Abstract
The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has highlighted the pivotal role of the immune response in determining the progression and severity of viral infections. In this paper, we review the most recent studies on the complicated dynamics between SARS-CoV-2 and the host immune system, highlight the importance of understanding these dynamics in developing effective treatments and formulate potent management strategies for COVID-19. We describe the activation of the host's innate immunity and the subsequent adaptive immune response following infection with SARS-CoV-2. In addition, the review emphasizes the immune evasion strategies of the SARS-CoV-2, including inhibition of interferon production and induction of cytokine storms, along with the resulting clinical outcomes. Finally, we assess the efficacy of current treatment strategies, including antiviral drugs, monoclonal antibodies, and anti-inflammatory treatments, and discuss their role in providing immunity and preventing severe disease.
Keywords: COVID-19, immune response, clinical intervention strategies
Introduction
Over the past two decades, coronaviruses such as severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) have emerged as major threats to global health, fundamentally altering our understanding of viral epidemiology and host–pathogen dynamics. In December 2019, a novel β-coronavirus emerged, initially named 2019-nCoV and later named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), precipitating the coronavirus disease 2019 (COVID-19) [1]. Although most patients experience mild symptoms and recover successfully, a notable minority suffer from severe complications. Acute respiratory distress syndrome (ARDS), driven by a dysregulated cytokine milieu or ‘cytokine storm’, has been the primary cause of mortality among these severe cases. The initial encounter between SARS-CoV-2 and the host triggers a broad-spectrum innate immune response, including the activation of physical barriers, cellular defenses, and the release of cytokines and chemokines. These early responses are crucial for containing the viral spread and orchestrating the subsequent engagement of the adaptive immune system. The complexity of the immune response to SARS-CoV-2 results in a spectrum of clinical outcomes, from life-threatening conditions to long covid. Understanding this variability is crucial for dissecting the host response mechanisms, comprehending the pathogenesis of COVID-19, and identifying potential therapeutic targets and providing precision therapeutic strategies.
SARS-CoV-2 interactions with host cell
Angiotensin-converting enzyme 2 (ACE2) is the primary receptor for SARS-CoV-2 entry into cells. Cells that express ACE2, except for certain mouse variants, are susceptible to SARS-CoV-2 infection. Conversely, cells lacking ACE2 are not infected, highlighting ACE2’s crucial role. Additionally, other cellular proteins, such as transmembrane serine protease 2 (TMPRSS2) and endosomal cysteine proteases B and L (CatB/L), are involved in the infection process. TMPRSS2 activates the spike protein, facilitating viral entry, while CatB/L also aids in this process. By inhibiting TMPRSS2 activity with camostat mesylate or suppressing CatB/L with ammonium chloride, cellular entry of SARS-CoV-2 could be partially blocked in the presence of another enzyme's activity [2]. When using a combination of camostat mesylate and E-64d (another CatB/L inhibitor), the viral infection was completely blocked. These studies suggest that both TMPRSS2 and CatB/L play a role in activating SARS-CoV-2’s S protein. It is intriguing to note that TMPRSS2 appears to have a more pivotal role in the viral entry than CatB/L, implying the possibility of other routes for viral infection.
The neuropilin-1 (NRP-1) receptor, a transmembrane receptor that is highly expressed in respiratory and olfactory epithelium but lacks a cytoplasmic protein kinase domain, has also been implicated in this entry process. S1 protein of SARS-CoV-2 can bind to the b1b2 domain of NRP-1. The polybasic amino acid sequence (682RRAR685) on the S1 protein promotes its interaction with NRP-1 [3, 4].
Furthermore, the entry mechanism of SARS-CoV-2 into cells might also be associated with the neutral amino acid transporter B0AT1, or solute carrier family 6 member 19 (SLC6A19). B0AT1 is a membrane-bound transporter protein primarily responsible for the transport of specific neutral amino acids. It forms a complex with ACE2, participating in various biological processes, including the absorption of amino acids. This interaction with ACE2 may significantly influence how the virus utilizes ACE2 as a gateway for entry into host cells. ACE2 may function as the membrane transport companion of B0AT1, controlling the intake of neutral amino acids into intestinal cells. Cryo-electron microscopy analysis indicates that ACE2 can form dimeric or heterodimeric structures with B0AT1 [5]. Residues 697 to 716 are located at the C-terminus of ACE2, where the TMPRSS2 protease can facilitate virus entry and participate in the dimerization process. Hence, the presence of B0AT1 might inhibit SARS-CoV-2 infection by restricting TMPRSS2’s access to the ACE2 cleavage site. Notably, B0AT1 is predominantly found in the kidney and intestine, and its expression is less than that of ACE2. In scenarios where B0AT1 is absent, ACE2 might preferentially form homodimers, as B0AT1 would not interfere. The exact role of B0AT1 in SARS-CoV-2 infections within the intestines remains an open question and warrants further investigation.
Mechanisms of SARS-CoV-2 immune evasion
In exploring the role of antiviral immunity in the host response to SARS-CoV-2, it is imperative to recognize that this encompasses a sophisticated and coordinated action between both the innate and adaptive branches of the immune system. Innate immunity acts as the body's first line of defense, employing a broad array of cellular and molecular mechanisms to rapidly confront and curb viral spread. This sets the stage for a more targeted and enduring response. Following this, the adaptive immune system, characterized by its specificity and memory, mounts a tailored attack against the virus. It involves the activation and proliferation of T cells and B cells. These two arms of the immune system work in concert, not only to neutralize and eliminate the virus but also to establish a state of immune memory that prepares the host for potential future encounters with the pathogen. This interplay between innate and adaptive immunity is crucial for a comprehensive understanding of the host's antiviral defense mechanisms and underscores the complexity of developing effective therapeutic interventions.
The role of innate immunity in SARS-CoV-2 infection
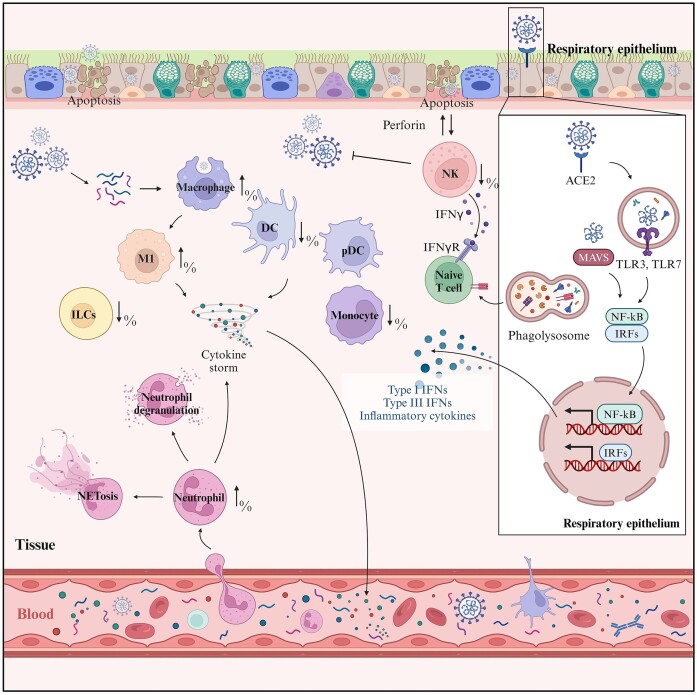
Innate immunity serves as the primary defense against pathogens, including SARS-CoV-2 (Fig. 1). It restricts viral activities such as entry, translation, replication, and assembly. Additionally, innate immunity facilitates the recognition and clearance of infected cells and speeds up the initiation of adaptive immunity. Key players in this defense mechanism against SARS-CoV-2 are cells like macrophages, basophils, eosinophils, neutrophils, dendritic cells, natural killer (NK) cells, and innate lymphoid cells (ILCs). The spike protein of SARS-CoV-2 primarily promotes viral invasion by binding to the ACE2 receptor, but it can also interact with proteins such as NRP-1 [6].
Figure 1.
Innate immune response mechanisms during SARS-CoV-2 infection. Innate immunity serves as the primary defense against SARS-CoV-2. When infected with SARS-CoV-2, the innate immune cells, including macrophages, neutrophils, dendritic cells (DCs), and plasmacytoid dendritic cells (pDCs), produce various pro-inflammatory cytokines and chemokines, contributing to the elimination of the infected cells. However, as a result of extensive cytokine release by the immune cells, neutrophil activation and degranulation as well as NETosis, a process where neutrophils expel their DNA to trap pathogens, could be observed. The cascade of signaling pathways involving toll-like receptor 3 (TLR3), toll-like receptor 7 (TLR7), mitochondrial antiviral-signaling protein (MAVS), and the transcription factors nuclear factor-kappa B (NF-κB) and interferon regulatory factors (IRFs) could be activated and further amplify the response, producing type I and type III interferons and other inflammatory cytokines, leading to exacerbated tissue damage. The innate immune response exhibits a multifaceted nature, including both protective and pathological outcomes in the context of COVID-19. (Created with BioRender.com).
Upon binding of the S protein to the receptor, the TMPRSS2 protease is instrumental in facilitating the proteolytic processing and entry of the S protein [2]. Other host proteins like neuropilin-1, heparan sulfate proteoglycan, C-type lectin, or furin protease can be co-factors for the virus's entry [4, 7]. After the virus binds to a cell, it may merge directly with the cell membrane, releasing its RNA into the cytoplasm, or enter certain cells, leading to a fusion of the viral and cellular membranes in a low pH environment [8].
Once inside the cytoplasm, SARS-CoV-2 is believed to operate similarly to other coronaviruses. Its genomic RNA translates into large polyproteins, pp1a and pp1ab, which produce 16 non-structural proteins. These proteins facilitate the formation of the virus's replication–transcription complex, converting the viral RNA into an antisense negative-strand template. The innate immune response to SARS-CoV-2 begins when pathogen-associated molecular patterns (PAMPs) of the virus are detected by pattern recognition receptors (PRRs) on immune cells. These receptors also recognize damage-associated molecular patterns (DAMPs) from infected tissues. Once PRRs bind to PAMPs or DAMPs, they activate signaling pathways inside the cells. This activation leads to the production of cytokines and chemokines, such as interferons (IFNs). The release of these cytokines not only suppresses pathogen proliferation but also bridges the gap to adaptive immunity activation.
Among the PRRs, those notably associated with RNA virus detection include toll-like receptors (TLRs) and RIG-I-like receptors (RLRs). TLRs, a diverse family of nine receptors, each recognize distinct PAMPs. Some TLRs, like TLR1, TLR2, TLR4, TLR5, and TLR6, are expressed on the cell surface, detecting extracellular pathogens. In contrast, TLRs like TLR3, TLR7, TLR8, and TLR9 are intracellular and recognize pathogens within cells. RLRs, on the other hand, are cytosolic receptors primarily involved in detecting viral infections. Key RLR members include melanoma differentiation-associated protein 5 (MDA-5), retinoic acid-inducible gene 1 (RIG-I), and laboratory of genetics and physiology 2 (LGP2)—RNA helicases present in the cytoplasm [9]. While they remain inactive in uninfected cells, the presence of viral RNA activates them, triggering interferon production to counteract the infection. For SARS-CoV-2 detection, TLRs stimulate transcription factors such as nuclear factor-kappa B (NF-κB), interferon regulatory factor 3 (IRF3), and IRF7. Subsequently, these activated factors migrate to the cell nucleus, promoting inflammasome assembly and amplifying the release of pro-inflammatory cytokines, and type I interferons. This cascade further propels the activation of additional innate immune cells.
Monocytes and macrophages play a crucial role in regulating inflammation and recruiting immune cells. As antigen-presenting cells (APCs), they are pivotal in rapidly responding to pathogens during acute infections. In the context of severe COVID-19 afflictions, monocytes contribute to amplified inflammatory surges and subsequent cytokine storms [10]. Compared to healthy controls, patients with severe COVID-19 display a decrease in total blood cells and CD16-typical monocytes, while the inflammatory monocyte subpopulation increases [11]. Lung-resident alveolar macrophages maintain pulmonary equilibrium and respond to SARS-CoV-2 by recognizing viral proteins through TLR receptors, leading to M1 polarization and contributing to macrophage activation syndrome (MAS) and acute respiratory distress syndrome (ARDS) [12, 13]. Analysis of single-cell RNA sequencing data from the bronchoalveolar lavage fluid of both mild and severe COVID-19 patients has shown an increase in inflammatory macrophage subgroups and a downregulation of type I IFN genes [14], highlighting macrophages' dual role in defense and pathology.
Dendritic cells (DCs), serve as the pivotal APCs in activating both the innate and adaptive arms of the immune arsenal. Conventional myeloid DCs (cDCs) are known for their effectiveness as APCs, and plasmacytoid DCs (pDCs) are renowned for their ability to produce type I IFN. In the context of SARS-CoV-2, these cells become a gateway through receptors like ACE2, CD147, and notably, DC-SIGN, with interstitial lung DCs emerging as the principal sites of viral entry. Severe COVID-19 patients exhibit decreased cDCs and pDCs, correlating with disease severity [15]. When stimulated by SARS-CoV-2, pDCs generate substantial amounts of type I and III IFNs through the toll-like receptor 7 (TLR-7) pathway, yet they refrain from releasing the inflammatory agents Tumor necrosis factor-α (TNF-α) and interleukin 6 (IL-6) [16], indicating a complex regulatory role in the immune response.
SARS-CoV-2 infection leads to a significant migration of neutrophils into the lung capillaries and alveoli, causing acute capillaritis and fibrin accumulation. Notably, patients with severe SARS-CoV-2 infection exhibit an increased neutrophil to lymphocyte ratio (NLR), indicating heightened infection and inflammation [17]. Elevated markers of neutrophil activation, such as myeloperoxidase (MPO) and neutrophil extracellular traps (NETs), are observed in severe cases [18]. These factors contribute to excessive cytokine release and respiratory failure, exacerbating the severity of the disease [19].
In the fight against viruses, NK cells serve as vital components of the innate immune defense. These cells not only induce target-cell lysis directly but also secrete inflammatory cytokines, mediate antibody-dependent cellular cytotoxicity (ADCC), and collaborate with other immune cells, such as monocytes. SARS-CoV-2 infections reduce the number and function of NK cells, particularly the CD56dimCD16+ subset, crucial for cytolytic activity [20]. Increased expression of inhibitory receptors like NKG2A and diminished activation markers such as CD107a and IFN-γ in NK cells are noted in severe cases [21]. Furthermore, elevated circulating IL-6 levels, especially prominent in deceased patients, show a strong association with decreased NK cell activity and heightened activation of other innate immune cells, culminating in escalated pro-inflammatory cytokine production and severe disease outcomes [22]. Collectively, these findings imply that the marked impairment in innate immunity, especially the decline in NK cell functionality seen in severe COVID-19 cases, is largely ascribed to the SARS-CoV-2 infection [23].
The role of adaptive immunity in SARS-CoV-2 infection
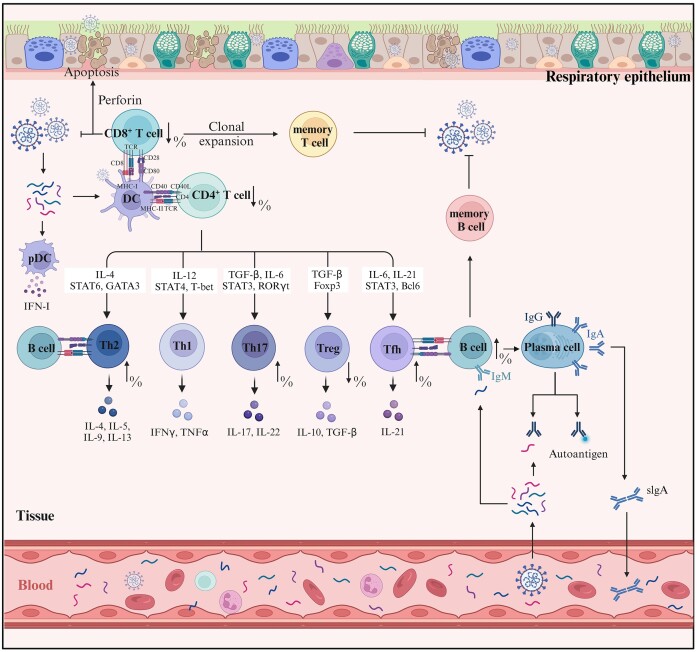
Adaptive response is characterized by its specificity to the SARS-CoV-2 antigen and the development of immunological memory, offering long-term protection against the virus (Fig. 2). The response begins when APCs, mainly dendritic cells, present viral antigens to T cells using major histocompatibility complex (MHC) molecules. This presentation activates T cells, leading to the differentiation of CD8+ cytotoxic T cells and CD4+ helper T cells. CD4+ T cells help activate and differentiate B cells into antibody-producing plasma cells and memory B cells, culminating in the production of virus-specific antibodies.
Figure 2.
T cell- and B cell-mediated immune responses during SARS-CoV-2 infection. During a SARS-CoV-2 infection, pDCs secrete IFN-I to activate CD8+ and CD4+ T cells. These T cells undergo clonal expansion with CD8+ T cells contributing to apoptosis of infected cells via perforin, and CD4+ T cells differentiating into various subtypes including Th1, Th2, Th17, Treg, and Tfh, based on the cytokine environment and transcription factors like STAT6, GATA3, T-bet, RORγt, and Foxp3. Among the cell subpopulations, Tfh cells facilitate the differentiation of B cells into memory B cells and plasma cells, which produce specific immunoglobulins (IgM, IgG, IgA) aimed at neutralizing the virus, contributing to the immune defense and potential immunopathology associated with COVID-19. (Created with BioRender.com).
Within ∼1 week post the onset of COVID-19 symptoms, an adaptive response, marked by T-cell and B-cell reactivity to SARS-CoV-2, becomes discernible in the bloodstream [24]. The disease trajectory illuminated by adaptive immune responses during COVID-19 infection unveils a landscape of complexity.
Serving as cornerstones in the adaptive immune milieu, T cells not only possess the capacity to eliminate virus-infected cells but also modulate the broader immune landscape via secreting cytokines. Historical encounters with highly pathogenic coronaviruses provide insights into the T cell dynamics. For instance, during the early phases of SARS-CoV and MERS-CoV infections, patients predominantly exhibited either normal or marginally decreased white blood cell and lymphocyte counts. As the disease progressed, these counts further dropped, revealing an inverse association with mortality rates [25, 26]. An interesting facet of SARS-CoV infections lies in the varied duration of immune responses. Although antibody responses decline within 1–2 years post-infection, T cell responses remain resilient, enduring for up to 17 years [27]. In the context of COVID-19, notable observations include the decline in CD4+ and CD8+ T cell populations and evident immune dysregulation [28–30]. These underscore the central role of T cells, not merely in direct antiviral defense, but also in orchestrating wider immune system interplay.
Dynamics of CD4+ T cells in SARS-CoV-2 infection
Observational studies emphasize the ubiquity of CD4+ T cell responses in COVID-19 afflicted individuals [28–30]. Intriguingly, patients bearing the brunt of the disease showcase amplified activation dynamics: elevated levels of CD4+ T cell aging, apoptosis, and metabolic perturbations. In contrast, individuals with milder clinical presentations illustrate a diversified spectrum of CD4+ T cell responses. This disparity hints at a potential correlation between CD4+ T cell reactions and the clinical manifestations of the disease. CD4+ T cells exhibit a robust response to the S protein of SARS-CoV-2, especially the N and C terminal regions of the S protein [31, 32], correlating with anti-SARS-CoV-2 IgG and IgA antibody titers [31].
On the other hand, longevity insights from the SARS-CoV epidemic underscore the persistence of memory T cells, lasting upwards of 2 years post-infection [27, 33]. These cells, upon subsequent encounters with either SARS-CoV or SARS-CoV-2, could potentially mount an efficacious immune defense. Diverse and frequent SARS-CoV-2 specific memory T cells, discernible in convalescent patients, seem to harbor recognition capacities for SARS-CoV-2 structural proteins and share a positive relationship with antibody titers [34]. From mild to severe cases, CD4+ T cell responses exhibit commendable persistence, enduring for at least 1 year post-infection, underscoring their presumptive prolonged protective capacities [35].
The ability of Th1 cells to clear viruses is attributed to their capacity to enhance cytotoxic T cells (CTLs) and macrophage phagocytosis. Additionally, their pro-inflammatory aptitude assists in robust viral elimination and fortifying the immunological bulwark. Th2 cells, essential for humoral responses against extracellular pathogens and allergens, are critical for antibody production dynamics. In SARS-CoV-2 infection, Th2 cells promote antibody production by activating B cells and modulating inflammatory landscapes, aiding in viral eradication and immune defense fortification.
The Th1/Th2 equilibrium plays a defining role across a spectrum of diseases, with disruptions in this balance being linked to severe clinical repercussions, such as persistent infections, tissue damage, allergic reactions, and tumor cell evasion. In the context of COVID-19, the immunological narrative highlights a notable Th1/Th2 imbalance. A common pattern among affected individuals is the decrease in Th1 cell representation concomitant with an increase in Th2 cell frequencies, leading to a Th2/Th1 cytokine imbalance [36]. This shift can impair immune efficacy and exacerbate inflammation. Further investigations have revealed a concurrent decrease in Th1 and Th17 cell proportions, along with an increase in activated Th2 cell numbers in COVID-19 patients [37]. The posthumous immune profiles also indicate a significant enrichment of senescent Th2 cells (characterized as PD1+/ICOS−). The elevated lymphocyte landscape, combined with a rise in senescent Th2 cells, is a concerning marker of mortality [38].
Th17 cells have been associated with chronic inflammatory and autoimmune diseases, and their relevance has been validated in the context of viral infections, especially COVID-19. Elevated Th17-mediated cytokine, such as IL-17, has been observed in COVID-19 patients [39], correlating with inflammation and tissue injuries. Reflecting on past epidemics, discernible Th17 reactions were similarly mapped in MERS-CoV- and SARS-CoV-afflicted individuals [40, 41]. One cannot ignore the conspicuous association of IL-17 with ARDS pathogenesis [42], a phenomenon potentially resulting from IL-17’s pivotal role in neutrophil mobilization and activation, along with its intricate interplay with cytokines such as granulocyte colony-stimulating factor (G-CSF), TNF-α, IL-1β, and IL-6 [43]. Hence, in the COVID-19 context, the abundance of neutrophils and Th17 cells correlates positively with both viral titers and clinical severity, with their presence in pulmonary tissues exacerbating inflammation and lung damage [44].
Regulatory T cells (Tregs) attenuate cytokine storms triggered by respiratory viruses, paving the path for improved patient prognosis. In the context of COVID-19, Treg dynamics differ: severe cases show decreased Tregs, while mild or latent cases show increased Tregs [17, 45, 46]. This relationship indicates that Treg prevalence correlates with disease severity and respiratory distress susceptibility, highlighting Tregs as potential therapeutic targets. Higher pulmonary Treg presence is linked to milder symptoms, suggesting that Treg concentrations influence clinical outcomes [47]. However, compromised Treg functions can trigger excessive immune reactions against SARS-CoV-2, leading to inflammation [48]. Treg scarcity can destabilize immune balance, allowing the unchecked proliferation of immune cells, including NK cells, Th1 cells, and Th17 cells to secrete pro-inflammatory cytokines, TNF-α, IL-6, and IL-1β. Accentuating this paradigm, patients severely afflicted with COVID-19 exhibit increased Th17 populations concurrent with a Treg recession [48]. Hence, Tregs might exhibit a dual role during both the initial and advanced stages throughout the course of COVID-19: initially reducing inflammation and cytokine storms, but later, their deficit can provoke pro-inflammatory responses, leading to complications such as acute lung injury, ARDS, and mortality.
During the acute phase of SARS-CoV-2 infection, T follicular helper (Tfh) cells and their memory cells develop to support the immune response. There is also a noted increase in these specific Tfh cells among hospitalized COVID-19 patients [45], which may result from the Tfh cells' potential to target B cells and inhibit germinal center reactions [49]. However, other studies demonstrate a reduced prevalence of Tfh cells in individuals recovering from COVID-19, accompanied by shifts in circulating T follicular helper (cTfh) subpopulations [50]. Convalescent sera contain IgG and IgM antibodies against SARS-CoV-2’s nucleocapsid protein (NP) and spike protein (SP), with IgG levels positively correlating with Tfh cell counts. Notably, those recovering from severe COVID-19 display enhanced neutralizing antibody titers, rapid lymphocyte rebound, and increased CXCR3+ Tfh levels—these latter levels are directly correlated to the concentrations of neutralizing antibodies. Therefore, SARS-CoV-2-specific cTfh cells are characterized by their unique specificity and function, which are associated with milder disease outcomes [51, 52].
The multifaceted role of CD8+ T cells in SARS-CoV-2 infection
CD8+ T cells, key effectors in adaptive immunity, play a pivotal role in the direct neutralization of pathogens and tumor cells. In the landscape of COVID-19 immunobiology, the response of CD8+ T cells against specific SARS-CoV-2 proteins, including S, M, N, nsp6, and ORF3a, has been elucidated [31, 53]. Notably, the concentration of SARS-CoV-2-specific CD8+ T cells is profoundly linked to clinical disease progression [51]. Intriguingly, one study highlighted the rapid proliferation of these T cells from the day symptoms appeared, even if they were imperceptible pre-infection [54]. Functionally, the memory CD8+ T cells targeting SARS-CoV-2 exhibit striking parallels to those against influenza, persisting even in patients who have cleared their SARS-CoV-2 infection but lack detectable levels of spike (S) and nucleocapsid (N) antibodies [54].
Further insights reveal that the magnitude of the SARS-CoV-2-specific CD8+ T cell response is associated with disease severity and often diminishes in more severe cases [55]. Even with high antibody concentrations, a weakened CD8+ T cell response might exacerbate acute COVID-19 symptoms [56]. Compared to healthy individuals, COVID-19 patients’ CD8+ T cells show increased levels of the degranulation marker CD107a, IL-2, and IL-17 upon anti-CD3/CD28 stimulation [39]. However, some studies describe a diminished cytokine-production capability in CD8+ T cells from COVID-19 patients, independent of any SARS-CoV-2 antigenic stimulation [22].
Emerging evidence also shows a significant increase in exhaustion markers such as CTLA-4, PD-1, LAG-3, TIM-3, NKG2A, and CD39 on circulating CD8+ T cells in severe COVID-19 cases, potentially contributing to impaired cellular immune responses. Notably, PD-1 expression on CD8+ T cells is inversely proportional to CD38, a marker of cellular activation [21, 57]. This increase in PD-1 expression is particularly pronounced in intensive care unit-admitted patients [58]. Collectively, these findings suggest a dual role of CD8+ T cells in COVID-19: they may protect during the early stages of the disease but contribute to its pathogenesis in advanced stages due to compromised cytotoxic capabilities and unchecked cytokine release [59].
Following the initial manifestation of COVID-19 symptoms, memory CD8+ T cells emerge in the bloodstream between 20 to 50 days, displaying a noteworthy half-life of ∼225 days. This extended persistence indicates a potential for durable immune memory against SARS-CoV-2 [60].
Another study suggests the indispensable role of memory CD8+ T cells in the effective clearance of SARS-CoV-2, especially evident in scenarios depicting T cell exhaustion, as observed in convalescent rhesus macaques [61]. Furthermore, certain vaccines, like Pfizer-BioNTech's BNT162b2, robustly stimulate CD8+ T cell populations, fortifying the immune system against SARS-CoV-2 reinfections [62]. Post-vaccination, increased CD8+IFN-γ and TNF-α are discernible within a week, implying robust T cell activation. Booster shots induce highly differentiated CD8+ T cells with functional profiles reminiscent of early memory T cells spurred by primary vaccine doses [63]. Hence, exploring the consequences of these overlapping functional behaviors, especially in the realm of sustained immunity, is a promising direction for further study.
The functions of B cells in SARS-CoV-2 infection
The humoral immune response against SARS-CoV-2 activates after the onset of COVID-19 symptoms and involves two crucial phases in the establishment of immune memory. Initially, upon exposure to the virus, a plasma cell response is triggered, leading to the induction of antigen-specific B cells with relatively low affinity. Subsequently, within secondary lymphoid tissues, CD4+ follicular helper T (Tfh) cells collaborate with B cells in a multifaceted process that promotes both antibody affinity maturation and isotype switching, essential for long-term immune protection [64].
Antibodies are immunoproteins produced by B lymphocytes in response to antigen exposure, and primarily circulate in human blood and tissues. Only specific antibodies can recognize and neutralize invading microbes, thus earning the name “neutralizing antibodies”. The emergence of these neutralizing antibodies aligns with seroconversion, possibly influenced by cytokines promoting antibody isotype switching. Notably, B cells can produce neutralizing antibodies relatively quickly and easily, existing primarily as heavy and light chains, with somatic hypermutations occurring infrequently.
Unlike previous viral infections like dengue fever and Zika virus, IgG serological responses to SARS-CoV-2 appear almost concurrently with IgM and IgA responses, generally within 7–10 days post-infection [65]. The presence of IgG antibodies targeting the receptor binding domain (RBD) positively correlates with anti-S neutralizing antibody titers. Most individuals stabilize these levels within 75 days after the onset of COVID-19 symptoms [66]. Within the subsequent 5 months, these IgG antibody titers remain relatively constant, playing a pivotal role in lowering the risk of reinfection [67]. However, a notable decline in IgG antibodies begins between weeks 5–7 and persists through weeks 34–42. During the period of weeks 8–11, 16.7% of patients exhibit a loss of IgM antibodies [68].
The consistency of antibody levels in the bloodstream can be influenced by several factors, such as the intensity and longevity of the immune response, distinct antibody isotypes, and the balance between short-lived and long-lived plasma cells. Additionally, higher viral antigen levels in the body often induce higher antibody titers, and patients with more severe symptoms usually have higher peak neutralizing antibody titers. However, some patients have notably low antibody levels, suggesting varying strategies the virus might adopt within the adaptive immune framework. Another intriguing observation is that symptomatic patients tend to lose detectable antibodies earlier than their asymptomatic counterparts [69]. A deeper exploration is warranted to elucidate the dynamics of antibody responses and the differences in immune retention between symptomatic and asymptomatic patients.
B cells play a multifaceted role in antiviral immune responses. Beyond their primary function of producing neutralizing antibodies against a plethora of viruses, including SARS, MERS, HIV, and Ebola, they orchestrate antiviral defenses through the secretion of inflammatory mediators and the activation of T cells. By binding directly to viruses, B cells inhibit their entry into host cells. Furthermore, antibodies possess the capability to activate the complement system and execute infected cells via ADCC. In certain scenarios, antibodies can enhance the uptake of viral particles either through Fc receptors or complement receptors. However, this can sometimes backfire, resulting in an intensified viral infection, a phenomenon known as antibody-dependent enhancement (ADE). ADE has been documented in several viral infections, including dengue, HIV, influenza, RSV, Ebola, and notably, SARS-CoV-2. ADE might be one of the factors leading to severe disease and death in patients, as ADE in monocytes and macrophages could result in excessive inflammation and severity of the disease. Recent findings suggest that certain approved monoclonal antibodies designed to combat COVID-19 might inadvertently promote ADE [70]. Given this backdrop, it is imperative for ongoing research to meticulously assess ADE risks, especially when confronted with emerging SARS-CoV-2 variants, such as Omicron.
Role of cytokines in SARS-CoV-2 infection
Production of interferons by the innate immune system against SARS-CoV-2
Interferons are vital cytokines integral to the immune response during viral infections, especially in terms of cytokine production and signal transduction. They are categorized into three main families: IFN-I, IFN-II, and IFN-III. Each family plays a significant role in countering microbial pathogens [71]. When infected by respiratory coronaviruses, such as CoV, the immune system activates a cascade of signaling pathways, leading to the synthesis of IFNs. These IFNs, in turn, amplify the immune response by enhancing IFN signaling, either via autocrine or paracrine pathways.
In the respiratory system, major sources of IFN during viral invasions include epithelial cells, endothelial cells, alveolar macrophages, natural killer cells, dendritic cells, and inflammatory monocyte macrophages (IMMs). Noteworthy is the role of pDCs during infections by coronaviruses like SARS-CoV, as they are considered primary producers of IFN-I. When a viral invasion occurs, specific subgenomic RNA (sgRNA) can interact with genome templates, forming double-stranded RNA (dsRNA). MDA5 then recognizes and clusters around these dsRNA formations, subsequently binding to the mitochondrial antiviral signaling protein (MAVS). This association kickstarts a sequence of molecular interactions, ultimately activating key transcription factors, such as NF-κB and interferon regulatory factors. Upon encountering specific viral ligands, TLRs engage adaptor proteins like myeloid differentiation primary response 88 (MyD88) or TIR-domain-containing adapter-inducing interferon-β (TRIF). This engagement triggers a signaling cascade, activating NF-κB and IRF [72].
Once activated, these factors form a complex termed the “enhancer”. This enhancer complex binds near the transcription start sites for both IFN-I (particularly IFNβ) and IFN-III (e.g. IFNλ-1, IFNλ-2, IFNλ-3), driving antiviral responses. Although both IFN-I and IFN-III lead to analogous transcriptional effects, they have distinct biological properties. These differences span the composition and distribution of their respective cell surface receptors, protein stability, and the magnitude and anatomical locale of their responses. Notably, IFN-I can mediate an antiviral state in nearly all cells. In contrast, IFN-III's action is more focused, primarily targeting epithelial barrier tissues, such as those in the respiratory and gastrointestinal tracts, and the blood–brain barrier. Both IFN types amplify their signaling via autocrine and paracrine pathways, spurring the increased expression of interferon-stimulated genes (ISGs). These ISGs provide antiviral defenses, either directly or indirectly, by inhibiting viral replication, reducing viral transcription/translation, or degrading viral genetic material.
IFN signaling is crucial for the host's ability to respond to coronaviruses. This viewpoint was first confirmed when research found that animals lacking the IFN-I receptor (IFNAR1−/−) exhibited more severe disease manifestations after infection with the mouse hepatitis virus (MHV) [73, 74]. This observation underscores the protective nature of the IFN-I response within the host, safeguarding distant organs from subsequent infections [73, 74]. An absence of this defense renders peripheral tissues susceptible to infiltration by circulating viruses, a phenomenon that can intensify disease severity. Notably, this pattern has been found in younger COVID-19 patients with specific genetic aberrations in IFNAR1 or TLRs [75], while certain older individuals exhibit autoantibodies against IFN-I [76]. During infection, variations in the expression of IFN-I, IFN-III, ISGs, and PRRs in relation to age have been linked to disease severity among COVID-19 patients [77].
Immune evasion by SARS-CoV-2 through inhibition of interferon production
As mentioned, the innate immune system typically springs into action, producing interferons and cytokines to counteract the viral threat. However, like other viruses, SARS-CoV-2 deftly evades immune surveillance to curtail IFN output through various cunning strategies, such as dampening PAMP and hampering IFN signaling [78, 79].
It was reported that SARS-CoV-2’s papain-like protease (PLpro) thwarts activation by stripping MDA5 of its ISG15 modification—a process that is fundamental post-RNA viral invasion [80]. Additionally, proteins like open reading frame 6 (ORF6), open reading frame 8 (ORF8), and nucleocapsid (N) of SARS-CoV-2 are known to impede the production of IFN-β by targeting the RIG-I and MDA5 signaling routes [81]. Furthermore, ORF6 not only disrupts the signal transduction pathways of TLR and RLR but also blocks transcription activator 1 (STAT1) from entering the nucleus, a crucial step for IFN synthesis [82]. These studies suggest that SARS-CoV-2 hinders the production of these interferons by either inhibiting mRNA release from transcription sites or accelerating its nuclear degradation [83].
Furthermore, numerous proteins dictated by SARS-CoV-2 disrupt both RLR sensors and the orchestration and transmission of IFN cues [84]. The N and membrane (M) proteins of SARS-CoV-2 inhibit IFN signaling, while ORF9b suppresses the TLR3–TRIF signaling machinery [85]. Interestingly, ORF3b of SARS-CoV-2 obstructs type I IFN synthesis more proficiently than its counterpart in SARS-CoV [86]. Non-structural proteins of the virus, such as NSP1 and NSP14, suppress cellular translation, which in turn affects the proteins involved in the IFN signaling cascade [87].
In line with the findings, decreased levels of Type I and Type III interferons in the serum of mild to moderate COVID-19 patients suggest that SARS-CoV-2 may employ mechanisms to evade the host's immune response [88]. These findings indicate that gaining a comprehensive understanding of the interaction between SARS-CoV-2 and the host immune system illuminates the way forward for therapeutic advancements.
Cytokine storm induced by SARS-CoV-2
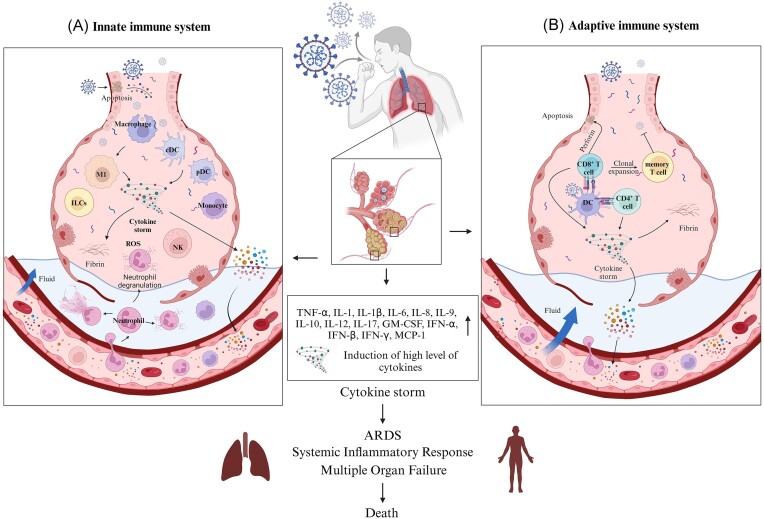
The severity of COVID-19 is intricately linked to the interplay between the virus-induced cellular pathologies and its evasion of the host immune response. In COVID-19 patients, the immune response might result in a potent inflammatory reaction known as cytokine release syndrome (CRS). Similar outcomes were observed in previous infections like SARS-CoV and MERS, both known to trigger cytokine storms. Consequently, the cytokine storm induced after SARS-CoV-2 infection is considered a key factor in severe COVID-19 cases (Fig. 3).
Figure 3.
Mechanisms of the cytokine storm in COVID-19 and its impact on immune response. (A) Initial reaction of innate immune system in COVID-19. SARS-CoV-2 triggers activation of macrophages, dendritic cells, and innate lymphoid cells to release pro-inflammatory cytokines and chemokines. This release enhances vascular permeability and recruits further immune cells, resulting in tissue damage and the onset of ARDS. (B) Upon interaction with antigen-presenting cells, the adaptive immune response, featuring the clonal expansion of specific T cells and B cells, amplifies cytokine release, and intensifies the cytokine storm which induces systemic inflammatory responses, leading to ARDS, multiple organ failure and even death. (Created with BioRender.com).
Numerous studies have shown elevated levels of various inflammatory cytokines in COVID-19 patients, including IL-1β, IL-2, IL-6, IL-10, IL-17, TNF-α, IFN-γ, IFN-γ induced protein 10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), and granulocyte-macrophage colony-stimulating factor (GM-CSF), and these cytokines correlate with the severity of the disease [89–92]. Be it in biopsy or post-mortem samples, multiple studies have verified inflammatory infiltration across diverse tissues in COVID-19 patients [93].
As previously described, interferons are critical in combating viruses and lead to the release of chemokines attracting neutrophils and macrophages [88]. An insufficient interferon response has been noted in infected cells. It has been found that the presence of autoantibodies against many immune-regulating proteins in patients, and the association of anti-type I interferon antibodies with severity and mortality is particularly noteworthy [94, 95]. Hence, the innate immune response might falter in clearing infected cells, potentially aiding viral replication during the early stages of infection. This theory is supported by the observation of SARS-CoV-2’s survival and multiplication within macrophages [96]. As the infection progresses, the immune response becomes more robust. The unchecked initial replication of the virus causes the response to be over-aggressive, which contributes to the cytokine storm and CRS.
Clinical intervention strategies for COVID-19
Based on our previous discussions, the selection of the most effective treatment strategy against different phases of COVID-19 is crucial [97]. In the case of asymptomatic SARS-CoV-2 carriers, isolation and observation are more suitable since antiviral drugs may not significantly improve virus clearance [98]. Additionally, these medications can have adverse effects, including potential liver damage [99]. For patients with severe symptoms, numerous immunotherapeutic methods are being explored to determine the optimal treatment protocols.
Therapeutic strategies for targeting inflammatory cytokines
The role of biological drugs in managing COVID-19-associated cytokine storm syndrome
The progression of biopharmaceutical technology has enabled the effective use of various cytokine inhibitors to treat immune rheumatological disorders such as cytokine storm syndrome (CSS). Given the suspected pivotal role of cytokine storms in the severity of COVID-19, several of these biologic drugs have been tested in patients infected with SARS-CoV-2 [100]. One of the primary culprits in cytokine storms is IL-1, which usually emerges following the activation of the innate immune response. In a retrospective study with 22 severe COVID-19 patients, treatment with Anakinra (an IL-1 receptor antagonist) over 8 days resulted in reduced need for mechanical ventilation, lowered serum C-reactive protein (CRP) levels, and improved clinical outcomes compared to a control group [101]. Additionally, Anakinra administration led to significant reductions in body temperature, white blood cell count, and plasma levels of several biomarkers [102]. Canakinumab, an IL-1β antagonist that prevents IL-1β from interacting with its receptor, exhibited potential in reducing mechanical ventilation in severe COVID-19 cases, although its impact on mortality rates remains inconclusive due to limited sample sizes [103]. However, the evidence for Canakinumab is currently constrained by limited sample sizes, necessitating more extensive clinical trials.
IL-6 has been identified as another potential therapeutic target, due to its prominent role in cytokine storm pathology. Tocilizumab was approved by the Food and Drug Administration (FDA) in 2017 for treating cytokine release syndrome (CRS) in certain conditions by binding to both circulating and membrane-bound IL-6 receptors, and in turn inhibiting its downstream signaling [104]. Multiple pieces of evidence have demonstrated its efficacy in treating CRS induced by COVID-19, notably reducing the mortality rate [105–107]. Thus, tocilizumab is considered one of the effective drugs for treating CRS induced by COVID-19.
In line with the findings, another monoclonal antibody, sarilumab, could inhibit both membrane-bound and soluble IL-6 receptors, promoting recovery in severe COVID-19 patients and reducing serum CRP levels [108]. Siltuximab, is an antagonist of IL-6 [109] and the impact of siltuximab in CRS induced by COVID-19 also has been investigated. Although it showed a promising effect in relieving symptoms and minimizing mechanical ventilation, comprehensive data on its safety and effectiveness in COVID-19-induced CRS are still lacking [109]. As such, tocilizumab has been recommended as the primary treatment, and siltuximab serves as an alternative choice.
The efficacy of anti-inflammatory drugs in managing severe COVID-19 symptoms
Excessive inflammation and cytokine storms induced by COVID-19 have been identified as the primary culprits behind severe interstitial pneumonia, ARDS, and coagulopathy. Urgent treatments targeting both the virus and its excessive inflammatory response were desired, and machine learning has been applied to develop anti-viral drug [110].
A large randomized controlled trial with 6 425 participants indicated dexamethasone's efficacy in reducing mortality in those requiring respiratory support, but not in those who didn not [111]. A meta-analysis involving 1 703 severe COVID-19 patients further supported dexamethasone's potential in reducing mortality rates [112]. Dexamethasone or other equivalent systemic corticosteroids have been endorsed for COVID-19 patients who require oxygen support or mechanical ventilation [113]. Intravenous immunoglobulin (IVIG), sourced from the serum of healthy donors, primarily consists of subclasses IgG1 and IgG2. These immunoglobulins harbor autoantibodies against cytokines, which might contribute to their anti-inflammatory properties [114]. As IVIG from healthy individuals contains these autoantibodies against cytokines such as IL-1, IL-6, and IFN-γ, their presence might elucidate the anti-inflammatory effects of IVIGs observed in inflammatory and autoimmune conditions [115]. In previous pandemics, such as SARS and H1N1 influenza, IVIG was effectively applied to treat the virus infection [116]. Recent studies suggested that early-stage COVID-19 treatment with IVIG could be beneficial, and there is a potential correlation between IVIG and a reduction in severe patient mortality rates [117]. However, some studies explored that the combined regimen of IVIG, hydroxychloroquine, and lopinavir/ritonavir might not be effective for severe cases [117].
Janus kinases (including JAK1, JAK2, JAK3, and Tyk2) are members of the receptor-associated tyrosine kinase family. By regulating immune responses, these kinases respond to extracellular signals like cytokines and interferons [118]. JAK inhibitors, including baricitinib and tofacitinib, have been employed in treating rheumatoid arthritis, and can potentially be repurposed to curtail the inflammatory response correlated to COVID-19 [119], highlighting their potential in inhibiting certain kinases responsible for viral endocytosis [120]. Similarly, tofacitinib, another JAK inhibitor, was found to reduce the risk of respiratory failure or death in hospitalized COVID-19 patients [121]. Colchicine, a gout treatment, restricts inflammatory responses to urate crystals and limits leukocyte migration [122]. By suppressing certain inflammasomes and cytokines, colchicine emerges as a prospective solution against COVID-19-induced inflammation, specifically 3–6 days after symptom onset, where it can potentially reduce fevers in a 72-h timeframe [123]. Furthermore, synergistic treatments combining colchicine with drugs like hydroxychloroquine and azithromycin have been shown to decrease mortality rates and hospitalization duration [122].
Clinical interventions regulating adaptive immune responses
Development and efficacy of COVID-19 vaccines
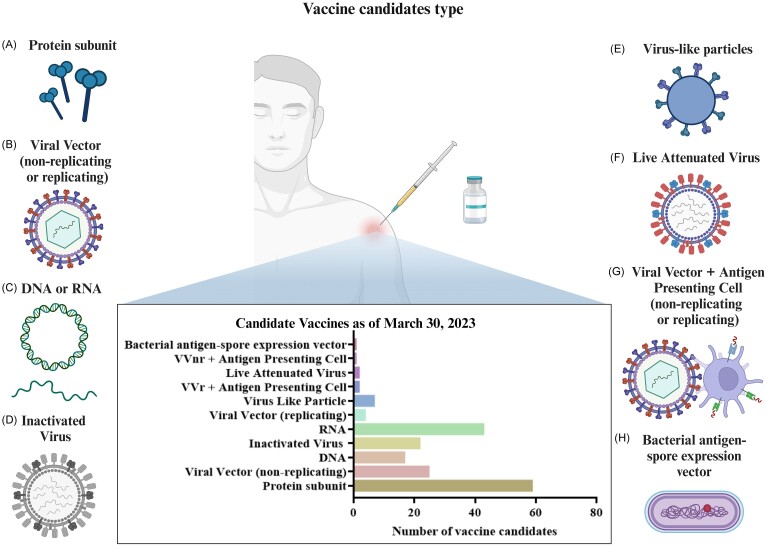
Vaccines have proven to be safe, effective, and lifesaving for multiple infectious diseases. Since the pandemic's onset, >273 vaccines have transitioned to preclinical or clinical evaluation (Fig. 4). Vaccines not only offer individual protection through acquired immunity but also pave the way for societal normalization. This immunity is achieved when CD4+ T cells, upon recognizing antigen-presenting major histocompatibility complex (AP-MHC) class II molecules, differentiate into Th cells, and CD8+ T cells evolve into CTLs by identifying AP-MHC class I molecules. Assisted by Th cells, B cells activate and produce antibodies, while subsequent antigen exposure results in long-lived memory cells, offering long-term protection against the same pathogens. Of the myriad SARS-CoV-2 vaccine candidates being explored, >10 have reached advanced clinical stages or secured emergency use clearance. These encompass diverse platforms: viral vectors, protein subunits, nucleic acids (e.g. mRNA, DNA), and virus-like particles [124]. Recent studies show that the effectiveness of mRNA vaccines against the XBB.1.5 Omicron lineage has significantly decreased [125]. Therefore, XBB.1.5 can significantly evade vaccine-induced humoral immunity, while bivalent vaccines can enhance the humoral immune response against the currently prevalent SARS-CoV-2 VOI XBB.1.5 lineage [126]. Additionally, the monovalent XBB.1.5 BNT162b2 COVID-19 vaccine has been approved as a single-dose vaccine for all individuals aged ≥6 months [127]. Recent studies have shown that booster shots of these updated vaccines significantly enhance immune responses and provide strong protection against severe diseases caused by these variants. These vaccines hold promise in curbing SARS-CoV-2 transmission and mitigating COVID-19’s impact.
Figure 4.
Schematic summary of the various types of vaccine candidates developed against SARS-CoV-2. The vaccine types are illustrated and categorized into several key technologies. (A) Protein subunit—vaccine candidates that use pieces of the virus protein to elicit an immune response without using live components. (B) Viral vector (non-replicating or replicating)—vaccines. The virus is modified to deliver genetic material of SARS-CoV-2 to induce an immune response. (C) DNA or RNA—vaccines. Genetic material is applied to encode antigenic parts of the virus that are expressed in the host to induce immunity. (D) Inactivated virus. These vaccines contain virus particles that have been killed or inactivated but still elicit an immune response. (E) Virus-like particles—mimic the virus structure but lack viral genetic material, typically focusing on the virus's outer protein coat. (F) Live attenuated virus—vaccines that use a weakened form of the virus that can still replicate without causing illness. (G) Viral vector plus antigen presenting cell (non-replicating or replicating)—combines viral vector technology with cells that effectively present antigens to the immune system. (H) Bacterial antigen-spore expression vector—employs bacterial spores to express the SARS-CoV-2 antigen, presenting a novel vaccination strategy. (Created with BioRender.com).
Monoclonal and polyclonal antibodies against SARS-CoV-2
Monoclonal antibodies (mAbs) are synthesized entities that, with high affinity, bind to viral antigens, replicating host immune defenses [128]. For example, LY-CoV555 (Bamlanivimab) targets multiple epitopes of the SARS-CoV-2 spike protein, correlating with reduced viral loads and hospitalizations [129]. LY-CoV1404 (Bebtelovimab), another mAb, neutralizes all SARS-CoV-2 variants by focusing on the spike protein RBD [130]. Unlike mAbs that recognize a single epitope, polyclonal antibodies (pAbs) originate from diverse B-cell lines, enabling them to detect multiple epitopes of an antigen. This broad recognition presents benefits in addressing therapeutic gaps in COVID-19 [131]. pAbs outstrip other treatments in terms of cost-effectiveness, rapid production, superior therapeutic potential against multiple viral epitopes, and minimized risks of cross-infection or adverse immune reactions. Their capability to target numerous epitopes also deters the rise of viral variants, amplifying neutralization. Several companies are advancing in this arena [132, 133]. Emergent BioSolutions, for instance, introduced the COVID-EIG/HIG hyperimmune antibody derived from human or horse plasma. Similarly, Regen Company's offerings, such as REGN3048-3051, are either patient-sourced or procured from genetically-altered mice, targeting the SARS-CoV-2 spike protein [132, 134, 135].
Using immunological memory to prevent cytokine storms and recurrence of SARS-CoV-2 infection
The escalating global vaccination effort offers a beacon of hope; however, the evolutionary agility of SARS-CoV-2 presents hurdles to achieving lasting epidemic containment. Recent data identified at least five predominant variants of concern: Alpha, Beta, Gamma, Delta, and Omicron [136]. These variants owe their emergence to modifications in the spike protein, a pivotal interface enabling viral entry into host cells. Intriguingly, these alterations may provide the virus with an evolutionary advantage, potentially enabling immune evasion [137]. Despite the challenges posed by these variants, current evidence confirms that existing COVID-19 vaccines offer considerable protection against severe outcomes post-infection [138]. It is worth noting that the humoral response, characterized by the presence of SARS-CoV-2 neutralizing antibodies, exhibits declining potency over time [139]. A marked decline is observed within 6 months post-vaccination, with immunosenescent and immunocompromised individuals being particularly susceptible [140].
Given these dynamics, global health consortia advocate for periodic booster immunizations to strengthen defenses against emerging variants. Recent studies indicate that booster doses significantly enhance the durability and scope of the immune response. For instance, booster shots with updated formulations aimed at specific variants, such as Omicron, have demonstrated improved efficacy in preventing severe disease and hospitalization. Additionally, the role of cellular immunity, particularly that of memory T cells, is increasingly recognized for its contribution to long-term protection. These memory T cells, formed during the initial vaccination or infection, remain alert and can rapidly respond to subsequent exposures to the virus, thereby preventing severe disease and reducing the incidence of cytokine storms. Utilizing immunological memory through booster programs and innovative vaccine designs is critical in the continued effort to manage COVID-19 as it transitions from a pandemic to an endemic phase. Recent studies emphasize the importance of adaptive immunity in long-term protection. Recent studies have highlighted the synergistic effect of combining booster doses with other therapeutic strategies, such as monoclonal antibodies and antiviral drugs, to enhance overall disease management.
Long COVID-19 and precision therapeutic strategies
Long COVID-19, also termed persistent symptomatic COVID-19 or post-acute sequelae of COVID-19, is defined by symptoms that persist for more than 3 months following a COVID-19 diagnosis [141, 142]. These symptoms, which include fatigue, respiratory difficulties, cognitive dysfunction, and cardiovascular issues, can significantly impact quality of life [143].
Managing long COVID requires a comprehensive approach that includes pharmacological treatments, rehabilitation programs, and continuous monitoring to address its complex and varied manifestations. Pharmacological treatments play a pivotal role in managing long COVID, particularly targeting inflammation and immune dysregulation. Common treatments include acetaminophen or nonsteroidal anti-inflammatory drugs (NSAIDs) for pain relief and corticosteroids to reduce inflammation. Additionally, antidepressants or anti-anxiety medications may be used to address emotional issues [144, 145].
The diverse symptoms and underlying mechanisms of long COVID necessitate a precision medicine approach, which customizes treatments based on individual patient characteristics [146]. Precision medicine aims to develop targeted therapies that address the specific pathways and mechanisms implicated in an individual's symptoms, such as therapies targeting persistent inflammation with specialized anti-inflammatory agents or modulating immune responses with immunotherapies.
In the context of long COVID, precision medicine involves using detailed clinical, genetic, and biomarker profiles to guide healthcare decisions. Identifying specific biomarkers that predict susceptibility and the varied manifestations of long COVID, including genetic predispositions, immune signatures, and metabolic profiles, is fundamental. This approach contrasts with traditional one-size-fits-all models and allows for more personalized treatment plans. Patient stratification is another critical component of precision medicine, where patients are classified into subgroups based on their biomarker profiles and clinical presentations. This stratification facilitates the development of tailored treatment plans that are more likely to be effective for each subgroup.
Recent advances in omics technologies—such as genomics, proteomics, and metabolomics—combined with computational biology, have significantly improved the implementation of precision medicine in clinical settings. Advanced diagnostic tools and wearable technologies also play a crucial role by enabling continuous monitoring of a patient's response to treatments, allowing for real-time adjustments and optimization of therapeutic strategies.
By integrating precision medicine into the management of long COVID, healthcare providers can enhance patient outcomes, alleviate the burden of chronic symptoms, and improve the overall quality of life for those affected. This personalized approach not only addresses the current challenges posed by long COVID but also sets a foundation for managing other emerging infectious diseases with similar post-acute conditions. As the virus continues to evolve, maintaining a dynamic and personalized treatment strategy is essential for effective disease management.
Summary and perspective
Since the emergence of SARS-CoV-2, the complex interaction between the virus's virulence factors and the host immune response has become a focal point in virology and immunology research. The viral incursion triggers a coordinated immune defense, encompassing both innate and adaptive mechanisms, which dictate the progression of the disease. A subset of patients demonstrates an exacerbated immune response, leading to a "cytokine storm" characterized by excessive production of pro-inflammatory cytokines. This overreaction can progress to severe pulmonary injury and systemic organ dysfunction. A critical aspect of SARS-CoV-2 pathogenesis is its capacity to modulate the host's immune system. Recent studies spotlight its potential to interfere with the type I interferon (IFN) pathway, a primary antiviral defense. This strategic evasion permits the virus an extended window for replication, potentially enhancing transmission. In the therapeutic arsenal against COVID-19, options range from antivirals to sophisticated modulators of the host immune response. A particularly promising avenue involves intercepting the inflammatory cascade, given its association with severe disease phenotypes. Deploying anti-inflammatory cytokine antibodies and specific molecular inhibitors could be transformative for critically ill patients. As the virus continues to evolve, refining its genetic repertoire, the dynamism of our immune response requires perpetual scrutiny. Adaptation and innovation in therapeutic modalities are imperative. Deepening our understanding of SARS-CoV-2, its emergent variants, and the intricacies of human immunity will not only prepare us against the current pandemic but also strengthen our defenses against future viral threats.
Acknowledgement
This work was supported by The Science and Technology Development Fund, Macau SAR (Grant Nos. 0058/2020/A, 0018/2023/AMJ).
Contributor Information
Zheng-yang Guo, State Key Laboratory of Quality Research in Chinese Medicines, Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau 999078, China.
Yan-qing Tang, State Key Laboratory of Quality Research in Chinese Medicines, Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau 999078, China.
Zi-bo Zhang, State Key Laboratory of Quality Research in Chinese Medicines, Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau 999078, China.
Juan Liu, State Key Laboratory of Quality Research in Chinese Medicines, Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau 999078, China.
Yu-xin Zhuang, State Key Laboratory of Quality Research in Chinese Medicines, Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau 999078, China.
Ting Li, State Key Laboratory of Quality Research in Chinese Medicines, Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau 999078, China.
Author contributions
Conceptualization, Writing-Review & Editing, Funding Acquisition, T.L.; Writing-Original, Draft Preparation, Visualization, Data Curation, Z.-Y.G. , Y.-Q.T. and Z.-B.Z.; Writing-Review & Editing, J.L., Y.-X.Z. All authors gave final approval for the submission and agree to be accountable for all aspects of the work. We have obtained the relevant permissions for BioRender (www.biorender.com) and have used the correct permission text as required by the copyright holders.
Conflict of interest
The authors declare no conflict of interest.
References
- 1. Tan W, Zhao X, Ma X et al. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019-2020. China CDC weekly. 2020;2:61–2. 10.46234/ccdcw2020.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoffmann M, Kleine-Weber H, Schroeder S et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chekol Abebe E, Mengie Ayele T, Tilahun Muche Z et al. Neuropilin 1: a novel entry factor for SARS-CoV-2 infection and a potential therapeutic target. Biologics: Targets and Therapy. 2021;15:143–52. 10.2147/BTT.S307352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cantuti-Castelvetri L, Ojha R, Pedro LD et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–60. 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan R, Zhang Y, Li Y et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–8. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kyrou I, Randeva HS, Spandidos DA et al. Not only ACE2-the quest for additional host cell mediators of SARS-CoV-2 infection: Neuropilin-1 (NRP1) as a novel SARS-CoV-2 host cell entry mediator implicated in COVID-19. Signal Transduction and Targeted Therapy. 2021;6:21. 10.1038/s41392-020-00460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ou X, Liu Y, Lei X et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bayati A, Kumar R, Francis V et al. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J Biol Chem. 2021;296:100306. 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jung HE, Lee HK Current understanding of the innate control of toll-like receptors in response to SARS-CoV-2 infection. Viruses. 2021;13:2132. 10.3390/v13112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martinez FO, Combes TW, Orsenigo F et al. Monocyte activation in systemic Covid-19 infection: assay and rationale. EBioMedicine. 2020;59:102964. 10.1016/j.ebiom.2020.102964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qin S, Jiang Y, Wei X et al. Dynamic changes in monocytes subsets in COVID-19 patients. Hum Immunol. 2021;82:170–6. 10.1016/j.humimm.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choudhury A, Das NC, Patra R et al. In silico analyses on the comparative sensing of SARS-CoV-2 mRNA by the intracellular TLRs of humans. J Med Virol. 2021;93:2476–86. 10.1002/jmv.26776. [DOI] [PubMed] [Google Scholar]
- 13. Jafarzadeh A, Chauhan P, Saha B et al. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257:118102. 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bost P, Giladi A, Liu Y et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–88. 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kulkarni-Munje A, Palkar S, Shrivastava S et al. Disease-duration based comparison of subsets of immune cells in SARS CoV-2 infected patients presenting with mild or severe symptoms identifies prognostic markers for severity. Immunity, Inflammation and Disease. 2021;9:419–34. 10.1002/iid3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Onodi F, Bonnet-Madin L, Meertens L et al. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. J Exp Med. 2021;218:e20201387. 10.1084/jem.20201387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qin C, Zhou L, Hu Z et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–8. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Middleton EA, He XY, Denorme F et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–79. 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laforge M, Elbim C, Frère C et al. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol. 2020;20:515–6. 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang F, Nie J, Wang H et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221:1762–9. 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng M, Gao Y, Wang G et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cellular & Molecular Immunology. 2020;17:533–5. 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mazzoni A, Salvati L, Maggi L et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694–703. 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Market M, Angka L, Martel AB et al. Flattening the COVID-19 curve with natural killer cell based immunotherapies. Front Immunol. 2020;11:1512. 10.3389/fimmu.2020.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tay MZ, Poh CM, Renia L et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–74. 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Wit E, van Doremalen N, Falzarano D et al. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–34. 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chien JY, Hsueh PR, Cheng WC et al. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirol. 2006;11:715–22. 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Le Bert N, Tan AT, Kunasegaran K et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–62. 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 28. Shahridan Faiez T, Singanayagam A Down to a T: the functional importance of lymphopenia in severe COVID-19. Am J Respir Crit Care Med. 2022;205:1370–2. 10.1164/rccm.202203-0526ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang M, Lin C, Wang Y et al. Cytokine storm promoting T cell exhaustion in severe COVID-19 revealed by single cell sequencing data analysis. Precision Clinical Medicine. 2022;5:pbac014. 10.1093/pcmedi/pbac014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tarke A, Potesta M, Varchetta S et al. Early and polyantigenic CD4 T cell responses correlate with mild disease in acute COVID-19 donors. Int J Mol Sci. 2022;23:7155. 10.3390/ijms23137155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grifoni A, Weiskopf D, Ramirez SI et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–501. e15. 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zlei M, Sidorov IA, Joosten SA et al. Immune determinants of viral clearance in hospitalised COVID-19 patients: reduced circulating naive CD4+ T cell counts correspond with delayed viral clearance. Cells. 2022;11:2743. 10.3390/cells11172743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peng H, Yang LT, Li J et al. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 2006;8:2424–31. 10.1016/j.micinf.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng Y, Mentzer AJ, Liu G et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336–45. 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taeschler P, Adamo S, Deng Y et al. T-cell recovery and evidence of persistent immune activation 12 months after severe COVID-19. Allergy. 2022;77:2468–81. 10.1111/all.15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aleebrahim-Dehkordi E, Molavi B, Mokhtari M et al. T helper type (Th1/Th2) responses to SARS-CoV-2 and influenza A (H1N1) virus: from cytokines produced to immune responses. Transpl Immunol. 2022;70:101495. 10.1016/j.trim.2021.101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gil-Etayo FJ, Suàrez-Fernández P, Cabrera-Marante O et al. T-helper cell subset response is a determining factor in COVID-19 progression. Front Cell Infect Microbiol. 2021;11:624483. 10.3389/fcimb.2021.624483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fathi F, Sami R, Mozafarpoor S et al. Immune system changes during COVID-19 recovery play key role in determining disease severity. Int J Immunopathol Pharmacol. 2020;34:2058738420966497. 10.1177/2058738420966497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Biasi S, Meschiari M, Gibellini L et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11:3434. 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mahallawi WH, Khabour OF, Zhang Q et al. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Josset L, Menachery VD, Gralinski LE et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013;4:e00165–13. 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim TO, Park KJ, Cho YN et al. Altered distribution, activation and increased IL-17 production of mucosal-associated invariant T cells in patients with acute respiratory distress syndrome. Thorax. 2022;77:865–72. 10.1136/thoraxjnl-2021-217724. [DOI] [PubMed] [Google Scholar]
- 43. Becerril-Rico J, Alvarado-Ortiz E, Toledo-Guzmán ME et al. The cross talk between gastric cancer stem cells and the immune microenvironment: a tumor-promoting factor. Stem Cell Research & Therapy. 2021;12:1–14. 10.1186/s13287-021-02562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choto TA, Makupe I, Cakana AZ et al. Excessive neutrophil recruitment promotes typical T-helper 17 responses in Coronavirus disease 2019 patients. PLoS One. 2022;17:e0273186. 10.1371/journal.pone.0273186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meckiff BJ, Ramirez-Suastegui C, Fajardo V et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183:1340–53. e16. 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kalfaoglu B, Almeida-Santos J, Tye CA et al. T-cell hyperactivation and paralysis in severe COVID-19 infection revealed by single-cell analysis. Front Immunol. 2020;11:589380. 10.3389/fimmu.2020.589380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taefehshokr N, Taefehshokr S, Heit B Mechanisms of dysregulated humoral and cellular immunity by SARS-CoV-2. Pathogens. 2020:9:1027. 10.3390/pathogens9121027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dhawan M, Rabaan AA, Fawarah MMA et al. Updated insights into the T cell-mediated immune response against SARS-CoV-2: a step towards efficient and reliable vaccines. Vaccines. 2023;11:101. 10.3390/vaccines11010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie MM, Fang S, Chen Q et al. Follicular regulatory T cells inhibit the development of granzyme B–expressing follicular helper T cells. JCI insight. 2019;4:16. 10.1172/jci.insight.128076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gong F, Dai Y, Zheng T et al. Peripheral CD4+ T cell subsets and antibody response in COVID-19 convalescent individuals. J Clin Invest. 2020;130:6588–99. 10.1172/JCI141054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moderbacher CR, Ramirez SI, Dan JM et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang J, Wu Q, Liu Z et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat Microbiol. 2021;6:51–8. 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- 53. Dong Y, Dai T, Wei Y et al. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduction and Targeted Therapy. 2020;5:237. 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schulien I, Kemming J, Oberhardt V et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med. 2021;27:78–85. 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 55. Sekine T, Perez-Potti A, Rivera-Ballesteros O et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–68. 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou R, To KK, Wong YC et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–77. 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Song JW, Zhang C, Fan X et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat Commun. 2020;11:3410. 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Diao B, Wang C, Tan Y et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol. 2020;11:827. 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen G, Wu D, Guo W et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–9. 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dan JM, Mateus J, Kato Y et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McMahan K, Yu J, Mercado NB et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–4. 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reinscheid M, Luxenburger H, Karl V et al. COVID-19 mRNA booster vaccine induces transient CD8+ T effector cell responses while conserving the memory pool for subsequent reactivation. Nat Commun. 2022;13:4631. 10.1038/s41467-022-32324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oberhardt V, Luxenburger H, Kemming J et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–73. 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Khanolkar A Elucidating T cell and B cell responses to SARS-CoV-2 in humans: gaining insights into protective immunity and immunopathology. Cells. 2021;11:67. 10.3390/cells11010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mansourabadi AH, Aghamajidi A, Dorfaki M et al. B lymphocytes in COVID-19: a tale of harmony and discordance. Arch Virol. 2023;168:148. 10.1007/s00705-023-05773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Iyer AS, Jones FK, Nodoushani A et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. 2020;5:eabe0367. 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Havervall S, Ng H, Jernbom Falk A et al. Robust humoral and cellular immune responses and low risk for reinfection at least 8 months following asymptomatic to mild COVID-19. J Intern Med. 2022;291:72–80. 10.1111/joim.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cheng ML, Liu HY, Zhao H et al. Longitudinal dynamics of antibody responses in recovered COVID-19 patients. Signal Transduction and Targeted Therapy. 2021;6:137. 10.1038/s41392-021-00559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Choe PG, Kang CK, Suh HJ et al. Waning antibody responses in asymptomatic and symptomatic SARS-CoV-2 infection. Emerg Infect Dis. 2021;27:327–9. 10.3201/eid2701.203515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mu S, Song S, Hao Y et al. Neutralizing antibodies from the rare convalescent donors elicited antibody-dependent enhancement of SARS-CoV-2 variants infection. Front Med (Lausanne). 2022;9:952697. 10.3389/fmed.2022.952697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dowling JW, Forero A Beyond good and evil: molecular mechanisms of type I and III IFN functions. J Immunol. 2022;208:247–56. 10.4049/jimmunol.2100707. [DOI] [PubMed] [Google Scholar]
- 72. Fitzgerald KA, Kagan JC Toll-like receptors and the control of immunity. Cell. 2020;180:1044–66. 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cervantes-Barragán L, Kalinke U, Zust R et al. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J Immunol. 2009;182:1099–106. 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- 74. Ireland DD, Stohlman SA, Hinton DR et al. Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J Virol. 2008;82:300–10. 10.1128/JVI.01794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pairo-Castineira E, Clohisey S, Klaric L et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–8. 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 76. Bastard P, Rosen LB, Zhang Q et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Loske J, Rohmel J, Lukassen S et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol. 2022;40:319–24. 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 78. Min YQ, Huang M, Sun X et al. Immune evasion of SARS-CoV-2 from interferon antiviral system. Comput Struct Biotechnol J. 2021;19:4217–25. 10.1016/j.csbj.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Minkoff JM, tenOever B Innate immune evasion strategies of SARS-CoV-2. Nat Rev Microbiol. 2023;21:178–94. 10.1038/s41579-022-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu G, Lee J-H, Parker ZM et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat Microbiol. 2021;6:467–78. 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li JY, Liao CH, Wang Q et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Miorin L, Kehrer T, Sanchez-Aparicio MT et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci USA. 2020;117:28344–54. 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Burke JM, St Clair LA, Perera R et al. SARS-CoV-2 infection triggers widespread host mRNA decay leading to an mRNA export block. RNA. 2021;27:1318–29. 10.1261/rna.078923.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kasuga Y, Zhu B, Jang KJ et al. Innate immune sensing of coronavirus and viral evasion strategies. Exp Mol Med. 2021;53:723–36. 10.1038/s12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Han L, Zhuang MW, Deng J et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5–MAVS, TLR3–TRIF, and cGAS–STING signaling pathways. J Med Virol. 2021;93:5376–89. 10.1002/jmv.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Konno Y, Kimura I, Uriu K et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:12. 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hsu JC, Laurent-Rolle M, Pawlak JB et al. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc Natl Acad Sci USA. 2021;118:e2101161118. 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Blanco-Melo D, Nilsson-Payant BE, Liu WC et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–45. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ruan Q, Yang K, Wang W et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–8. 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Del Valle DM, Kim-Schulze S, Huang HH et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–43. 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kapten K, Orczyk K, Smolewska E Immunity in SARS-CoV-2 infection: clarity or mystery? A broader perspective in the third year of a worldwide pandemic. Arch Immunol Ther Exp (Warsz). 2023;71:7. 10.1007/s00005-023-00673-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hasanvand A COVID-19 and the role of cytokines in this disease. Inflammopharmacology. 2022;30:789–98. 10.1007/s10787-022-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Manetti AC, Maiese A, Paolo MD et al. MicroRNAs and sepsis-induced cardiac dysfunction: A systematic review. Int J Mol Sci. 2020;22:321. 10.3390/ijms22010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang EY, Mao T, Klein J et al. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595:283–8. 10.1038/s41586-021-03631-y. [DOI] [PubMed] [Google Scholar]
- 95. Bastard P, Gervais A, Le Voyer T et al. Autoantibodies neutralizing type I IFNs are present in∼ 4% of uninfected individuals over 70 years old and account for∼ 20% of COVID-19 deaths. Sci Immunol. 2021;6:eabl4340. 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lv J, Wang Z, Qu Y et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov. 2021;7:24. 10.1038/s41421-021-00258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li G, Hilgenfeld R, Whitley R et al. Therapeutic strategies for COVID-19: progress and lessons learned. Nat Rev Drug Discov. 2023;22:449–75. 10.1038/s41573-023-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gao Z, Xu Y, Sun C et al. A systematic review of asymptomatic infections with COVID-19. J Microbiol Immunol Infect. 2021;54:12–6. 10.1016/j.jmii.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Luo SH, Liu W, Liu ZJ et al. A confirmed asymptomatic carrier of 2019 novel coronavirus. Chin Med J (Engl). 2020;133:1123–5. 10.1097/CM9.0000000000000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gavriatopoulou M, Ntanasis-Stathopoulos I, Korompoki E et al. Emerging treatment strategies for COVID-19 infection. Clin Exp Med. 2021;21:167–79. 10.1007/s10238-020-00671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cauchois R, Koubi M, Delarbre D et al. Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc Natl Acad Sci USA. 2020;117:18951–3. 10.1073/pnas.2009017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kooistra EJ, Waalders NJB, Grondman I et al. Anakinra treatment in critically ill COVID-19 patients: a prospective cohort study. Crit Care. 2020;24:688. 10.1186/s13054-020-03364-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Caricchio R, Abbate A, Gordeev I et al. Effect of canakinumab vs placebo on survival without invasive mechanical ventilation in patients hospitalized with severe COVID-19: a randomized clinical trial. JAMA. 2021;326:230–9. 10.1001/jama.2021.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tanaka T, Narazaki M, Kishimoto T Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy. 2016;8:959–70. 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 105. Luo P, Liu Y, Qiu L et al. Tocilizumab treatment in COVID-19: a single center experience. J Med Virol. 2020;92:814–8. 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sciascia S, Aprà F, Baffa A et al. Pilot prospective open, single-arm multicentre study on off-label use of tocilizumab in patients with severe COVID-19. Clin Exp Rheumatol. 2020;38:529–32., [PubMed] [Google Scholar]
- 107. Boregowda U, Perisetti A, Nanjappa A et al. Addition of Tocilizumab to the standard of care reduces mortality in severe COVID-19: A systematic review and meta-analysis. Front Med (Lausanne). 2020;7:586221. 10.3389/fmed.2020.586221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Benucci M, Giannasi G, Cecchini P et al. COVID-19 pneumonia treated with sarilumab: a clinical series of eight patients. J Med Virol. 2020;92:2368. 10.1002/jmv.26062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mahmoudjafari Z, Hawks KG, Hsieh AA et al. American Society for Blood and Marrow Transplantation Pharmacy Special Interest Group Survey on chimeric antigen receptor T cell therapy administrative, logistic, and toxicity management practices in the United States. Biol Blood Marrow Transplant. 2019;25:26–33. 10.1016/j.bbmt.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 110. Tao S, Chen Y, Wu J et al. VDDB: A comprehensive resource and machine learning tool for antiviral drug discovery. MedComm–Future Medicine. 2023;2:e32. 10.1002/mef2.32. [DOI] [Google Scholar]
- 111. Group RC Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384:693–704. 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sterne JA, Murthy S, Diaz JV et al. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. JAMA. 2020;324:1330–41. 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ning W, Lei S, Yang J et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat Biomed Eng. 2020;4:1197–207. 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Schwab I, Nimmerjahn F Intravenous immunoglobulin therapy: how does IgG modulate the immune system?. Nat Rev Immunol. 2013;13:176–89. 10.1038/nri3401. [DOI] [PubMed] [Google Scholar]
- 115. Ross C, Svenson M, Hansen MB et al. High avidity IFN-neutralizing antibodies in pharmaceutically prepared human IgG. J Clin Invest. 1995;95:1974–8. 10.1172/JCI117881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hung IFN, To KKW, Lee CK et al. Hyperimmune IV immunoglobulin treatment: a multicenter double-blind randomized controlled trial for patients with severe 2009 influenza A(H1N1) infection. Chest. 2013;144:464–73. 10.1378/chest.12-2907. [DOI] [PubMed] [Google Scholar]
- 117. Tabarsi P, Barati S, Jamaati H et al. Evaluating the effects of intravenous Immunoglobulin (IVIg) on the management of severe COVID-19 cases: A randomized controlled trial. Int Immunopharmacol. 2021;90:107205. 10.1016/j.intimp.2020.107205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Philips RL, Wang Y, Cheon H et al. The JAK-STAT pathway at 30: much learned, much more to do. Cell. 2022;185:3857–76., 10.1016/j.cell.2022.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Stebbing J, Sanchez Nievas G, Falcone M et al. JAK inhibition reduces SARS-CoV-2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci Adv. 2021;7:eabe4724. 10.1126/sciadv.abe4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Richardson P, Griffin I, Tucker C et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet North Am Ed. 2020;395:e30–e1. 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Guimarães PO, Quirk D, Furtado RH et al. Tofacitinib in patients hospitalized with Covid-19 pneumonia. N Engl J Med. 2021;385:406–15. 10.1056/NEJMoa2101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Brunetti L, Diawara O, Tsai A et al. Colchicine to weather the Cytokine storm in hospitalized patients with COVID-19. J Clin Med. 2020;9:2961. 10.3390/jcm9092961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hariyanto TI, Halim DA, Jodhinata C et al. Colchicine treatment can improve outcomes of coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. Clin Exp Pharmacol Physiol. 2021;48:823–30. 10.1111/1440-1681.13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Liu C, Zhou Q, Li Y et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related human Coronavirus Diseases. Washington DC: ACS Publications, 2020. 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Qiu Y, Li Z, Lin F et al. Comparison of the disease severity with infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) delta and Omicron variants: A meta-analysis. MedComm–Future Medicine. 2023;2:e39. 10.1002/mef2.39. [DOI] [Google Scholar]
- 126. Uraki R, Ito M, Kiso M et al. Antiviral and bivalent vaccine efficacy against an omicron XBB. 1.5 isolate. Lancet Infect Dis. 2023;23:402–3. 10.1016/S1473-3099(23)00070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ghasemiyeh P, Mohammadi-Samani S Lessons we learned during the past four challenging years in the COVID-19 era: pharmacotherapy, long COVID complications, and vaccine development. Virol J. 2024;21:98. 10.1186/s12985-024-02370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Xu YG, Weng MZ, Zhang JY et al. Combination of N-(3'4'-dimethoxycinnamoyl) anthranilic acid with cyclosporin A treatment preserves immunosuppressive effect and reduces the side effect of cyclosporin A in rat. Eur J Pharmacol. 2014;728:16–23. 10.1016/j.ejphar.2014.01.055. [DOI] [PubMed] [Google Scholar]
- 129. Chen P, Nirula A, Heller B et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021;384:229–37. 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Westendorf K, Žentelis S, Wang L et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:7. 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ali MG, Zhang Z, Gao Q et al. Recent advances in therapeutic applications of neutralizing antibodies for virus infections: an overview. Immunol Res. 2020;68:325–39. 10.1007/s12026-020-09159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Owji H, Negahdaripour M, Hajighahramani N Immunotherapeutic approaches to curtail COVID-19. Int Immunopharmacol. 2020;88:106924. 10.1016/j.intimp.2020.106924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Pinto D, Park YJ, Beltramello M et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–5. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 134. Liu Y, Wang K, Massoud TF et al. SARS-CoV-2 vaccine development: an overview and perspectives. ACS Pharmacol Transl Sci. 2020;3:844–58. 10.1021/acsptsci.0c00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sheikhshahrokh A, Ranjbar R, Saeidi E et al. Frontier Therapeutics and Vaccine Strategies for SARS-CoV-2 (COVID-19): A review. Iran J Public Health. 2020;49:18–29. 10.18502/ijph.v49iS1.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Haque A, Pant AB Mitigating Covid-19 in the face of emerging virus variants, breakthrough infections and vaccine hesitancy. J Autoimmun. 2022;127:102792. 10.1016/j.jaut.2021.102792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Callaway E The Coronavirus Is Mutating—Does It Matter?. London, UK: Nature Publishing Group, 2020. [DOI] [PubMed] [Google Scholar]
- 138. Shekhar R, Garg I, Pal S et al. COVID-19 vaccine booster: to boost or not to boost. Infect Dis Rep. 2021;13:924–9. 10.3390/idr13040084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Thomas SJ, Moreira ED Jr, Kitchin N et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 months. N Engl J Med. 2021;385((19):):1761–73. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Dolgin E COVID vaccine immunity is waning—How much does that matter?. Nature. 2021;597:606–7. 10.1038/d41586-021-02532-4. [DOI] [PubMed] [Google Scholar]
- 141. Venkatesan P NICE guideline on long COVID. The Lancet Respiratory medicine. 2021;9:129. 10.1016/S2213-2600(21)00031-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Shah W, Hillman T, Playford ED et al. Managing the long term effects of covid-19: summary of NICE, SIGN, and RCGP rapid guideline. BMJ. 2021;372:n136. 10.1136/bmj.n136. [DOI] [PubMed] [Google Scholar]
- 143. Marjenberg Z, Leng S, Tascini C et al. Risk of long COVID main symptoms after SARS-CoV-2 infection: a systematic review and meta-analysis. Sci Rep. 2023;13:15332. 10.1038/s41598-023-42321-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Seo J-W, Kim SE, Kim Y et al. Updated clinical practice guidelines for the diagnosis and management of long COVID. Infection & Chemotherapy. 2024;56:122. 10.3947/ic.2024.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bartone PT, McDonald K, Hansma BJ et al. Hardiness moderates the effects of COVID-19 stress on anxiety and depression. J Affect Disord. 2022;317:236–44. 10.1016/j.jad.2022.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fernández-de-Las-Peñas C, Raveendran AV, Giordano R et al. Long COVID or Post-COVID-19 condition: past, present and future research directions. Microorganisms. 2023;11:2959. 10.3390/microorganisms11122959. [DOI] [PMC free article] [PubMed] [Google Scholar]