ABSTRACT
Autism spectrum disorders (ASD) are highly heritable, heterogeneous neurodevelopmental disorders characterized by clinical presentation of atypical social, communicative, and repetitive behaviors. Over the past 25 years, hundreds of ASD risk genes have been identified. Many converge on key molecular pathways, from translational control to those regulating synaptic structure and function. Despite these advances, therapeutic approaches remain elusive. Emerging data unearthing the relationship between genetics, microbes, and immunity in ASD suggest an integrative physiology approach could be paramount to delivering therapeutic breakthroughs. Indeed, the advent of large-scale multi-OMIC data acquisition, analysis, and interpretation is yielding an increasingly mechanistic understanding of ASD and underlying risk factors, revealing how genetic susceptibility interacts with microbial genetics, metabolism, epigenetic (re)programming, and immunity to influence neurodevelopment and behavioral outcomes. It is now possible to foresee exciting advancements in the treatment of some forms of ASD that could markedly improve quality of life and productivity for autistic individuals. Here, we highlight recent work revealing how gene X maternal exposome interactions influence risk for ASD, with emphasis on the intrauterine environment and fetal neurodevelopment, host–microbe interactions, and the evolving therapeutic landscape for ASD.
KEYWORDS: Host–microbe interactions, gut-microbiota-brain axis, autism spectrum disorder, neurodevelopmental disorders, hologenome, early life, neuroimmune
Introduction
Autism spectrum disorders (ASD) are characterized by heterogeneous presentation of social, communication, behavioral, and cognitive deficits. In 2023, the Autism and Developmental Disabilities Monitoring (ADDM) Network estimated that 1 in 25 boys and 1 in 100 girls eight years of age (or 1 in 36 children) in the United States is autistic,1 a marked increase from its most recent estimate of 1 in 44 children.2 Despite its prevalence and identification of molecular-to-neural circuit-level mechanisms contributing to syndromic and other genetically defined ASD populations,3,4 risk profiles for ASD remain poorly defined, most cases are of unknown etiology, and effective preventative and therapeutic measures remain limited.
The genetic landscape of ASD is heterogenous and multifaceted, encompassing (1) syndromic forms, such as that associated with Fragile X Syndrome,5 (2) single nucleotide polymorphisms like mutations in the gene encoding voltage gated sodium channel 1.2 (Nav1.2), SCN2A,6 and (3) copy number variants (CNVs), such as 15q11.2q13 deletions or duplications.7 This genetic complexity is reflected in the wide range of features, symptoms, and severity which characterize ASD (the ‘spectrum’). Despite these relatively well-characterized disorders, the underlying cause of most cases remains completely unknown. Given the high heritability of ASD, most studies have rationally focused on understanding how genetic variants contribute to disease risk and the underlying pathology. However, it is increasingly evident that environmental factors can modify ASD risk.8,9 The concept that early-life environmental exposures that modify gene expression through epigenetic reprogramming could result in a phenotype indistinguishable from that of genomic variants is gaining traction, as the burgeoning field of exposomics attempts to capture how the full array of environmental exposures experienced from fetal development onward contribute to disease risk and outcomes, even across generations.
The “exposome,” a term coined by cancer biologist Christopher P. Wild in 2005,10 encompasses the myriad environmental factors to which an individual is exposed throughout life. Notably, a growing body of evidence highlights the role of the early life exposome in setting the stage for disease risk later in life. Beyond cancer, the interplay between genetics and the exposome appears to be particularly relevant to neurodevelopmental disorders, which typically emerge during early childhood.11 Detrimental events, such as in utero exposure to valproic acid (VPA),12 occurring during developmental critical periods can disrupt highly orchestrated changes in gene expression and thereby affect the formation of behaviorally relevant neural circuits. While investigation into the relationship between the exposome and neurodevelopmental health is in its infancy, exposure science is on the radar of major biomedical funding agencies, including the National Institute of Environmental Health Science (NIEHS).
Most environmental exposures that influence early life neurodevelopment likewise act on the maternal and offspring gut microbiome, as well as the host immune system.13–15 Microbiota-immune interactions occur both in utero and during early-postnatal life, underscoring the importance of the maternal exposome in offspring risk for neurodevelopmental disorders. Maternal diet, infections, and medications can significantly alter the microbiome and the maternal inflammatory response. Each has been linked to risk for neurodevelopmental disorders in both preclinical and clinical studies.16
Multi-OMICs-based studies are beginning to unravel the complex relationship between the microbiome and its associated metabolome with the host immunome, epigenome, and, ultimately, the tissue-specific transcriptome. Intriguingly, some preclinical studies exploit a key feature of environmentally driven epigenetic alterations: their reversibility.17,18 Alongside host genome-targeted approaches,19 modulation of the gut microbiome and the immune system during early development are being explored as innovative strategies for ASD. Here, we review recent evidence for complex genome–metagenome interactions in ASD and how early life environmental exposures, particularly those that affect the maternal and intrauterine environments, contribute to risk for ASD by influencing development of the brain and the immune system, and their interactions.
Autism spectrum disorder: a complex condition with increasing prevalence
Autism spectrum disorder (ASD) is characterized by presentation of persistent deficits in communication and social interaction as well as restricted and repetitive pattern of interests or activities which lead to significant impairment in social and occupational functioning.20 The core features of ASD often emerge in early development, but the age of diagnosis varies. ‘Spectrum’ reflects the heterogeneous clinical manifestation of ASD, which can differ according to severity of the disorder as well as the developmental stage, biological sex, and age of the subject. While early detection and intervention, ideally in infancy, is associated with better outcomes,21 most cases are diagnosed after age three.
The prevalence and demographics of children diagnosed with ASD in the United States are changing.22 Biological sex is a well-established risk factor for ASD, with the DSM-V reporting that “[ASD] is diagnosed four times more often in males than in females.” Consistent with much of the human literature, our group23 and others have reported increased severity of autism-like phenotypes among male animals in preclinical models for ASD. However, in a 2009 meta-review of 43 studies on the prevalence of ASD published since 1966,24 the CDC concluded that this gap is narrowing, as the ratio between boys and girls has decreased in the autism and developmental disabilities monitoring (ADDM) network overall. Girls affected by ASD are more likely to be misdiagnosed25 or late-identified26 given that female autism phenotypes diverge from the classically established diagnostic criteria for ASD.27,28 More recent studies implementing improved diagnostic assessments of social communication and restricted and repetitive behaviors that adjust for bias in sex-related measurements and account for sex-specific symptom trajectories identify a more equivalent ASD prevalence among boys and girls.29 Similarly, classical behavioral analyses in preclinical models for ASD were designed to assess male-dominant behavioral traits;30 thus, there is a historical gap assessing and reporting on female-specific behaviors, like maternal behavior and alloparenting,31 in the preclinical literature. Nonetheless, biological factors such as sex hormones and sex-based genetic risk, areas of active investigation, might be responsible for the apparent increase in vulnerability to ASD among males.32–34
The etiology of non-syndromic ASD, like other neurodevelopmental and neuropsychiatric disorders, is now thought to be multifactorial, involving complex interactions between risk genes and environmental insults35,36 incurred in early development. ASD etiology has been under intense investigation for decades. In the 1960s, a popular, but errant, theory that lack of parental warmth as a determinant of ASD37 emerged in opposition to competing theories focused on biological factors like brain development.38 In the following decades, epidemiologic, genetic, cytogenetic, and neuroimaging studies39,40 provided causal links between ASD and altered brain development. The resulting data definitively characterized autism as a disorder of biological origin. This ushered in a new era of investigation into ASD anchored by the concept that gene mutations were the primary driver of autistic phenotypes, leading to the classification of ASD into syndromic and non-syndromic forms.41 Syndromic autism occurs in subjects with other neurological conditions, such tuberous sclerosis complex (TSC),42 Rett syndrome (RTT),43 fragile X syndrome (FXS),44 and phosphatase and tensin homologue (PTEN) macrocephaly syndrome.45 It is determined by a mutation in a specific gene or group of genes. Non-syndromic autism, which accounts for most cases, is not linked to other defined conditions and cannot be traced to mutations in a single gene or specific chromosomic aberrations. Nonetheless, investigation of syndromic ASD can aid in understanding non-syndromic autism.46
Several nongenetic – or, environmental – factors can interfere with fetal brain development in ways that converge onto the molecular and cellular pathways implicated in syndromic autism and are, thereby, proposed to contribute to either risk for or the severity of autistic phenotypes.47,48 While there is a significant amount of evidence to support that the early life exposome is relevant to ASD pathology, the causal relationship between environmental exposures and ASD remains an area of intense investigation and equally intense debate in the field.
Pregnancy and the periconceptional period have been identified as developmental critical periods during which environmental exposures can influence child health outcomes by longitudinal population-based birth cohort studies, such as the Human Early Life Exposome (HELIX) study.49 The environmental factors – chemical or physical agents, nutrition, psychological and social conditions, and infectious disease, among others – to which we are exposed throughout life constitute the ‘exposome.50,51 The idea that the pregnancy exposome exerts a considerable impact on fetal development originates from David Barker’s “Fetal Programming Hypothesis”52 that chronic disorders manifesting in postnatal/adult life may, in part, result from environmental insults in utero. This influential theory is also known as the “developmental origins of health and disease (DOHaD)” hypothesis.
In 2003, the CHARGE (Childhood Autism Risks from Genetics and the Environment) study was launched to comprehensively assess the contribution of environmental factors to ASD.53 CHARGE identified increased risk for ASD associated with maternal occupational exposure to solvents and postulated that other chemicals might have a similar effect on neurodevelopment.54 Human epidemiological55 and animal studies suggest that other maternal factors, such as infection and diet/metabolic status during pregnancy can likewise contribute to ASD risk in offspring. The underlying mechanisms by which discrete environmental exposures increase risk for ASD, among other non-communicable chronic disorders, are an area of active investigation. One unifying theory is that they induce epigenetic reprogramming of various systems (e.g., the immune and/or nervous systems) during developmental critical periods, which depend on temporally strict and spatially precise regulation of gene expression. The hypothesis that environmentally induced reprogramming of the fetal epigenome can trigger fetal programming of disease risk has gained ground in the scientific and medical communities, setting the stage for large-scale epigenomic studies which have the potential to greatly expand upon current screening and therapeutic approaches for ASD, among other diseases.56,57
Reimagining ASD as a disease at the crossroad between genes and the environment, in which environmental exposures can serve as a “tipping point” toward disease manifestation in genetically susceptible individuals has facilitated the development of new and innovative theories and investigation into the determinants of ASD. In this context, a growing body of evidence suggests that gut microbiome-immune interactions play a critical role in mediating the effects of early life environmental exposures on long-term neurodevelopmental health outcomes.
Host and microbial genetics in ASD
In recent decades, rodent and non-human primate models for ASD have been developed to facilitate mechanistic understanding of ASD pathology and spur the discovery of novel therapeutics. These models target human risk variants, obtained by genetic engineering,58,59 and include models for single-gene mutations associated with syndromic ASD or non-syndromic ASD and models of copy number variations (CNVs).
Mecp2 mutant mice60 and macaques,61 for instance, reproduce genetic mutations associated with Rett syndrome (RTT), a neurodevelopmental disorder caused by mutation of the gene encoding X-linked methyl-CpG binding protein 2 (MECP2), which regulates transcription62 and RNA splicing.63 Similarly, Tsc1/Tsc2 mutant mice model Tuberous sclerosis complex (TSC), an autosomal dominant neurodevelopmental syndrome resulting from mutation in TSC1 or TSC2, which encode proteins hamartin (Tsc1) and tuberin (Tsc2), respectively, inhibitors of the mTORC1 translational control pathway.64 About 50% of TSC individuals are also diagnosed with ASD, and TSC genetic alterations account for about 1–4% of ASD cases.65 Tsc1 heterozygous mice show increased anxiety-like behavior, impaired learning and memory, and reduced social interaction, which is rescued by rapamycin administration.66,67
Mouse models of single-gene mutations associated with non-syndromic ASD, like those encoding neurexins (NRXN), neuroligins (NLGN), and SHANK proteins, have uncovered a significant and valuable portion of ASD biology.68 However, modeling the contribution of a single gene in an animal model, particularly when using Cre-drivers to investigate cell type-specific roles of the gene, does not yield full insight into the role of that gene in the pathophysiology of the disease in genetically heterogeneous human patient populations. To overcome these limitations, functional studies have been developed to identify and manipulate one or more cellular and molecular pathways on which common and rare variants associated with ASD converge.69,70
Most human ASD risk genes belong to at least two main clusters: (1) regulation of mRNA translation and protein synthesis71 and (2) regulation of synaptic structure and function.72 Investigation of syndromic ASD strongly supports a mechanistic link between synaptic dysfunction and dysregulated translational control. An intriguing hypothesis is that non-syndromic ASD is driven by a similar convergence.46 We propose that this could also true of some cases associated with specific environmental exposures, particularly those that affect the intrauterine environment.
Epigenetic modifications in ASD
Despite its multifactorial etiology, ASD aggregates in families and is highly heritable.73 However, the concordance rate for ASD in MZ twin pairs ranges from 36% to 96%,74,75 implicating nongenetic disease liability.76,77 The emerging hypothesis that epigenetic mechanisms can causally contribute to ASD risk78,79 provides a rational explanation for disease-discordant MZ twin pairs. Epigenetic modifications, such as DNA methylation, histone modification and regulation, and transcriptional gene silencing by means of long non-coding RNAs (lncRNAs) and small non-coding RNAs (sncRNAs), influence chromatin architecture and conformation, the accessibility of genes to transcriptional complexes, and gene expression.80 Epigenetic programming is complex. It can be the result of primary stochastic phenomena, environmental factors, or DNA mutations.81 Given that precise spatial and temporal regulation of gene expression is crucial for the establishment of proper excitatory and inhibitory synaptic connections, activity-dependent responses, and neuronal specification, epigenetic reprogramming driven by environmental exposures has significant ramifications for brain development, function, and disease risk.
Mutations in genes encoding chromatin remodeling enzymes are implicated in ASD and other neurodevelopmental disorders.82–84 Significant epigenetic variability has been reported between MZ twins.85,86 Consistently, considerable differences in DNA methylation have been found in MZ twins discordant for phenotypically complex disorders, like schizophrenia and bipolar disorder.87 Analysis of lymphoblastoid cell lines derived from ASD-discordant MZ twin pairs’ peripheral blood lymphocytes revealed many ASD-relevant loci with differential methylation profiles.88 Moreover, a genome-wide analysis revealed that DNA methylation at specific CpG sites varied significantly within ASD-discordant MZ twin pairs.89 Further studies90–92 identified differential DNA methylation patterns in autistic versus non-affected individuals. Some autistic individuals carry mutations in genes encoding proteins involved in epigenetic modification. Indeed, a de novo mutation in the HIST1H1E gene, which encodes for the linker histone H, was reported to disrupt chromatin organization and downregulate protein expression in ASD patients.93 Intriguingly, in the same study, a review of SFARI GENE (https://gene.sfari.org/),94 a curated database of candidate ASD risk genes, determined that almost 20% of risk genes encode proteins involved in epigenetic regulation and chromatin remodeling. For instance, SETD5, a member of the SET-domain family encoding for histone methyltransferase (HMT) which regulates gene expression during early development and is implicated in both synaptic plasticity and cell fate determination, is linked to both ASD and ID.95–97
Beyond non-coding RNAs, DNA methylation, and chromatin remodeling, short stretches of RNA known to regulate mRNA translation and degradation, microRNAs (miRNA), are implicated in ASD pathology. The contributions of miRNAs to ASD pathoetiology have been recently well reviewed elsewhere.98
The gut microbiome in ASD
Large-scale studies aimed at characterizing the human microbiome (i.e., the NIH-funded Human Microbiome Project (HMP)99 and the European Metagenomics of the Human Intestinal Tract (MetaHIT))100 contributed to technological and computational advances that dramatically increased accessibility to the field and, together, drove a consequent boom in the number of publications on the gut microbiome in human physiology and pathology. It is now well established that the gut microbiome plays key roles in both health and disease, from cancer to autoimmune disorders and neuropsychiatric disorders.101
The gut microbiome is a dynamic ecological system that varies between individuals and within the same individual across the lifespan. Community dynamism occurs over even shorter timescales, as the relative abundance of certain taxa can ebb and flow according to circadian rhythms.102 Host factors, including diet, drugs, toxins, pathogens, the immune system, and physical and psychological conditions together drive a constant reshaping of gut microbiome composition (Figure 1), which can evolve into a detrimental state – here referred to as, “dysbiosis.”103 Powerful stressors can trigger a cascade of events that ultimately lead to a decrease in microbial diversity while simultaneously creating a permissive environment for the growth of opportunistic pathogenic taxa.104,105 These events, in turn, alter the pool of metabolites produced by the microorganisms, and consequently, interactions between the microbiome and host, often with adverse consequences for the host.
Figure 1.
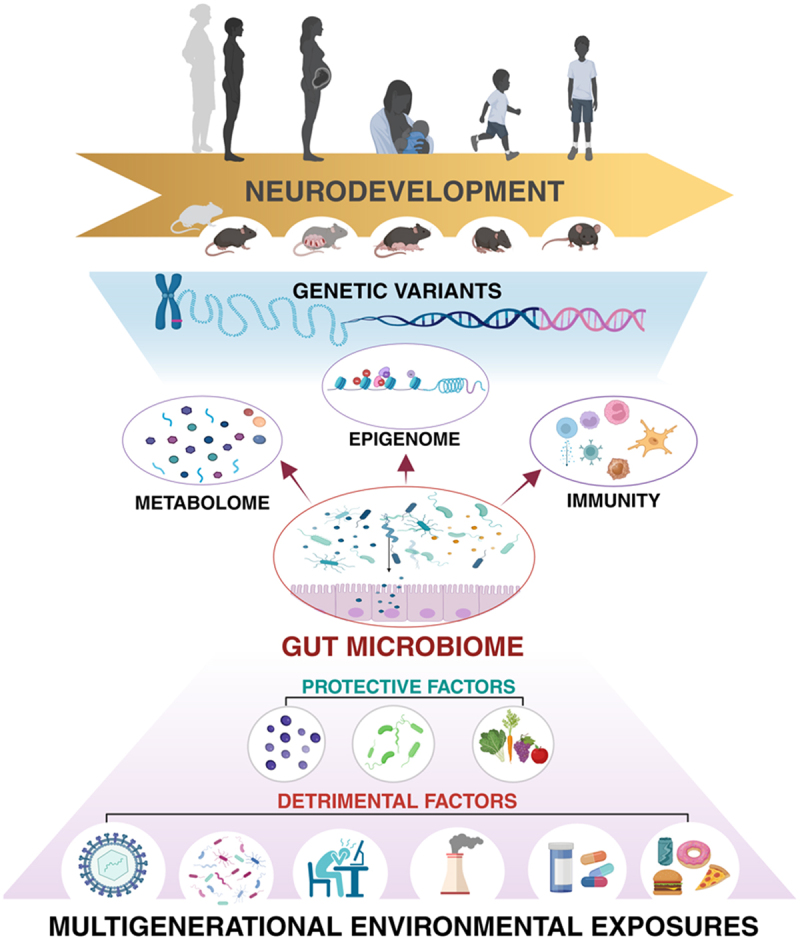
As a key intermediary between the exposome and genetic susceptibility, the gut microbiome is poised to influence risk for neurodevelopmental disorders.
Classically, mutations in one or more genes involved in regulating brain development and function were thought to be the exclusive drivers of neurodevelopmental disorders; however, a growing body of preclinical and clinical research is revealing a critical role for gene X environment interactions in determining predisposition to and the severity of neurodevelopmental disorders. Environmental exposures – from infection to diet to air quality – that influence the functional composition of the maternal and early life infant gut microbiome can alter the diverse pool of microbially associated metabolites available to the host, which can then affect maturation and function of the immune system and drive cell type-specific epigenetic reprogramming that influences neurobehavioral outcomes. Notably, environmental exposures incurred across multiple generations can affect early life neurodevelopment and disease risk through inherited patterns of microbiome alterations and epigenetic modifications. Given its strategic position between the host and its environment, the gut microbiome presents an intriguing duality as both a potential contributor to and a therapeutic target for reducing risk for neurodevelopmental disorders in children.
Investigations at the intersection of neuroscience and microbiology have begun to unravel the contributions of gut resident microorganisms to the development and homeostasis of host brain structure and function. The microbiome-gut-brain axis (MGBA) encompasses the mechanisms involved in mediating the interplay between the gut and the brain, which include anatomical (e.g., the vagus nerve), immunological, metabolic, neuronal, and chemical pathways.106 Early evidence for the existence of the MGBA was provided by experiments using germ-free (GF) animals,107 fecal microbiota transplantation (FMT), and antibiotic-driven changes in the gut microbiome composition108 in the context of brain disorders.109–113 These pioneering studies identified key contributions of gut microbiota to brain function and behavior,114 making evident that elucidating the mechanisms by host–microbe interactions affect brain development, function, and behavior is imperative to understanding the physiopathology of many neuropsychiatric conditions and to the development of new microbiota-based and -targeted treatments.115
Among the multiple comorbidities associated with ASD, gastrointestinal (GI) dysfunction – manifesting as diarrhea, constipation, and abdominal pain116 – is one of the most frequently reported,117,118 though different studies show varying prevalence,119 likely due to the genetic and epigenetic heterogeneity of ASD. The presence of GI symptoms is associated with more severe ASD120 and is heavily cited in surveys of parents of autistic children.118 Several studies report substantial differences in the composition of the gut microbiome of autistic children compared to neurotypical controls,121–123 although a direct causal relationship remains to be demonstrated. Furthermore, the directionality of a relationship – whether ASD pathology alters the gut microbiome or changes in the gut microbiome could causally contribute to ASD – is an area of debate, with limited investigations demonstrating that divergent and limited dietary preferences in autistic individuals may be the driver of any microbiome changes observed in autistic populations when compared to neurotypical controls124 (but see,125 with which we side). Despite limited reproducibility of taxa-specific signatures across studies of gut microbiome composition among children with ASD, recent investigations coupling intestinal metagenomics with metabolomic analyses have identified metabolic signatures reflecting functional alterations in gut microbial ecology of affected individuals.126–128 In an effort to reconcile the notorious inconsistency comparing microbiome composition among an autistic population versus controls, Morton et al.127 applied a Bayesian differential ranking algorithm to identify commonalities among 10 cross-sectional microbiome datasets and 15 others. The authors identified distinct patterns between children with ASD and age- and sex-matched counterparts when accounting for microbiome (16S rRNA and whole-genome shotgun metagenomic sequencing) and the human transcriptome (RNA-seq). They also revealed positive correlations between pro-inflammatory cytokines, read immune dysregulation, and the global microbial log fold changes between ASD and control pairs – interestingly, canonical pro-inflammatory cytokine IL-6 was only linked to a few taxa, whereas TGF-β levels were linked to many taxa. Moreover, a recent study129 comparing microbiome composition between children and adolescents diagnosed with ASD, attention deficit hyperactivity disorder (ADHD), and comorbid ASD/ADHD,130,131 was the first to show the presence of shared microbiome signature in children with ASD and ADHD which are distinct from non-related controls as well as similarities in altered immune markers and an increase in gut permeability indicators. Here, the authors report lower bacterial richness among youth with both disorders compared to non-related controls, including a specific decrease in the relative abundance of Coprobacter and Howardella; in contrast, Eggerthella – a taxa previously associated with developmental delays in children132 – Hungatelle, and Ruminococcus gnavus group were found to be enriched in both the ASD and ADHD groups. While greater microbial variability was reported between children with ASD, as compared to ADHD, and controls, relatively few variations were observed between the gut microbiota of youth with ASD and ADHD. Together, these findings suggest a common, microbiome-mediated mechanism might contribute to the overlapping clinical features of ASD and ADHD.
Gut microbiota produce metabolites that can have a strong impact on behavior and the underlying neural correlates via the gut-brain-axis.133–135 Consequently, modulating gut microbiome composition represents a novel and innovative strategy for treating ASD. An open-label study of Microbiota Transfer Therapy (MTT)136 was among the first to investigate the effects of fecal microbiota transplant (FMT) in children with ASD and chronic gastrointestinal disturbances. A two-year follow-up study137 aimed at assessing the long-term results of MTT showed significant improvements not only gastrointestinal symptoms but also in core autism symptoms for all 18 subjects. MTT specifically increased microbial diversity and restored microbial metabolic capability to a similar level to the typically developing (TD) children.138 These promising results prompted the FDA to grant fast track status to MTT for autistic children in 2019. More recently, oral delivery of AB-2004, a small-molecule sequestrant targeting microbially derived metabolites, was shown to significantly reduce irritability in children with ASD.139 Additionally, a double-blind randomized placebo-controlled trial of precision treatment with a microbe shown to reverse social dysfunction in multiple mouse models for ASD133,140,141 was found to specifically enhance social behavior in children with ASD, consistent with its behavior-specific effects in the preclinical experiments.142
Microbial communities can serve as a source of epigenetic modifiers influencing gene expression.143 Microbially derived metabolites have been shown to both directly and indirectly impact the activity of enzymes involved in regulating epigenetic pathways, including those orchestrating DNA methylation and histone modification.144 Microbial regulation of host chromatin modification states and associated transcriptional responses is strictly dependent on host dietary patterns, in particular fiber content. Microbial anaerobic fermentation of insoluble dietary fiber produces short-chain fatty acids (SCFAs),145 organic monocarboxylic acids that can cross the intestinal barrier through monocarboxylate transporters (MCTs), travel through systemic circulation, and reach distal organs.146 Here, they can be metabolized as an energy source via the Krebs cycle but also play multiple signaling roles as SCFAs bind to the G protein-coupled receptors (GPCR) GPR43 and GPR41, later renamed free fatty acid receptor 2 (FFAR2) and 3 (FFAR3).147 SCFAs promote intestinal barrier integrity,148 counteract intestinal inflammation,149 and modulate gastrointestinal motility, adipogenesis, and glucose homeostasis.150 They have been shown to modulated intestinal mucosal immunity151 and are thought to also affect the peripheral immune system. CD4+ regulatory T cells, in particular Th17 cells,152 are regulated by SCFAs and their differentiation is impaired in GF mice.153 Similarly, SCFAs are required for CD8+ cytotoxic T cell transition in memory cells.154 SCFA oral administration promoted peripheral regulatory T cell differentiation in mice,155 and FFAR agonists have been shown to modulate the human monocyte inflammatory pathway by decreasing the release of pro-inflammatory cytokines.156 Such regulation of peripheral immunity might be key to their impact on brain function.
SCFAs facilitate MGBA communication and affect brain physiology through multiple mechanisms, as reviewed in Dalile et al. (2019).157 SCFA receptors are expressed by neurons in both the peripheral and central nervous systems.158 Recent work showed an association between SCFA production in the gut and regulation of feeding behavior via direct hypothalamic regulation.159 Additionally, SCFAs maintain blood–brain barrier (BBB) integrity.160 Propionate-induced FFAR3 activation of vagal fibers has been shown to increase the activity of the dorsal vagal complex, parabrachial nuclei, and hypothalamus,161 thus suggesting that SCFAs can directly influence brain activity through vagal signaling. Another mechanism by which SCFAs influence brain activity is via enteroendocrine signaling. FFAR activation in gut enteroendocrine L cells determines the production of hormones released in response to food, GLP-1 and PYY, into circulation162,163 to regulate appetite and nutrient intake.164,165 Animal studies have shown GLP-1 can further promote learning and memory166 and improve neuroplasticity while reducing microglial activation in the hippocampus.167 Furthermore, microbially derived SCFAs influence hippocampal neurogenesis by acting on monocytes.168 SCFAs are also implicated in the modulation of anxiety- and depressive-like behavior following psychosocial stress, which is associated with alterations in the gut microbiome.169
SCFAs can directly modulate the activity of histone deacetylase enzymes (HDACs), which regulate chromatin accessibility and gene expression. Acetate, butyrate, and propionate exert an inhibitory effect on HDACs,170 dysregulation of which is linked to neurodegenerative and neuropsychiatric disorders, including schizophrenia.171 They are proposed to mediate hippocampal long-term potentiation by enhancing histone acetylation, a process required for long-term memory formation.172 Additionally, SCFAs modulate epigenetic modifications in neuro-, peripheral, and enteric immune systems.173,174 Acetate was recently shown to regulate microglial metabolism and function through modifying histone methylation on genes related to microglial proliferation, morphology, and activation.175 Finally, SCFAs, particularly propionate, decrease IL-17 production by human and mouse γδ intraepithelial lymphocytes in a HDAC-dependent manner.176
Several recent studies investigated the association between altered SCFAs and neurodevelopmental disorders in children, including ASD;177,178 however, whether SCFAs contribute to or relieve ASD pathology and symptom severity is controversial. While some studies report elevated SCFAs in fecal samples isolated from autistic individuals compared to controls, others report lower levels. Serum SCFA concentration is less commonly reported but this could be an important data point for teasing out the effects of SCFA concentration on host brain function and behavior in the context of ASD.179,180 Higher levels of SCFAs have been found in a valproic acid mouse model for ASD,181 while a beneficial effect of butyrate on social dysfunction has been reported in the BTBR mouse model for idiopathic ASD.182 Hence, it is likely that the impact of SCFAs on host health is context-dependent.
Taken together, the studies above demonstrate that gut microbiota influence host gene expression and can thereby contribute to disease risk and outcomes. This intricate interplay between host and microbial factors could contribute to the huge variability characteristic of nonsyndromic ASD. Heritability of the gut microbiome, another form of genetic inheritance, could furthermore influence disease risk across generations, as we have found in an animal model for maternal overnutrition.23 However, such heritability appears to be responsive to environmental forces.183 Further studies with large sample sizes and high resolution longitudinal multi-OMICs-based assessments are required to elucidate the extent to which gut microbiota impact health outcomes across multiple generations. Nonetheless, increasing evidence suggests that antenatal maternal gut microbial communities play a particularly pivotal role in offspring neurodevelopment, especially during critical developmental stages such fetal and early-post natal life.
Neurodevelopmental disorders and the maternal exposome
In recent years, the pregnancy exposome 184 has garnered increased attention in the investigation of risk factors occurring in early development that may permanently affect vulnerability to disease later in life. Given that the maternal environment is, to an extent, modifiable, it has also become a target for the development of therapeutic interventions. Multiple investigations highlight the connection between the pregnancy exposome and fetal programming of disease, particularly metabolic impairments and cardiovascular disorders.185–188 We and others propose that early life programming is likely to influence risk for neurodevelopmental disorders, including ASD.189–191
Intrauterine brain development involves a precise succession of events orchestrated by lineage-specific gene expression programs.192–194 Environmental insults can determine cell type-specific epigenetic programming at different stages of neurodevelopment,195,196 including of neurons, glia, and immune cells.197 Epigenetic reprogramming is implicated in synaptic formation,198,199 and environmental exposures incurred during the third trimester of human development can disrupt synaptogenesis, a key process dysregulated in ASD. Multiple nongenetic factors, such as maternal infection during pregnancy, maternal diet/metabolic status, and maternal chemical exposure, incurred at various pregnancy stages have been proposed to interfere with the developing human brain and, thereby, contribute to autistic phenotypes.47,48 Below, we consider the relationship between environmental exposures, the maternal gut microbiome, and offspring outcomes.
Maternal infection
In utero exposure to maternal viral infections, particularly those requiring hospitalization, is associated with increased risk for ASD. In the 1970s, an American child psychiatrist, Stella Chess, diagnosed symptoms of ASD in a group of pediatric patients with congenital rubella syndrome, resulting from the 1963–1964 rubella epidemic in New York. The reported prevalence was 200 times higher than that of the general population in the US.200,201 Subsequent studies revealed similar findings, not only in relation to rubella infection but also in response to maternal infection during pregnancy with other viruses, such as cytomegalovirus202,203 and influenza.204,205 A great body of work now demonstrates that damage to the developing brain results from maternal immune activation (MIA) and related inflammatory responses independent of the specific class of pathogen (e.g., viral versus bacterial).206–208 Relatedly, multiple studies identify an association between ASD and dysregulation of the inflammatory response: increased activation of microglia and astroglia,209 upregulation of markers of inflammation,210 and alterations in genes involved in immune function211 have been identified in autistic patient populations. Consequently, cytokine profiles have been proposed as biomarkers of immune dysfunction in autistic individuals.212 Indeed, human studies have identified a significant increase in the levels of pro-inflammatory cytokines, including IL-6,210 TNFα,213 IFN-γ,214 IL-17,215 in the brain and in biological fluids, including serum and cerebrospinal fluid, of autistic individuals compared to controls. In contrast, levels of anti-inflammatory, regulatory cytokines such as IL-23216 and TGF-β217 have been found to be downregulated in autistic individuals. Similarly, analyses of serum218,219 and amniotic fluid206,220 from mothers who gave birth to autistic children reveal an increase in proinflammatory cytokine and chemokine levels, when compared to control subjects.
The causal relationship between gestational MIA and ASD and the biological mechanisms by which MIA interferes with fetal neurodevelopment has been investigated using preclinical animal models221 including: (1) maternal exposure to pathogens during pregnancy, such as influenza virus,205,222,223 (2) exposure to agents which stimulate the innate immune system, such as the bacterial endotoxin lipopolysaccharide (LPS), polyinosinic-polycytidylic acid (poly(I:C)),224 a synthetic analog of double-stranded RNA (dsRNA), or the soluble tachyzoite antigen of the protozoan Toxoplasma gondii 225 (3) stimulation of the immune system by pro-inflammatory cytokines, such as IL-6,226 and (4) administration of immunological factors linked with the pathogenesis of the disorder,227 as in the case of ASD-associated maternal autoantibodies.228
The effects of MIA on fetal neurodevelopment differ according to pregnancy stage at the time of exposure. Studies in LPS/Poly(I:C)-stimulated models have shown that the development of the dopaminergic (DA) system, a neuronal network strongly implicated in ASD,229 is selectively impaired when MIA is triggered during early gestation.230 In LPS/Poly(I:C) models, stimulants do not reach the fetus directly; instead, their effects on neurodevelopment are mediated by maternal cytokines released in the circulation and transmitted to the fetus through the placenta.231 Cytokines modulate neurodevelopment,232,233 however an imbalance in maternal pro- and anti-inflammatory cytokines can have detrimental effects on the fetal brain. The relationship between maternal cytokine imbalance and disruption of neurodevelopment has also been investigated in the context of ASD.234 Offspring of mice stimulated with Poly(I:C) or LPS during gestation display core ASD-like behavioral impairments:235 repetitive self-grooming and stereotypies, restricted interests/cognitive inflexibility, and decreased sociability.134,236–240 In this context, IL-6,241 IL-17a,240 and IL-1β242 have been implicated in MIA phenotypes. However, the precise mechanisms by which pro-inflammatory cytokines impact fetal brain development, brain function, and behavior remain mostly unknown.
Another mechanism linking immune and brain function (or dysfunction, in the case of MIA) converges on mammalian target of rapamycin (mTOR) complexes one and two (mTORC1, mTORC2), powerful regulators of mRNA translation and actin (and, thereby, synaptic) remodeling, respectively.243 mTORC1 and mTORC2 are involved in the differentiation of Th1 and T helper 17 (Th17) and Th2 cells, respectively.244 mTOR complex inhibition drives T cells to differentiate into Treg cells.245 Thus, an interesting hypothesis suggests that mTOR hyperactivity along the gut-brain-immune axis, one of the pathological mechanisms involved in ASD, might lead to a decrease in Treg cell-associated anti-inflammatory cytokines, including IL-10 and TGF-β.246
Multiple studies highlight a role for the gut microbiome in the interplay between the immune system and neurodevelopment in the context of MIA.247 A seminal study in the field demonstrated that supplementation of MIA offspring with a probiotic species shown to contribute to host immune maturation and protect against Helicobacter hepaticus-driven colitis in mice,248 Bacteroides fragilis, could rescue many ASD-like behaviors including aberrant communication, stereotypy, anxiety-like, and hyperactivity, but notably did not rescue social dysfunction.134 While the gut microbiome influences the development of both the adaptive and innate immune systems both locally and systemically, the immune system finely tunes the symbiotic host–microbe relationship to avoid microorganism overgrowth while simultaneously allowing tolerance.249,250 For instance, a MIA-associated spike in IL-17a in maternal circulation is a consequence of Th17 cell expansion in the gut and depends on segmented filamentous bacteria (SFB). Mice devoid of SFB are protected from the pathogenic release of IL-17a and do not display the associated phenotypical aberrations typical of the MIA model.135,240 Immunotherapy-mediated blockade of pathological IL-17a pathway activation likewise prevented ASD-like symptoms in the poly(I:C) MIA model.240
Maternal infections during gestation could directly affect fetal microglia, which could contribute to behavioral dysfunction observed in MIA offspring.251 Microglia are innate sentinel immune cells which regulate inflammatory processes in the brain by the release of pro- and anti-inflammatory cytokines and chemokines. In addition to immune surveillance, CNS microglia also regulate CNS maturation and synaptic plasticity. Microglial abnormalities have been reported in postmortem analysis of brains of autistic individuals.209 Microglial maturation and differentiation of microglia are partially regulated by gut microbes.174,252 The maternal microbiome in particular influences the activity of microglia during prenatal life, as microglial homeostasis was shown to be disrupted in mice born to germ-free dams.253 Microglia also mobilize monocytes from the periphery to enter the brain, a process mediated by systemic TNF-α signaling, which leads to microglial activation and subsequent recruitment of activated monocytes.254,255 Interestingly, the trafficking of monocytes from the spleen might be modulated by microbiota-produced SCFAs, which bind to free fatty acid receptor type 2 (FFAR2) expressed on peripheral lymphocytes.256
Recent work highlighted the effect of SCFA supplementation, via high-fiber diet, on the inhibition of inflammatory microglia by downregulation of HDAC activity, NF-κB activity, and inflammation caused by LPS stimulation.257 Inhibition of HDACs, which leads to transcriptional repression, has been proposed as a primary downstream action of SCFAs toward prevention of neuroinflammation.258 A recent study conducted in the BTBR mouse model for idiopathic autism, which is characterized by systemic immune dysregulation and comorbid gut dysbiosis, traced the immune abnormalities back to the embryonic stages of the yolk sac where macrophages (microglia) and peripheral immune cells differentiate.259 The underlying mechanism involved transcriptional regulation by HDAC1. The epigenetic abnormalities, associated with increased proinflammatory cytokines and microglia activation, were successfully reversed upon administration of sodium butyrate, which inhibits HDAC1 activity. This study reveals a key role for epigenetic reprogramming of immune function as common etiology between environmental risk factors for ASD and highlights the potential for correcting postnatal immune dysregulation at the embryonic stage through maternal microbiome-targeted therapies.
Maternal diet
Maternal nutrition and metabolism exert a major impact on offspring fetal and early-postnatal development, including on gamete maturation and placental growth.260 Furthermore, maternal intake of micro- and macronutrients-alike has been shown to be crucial for successful development of offspring organs and systems, including the nervous and immune systems, which has significant implications for neurodevelopment and risk for neurodevelopmental disorders.47,261
Recent epigenome-wide studies of low-income populations262,263 suggest that epigenetic alterations contribute to the detrimental consequences of micronutrient deficiency on neurodevelopment. Vitamin D and folate deficiency have been extensively investigated in the context of ASD and other NDDs.264 Vitamin D plays a crucial role in many biological processes,265 with numerous studies highlighting its importance in pregnancy and fetal growth and development.266 Suboptimal levels of circulating vitamin D in pregnant women267 are associated with increased pregnancy complications, such as miscarriage,268 hypertensive disorders,269 and gestational diabetes.270 Notably, low maternal vitamin D also increases risk for developmental deficits, including ASD, in offspring.271–273 To this end, vitamin D supplementation is indicated when vitamin D deficiency is identified during pregnancy.274 Interestingly, it has been reported that maternal depletion of vitamin D is associated with alterations of the epigenetic landscape, specifically in DNA methylation, across multiple generations.275 Recent studies in human populations276 have reported significant associations between vitamin D and changes microbiome composition in the context of autoimmune disorders. These findings suggest a link between maternal vitamin D, microbiome composition, and immune function with potential effects on fetal epigenetic programming. However, further studies are required to test this hypothesis. Similarly, folate (vitamin B-9) is a crucial nutrient during early pregnancy for reducing risk for birth defects, notably neural tube defects (NTDs).277 The neuroprotective effect of folate is likely mediated by genome-wide modification of methylation patterns in neural target genes.278,279 While mammalian cells are unable to produce folate, and therefore exogenous intake is required, there is substantial evidence that colonic bacteria produce a considerable amount of folate, as well as other B-vitamins. While their production can be enhanced by prebiotic supplementation,280 there is no evidence for an essential role for bacterial folate biosynthesis in early development.
Beyond micronutrient supplementation, large preclinical and clinical studies on the effects of macronutrient intake – general dietary habits considering the ratio of protein to fat to carbohydrates consumed – such as Western Pattern Diet (WPD),281 also known as the Standard American Diet (SAD), Mediterranean Diet (MedDiet),282 and ketogenic diet (KD),283 and associated metabolic conditions, are beginning to reveal how maternal diet could contribute to risk for ASD and other NDDs. As introduced above, the developmental origins of health and disease (DOHaD) hypothesis suggests that maternal diet, in particular maternal undernutrition, has a causal role in the incidence of various disorders in adulthood. While initial observations focused on alterations in cardiovascular and metabolic function in subsequent generations of populations impacted by famine, this theory was later extended to include risk for mental disorders.284 Increased prevalence of major affective disorder,285 antisocial personality disorder,286 schizophrenia spectrum disorder,287 impaired cognitive performances,288 and substance addiction289 have each been correlated with prenatal exposure to extreme caloric restriction in epidemiological studies. Notably, further epidemiological data suggests specific correlations between the trimester in which mothers are exposed to dietary restrictions and the specific disorder induced in the offspring,290,291 thus highlighting the relevance of timing in developmental programming of future disorders, and suggest a critical role for exposure during early gestation.
Operating as the interface between the maternal and fetal blood circulation and regulating the nutrient and oxygen transfer from the mother to the fetus, the placenta directly contributes to intrauterine fetal programming. It integrates maternal and fetal signals and constantly balances fetal needs with maternal supply. Perturbations in the maternal compartment are sensed by the placenta, which in turn modulates blood flow and nutrient supply and adaptively modifies hormonal release through epigenetic changes in placental cells.260,292 Excessive deprivation (undernourishment) or abnormal increase (overnutrition) in maternal nutrient intake at conception and throughout pregnancy impact the ability of the placenta to properly allocate necessary resources for fetal growth.293 Nutrient sensing in the placenta occurs by means of multiple mechanisms, including one involving the mTORC1 translational control pathway294 in the syncytiotrophoblast, which is regulated by several factors, including maternal adipokines,295 IL-6,296 TNF-α,297 leptin,298 adiponectin.299 Maternal undernourishment causes a decrease placental amino acid transport, which in turn drives intrauterine growth restriction (IUGR).300,301 Maternal obesity and diabetes drive excess of nutrient supply to the placenta which results in fetal overgrowth302,303 and increased risk for the infants to develop obesity and metabolic dysfunction in adulthood.304,305 This evidence suggests a U-shaped relationship between maternal nutritional imbalances, either maternal malnutrition306,307 or obesity,306–308 with offspring risk for metabolic disorders.
Maternal diet and metabolism are also implicated in offspring mental health outcomes. Preclinical and human studies suggested a role for maternal nutrition in the etiology of neuropsychiatric disorders,309–312 neurodevelopmental disorders,313,314 and cognitive function.315,316 As we recently reviewed in Di Gesu et al. (2021),281 maternal obesity, overweight, and associated metabolic disorders increase odds ratios for neurodevelopmental disorders, including ASD and ADHD, in children.317–319 Accumulating evidence provided by epidemiological studies suggests a strong correlation between maternal obesity320–323 and diabetes323,324 and an increased risk for ASD among children exposed to maternal obesity and diabetes in utero. Animal models325 of diet-induced obesity have been used to investigate the mechanisms underlying the detrimental effects maternal obesity on offspring neurodevelopment in neuropsychiatric disorders. Maternal high-fat diet (MHFD) has been shown to impair synaptic plasticity,326 social behavior,23,133,327 learning and memory328,329 and neurogenesis330,331 in offspring. Yet, the precise mechanisms underlying the impact of maternal diet on offspring neurodevelopment remain to be determined.
A growing body of evidence suggests that dietary challenges, as in the case of WPD, and associated dysbiosis of the gut microbiome have the potential to reprogram the host epigenome in a tissue-specific fashion due to alterations in the intestinal metabolite pool either produced or transformed by microbiome enzymatic activity.145,332 Epigenetic alterations are associated with other maternal nutritional conditions, such as maternal starvation333 and maternal deficiency of vitamins and cofactors,334,335 and are associated with ASD and other NDDs.79,336 Modifications in the fetal epigenome are associated with maternal obesity337,338 and maternal diabetes.339–341 For instance, MHFD offspring display alterations in histone binding and expression of the oxytocin receptor (OXT-R) in the hippocampus342 which plays an important role in social behavior,343 hypermethylation in the regulatory regions of hypothalamic POMC gene344,345 which is involved in regulation of food intake,346 and alteration of histone modifications and expression in the hippocampal leptin receptor (Lepr),347 which is involved in synaptogenesis and neural circuit maturation.348
Maternal high-fat diet (MHFD)-induced changes in both the maternal and offspring immune system could, likewise, causally contribute to increased risk for NDDs. Interestingly, in perinatal MHFD mouse models, changes in the expression of several epigenetic regulators in the offspring developing brains were found in association with an anxiety-like phenotype.349 One intriguing possibility is that, similar to what has been observed in MIA offspring, one of the causal mechanisms underlying social dysfunction in MHFD offspring might depend on maternal microbiota-dependent pathological activation of pro-inflammatory pathways, such as the IL-17a pathway, given that HFD regimen also results in a TH17 bias,350 and MHFD drives microbiota-dependent IL-17–producing type 3 innate lymphoid cell expansion (ILC3s).351 Of note, L. reuteri supplementation has been shown to promote differentiation of CD4+CD8aa+ T precursor cells into immunoregulatory T cells (Tregs), as opposed to a TH17 cell fate,352 an effect which might be mediated by probiotic-dependent increase in SCFA levels.
More recently, birth cohort studies have investigated the potential beneficial effects of a Mediterranean-style diet, rich in fruits, vegetables, and polyunsaturated fatty acids (PUFAs), and low in ultra-processed foods and saturated fatty acids, during pregnancy in reducing risk for neurodevelopmental disabilities in offspring.282,353,354 While adherence to a MedDiet has been linked to lower mortality and decreased prevalence of obesity, diabetes, low-grade inflammation, cancer, neurodegenerative disorders, and depression,355,356 the underlying molecular and cellular mechanisms of action are not yet fully elucidated. Recently, a link between the MedDiet and the gut microbiota in disease risk was proposed.357 In randomized controlled studies, a MedDiet regimen in obese subjects lowered plasma cholesterol and was associated with changes in the gut microbiome and systemic metabolome independent of caloric intake.358 A similar study in elderly populations showed that MedDiet increased the abundance of microbial taxa associated with SCFA production, lowered inflammation, and improved cognitive performance.359
The effects of adherence to Mediterranean dietary patterns during gestation on both maternal and offspring health outcomes have been investigated in contrast to obesity-associated WPD,360 however the relationship between maternal MedDiet and offspring neurodevelopment has yet to be fully explored. Interestingly, a recent study including mother and infant dyads enrolled in the Newborn Epigenetics STudy (NEST), showed that MedDiet in early gestation was associated with favorable neurobehavioral outcomes and sex-dependent changes in methylation patterns of imprinted genes in offspring.361 As mentioned above, the MedDiet is described as rich in polyunsaturated fatty acids (PUFAs), which are classified as n-6 PUFAs and n-3 PUFAs and considered essential nutrients given the absence of specific enzymes required for their synthesis in mammals.362 Intriguingly, low levels of n-3 PUFAs (mainly eicosapentaenoic acid (EPA) and docosahexaenoic (DHA) acids) in the plasma and the brain of autistic individuals have reported in epidemiological studies.363,364 Randomized controlled trials reported behavioral improvements in ASD children treated with n-3 fatty acid dietary supplementation.365 Animal studies corroborated epidemiological evidence showing that n-3 PUFA deficiency to be associated with alterations in GABAergic and dopaminergic neurotransmission as well as ASD-like behavioral impairment in rodents.366,367
Studies in MIA models report a positive effect of n-3 PUFA-enriched diet in reducing ASD-like symptoms.368 Given that n-3 PUFAs can regulate neuroinflammatory processes and microglial activity,369 their protective effects stem from anti-inflammatory activity in the developing brain. In further support of the beneficial role of these fat molecules in preventing ASD-like symptoms in animal models, a recent study showed that n-3 PUFA supplementation prevented the behavioral, cellular, and molecular ASD-related disturbances in the VPA mouse model for ASD, especially in female offspring.370 Interestingly, n-3 PUFAs have been shown to modulate the composition of the gut microbiome,371 and increase the abundance of probiotic taxa, such as Lactobacilli and Bifidobacterium, and SCFA-producing species.372 Conversely, gut microbiota can also affect the metabolism and absorption of n-3 PUFAs.373 While the interplay between gut microbes and this class of polyunsaturated fats remains to be fully understood, recent research demonstrated PUFA supplementation ameliorated autistic phenotypes and GI dysfunction in Fmr1 knockout (KO) mice, a genetic model for fragile X syndrome (FXS), by altering the gut microbiome.374 Epidemiological studies show that maternal consumption of n-3 PUFAs is associated with lower risk of impairments in social development scores and motor and communication skills in children.375 While preclinical studies showed that exposure to a diet rich in n-3 PUFAs during pregnancy modulate the gut microbiome of the offspring and prevents metabolic alterations induced by HFD, further investigation is required to elucidate how dietary PUFAs act on maternal microbial ecology and contribute to the development of ASD in offspring.376 Given the ability of PUFAs to positively modulate epigenetic modifications in both the placenta and the fetal brain,377–379 it is possible that maternal gut microbiota mediate the effects of PUFAs in offspring via epigenetic programming of neurodevelopment. Consistently, the relationship between maternal PUFA consumption, associated changes in the gut microbiome, and offspring risk for neurodevelopmental disorders, including ASD, is of great interest in the research community and we anticipate increasingly mechanistic work to come out on this topic.380
Another dietary pattern which has gained popularity in recent years is the ketogenic diet (KD), which is characterized by a high proportion (modeled at 74% kcal from fat) of fats and proteins with low intake of carbohydrates. KD enhances the production of ketone bodies (KBs), which can substitute for glucose as the primary energy source, especially in the brain where KBs are used as substrates for oxidative metabolic processes. KBs are considered beneficial in certain neurological conditions given their role in various in multiple brain processes, including neuroinflammation, neuroplasticity, synaptic transmission, and cellular energetics and metabolism.381 The beneficial effects of KD have been proposed to be mediated by changes in the gut microbiome. Importantly, despite the high intake of fats characterizing the KD, these alterations seem to be distinct from those induced by HFD regimen, probably due to the concomitant production of KBs by the host. KBs have been shown to reduce the abundance of Bifidobacterium, with a concomitant decrease in Th17-mediated immune response,382 a pathway implicated in ASD pathogenesis.240,383 Multiple studies have demonstrated the ability of a KD to mitigate some of the behavioral symptoms displayed in animal models of ASD.384,385 Similar effects were also observed in autistic children following a KD regimen, including amelioration of hyperactivity, social deficits,386 and seizure frequency.387 Preclinical evidence suggests that the antiseizure effects of KD occurs via increases in the relative abundance of Akkermansia and Parabacteroides in mice.388 Additionally, studies in the BTBR model of ASD showed that microbiome remodeling was crucial in the modulation of neurobehavioral symptoms.389,390 While these data suggest that the KD and associated effects on the gut microbiome may be a new therapeutic approach in autistic patients, there is a paucity of investigations on alterations of the gut microbiota in children treated with a KD and caution is warranted when proposing KD in autistic populations. KD is an extreme dietary regimen which could be difficult to implement in children affected with ASD, who often display signs of food aversion and might therefore lead to macronutrient and micronutrient deficiencies. Similar considerations should be made when contemplating maternal nutritional interventions based on KD, given limited evidence for beneficial effects on offspring neurodevelopment. Notably, a recent study investigating the effects of KD on the course of gestation and fetal development in rats showed both metabolic impairments and delays in neurological development, particularly in female offspring.391
The studies summarized here highlight the profound effects of maternal nutrition – from the periconceptional period throughout gestation and lactation – on offspring neurodevelopment and provide compelling evidence for a complex interplay between maternal nutrition, the gut microbiome, and the immune response, with epigenetic modifications as a consequential bridge between these environmental cues and early life programming of neurodevelopmental disorders.
Maternal chemical exposure
Exposure to even small amounts of toxic chemicals, such as lead, methylmercury, polychlorinated biphenyls [PCBs], arsenic, and toluene, relatively harmless in adult individuals, during pre- and early postnatal neurodevelopment392,393 can result in neurodevelopmental toxicity and brain damage.394 The placenta is permeable to a large number of environmental toxins,395 therefore the fetus has little-to-no protection against these agents, many of which can easily cross the blood–brain barrier.396–398 The detrimental effects of exposure to toxic substances are not limited to the intrauterine period, but instead extend across many years.192
PCBs (polychlorinated biphenyls), a class of compounds utilized as additives for pesticides, insulators, paints, and glues, are internationally recognized as hazardous. While industrial exposure to PCBs showed mild toxicity in adult individuals, severe behavioral impairments, as well as hormonal and immune dysfunction were observed in children born to exposed mothers.399–401 The first evidence for neurotoxicity of PCBs dates back to the 1970s, when babies born to women who ingested PCB-contaminated cooking oil during pregnancy in Japan and Taiwan suffered from significantly higher rates and more severe forms of cognitive and psychomotor impairments.402 However, subsequent epidemiological investigations revealed that maternal exposures to even lower, yet environmentally relevant, levels of PCBs poised significant risk for neurotoxicity in offspring,403 and was associated with higher risk of neurodevelopmental disorders, including ADHD and ASD.404,405 Mechanisms by which PCBs have been proposed to disrupt neurodevelopment include interference with thyroid hormone (TH), altered signaling of γ-aminobutyric acid (GABA), and disruption of intracellular calcium ion (Ca2+) dynamics.406
Pesticides, including insecticides and fungicides, are widely used in agriculture and their residues can be found in and on fruits, vegetables, and other food products,407 with a large number of these being specifically designed to produce neurotoxic effects in targeted pests.408,409 Prenatal exposure to organophosphates, which are the most widely used class of pesticides, has been associated with neurodevelopmental deficits, including cognitive dysfunction and attention deficits.410–413 Interestingly, maternal intake of high dose folic acid preceding pregnancy has been shown to reduce ASD risk arising from prenatal exposure to pesticides,414 suggesting a protective role for folic acid against harmful chemical exposures.
Fortunately, a growing number of countries, including the US, have begun to implement environmental regulations to limit exposures to known toxicants. Among these, the Toxic Substances Control Act (TSCA) of 1976 provided the US Environmental Protection Agency (EPA) with the authority to require rigorous recording and reporting of chemical substances and/or mixtures released into the environment; however, most food, drugs, cosmetics, and pesticides were notably excluded from the chemicals covered by this act. After these regulatory initiatives, the environmental concentrations of PCBs in commercial mixtures gradually declined. Nevertheless, the persistence of legacy PCBs pose a health hazard to humans,415 with ongoing utilization of old equipment containing PCBs and their release from aging construction components.
Despite well-intended regulation, the enactment, enforcement, and achievement of strict environmental quality standards remains a challenge, given the inevitable lag between the production and broad commercial application of new classes of synthetic chemicals and research efforts to objectively perform a comprehensive assessment of their impacts on short- and long-term health outcomes. While PCBs came to the forefront of environmental policy when the EPA developed regulations under the TSCA, PFAS (Per- and Polyfluoroalkyl Substances) were subject to relatively less strict regulations, with scientists mainly concentrating on offering guidance on acceptable levels of PFAS in drinking water and groundwater. However, recent studies indicate that PFAS are building up in the environment and exhibit long-lasting stability, to the point that are commonly referred to as “forever” chemicals.416 This accumulation of PFAS has increasingly adverse effects on both human health and the environment. Consequently, PFAS are becoming as widely recognized among the public as PCBs. PubMed citations including “PFAS” grew from 1 in 2002 to >1,200 in 2023. They are accompanied by a growing number of articles published by popular press outlets, including news and lifestyle magazines, highlighting links between PFAS and health risks, as well as a nuanced consideration of the danger presented by how little is known about them and the consequences of cumulative exposures.
Epidemiological studies have highlighted associations between exposure to specific PFAS, which find extensive application in both industrial and consumer goods, including coatings applied to fabrics, carpets, paper goods, and as nonstick coatings on cookware, and multiple health outcomes, including alterations in immune function, liver and kidney disease, metabolic dysregulation, adverse reproductive and developmental effects, and cancer.417 As mentioned in previous sections, the CHARGE population-based case–control study investigated the association between environmental factors, including prenatal maternal exposure to PFAS, and risk for autism and developmental delay in 1,800 children and their families. Results from this study revealed that modeled prenatal exposure to perfluorohexane sulfonate (PFHxS) and perfluorooctane sulfonate (PFOS), but not other PFAS, was linked to higher odds of a child being diagnosed with ASD.418 However, the authors concluded that additional studies in which PFAS concentrations are prospectively measured in mothers and children at multiple developmental stages were required to corroborate these findings. Another study suggested that prenatal serum levels of Perfluorononanoic acid (PFNA), a type of PFAS, might be linked to slight increases in autism-related traits in children.419 However, future research is warranted to investigate the correlation between maternal exposure to both established and newly emerging PFAS and various quantitative measures of autism-related health outcomes in offspring.
Intriguingly, recent evidence suggests that the gut microbiome might be a major player in the toxicity of environmental pollutants, including pesticides and PFAS.420 Exposure to such toxicants might increase risk for psychiatric or neurological disorders through perturbations of the microbiome-gut-brain axis.421 Maternal exposure to environmental pollutants, including metals, PFAS, and pesticides, was associated with alterations in developmental trajectory of the gut microbiome in infants, especially breastfed infants, in a recent birth cohort study.422 Additional evidence of maternal PFAS exposure on microbiota composition in mother-infant dyads in Finland revealed alterations in the maternal gut microbiome and in the levels of certain metabolites, such as bile acid glycoursodeoxycholic acid (GUDCA) and cholic acid (CA).423 Further studies are required to determine the full extent of consequences of maternal exposures to individual environmental chemicals on offspring neurodevelopment, their impact of the risk neurodevelopmental disorders, and the potential involvement of the gut microbiome in mediating neurotoxic effects of such compounds.
Therapeutic targeting of the maternal gut microbiome to reduce ASD risk
Pregnancy is characterized by significant remodeling of the maternal gut microbiome, even in the absence of environmental insults.424–426 This remodeling is driven by changes in hormone levels, immunity, and metabolic function required to support fetal development. In parallel, microbiota are poised to modulate immune and metabolic adaptations during pregnancy, as well as gut barrier function, by means of bioactive microbially derived metabolites. In the third trimester, total bacterial load is increased while microbial richness is decreased in the maternal gut microbiome. A concomitant increase in the proportion of opportunistic pathogens is thought to promote in the development of the offspring immune system.427 A surge in maternal progesterone during the third trimester increases the abundance of Bifidobacteria abundance,428 which play an important role in the developing infant microbiota.429 Human studies suggest that pregnancy-specific microbiota rearrangements are most pronounced in the third trimester, regardless of maternal pre-pregnancy BMIs and gestational diabetes status.424 Intriguingly, transplantation of microbial strains from women in the third trimester induced greater weight gain and inflammation in germ-free recipient female mice than those isolated from women in their first trimester, which resemble increased adiposity and insulin resistance430 as well as low-grade inflammation431 observed in the latter stages of gestation in healthy women. While such changes are typically associated with metabolic disorders in non-pregnant individuals, in the context of gestation they are thought to be beneficial metabolic adaptations required ensure proper fetal growth and development.432 The precise mechanisms by which pregnancy shapes the maternal microbiome, how microbiota modulate the maternal environment, and the consequences of environmental exposures that disrupt the maternal gut microbiome and its remodeling on offspring development have yet to be fully elucidated. Further studies incorporating multi-OMIC approaches are warranted.433
Earlier, we introduced how maternal environmental factors, such as nutrition, shape both the maternal and offspring microbiome, with ramifications for development and long-term health outcomes in offspring. High-fat diet (HFD) exposures leading up to and throughout gestation increase the abundance of taxa involved in the biosynthesis of ketone bodies, fatty acid, vitamins, and bile acids.434 Additionally, HFD reduces the abundance of short-chain fatty acids (SCFAs) and SCFA-producing bacteria, while increasing pro-inflammatory markers lipopolysaccharides (LPS) and tumor necrosis factor (TNF), and promoting loss of intestinal epithelial barrier integrity, and placental hypoxia and inflammation.435,436 Intriguingly, human studies show that overweight pregnant women harbor distinct microbiota compared to those of normal weight437 associated with alterations in several metabolic hormones and pregnancy metabolism.438 Taken together, these studies highlight the crucial role of the maternal microbiome in pregnancy-associated metabolic adaptations and make the case for more extensive investigation into how environmentally induced alterations in maternal gut microbial ecology influence physiological changes in offspring and their consequences for fetal development and programming of disease, including neurodevelopmental disorders.
We propose that precision targeting the maternal gut microbiome could provide for a healthier in utero environment, thus facilitating typical fetal development and reducing risk for adverse health outcomes. Select probiotics have the potential to remedy dysbiosis of the gut microbiome through a variety of mechanisms439 and are generally regarded as safe (GRAS) to administer during pregnancy.440,441 In a recent study, maternal probiotic supplementation during gestation and lactation was found to promote intestinal barrier integrity and reduce inflammation.442 A limited number of studies have begun to investigate the impacts of daily probiotic administration in obese pregnant women or animal models of diet-induced obesity443 with some of them showing increased gut microbial diversity,444 albeit small effects on metabolic parameters.445 Promisingly, a randomized double-blinded Danish study investigating the efficacy of a multi-strain probiotic versus placebo-control to regulate blood glucose, gestational weight gain, and reduce risk for gestational diabetes mellitus in obese pregnant women demonstrated >80% adherence to the probiotic regimen and an increase in alpha diversity of the gut microbiome of group receiving the probiotic intervention over time. In contrast, no increase in alpha diversity was observed in the placebo control group.
Both single- and multi-strain probiotic treatments have been used to target gut-brain-behavior interactions and ameliorate or prevent neuropsychiatric outcomes in human and animal studies.446,447 Clinical trials have reported ameliorations of core and comorbid symptoms in autistic children after rebalancing the composition of the gut microbiome through probiotic interventions.448,449 However, the efficacy of maternal probiotic supplementation to counteract the detrimental effects of diet-induced dysbiosis of gut microbiome on offspring neurodevelopment has not yet been explored. A recent study in CD-1 IGS mice450 showed that multi-strain probiotic (Bio-Kult Advanced® containing Bifidobacterium spp. and Lactobacillus spp.) exposure during the perinatal period reduced anxiety-like behaviors associated with maternal obesity, modulated the expression of genes involved in synaptic plasticity in the prefrontal cortex of offspring, and increased brain lactate and SCFA levels, which are known to regulate gene expression.146 Additionally, the multi-strain probiotic decreased inflammation, as indicated by a reduction in circulating levels of pro-inflammatory cytokine interleukin-6 (IL-6) and increased SCFA production in obese dams treated with probiotics.450 This study also provided evidence of a critical role for probiotic species and their metabolites in the regulation of mood and behavior through changes in the expression of synaptic plasticity-related genes. In another study in which mouse dams were exposed to a pregnancy-specific dietary regimen, single-strain probiotic administration during the second trimester of pregnancy decreased anxiety-like behavior and modified cortical cytoarchitecture, with differential effects on male versus female offspring.451 In the context of MIA models, a combination of pre- and probiotics (Bifidobacteria and Lactobacillus combined with fructooligosaccharides and maltodextrin) administered to pregnant dams prevented MIA-induced depression-like and ASD-like behaviors in adult offspring, while also reducing the abundance of proinflammatory cytokines levels in the fetal brain.452
The potential for early life probiotic interventions to prevent or treat ASD-associated phenotypes in nonsyndromic autism is further supported by successful intervention in models for genetic and idiopathic ASD. In the BTBR model, administration of probiotic Lactobacillus (L.) rhamnosus rescued social deficits and modulated the composition of the gut microbiome by increasing microbial richness and the abundance of SCFA-producing taxa. Notably, however, in a randomized, placebo-controlled, cross-over study in human males, the JB-1 strain of L. rhamnosus failed to modulate stress or cognitive performance.453 Not long after, treatment with the probiotic Limosilactobacillus reuteri ATTC-PTA-6475 in the Cntnap2−/− 140 and Shank3B−/− mouse models for disorders of social dysfunction was shown to rescue ASD-like behavior and underlying deficits in synaptic plasticity.141 Excitingly, in a recent double-blind, randomized, placebo-controlled clinical trial, this same human-derived strain selectively reversed social deficits in children with ASD when given in combination with its parent strain, Limosilactobacillus reuteri DSM-17938.142 Thus, the promise of preclinical discoveries that precision targeting of the gut microbiome can relieve some core ASD symptoms is beginning to be realized in human patients. Therapeutic targeting of the gut microbiome for the prevention and treatment of neurodevelopmental disorders is a frontier ripe for discovery, innovation, and implementation.
Concluding perspective
Investigative teams around the world are beginning to elucidate the relative contribution of genetics and the environment, and their interactions, to the etiology of ASD and other neurodevelopmental disorders. Genetics are undoubtedly at the core of ASD pathology, and promising treatment options are emerging for monogenic ASDs, such as anti-sense oligonucleotides (ASOs).19 However, in many cases, genetic variants could establish baseline risk for ASD, while environmental factors modify the phenotypic expression of genetic determinants, influencing disease severity and clinical manifestation. Therapeutically, this is very exciting given that the environment is modifiable and some changes can even be reversible. Environmental exposure-related epigenetic reprogramming and remodeling of the maternal gut microbiome provide new perspectives from which to investigate the mechanisms underlying some forms of neurodevelopmental disorders and could spur innovation in the development of therapeutic approaches. For instance, integrating microbiome-targeted (prebiotic, probiotic, and/or symbiotic) treatments for expectant mothers and/or infants could influence the maternal immune landscape and thereby facilitate healthy brain development in children. New tools for precision sampling and delivery of gut microbiota will likely be key to advancing diagnostics and developing breakthrough therapeutic approaches in coming years. Multidisciplinary investigations exploiting the power of integrated multi-OMICs in preclinical animal models for ASD and carefully stratified patient populations are warranted to advance our mechanistic understanding of how early life environmental exposures contribute to risk for ASD and could be key to the development of innovative diagnostics and interventions for ASD.
Acknowledgments
This work was supported by The McNair Medical Institute of The Robert & Janice McNair Foundation, The John S. Dunn Foundation, and the National Institutes of Health [National Institute of Child Health & Human Development (NICHD) R01 HD109095 and R01 HD109780] awards to S.A.B. Further support was provided by the Gulf Coast Center for Precision Environmental Health (GC-CPEH) through the National Institute for Environmental Health Science Core Center Program P30 ES030285.
Funding Statement
The work was supported by the National Institute of Child Health and Human Development [R01 HD109095, R01 HD109780]; National Institute of Environmental Health Sciences [P30 ES030285]; Robert and Janice McNair Foundation; The John S. Dunn Foundation.
Disclosure statement
S.A.B. is an inventor on a patent granted to Baylor College of Medicine related to the use of Limosilactobacillus reuteri for treating disorders characterized by social dysfunction (US Patent No. 11135252). The authors declare no other competing interests.
References
- 1.Maenner MJ, Warren Z, Williams AR, Amoakohene E, Bakian AV, Bilder DA, Durkin MS, Fitzgerald RT, Furnier SM, Hughes MM, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years — autism and developmental disabilities monitoring network, 11 sites, United States, 2020. MMWR Surveill Summ. 2023;72(2):1–39. doi: 10.15585/mmwr.ss7202a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maenner MJ, Shaw KA, Bakian AV, Bilder DA, Durkin MS, Esler A, Furnier SM, Hallas L, Hall-Lande J, Hudson A, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years — autism and developmental disabilities monitoring network, 11 sites, United States, 2018. MMWR Surveill Summ. 2021;70(11):1–16. doi: 10.15585/mmwr.ss7011a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buch AM, Vertes PE, Seidlitz J, Kim SH, Grosenick L, Liston C.. Molecular and network-level mechanisms explaining individual differences in autism spectrum disorder. Nat Neurosci. 2023;26(4):650–663. doi: 10.1038/s41593-023-01259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donovan AP, Basson MA. The neuroanatomy of autism – a developmental perspective. J Anat. 2017;230(1):4–15. doi: 10.1111/joa.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufmann WE, Kidd SA, Andrews HF, Budimirovic DB, Esler A, Haas-Givler B, Stackhouse T, Riley C, Peacock G, Sherman SL, et al. Autism spectrum disorder in fragile X syndrome: cooccurring conditions and current treatment. Pediatr. 2017;139(Suppl 3):S194–S206. doi: 10.1542/peds.2016-1159F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruth KA, Grisolano TM, Ahern CA, Williams AJ. SCN2A channelopathies in the autism spectrum of neuropsychiatric disorders: a role for pluripotent stem cells? Mol Autism. 2020;11(1):23. doi: 10.1186/s13229-020-00330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38(2):181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheroni C, Caporale N, Testa G. Autism spectrum disorder at the crossroad between genes and environment: contributions, convergences, and interactions in ASD developmental pathophysiology. Mol Autism. 2020;11(1):69. doi: 10.1186/s13229-020-00370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strathearn L, Momany A, Kovacs EH, Guiler W, Ladd-Acosta C. The intersection of genome, epigenome and social experience in autism spectrum disorder: exploring modifiable pathways for intervention. Neurobiol Learn Mem. 2023;202:107761. doi: 10.1016/j.nlm.2023.107761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarker Prev. 2005;14(8):1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 11.Riglin L, Wootton RE, Thapar AK, Livingston LA, Langley K, Collishaw S, Tagg J, Smith GD, Stergiakouli E, Tilling K, et al. Variable emergence of autism spectrum disorder symptoms from childhood to early adulthood. Am J Psychiatry. 2021;178(8):752–760. doi: 10.1176/appi.ajp.2020.20071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roullet FI, Lai JK, Foster JA. In utero exposure to valproic acid and autism — a current review of clinical and animal studies. Neurotoxicol Teratol. 2013;36:47–56. doi: 10.1016/j.ntt.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Yu LW, Agirman G, Hsiao EY. The gut microbiome as a regulator of the neuroimmune landscape. Annu Rev Immunol. 2022;40(1):143–167. doi: 10.1146/annurev-immunol-101320-014237. [DOI] [PubMed] [Google Scholar]
- 14.Begum N, Mandhare A, Tryphena KP, Srivastava S, Shaikh MF, Singh SB, Khatri DK. Epigenetics in depression and gut-brain axis: a molecular crosstalk. Front Aging Neurosci. 2022;14:1048333. doi: 10.3389/fnagi.2022.1048333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forssberg H. Microbiome programming of brain development: implications for neurodevelopmental disorders. Dev Med Child Neurol. 2019;61(7):744–749. doi: 10.1111/dmcn.14208. [DOI] [PubMed] [Google Scholar]
- 16.Courchesne E, Pramparo T, Gazestani VH, Lombardo MV, Pierce K, Lewis NE. The ASD living biology: from cell proliferation to clinical phenotype. Mol Psychiatry. 2019;24(1):88–107. doi: 10.1038/s41380-018-0056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiele MA, Gottschalk MG, Domschke K. The applied implications of epigenetics in anxiety, affective and stress-related disorders - a review and synthesis on psychosocial stress, psychotherapy and prevention. Clin Psychol Rev. 2020;77:101830. doi: 10.1016/j.cpr.2020.101830. [DOI] [PubMed] [Google Scholar]
- 18.Kubota T, Miyake K, Hirasawa T. Epigenetic understanding of gene-environment interactions in psychiatric disorders: a new concept of clinical genetics. Clin Epigenet. 2012;4(1):1. doi: 10.1186/1868-7083-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Y, Sztainberg Y, Wang Q, Bajikar SS, Trostle AJ, Wan YW, Jafar-Nejad P, Rigo F, Liu Z, Tang J, et al. Antisense oligonucleotide therapy in a humanized mouse model of MECP2 duplication syndrome. Sci Transl Med. 2021;13(583). doi: 10.1126/scitranslmed.aaz7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Battle DE. Diagnostic and statistical manual of mental disorders (DSM). Codas. 2013;25(2):191–192. doi: 10.1590/s2317-17822013000200017. [DOI] [PubMed] [Google Scholar]
- 21.Elder JH, Kreider CM, Brasher SN, Ansell M. Clinical impact of early diagnosis of autism on the prognosis and parent-child relationships. Psychol Res Behav Manag. 2017;10:283–292. doi: 10.2147/PRBM.S117499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw KA, Bilder DA, McArthur D, Williams AR, Amoakohene E, Bakian AV, Durkin MS, Fitzgerald RT, Furnier SM, Hughes MM, et al. Early identification of autism spectrum disorder among children aged 4 years — autism and developmental disabilities monitoring network, 11 sites, United States, 2020. MMWR Surveill Summ. 2023;72(1):1–15. doi: 10.15585/mmwr.ss7201a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Gesu CM, Matz LM, Bolding IJ, Fultz R, Hoffman KL, Gammazza AM, Petrosino JF, Buffington SA. Maternal gut microbiota mediate intergenerational effects of high-fat diet on descendant social behavior. Cell Rep. 2022;41(2):111461. doi: 10.1016/j.celrep.2022.111461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr Res. 2009;65(6):591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 25.Bargiela S, Steward R, Mandy W. The experiences of late-diagnosed women with autism spectrum conditions: an investigation of the female autism phenotype. J Autism Dev Disord. 2016;46(10):3281–3294. doi: 10.1007/s10803-016-2872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giarelli E, Wiggins LD, Rice CE, Levy SE, Kirby RS, Pinto-Martin J, Mandell D. Sex differences in the evaluation and diagnosis of autism spectrum disorders among children. Disabil Health J. 2010;3(2):107–116. doi: 10.1016/j.dhjo.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai M-C, Lombardo MV, Auyeung B, Chakrabarti B, Baron-Cohen S. Sex/gender differences and autism: setting the scene for future research. J Am Acad Child & Adolesc Psychiatry. 2015;54(1):11–24. doi: 10.1016/j.jaac.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dworzynski K, Ronald A, Bolton P, Happé F. How different are girls and boys above and below the diagnostic threshold for autism spectrum disorders? J Am Acad Child & Adolesc Psychiatry. 2012;51(8):788–797. doi: 10.1016/j.jaac.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 29.Burrows CA, Grzadzinski RL, Donovan K, Stallworthy IC, Rutsohn J, St John T, Marrus N, Parish-Morris J, MacIntyre L, Hampton J, et al. A data driven approach in an unbiased sample reveals equivalent sex ratio of autism spectrum disorder–associated impairment in early childhood. Biol Psychiatry. 2022;92(8):654–662. doi: 10.1016/j.biopsych.2022.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11(7):490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carcea I, Caraballo NL, Marlin BJ, Ooyama R, Riceberg JS, Mendoza Navarro JM, Opendak M, Diaz VE, Schuster L, Alvarado Torres MI, et al. Oxytocin neurons enable social transmission of maternal behaviour. Nature. 2021;596(7873):553–557. doi: 10.1038/s41586-021-03814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron-Cohen S. Empathizing, systemizing, and the extreme male brain theory of autism. Prog Brain Res. 2010;186:167–175. [DOI] [PubMed] [Google Scholar]
- 33.Baron-Cohen S. The extreme male brain theory of autism. Trends Cogn Sci. 2002;6(6):248–254. doi: 10.1016/S1364-6613(02)01904-6. [DOI] [PubMed] [Google Scholar]
- 34.Jacquemont S, Coe Bradley P, Hersch M, Duyzend Michael H, Krumm N, Bergmann S, Beckmann J, Rosenfeld J, Eichler E. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. The Am J Hum Genet. 2014;94(3):415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Felice A, Ricceri L, Venerosi A, Chiarotti F, Calamandrei G. Multifactorial origin of neurodevelopmental disorders: approaches to understanding complex etiologies. Toxics. 2015;3(1):89–129. doi: 10.3390/toxics3010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martens G, van Loo K. Genetic and environmental factors in complex neurodevelopmental disorders. CG. 2007;8(7):429–444. doi: 10.2174/138920207783591717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettelheim B. The empty fortress: infantile autism and the birth of the self. New York (NY): Free Press; 1967. [Google Scholar]
- 38.Rimland B. Infantile autism. East Norwalk (CT), US: Appleton-Century-Crofts; 1964. [Google Scholar]
- 39.Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. Child Phychol Psychiatry. 1977;18(4):297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 40.Piven J, Berthier ML, Starkstein SE, Nehme E, Pearlson G, Folstein S. Magnetic resonance imaging evidence for a defect of cerebral cortical development in autism. Am J Psychiatry. 1990;147:734–739. [DOI] [PubMed] [Google Scholar]
- 41.Ziats CA, Patterson WG, Friez M. Syndromic autism revisited: review of the literature and lessons learned. Pediatr Neurol. 2021;114:21–25. doi: 10.1016/j.pediatrneurol.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Fryer AE, Connor JM, Povey S, Yates JRW, Chalmers A, Fraser I, Yates AD, Osborne JP. Evidence that the gene for tuberous sclerosis is on chromosome 9. The Lancet. 1987;329(8534):659–661. doi: 10.1016/S0140-6736(87)90416-8. [DOI] [PubMed] [Google Scholar]
- 43.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 44.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905–914. doi: 10.1016/0092-8674(91)90397-H. [DOI] [PubMed] [Google Scholar]
- 45.Butler MG. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42(4):318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sztainberg Y, Zoghbi HY. Lessons learned from studying syndromic autism spectrum disorders. Nat Neurosci. 2016;19(11):1408–1417. doi: 10.1038/nn.4420. [DOI] [PubMed] [Google Scholar]
- 47.Hertz-Picciotto I, Schmidt RJ, Krakowiak P. Understanding environmental contributions to autism: causal concepts and the state of science. Autism Res. 2018;11(4):554–586. doi: 10.1002/aur.1938. [DOI] [PubMed] [Google Scholar]
- 48.Bolte S, Girdler S, Marschik PB. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell Mol Life Sci. 2019;76(7):1275–1297. doi: 10.1007/s00018-018-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maitre L, de Bont J, Casas M, Robinson O, Aasvang GM, Agier L, Andrušaitytė S, Ballester F, Basagaña X, Borràs E, et al. Human early life exposome (HELIX) study: a European population-based exposome cohort. BMJ Open. 2018;8(9):e021311. doi: 10.1136/bmjopen-2017-021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarker & Prev. 2005;14(8):1847. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 51.Lein PJ. Chapter 1 - Overview of the role of environmental factors in neurodevelopmental disorders. In: Aschner M, Costa L. editors. Environmental factors in neurodevelopmental and neurodegenerative disorders. Boston: Academic Press; 2015. p. 3–20. [Google Scholar]
- 52.Barker DJ, Osmond C, Simmonds SJ, Wield GA. The relation of small head circumference and thinness at birth to death from cardiovascular disease in adult life. BMJ. 1993;306(6875):422–426. doi: 10.1136/bmj.306.6875.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hertz-Picciotto I, Croen LA, Hansen R, Jones CR, van de Water J, Pessah IN. The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect. 2006;114(7):1119–1125. doi: 10.1289/ehp.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCanlies EC, Ma CC, Gu JK, Fekedulegn D, Sanderson WT, Ludena-Rodriguez YJ, Hertz-Picciotto I. The CHARGE study: an assessment of parental occupational exposures and autism spectrum disorder. Occup Environ Med. 2019;76(9):644–651. doi: 10.1136/oemed-2018-105395. [DOI] [PubMed] [Google Scholar]
- 55.Santos JX, Sampaio P, Rasga C, Martiniano H, Faria C, Cafe C, Oliveira A, Duque F, Oliveira G, Sousa L, et al. Evidence for an association of prenatal exposure to particulate matter with clinical severity of autism spectrum disorder. Environ Res. 2023;228:115795. doi: 10.1016/j.envres.2023.115795. [DOI] [PubMed] [Google Scholar]
- 56.Rizzo HE, Escaname EN, Alana NB, Lavender E, Gelfond J, Fernandez R, Hibbs MA, King JM, Carr NR, Blanco CL, et al. Maternal diabetes and obesity influence the fetal epigenome in a largely Hispanic population. Clin Epigenet. 2020;12(1):34. doi: 10.1186/s13148-020-0824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LaSalle JM. Epigenomic signatures reveal mechanistic clues and predictive markers for autism spectrum disorder. Mol Psychiatry. 2023;28(5):1890–1901. doi: 10.1038/s41380-022-01917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bey AL, Jiang Y-H. Overview of mouse models of autism spectrum disorders. Curr Protoc Pharmacol. 2014;66(1):66:5.66.1–5.26. doi: 10.1002/0471141755.ph0566s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hulbert SW, Jiang YH. Monogenic mouse models of autism spectrum disorders: common mechanisms and missing links. Neurosci. 2016;321:3–23. doi: 10.1016/j.neuroscience.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong DL, Paylor R, Zoghbi HY. Mice with truncated MeCP2 recapitulate many rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35(2):243–254. doi: 10.1016/S0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 61.Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, et al. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell STEM Cell. 2014;14(3):323–328. doi: 10.1016/j.stem.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13(15):1259–1268. doi: 10.1016/S0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 65.Caglayan AO. Genetic causes of syndromic and non-syndromic autism. Develop Med Child Neuro. 2010;52(2):130–138. doi: 10.1111/j.1469-8749.2009.03523.x. [DOI] [PubMed] [Google Scholar]
- 66.Sato A, Kasai S, Kobayashi T, Takamatsu Y, Hino O, Ikeda K, Mizuguchi M. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat Commun. 2012;3(1):1292. doi: 10.1038/ncomms2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1 +/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62(6):648–655. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- 68.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22(4):345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lord C, Brugha TS, Charman T, Cusack J, Dumas G, Frazier T, Jones EJH, Jones RM, Pickles A, State MW, et al. Autism spectrum disorder. Nat Rev Dis Primers. 2020;6(1):5. doi: 10.1038/s41572-019-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind D. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelleher RJ 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135(3):401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 72.Guang S, Pang N, Deng X, Yang L, He F, Wu L, Chen C, Yin F, Peng J. Synaptopathology involved in autism spectrum disorder. Front Cell Neurosci. 2018;12:470. doi: 10.3389/fncel.2018.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. The heritability of autism spectrum disorder. JAMA. 2017;318(12):1182–1184. doi: 10.1001/jama.2017.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet. 2011;156(3):255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- 75.Hallmayer J. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68(11):1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46(8):881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tick B, Bolton P, Happe F, Rutter M, Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J Child Psychol Psychiatry. 2016;57(5):585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mbadiwe T, Millis RM. Epigenetics and autism. Autism Res And Treat. 2013;2013:826156. doi: 10.1155/2013/826156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wiśniowiecka-Kowalnik B, Nowakowska BA. Genetics and epigenetics of autism spectrum disorder—current evidence in the field. J Appl Genet. 2019;60(1):37–47. doi: 10.1007/s13353-018-00480-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robertson KD. Epigenetic mechanisms of gene regulation. In: Szyf M. editor. DNA methylation and cancer therapy. Boston (MA): Springer US; 2005. p. 13–30. [Google Scholar]
- 81.Grafodatskaya D, Chung B, Szatmari P, Weksberg R. Autism spectrum disorders and epigenetics. J Am Acad Child Adolesc Psychiatry. 2010;49(8):794–809. doi: 10.1016/j.jaac.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 82.Pilarowski GO, Vernon HJ, Applegate CD, Boukas L, Cho MT, Gurnett CA, Benke PJ, Beaver E, Heeley JM, Medne L, et al. Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability. J Med Genet. 2018;55(8):561–566. doi: 10.1136/jmedgenet-2017-104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodman JV, Bonni A. Regulation of neuronal connectivity in the mammalian brain by chromatin remodeling. Curr Opin Neurobiol. 2019;59:59–68. doi: 10.1016/j.conb.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout A, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158(2):263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong CCY, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Moffitt TE, Mill J. A longitudinal study of epigenetic variation in twins. Epigenet. 2010;5(6):516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, Andronikos R, Cruickshank MN, Conneely KN, Smith AK, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22(8):1395–1406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, Kalidindi S, Picchioni M, Kravariti E, Toulopoulou T, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20(24):4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB j. 2010;24(8):3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wong CC, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, Plomin R, Mill J. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2014;19(4):495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry. 2014;19(8):862–871. doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellis SE, Gupta S, Moes A, West AB, Arking DE. Exaggerated CpH methylation in the autism-affected brain. Mol Autism. 2017;8(1):6. doi: 10.1186/s13229-017-0119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berko ER, Suzuki M, Beren F, Lemetre C, Alaimo CM, Calder RB, Ballaban-Gil K, Gounder B, Kampf K, Kirschen J, et al. Mosaic epigenetic dysregulation of ectodermal cells in autism spectrum disorder. PLoS Genet. 2014;10(5):e1004402. doi: 10.1371/journal.pgen.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duffney LJ, Valdez P, Tremblay MW, Cao X, Montgomery S, McConkie-Rosell A, Jiang Y-H. Epigenetics and autism spectrum disorder: a report of an autism case with mutation in H1 linker histone HIST1H1E and literature review. Am J Of Med Genet Pt B. 2018;177(4):426–433. doi: 10.1002/ajmg.b.32631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism. 2013;4(1):36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fernandes IR, Cruz ACP, Ferrasa A, Phan D, Herai RH, Muotri AR. Genetic variations on SETD5 underlying autistic conditions. Dev Neurobiol. 2018;78(5):500–518. doi: 10.1002/dneu.22584. [DOI] [PubMed] [Google Scholar]
- 96.Grozeva D, Carss K, Spasic-Boskovic O, Parker MJ, Archer H, Firth HV, Park S-M, Canham N, Holder S, Wilson M, et al. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet. 2014;94(4):618–624. doi: 10.1016/j.ajhg.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deliu E, Arecco N, Morandell J, Dotter CP, Contreras X, Girardot C, Käsper E-L, Kozlova A, Kishi K, Chiaradia I, et al. Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat Neurosci. 2018;21(12):1717–1727. doi: 10.1038/s41593-018-0266-2. [DOI] [PubMed] [Google Scholar]
- 98.Li J, Xu X, Liu J, Zhang S, Tan X, Li Z, Zhang J, Wang Z. Decoding microRNAs in autism spectrum disorder. Mol Ther Nucleic Acids. 2022;30:535–546. doi: 10.1016/j.omtn.2022.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Human Microbiome Project C. A framework for human microbiome research. Nature. 2012;486:215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kho ZY, Lal SK. The human ct microbiome – a potential controller of wellness and disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26(8):26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frost F, Kacprowski T, Ruhlemann M, Pietzner M, Bang C, Franke A, Nauck M, Völker U, Völzke H, Dörr M, et al. Long-term instability of the intestinal microbiome is associated with metabolic liver disease, low microbiota diversity, diabetes mellitus and impaired exocrine pancreatic function. Gut. 2021;70(3):522–530. doi: 10.1136/gutjnl-2020-322753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis. 2016;22(5):1137–1150. doi: 10.1097/MIB.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Forsythe P, Bienenstock J, Kunze WA. Vagal pathways for microbiome-brain-gut axis communication. Adv Exp Med Biol. 2014;817:115–133. [DOI] [PubMed] [Google Scholar]
- 107.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol. 2004;558(1):263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterol. 2011;141(2):599–609.e3. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 109.Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J Psychiatr Res. 2008;43(2):164–174. doi: 10.1016/j.jpsychires.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 110.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23(3):255–e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 111.Arentsen T, Raith H, Qian Y, Forssberg H, Diaz Heijtz R. Host microbiota modulates development of social preference in mice. Microb Ecol Health Dis. 2015;26(8):29719. doi: 10.3402/mehd.v26.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, MacQueen G, Sherman PM. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60(3):307–317. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- 113.Mohajeri MH, La Fata G, Steinert RE, Weber P. Relationship between the gut microbiome and brain function. nutr Rev. 2018;76(7):481–496. doi: 10.1093/nutrit/nuy009. [DOI] [PubMed] [Google Scholar]
- 114.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 2011;108(7):3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim YK, Shin C. The microbiota-gut-brain axis in neuropsychiatric disorders: pathophysiological mechanisms and novel treatments. Curr Neuropharmacol. 2018;16(5):559–573. doi: 10.2174/1570159X15666170915141036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McElhanon BO, McCracken C, Karpen S, Sharp WG. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics. 2014;133(5):872–883. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- 117.Madra M, Ringel R, Margolis KG. Gastrointestinal issues and autism spectrum disorder. Child Adolesc Psychiatr Clin N Am. 2020;29(3):501–513. doi: 10.1016/j.chc.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gorrindo P, Williams KC, Lee EB, Walker LS, McGrew SG, Levitt P. Gastrointestinal dysfunction in autism: parental report, clinical evaluation, and associated factors. Autism Res. 2012;5(2):101–108. doi: 10.1002/aur.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Buie T, Campbell DB, Fuchs GJ, Furuta GT, Levy J, Vandewater J, Whitaker AH, Atkins D, Bauman ML, Beaudet AL, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125(Suppl 1):SS1–S18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- 120.Wang LW, Tancredi DJ, Thomas DW. The prevalence of gastrointestinal problems in children across the United States with autism spectrum disorders from families with multiple affected members. J Dev Behav Pediatr. 2011;32(5):351–360. doi: 10.1097/DBP.0b013e31821bd06a. [DOI] [PubMed] [Google Scholar]
- 121.Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, Youn E, Summanen PH, Granpeesheh D, Dixon D, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16(4):444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 122.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni ME, Gobbetti M, Francavilla R, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8(10):e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gondalia SV, Palombo EA, Knowles SR, Cox SB, Meyer D, Austin DW. Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 2012;5(6):419–427. doi: 10.1002/aur.1253. [DOI] [PubMed] [Google Scholar]
- 124.Yap CX, Henders AK, Alvares GA, Wood DLA, Krause L, Tyson GW, Restuadi R, Wallace L, McLaren T, Hansell NK, et al. Autism-related dietary preferences mediate autism-gut microbiome associations. Cell. 2021;184(24):5916–5931.e17. doi: 10.1016/j.cell.2021.10.015. [DOI] [PubMed] [Google Scholar]
- 125.Ozcan E, Hsiao EY. Are changes in the gut microbiome a contributor or consequence of autism—why not both? Cell Rep Med. 2022;3(1):100505. doi: 10.1016/j.xcrm.2021.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu M, Xu X, Li J, Li F. Association between gut microbiota and autism spectrum disorder: a systematic review and meta-analysis. Front. Psychiatry. 2019;10:473. doi: 10.3389/fpsyt.2019.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Morton JT, Jin DM, Mills RH, Shao Y, Rahman G, McDonald D, Zhu Q, Balaban M, Jiang Y, Cantrell K, et al. Multi-level analysis of the gut–brain axis shows autism spectrum disorder-associated molecular and microbial profiles. Nat Neurosci. 2023;26(7):1208–1217. doi: 10.1038/s41593-023-01361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wan Y, Zuo T, Xu Z, Zhang F, Zhan H, Chan D, Leung T-F, Yeoh YK, Chan FKL, Chan R, et al. Underdevelopment of the gut microbiota and bacteria species as non-invasive markers of prediction in children with autism spectrum disorder. Gut. 2022;71(5):910–918. doi: 10.1136/gutjnl-2020-324015. [DOI] [PubMed] [Google Scholar]
- 129.Bundgaard-Nielsen C, Lauritsen MB, Knudsen JK, Rold LS, Larsen MH, Hindersson P, Villadsen AB, Leutscher PDC, Hagstrøm S, Nyegaard M, et al. Children and adolescents with attention deficit hyperactivity disorder and autism spectrum disorder share distinct microbiota compositions. Gut Microbes. 2023;15(1):2211923. doi: 10.1080/19490976.2023.2211923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lai MC, Kassee C, Besney R, Bonato S, Hull L, Mandy W, Szatmari P, Ameis SH. Prevalence of co-occurring mental health diagnoses in the autism population: a systematic review and meta-analysis. Lancet Psychiatry. 2019;6(10):819–829. doi: 10.1016/S2215-0366(19)30289-5. [DOI] [PubMed] [Google Scholar]
- 131.Lugo-Marín J, Magán-Maganto M, Rivero-Santana A, Cuellar-Pompa L, Alviani M, Jenaro-Rio C. Prevalence of psychiatric disorders in adults with autism spectrum disorder: a systematic review and meta-analysis. Res In Autism Spectr Disord. 2019;59:22–33. doi: 10.1016/j.rasd.2018.12.004. [DOI] [Google Scholar]
- 132.Zhang W, Sun Z, Zhang Q, Sun Z, Su Y, Song J, Wang B, Gao R. Preliminary evidence for an influence of exposure to polycyclic aromatic hydrocarbons on the composition of the gut microbiota and neurodevelopment in three-year-old healthy children. BMC Pediatr. 2021;21(1):86. doi: 10.1186/s12887-021-02539-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165(7):1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli J, Chow J, Reisman S, Petrosino J, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman RS, Honda K, Littman DR, Choi GB, et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549(7673):528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, Khoruts A, Geis E, Maldonado J, McDonough-Means S, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5(1):10. doi: 10.1186/s40168-016-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kang DW, Adams JB, Coleman DM, Pollard EL, Maldonado J, McDonough-Means S, Caporaso JG, Krajmalnik-Brown R. Long-term benefit of microbiota transfer therapy on autism symptoms and gut microbiota. Sci Rep. 2019;9(1):5821. doi: 10.1038/s41598-019-42183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nirmalkar K, Qureshi F, Kang DW, Hahn J, Adams JB, Krajmalnik-Brown R. Shotgun metagenomics study suggests alteration in sulfur metabolism and oxidative stress in children with autism and improvement after microbiota transfer therapy. Int J Mol Sci. 2022;23(21):23. doi: 10.3390/ijms232113481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stewart Campbell A, Needham BD, Meyer CR, Tan J, Conrad M, Preston GM, Bolognani F, Rao SG, Heussler H, Griffith R, et al. Safety and target engagement of an oral small-molecule sequestrant in adolescents with autism spectrum disorder: an open-label phase 1b/2a trial. Nat Med. 2022;28(3):528–534. doi: 10.1038/s41591-022-01683-9. [DOI] [PubMed] [Google Scholar]
- 140.Buffington SA, Dooling SW, Sgritta M, Noecker C, Murillo OD, Felice DF, Turnbaugh PJ, Costa-Mattioli M. Dissecting the contribution of host genetics and the microbiome in complex behaviors. Cell. 2021;184(7):1740–1756.e16. doi: 10.1016/j.cell.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sgritta M, Dooling SW, Buffington SA, Momin EN, Francis MB, Britton RA, Costa-Mattioli M. Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron. 2019;101(2):246–259.e6. doi: 10.1016/j.neuron.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mazzone L, Dooling SW, Volpe E, Uljarevic M, Waters JL, Sabatini A, Arturi L, Abate R, Riccioni A, Siracusano M, et al. Precision microbial intervention improves social behavior but not autism severity: a pilot double-blind randomized placebo-controlled trial. Cell Host & Microbe. 2024;32(1):106–116.e6. doi: 10.1016/j.chom.2023.11.021. [DOI] [PubMed] [Google Scholar]
- 143.Woo V, Alenghat T. Epigenetic regulation by gut microbiota. Gut Microbes. 2022;14(1):2022407. doi: 10.1080/19490976.2021.2022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Miro-Blanch J, Yanes O. Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front Genet. 2019;10:638. doi: 10.3389/fgene.2019.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64(5):982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van der Hee B, Wells JM. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021;29(8):700–712. doi: 10.1016/j.tim.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 147.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278(13):11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 148.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139(9):1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lewis K, Lutgendorff F, Phan V, Soderholm JD, Sherman PM, McKay DM. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate†. Inflamm Bowel Dis. 2010;16(7):1138–1148. doi: 10.1002/ibd.21177. [DOI] [PubMed] [Google Scholar]
- 150.Muller M, Hernandez MAG, Goossens GH, Reijnders D, Holst JJ, Jocken JWE, van Eijk H, Canfora EE, Blaak EE. Circulating but not faecal short-chain fatty acids are related to insulin sensitivity, lipolysis and GLP-1 concentrations in humans. Sci Rep. 2019;9(1):12515. doi: 10.1038/s41598-019-48775-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin & Trans Imm. 2016;5(4):e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Omenetti S, Bussi C, Metidji A, Iseppon A, Lee S, Tolaini M, Li Y, Kelly G, Chakravarty P, Shoaie S, et al. The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity. 2019;51(1):77–89.e6. doi: 10.1016/j.immuni.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dähling S, Kastenmüller W, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity. 2019;51(2):285–297.e5. doi: 10.1016/j.immuni.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 155.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ang Z, Er JZ, Tan NS, Lu J, Liou YC, Grosse J, Ding JL. Human and mouse monocytes display distinct signalling and cytokine profiles upon stimulation with FFAR2/FFAR3 short-chain fatty acid receptor agonists. Sci Rep. 2016;6(1):34145. doi: 10.1038/srep34145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat Rev Gastroenterol Hepatol. 2019;16(8):461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 158.Falomir-Lockhart LJ, Cavazzutti GF, Gimenez E, Toscani AM. Fatty acid signaling mechanisms in neural cells: fatty acid receptors. Front Cell Neurosci. 2019;13:162. doi: 10.3389/fncel.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li Z, Yi CX, Katiraei S, Kooijman S, Zhou E, Chung CK, Gao Y, van den Heuvel JK, Meijer OC, Berbée JFP, et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut. 2018;67(7):1269–1279. doi: 10.1136/gutjnl-2017-314050. [DOI] [PubMed] [Google Scholar]
- 160.Hoyles L, Snelling T, Umlai UK, Nicholson JK, Carding SR, Glen RC, McArthur S. Microbiome–host systems interactions: protective effects of propionate upon the blood–brain barrier. Microbiome. 2018;6(1):55. doi: 10.1186/s40168-018-0439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 162.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-Protein–coupled receptor FFAR2. Diabetes. 2012;61(2):364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Larraufie P, Martin-Gallausiaux C, Lapaque N, Dore J, Gribble FM, Reimann F, Blottiere HM. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci Rep. 2018;8(1):74. doi: 10.1038/s41598-017-18259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alvarez E, Martinez MD, Roncero I, Chowen JA, Garcia-Cuartero B, Gispert JD, Sanz C, Vázquez P, Maldonado A, De Cáceres J, et al. The expression of GLP-1 receptor mRNA and protein allows the effect of GLP-1 on glucose metabolism in the human hypothalamus and brainstem. J Neurochem. 2005;92(4):798–806. doi: 10.1111/j.1471-4159.2004.02914.x. [DOI] [PubMed] [Google Scholar]
- 165.Nonaka N, Shioda S, Niehoff ML, Banks WA. Characterization of blood-brain barrier permeability to PYY3-36 in the mouse. J Pharmacol Exp Ther. 2003;306(3):948–953. doi: 10.1124/jpet.103.051821. [DOI] [PubMed] [Google Scholar]
- 166.During MJ, Cao L, Zuzga DS, Francis JS, Fitzsimons HL, Jiao X, Bland RJ, Klugmann M, Banks WA, Drucker DJ, et al. Glucagon-like peptide-1 receptor is involved in learning and neuroprotection. Nat Med. 2003;9(9):1173–1179. doi: 10.1038/nm919. [DOI] [PubMed] [Google Scholar]
- 167.McClean PL, Parthsarathy V, Faivre E, Holscher C. The diabetes drug liraglutide prevents degenerative processes in a mouse model of Alzheimer’s disease. J Neurosci. 2011;31:6587–6594. doi: 10.1523/JNEUROSCI.0529-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mohle L, Mattei D, Heimesaat MM, Bereswill S, Fischer A, Alutis M, French T, Hambardzumyan D, Matzinger P, Dunay I, et al. Ly6Chi monocytes provide a link between antibiotic-induced changes in gut microbiota and adult hippocampal neurogenesis. Cell Rep. 2016;15(9):1945–1956. doi: 10.1016/j.celrep.2016.04.074. [DOI] [PubMed] [Google Scholar]
- 169.van de Wouw M, Boehme M, Lyte JM, Wiley N, Strain C, O’Sullivan O, Clarke G, Stanton C, Dinan TG, Cryan JF. Short-chain fatty acids: microbial metabolites that alleviate stress-induced brain–gut axis alterations. J Physiol. 2018;596(20):4923–4944. doi: 10.1113/JP276431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Soliman ML, Rosenberger TA. Acetate supplementation increases brain histone acetylation and inhibits histone deacetylase activity and expression. Mol Cell Biochem. 2011;352(1–2):173–180. doi: 10.1007/s11010-011-0751-3. [DOI] [PubMed] [Google Scholar]
- 171.Volmar C-H, Wahlestedt C. Histone deacetylases (HDACs) and brain function. Neuroepigenet. 2015;1:20–27. doi: 10.1016/j.nepig.2014.10.002. [DOI] [Google Scholar]
- 172.Garcez ML, de Carvalho CA, Mina F, Bellettini-Santos T, Schiavo GL, da Silva S, Campos ACBF, Varela RB, Valvassori SS, Damiani AP, et al. Sodium butyrate improves memory and modulates the activity of histone deacetylases in aged rats after the administration of d-galactose. Exp Gerontol. 2018;113:209–217. doi: 10.1016/j.exger.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 173.Mihaylova MM, Stratton MS. Short chain fatty acids as epigenetic and metabolic regulators of neurocognitive health and disease. In: Ferguson BS. editor. Nutritional epigenomics. 1st ed. Academic Press, Elsevier; 2019. p. 381–397, 478. [Google Scholar]
- 174.Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18(7):965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Erny D, Dokalis N, Mezo C, Castoldi A, Mossad O, Staszewski O, Frosch M, Villa M, Fuchs V, Mayer A, et al. Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab. 2021;33(11):2260–2276.e7. doi: 10.1016/j.cmet.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 176.Dupraz L, Magniez A, Rolhion N, Richard ML, Da Costa G, Touch S, Mayeur C, Planchais J, Agus A, Danne C, et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. 2021;36(1):109332. doi: 10.1016/j.celrep.2021.109332. [DOI] [PubMed] [Google Scholar]
- 177.Liu S, Li E, Sun Z, Fu D, Duan G, Jiang M, Yu Y, Mei L, Yang P, Tang Y, et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci Rep. 2019;9(1):287. doi: 10.1038/s41598-018-36430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bojovic K, Ignjatovic Eth I, Sokovic Bajic S, Vojnovic Milutinovic D, Tomic M, Golic N, Tolinački M. Gut microbiota dysbiosis associated with altered production of short chain fatty acids in children with neurodevelopmental disorders. Front Cell Infect Microbiol. 2020;10:223. doi: 10.3389/fcimb.2020.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11(1):22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig Dis Sci. 2012;57(8):2096–2102. doi: 10.1007/s10620-012-2167-7. [DOI] [PubMed] [Google Scholar]
- 181.de Theije CG, Wopereis H, Ramadan M, van Eijndthoven T, Lambert J, Knol J, Garssen J, Kraneveld AD, Oozeer R. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 2014;37:197–206. doi: 10.1016/j.bbi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 182.Kratsman N, Getselter D, Elliott E. Sodium butyrate attenuates social behavior deficits and modifies the transcription of inhibitory/excitatory genes in the frontal cortex of an autism model. Neuropharmacol. 2016;102:136–145. doi: 10.1016/j.neuropharm.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 183.Grieneisen L, Dasari M, Gould TJ, Bjork JR, Grenier JC, Yotova V, Jansen D, Gottel N, Gordon JB, Learn NH, et al. Gut microbiome heritability is nearly universal but environmentally contingent. Sci. 2021;373(6551):181–186. doi: 10.1126/science.aba5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Robinson O, Vrijheid M. The pregnancy exposome. Curr Envir Health Rpt. 2015;2(2):204–213. doi: 10.1007/s40572-015-0043-2. [DOI] [PubMed] [Google Scholar]
- 185.Gillman MW, Rifas-Shiman SL, Kleinman K, Oken E, Rich-Edwards JW, Taveras EM. Developmental origins of childhood overweight: potential public health impact. Obes (Silver Spring). 2008;16(7):1651–1656. doi: 10.1038/oby.2008.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351(9097):173–177. doi: 10.1016/S0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 187.Crowther NJ, Cameron N, Trusler J, Gray IP. Association between poor glucose tolerance and rapid post natal weight gain in seven-year-old children. Diabetologia. 1998;41(10):1163–1167. doi: 10.1007/s001250051046. [DOI] [PubMed] [Google Scholar]
- 188.Jornayvaz FR, Selz R, Tappy L, Theintz GE. Metabolism of oral glucose in children born small for gestational age: evidence for an impaired whole body glucose oxidation. Metabol. 2004;53(7):847–851. doi: 10.1016/j.metabol.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 189.Courchesne E, Gazestani VH, Lewis NE. Prenatal origins of ASD: The When, What, and How of ASD development. Trends Neurosci. 2020;43(5):326–342. doi: 10.1016/j.tins.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Szatmari P. Is autism, at least in part, a disorder of fetal programming? Arch Gen Psychiatry. 2011;68(11):1091–1092. doi: 10.1001/archgenpsychiatry.2011.99. [DOI] [PubMed] [Google Scholar]
- 191.Bonnet-Brilhault F, Rajerison TA, Paillet C, Guimard-Brunault M, Saby A, Ponson L, Tripi G, Malvy J, Roux S. Autism is a prenatal disorder: evidence from late gestation brain overgrowth. Autism Res. 2018;11(12):1635–1642. doi: 10.1002/aur.2036. [DOI] [PubMed] [Google Scholar]
- 192.Rice D, Barone S. Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ Health Perspect. 2000;108(Suppl 3):511–533. doi: 10.1289/ehp.00108s3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dobbing J. Vulnerable periods of brain development. In: lipids, malnutrition & the developing brain. Ciba Found Symp. 1971; 9–29. PMID: 4949882. [PubMed] [Google Scholar]
- 194.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20(4):327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Salinas RD, Connolly DR, Song H. Invited review: epigenetics in neurodevelopment. Neuropathol Appl Neurobio. 2020;46(1):6–27. doi: 10.1111/nan.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stroud H, Su SC, Hrvatin S, Greben AW, Renthal W, Boxer LD, Nagy MA, Hochbaum DR, Kinde B, Gabel HW, et al. Early-life gene expression in neurons modulates lasting epigenetic states. Cell. 2017;171(5):1151–1164.e16. doi: 10.1016/j.cell.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Komada M, Nishimura Y. Epigenetics and neuroinflammation associated with neurodevelopmental disorders: a microglial perspective. Front Cell Dev Biol. 2022;10:852752. doi: 10.3389/fcell.2022.852752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nayak M, Das D, Pradhan J, Ahmed RG, Laureano-Melo R, Dandapat J. Epigenetic signature in neural plasticity: the journey so far and journey ahead. Heliyon. 2022;8(12):e12292. doi: 10.1016/j.heliyon.2022.e12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chess S. Autism in children with congenital rubella. J Autism Dev Disord. 1971;1(1):33–47. doi: 10.1007/BF01537741. [DOI] [PubMed] [Google Scholar]
- 201.Chess S. Follow-up report on autism in congenital rubella. J Autism Dev Disord. 1977;7(1):69–81. doi: 10.1007/BF01531116. [DOI] [PubMed] [Google Scholar]
- 202.Maeyama K, Tomioka K, Nagase H, Yoshioka M, Takagi Y, Kato T, Mizobuchi M, Kitayama S, Takada S, Nagai M, et al. Congenital cytomegalovirus infection in children with autism spectrum disorder: systematic review and meta-analysis. J Autism Dev Disord. 2018;48(5):1483–1491. doi: 10.1007/s10803-017-3412-x. [DOI] [PubMed] [Google Scholar]
- 203.Stubbs EG, Ash E, Williams CP. Autism and congenital cytomegalovirus. J Autism Dev Disord. 1984;14(2):183–189. doi: 10.1007/BF02409660. [DOI] [PubMed] [Google Scholar]
- 204.Brown AS, Begg MD, Gravenstein S, Schaefer CA, Wyatt RJ, Bresnahan M, Babulas VP, Susser ES. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry. 2004;61(8):774–780. doi: 10.1001/archpsyc.61.8.774. [DOI] [PubMed] [Google Scholar]
- 205.Shi L, Fatemi SH, Sidwell RW, Patterson PH. Maternal influenza infection causes marked behavioral and pharmacological changes in the offspring. J Neurosci. 2003;23(1):297–302. doi: 10.1523/JNEUROSCI.23-01-00297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Abdallah MW, Larsen N, Grove J, Nørgaard-Pedersen B, Thorsen P, Mortensen EL, Hougaard DM. Amniotic fluid chemokines and autism spectrum disorders: an exploratory study utilizing a Danish historic birth cohort. Brain Behav Immun. 2012;26(1):170–176. doi: 10.1016/j.bbi.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 207.Jiang HY, Xu LL, Shao L, Xia RM, Yu ZH, Ling ZX, Yang F, Deng M, Ruan B. Maternal infection during pregnancy and risk of autism spectrum disorders: a systematic review and meta-analysis. Brain Behav Immun. 2016;58:165–172. doi: 10.1016/j.bbi.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 208.Brown AS, Sourander A, Hinkka-Yli-Salomaki S, McKeague IW, Sundvall J, Surcel HM. Elevated maternal C-reactive protein and autism in a national birth cohort. Mol Psychiatry. 2014;19(2):259–264. doi: 10.1038/mp.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, Courchesne E, Everall IP. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. 2010;68(4):368–376. doi: 10.1016/j.biopsych.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 210.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57(1):67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 211.Siniscalco D, Schultz S, Brigida AL, Antonucci N. Inflammation and neuro-immune dysregulations in autism spectrum disorders. Pharmaceut (Basel). 2018;11(2):11. doi: 10.3390/ph11020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu N, Li X, Zhong Y. Inflammatory cytokines: potential biomarkers of immunologic dysfunction in autism spectrum disorders. Mediators Inflamm. 2015;2015:1–10. doi: 10.1155/2015/531518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chez MG, Dowling T, Patel PB, Khanna P, Kominsky M. Elevation of tumor necrosis factor-alpha in cerebrospinal fluid of autistic children. Pediatr Neurol. 2007;36(6):361–365. doi: 10.1016/j.pediatrneurol.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 214.Croonenberghs J, Bosmans E, Deboutte D, Kenis G, Maes M. Activation of the inflammatory response system in autism. Neuropsychobiol. 2002;45(1):1–6. doi: 10.1159/000048665. [DOI] [PubMed] [Google Scholar]
- 215.Al-Ayadhi LY, Mostafa GA. Elevated serum levels of interleukin-17A in children with autism. J Neuroinflammat. 2012;9(1):158. doi: 10.1186/1742-2094-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Enstrom A, Onore C, Hertz-Picciotto I, Hansen R, Croen L, Van de Water J, Ashwood P. Detection of IL-17 and IL-23 in plasma samples of children with autism. Am J Biochem Biotechnol. 2008;4(2):114–120. doi: 10.3844/ajbbsp.2008.114.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Okada K, Hashimoto K, Iwata Y, Nakamura K, Tsujii M, Tsuchiya KJ, Sekine Y, Suda S, Suzuki K, Sugihara G-I, et al. Decreased serum levels of transforming growth factor-β1 in patients with autism. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31(1):187–190. doi: 10.1016/j.pnpbp.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 218.Goines PE, Croen LA, Braunschweig D, Yoshida CK, Grether J, Hansen R, Kharrazi M, Ashwood P, Van de Water J. Increased midgestational IFN-γ, IL-4 and IL-5 in women bearing a child with autism: a case-control study. Mol Autism. 2011;2(1):13. doi: 10.1186/2040-2392-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jones KL, Croen LA, Yoshida CK, Heuer L, Hansen R, Zerbo O, DeLorenze GN, Kharrazi M, Yolken R, Ashwood P, et al. Autism with intellectual disability is associated with increased levels of maternal cytokines and chemokines during gestation. Mol Psychiatry. 2017;22(2):273–279. doi: 10.1038/mp.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Abdallah MW, Larsen N, Grove J, Norgaard-Pedersen B, Thorsen P, Mortensen EL, Hougaard DM. Amniotic fluid inflammatory cytokines: potential markers of immunologic dysfunction in autism spectrum disorders. World J Biol Psychiatry. 2013;14(7):528–538. doi: 10.3109/15622975.2011.639803. [DOI] [PubMed] [Google Scholar]
- 221.Meyer U, Feldon J, Fatemi SH. In-vivo rodent models for the experimental investigation of prenatal immune activation effects in neurodevelopmental brain disorders. Neurosci Biobehav Rev. 2009;33(7):1061–1079. doi: 10.1016/j.neubiorev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 222.Fatemi SH, Folsom TD, Reutiman TJ, Abu-Odeh D, Mori S, Huang H, Oishi K. Abnormal expression of myelination genes and alterations in white matter fractional anisotropy following prenatal viral influenza infection at E16 in mice. Schizophr Res. 2009;112(1–3):46–53. doi: 10.1016/j.schres.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Fatemi SH, Reutiman TJ, Folsom TD, Huang H, Oishi K, Mori S, Smee DF, Pearce DA, Winter C, Sohr R, et al. Maternal infection leads to abnormal gene regulation and brain atrophy in mouse offspring: implications for genesis of neurodevelopmental disorders. Schizophr Res. 2008;99(1–3):56–70. doi: 10.1016/j.schres.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Garay PA, Hsiao EY, Patterson PH, McAllister AK. Maternal immune activation causes age- and region-specific changes in brain cytokines in offspring throughout development. Brain Behav Immun. 2013;31:54–68. doi: 10.1016/j.bbi.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xu Z, Zhang X, Chang H, Kong Y, Ni Y, Liu R, Zhang X, Hu Y, Yang Z, Hou M, et al. Rescue of maternal immune activation-induced behavioral abnormalities in adult mouse offspring by pathogen-activated maternal treg cells. Nat Neurosci. 2021;24(6):818–830. doi: 10.1038/s41593-021-00837-1. [DOI] [PubMed] [Google Scholar]
- 226.Lim AI, McFadden T, Link VM, Han SJ, Karlsson RM, Stacy A, Farley TK, Lima-Junior DS, Harrison OJ, Desai JV, et al. Prenatal maternal infection promotes tissue-specific immunity and inflammation in offspring. Sci. 2021;373(6558). doi: 10.1126/science.abf3002. [DOI] [PubMed] [Google Scholar]
- 227.Braunschweig D, Duncanson P, Boyce R, Hansen R, Ashwood P, Pessah IN, Hertz-Picciotto I, Van de Water J. Behavioral correlates of maternal antibody status among children with autism. J Autism Dev Disord. 2012;42(7):1435–1445. doi: 10.1007/s10803-011-1378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Martinez-Cerdeno V, Camacho J, Fox E, Miller E, Ariza J, Kienzle D, Plank K, Noctor SC, Van de Water J. Prenatal exposure to autism-specific maternal autoantibodies alters proliferation of cortical neural precursor cells, enlarges brain, and increases neuronal size in adult animals. Cereb Cortex. 2016;26(1):374–383. doi: 10.1093/cercor/bhu291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Paval D. A dopamine hypothesis of autism spectrum disorder. Dev Neurosci. 2017;39(5):355–360. doi: 10.1159/000478725. [DOI] [PubMed] [Google Scholar]
- 230.Meyer U, Yee BK, Feldon J. The neurodevelopmental impact of prenatal infections at different times of pregnancy: the earlier the worse? Neuroscientist. 2007;13(3):241–256. doi: 10.1177/1073858406296401. [DOI] [PubMed] [Google Scholar]
- 231.Urakubo A, Jarskog LF, Lieberman JA, Gilmore JH. Prenatal exposure to maternal infection alters cytokine expression in the placenta, amniotic fluid, and fetal brain. Schizophr Res. 2001;47(1):27–36. doi: 10.1016/S0920-9964(00)00032-3. [DOI] [PubMed] [Google Scholar]
- 232.Burns TM, Clough JA, Klein RM, Wood GW, Berman NE. Developmental regulation of cytokine expression in the mouse brain. Growth Factors. 1993;9(4):253–258. doi: 10.3109/08977199308991585. [DOI] [PubMed] [Google Scholar]
- 233.Deverman BE, Patterson PH. Cytokines and CNS development. Neuron. 2009;64(1):61–78. doi: 10.1016/j.neuron.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 234.Brynge M, Gardner RM, Sjoqvist H, Lee BK, Dalman C, Karlsson H. Maternal levels of cytokines in early pregnancy and risk of autism spectrum disorders in offspring. Front Publ Health. 2022;10:917563. doi: 10.3389/fpubh.2022.917563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Careaga M, Murai T, Bauman MD. Maternal immune activation and autism spectrum disorder: from rodents to nonhuman and human primates. Biol Psychiatry. 2017;81(5):391–401. doi: 10.1016/j.biopsych.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kirsten TB, Bernardi MM. Prenatal lipopolysaccharide induces hypothalamic dopaminergic hypoactivity and autistic-like behaviors: repetitive self-grooming and stereotypies. Behav Brain Res. 2017;331:25–29. doi: 10.1016/j.bbr.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 237.Malkova NV, Yu CZ, Hsiao EY, Moore MJ, Patterson PH. Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav Immun. 2012;26(4):607–616. doi: 10.1016/j.bbi.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hsiao EY, McBride SW, Chow J, Mazmanian SK, Patterson PH. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc Natl Acad Sci USA. 2012;109(31):12776–12781. doi: 10.1073/pnas.1202556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Schwartzer JJ, Careaga M, Onore CE, Rushakoff JA, Berman RF, Ashwood P. Maternal immune activation and strain specific interactions in the development of autism-like behaviors in mice. Transl Psychiatry. 2013;3(3):e240. doi: 10.1038/tp.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, Huh JR. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351(6276):933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007;27(40):10695–10702. doi: 10.1523/JNEUROSCI.2178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Girard S, Tremblay L, Lepage M, Sebire G. IL-1 receptor antagonist protects against placental and neurodevelopmental defects induced by maternal inflammation. J Immunol. 2010;184(7):3997–4005. doi: 10.4049/jimmunol.0903349. [DOI] [PubMed] [Google Scholar]
- 243.Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci. 2013;16:1537–1543. doi: 10.1038/nn.3546. [DOI] [PubMed] [Google Scholar]
- 244.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12(4):295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32(6):743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.van Sadelhoff JHJ, Perez Pardo P, Wu J, Garssen J, van Bergenhenegouwen J, Hogenkamp A, Hartog A, Kraneveld AD. The gut-immune-brain axis in autism spectrum disorders; a focus on amino acids. Front Endocrinol (Lausanne). 2019;10:247. doi: 10.3389/fendo.2019.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Fung TC. The microbiota-immune axis as a central mediator of gut-brain communication. Neurobiol Dis. 2020;136:104714. doi: 10.1016/j.nbd.2019.104714. [DOI] [PubMed] [Google Scholar]
- 248.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 249.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14(7):668–675. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bilbo SD, Block CL, Bolton JL, Hanamsagar R, Tran PK. Beyond infection - maternal immune activation by environmental factors, microglial development, and relevance for autism spectrum disorders. Exp Neurol. 2018;299:241–251. doi: 10.1016/j.expneurol.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cryan JF, Dinan TG. Microbiota and neuroimmune signalling—metchnikoff to microglia. Nat Rev Gastroenterol Hepatol. 2015;12(9):494–496. doi: 10.1038/nrgastro.2015.127. [DOI] [PubMed] [Google Scholar]
- 253.Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, Blecher R, Ulas T, Squarzoni P, Hoeffel G, et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. 2018;172(3):500–516.e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.D’Mello C, Ronaghan N, Zaheer R, Dicay M, Le T, MacNaughton WK, Surrette MG, Swain MG. Probiotics improve inflammation-associated sickness behavior by altering communication between the peripheral immune system and the brain. J Neurosci. 2015;35(30):10821–10830. doi: 10.1523/JNEUROSCI.0575-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.D’Mello C, Le T, Swain MG. Cerebral microglia recruit monocytes into the brain in response to tumor necrosis factorα signaling during peripheral organ inflammation. J Neurosci. 2009;29(7):2089–2102. doi: 10.1523/JNEUROSCI.3567-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nilsson NE, Kotarsky K, Owman C, Olde B. Identification of a free fatty acid receptor, FFA2R, expressed on leukocytes and activated by short-chain fatty acids. Biochem Biophys Res Commun. 2003;303(4):1047–1052. doi: 10.1016/S0006-291X(03)00488-1. [DOI] [PubMed] [Google Scholar]
- 257.Caetano-Silva ME, Rund L, Hutchinson NT, Woods JA, Steelman AJ, Johnson RW. Inhibition of inflammatory microglia by dietary fiber and short-chain fatty acids. Sci Rep. 2023;13(1):2819. doi: 10.1038/s41598-022-27086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Reddy DS, Wu X, Golub VM, Dashwood WM, Dashwood RH. Measuring histone deacetylase inhibition in the brain. Curr Protoc Pharmacol. 2018;81(1):e41. doi: 10.1002/cpph.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lin CW, Septyaningtrias DE, Chao HW, Konda M, Atarashi K, Takeshita K, Tamada K, Nomura J, Sasagawa Y, Tanaka K, et al. A common epigenetic mechanism across different cellular origins underlies systemic immune dysregulation in an idiopathic autism mouse model. Mol Psychiatry. 2022;27(8):3343–3354. doi: 10.1038/s41380-022-01566-y. [DOI] [PubMed] [Google Scholar]
- 260.King JC. A summary of pathways or mechanisms linking preconception maternal nutrition with birth outcomes. J Nutr. 2016;146(7):1437S–1444S. doi: 10.3945/jn.115.223479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Li M, Francis E, Hinkle SN, Ajjarapu AS, Zhang C. Preconception and prenatal nutrition and neurodevelopmental disorders: a systematic review and meta-analysis. Nutr. 2019;11(7):11. doi: 10.3390/nu11071628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Darnton-Hill I, Mkparu UC. Micronutrients in pregnancy in low- and middle-income countries. Nutr. 2015;7(3):1744–1768. doi: 10.3390/nu7031744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Saffari A, Shrestha S, Issarapu P, Sajjadi S, Betts M, Sahariah SA, Tomar AS, James P, Dedaniya A, Yadav DK, et al. Effect of maternal preconceptional and pregnancy micronutrient interventions on children’s DNA methylation: findings from the EMPHASIS study. The Am J Clin Nutr. 2020;112(4):1099–1113. doi: 10.1093/ajcn/nqaa193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhong C, Tessing J, Lee BK, Lyall K. Maternal dietary factors and the risk of autism spectrum disorders: a systematic review of existing evidence. Autism Res. 2020;13(10):1634–1658. doi: 10.1002/aur.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Eyles DW, Burne TH, McGrath JJ. Vitamin D, effects on brain development, adult brain function and the links between low levels of vitamin D and neuropsychiatric disease. Front Neuroendocrinol. 2013;34(1):47–64. doi: 10.1016/j.yfrne.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 266.Heyden EL, Wimalawansa SJ. Vitamin D: effects on human reproduction, pregnancy, and fetal well-being. J Steroid Biochem Mol Biol. 2018;180:41–50. doi: 10.1016/j.jsbmb.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 267.Fernell E, Barnevik-Olsson M, Bagenholm G, Gillberg C, Gustafsson S, Saaf M. Serum levels of 25-hydroxyvitamin D in mothers of Swedish and of Somali origin who have children with and without autism. Acta Paediatr. 2010;99(5):743–747. doi: 10.1111/j.1651-2227.2010.01755.x. [DOI] [PubMed] [Google Scholar]
- 268.Hou W, Yan XT, Bai CM, Zhang XW, Hui LY, Yu XW. Decreased serum vitamin D levels in early spontaneous pregnancy loss. Eur J Clin Nutr. 2016;70(9):1004–1008. doi: 10.1038/ejcn.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Purswani JM, Gala P, Dwarkanath P, Larkin HM, Kurpad A, Mehta S. The role of vitamin D in pre-eclampsia: a systematic review. BMC Pregnancy Childbirth. 2017;17(1):231. doi: 10.1186/s12884-017-1408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Rizzo G, Garzon S, Fichera M, Panella MM, Catena U, Schiattarella A, de Franciscis P, Vilos G, Tesarik J, Török P, et al. Vitamin D and gestational diabetes mellitus: Is there a link? Antioxid (Basel). 2019;8(11):511. doi: 10.3390/antiox8110511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Garcia-Serna AM, Morales E. Neurodevelopmental effects of prenatal vitamin D in humans: systematic review and meta-analysis. Mol Psychiatry. 2020;25(10):2468–2481. doi: 10.1038/s41380-019-0357-9. [DOI] [PubMed] [Google Scholar]
- 272.Lee BK, Eyles DW, Magnusson C, Newschaffer CJ, McGrath JJ, Kvaskoff D, Ko P, Dalman C, Karlsson H, Gardner RM, et al. Developmental vitamin D and autism spectrum disorders: findings from the Stockholm youth cohort. Mol Psychiatry. 2019;26(5):1578–1588. doi: 10.1038/s41380-019-0578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Magnusson C, Lundberg M, Lee BK, Rai D, Karlsson H, Gardner R, Karlsson H, Gardner R, Arver S. Maternal vitamin D deficiency and the risk of autism spectrum disorders: population-based study. BJPsych Open. 2016;2(2):170–172. doi: 10.1192/bjpo.bp.116.002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wagner CL, Greer FR. American Academy of Pediatrics section on B, American Academy of Pediatrics Committee on N. Prevention of rickets and vitamin D deficiency in infants, children, and adolescents. Pediatr. 2008;122(5):1142–1152. doi: 10.1542/peds.2008-1862. [DOI] [PubMed] [Google Scholar]
- 275.Xue J, Schoenrock SA, Valdar W, Tarantino LM, Ideraabdullah FY. Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations. Clin Epigenet. 2016;8(1):107. doi: 10.1186/s13148-016-0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Yamamoto EA, Jorgensen TN. Relationships between vitamin D, gut microbiome, and systemic autoimmunity. Front Immunol. 2019;10:3141. doi: 10.3389/fimmu.2019.03141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Sato K. Why is folate effective in preventing neural tube closure defects? Med Hypotheses. 2020;134:109429. doi: 10.1016/j.mehy.2019.109429. [DOI] [PubMed] [Google Scholar]
- 278.Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr. 2012;3:21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ondicova M, Irwin RE, Thursby SJ, Hilman L, Caffrey A, Cassidy T, McLaughlin M, Lees-Murdock DJ, Ward M, Murphy M, et al. Folic acid intervention during pregnancy alters DNA methylation, affecting neural target genes through two distinct mechanisms. Clin Epigenet. 2022;14(1):63. doi: 10.1186/s13148-022-01282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Asrar FM, O’Connor DL. Bacterially synthesized folate and supplemental folic acid are absorbed across the large intestine of piglets. J Nutr Biochem. 2005;16(10):587–593. doi: 10.1016/j.jnutbio.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 281.Di Gesu CM, Matz LM, Buffington SA. Diet-induced dysbiosis of the maternal gut microbiome in early life programming of neurodevelopmental disorders. Neurosci Res. 2021;168:3–19. doi: 10.1016/j.neures.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Vecchione R, Wang S, Rando J, Chavarro JE, Croen LA, Fallin MD, Hertz-Picciotto I, Newschaffer CJ, Schmidt RJ, Lyall K, et al. Maternal dietary patterns during pregnancy and child autism-related traits: results from two US cohorts. Nutr. 2022;14(13):14. doi: 10.3390/nu14132729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ruskin DN, Murphy MI, Slade SL, Masino SA, Chapouthier G. Ketogenic diet improves behaviors in a maternal immune activation model of autism spectrum disorder. PLoS One. 2017;12(2):e0171643. doi: 10.1371/journal.pone.0171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.van den Broek T, Fleischmann M. Prenatal famine exposure and mental health in later midlife. Aging Ment Health. 2019;23(2):166–170. doi: 10.1080/13607863.2017.1402293. [DOI] [PubMed] [Google Scholar]
- 285.Brown AS, van Os J, Driessens C, Hoek HW, Susser ES. Further evidence of relation between prenatal famine and major affective disorder. Am J Psychiatry. 2000;157(2):190–195. doi: 10.1176/appi.ajp.157.2.190. [DOI] [PubMed] [Google Scholar]
- 286.Neugebauer R, Hoek HW, Susser E. Prenatal exposure to wartime famine and development of antisocial personality disorder in early adulthood. JAMA. 1999;282(5):455–462. doi: 10.1001/jama.282.5.455. [DOI] [PubMed] [Google Scholar]
- 287.Hoek HW, Brown AS, Susser E. The Dutch famine and schizophrenia spectrum disorders. Soc Psychiatry Psychiatr Epidemiol. 1998;33(8):373–379. doi: 10.1007/s001270050068. [DOI] [PubMed] [Google Scholar]
- 288.de Rooij SR, Wouters H, Yonker JE, Painter RC, Roseboom TJ. Prenatal undernutrition and cognitive function in late adulthood. Proc Natl Acad Sci USA. 2010;107(39):16881–16886. doi: 10.1073/pnas.1009459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Franzek EJ, Sprangers N, Janssens AC, Van Duijn CM, Van De Wetering BJ. Prenatal exposure to the 1944–45 Dutch ‘hunger winter’ and addiction later in life. Addict. 2008;103(3):433–438. doi: 10.1111/j.1360-0443.2007.02084.x. [DOI] [PubMed] [Google Scholar]
- 290.Roseboom T, de Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum Dev. 2006;82(8):485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 291.Susser M, Stein Z. Timing in prenatal nutrition: a reprise of the Dutch famine study. Nutr Rev. 2009;52(3):84–94. doi: 10.1111/j.1753-4887.1994.tb01395.x. [DOI] [PubMed] [Google Scholar]
- 292.Cox B, Leavey K, Nosi U, Wong F, Kingdom J. Placental transcriptome in development and pathology: expression, function, and methods of analysis. Am J Obstet Gynecol. 2015;213(4):S138–51. doi: 10.1016/j.ajog.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 293.Jansson T, Powell TL. Role of the placenta in fetal programming: underlying mechanisms and potential interventional approaches. Clin Sci (Lond). 2007;113(1):1–13. doi: 10.1042/CS20060339. [DOI] [PubMed] [Google Scholar]
- 294.Jansson T, Aye IL, Goberdhan DC. The emerging role of mTORC1 signaling in placental nutrient-sensing. Placenta. 2012;33(Suppl 2):e23–9. doi: 10.1016/j.placenta.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Dos Santos E, Duval F, Vialard F, Dieudonné MN. The roles of leptin and adiponectin at the fetal-maternal interface in humans. Horm Mol Biol Clin Investig. 2015;24(1):47–63. doi: 10.1515/hmbci-2015-0031. [DOI] [PubMed] [Google Scholar]
- 296.Friis CM, Paasche Roland MC, Godang K, Ueland T, Tanbo T, Bollerslev J, Henriksen T. Adiposity-related inflammation: effects of pregnancy. Obes (Silver Spring). 2013;21(1):E124–30. doi: 10.1002/oby.20120. [DOI] [PubMed] [Google Scholar]
- 297.Siwetz M, Blaschitz A, El-Heliebi A, Hiden U, Desoye G, Huppertz B, Gauster M. TNF-α alters the inflammatory secretion profile of human first trimester placenta. Lab Invest. 2016;96(4):428–438. doi: 10.1038/labinvest.2015.159. [DOI] [PubMed] [Google Scholar]
- 298.Tessier DR, Ferraro ZM, Gruslin A. Role of leptin in pregnancy: consequences of maternal obesity. Placenta. 2013;34(3):205–211. doi: 10.1016/j.placenta.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 299.Duval F, Santos ED, Poidatz D, Serazin V, Gronier H, Vialard F, Dieudonné M-N. Adiponectin inhibits nutrient transporters and promotes apoptosis in human villous cytotrophoblasts: involvement in the control of fetal growth. Biol Reprod. 2016;94(5):111. doi: 10.1095/biolreprod.115.134544. [DOI] [PubMed] [Google Scholar]
- 300.Pantham P, Rosario FJ, Weintraub ST, Nathanielsz PW, Powell TL, Li C, Jansson T. Down-regulation of placental transport of amino acids precedes the development of intrauterine growth restriction in maternal nutrient restricted baboons. Biol Reprod. 2016;95(5):98. doi: 10.1095/biolreprod.116.141085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Jansson N, Pettersson J, Haafiz A, Ericsson A, Palmberg I, Tranberg M, Ganapathy V, Powell TL, Jansson T. Down-regulation of placental transport of amino acids precedes the development of intrauterine growth restriction in rats fed a low protein diet. J Physiol. 2006;576(3):935–946. doi: 10.1113/jphysiol.2003.550004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Jansson N, Rosario FJ, Gaccioli F, Lager S, Jones HN, Roos S, Jansson T, Powell TL. Activation of placental mTOR signaling and amino acid transporters in obese women giving birth to large babies. J Clin Endocrinol Metab. 2013;98(1):105–113. doi: 10.1210/jc.2012-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Acosta O, Ramirez VI, Lager S, Gaccioli F, Dudley DJ, Powell TL, Jansson T. Increased glucose and placental GLUT-1 in large infants of obese nondiabetic mothers. Am J Obstet Gynecol. 2015;212(2):.e227.1–.e227.7. doi: 10.1016/j.ajog.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 304.Howell KR, Powell TL. Effects of maternal obesity on placental function and fetal development. Reproduct. 2017;153(3):R97–R108. doi: 10.1530/REP-16-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Desoye G, van Poppel M. The feto-placental dialogue and diabesity. Best Pract Res Clin Obstet Gynaecol. 2015;29(1):15–23. doi: 10.1016/j.bpobgyn.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 306.Bellinger L, Lilley C, Langley-Evans SC. Prenatal exposure to a maternal low-protein diet programmes a preference for high-fat foods in the young adult rat. Br J Nutr. 2004;92(3):513–520. doi: 10.1079/BJN20041224. [DOI] [PubMed] [Google Scholar]
- 307.Zambrano E, Bautista CJ, Deás M, Martínez-Samayoa PM, González-Zamorano M, Ledesma H, Morales J, Larrea F, Nathanielsz PW. A low maternal protein diet during pregnancy and lactation has sex- and window of exposure-specific effects on offspring growth and food intake, glucose metabolism and serum leptin in the rat. J Physiol. 2006;571(1):221–230. doi: 10.1113/jphysiol.2005.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Samuelsson AM, Matthews PA, Argenton M, Christie MR, McConnell JM, Jansen EH, Piersma AH, Ozanne SE, Twinn DF, Remacle C, et al. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: a novel murine model of developmental programming. Hypertens. 2008;51(2):383–392. doi: 10.1161/HYPERTENSIONAHA.107.101477. [DOI] [PubMed] [Google Scholar]
- 309.Gawlinska K, Gawlinski D, Korostynski M, Borczyk M, Frankowska M, Piechota M, Filip M, Przegaliński E. Maternal dietary patterns are associated with susceptibility to a depressive-like phenotype in rat offspring. Dev Cogn Neurosci. 2021;47:100879. doi: 10.1016/j.dcn.2020.100879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Schaefer CA, Brown AS, Wyatt RJ, Kline J, Begg MD, Bresnahan MA, Susser ES. Maternal prepregnant body mass and risk of schizophrenia in adult offspring. Schizophr Bull. 2000;26(2):275–286. doi: 10.1093/oxfordjournals.schbul.a033452. [DOI] [PubMed] [Google Scholar]
- 311.Kawai M, Minabe Y, Takagai S, Ogai M, Matsumoto H, Mori N, Takei N. Poor maternal care and high maternal body mass index in pregnancy as a risk factor for schizophrenia in offspring. Acta Psychiatr Scand. 2004;110(4):257–263. doi: 10.1111/j.1600-0447.2004.00380.x. [DOI] [PubMed] [Google Scholar]
- 312.Rodriguez A. Maternal pre-pregnancy obesity and risk for inattention and negative emotionality in children. J Child Psychol Psychiatry. 2010;51(2):134–143. doi: 10.1111/j.1469-7610.2009.02133.x. [DOI] [PubMed] [Google Scholar]
- 313.Kong L, Chen X, Gissler M, Lavebratt C. Relationship of prenatal maternal obesity and diabetes to offspring neurodevelopmental and psychiatric disorders: a narrative review. Int J Obes (Lond). 2020;44(10):1981–2000. doi: 10.1038/s41366-020-0609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.de Lauzon-Guillain B, Marques C, Kadawathagedara M, Bernard JY, Tafflet M, Lioret S, Charles MA. Maternal diet during pregnancy and child neurodevelopment up to age 3.5 years: the nationwide Étude longitudinale Française depuis l’Enfance (ELFE) birth cohort. Am J Clin Nutr. 2022;116(4):1101–1111. doi: 10.1093/ajcn/nqac206. [DOI] [PubMed] [Google Scholar]
- 315.Tanda R, Salsberry PJ, Reagan PB, Fang MZ. The impact of prepregnancy obesity on children’s cognitive test scores. Matern Child Health J. 2013;17(2):222–229. doi: 10.1007/s10995-012-0964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Mahmassani HA, Switkowski KM, Scott TM, Johnson EJ, Rifas-Shiman SL, Oken E, Jacques PF. Maternal diet quality during pregnancy and child cognition and behavior in a US cohort. Am J Clin Nutr. 2022;115(1):128–141. doi: 10.1093/ajcn/nqab325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Márquez-Valadez B, Valle-Bautista R, García-López G, Díaz NF, Molina-Hernández A. Maternal diabetes and fetal programming toward neurological diseases: beyond neural tube defects. Front Endocrinol. 2018;9:664. doi: 10.3389/fendo.2018.00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Rodriguez A, Miettunen J, Henriksen TB, Olsen J, Obel C, Taanila A, Ebeling H, Linnet KM, Moilanen I, Järvelin M-R, et al. Maternal adiposity prior to pregnancy is associated with ADHD symptoms in offspring: evidence from three prospective pregnancy cohorts. Int J Obes. 2008;32(3):550–557. doi: 10.1038/sj.ijo.0803741. [DOI] [PubMed] [Google Scholar]
- 319.Chen Q, Sjolander A, Langstrom N, Rodriguez A, Serlachius E, D’Onofrio BM, Lichtenstein P, Larsson H. Maternal pre-pregnancy body mass index and offspring attention deficit hyperactivity disorder: a population-based cohort study using a sibling-comparison design. Int J Epidemiol. 2014;43(1):83–90. doi: 10.1093/ije/dyt152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Krakowiak P, Walker CK, Bremer AA, Baker AS, Ozonoff S, Hansen RL, Hertz-Picciotto I. Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatr. 2012;129(5):e1121–8. doi: 10.1542/peds.2011-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Shen Y, Dong H, Lu X, Lian N, Xun G, Shi L, Xiao L, Zhao J, Ou J. Associations among maternal pre-pregnancy body mass index, gestational weight gain and risk of autism in the Han Chinese population. BMC Psychiatry. 2018;18(1):11. doi: 10.1186/s12888-018-1593-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Reynolds LC, Inder TE, Neil JJ, Pineda RG, Rogers CE. Maternal obesity and increased risk for autism and developmental delay among very preterm infants. J Perinatol. 2014;34(9):688–692. doi: 10.1038/jp.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Wang Y, Tang S, Xu S, Weng S, Liu Z. Maternal body mass index and risk of autism spectrum disorders in offspring: a meta-analysis. Sci Rep. 2016;6(1):34248. doi: 10.1038/srep34248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Xu G, Jing J, Bowers K, Liu B, Bao W. Maternal diabetes and the risk of autism spectrum disorders in the offspring: a systematic review and meta-analysis. J Autism Dev Disord. 2014;44(4):766–775. doi: 10.1007/s10803-013-1928-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Hariri N, Thibault L. High-fat diet-induced obesity in animal models. Nutr Res Rev. 2010;23(2):270–299. doi: 10.1017/S0954422410000168. [DOI] [PubMed] [Google Scholar]
- 326.Ding Q, Zhao Y, Yang Y, Chen Z. Cognitive impairment due to leptin withdrawal in rat offspring of dams with maternal diet-induced obesity. Med Sci Monit. 2018;24:6208–6217. doi: 10.12659/MSM.911906. PMID: 30187895; PMCID: PMC6139113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Kang SS, Kurti A, Fair DA, Fryer JD. Dietary intervention rescues maternal obesity induced behavior deficits and neuroinflammation in offspring. J Neuroinflammat. 2014;11(1):156. doi: 10.1186/s12974-014-0156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Mucellini AB, Laureano DP, Silveira PP, Sanvitto GL. Maternal and post-natal obesity alters long-term memory and hippocampal molecular signaling of male rat. Brain Res. 2019;1708:138–145. doi: 10.1016/j.brainres.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 329.Wolfrum C, Peleg-Raibstein D. Maternal overnutrition leads to cognitive and neurochemical abnormalities in C57BL/6 mice. Nutr Neurosci. 2019;22(10):688–699. doi: 10.1080/1028415X.2018.1432096. [DOI] [PubMed] [Google Scholar]
- 330.Tozuka Y, Wada E, Wada K. Diet-induced obesity in female mice leads to peroxidized lipid accumulations and impairment of hippocampal neurogenesis during the early life of their offspring. FASEB J. 2009;23(6):1920–1934. doi: 10.1096/fj.08-124784. [DOI] [PubMed] [Google Scholar]
- 331.Stachowiak EK, Srinivasan M, Stachowiak MK, Patel MS. Maternal obesity induced by a high fat diet causes altered cellular development in fetal brains suggestive of a predisposition of offspring to neurological disorders in later life. Metab Brain Dis. 2013;28(4):721–725. doi: 10.1007/s11011-013-9437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, Chrysovergis K, Wade PA. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19(1):7. doi: 10.1186/s13059-018-1389-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Shen L, Li C, Wang Z, Zhang R, Shen Y, Miles T, Wei J, Zou Z. Early-life exposure to severe famine is associated with higher methylation level in the IGF2 gene and higher total cholesterol in late adulthood: the genomic research of the Chinese famine (GRECF) study. Clin Epigenet. 2019;11(1):88. doi: 10.1186/s13148-019-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Chango A, Pogribny IP. Considering maternal dietary modulators for epigenetic regulation and programming of the fetal epigenome. Nutri. 2015;7(4):2748–2770. doi: 10.3390/nu7042748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Bastaki KN, Alwan S, Zahir FR. Maternal prenatal exposures in pregnancy and autism spectrum disorder: an insight into the epigenetics of drugs and diet as key environmental influences. Adv Neurobiol. 2020;24:143–162. [DOI] [PubMed] [Google Scholar]
- 336.Kubota T, Mochizuki K. Epigenetic effect of environmental factors on autism spectrum disorders. Int J Environ Res Public Health. 2016;13(5):504. doi: 10.3390/ijerph13050504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Panchenko PE, Voisin S, Jouin M, Jouneau L, Prezelin A, Lecoutre S, Breton C, Jammes H, Junien C, Gabory A, et al. Expression of epigenetic machinery genes is sensitive to maternal obesity and weight loss in relation to fetal growth in mice. Clin Epigenet. 2016;8(1):22. doi: 10.1186/s13148-016-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, Lane RH. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41(2):91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Masuyama H, Hiramatsu Y. Effects of a high-fat diet exposure in utero on the metabolic syndrome-like phenomenon in mouse offspring through epigenetic changes in adipocytokine gene expression. Endocrinol. 2012;153(6):2823–2830. doi: 10.1210/en.2011-2161. [DOI] [PubMed] [Google Scholar]
- 340.Ornoy A, Reece EA, Pavlinkova G, Kappen C, Miller RK. Effect of maternal diabetes on the embryo, fetus, and children: congenital anomalies, genetic and epigenetic changes and developmental outcomes. Birth Defects Res Embryo Today. 2015;105(1):53–72. doi: 10.1002/bdrc.21090. [DOI] [PubMed] [Google Scholar]
- 341.Salbaum JM, Kappen C. Responses of the embryonic epigenome to maternal diabetes. Birth Defects Res Part A: Clin And Mol Teratol. 2012;94(10):770–781. doi: 10.1002/bdra.23035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Glendining KA, Jasoni CL. Maternal high fat diet-induced obesity modifies histone binding and expression of oxtr in offspring hippocampus in a sex-specific manner. Int J Mol Sci. 2019;20(2):20. doi: 10.3390/ijms20020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Fineberg SK, Ross DA. Oxytocin and the social brain. Biol Psychiatry. 2017;81(3):e19–e21. doi: 10.1016/j.biopsych.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Gali Ramamoorthy T, Allen TJ, Davies A, Harno E, Sefton C, Murgatroyd C, White A. Maternal overnutrition programs epigenetic changes in the regulatory regions of hypothalamic pomc in the offspring of rats. Int J Obes (Lond). 2018;42(8):1431–1444. doi: 10.1038/s41366-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Zheng J, Xiao X, Zhang Q, Yu M, Xu J, Wang Z, Qi C, Wang T. Maternal and post-weaning high-fat, high-sucrose diet modulates glucose homeostasis and hypothalamic POMC promoter methylation in mouse offspring. Metab Brain Dis. 2015;30(5):1129–1137. doi: 10.1007/s11011-015-9678-9. [DOI] [PubMed] [Google Scholar]
- 346.Coll AP. Effects of pro-opiomelanocortin (POMC) on food intake and body weight: mechanisms and therapeutic potential? Clin Sci (Lond). 2007;113(4):171–182. doi: 10.1042/CS20070105. [DOI] [PubMed] [Google Scholar]
- 347.Glendining KA, Higgins MBA, Fisher LC, Jasoni CL. Maternal obesity modulates sexually dimorphic epigenetic regulation and expression of leptin receptor in offspring hippocampus. Brain Behav Immun. 2020;88:151–160. doi: 10.1016/j.bbi.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 348.Bouret SG, Gorski JN, Patterson CM, Chen S, Levin BE, Simerly RB. Hypothalamic neural projections are permanently disrupted in diet-induced obese rats. Cell Metab. 2008;7(2):179–185. doi: 10.1016/j.cmet.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Glendining KA, Fisher LC, Jasoni CL. Maternal high fat diet alters offspring epigenetic regulators, amygdala glutamatergic profile and anxiety. Psychoneuroendocrinol. 2018;96:132–141. doi: 10.1016/j.psyneuen.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 350.Winer S, Paltser G, Chan Y, Tsui H, Engleman E, Winer D, Dosch H-M. Obesity predisposes to Th17 bias. Eur J Immunol. 2009;39(9):2629–2635. doi: 10.1002/eji.200838893. [DOI] [PubMed] [Google Scholar]
- 351.Babu ST, Niu X, Raetz M, Savani RC, Hooper LV, Mirpuri J. Maternal high-fat diet results in microbiota-dependent expansion of ILC3s in mice offspring. JCI Insight. 2018;3(19):3. doi: 10.1172/jci.insight.99223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, et al. Lactobacillus reuteri induces gut intraepithelial CD4 + CD8αα + T cells. Sci. 2017;357(6353):806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Dai FC, Wang P, Li Q, Zhang L, Yu LJ, Wu L, Tao R-X, Zhu P. Mediterranean diet during pregnancy and infant neurodevelopment: a prospective birth cohort study. Front Nutr. 2023;9:1078481. doi: 10.3389/fnut.2022.1078481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Crovetto F, Nakaki A, Arranz A, Borras R, Vellve K, Paules C, Boutet ML, Castro-Barquero S, Freitas T, Casas R, et al. Effect of a Mediterranean diet or mindfulness-based stress reduction during pregnancy on child neurodevelopment: a prespecified analysis of the IMPACT BCN randomized clinical trial. JAMA Netw Open. 2023;6(8):e2330255. doi: 10.1001/jamanetworkopen.2023.30255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Sofi F, Cesari F, Abbate R, Gensini GF, Casini A. Adherence to Mediterranean diet and health status: meta-analysis. BMJ. 2008;337(2):a1344. doi: 10.1136/bmj.a1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Mentella MC, Scaldaferri F, Ricci C, Gasbarrini A, Miggiano GAD. Cancer and Mediterranean diet: a review. Nutr. 2019;11(9):11. doi: 10.3390/nu11092059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Cani PD, Van Hul M. Mediterranean diet, gut microbiota and health: when age and calories do not add up! Gut. 2020;69(7):1167–1168. doi: 10.1136/gutjnl-2020-320781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Meslier V, Laiola M, Roager HM, De Filippis F, Roume H, Quinquis B, Giacco R, Mennella I, Ferracane R, Pons N, et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut. 2020;69(7):1258–1268. doi: 10.1136/gutjnl-2019-320438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, Giampieri E, Jennings A, Candela M, Turroni S, et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut. 2020;69(7):1218–1228. doi: 10.1136/gutjnl-2019-319654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Amati F, Hassounah S, Swaka A. The impact of Mediterranean dietary patterns during pregnancy on maternal and offspring health. Nutr. 2019;11(5):11. doi: 10.3390/nu11051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.House JS, Mendez M, Maguire RL, Gonzalez-Nahm S, Huang Z, Daniels J, Murphy SK, Fuemmeler BF, Wright FA, Hoyo C, et al. Periconceptional maternal Mediterranean diet is associated with favorable offspring behaviors and altered CpG methylation of imprinted genes. Front Cell Dev Biol. 2018;6:107. doi: 10.3389/fcell.2018.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wiktorowska-Owczarek A, Berezinska M, Nowak JZ. PUFAs: structures, metabolism and functions. Adv Clin Exp Med. 2015;24(6):931–941. doi: 10.17219/acem/31243. [DOI] [PubMed] [Google Scholar]
- 363.Jory J. Abnormal fatty acids in Canadian children with autism. Nutr. 2016;32(4):474–477. doi: 10.1016/j.nut.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 364.Mostafa GA, El-Khashab HY, Al-Ayadhi LY. A possible association between elevated serum levels of brain-specific auto-antibodies and reduced plasma levels of docosahexaenoic acid in autistic children. J Neuroimmunol. 2015;280:16–20. doi: 10.1016/j.jneuroim.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 365.Bent S, Bertoglio K, Ashwood P, Bostrom A, Hendren RL. A pilot randomized controlled trial of omega-3 fatty acids for autism spectrum disorder. J Autism Dev Disord. 2011;41(5):545–554. doi: 10.1007/s10803-010-1078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Fedorova I, Alvheim AR, Hussein N, Salem N. Deficit in prepulse inhibition in mice caused by dietary n-3 fatty acid deficiency. Behav Neurosci. 2009;123(6):1218–1225. doi: 10.1037/a0017446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Madore C, Leyrolle Q, Lacabanne C, Benmamar-Badel A, Joffre C, Nadjar A, Layé S. Neuroinflammation in autism: plausible role of maternal inflammation, dietary Omega 3, and microbiota. Neural Plast. 2016;2016:1–15. doi: 10.1155/2016/3597209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Li Q, Leung YO, Zhou I, Ho LC, Kong W, Basil P, Wei R, Lam S, Zhang X, Law ACK, et al. Dietary supplementation with n-3 fatty acids from weaning limits brain biochemistry and behavioural changes elicited by prenatal exposure to maternal inflammation in the mouse model. Transl Psychiatry. 2015;5(9):e641. doi: 10.1038/tp.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Delpech JC, Thomazeau A, Madore C, Bosch-Bouju C, Larrieu T, Lacabanne C, Remus-Borel J, Aubert A, Joffre C, Nadjar A, et al. Dietary n-3 PUFAs deficiency increases vulnerability to inflammation-induced spatial memory impairment. Neuropsychopharmacol. 2015;40(12):2774–2787. doi: 10.1038/npp.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Turpin V, Schaffhauser M, Thabault M, Aubert A, Joffre C, Balado E, Longueville J-E, Francheteau M, Burucoa C, Pichon M, et al. Mice prenatally exposed to valproic acid do not show autism-related disorders when fed with polyunsaturated fatty acid-enriched diets. Sci Rep. 2023;13(1):11235. doi: 10.1038/s41598-023-38423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Pusceddu MM, El Aidy S, Crispie F, O’Sullivan O, Cotter P, Stanton C, Kelly P, Cryan JF, Dinan TG, et al. N-3 polyunsaturated fatty acids (PUFAs) reverse the impact of early-life stress on the gut microbiota. PLoS One. 2015;10(10):e0139721. doi: 10.1371/journal.pone.0139721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Watson H, Mitra S, Croden FC, Taylor M, Wood HM, Perry SL, Spencer JA, Quirke P, Toogood GJ, Lawton CL, et al. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut. 2018;67(11):1974–1983. doi: 10.1136/gutjnl-2017-314968. [DOI] [PubMed] [Google Scholar]
- 373.Schoeler M, Ellero-Simatos S, Birkner T, Mayneris-Perxachs J, Olsson L, Brolin H, Loeber U, Kraft JD, Polizzi A, Martí-Navas M, et al. The interplay between dietary fatty acids and gut microbiota influences host metabolism and hepatic steatosis. Nat Commun. 2023;14(1):5329. doi: 10.1038/s41467-023-41074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Guo P, Yang X, Guo X, Yang H, Pan J, Li Y. Dietary fish oil improves autistic behaviors and gut homeostasis by altering the gut microbial composition in a mouse model of fragile X syndrome. Brain Behav Immun. 2023;110:140–151. doi: 10.1016/j.bbi.2023.02.019. [DOI] [PubMed] [Google Scholar]
- 375.Hibbeln JR, Davis JM, Steer C, Emmett P, Rogers I, Williams C, Golding J. Maternal seafood consumption in pregnancy and neurodevelopmental outcomes in childhood (ALSPAC study): an observational cohort study. Lancet. 2007;369(9561):578–585. doi: 10.1016/S0140-6736(07)60277-3. [DOI] [PubMed] [Google Scholar]
- 376.Robertson RC, Kaliannan K, Strain CR, Ross RP, Stanton C, Kang JX. Maternal omega-3 fatty acids regulate offspring obesity through persistent modulation of gut microbiota. Microbiome. 2018;6(1):95. doi: 10.1186/s40168-018-0476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Basak S, Duttaroy AK. Maternal PUFAs, placental epigenetics, and their relevance to fetal growth and brain development. Reprod Sci. 2023;30(2):408–427. doi: 10.1007/s43032-022-00989-w. [DOI] [PubMed] [Google Scholar]
- 378.Basil P, Li Q, Gui H, Hui TCK, Ling VHM, Wong CCY, Mill J, McAlonan GM, Sham P-C. Prenatal immune activation alters the adult neural epigenome but can be partly stabilised by a n-3 polyunsaturated fatty acid diet. Transl Psychiatry. 2018;8(1):125. doi: 10.1038/s41398-018-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Basak S, Vilasagaram S, Duttaroy AK. Maternal dietary deficiency of n-3 fatty acids affects metabolic and epigenetic phenotypes of the developing fetus. Prostaglandins Leukot Essent Fat Acids. 2020;158:102109. doi: 10.1016/j.plefa.2020.102109. [DOI] [PubMed] [Google Scholar]
- 380.Rodrigues EL, Figueiredo PS, Marcelino G, de Cassia Avellaneda Guimaraes R, Pott A, Santana LF, Hiane PA, Do Nascimento VA, Bogo D, de Cássia Freitas K, et al. Maternal intake of polyunsaturated fatty acids in autism spectrum etiology and its relation to the gut microbiota: What do we know? Nutr. 2023;15(7):15. doi: 10.3390/nu15071551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Field R, Field T, Pourkazemi F, Rooney K. Ketogenic diets and the nervous system: a scoping review of neurological outcomes from nutritional ketosis in animal studies. Nutr Res Rev. 2022;35(2):268–281. doi: 10.1017/S0954422421000214. [DOI] [PubMed] [Google Scholar]
- 382.Ang QY, Alexander M, Newman JC, Tian Y, Cai J, Upadhyay V, Turnbaugh JA, Verdin E, Hall KD, Leibel RL, et al. Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell. 2020;181(6):1263–1275.e16. doi: 10.1016/j.cell.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Moaaz M, Youssry S, Elfatatry A, El Rahman MA. Th17/Treg cells imbalance and their related cytokines (IL-17, IL-10 and TGF-β) in children with autism spectrum disorder. J Neuroimmunol. 2019;337:577071. doi: 10.1016/j.jneuroim.2019.577071. [DOI] [PubMed] [Google Scholar]
- 384.Verpeut JL, DiCicco-Bloom E, Bello NT. Ketogenic diet exposure during the juvenile period increases social behaviors and forebrain neural activation in adult engrailed 2 null mice. Physiol Behav. 2016;161:90–98. doi: 10.1016/j.physbeh.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 385.Castro K, Baronio D, Perry IS, Riesgo RDS, Gottfried C. The effect of ketogenic diet in an animal model of autism induced by prenatal exposure to valproic acid. Nutr Neurosci. 2017;20(6):343–350. doi: 10.1080/1028415X.2015.1133029. [DOI] [PubMed] [Google Scholar]
- 386.Zarnowska I, Chrapko B, Gwizda G, Nocun A, Mitosek-Szewczyk K, Gasior M. Therapeutic use of carbohydrate-restricted diets in an autistic child; a case report of clinical and 18FDG PET findings. Metab Brain Dis. 2018;33(4):1187–1192. doi: 10.1007/s11011-018-0219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Herbert MR, Buckley JA. Autism and dietary therapy: case report and review of the literature. J Child Neurol. 2013;28(8):975–982. doi: 10.1177/0883073813488668. [DOI] [PubMed] [Google Scholar]
- 388.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell. 2018;173(7):1728–1741.e13. doi: 10.1016/j.cell.2018.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Newell C, Bomhof MR, Reimer RA, Hittel DS, Rho JM, Shearer J. Ketogenic diet modifies the gut microbiota in a murine model of autism spectrum disorder. Mol Autism. 2016;7(1):37. doi: 10.1186/s13229-016-0099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Olivito I, Avolio E, Minervini D, Soda T, Rocca C, Angelone T, Iaquinta FS, Bellizzi D, De Rango F, Bruno R, et al. Ketogenic diet ameliorates autism spectrum disorders-like behaviors via reduced inflammatory factors and microbiota remodeling in BTBR T+ Itpr3tf/J mice. Exp Neurol. 2023;366:114432. doi: 10.1016/j.expneurol.2023.114432. [DOI] [PubMed] [Google Scholar]
- 391.Kosiek W, Rauk Z, Szulc P, Cichy A, Rugiel M, Chwiej J, Janeczko K, Setkowicz Z. Ketogenic diet impairs neurological development of neonatal rats and affects biochemical composition of maternal brains: evidence of functional recovery in pups. Brain Struct Funct. 2022;227(3):1099–1113. doi: 10.1007/s00429-021-02450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Rodier PM. Developing brain as a target of toxicity. Environ Health Perspect. 1995;103(Suppl 6):73–76. doi: 10.1289/ehp.95103s673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Bellinger DC. Prenatal exposures to environmental chemicals and children’s neurodevelopment: an update. Saf Health Work. 2013;4(1):1–11. doi: 10.5491/SHAW.2013.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Grandjean P, Landrigan PJ. Developmental neurotoxicity of industrial chemicals. The Lancet. 2006;368(9553):2167–2178. doi: 10.1016/S0140-6736(06)69665-7. [DOI] [PubMed] [Google Scholar]
- 395.Needham LL, Grandjean P, Heinzow B, Jørgensen PJ, Nielsen F, Patterson DG, Sjödin A, Turner WE, Weihe P. Partition of environmental chemicals between maternal and fetal blood and tissues. Environ Sci Technol. 2011;45(3):1121–1126. doi: 10.1021/es1019614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Saunders NR, Dziegielewska KM, Mollgard K, Habgood MD. Recent developments in understanding barrier mechanisms in the developing brain: drugs and drug transporters in pregnancy, susceptibility or protection in the fetal brain? Annu Rev Pharmacol Toxicol. 2019;59(1):487–505. doi: 10.1146/annurev-pharmtox-010818-021430. [DOI] [PubMed] [Google Scholar]
- 397.Saunders NR, Liddelow SA, Dziegielewska KM. Barrier mechanisms in the developing brain. Front Pharmacol. 2012;3:46. doi: 10.3389/fphar.2012.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Adinolfi M. The development of the human blood-CSF-brain barrier. Dev Med Child Neurol. 1985;27(4):532–537. doi: 10.1111/j.1469-8749.1985.tb04581.x. [DOI] [PubMed] [Google Scholar]
- 399.Chen YJ, Hsu CC. Effects of prenatal exposure to PCBs on the neurological function of children: a neuropsychological and neurophysiological study. Develop Med Child Neuro. 1994;36(4):312–320. doi: 10.1111/j.1469-8749.1994.tb11851.x. [DOI] [PubMed] [Google Scholar]
- 400.Gladen BC, Rogan WJ, Hardy P, Thullen J, Tingelstad J, Tully M. Development after exposure to polychlorinated biphenyls and dichlorodiphenyl dichloroethene transplacentally and through human milk. J Pediatr. 1988;113(6):991–995. doi: 10.1016/S0022-3476(88)80569-9. [DOI] [PubMed] [Google Scholar]
- 401.Jacobson JL, Jacobson SW. Intellectual impairment in children exposed to polychlorinated biphenyls in utero. N Engl J Med. 1996;335(11):783–789. doi: 10.1056/NEJM199609123351104. [DOI] [PubMed] [Google Scholar]
- 402.Mitoma C, Uchi H, Tsukimori K, Yamada H, Akahane M, Imamura T, Utani A, Furue M. Yusho and its latest findings—a review in studies conducted by the yusho group. Environ Int. 2015;82:41–48. doi: 10.1016/j.envint.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 403.Pessah IN, Lein PJ, Seegal RF, Sagiv SK. Neurotoxicity of polychlorinated biphenyls and related organohalogens. Acta Neuropathol. 2019;138(3):363–387. doi: 10.1007/s00401-019-01978-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Granillo L, Sethi S, Keil KP, Lin Y, Ozonoff S, Iosif AM, Puschner B, Schmidt RJ. Polychlorinated biphenyls influence on autism spectrum disorder risk in the MARBLES cohort. Environ Res. 2019;171:177–184. doi: 10.1016/j.envres.2018.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Sagiv SK, Thurston SW, Bellinger DC, Tolbert PE, Altshul LM, Korrick SA. Prenatal organochlorine exposure and behaviors associated with attention deficit hyperactivity disorder in school-aged children. Am J Epidemiol. 2010;171(5):593–601. doi: 10.1093/aje/kwp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Klocke C, Sethi S, Lein PJ. The developmental neurotoxicity of legacy vs. contemporary polychlorinated biphenyls (PCBs): similarities and differences. Environ Sci Pollut ResearchInt. 2020;27(9):8885–8896. doi: 10.1007/s11356-019-06723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Schafer KS. Persistent toxic chemicals in the US food supply. J Epidemiol Community Health. 2002;56(11):813–817. doi: 10.1136/jech.56.11.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Voorhees JR, Rohlman DS, Lein PJ, Pieper AA. Neurotoxicity in preclinical models of occupational exposure to organophosphorus compounds. Front Neurosci. 2017;10:590. doi: 10.3389/fnins.2016.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Banks CN, Lein PJ. A review of experimental evidence linking neurotoxic organophosphorus compounds and inflammation. Neurotoxicol. 2012;33(3):575–584. doi: 10.1016/j.neuro.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Engel SM, Wetmur J, Chen J, Zhu C, Barr DB, Canfield RL, Wolff MS. Prenatal exposure to organophosphates, paraoxonase 1, and cognitive development in childhood. Environ Health Perspect. 2011;119(8):1182–1188. doi: 10.1289/ehp.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Ruckart PZ, Kakolewski K, Bove FJ, Kaye WE. Long-term neurobehavioral health effects of methyl parathion exposure in children in Mississippi and Ohio. Environ Health Perspect. 2004;112(1):46–51. doi: 10.1289/ehp.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Young JG, Eskenazi B, Gladstone EA, Bradman A, Pedersen L, Johnson C, Barr DB, Furlong CE, Holland NT. Association between in utero organophosphate pesticide exposure and abnormal reflexes in neonates. Neurotoxicology. 2005;26(2):199–209. doi: 10.1016/j.neuro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 413.Guodong D, Pei W, Ying T, Jun Z, Yu G, Xiaojin W, Rong S, Guoquan W, Xiaoming S. Organophosphate pesticide exposure and neurodevelopment in young Shanghai children. Environ Sci Technol. 2012;46(5):2911–2917. doi: 10.1021/es202583d. [DOI] [PubMed] [Google Scholar]
- 414.Schmidt RJ, Kogan V, Shelton JF, Delwiche L, Hansen RL, Ozonoff S, Ma CC, McCanlies EC, Bennett DH, Hertz-Picciotto I, et al. Combined prenatal pesticide exposure and folic acid intake in relation to autism spectrum disorder. Environ Health Perspect. 2017;125(9):097007. doi: 10.1289/EHP604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Montano L, Pironti C, Pinto G, Ricciardi M, Buono A, Brogna C, Venier M, Piscopo M, Amoresano A, Motta O. Polychlorinated biphenyls (PCBs) in the environment: occupational and exposure events, effects on human health and fertility. Toxics. 2022;10(7):10. doi: 10.3390/toxics10070365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Langenbach B, Wilson M. Per- and polyfluoroalkyl substances (PFAS): significance and considerations within the regulatory framework of the USA. Int J Environ Res Public Health. 2021;18(21):18. doi: 10.3390/ijerph182111142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Fenton SE, Ducatman A, Boobis A, DeWitt JC, Lau C, Ng C, Smith JS, Roberts SM. Per- and polyfluoroalkyl substance toxicity and human health review: current state of knowledge and strategies for informing future research. Enviro Toxic And Chem. 2021;40(3):606–630. doi: 10.1002/etc.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Shin HM, Bennett DH, Calafat AM, Tancredi D, Hertz-Picciotto I. Modeled prenatal exposure to per- and polyfluoroalkyl substances in association with child autism spectrum disorder: a case-control study. Environ Res. 2020;186:109514. doi: 10.1016/j.envres.2020.109514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Ames JL, Burjak M, Avalos LA, Braun JM, Bulka CM, Croen LA, Dunlop AL, Ferrara A, Fry RC, Hedderson MM, et al. Prenatal exposure to per- and polyfluoroalkyl substances and childhood autism-related outcomes. Epidemiology. 2023;34(3):450–459. doi: 10.1097/EDE.0000000000001587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Milan M, Carraro L, Fariselli P, Martino ME, Cavalieri D, Vitali F, Boffo L, Patarnello T, Bargelloni L, Cardazzo B, et al. Microbiota and environmental stress: how pollution affects microbial communities in manila clams. Aquat Toxicol. 2018;194:195–207. doi: 10.1016/j.aquatox.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 421.Singh S, Sharma P, Pal N, Kumawat M, Shubham S, Sarma DK, Tiwari RR, Kumar M, Nagpal R. Impact of environmental pollutants on gut microbiome and mental health via the gut–brain axis. Microorganisms. 2022;10(7):10. doi: 10.3390/microorganisms10071457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Naspolini NF, Meyer A, Moreira JC, Sun H, Froes-Asmus CIR, Dominguez-Bello MG. Environmental pollutant exposure associated with altered early-life gut microbiome: results from a birth cohort study. Environ Res. 2022;205:112545. doi: 10.1016/j.envres.2021.112545. [DOI] [PubMed] [Google Scholar]
- 423.Lamichhane S, Harkonen T, Vatanen T, Hyotylainen T, Knip M, Oresic M. Impact of exposure to per- and polyfluoroalkyl substances on fecal microbiota composition in mother-infant dyads. Environ Int. 2023;176:107965. doi: 10.1016/j.envint.2023.107965. [DOI] [PubMed] [Google Scholar]
- 424.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner J, Angenent L, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Turjeman S, Collado MC, Koren O. The gut microbiome in pregnancy and pregnancy complications. Curr Opin In Endocr And Metabolic Res. 2021;18:133–138. doi: 10.1016/j.coemr.2021.03.004. [DOI] [Google Scholar]
- 426.Yang H, Guo R, Li S, Liang F, Tian C, Zhao X, Long Y, Liu F, Jiang M, Zhang Y, et al. Systematic analysis of gut microbiota in pregnant women and its correlations with individual heterogeneity. NPJ Biofilms Microbiomes. 2020;6(1):32. doi: 10.1038/s41522-020-00142-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host & Microbe. 2018;24(1):133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Nuriel-Ohayon M, Neuman H, Ziv O, Belogolovski A, Barsheshet Y, Bloch N, Uzan A, Lahav R, Peretz A, Frishman S, et al. Progesterone increases bifidobacterium relative abundance during late pregnancy. Cell Rep. 2019;27(3):730–736.e3. doi: 10.1016/j.celrep.2019.03.075. [DOI] [PubMed] [Google Scholar]
- 429.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Sonagra AD. Normal pregnancy- a state of insulin resistance. J Clin Diagn Res. 2014;8:CC01–3. doi: 10.7860/JCDR/2014/10068.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Mor G, Cardenas I. REVIEW ARTICLE: the immune system in pregnancy: a unique complexity. Am J Reprod Immunol. 2010;63(6):425–433. doi: 10.1111/j.1600-0897.2010.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Parrettini S, Caroli A, Torlone E. Nutrition and metabolic adaptations in physiological and complicated pregnancy: focus on obesity and gestational diabetes. Front Endocrinol (Lausanne). 2020;11:611929. doi: 10.3389/fendo.2020.611929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Ghaemi MS, DiGiulio DB, Contrepois K, Callahan B, Ngo TTM, Lee-McMullen B, Lehallier B, Robaczewska A, Mcilwain D, Rosenberg-Hasson Y, et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformat. 2019;35(1):95–103. doi: 10.1093/bioinformatics/bty537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Gohir W, Whelan FJ, Surette MG, Moore C, Schertzer JD, Sloboda DM. Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother’s periconceptional diet. Gut Microbes. 2015;6:310–320. doi: 10.1080/19490976.2015.1086056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Gohir W, Kennedy KM, Wallace JG, Saoi M, Bellissimo CJ, Britz-McKibbin P, Petrik JJ, Surette MG, Sloboda DM. High-fat diet intake modulates maternal intestinal adaptations to pregnancy and results in placental hypoxia, as well as altered fetal gut barrier proteins and immune markers. J Physiol. 2019;597(12):3029–3051. doi: 10.1113/JP277353. [DOI] [PubMed] [Google Scholar]
- 436.Wallace JG, Bellissimo CJ, Yeo E, Fei Xia Y, Petrik JJ, Surette MG, Bowdish DME, Sloboda DM. Obesity during pregnancy results in maternal intestinal inflammation, placental hypoxia, and alters fetal glucose metabolism at mid-gestation. Sci Rep. 2019;9(1):17621. doi: 10.1038/s41598-019-54098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88(4):894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 438.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker Nitert M, et al. Connections between the gut microbiome and metabolic hormones in early pregnancy in overweight and obese women. Diabetes. 2016;65(8):2214–2223. doi: 10.2337/db16-0278. [DOI] [PubMed] [Google Scholar]
- 439.van Zyl WF, Deane SM, Dicks LMT. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes. 2020;12:1831339. doi: 10.1080/19490976.2020.1831339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Allen SJ, Jordan S, Storey M, Thornton CA, Gravenor M, Garaiova I, Plummer SF, Wang D, Morgan G. Dietary supplementation with lactobacilli and bifidobacteria is well tolerated and not associated with adverse events during late pregnancy and early infancy. J Nutr. 2010;140(3):483–488. doi: 10.3945/jn.109.117093. [DOI] [PubMed] [Google Scholar]
- 441.Sheyholislami H, Connor KL. Are probiotics and prebiotics safe for use during pregnancy and lactation? A systematic review and meta-analysis. Nutrients. 2021;13(7):2382. doi: 10.3390/nu13072382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Yu Y, Lu J, Oliphant K, Gupta N, Claud K, Lu L, Aguila MB. Maternal administration of probiotics promotes gut development in mouse offsprings. PLoS One. 2020;15(8):e0237182. doi: 10.1371/journal.pone.0237182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Wiedmer EB, Herter-Aeberli I. The potential of prebiotic and probiotic supplementation during obese pregnancy to improve maternal and offspring’s metabolic health and reduce obesity risk—a narrative review. Front Nutr. 2022;9:819882. doi: 10.3389/fnut.2022.819882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Halkjaer SI, de Knegt VE, Lo B, Nilas L, Cortes D, Pedersen AE, Mirsepasi-Lauridsen HC, Andersen LO, Nielsen HV, Stensvold CR, et al. Multistrain probiotic increases the gut microbiota diversity in obese pregnant women: results from a randomized, double-blind placebo-controlled study. Curr Dev Nutr. 2020;4(7):nzaa095. doi: 10.1093/cdn/nzaa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Callaway LK, McIntyre HD, Barrett HL, Foxcroft K, Tremellen A, Lingwood BE, Tobin JM, Wilkinson S, Kothari A, Morrison M, et al. Probiotics for the prevention of gestational diabetes mellitus in overweight and obese women: findings from the SPRING double-blind randomized controlled trial. Diabetes Care. 2019;42(3):364–371. doi: 10.2337/dc18-2248. [DOI] [PubMed] [Google Scholar]
- 446.Joseph JM, Law C. Cross-species examination of single- and multi-strain probiotic treatment effects on neuropsychiatric outcomes. Neurosci Biobehav Rev. 2019;99:160–197. doi: 10.1016/j.neubiorev.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Le Morvan de Sequeira C, Hengstberger C, Enck P, Mack I. Effect of probiotics on psychiatric symptoms and central nervous system functions in human health and disease: a systematic review and meta-analysis. Nutrients. 2022;14(3):14. doi: 10.3390/nu14030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Lu C, Rong J, Fu C, Wang W, Xu J, Ju XD. Overall rebalancing of gut microbiota is key to autism intervention. Front Psychol. 2022;13:862719. doi: 10.3389/fpsyg.2022.862719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.He X, Liu W, Tang F, Chen X, Song G. Effects of probiotics on autism spectrum disorder in children: a systematic review and meta-analysis of clinical trials. Nutr. 2023;15(6):15. doi: 10.3390/nu15061415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Radford-Smith DE, Probert F, Burnet PWJ, Anthony DC. Modifying the maternal microbiota alters the gut–brain metabolome and prevents emotional dysfunction in the adult offspring of obese dams. Proc Natl Acad Sci USA. 2022;119(9):119. doi: 10.1073/pnas.2108581119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Surzenko N, Pjetri E, Munson CA, Friday WB, Hauser J, Mitchell ES, Homberg J. Prenatal exposure to the probiotic Lactococcus lactis decreases anxiety-like behavior and modulates cortical cytoarchitecture in a sex specific manner. PLoS One. 2020;15(7):e0223395. doi: 10.1371/journal.pone.0223395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Wang X, Yang J, Zhang H, Yu J, Yao Z. Oral probiotic administration during pregnancy prevents autism-related behaviors in offspring induced by maternal immune activation via anti-inflammation in mice. Autism Res. 2019;12(4):576–588. doi: 10.1002/aur.2079. [DOI] [PubMed] [Google Scholar]
- 453.Kelly JR, Allen AP, Temko A, Hutch W, Kennedy PJ, Farid N, Murphy E, Boylan G, Bienenstock J, Cryan JF, et al. Lost in translation? The potential psychobiotic lactobacillus rhamnosus (JB-1) fails to modulate stress or cognitive performance in healthy male subjects. Brain Behav Immun. 2017;61:50–59. doi: 10.1016/j.bbi.2016.11.018. [DOI] [PubMed] [Google Scholar]


