Abstract
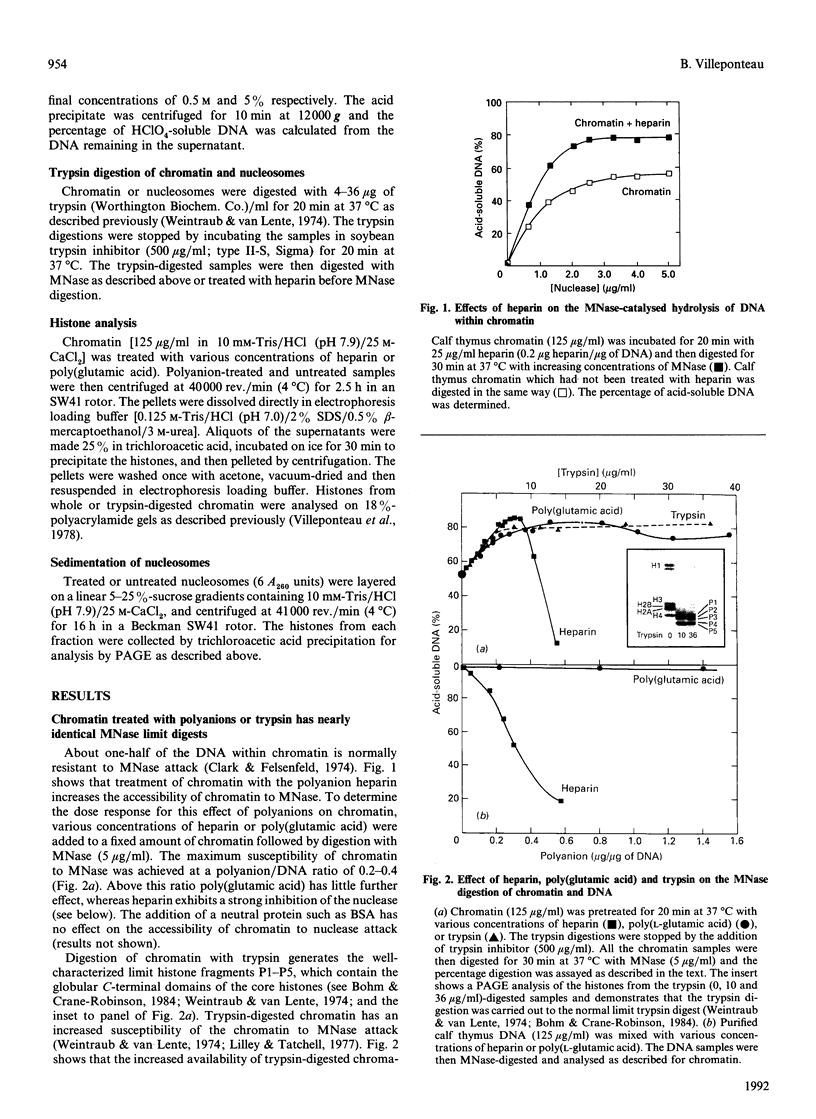
Recent evidence indicates that chromatin accessibility to transcription factors is of regulatory significance. The polyanion heparin is known to increase chromatin accessibility to DNAase I and to stimulate both RNA and DNA synthesis. In the present study, chromatin structure and its modification by polyanions were examined by using trypsin and micrococcal nuclease as probes. Both heparin and poly(glutamic acid) were found to be equivalent to trypsin digestion of histones in their ability to increase nuclease accessibility in chromatin. However, no increase in nuclease accessibility was observed when trypsin-digested chromatin was further treated with heparin, indicating that polyanions and trypsin are not additive in their effects on chromatin accessibility. Moreover, sucrose-gradient analysis demonstrated that heparin binds tightly to intact nucleosomes but not to trypsin-digested nucleosomes. These data suggest that polyanions interact predominantly with the trypsin-sensitive lysine and arginine residues in histone H1 and the N-terminal segments of the core histones. The possible relevance of these results to the chromatin structure of actively transcribed regions is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloom K. S., Anderson J. N. Conformation of ovalbumin and globin genes in chromatin during differential gene expression. J Biol Chem. 1979 Oct 25;254(20):10532–10539. [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Fractionation of hen oviduct chromatin into transcriptionally active and inactive regions after selective micrococcal nuclease digestion. Cell. 1978 Sep;15(1):141–150. doi: 10.1016/0092-8674(78)90090-9. [DOI] [PubMed] [Google Scholar]
- Brotherton T. W., Jagannadham M. V., Ginder G. D. Heparin binds to intact mononucleosomes and induces a novel unfolded structure. Biochemistry. 1989 Apr 18;28(8):3518–3525. doi: 10.1021/bi00434a055. [DOI] [PubMed] [Google Scholar]
- Böhm L., Crane-Robinson C. Proteases as structural probes for chromatin: the domain structure of histones. Biosci Rep. 1984 May;4(5):365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Cary P. D., Moss T., Bradbury E. M. High-resolution proton-magnetic-resonance studies of chromatin core particles. Eur J Biochem. 1978 Sep 1;89(2):475–482. doi: 10.1111/j.1432-1033.1978.tb12551.x. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Chemical probes of chromatin structure. Biochemistry. 1974 Aug 13;13(17):3622–3628. doi: 10.1021/bi00714a034. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. Chromatin structure and gene activity. Curr Opin Cell Biol. 1990 Jun;2(3):437–445. doi: 10.1016/0955-0674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988 Aug 25;334(6184):721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Goldblatt D., Bustin M. Exposure of histone antigenic determinants in chromatin. Biochemistry. 1975 Apr 22;14(8):1689–1695. doi: 10.1021/bi00679a022. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Partington G. A. Distribution of messenger RNA-coding sequences in fractionated chromatin. Cell. 1977 Dec;12(4):953–962. doi: 10.1016/0092-8674(77)90160-x. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988 Dec 23;55(6):1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Levy-Wilson B., Dixon G. H. Limited action of micrococcal nuclease on trout testis nuclei generates two mononucleosome subsets enriched in transcribed DNA sequences. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1682–1686. doi: 10.1073/pnas.76.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Tatchell K. Chromatin core particle unfolding induced by tryptic cleavage of histones. Nucleic Acids Res. 1977 Jun;4(6):2039–2055. doi: 10.1093/nar/4.6.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundell M., Martinson H. G. The DNase I sensitive state of "active" globin gene chromatin resists trypsin treatments which disrupt chromatin higher order structure. Biochemistry. 1989 Dec 12;28(25):9757–9765. doi: 10.1021/bi00451a032. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Converting a eukaryotic transcriptional inhibitor into an activator. Cell. 1988 Nov 4;55(3):443–446. doi: 10.1016/0092-8674(88)90030-x. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., True R., Burch J. B., Kunkel G. Semihistone protein A24 replaces H2A as an integral component of the nucleosome histone core. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1030–1034. doi: 10.1073/pnas.76.3.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Smith M. R., Cook R. T. Mechanisms of polyanion stimulation of nuclear DNA synthesis. Examination of thermal destabilization. Exp Cell Res. 1977 Nov;110(1):15–23. doi: 10.1016/0014-4827(77)90264-6. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Stein A., Townsend T. HMG 14/17 binding affinities and DNAase I sensitivities of nucleoprotein particles. Nucleic Acids Res. 1983 Oct 11;11(19):6803–6819. doi: 10.1093/nar/11.19.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteaux B., Lasky L., Harary I. Lysine-rich histones and the selective digestion of the globin gene in avian red blood cells. Biochemistry. 1978 Dec 12;17(25):5532–5536. doi: 10.1021/bi00618a031. [DOI] [PubMed] [Google Scholar]
- Walker J. M., Hastings J. R., Johns E. W. A novel continuous sequence of 41 aspartic and glutamic residues in a non-histone chromosomal protein. Nature. 1978 Jan 19;271(5642):281–282. doi: 10.1038/271281a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Van Lente F. Dissection of chromosome structure with trypsin and nucleases. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4249–4253. doi: 10.1073/pnas.71.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Localization of the sites along nucleosome DNA which interact with NH2-terminal histone regions. J Biol Chem. 1977 Sep 25;252(18):6516–6520. [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Stein A. Folding of DNA by histones which lack their NH2-terminal regions. J Biol Chem. 1978 Jun 10;253(11):3857–3861. [PubMed] [Google Scholar]
- van Holde K. E., Lohr D. E., Robert C. What happens to nucleosomes during transcription? J Biol Chem. 1992 Feb 15;267(5):2837–2840. [PubMed] [Google Scholar]