stephen Elledge, and Manfred Schubert for generously providing various reagents. We also thank Guangxia Gao, Marion Dorsch, and Matthew Evans for helpful discussions and Sharon Boast and Kenia de los Santos for technical assistance.
This work was supported by PHS grant CA 30488 from the National Cancer Institute. E.B. and M.O. are Associates, and S.P.G. is an Investigator, of the Howard Hughes Medical Institute.
REFERENCES
- 1.Accola M A, Hoglund S, Gottlinger H G. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola M A, Strack B, Gottlinger H G. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alin K, Goff S P. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology. 1996;222:339–351. doi: 10.1006/viro.1996.0431. [DOI] [PubMed] [Google Scholar]
- 4.Alin K, Goff S P. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology. 1996;216:418–424. doi: 10.1006/viro.1996.0078. [DOI] [PubMed] [Google Scholar]
- 5.Bacharach E, Goff S P. Binding of the human immunodeficiency virus type 1 Gag protein to the viral RNA encapsidation signal in the yeast three-hybrid system. J Virol. 1998;72:6944–6949. doi: 10.1128/jvi.72.8.6944-6949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel P L, Fields S, editors. The yeast two-hybrid system. Oxford, United Kingdom: Oxford University Press; 1997. [Google Scholar]
- 7.Boone L R, Myer F E, Yang D M, Ou C Y, Koh C K, Roberson L E, Tennant R W, Yang W K. Reversal of Fv-1 host range by in vitro restriction endonuclease fragment exchange between molecular clones of N-tropic and B-tropic murine leukemia virus genomes. J Virol. 1983;48:110–119. doi: 10.1128/jvi.48.1.110-119.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourbon H M, Lapeyre B, Amalric F. Structure of the mouse nucleolin gene. The complete sequence reveals that each RNA binding domain is encoded by two independent exons. J Mol Biol. 1988;200:627–638. doi: 10.1016/0022-2836(88)90476-7. [DOI] [PubMed] [Google Scholar]
- 9.Bouvet P, Jain C, Belasco J G, Amalric F, Erard M. RNA recognition by the joint action of two nucleolin RNA-binding domains: genetic analysis and structural modeling. EMBO J. 1997;16:5235–5246. doi: 10.1093/emboj/16.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bugler B, Bourbon H, Lapeyre B, Wallace M O, Chang J H, Amalric F, Olson M O. RNA binding fragments from nucleolin contain the ribonucleoprotein consensus sequence. J Biol Chem. 1987;262:10922–10925. [PubMed] [Google Scholar]
- 11.Burniston M T, Cimarelli A, Colgan J, Curtis S P, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callebaut C, Blanco J, Benkirane N, Krust B, Jacotot E, Guichard G, Seddiki N, Svab J, Dam E, Muller S, Briand J P, Hovanessian A G. Identification of V3 loop-binding proteins as potential receptors implicated in the binding of HIV particles to CD4(+) cells. J Biol Chem. 1998;273:21988–1997. doi: 10.1074/jbc.273.34.21988. [DOI] [PubMed] [Google Scholar]
- 13.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimarelli A, Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1999;73:5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 17.Crawford S, Goff S P. Mutations in Gag proteins P12 and P15 of Moloney murine leukemia virus block early stages of infection. J Virol. 1984;49:909–917. doi: 10.1128/jvi.49.3.909-917.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creancier L, Prats H, Zanibellato C, Amalric F, Bugler B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol Biol Cell. 1993;4:1239–1250. doi: 10.1091/mbc.4.12.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DesGroseillers L, Jolicoeur P. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J Virol. 1983;48:685–696. doi: 10.1128/jvi.48.3.685-696.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson C, Eisenman R, Fan H, Hunter E, Teich N. Protein biosynthesis and assembly. In: Weiss R, Teich T, Varmus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 513–648. [Google Scholar]
- 22.Dorsch M, Fan P D, Danial N N, Rothman P B, Goff S P. The thrombopoietin receptor can mediate proliferation without activation of the Jak-STAT pathway. J Exp Med. 1997;186:1947–1955. doi: 10.1084/jem.186.12.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 24.Fang S H, Yeh N H. The self-cleaving activity of nucleolin determines its molecular dynamics in relation to cell proliferation. Exp Cell Res. 1993;208:48–53. doi: 10.1006/excr.1993.1221. [DOI] [PubMed] [Google Scholar]
- 25.Franke E K, Yuan H E, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 26.Gao G, Goff S P. Replication defect of Moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J Virol. 1998;72:5905–5911. doi: 10.1128/jvi.72.7.5905-5911.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 28.Ghisolfi L, Kharrat A, Joseph G, Amalric F, Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur J Biochem. 1992;209:541–548. doi: 10.1111/j.1432-1033.1992.tb17318.x. [DOI] [PubMed] [Google Scholar]
- 29.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 30.Goff S P. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell. 1996;86:691–693. doi: 10.1016/s0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 31.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granowitz C, Goff S P. Substitution mutations affecting a small region of the Moloney murine leukemia virus MA gag protein block assembly and release of virion particles. Virology. 1994;205:336–344. doi: 10.1006/viro.1994.1650. [DOI] [PubMed] [Google Scholar]
- 34.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 35.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 36.Heine M A, Rankin M L, DiMario P J. The Gly/Arg-rich (GAR) domain of Xenopus nucleolin facilitates in vitro nucleic acid binding and in vivo nucleolar localization. Mol Biol Cell. 1993;4:1189–1204. doi: 10.1091/mbc.4.11.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu H W, Schwartzberg P, Goff S P. Point mutations in the P30 domain of the gag gene of Moloney murine leukemia virus. Virology. 1985;142:211–214. doi: 10.1016/0042-6822(85)90435-0. [DOI] [PubMed] [Google Scholar]
- 38.Issinger O G, Martin T, Richter W W, Olson M, Fujiki H. Hyperphosphorylation of N-60, a protein structurally and immunologically related to nucleolin after tumour-promoter treatment. EMBO J. 1988;7:1621–1626. doi: 10.1002/j.1460-2075.1988.tb02988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalpana G V, Goff S P. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim W, Tang Y, Okada Y, Torrey T A, Chattopadhyay S K, Pfleiderer M, Falkner F G, Dorner F, Choi W, Hirokawa N, Morse H C., III Binding of murine leukemia virus Gag polyproteins to KIF4, a microtubule-based motor protein. J Virol. 1998;72:6898–6901. doi: 10.1128/jvi.72.8.6898-6901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lama J, Trono D. Human immunodeficiency virus type 1 matrix protein interacts with cellular protein HO3. J Virol. 1998;72:1671–1676. doi: 10.1128/jvi.72.2.1671-1676.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Yuan B, Goff S P. Genetic analysis of interactions between Gag proteins of Rous sarcoma virus. J Virol. 1997;71:5624–5630. doi: 10.1128/jvi.71.7.5624-5630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lischwe M A, Cook R G, Ahn Y S, Yeoman L C, Busch H. Clustering of glycine and NG,NG-dimethylarginine in nucleolar protein C23. Biochemistry. 1985;24:6025–6028. doi: 10.1021/bi00343a001. [DOI] [PubMed] [Google Scholar]
- 44.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 45.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 46.Maldarelli F, Martin M A, Strebel K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDermott J, Karanjia S, Love Z, Barklis E. Crosslink analysis of N-terminal, C-terminal, and N/B determining regions of the Moloney murine leukemia virus capsid protein. Virology. 2000;269:190–200. doi: 10.1006/viro.2000.0212. [DOI] [PubMed] [Google Scholar]
- 48.Meric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olson M O, Kirstein M N, Wallace M O. Limited proteolysis as a probe of the conformation and nucleic acid binding regions of nucleolin. Biochemistry. 1990;29:5682–5686. doi: 10.1021/bi00476a006. [DOI] [PubMed] [Google Scholar]
- 50.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder II R C, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ou C Y, Boone L R, Koh C K, Tennant R W, Yang W K. Nucleotide sequences of gag-pol regions that determine the Fv-1 host range property of BALB/c N-tropic and B-tropic murine leukemia viruses. J Virol. 1983;48:779–784. doi: 10.1128/jvi.48.3.779-784.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinter A, Honnen W J, Tung J S, O'Donnell P V, Hammerling U. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology. 1982;116:499–516. doi: 10.1016/0042-6822(82)90143-x. [DOI] [PubMed] [Google Scholar]
- 54.Qiu J, Brown K E. A 110-kDa nuclear shuttle protein, nucleolin, specifically binds to adeno-associated virus type 2 (AAV-2) capsid. Virology. 1999;257:373–382. doi: 10.1006/viro.1999.9664. [DOI] [PubMed] [Google Scholar]
- 55.Reicin A S, Ohagen A, Yin L, Hoglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reiser J, Harmison G, Kluepfel-Stahl S, Brady R O, Karlsson S, Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 60.Sapp M, Richter A, Weisshart K, Caizergues-Ferrer M, Amalric F, Wallace M O, Kirstein M N, Olson M O. Characterization of a 48-kDa nucleic-acid-binding fragment of nucleolin. Eur J Biochem. 1989;179:541–548. doi: 10.1111/j.1432-1033.1989.tb14581.x. [DOI] [PubMed] [Google Scholar]
- 61.Schwartzberg P, Colicelli J, Gordon M L, Goff S P. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virions and reverse transcriptase. J Virol. 1984;49:918–924. doi: 10.1128/jvi.49.3.918-924.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shields A, Witte W N, Rothenberg E, Baltimore D. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell. 1978;14:601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- 63.Shioda T, Shibuta H. Production of human immunodeficiency virus (HIV)-like particles from cells infected with recombinant vaccinia viruses carrying the gag gene of HIV. Virology. 1990;175:139–148. doi: 10.1016/0042-6822(90)90194-v. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki T, Suzuki N, Hosoya T. Limited proteolysis of rat liver nucleolin by endogenous proteases: effects of polyamines and histones. Biochem J. 1993;289:109–115. doi: 10.1042/bj2890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 66.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 67.Tang Y, Winkler U, Freed E O, Torrey T A, Kim W, Li H, Goff S P, Morse H C., III Cellular motor protein KIF-4 associates with retroviral Gag. J Virol. 1999;73:10508–10513. doi: 10.1128/jvi.73.12.10508-10513.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Telesnitsky A, Blain S, Goff S P. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 69.Tuteja N, Huang N W, Skopac D, Tuteja R, Hrvatic S, Zhang J, Pongor S, Joseph G, Faucher C, Amalric F, et al. Human DNA helicase IV is nucleolin, an RNA helicase modulated by phosphorylation. Gene. 1995;160:143–148. doi: 10.1016/0378-1119(95)00207-m. [DOI] [PubMed] [Google Scholar]
- 70.Vidal M, Brachmann R K, Fattaey A, Harlow E, Boeke J D. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidal M, Braun P, Chen E, Boeke J D, Harlow E. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc Natl Acad Sci USA. 1996;93:10321–10326. doi: 10.1073/pnas.93.19.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 73.Yu X, Yu Q C, Lee T H, Essex M. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J Virol. 1992;66:5667–5670. doi: 10.1128/jvi.66.9.5667-5670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan B, Campbell S, Bacharach E, Rein A, Goff S P. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan B, Li X, Goff S P. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaidi S H, Malter J S. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J Biol Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]