Abstract
Figs (Ficus spp., Moraceae) and their pollinating wasps (Agaonidae, Chalcidoidea) constitute perhaps the most tightly integrated pollination mutualism that is known. Figs are characterized by extraordinarily high global and local species diversity. It has been proposed that the diversification of this mutualism has occurred through strict-sense coadaptation and cospeciation between pairs of fig and wasp species that are associated in highly specific one-to-one relationships. However, existing studies cast doubt on the generality of this proposition. Here, we review our current knowledge of the evolutionary history of the fig/fig-wasp mutualism. We critically examine the idea that codivergence between figs and their pollinators has been dominated by strict-sense cospeciation. We present phylogenetic and population genetic data from neotropical fig and fig wasp species that suggest that a more accurate model for diversification in this mutualism is that of groups of genetically well defined wasp species coevolving with genetically less well defined (frequently hybridizing) groups of figs. Last, we use our results to assess previously proposed hypotheses on models of speciation in this mutualism.
Both ecologically and evolutionarily, mutualisms represent one of the most influential of all biological interactions, with fundamental consequences for the evolution and maintenance of biotic diversity (1-7). The long-term stability of mutualisms poses a considerable and, as yet, not fully resolved challenge to evolutionary theory. However, the obvious fact of long-term stability coupled with the proliferation and diversification of many mutualisms raises a set of interesting questions concerning coadaptation and speciation among the partners in the interaction. The obligate mutualisms between flowering plants and their insect pollinators (8-11) constitute fascinating extreme cases of interspecific mutualisms. Most obligate plant-pollinator mutualisms show high levels of reciprocal species or taxon specificity. Usually, the insect requires the plant for food or other substances to complete its life cycle successfully, and the plant requires the insect for pollination. Further, it is the insect's recognition and choice of hosts that determine the patterns of host gene flow. Although there are relatively few cases of obligate pollination mutualisms (8-11), these few cases are often marked by high to extreme speciation and diversification in both partners, raising the question of how host specificity and control of gene flow affects patterns of speciation in one or both partners.
Figs (Ficus spp., Moraceae) and their pollinating wasps (Agaonidae, Chalcidoidea) constitute perhaps the most tightly integrated pollination mutualism that is known (8, 9, 12-16). Ficus is one of the most diverse genera of flowering plants in number of species and growth and life forms (17, 18). The nearly 750 described species of Ficus (19) occur worldwide in tropical and subtropical regions, and they are considered “keystone” species in tropical forests because of their year-round production of fruit that is essential to a large number of frugivores (20-23). Figs depend on minute, pollen-bearing female wasps to pollinate the flowers and thereby initiate seed production (8, 12, 24-28). The mated female wasps, in turn, depend on the developing fig inflorescence for the production of their offspring, because each wasp larva consumes the contents of one would-be seed. The cycle begins when mated female wasps locate a receptive tree and enter the enclosed fig inflorescences (Syconia). As the females search for oviposition sites, they pollinate the flowers. Usually the foundresses die inside the syconium, and then both their offspring and the seeds begin to develop (24-28). Last, after maturation, the offspring mate and the mated females collect pollen and fly off to find a receptive tree and begin the cycle anew.
Results from morphological studies and the notion of the high (one-to-one) species specificity of the interaction led to the proposal of strict-sense coevolution and tight cospeciation between the two groups (17, 19, 24, 29, 30). Recent molecular phylogenetic studies (31-36) have provided some support for the proposition of cocladogenesis and coadaptation between recognized genera of pollinating wasps and their respective fig sections (species groups within fig subgenera). Results from those studies (as well as earlier morphological studies) have led some authors to suggest that finer-scale cospeciation of individual fig and wasp species should be widespread (9, 15, 17, 24).
However, even at this fairly coarse level of wasp genera and fig sections, clear incongruencies among their phylogenies have been detected, directly undermining the empirical basis of strict-sense cospeciation (32, 33, 35, 37). Also, existing phylogenetic studies are not adequate to clearly distinguish between strict-sense cospeciation between individual pairs of species-specific wasps and figs, and a much less specific form of broad-sense coevolution between related groups of wasps with related groups of figs. This difference is crucial with respect to understanding the actual mechanisms of coadaptation and cospeciation in the mutualism. If strict-sense cospeciation were the rule, then the evolutionary dynamics of coadaptation would be expected to take place independently in a series of genetically isolated, tightly coupled pairs of fig and pollinator wasp species. If strict-sense cospeciation were not the rule, then the dynamics of coadaptation would be quite different. Specifically, under this latter scenario, we expect a much looser coevolutionary coupling between particular pairs of figs and wasps, with host colonization, hybridization, and introgression of relatively novel genetic material across different fig species contributing to the generation of phenotypic diversity on which selection can act.
Here, we evaluate the hypothesis that strict-sense cospeciation has dominated the evolutionary history of figs and their pollinators. We review the current knowledge on the evolutionary history of the mutualism, focusing on the available evidence suggesting cocladogenesis at high taxonomic levels (i.e., between different sections of Ficus and their associated genera of pollinating wasps) and cospeciation at finer taxonomic scales. We present data from neotropical fig and fig wasp species that suggest an alternative view. Specifically, we find that groups of genetically well defined pollinator wasp species coevolve in association with groups of genetically poorly defined (hybridizing) figs. Last, we link our results to previously proposed models of speciation in this mutualism that bear directly on the question of the origin of the high species diversity in these organisms.
The Coevolutionary History of the Mutualism
The fig-wasp mutualism is ancient and diverse, originating ≈80-90 million years ago (32, 38). The outgroup to figs is not yet clear and includes both New and Old World taxa as possibilities (38). However, the hypothesis of a Gondwanan origin of the mutualism (39, 40) is supported by the observations that the most basal group to extant figs and pollinator wasp are, respectively, the New World Ficus subgenus Pharmacosycea and their associated pollinators in the genus, Tetrapus (19, 24, 31, 32, 35, 37). The idea that extant figs and wasps have radiated from these basal New World groups is supported further by the observation that the estimated divergence times among the pollinator genera and their current geographical distributions correspond well with several features of the break up of the southern continents during the late Cretaceous period (32).
The nearly 750 described species of Ficus have been classified in four subgenera (Pharmacosycea, Urostigma, Sycomorus, and Ficus) and 18 sections (species groups within subgenera) (17, 19, 41). The pollinators of figs, minute chalcid wasps from family Agaonidae, have been assigned to 20 different genera (17, 42), with >300 described species (15). With two exceptions (Ceratosolen and Wiebesia), each recognized pollinator genus is generally restricted to a single taxonomic group of Ficus (subgenus or section). Molecular data show that fig-pollinating wasps form a clearly monophyletic group within the Chalcidoidea that is separate from other fig wasps (a large number of nonpollinating wasps are also associated with figs), supporting the idea that the pollination syndrome evolved only once during the evolution of the mutualism (43, 44).
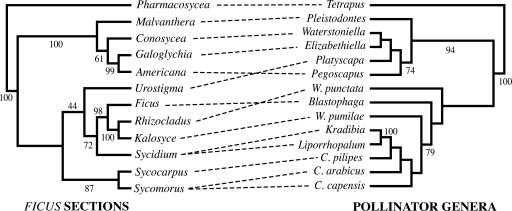
The original taxonomic studies of the fig-pollinating wasps using morphological characters (24, 29, 42, 45) showed that related species of wasps pollinate related species of figs. This result led to the hypothesis of strict-sense coevolution between the mutualists. Also, the almost invariable finding at that time of one species of pollinator per host fig species reinforced this idea, and it led to the inference that cospeciation should, therefore, be the dominant pattern of codivergence at both coarser (e.g., sections of figs and genera of wasps) and finer (species of both) taxonomic scales (9, 17, 24, 46). Recent molecular phylogenetic studies of the major groups of fig-pollinating wasps (32, 33) and figs (31, 35, 37) have allowed mapping the morphological classification of figs into the fig-wasp phylogeny and vice versa. These studies have provided some support to the original insight from taxonomy about the strong conservation of host associations and the preponderance of cocladogenesis during the diversification of wasp genera and associated fig subgenera and sections. However, these studies have also revealed clear incongruencies between the phylogenies of figs and those of their pollinators (Fig. 1) (32, 33, 35, 37).
Fig. 1.
Tanglegram comparison of the phylogenies of sections of Ficus and their associated genera of pollinating wasps. Both phylogenies correspond to pruned trees from larger phylogenetic studies (32, 35, 37). In the pollinator phylogeny, C stands for Ceratosolen and W stands for Wiebesia, the only genera of pollinating wasps associated with more than one Ficus section. Numbers associated with branches are bootstrap values (>40%) that were taken directly from the studies in which the complete phylogenies were presented originally (32, 35, 37). They are shown to indicate the level of support for each node in the original phylogenetic studies. The two cladograms are significantly incongruent (see text). treemap 1.0 was used to create this image (47) (see text for details).
In Fig. 1, we present a pollinator phylogeny based on cytochrome oxidase I (COI) data from ref. 32. This tree is largely consistent with results for the pollinators obtained by Weiblen (33). The corresponding fig phylogeny is a strict consensus from maximum-parsimony analyses of combined sequences of the internal and external transcribed spacers (ITS and ETS) from a large world-wide sample of Ficus, and is largely consistent with results from Jousselin et al. (35). This phylogeny is also largely consistent with results from a different study on Indo-Australian Ficus taxa from Weiblen using ITS sequences (37). Specifically, the well resolved relationships among sections Ficus, Sycidium, Rhizocladus, and Kalosyce were incorporated from that study (37). The degree of congruency between the two cladograms shown in Fig. 1 is not significant (P = 0.85) (47), a result that is not consistent with previous assertions about the preponderance of cocladogenesis. However, this result should be viewed cautiously because of the lack of resolution and uncertainty of relationships of several fig sections (Urostigma and Oreosycea) and pollinator genera (Wiebesia, Blastophaga, and Platyscapa) (31-33, 35, 37), and to the partial sampling of wasp genera (11 of 20) and fig sections (12 of 18) in the phylogenies.
Nonetheless, despite these caveats, the phylogenetic reconstructions show that ancestral host switches have occurred at different times during the evolution of the mutualism, even when considering the strongly supported clades. For example, wasps that pollinate the figs in subgenera Ficus and Sycidium are not close relatives (32, 33), although both morphological (17, 19) and molecular (31, 35, 37) studies indicate that Ficus and Sycidium are sister groups. Further, molecular data suggest that the pollinators of Ficus are more closely related to the pollinators of Urostigma (33). Because figs from the subgenera Ficus and Sycidium are supported to be related more closely to Sycomorus than to Urostigma, the most likely scenario is that the ancestor of Ficus figs was colonized by the ancestor of wasps associated currently with Urostigma figs and that this new combination then jointly diversified. Thus, even at high taxonomic scales, the evidence indicates at least one breakdown (and possibly more) in strict cocladogenesis in the mutualism.
Finer-Scale Coevolution: Is There Evidence for Strict-Sense Cospeciation?
A major weakness of existing molecular phylogenetic studies in addressing the degree to which strict-sense cospeciation dominates the evolutionary dynamics of this mutualism is that they typically have concentrated on a small number of taxa that represent very ancient, distantly related taxonomic subdivisions within the genus Ficus and their associated genera of pollinator wasps (31-33, 35). Also, these studies have also tended to concentrate on analyses of one gene (or a few genes) from a few individuals of each species. Such sampling differentially emphasizes the products of ancient processes and inevitably tends to bias interpretation toward cospeciation, without providing a real and rigorous test of the hypothesis. Specifically, even if there were a perfect congruence of fig and wasp phylogenies at coarse scales (and available evidence suggests that there is not), such a pattern would not necessarily result from strict-sense, one-to-one cospeciation of fig and wasp species. Also, the sampling of multiple loci from multiple individuals within a species is essential to detect potentially important ongoing processes such as hybridization and introgression. Therefore, to find compelling evidence for, or against, strict-sense cospeciation as a real, ongoing process, detailed genetic sampling of multiple individuals from multiple, relatively closely related species of the same section of fig and genus of pollinator is required.
The only published coevolutionary study that has attempted to survey relationships at a finer (within section/genus) taxonomic scale (48), reconstructed molecular phylogenies for 17 Indo-Australian and two African fig species from subgenus Sycomorus and their pollinators (Ceratosolen sp.). Significant congruency between the phylogenies of the two groups supported the cospeciation hypothesis (48). However, although some of the sampled species appear to be close relatives based on their taxonomic associations (e.g., they are part of the same Ficus section), most species included in this sample are very distinct genetically, and speciation events appear to be ancient (32). Also, within species genetic sampling was minimal or not reported. Therefore, although that study represents a step in the right direction, it does not provide the kind of information necessary to understand species-level processes of diversification in the mutualism and to adequately test coevolutionary hypotheses.
We have begun to fill that gap by gathering detailed genetic data from a group of 17 sympatric species of figs and their associated pollinators found in the vicinity of the Panama Canal. In particular, we have collected information from multiple loci across multiple individuals within these taxa. These figs and wasps also represent two of the most divergent lineages of Ficus and their pollinators (19), and a series of extensive, long-term studies have made them the ecologically and genetically best characterized group of species from this mutualism (26, 49-52). Free-standing neotropical monoecious fig trees (subgenus Pharmacosycea, section Pharmacosycea) are the most basal of the extant fig lineages, with 20 described species (19). They are pollinated by wasps of the genus Tetrapus, the most basal group of pollinators (Fig. 1). The second lineage is a derived section of strangling monoecious figs (subgenus Urostigma, section Americana) and their pollinating fig wasps (Pegoscapus sp.), with ≈100-120 described species (19). Of the 17 Panamanian fig species we have studied, 4 belong to section Pharmacosycea and 13 belong to section Americana.
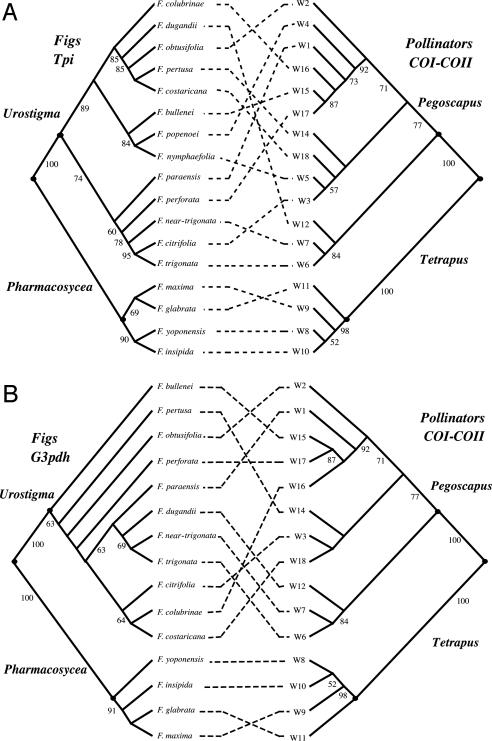
We reconstructed molecular phylogenies of the 17 species of Panamanian figs and their pollinators (Fig. 2) using 1,587 bp of COI-COII from the pollinators and the following two nuclear regions from the figs: 530 bp from an intron of the triosephosphate isomerase gene (Tpi) and 723 bp of sequence encompassing four introns and three exons from the glycerol-3-phosphate dehydrogenase gene (G3pdh). The primers used for PCR and sequencing have been described elsewhere (32, 53, 54). PCR products were sequenced directly in both directions by using standard protocols. If the Tpi or G3pdh sequence from an individual was polymorphic, the PCR product was cloned with the TOPO TA cloning kit (Invitrogen), and 8-10 cloned PCR fragments sequenced to identify the different haplotypes. Multiple individuals per species were sampled (see below), but the phylogenies used for reconciliation analyses (47) were reconstructed by using either the most common haplotype per species or a randomly chosen individual sequence (in a few cases, all haplotypes had the same frequency). Results from the reconciliation analyses are similar if different combinations of haplotypes are used (data not shown). Phylogenies were reconstructed using maximum-likelihood (ML) methods and the general reversible model with rate heterogeneity (REV+Γ) using of paup* (version 4.0b10) (55). Topological comparisons were conducted by using the Shimodaira-Hasegawa test (one-tailed test) (56) with Resampling of Estimated Log Likelihoods (RELL) bootstrap (1,000 replications). The degree of match between the Tpi ML fig phylogeny [-log (L) = 1,095.06375; α = ∞] and the COI-COII pollinator ML phylogeny [-log (L) = 8,236.58884; α = 0.572] is not significantly different from what would be expected by chance (P = 0.25; Fig. 2 A). Also, forcing the fig Tpi phylogeny to match the pollinator phylogeny generates a tree that is significantly worse than the ML tree [Δ -log (L) = 141.9446; P < 0.001]. The G3pdh ML phylogeny reconstructed with the REV+Γ model [-log (L) = 1,385.80552; α = 0.847] and the corresponding COI-COII pollinator phylogenies are shown in Fig. 2B. The G3pdh and COI-COII phylogenies are incongruent (P = 0.09), and forcing the topology of the G3pdh phylogeny to match the pollinator phylogeny also generates a tree that is significantly worse than the ML tree (Δ -log (L) = 28.29731, P = 0.025). Therefore, despite the low resolution for several nodes in the fig phylogenies (Fig. 2), the phylogenies of two different nuclear genes show significant incongruency with the phylogenetic history of the associated pollinators.
Fig. 2.
Tanglegram comparisons of molecular phylogenies from neotropical Ficus and their pollinators. Species names for the wasps are omitted for brevity and are shown as codes, with each number corresponding to a different species. All phylogenies are ML trees (see text for details). The individual sequences per species used in these reconstructions correspond to the most common haplotypes found for each species. Fig wasp sequences from fig species with multiple pollinators are from the most common species associated with a given fig host. The pollinator phylogeny compared with the G3pdh phylogeny is a pruned version of the larger COI-COII tree compared with the Tpi phylogeny. Numbers below branches are bootstrap values based on 1,000 replications. The fig and pollinator phylogenies are significantly incongruent (see text).
These results suggest that cospeciation is not the dominant pattern of codivergence in these groups of sympatric neotropical figs and their pollinators. We suggest that the most likely explanation for the lack of congruency between the molecular phylogenies of the two mutualists is the recently documented presence of multiple species of pollinator per host in neotropical figs (52), which would generate incongruent phylogenetic histories because of pollinator host switches, with the resultant hybridization and possible genetic introgression across different fig species (see below).
A Complex Mutualism Unveiled? Multiple Pollinators per Host Are Not the Exception
The degree to which tight specificity of a particular wasp species to a given host species breaks down will influence the possibility of naturally occurring hybridization among different host species and affect the trajectories of fig and wasp coadaptation. The notion that the vast majority of species of fig are pollinated by its own highly specific species of pollinator (“the one-to-one rule”) has long been claimed as one of the most extraordinary aspects of this mutualism (9, 15, 16). Initial documented cases of breakdown of species specificity were thought to represent taxonomic artifacts (57). Nonetheless, to determine the extent to which tight cospecificity reflects a real and predominant pattern rather than an assumption requires detailed taxonomic and genetic studies.
Several recent and detailed studies have confirmed that this rule is often broken (15-17, 42, 52, 58). Recent studies in Africa and Australia have shown that breakdowns of specificity are relatively common, occurring in approximately one third of the studied cases (24 species of Ficus) (16, 59, 60). In some cases, different wasp species are associated only with a host fig in different parts of its geographic range (58, 61, 62). Importantly, it has been demonstrated in other cases that multiple pollinators associate routinely and successfully with a given host in sympatry (13, 42, 52, 59, 60, 63-65).
Moreover, several independent observations by different researchers in different geographic regions suggest the routine occurrence of interspecific hybridization in figs. Frequent cases of adult figs with hybrid morphological characteristics have been observed in Panama and South America (19) (W. Ramirez, personal communication; C.C. Berg, personal communication; E.A.H, unpublished data), Africa (S. Compton, personal communication; G. Michaloud, personal communication), and Indonesia (66). Reports of local pollinators breeding in introduced fig species have been reported in Florida (67), where hybrid viable seedlings were also observed. Also, Parrish et al. (66) recently reported genetic evidence of hybridization among three dioecious fig species found in three Indonesian islands. The observation of multiple individuals with hybrid genetic composition at several amplified fragment length polymorphism loci was interpreted to be caused by a breakdown of pollinator specificity in the isolated island populations due to pollinator limitation. Last, a detailed study from South Africa (64) found that a fig tree (Ficus lutea) growing >100 km out of its normal range was pollinated by several different species of wasps that are not normally associated with it. Likely relevant to understanding the broader mechanisms that generate diversity in figs, this study found that the species that were most closely related to the normal pollinator of this host species showed a higher capacity to reproduce and produce seeds that would germinate (64, 68, 69).
Of all documented violations of the one-to-one rule, the most thorough sampling of genetic data comes from a group of sympatric New World fig species (52). Extensive genetic sampling of pollinators associated with a subset of the 17 sympatric neotropical fig species from Fig. 2, shows that the rule is broken in ≈60% of the cases (52) (Table 1). In some cases, the species pairs associated with the same host are each other's closest relatives, a finding that is consistent with long-term coexistence on a single host. In other cases, however, the species pairs associated with the same host are not each other's closest relatives, indicating a host switch. Importantly, wasps that are genetically indistinguishable regularly pollinate different host fig species (Table 1). The molecular phylogeny of the neotropical pollinators presented in Fig. 2, was reconstructed by using the most common haplotype of each wasp species found in our survey and, therefore, represents the phylogeny of the most common pollinators of the 17 neotropical figs included, based on our genetic sampling. Inclusion of the multiple pollinators in the cophylogenetic analyses does not generate a better fit between the fig and pollinator phylogenies (Tpi, P = 0.33; G3pdh, P = 0.15).
Table 1. Genetically confirmed cases of multiple pollinators per Ficus in Neotropical species.
| Ficus species | No. of pollinators | Shared pollinators |
|---|---|---|
| F. bullenei† | 4 | F. popenoei, F. near-trigonata |
| F. near-trigonata† | 3 | F. popenoei, F. bullenei |
| F. popenoei† | 2 | F. near-trigonata, F. bullenei |
| F. perforata† | 2 | F. colubrinae |
| F. obtusifolia† | 2 | — |
| F. glabrata‡ | 2 | — |
| F. maxima‡ | 2 | — |
Other species have been sampled (>10 individuals) without revealing the presence of multiple pollinators: F. citrifolia, F. nymphaefolia, F. trigonata, F. dugandii, and F. colubrinae.
Ficus from subgenus Urostigma.
Ficus from the subgenus Pharmacosycea. Sample sizes for those two species are small (<10 individuals).
These findings reveal previously unsuspected levels of complexity in the mutualism, and they seriously challenge the idea that strict-sense cospeciation should be the dominant pattern of codivergence among figs and their pollinators. The inferred host switches and the observation of different fig hosts sharing identical pollinators suggests the potential for hybridization and genetic introgression between host fig species (see below), a situation that could generate incongruent histories of divergence between the mutualists. Thus, our observations provide a clear mechanism for explaining the incongruent phylogenetic histories of neotropical figs and their pollinators (Fig. 2), and they also provide a potential mechanism to explain phylogenetic incongruencies at higher taxonomic levels (Fig. 1).
Assessing the Evidence of Hybridization and Introgression Among Neotropical Fig Species
To test for evidence of introgression in neotropical figs, we conducted a divergence population genetic study (70, 71) in three species of neotropical Urostigma figs: Ficus popenoei, Ficus bullenei, and Ficus near-trigonata. These three species share one pollinator, and also, each species has one or two additional pollinators that, based on our current sample sizes, are not shared with other fig species (Table 1). Thus, we expect to observe some evidence of past or current introgression between these species pairs. We collected DNA polymorphism data for these three species (Table 2) by using the two nuclear loci described above (Tpi and G3pdh) and the locus C2E19, an EST isolated from a cDNA library made from leaf tissue of a neotropical Urostigma fig species (Ficus citrifolia). The locus C2E19 has no clear homologues in GenBank, and therefore, we consider it here to be an ORFan (an expressed gene with unknown function). PCR products were directly sequenced in both directions by using standard protocols, and polymorphic sequences were cloned to distinguish haplotypes.
Table 2. Polymorphism statistics for three nuclear loci sequenced in three species of neotropical Ficus (sect. Urostigma).
| Locus | Species | n† | L‡ | S§ |
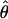 ¶ ¶
|
π∥ | D†† |
|---|---|---|---|---|---|---|---|
| Tpi | F. popenoei | 16 | 517 | 2 | 0.00117 | 0.00193 | 1.6871 |
| F. near-trigonata | 10 | 521 | 30 | 0.02034 | 0.01858 | −0.4146 | |
| F. bullenei | 10 | 517 | 14 | 0.00957 | 0.00606 | −1.6802 | |
| G3pdh | F. bullenei | 10 | 719 | 1 | 0.00049 | 0.00049 | 0.0150 |
| F. near-trigonata | 12 | 719 | 3 | 0.00138 | 0.00148 | 0.2219 | |
| F. bullenei | 8 | 720 | 2 | 0.00107 | 0.00070 | −1.3101 | |
| C2E19‡‡ | F. popenoei | 9 | 704 | 13 | 0.00679 | 0.00458 | −1.5570 |
| F. near-trigonata | 8 | 704 | 13 | 0.00712 | 0.01040 | 2.3306§§ | |
| F. bullenei | 11 | 704 | 3 | 0.00145 | 0.00232 | 2.0454§§ |
No. of alleles sequenced.
Average length (bp) of the sequences from each species.
No. of polymorphic sites.
Estimate of 4Nμ per bp using the no. of polymorphic sites (76).
Estimate of 4Nμ by using the average no. of nucleotide differences per site (95).
Tajima's statistic (74).
ORFan locus isolated from a cDNA library.
Significantly different from zero (P < 0.01).
The polymorphism data were fitted to a null model of speciation with no gene flow (isolation model) by using the method described by Wang, Wakeley, and Hey (WWH) (72). In addition to the WWH method, we also determined the fit of the fig nucleotide data to the isolation model by comparing observed to expected values of different polymorphism types (shared, exclusive, and fixed differences) by using a χ2 statistic (70). Under the isolation model, two very recently diverged species are expected to share polymorphisms that are present in their ancestor. As the species diverge, shared polymorphisms are lost, exclusive polymorphisms arise, and fixed genetic differences accumulate in each lineage through genetic drift. With observations from a number of loci, one expects to find a consistent negative correlation between fixed differences and shared polymorphisms across loci. However, if the null model of speciation is not correct and gene flow has occurred during species divergence, the divergence of a given locus will be slowed down because gene flow removes and prevents the accumulation of fixed differences at the same time as it introduces shared polymorphisms. Gene flow will elevate the numbers of shared polymorphisms and reduce both the number of exclusive polymorphisms and the number of fixed differences between taxa. More importantly, if gene flow occurs at some loci but not at others, it will elevate the variance among loci in the numbers of shared polymorphisms and fixed differences. These ideas form the basis of the WWH method (72). This method is conservative and takes into account the number of exclusive polymorphisms for each species, and the numbers of shared polymorphisms and fixed differences between the species. These polymorphism counts are used to fit the isolation model to the data and estimate the four parameters of the model: the three population mutation rates (one for the ancestral population and one for each descendant), and the time since divergence. By using these four parameters and estimates of the recombination rate for each locus, it is then possible to conduct coalescent simulations to determine the significance of the WWH (72) and χ2 (70) test statistics.
The fit of the fig polymorphism data to the isolation model is poor (Table 3), and significantly so for the F. popenoei/F. bullenei and F. popenoei/F. near-trigonata comparisons. These results suggest the occurrence of gene flow among these two species pairs during their divergence. Based on this small data set, we can infer that gene flow has occurred but has not occurred recently because we see no evidence of shared identical haplotypes across species at any of the three studied loci.
Table 3. Fitting Ficus sequence data to an isolation model of species divergence.
| Species 1 | Species 2 | 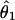 |
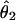 |
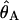 |
T | χ2 | Pχ2 | WWH | PWWH |
|---|---|---|---|---|---|---|---|---|---|
| F. popenoei | F. near-trigonata | 15.91 (4.84-59.57) | 5.37 (1.95-10.78) | 15.561 (0-33.34) | 0.436 (0.11-1.42) | 70.02 | <0.001 | 11 | 0.079 |
| F. popenoei | F. bullenei | 5.512 (2.38-2.03) | 6.828 (3.25-22.67) | 0 (0-0.002) | 2.442 (0.49-2.82) | 34.33 | 0.001 | 9 | 0.016 |
| F. near-trigonata | F. bullenei | 9.377 (0-37.51) | 3.560 (0-10.45) | 27.354 (0-47.51) | 0.397 (0.14-1.82) | 36.03 | 0.088 | 11 | 0.333 |
The 95% confidence intervals are shown in parentheses. 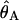 is the estimate of the population mutation parameter for the ancestor of species 1 and 2. T is the estimated time of divergence between the two species in 2N1 generation units (where N1 is the estimate of the effective population size of species 1). The P values for both the χ2 and the WWH test statistics are the proportion of simulated values greater than or equal to the observed value.
is the estimate of the population mutation parameter for the ancestor of species 1 and 2. T is the estimated time of divergence between the two species in 2N1 generation units (where N1 is the estimate of the effective population size of species 1). The P values for both the χ2 and the WWH test statistics are the proportion of simulated values greater than or equal to the observed value.
An assumption of these tests is that the DNA sequences should be evolving neutrally. We first determined whether the amounts of polymorphism and divergence across loci are correlated, as expected under neutrality, by using the Hudson-Kreitman-Aguade (HKA) test (73). The significance of the χ2 statistic from this test was evaluated by using a distribution generated from 1,000 coalescent simulations. HKA tests were applied to each of the three taxa, in each case by using a single sequence from F. citrifolia as an outgroup. The data fit the neutral model in all three species: F. popenoei, χ2 = 1.40 and P = 0.212; F. near-trigonata, χ2 = 2.23 and P = 0.103; and F. bullenei, χ2 = 1.73 and P = 0.128. A second type of neutrality tests evaluates the fit of the data to the neutral model by using DNA polymorphism data from a single locus (74, 75). These tests evaluate whether the frequency spectrum of mutations is significantly different from that expected under neutrality. Tajima's D statistic (74) is proportional to the difference between two estimates of the population mutation parameter 4Nμ, the mean pairwise difference between the sampled sequences (π) and Watterson's estimator (θ) (76). Under a neutral model with constant population size, both estimators have the same expected value, and therefore, the value of Tajima's D under neutrality is zero. The observed values of Tajima's D for the C2E19 sequences of F. near-trigonata (D = 2.3306) and F. bullenei (D = 2.0454) are significantly positive (Table 2). Significant positive values of Tajima's D are usually interpreted as evidence of balancing selection (74). However, in the case of F. near-trigonata, that result is likely to be caused by introgression: groups of F. near-trigonata C2E19 alleles are more closely related to F. popenoei alleles than to other conspecific alleles (data not shown), a result consistent with the poor fit of the data from that species pair to a strict isolation model of species divergence. Introgression generates patterns of polymorphism that mimic those produced by natural selection (77, 78). In the case of F. bullenei, balancing selection could be the explanation for the large positive value of Tajima's D. However, we question that inference because an alternative test of neutrality that also uses DNA polymorphism from a single locus, Fu and Li's D (75), is not significant (D = 1.1284).
Our findings on the pollinators of these fig species contrast with those of their hosts. None of the available data from microsatellite loci (52, 79) or mitochondrial sequences (52) show any evidence of genetic introgression across different species of pollinator. Allele size distributions of microsatellites for different cryptic pollinator species are nonoverlapping (52, 79). Nucleotide sequences of mitochondrial genes (52) from different species are reciprocally monophyletic. These findings suggest that the process of divergence in figs and in the pollinators is very different. Thus, whereas figs may exchange genes during divergence because of nonspecific pollinators, the pollinators do not, and thus, they seem to be reproductively isolated species that satisfy a strict interpretation of the biological species concept.
A second potential reason could partially explain the observed phylogenetic incongruencies among neotropical figs and their pollinators (Fig. 2): the large effective population size (Ne) that is believed to characterize neotropical figs (50). Large Ne could interfere with species-level phylogenetic analyses because of the expected large amounts of ancestral polymorphism shared across recently diverged species. The slow lineage sorting of species-specific alleles into reciprocally monophyletic clades generates the well known “gene tree vs. species tree” problem (80, 81), leading to erroneous phylogenetic inference for recently diverged lineages and to low resolution of species-level phylogenies. However, we believe that this explanation is unlikely for three reasons. First, this issue is effectively considered by the test of the strict isolation model. The test seeks to determine whether the observed level of shared polymorphisms, exclusive polymorphisms, and fixed differences can be explained based on the estimates of the four basic parameters of the model (the population sizes of the two species and their ancestor, and the time of divergence). The lack of fit of the data to the model suggests that shared variation is the result of gene flow and not of ancestral polymorphism. Second, the data from G3pdh, used to test cospeciation (Fig. 2B), shows 7-10 fixed differences between the three species pairs and not a single shared polymorphism. Thus, the significant incongruency among the fig G3pdh phylogeny and the pollinator COI-COII phylogeny is clearly not due to unsorted ancestral polymorphisms. Third, although the nucleotide variability in neotropical fig species is expected to be high based on results from protein electrophoresis studies (50), Table 2 shows that the observed level of DNA polymorphism in these species is not uncommonly high for the three surveyed genes. In fact, DNA polymorphism in these three species is much lower than that observed in other outbred plant species. Average values of θ, Watterson's estimator of the population mutation rate (4Nμ) (76), for F. popenoei (0.00292), F. bullenei (0.00403), F. near-trigonata (0.00961), are approximately two to nine times lower than averages values at silent sites from the two outcrossing plants (Zea mays ssp. parviglumis and Arabidopsis lyrata ssp. petraea) that have the highest levels of nucleotide diversity that have been measured (82). However, our survey of variation is based on a small sample of genes and species, and thus, genetic studies that include more loci and species would help to resolve this last point.
Conclusions and Prospects: Multiple Pollinators and the Origin of Fig Diversity
In the past, it has been difficult to explain the origin of the remarkable global and local diversity of fig species given the assumption of strict-sense cospeciation and one-to-one pollinator species-specificity. As a result, there is no generally accepted view on the mechanisms of speciation in figs and their pollinators (15, 62). A few authors have proposed that speciation in these organisms happens by allopatric isolation (13, 14, 62). However, genetic studies using paternity analyses have shown that neotropical figs have the highest documented distances of gene flow of any tropical plant (50, 83). Despite growing at very low densities (1-10 individuals per km2) and having asynchronous flowering within populations, individual fig trees receive pollinating wasps (and pollen) from a large number of individual trees. Conservative estimates suggest that fig wasps routinely disperse pollen over distances of >10 km and that breeding populations of figs constitute hundreds of individuals spread over areas >100 km2 (50). Therefore, allopatric isolation in monoecious neotropical species is highly unlikely. Other authors have proposed that temporal isolation (allochrony) generated by flower asynchrony could lead to isolation in sympatry (84). However, population genetic studies of the pollinators reveal no evidence of population subdivision (79), an essential condition for the allochronic speciation model (84).
In a short essay published in 1961 (85), Baker proposed pollinator generalization (lack of species specificity) and hybridization among fig species as a potential explanation for the large diversity of recognized species of Ficus: “if hybridization is possible... it may be that only the natural eagerness of taxonomists to name new (fig) `species' has prevented the recognition of some of the intermediates between extreme forms as hybrids.” Given the scarce evidence of breakdowns to the one-to-one rule at that time, Baker's suggestion was not taken as seriously as more recent results now suggest it should be. Our preliminary genetic results, the findings on the large number of exceptions to the one-to-one rule (42, 52, 58-65), and the observation of hybrid (or backcross) figs in nature reviewed above, support Baker's suggestion and provide the foundations for developing an appropriate hypothesis for understanding the coevolutionary dynamics of this mutualism.
Here, we propose that hybridization and introgression due to pollinator host switches and pollinator host sharing may be a major factor underlying much of the tremendous diversity of fig species. Hybridization can lead to generation of new genotypic combinations that may then diversify and lead to the evolution of additional specialized pollinators. The process of divergence in the pollinators can be reinforced by their inbred population structure (86), and fine-tuning of the host recognition mechanisms would promote pollinator divergence and speciation. By this view, the coevolutionary history of the mutualism is that of semispecific wasps (that are good biological species) moving back and forth between figs that may not be good biological species. Although this hypothesis does not yet provide a detailed model for explaining speciation in figs and wasps, the recognition that wasps are not strictly host-specific and that interspecific hybridization in figs is not rare over both ecological and evolutionary time is almost certainly crucial for developing such a model. Minimally, breakdowns in specificity appear to explain the frequent observation of fig individuals with intermediate or mosaic morphologies, the existence of species complexes of figs (19), as well as the observed lack of congruence of fig and wasp phylogenies at various taxonomic scales. Also, host switching of pollinators followed by the introgression of complete gene complexes could help to explain several interesting cases of gains and losses of elaborate characters (e.g., passive or active pollination) (32, 87, 88).
Progress will depend fundamentally on improved population genetic and phylogenetic datasets for resolving the many outstanding questions on the coevolutionary dynamics of the mutualism both at local and regional geographic scales. Furthermore, we need to document in detail the reproductive consequences of pollinator “mistakes” (host switches and host sharing) for both partners in the mutualism. Understanding the evolutionary trajectories of ecologically important fig-associated characters that have been shown to influence reproductive success and host recognition in the wasps (flower number, seed size, seed number, and receptive volatiles) (26-28, 89-94) could allow us understand what allows some combinations of wasp fig-pollination mistakes or host-switches to occur more often than others. Understanding the consequence of pollination mistakes appears to be critical for understanding the processes that affect gene flow, coadaptation, and cospeciation at a fine taxonomic scale in the fig/fig-wasp mutualism.
Given the information now available to us, it appears that strict-sense cospeciation of one-to-one species specific figs and wasps should not be the default paradigm for formulating hypotheses to explain the extraordinary diversification of fig and wasp species. For cases in which appropriate studies have been conducted, most figs appear to be pollinated by more than one species of wasp, many of these wasps appear to colonize new species of figs, and interspecific hybridization and introgression appear to be widespread among figs. We propose that the best model for understanding evolutionary dynamics in this mutualism is one in which groups of genetically well defined species of wasps coevolve with groups of genetically less well defined (frequently hybridizing) groups of figs.
Acknowledgments
We thank Adalberto Gomez and Maritza Lopez for help with field work; Whitman Schofield, Erica Hudson, Zuleyka Mainard, and Jeovanna Lowe for help with molecular data collection; G. Michaloud, S. G. Compton, W. Ramirez, C. C. Berg, J. M. Cook, and M. Zavodna for discussion of their observations suggesting hybridization in nature and species complexes in figs; and an anonymous reviewer and E. Leigh, Jr., S. Van Bael, and J. Hey for comments that greatly improved the presentation. This work was supported by the Smithsonian Tropical Research Institute, National Science Foundation Grant DEB-0108475, and start-up research funds from the University of Arizona.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Systematics and the Origin of Species: On Ernst Mayr's 100th Anniversary,” held December 16-18, 2004, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: COI-II, cytochrome oxidase I/II; ML, maximum likelihood.
Data deposition: The nucleotide sequences reported in this paper have been deposited in the GenBank database (accession nos. AY967870-AY968016).
References
- 1.Boucher, D. H. (1985) The Biology of Mutualism: Ecology and Evolution (Croom Helm, London).
- 2.Margulis, L. & Fester, R. (1991) Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis (MIT Press, Cambridge, MA). [PubMed]
- 3.Douglas, A. E. (1994) Symbiotic Interactions (Oxford Univ. Press, Oxford).
- 4.Thompson, J. N. (1994) The Coevolutionary Process (Univ. of Chicago Press, Chicago).
- 5.Maynard Smith, J. & Szathmáry, E. (1995) The Major Transitions in Evolution (Freeman Spektrum, Oxford).
- 6.Herre, E. A., Knowlton, N., Mueller, U. G. & Rehner, S. A. (1999) Trends Ecol. Evol. 14, 49-53. [DOI] [PubMed] [Google Scholar]
- 7.Bronstein, J. L. (2001) in Evolutionary Ecology: Concepts and Case Studies, eds. Fox, C. W., Roff, D. A. & Fairbairn, D. J. (Oxford Univ. Press, Oxford), pp. 315-330.
- 8.Corner, E. J. H. (1952) Wayside Trees of Malaya (Government Printer Office, Singapore).
- 9.Wiebes, J. T. (1979) Annu. Rev. Ecol. Syst. 10, 1-12. [Google Scholar]
- 10.Pellmyr, O. (2003) Ann. Missouri Bot. Gard. 90, 35-55. [Google Scholar]
- 11.Kato, M., Takimura, A. & Kawakita, A. (2003) Proc. Natl. Acad. Sci. USA 100, 5264-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galil, J. & Eisikowitch, D. (1968) Ecology 49, 259-269. [Google Scholar]
- 13.Ramirez, W. (1970) Evolution (Lawrence, Kans.) 24, 681-691. [Google Scholar]
- 14.Janzen, D. H. (1979) Annu. Rev. Ecol. Syst. 10, 13-51. [Google Scholar]
- 15.Weiblen, G. D. (2002) Annu. Rev. Entomol. 47, 299-330. [DOI] [PubMed] [Google Scholar]
- 16.Cook, J. M. & Rasplus, J.-Y. (2003) Trends Ecol. Evol. 18, 241-248. [Google Scholar]
- 17.Berg, C. C. & Wiebes, J. T. (1992) African Fig Trees and Fig Wasps (North-Holland, Amsterdam).
- 18.Harrison, R. D. (2005) Bioscience, in press.
- 19.Berg, C. C. (1989) Experientia 45, 605-611. [Google Scholar]
- 20.Terborgh, J. (1986) in Conservation Biology: The Science of Scarcity and Diversity, ed. Soulé, M. E. (Sinauer, Sunderland, MA), pp. 330-344.
- 21.McKey, D. (1989) Experientia 45, 661-673. [Google Scholar]
- 22.Kalko, E. K. V., Herre, E. A. & Handley, C. O. (1996) J. Biogeogr. 23, 565-576. [Google Scholar]
- 23.Korine, C., Kalko, E. K. V. & Herre, E. A. (2000) Oecologia 123, 560-568. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez, W. (1974) Ann. Missouri Bot. Gard. 61, 770-780. [Google Scholar]
- 25.Galil, J. (1977) Endeavour 1, 52-56. [Google Scholar]
- 26.Herre, E. A. (1989) Experientia 45, 637-647. [Google Scholar]
- 27.Herre, E. A. & West, S. A. (1997) Proc. R. Soc. London Ser. B 264, 1501-1507. [Google Scholar]
- 28.Herre, E. A. (1999) in Levels of Selection in Evolution, ed. Keller, L. (Princeton Univ. Press, Princeton), pp. 209-237.
- 29.Wiebes, J. T. (1982) Neth. J. Zool. 32, 395-411. [Google Scholar]
- 30.Wiebes, J. T. (1987) in Systematics and Evolution: A Matter of Diversity, ed. Hovenkamp, P. (Utrecht Univ. Press, Utrecht, The Netherlands), pp. 309-314.
- 31.Herre, E. A., Machado, C. A., Bermingham, E., Nason, J. D., Windsor, D. M., McCafferty, S. S., VanHouten, W. & Bachmann, K. (1996) J. Biogeogr. 23, 521-530. [Google Scholar]
- 32.Machado, C. A., Jousselin, E., Kjellberg, F., Compton, S. G. & Herre, E. A. (2001) Proc. R. Soc. London Ser. B 268, 685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiblen, G. D. (2001) Syst. Biol. 50, 243-267. [PubMed] [Google Scholar]
- 34.Yokoyama, J. (1995) in Biodiversity and Evolution, eds. Arai, R., Kato, M. & Doi, Y. (The National Science Museum Foundation, Tokyo), pp. 115-130.
- 35.Jousselin, E., Rasplus, J.-Y. & Kjellberg, F. (2003) Evolution (Lawrence, Kans.) 57, 1255-1269. [DOI] [PubMed] [Google Scholar]
- 36.Weiblen, G. D. (2004) Syst. Biol. 53, 128-139. [DOI] [PubMed] [Google Scholar]
- 37.Weiblen, G. D. (2000) Am. J. Bot. 87, 1342-1357. [PubMed] [Google Scholar]
- 38.Datwyler, S. L. & Weiblen, G. D. (2004) Am. J. Bot. 91, 767-777. [DOI] [PubMed] [Google Scholar]
- 39.Murray, M. G. (1985) Biol. J. Linn. Soc. 26, 69-81. [Google Scholar]
- 40.Corner, E. J. H. (1958) Reinwardtia 4, 325-355. [Google Scholar]
- 41.Corner, E. J. H. (1965) Gdns. Bull. Singapore 21, 1-186. [Google Scholar]
- 42.Wiebes, J. T. (1994) The Indo-Australian Agaoninae: Pollinators of Figs (North-Holland, Amsterdam).
- 43.Machado, C. A., Herre, E. A., McCafferty, S. & Bermingham, E. (1996) J. Biogeogr. 23, 531-542. [Google Scholar]
- 44.Rasplus, J.-Y., Kerdelhué, C., Le Clainche, I. & Mondor, G. (1998) C. R. Acad. Sci. III 321, 517-527. [DOI] [PubMed] [Google Scholar]
- 45.Ramirez, W. (1991) Rev. Biol. Trop. 39, 87-95. [Google Scholar]
- 46.Corner, E. J. H. (1985) Biol. J. Linn. Soc. 25, 187-195. [Google Scholar]
- 47.Page, R. D. M. (1994) Cladistics 10, 155-173. [Google Scholar]
- 48.Weiblen, G. D. & Bush, G. L. (2002) Mol. Ecol. 11, 1573-1578. [DOI] [PubMed] [Google Scholar]
- 49.Herre, E. A. (1996) J. Biogeogr. 23, 593-607. [Google Scholar]
- 50.Nason, J. D., Herre, E. A. & Hamrick, J. L. (1998) Nature 391, 685-687. [Google Scholar]
- 51.Herre, E. A., Machado, C. A. & A., W. S. (2001) in Adaptationism and Optimality, eds. Orzack, S. & Sober, E. (Cambridge Univ. Press, New York), pp. 191-218.
- 52.Molbo, D., Machado, C. A., Sevenster, J. G., Keller, L. & Herre, E. A. (2003) Proc. Natl. Acad. Sci. USA 100, 5867-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H. & Flook, P. (1994) Ann. Entomol. Soc. Am. 87, 651-701. [Google Scholar]
- 54.Strand, A. E., Leebens-Mack, J. & Milligan, B. G. (1997) Mol. Ecol. 6, 113-118. [DOI] [PubMed] [Google Scholar]
- 55.Swofford, D. L. (1998) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 56.Shimodaira, H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16, 1114-1116. [Google Scholar]
- 57.Wiebes, J. T. (1963) Tijdschr. Entomol. 106, 1-112. [Google Scholar]
- 58.Rasplus, J.-Y. (1996) in The Biodiversity of African Plants, eds. van der Maesen, L. J. G., van der Burgt, X. M. & van Medenbach de Rooy, J. M. (Kluwer, Wageningen, The Netherlands), pp. 639-649.
- 59.Kerdelhué, C., Le Clainche, I. & Rasplus, J. Y. (1999) Mol. Phylogenet. Evol. 11, 401-414. [DOI] [PubMed] [Google Scholar]
- 60.Lopez-Vaamonde, C., Dixon, D. J., Cook, J. M. & Rasplus, J.-Y. (2002) Zool. J. Linn. Soc. 136, 637-683. [Google Scholar]
- 61.Michaloud, G., Michaloud-Pelletier, S., Wiebes, J. T. & Berg, C. C. (1985) Proc. K Ned. Akad. Wet. C 88, 93-119. [Google Scholar]
- 62.Michaloud, G., Carriere, S. & Kobbi, M. (1996) J. Biogeogr. 23, 513-520. [Google Scholar]
- 63.Ramirez, W. (1970) Univ. Kansas Sci. Bull. 49, 1-44. [Google Scholar]
- 64.Compton, S. G. (1990) S. Afr. J. Sci. 86, 39-40. [Google Scholar]
- 65.Kerdelhué, C., Hochberg, M. E. & Rasplus, J. Y. (1997) Biotropica 29, 69-75. [Google Scholar]
- 66.Parrish, T. L., Koelewijn, H. P. & van Dijk, P. J. (2003) Biotropica 35, 333-343. [Google Scholar]
- 67.Ramirez, W. (1994) Rev. Biol. Trop. 42, 339-342. [Google Scholar]
- 68.Ware, A. B. & Compton, S. G. (1992) Biotropica 24, 544-549. [Google Scholar]
- 69.Compton, S. G. (1993) Afr. Entomol. 1, 151-158. [Google Scholar]
- 70.Kliman, R. M., Andolfatto, P., Coyne, J. A., Depaulis, F., Kreitman, M., Berry, A. J., McCarter, J., Wakeley, J. & Hey, J. (2000) Genetics 156, 1913-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Machado, C. A., Kliman, R. M., Markert, J. A. & Hey, J. (2002) Mol. Biol. Evol. 19, 472-488. [DOI] [PubMed] [Google Scholar]
- 72.Wang, R. L., Wakeley, J. & Hey, J. (1997) Genetics 147, 1091-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hudson, R. R., Kreitman, M. & Aguade, M. (1987) Genetics 116, 153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tajima, F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu, Y. X. & Li, W. H. (1993) Genetics 133, 693-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watterson, G. A. (1975) Theor. Popul. Biol. 7, 256-276. [DOI] [PubMed] [Google Scholar]
- 77.Sweigart, A. L. & Willis, J. H. (2003) Evolution (Lawrence, Kans.) 57, 2490-2506. [DOI] [PubMed] [Google Scholar]
- 78.Wright, S. I., Lauga, B. & Charlesworth, D. (2003) Mol. Ecol. 12, 1247-1263. [DOI] [PubMed] [Google Scholar]
- 79.Molbo, D., Machado, C. A., Herre, E. A. & Keller, L. (2004) Mol. Ecol. 13, 1613-1623. [DOI] [PubMed] [Google Scholar]
- 80.Tajima, F. (1983) Genetics 105, 437-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pamilo, P. & Nei, M. (1988) Mol. Biol. Evol. 5, 568-583. [DOI] [PubMed] [Google Scholar]
- 82.Wright, S. I. & Gaut, B. S. (2005) Mol. Biol. Evol. 22, 506-519. [DOI] [PubMed] [Google Scholar]
- 83.Nason, J. D., Herre, E. A. & Hamrick, J. L. (1996) J. Biogeogr. 23, 501-512. [Google Scholar]
- 84.Kiester, A. R., Lande, R. & Schemske, D. W. (1984) Am. Nat. 124, 220-243. [Google Scholar]
- 85.Baker, H. G. (1961) Evolution (Lawrence, Kans.) 15, 378-379. [Google Scholar]
- 86.Askew, R. R. (1968) Evolution (Lawrence, Kans.) 22, 642-645. [DOI] [PubMed] [Google Scholar]
- 87.Cook, J. M., Bean, D., Power, S. A. & Dixon, D. J. (2004) J. Evol. Biol. 17, 238-246. [DOI] [PubMed] [Google Scholar]
- 88.Kjellberg, F., Jousselin, E., Bronstein, J.L., Patel, A., Yokoyama, J. & Rasplus, J. Y. (2001) Proc. R. Soc. London Ser. B 268, 1113-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barker, N. P. (1985) Bothalia 15, 607-611. [Google Scholar]
- 90.Ware, A. B., Kaye, P. T., Compton, S. & Van Noort, S. (1993) Plant Syst. Evol. 186, 147-156. [Google Scholar]
- 91.Van Noort, S., Ware, A. B. & Compton, S. G. (1989) S. Afr. J. Sci. 85, 323-324. [Google Scholar]
- 92.Hossaert-McKey, M., Gibernau, M. & Frey, J. E. (1994) Entomol. Exp. Appl. 70, 185-191. [Google Scholar]
- 93.Gibernau, M. & Hossaert-McKey, M. (1998) Ecoscience 5, 306-311. [Google Scholar]
- 94.Grison, L., Edwards, A. A. & Hossaert-McKey, M. (1999) Phytochemistry 52, 1293-1299. [Google Scholar]
- 95.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).