Abstract
Mammalian cells exhibit a complex response to DNA damage. The tumor suppressor BRCA1 and associated protein BARD1 are thought to play an important role in this response, and our previous work demonstrated that this includes transient inhibition of the pre-mRNA 3′ processing machinery. Here we provide evidence that this inhibition involves proteasomal degradation of a component necessary for processing, RNA polymerase II (RNAP II). We further show that RNAP IIO, the elongating form of the enzyme, is a specific in vitro target of the BRCA1/BARD1 ubiquitin ligase activity. Significantly, siRNA-mediated knockdown of BRCA1 and BARD1 resulted in stabilization of RNAP II after DNA damage. In addition, inhibition of 3′ cleavage induced by DNA damage was reverted in extracts of BRCA1-, BARD1-, or BRCA1/BARD1-depleted cells. We also describe corresponding changes in the nuclear localization and/or accumulation of these factors following DNA damage. Our results support a model in which a BRCA1/BARD1-containing complex functions to initiate degradation of stalled RNAP IIO, inhibiting the coupled transcription-RNA processing machinery and facilitating repair.
Keywords: BARD1, BRCA1, RNA polymerase II, polyadenylation
The cellular response to DNA damage involves changes in the properties of a number of nuclear proteins. One example is provided by the product of the breast and ovarian cancer susceptibility gene BRCA1 and the associated protein BARD1 (for reviews, see Baer and Ludwig 2002; Jasin 2002; Venkitaraman 2002; Starita and Parvin 2003). The subnuclear location of the BRCA1/BARD1 complex and the phosphorylation state of BRCA1 have been shown to change in response to DNA damage (e.g., Scully et al. 1997b). Although the functional significance of these changes has not been fully elucidated, they likely reflect proposed roles of BRCA1 in DNA repair and/or in transcription and RNA processing (Baer and Ludwig 2002; Jasin 2002; Venkitaraman 2002). Both proteins are large and likely multifunctional, capable of interacting with many other proteins. Additionally, both BARD1 and BRCA1 contain N-terminal RING domains that exhibit significant E3 ubiquitin (Ub) ligase activity in the context of the BRCA1/BARD1 heterodimer (Hashizume et al. 2001; Chen et al. 2002; Xia et al. 2003).
Moreover, this activity is disrupted by several breast cancer-associated mutations in BRCA1, suggesting a relationship between this function and breast cancer development. The Ub ligase activity of BRCA1 has also been shown to enhance cell survival following DNA damage (Dong et al. 2003). BRCA1/BARD1 undergoes autoubiquitination (Chen et al. 2002; Mallery et al. 2002), and is reported to either monoubiquitinate (Chen et al. 2002; Mallery et al. 2002; Xia et al. 2003; Starita et al. 2004) or polyubiquitinate (Sato et al. 2004) potential substrates in vitro.
mRNA transcription is transiently inhibited following DNA damage (e.g., Mayne and Lehmann 1982; Mullenders 1998) as part of the transcription-coupled repair (TCR) response. TCR is a process in which DNA damage is repaired more rapidly in transcriptionally active DNA than in the genome as a whole (for review, see Svejstrup 2002). As part of this response, for example, after UV irradiation, a fraction of the RNAP II largest subunit (RNAP II LS) is phosphorylated, ubiquitinated, and degraded by the proteasome (for reviews, see van den Boom et al. 2002; Muratani and Tansey 2003). The levels of both RNAP IIO, the hyperphosphorylated form of the enzyme that functions in transcription elongation, and RNAP IIA, the hypophosphorylated form that engages promoters, decrease in response to UV light (Ratner et al. 1998; Luo et al. 2001; Lee et al. 2002; Woudstra et al. 2002). RNAP IIO is ubiquitinated and degraded by the proteasome, while RNAP IIA appears to be depleted simply by phosphorylation and conversion to RNAP IIO (McKay et al. 2001). BRCA1 has also been implicated in TCR. In particular, BRCA1-deficient HCC1937 breast carcinoma cells display a block to RNAP II transcription at sites of DNA damage and an impaired ability to repair such damage, and these defects can be ameliorated by expression of exogenous BRCA1 (Abbott et al. 1999; Le Page et al. 2000). Consistent with this, BRCA1 can be found in a complex with proteins involved in the recognition and repair of aberrant DNA structures (Wang et al. 2000). Moreover, both BRCA1 and BARD1 can associate with the RNAP II holoenzyme, and this association seems to change following DNA damage (Scully et al. 1997a; Chiba and Parvin 2001, 2002). Indeed, recent data show that BRCA1 associates preferentially with RNAP IIO, the elongating form of RNAP II that is ubiquitinated and degraded during TCR (Krum et al. 2003).
Processing of mRNA precursors, and specifically 3′ end formation, is also affected by DNA damage. Specifically, pre-mRNA polyadenylation in cell extracts is strongly but transiently inhibited following treatment of cells with DNA damage-inducing agents (Kleiman and Manley 2001). The polyadenylation reaction consists of an endonucleolytic cleavage followed by synthesis of the poly(A) tail, and requires numerous protein factors (for review, see Zhao et al. 1999; Shatkin and Manley 2000). Highlighting the link between transcription and RNA processing, these factors include RNAP II, which plays a strong stimulatory role in vitro (Hirose and Manley 1998). Regulation of 3′ end formation can play significant roles in cell growth control (e.g., Takagaki et al. 1996; Takagaki and Manley 1998; Chuvpilo et al. 1999) and perhaps in disease, especially in tumor cells (for review, see Scorilas 2002). Cleavage stimulation factor (CstF) is one of the essential 3′ processing factors. Genetically modified chicken B cells deficient in CstF-64, one of the CstF subunits, undergo cell cycle arrest and apoptotic death (Takagaki and Manley 1998). Another subunit, CstF-50, has been shown to interact with the C-terminal domain of the RNAP II LS (CTD), likely facilitating the RNAP II-mediated activation of processing (McCracken et al. 1997; Hirose and Manley 1998). Moreover, 3′ processing can be repressed by direct interaction between CstF-50 and the BARD1 subunit of the BRCA1/BARD1 heterodimer (Kleiman and Manley 1999). The DNA damage-induced inhibition of polyadenylation correlates with increasing amounts of a BRCA1/BARD1/CstF-containing complex, supporting the involvement of BRCA1/BARD1 in the inhibition of 3′ RNA processing following DNA damage (Kleiman and Manley 2001).
These findings indicate a functional interplay between BRCA1/BARD1, CstF, and RNAP II following DNA damage. Here we provide unexpected insights into the underlying mechanisms. We show that the UV-induced inhibition of 3′ processing results not only from the BARD1–CstF interaction but also from proteasome-mediated degradation of RNAP II. Extending these results, we show that RNAP IIO, but not RNAP IIA, can be polyubiquitinated in vitro in a manner dependent upon the Ub ligase activity of BRCA1/BARD1. Using siRNA-mediated depletion of BRCA1 and BARD1 in human cells, we find that both proteins are necessary for degradation of RNAP II after DNA damage. We also determined that reduced expression of BARD1 and/or BRCA1 is sufficient to revert DNA damage-induced inhibition of 3′ cleavage. We show that these functional interactions correlate with transient changes in the nuclear localization/accumulation of all these factors following UV irradiation. Our results provide an explanation for the inhibition of transcription and 3′ processing that occurs in response to DNA damage and also identify RNAP IIO as an enzymatic target of the BRCA1/BARD1 heterodimer.
Results
Role of the CTD of RNAP II in UV-induced inhibition of 3′ processing
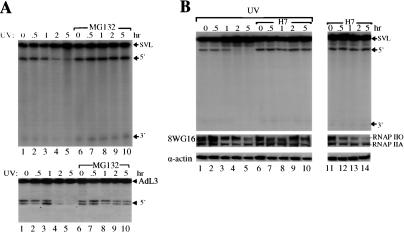
We previously provided evidence that an interaction between the BRCA1/BARD1 complex and CstF regulates UV-induced inhibition of mRNA 3′ end formation (Kleiman and Manley 2001). The simplest view of this repression might be that BARD1 binding to CstF-50 interferes with CstF function in cleavage, and indeed this may be a significant aspect of the mechanism. However, in light of the E3 ligase activity displayed by the BRCA1/BARD1 heterodimer, we decided to test whether proteasomal degradation might also be involved. To examine this, we first added the proteasome inhibitor MG132 to HeLa cells immediately after exposure to UV light, and then prepared nuclear extracts (NEs) at different times and assayed 3′ endonucleolytic cleavage by addition of a 32P-labeled SV40 RNA substrate (see Materials and Methods). Strikingly, we found that MG132 prevented UV-induced inhibition of processing (Fig. 1A, top, cf. lanes 1–5 and 6–10), suggesting that UV light induces proteasomal degradation of a protein(s) required for 3′ end formation. These effects were not specific to the SV40 RNA, as processing of an adenovirus RNA was similarly affected (Fig. 1A, bottom). Although not done exhaustively, Western blots with antibodies against several of the general required factors for 3′ processing (CstF, CPSF, PAP) failed to reveal any change in abundance of these proteins following UV treatment (data not shown).
Figure 1.
Role of protein turnover and RNAP II phosphorylation in UV-induced inhibition of 3′ processing. (A) An inhibitor of the proteasome rescues UV-induced inhibition of 3′ cleavage. HeLa cells were treated with UV irradiation and allowed to recover in the presence (lanes 1–5) or absence (lanes 6–10) of the proteasomal inhibitor MG132 for the times indicated. NEs prepared from these cells were used in cleavage reactions with an SV40 late (top) or adenovirus L3 (bottom) pre-mRNA. Positions of pre-mRNA and the 5′ and 3′ cleavage products are indicated. (B) The CTD-kinase inhibitor H7 blocks UV-induced inhibition of 3′ cleavage. HeLa cells were treated with UV irradiation and allowed to recover in the absence (lanes 1–5) or presence (lanes 6–10) of H7 for the times indicated. Cells were also treated with H7 only (lanes 11–14). NEs prepared from these cells were used in 3′ cleavage reactions as above. Positions of the SV40 pre-mRNA and 5′ and 3′ cleavage products are indicated. RNAP IIO, RNAP IIA, and actin protein levels in NEs from treated cells were monitored by Western blotting. Proteins were detected by immunoblotting with antibodies against a nonphosphorylated CTD epitope of RNAP II LS (8WG16) and actin.
An intriguing candidate for the proteasome-targeted protein is RNAP II. As described above, a fraction of the cell's RNAP IIO is ubiquitinated and degraded by the proteasome in response to UV treatment, and RNAP II, specifically the CTD, is necessary for efficient 3′ cleavage in vitro (Hirose and Manley 1998). Thus it is possible that degradation of RNAP II contributes to the inhibition of processing. To examine the possible role of RNAP II in UV-induced 3′ processing inhibition, we first analyzed the effects of CTD kinases inhibitors (H7 and DRB) on 3′ cleavage in vitro when added to cells immediately after UV treatment (Fig. 1B). The rationale for this was that reducing the levels of RNAP IIO would in turn limit UV-induced RNAP II destruction, and thus rescue processing. Strikingly, H7 prevented UV-induced inhibition of 3′ cleavage (Fig. 1B, cf. lanes 4–5 and 9–10). Similar results were obtained after treatment with DRB (data not shown). Although, as expected, treatment of cells with H7 alone decreased the amount of RNAP IIO compared with nontreated samples (Fig. 1B, cf. lanes 11–14 and 1), this did not have an effect on 3′ processing, likely reflecting the fact that either RNAP IIA or RNAP IIO can function in 3′ processing (Hirose and Manley 1998). Consistent with previous results (Bregman et al. 1995; Luo et al. 2001; McKay et al. 2001), Western blot analysis revealed that UV treatment decreased accumulation of both RNAP IIO and RNAP IIA forms (Fig. 1B, cf. lanes 3–5 and 1). Significantly, treatment with both UV and H7 reversed the decrease in the two RNAP II forms that resulted from UV treatment alone (Fig. 1B, cf. lanes 8–10 and 3–5). Stabilization of RNAP IIO by H7 suggests that ongoing transcription and/or CTD phosphorylation is necessary for UV-induced degradation, consistent with previous results (Luo et al. 2001).
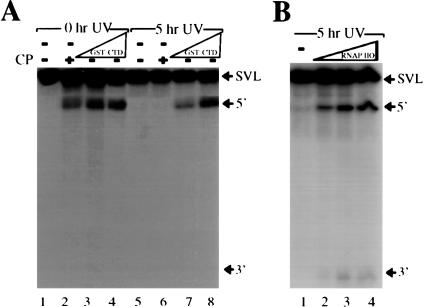
The above results are consistent with the idea that RNAP II plays a role in UV-induced inhibition of 3′ processing. To test directly the idea that RNAP II might become limiting for 3′ processing following DNA damage, we next analyzed the effect of adding increasing amounts of a recombinant GST-CTD fusion protein to NEs prepared from cells exposed to UV light (Fig. 2A). The cofactor creatine phosphate, which appears to mimic the CTD in 3′ processing (Hirose and Manley 1997, 1998), was omitted from reaction mixtures. Strikingly, addition of increasing amounts of GST-CTD to inhibited NEs (5 h post-UV treatment) effectively restored 3′ processing (Fig. 2A, lanes 5–8), while having no effect on untreated extracts (Fig. 2A, lanes 1–4). Although we showed previously that GST-CTD and RNAP II function similarly in reconstituting 3′ cleavage (Hirose and Manley 1998), we also analyzed the effect of adding increasing amounts of purified RNAP IIO to UV-inhibited NEs. The results (Fig. 2B) show that RNAP IIO also effectively restored 3′ cleavage. Together, the results support the idea that RNAP II becomes limiting in UV-treated extract and that this contributes to the inhibition of 3′ processing. Consistent with this, the time course over which RNAP II turnover is observed after DNA damage (e.g., Ratner et al. 1998) is similar to the time course of 3′ cleavage inhibition (Kleiman and Manley 2001).
Figure 2.
RNAP II reverse the inhibitory effect of DNA damage on 3′ processing. (A) Activation of cleavage by the addition of GST-CTD. NEs active (0-h UV treatment) and inactive (5-h UV treatment) for 3′ cleavage were preincubated with no addition (lanes 1,2,5,6) or with increasing amounts (100 and 200 ng) of recombinant GST-CTD (lanes 3,4,7,8). The cleavage reactions were performed in the absence (lanes 1,3–5,7,8) or presence (lanes 2,6) of creatine phosphate. Positions of pre-mRNA and the 5′ and 3′ cleavage products are indicated. (B) Activation of cleavage by the addition of RNAP IIO. NEs inactive (5-h UV treatment) for 3′ cleavage were preincubated with no addition (lane 1) or with increasing amounts (25, 50, and 100 ng) of purified RNAP IIO (lanes 2–4). Positions of pre-mRNA and the 5′ and 3′ cleavage products are indicated.
The BRCA1/BARD1 heterodimer ubiquitinates RNAP IIO but not RNAP IIA in vitro
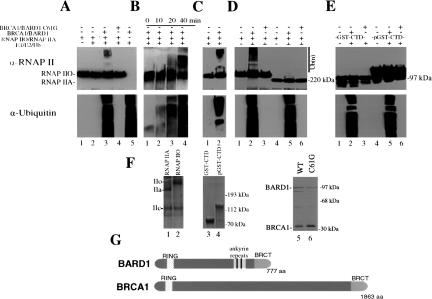
The above results implicate proteasome-mediated degradation of RNAP IIO in UV-induced inhibition of 3′ processing. Given the involvement of BRCA1/BARD1 in this process, an intriguing possibility is that BRCA1/BARD1 targets RNAP IIO for destruction by providing an E3 ligase activity necessary for polyubiquitination of RNAP IIO. To examine this hypothesis, we first purified a heterodimeric complex (ΔBRCA1/BARD1-wt) comprised of truncated BRCA1 (residues 1–304) and full-length BARD1 from bacterial cells and incubated it with purified RNAP IIO (SDS-PAGE of RNAP II proteins and the truncated BRCA1/BARD1 complex used in these experiments is shown in Fig. 3F and the proteins are depicted schematically in Fig. 3G), along with E1, its cognate E2 (UbcH5c), and His-HA-tagged Ub (see Materials and Methods). The reaction products were then analyzed by Western blotting with an anti-RNAP II LS antibody (Fig. 3A, top panel), as well as an anti-Ub antibody (Fig. 3A, lower panel) to measure total, nonspecific ubiquitination. As indicated by the low mobility species in Figure 3A, lane 3, RNAP IIO LS was ubiquitinated in the complete reaction. The very high molecular weight of these species indicates the addition of multiple Ub monomers, presumably in the form of polyubiquitin. The ubiquitination observed (both specific and nonspecific) was completely dependent on the presence of recombinant ΔBRCA1/BARD1 (Fig. 3A, lane 2), and the tumor-associated BRCA1 mutation C61G, which disrupts the RING domain, abolished both total ubiquitination and the RNAP IIO modification (Fig. 3A, lane 4). Kinetic analysis showed a slight reduction in unmodified RNAP IIO by 40 min, concomitant with an accumulation of the high-molecular-weight species (Fig. 3B, upper panel). The nonspecific ubiquitination activity displayed similar kinetics (Fig. 3B, lower panel). Significantly, a complex consisting of full-length BRCA1 and BARD1, purified from insect cells coinfected with recombinant baculoviruses (Wu-Baer et al. 2003), also polyubiquitinated RNAP IIO LS (Fig. 3C).
Figure 3.
RNAP IIO but not RNAP IIA is ubiquitinated by BRCA1/BARD1. (A) RNAP IIO was incubated with E1, E2, His-HA-Ub, and a purified heterodimer comprised of truncated BRCA1 and full-length BARD1 (ΔBRCA1/BARD1-wt) as indicated (lanes 1–3,5). Lane 4 shows a reaction containing the mutant heterodimer ΔBRCA1/BARD1-C61G. Reactions were terminated and proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-RNAP II LS and anti-Ub antibodies in the top and lower panels, respectively. Positions of the RNAP IIO and RNAP IIA forms are indicated on the left, and the polyubiquitinated forms of RNAP IIO and molecular-weight markers are indicated on the right. (B) Kinetics of RNAP IIO ubiquitination by ΔBRCA1/BARD1-wt. Ubiquitination reactions were performed and analyzed as in A, except that the incubation times were as indicated. (C) Ubiquitination of RNAP IIO with a heterodimeric complex comprised of full-length BRCA1 and BARD1. Reactions were performed and analyzed as in A. (D) ΔBRCA1/BARD1-wt stimulates polyubiquitination of RNAP IIO but not RNAP IIA. Ubiquitination reactions were performed as above except in presence of either RNAP IIO or RNAP IIA. (E) ΔBRCA1/BARD1 does not stimulate ubiquitination of either GST-CTD or phosphorylated GST-CTD. Ubiquitination reactions were done with nonphosphorylated (lanes 1–3) or in vitro phosphorylated (lanes 4–6) GST-CTD as in panel A. Positions of the nonphosphorylated and phosphorylated GST-CTD proteins are indicated on the left. (F) Concentration and purity of RNAP IIO and RNAP IIA (lanes1,2; only the largest subunits are shown), GST-CTD and pGST-CTD (lanes 3,4), and the heterodimeric complexes comprised of full-length BARD1 and truncated wild-type and C61G BRCA1 (lanes 5,6) were monitored by silver or Coomassie blue staining following SDS-PAGE. Positions of the three largest RNAP II subunits (IIa, IIo, and IIc) are indicated on the left, and positions of molecular-weight markers are indicated on the right.(G) Schematic diagrams of BRCA1 (1863 amino acids) and BARD1 (777 amino acids) showing the N-terminal RING domains (BRCA1 23–76, BARD1 49–100), the ankyrin repeats (BARD1 427–525), and C-terminal BRCT domains.
RNAP IIO, but not RNAP IIA, is specifically ubiquitinated and degraded following DNA damage (see Introduction). To determine whether the enzymatic activity of BRCA1/BARD1 is specific for RNAP IIO, we compared the ability of RNAP IIO and RNAP IIA to serve as substrates in the in vitro ubiquitination assay. Strikingly, while ΔBRCA1/BARD1 again catalyzed the ubiquitination of RNAP IIO LS, a corresponding modification of RNAP IIA LS was not detected (Fig. 3D, upper panel, cf. lanes 2 and 5). Nonspecific ubiquitination was not reduced in these reactions by RNAP IIA, indicating that the RNAP IIA used was not inhibitory (Fig. 3D, lower panel). Thus, the BRCA1/BARD1 heterodimer specifically catalyzes polyubiquitination of RNAP IIO, the form of RNAP II that is preferentially targeted for Ubmediated proteolysis during the response to UV damage.
To address further the specificity of the BRCA1/BARD1-mediated ubiquitination of RNAP IIO, we investigated whether phosphorylated and unphosphorylated GST-CTD fusion proteins were targets for in vitro ubiquitination. We found that neither protein was detectably modified, although nonspecific ubiquitination activity was again not affected (Fig. 3E). These results are consistent with previous studies that identified non-CTD lysines in RNAP IIO LS as targets for ubiquitination after UV irradiation in vivo (Ratner et el. 1998). The inactivity of the two forms of GST-CTD, as well as of RNAP IIA, is significant because it both highlights the specificity with which BRCA1/BARD1 targets RNAP IIO in vitro, and also reinforces the possible in vivo significance of our results.
RNAi-mediated knockdown of BRCA1 and BARD1 stabilizes RNAP II following DNA damage
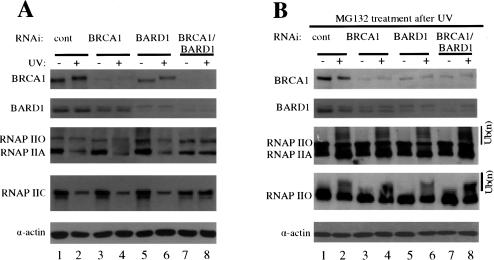
We next sought to determine whether the ability of the BRCA1/BARD1 heterodimer to specifically polyubiquitinate RNAP IIO in vitro reflects its activity in vivo. Specifically, are BRCA1 and/or BARD1 necessary for UV-induced degradation of RNAP II? To investigate this, we used siRNAs to knockdown expression of BRCA1, BARD1, or both in HeLa cells, and then determined the effect of these treatments on RNAP II stability following exposure to UV light. Cells were treated with siRNAs for 48 h, exposed to UV light, and allowed to recover for 2 h. NEs were then prepared and analyzed by Western blotting (Fig. 4A; see Materials and Methods). In the presence of a control siRNA (Fig. 4A, lanes 1,2), accumulation of neither BRCA1 nor BARD1 was affected by UV, although the electrophoretic mobility of BRCA1 was reduced, indicative of damage-induced phosphorylation (Scully et al. 1997b). RNAP II accumulation was measured by two anti-CTD antibodies, N-20, which detects both RNAP IIO and RNAP IIA LS, and H5, which detects primarily the Ser 2-phosphorylated, elongating form of the enzyme. Consistent with previous results and with our data shown above, UV treatment significantly reduced accumulation of both RNAP II isoforms, and this was unaffected by control siRNAs (Fig. 4A; data not shown). (Note that UV-induced degradation of RNAP IIO appeared somewhat greater when measured with H5, likely because H5 specifically recognizes the elongating form of RNAP II that would be targeted following UV.) Treatment of cells with BRCA1 siRNA resulted in substantial depletion of BRCA1 (50%–60%), both before and after UV treatment (Fig. 4A, lanes 3,4), and the BARD1 siRNA had similar effects on BARD1 expression (Fig. 4A, lanes 5,6). BRCA1 siRNA also reduced BARD1 accumulation, although to a lesser extent than BRCA1, and BARD1 siRNA had a similar effect on BRCA1, consistent with the idea that the two polypeptides stabilize each other (Hashizume et al. 2001). While siRNA-mediated depletion of either BRCA1 alone (Fig. 4A, lane 4) or BARD1 alone (Fig. 4A, lane 6) did not have a significant effect on RNAP II degradation following UV, a striking response was elicited by treating cells with both siRNAs simultaneously (Fig. 4A, lanes 7,8). Both BRCA1 and BARD1 accumulation was greatly reduced (by ∼90%) and more significantly, both isoforms of RNAP II were fully stabilized following UV, as observed with both N-20 and H5 antibodies. These results indicate that the BRCA1/BARD1 heterodimer is necessary for UV-induced destruction of RNAP II.
Figure 4.
siRNA knockdown of both BRCA1/BARD1 expression abolishes UV-induced degradation of RNAP II. (A) Protein levels of BRCA1, BARD1, actin, and RNAP II in NEs prepared from cells subjected to control (cont), BRCA1, BARD1, and BRCA1/BARD1 siRNA and UV irradiation. Nonirradiated and UV-irradiated samples were prepared 48 h after the addition of the siRNAs. Irradiated cells were allowed to recover 2 h after exposure to UV doses of 10 Jm–2. Panels depict blots using antibodies against BRCA1, BARD1, RNAP IIO (H5), RNAP II (N-20), and actin. Protein concentrations were equalized by immunostaining with antibodies against actin. The actin and the RNAP II blots correspond to the same gel. The positions of each protein are indicated in the corresponding panel. (B) An inhibitor of the proteasome prevents UV-induced degradation but not ubiquitination of RNAP II in siRNA-treated cells. siRNA-treated cells were irradiated with UV and allowed to recover in the presence of the proteasomal inhibitor MG132 for 2 h. BRCA1, BARD1, RNAP II, RNAP IIO, and actin protein levels in NEs from these cells were analyzed by Western blot as above.
To characterize further the BRCA1/BARD1-dependent degradation of RNAP II, we repeated the above experiments but added MG132 to the cells immediately after UV exposure (Fig. 4B). This prevented degradation of RNAP II not only in the control siRNA-treated cells but also in all the BRCA1 and/or BARD1 siRNA-treated cells, confirming that the observed degradation was mediated by the proteasome. With degradation blocked, we were also able to observe apparent ubiquitinated forms of RNAP IIO, which were detected as a high-molecular-weight smear that migrates above the RNAP IIO LS. Intriguingly, the BRCA1/BARD1 double knockdown that fully stabilized RNAP II did not detectably affect its ubiquitination. These results are consistent with the possibility that RNAP IIO is targeted for ubiquitination in vivo by more than one E3 ligase, as previously suggested (Mitsui and Sharp 1999; see also Discussion). Although our data indicate that only RNAP IIO is ubiquitinated by BRCA1/BARD1 in vitro (Fig. 3), these experiments do not rule out the possibility that RNAP IIA could also be a target in vivo. Nevertheless, our results establish that the BRCA1/BARD1 heterodimer is necessary for UV-induced proteasomal degradation of RNAP II.
RNAi-mediated knockdown of BRCA1 and BARD1 rescues UV-induced inhibition of polyadenylation
We next wished to determine whether depletion of BRCA1 and/or BARD1 was sufficient to rescue UV-induced inhibition of 3′ processing. To this end, HeLa cells were treated with control, BRCA1, and/or BARD1 siRNAs as described above, and NEs were prepared before or after 2-h UV treatment. Western blot analysis indicated effects on BARD1, BRCA1, and RNAP II essentially identical to those shown in Figure 4A. The NEs were then used to monitor 3′ cleavage of the SV40 pre-mRNA substrate as described above (Fig. 5A). Strikingly, 3′ processing was rescued not only in NEs from the double knockdown cells (Fig. 5A, lanes 7,8), which stabilized RNAP II, but also in both of the single knockdowns (Fig. 5A, lanes 3–6), which did not prevent RNAP II degradation.
Figure 5.
siRNA knockdown of both BRCA1/BARD1 abolishes the UV-induced inhibition of polyadenylation. (A) NEs were prepared from cells subjected to control, BRCA1, BARD1, and BRCA1/BARD1 siRNA treatment, and then used in cleavage reactions with the SV40 late substrate RNA. Positions of pre-mRNA and the 5′ and 3′ cleavage products are indicated. (B) NEs were prepared and used in processing reactions as in A. Reactions contained no addition (lanes 1–8), 10 ng full-length BRCA1/BARD1 (lanes 9–16), or 7 ng truncated BRCA1 (1–304)/BARD1 (1–202) complex (lanes 17–24).
The above results strongly suggest that depletion of BRCA1/BARD1 was by itself sufficient to prevent UV-induced inhibition of 3′ processing. To confirm this, we repeated the experiment in Figure 5A, except that we determined the effect of adding purified BRCA1/BARD1 heterodimers to the NEs (Fig. 5B). In the absence of added protein, results essentially identical to those shown in Figure 5A were obtained (lanes 1–8). Addition of full-length BRCA1/BARD1 to the NEs, however, had significant but extract-dependent effects (Fig. 5B, lanes 9–16). When added to the NEs from control siRNA-treated cells, the BRCA1/BARD1 complex inhibited 3′ cleavage even in the absence of UV treatment (Fig. 5B, cf. lanes 1 and 9). This is consistent with our initial finding that addition of BARD1 to HeLa NE inhibited 3′ processing by its interaction with CstF-50 (Kleiman and Manley 1999). A different pattern was seen when the heterodimer was added to the NEs from either the BRCA1 or BARD1 siRNA-treated cells (Fig. 5B, lanes 11–14). In both cases, added BRCA1/BARD1 did not significantly decrease cleavage in the NEs from non-UV-treated cells (Fig. 5B, cf. lanes 3 and 11 and lanes 5 and 13). Significantly however it restored cleavage inhibition to the extracts from the UV-treated cells (Fig. 5B, cf. lanes 4 and 12 and lanes 6 and 14), providing evidence that these extracts were resistant to UV-induced inhibition of processing due to reduced levels of BRCA1/BARD1. The inability of the exogenously added complex to inhibit processing in the NEs from non-UV-treated cells likely reflects the reduced levels of endogenous BRCA1/BARD1, such that the total concentration was insufficient to block cleavage. Consistent with this, addition of BRCA1/BARD1 to the NEs from cells treated with both BRCA1 and BARD1 siRNAs, which resulted in more complete depletion of the two proteins (see above), was without significant effect (Fig. 5B, lanes 15,16). Addition of a heterodimer consisting of truncated forms of BRCA1 and BARD1, which lacked the CstF-50 interaction domain in BARD1 (Kleiman and Manley 1999), did not significantly affect processing in any of the NEs (Fig. 5B, lanes 17–24).
Taken together, the above results indicate that UV-induced inhibition of 3′ processing can be rescued by stabilizing RNAP II and/or by reducing the concentration of the BRCA1/BARD1 heterodimer. BRCA1/BARD1 thus can block polyadenylation both by direct inhibitory interaction with the 3′ processing machinery, as shown previously (Kleiman and Manley 1999; 2001), and/or by causing proteasomal degradation of RNAP II. These may reflect redundant mechanisms to achieve the same result.
The nuclear distribution of BRCA1/BARD1, RNAP II, and polyadenylation factors changes after DNA damage
The cell nucleus contains distinct structural and functional domains, and their internal organization can vary in response to a variety of stimuli (for review, see Lamond and Spector 2003). For example, BRCA1/BARD1 can be found within discrete nuclear foci in S and G2 phase cells (Jin et al. 1997; Scully et al. 1997b), while RNAP II localizes to 20–50 discrete subnuclear domains as well as in a diffuse nucleoplasmic pattern (Bregman et al. 1996). CstF distributes diffusely throughout the nucleus as well, but also concentrates in a few small foci (cleavage bodies) (Schul et al. 1996). Upon DNA damage of S-phase cells, BRCA1/BARD1 disperses from its nuclear foci and associates with distinct nuclear structures that contain replicating DNA as well as the PCNA replication protein (Scully et al. 1997b).
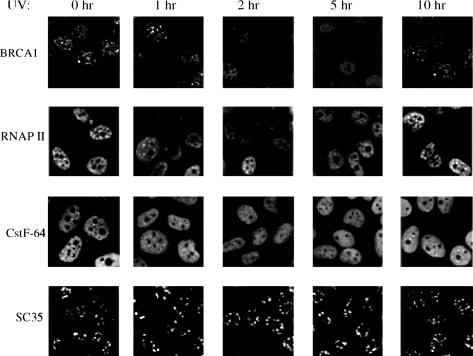
To examine the possible relationship between UV-induced inhibition of 3′ processing and the subcellular localization of the factors involved, we examined the nuclear distribution of these factors following UV irradiation by indirect immunofluorescence. Specifically, we analyzed both HeLa (data not shown) and MCF7 (Fig. 6) cells using antibodies that recognize BARD1, BRCA1, RNAP IIO, and subunits of the CPSF and CstF complexes (results with CPSF not shown). The results indicate that between 2 and 5 h after UV treatment not only was BRCA1/BARD1 dispersed from nuclear foci, but RNAP IIO and CstF also appeared to change their distribution. This was apparent more as a decrease in overall staining than as a relocalization to other discrete sites, which could reflect a more diffuse localization and/or turnover (i.e., of RNAP IIO). Significantly, these changes were reversible, because by 10 h after UV treatment the proteins displayed the patterns observed in untreated cells. These dynamic changes were not typical of all nuclear proteins, since the localization/abundance of two other RNA processing factors, SC35 and SRm300, was not affected by UV treatment (Fig. 6; results with SRm300 not shown). The timing of these events closely correlated with the observed inhibition of 3′ cleavage (Kleiman and Manley 2001), suggesting that the changes in nuclear distribution/accumulation and the inhibition of processing are different manifestations of the same cellular response.
Figure 6.
UV treatment transiently disperses BRCA1/BARD1 nuclear foci, RNAP IIO dots, and cleavage bodies, but not SC35 speckles. Asynchronous MCF7 cells were treated with UV and were fixed in 2% formaldehyde after the UV treatment at the times indicated. Fixed cells were immunostained with a BRCA1 mAb (MS110), BARD1 polyclonal antibody (669D), CstF-64 mAb, RNAP IIO LS mAb (H5), and an SC35 mAb. Immunostaining with monoclonal or polyclonal antibodies was visualized by using a goat anti-mouse or anti-rabbit IgG-fluorescein, respectively.
Discussion
We showed previously that the polyadenylation machinery is strongly but transiently inhibited following DNA damage, reflecting formation of a complex containing both BRCA1/BARD1 and CstF (Kleiman and Manley 1999, 2001). In this study, we analyzed the mechanism(s) of this inhibition, and provided evidence that degradation of RNAP IIO is a critical factor. Extending these results, we found that the large subunit of RNAP IIO, but not of RNAP IIA, is a specific in vitro target of the BRCA1/BARD1 E3 Ub ligase activity. Using siRNA-mediated depletion of BRCA1 and BARD1, we showed that efficient depletion of both proteins prevented degradation of RNAP II after DNA damage, while inhibition of 3′ cleavage was even more sensitive to levels of BRCA1/BARD1. These functional changes were found to correlate with alterations in nuclear organization and localization of the relevant factors. Our results thus indicate that BRCA1/BARD1 plays a significant role in targeting RNAP IIO for degradation following DNA damage, and that this could be sufficient to explain the UV-induced inhibition of 3′ processing. However, BRCA1/BARD1 also represses the 3′ processing machinery by its interaction with CstF-50 (Kleiman and Manley 1999), suggesting the existence of another, possibly redundant, mechanism to explain the inhibitory effect of UV irradiation.
Based on our results, we propose a model whereby BRCA1/BARD1 serves to help coordinate a ubiquitous response to DNA damage by a mechanism that includes interactions with RNAP II and components of the polyadenylation machinery. Following DNA damage, elongating RNAP IIO is stalled, causing premature termination or arrest of transcription. DNA lesions themselves could stall RNAP II, or arrest could occur via an interaction between components of the RNAP II holoenzyme and elements of the replication/repair machinery (for reviews, see van den Boom et al. 2002; Muratani and Tansey 2003). In any event, we suggest that BRCA1/BARD1-containing complexes are recruited to sites of DNA damage on active genes by interaction with RNAP IIO (see Chiba and Parvin 2001, 2002; Krum et al. 2003), resulting in BRCA/BARD1-mediated ubiquitination of RNAP IIO LS and inhibition of transcription by degradation/inactivation of RNAP IIO. These events would also inhibit the 3′ mRNA processing machinery, both by the BARD1/CstF-50 interaction and/or by RNAP IIO degradation. This mechanism allows for both the clearance of the damaged region for DNA repair and the elimination of prematurely terminated transcripts that could produce truncated, potentially deleterious, proteins.
Although the identity of the repair protein(s) that mediate(s) the damage-induced association between stalled RNAP IIO and the DNA repair machinery during TCR is not known, one candidate is PCNA. The association of PCNA with certain DNA repair proteins after UV treatment is dependent upon TCR (Balajee et al. 1998). PCNA also colocalizes with BRCA1/BARD1 at possible sites of DNA damage (Scully et al. 1997b; Wang et al. 2000) and, like BARD1, interacts with CstF-50 (Kleiman and Manley 1999). Furthermore, both CstF-50 and BRCA1/BARD1 can associate with the RNAP II holoenzyme (Scully et al. 1997a; McCracken et al. 1997; Chiba and Parvin 2002). Alternatively, BRCA1 by itself might signal sites of DNA damage, since BRCA1 can interact directly with DNA (Paull et al. 2001).
UV-induced ubiquitination and turnover of RNAP IIO is now well documented in both yeast (Beaudenon et al. 1999; Woudstra et al. 2002) and mammals (Bregman et al. 1996; Ratner et al. 1998; McKay et al. 2001; Luo et al. 2001). However, a number of key questions remain regarding, for example, its relationship to TCR (e.g., Lommel et al. 2000), how ubiquitous the process is in the DNA damage response, and, especially in mammals, the identities of the signaling factors. Woudstra et al. (2002) suggested that yeast cells may respond to RNAP II stalled following DNA damage in one of two ways. If the lesion is repaired rapidly, then RNAP II reengages and continues transcription, but if the damage persists, RNAP IIO is ubiquitinated and degraded. This is an attractive model because it offers an explanation for certain results suggesting that elongating RNAP II might not always be degraded at sites of DNA damage (Tornaletti et al. 1999).
Whether a similar situation occurs in mammalian cells is unclear, and there are significant differences in the response to DNA damage in the two systems. Most importantly in the current context, true orthologs of BRCA1 and BARD1 have not been identified in yeast (Baer and Ludwig 2002). Instead, UV-induced ubiquitination of RNAP II is brought about in yeast by Rsp5, a HECT domain E3 ligase (Huibregtse et al. 1997; Beaudenon et al. 1999). Interestingly, Rsp5 has also been implicated in the regulation of RNA processing in yeast (Neumann at al. 2003). It may be that Rpf1/Nedd4, a possible human homolog of Rsp5, participates in RNAP II ubiquitination, as it was shown to bind RNAP II in cell extracts and to ubiquitinate yeast RNAP II LS (Beaudenon et al. 1999). However, unlike our studies with BRCA1/BARD1, these experiments revealed no preference for ubiquitination of the RNAP IIO relative to the RNAP IIA isoform of RNAP II, which would appear inconsistent with the situation in vivo. It is possible that Rpf1/Nedd4 is responsible for the UV-induced ubiquitination of RNAP II we observed in the BRCA1/BARD1 double knockdown cells. Consistent with this, Mitsui and Sharp (1999) suggested that RNAP IIO is targeted for ubiquitination in vivo by more than one E3 ligase. It is not unprecedented that a single protein can be targeted by multiple E3 ligases. For example, p53 can be targeted in vitro and destabilized in vivo by several E3s (Dornan et al. 2004 and references therein). Also, BRCA1 itself can be polyubiquitinated in a manner independent of its own enzymatic activity, presumably by the action of other cellular E3 ligases (Choudhury et al. 2004).
In addition to providing insights into the BARD1-mediated inhibition of 3′ processing induced by DNA damage, our results have identified RNAP IIO as a potentially important target of the BRCA1/BARD1 E3 ligase activity. Although multiple studies have now shown that the BRCA1/BARD1 heterodimer, and specifically its N-terminal RING domains, possesses E3 ligase activity, only the autoubiquitination reaction has been well characterized (Chen et al. 2002). The nature of BRCA1/BARD1 autoubiquitination is unusual. It was initially suggested to involve the formation of polyubiquitin chains linked through Ub residues other than the canonical K48 (Chen et al. 2002; Xia et al. 2003) and more recently to occur via an unusual linkage involving K6 (Wu-Baer et al. 2003; Nishikawa et al. 2004). Significantly, this modification does not appear to target BRCA1/BARD1 for degradation and instead may enhance its stability and/or activity (Nishikawa et al. 2004). It will be important in the future to determine whether BRCA1/BARD1 forms K6-linked polyubiquitin chains on heterologous substrates (e.g., RNAP IIO), and whether K6-linked chains can target certain substrates for degradation. Given the uncertainties surrounding the nature and function of BRCA1/BARD1-catalyzed polyubiquitination, we must also entertain the possibility that the specific modification of RNAP IIO LS we have described here might have a function other than or in addition to signaling turnover of RNAP IIO, or might bring about RNAP II degradation indirectly, for example, by recruiting another E3 ligase. Nevertheless, by demonstrating that the BRCA1/BARD1 heterodimer is essential for UV-induced degradation of RNAP II and for inhibition of 3′ mRNA processing, our data establish a significant functional interaction between components of the transcription, 3′ processing and DNA repair machineries.
Materials and methods
Tissue culture methods
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-10% fetal bovine serum (FBS). 80% confluent cultures were exposed to UV, incubated with the indicated drugs, and then harvested at the stated times. Cells were treated as indicated with 2 μM MG132 (Sigma), 50 μM H7 (1-[5-isoquinolinesulfonyl]-2-methylpiperazine; Sigma), and 50 μM DRB (5,6-Dichloro-1-β-D-ribofuranosylbenzimi dazole; Calbiochem). UV doses of 10 Jm–2 were delivered in a single pulse using a Stratalinker (Stratagene). Prior to pulsing, medium was removed and replaced with medium containing the indicated drugs immediately after treatment.
Immunostaining and confocal microscopy
Coverslips with attached HeLa or MCF7 cells were rinsed once in phosphate-buffered saline (PBS) and incubated with 4% paraformaldehyde in PBS (pH 7.4) for 15 min at room temperature. After fixation, cells were rinsed three times for 10 min each time with PBS and permeabilized with 0.2% Triton X-100 plus 1% FBS in PBS for 5 min on ice. Cells subsequently were rinsed three times for 10 min each time in PBS/1% FBS. Fixed cells were incubated in the appropriate concentration of the appropriate primary antibody diluted in PBS/1% FBS for 1 h at room temperature. The following antibodies were used: BRCA1 mAb (MS110), BARD1 polyclonal antibody (669D), CstF-64 mAb (Kleiman and Manley 2001), RNAP IIO mAb (H5, Covance), SC35 mAb, and SRm 300 polyclonal antibody (kindly provided by Dr. Susan McCracken, University of Toronto, Toronto, Canada). Subsequently, cells were washed three times for 10 min each time in PBS/1% FBS and incubated with secondary antibodies diluted in PBS/1% FBS for 1 h. Labeled goat anti-mouse IgG and goat anti-rabbit IgG antibodies (Molecular Probes) were used. Finally cells were washed four times for 10 min each time in PBS and rinsed in water. Coverslips were mounted in one drop of mounting media (Polysciences Inc.).
NE preparation and immunoblotting analysis
After drug treatment, NEs were prepared from HeLa cells as described (Kleiman and Manley 2001). Sixty micrograms of the NEs were analyzed by immunoblotting with mAbs targeted against RNAP II (8WG16, Covance, and N-20, Santa Cruz), RNAP IIO (H5; Covance), BRCA1 (C-20, Santa Cruz), BARD1 (H-300, Santa Cruz), and actin (A 2066; Sigma).
3′ cleavage assays
32P-labelled pre-mRNA substrates were prepared as described (Kleiman and Manley 1999). Protein concentrations of the extracts were equalized by Bradford reaction (Bio-Rad) or by immunostaining with an α-actin antibody. Cleavage assays with NEs (5 μL) were carried out in reaction mixtures containing 0.2–0.5 ng labeled RNA, 250 ng tRNA, 0.25 U RNasin (Promega), 9.6 mM HEPES-NaOH (pH 7.9), 9.6% glycerol, 24 mM (NH4)2SO4, 0.24 mM DTT, 2.5% polyvinyl alcohol, 0.24 mM PMSF, 2 mM EDTA, and with or without 20 mM creatine phosphate as indicated. For the L3 pre-mRNA, the cleavage reaction was performed in the presence of 1 mM MgCl2 and 1 mM 3′ dATP. NE and added recombinant proteins were pre-incubated for 15 min at 30°C, after which the pre-mRNA was added and incubation continued for an additional 90 min. The in vitro phosphorylation of GST-CTD and purification and separation of the two RNAP II isoforms (IIA and IIO) from NE pellets was performed as described (Hirose and Manley 1998). Full-length and truncated BRCA1/BARD1 (BRCA1 1–304, BARD1 1–202) complexes were purified from SF9 insect cells coinfected with recombinant baculoviruses as described (Wu-Baer et al. 2003). RNA products were isolated and fractionated on 5% polyacrylamide, 8.3 M urea gels.
Ubiquitination assays
Purification and separation of the two RNAP II isoforms (IIA and IIO) from NE pellets was done as described (Hirose and Manley 1998). Heterodimeric complexes comprised of full-length BARD1 and truncated BRCA1 (ΔBRCA1/BARD1-wt and ΔBRCA1/BARD1-C61G) were generated by cotransfecting Escherichia coli cells with an expression plasmid (BARD1/pET28a) that encodes BARD1 with an N-terminal extension of 40 residues containing the 6xHis tag and a plasmid that encodes an untagged N-terminal segment of BRCA1 (residues 1–304) with either the wild-type BRCA1 sequence (BR304*-wt/pET14b) or harboring the C61G missense mutation (BR304*C61G/pET14b). The heterodimeric complexes were then purified from bacterial lysates by affinity chromatography on Ni-NTA agarose beads (Qiagen) as described (Wu-Baer et al. 2003). Full-length BRCA1/BARD1 (BRCA1/BARD1-wt) was isolated from Sf9 insect cells as described (Wu-Baer et al. 2003). E2-conjugating enzyme (His-UbcH5c) and His-HA-Ub were expressed in E. coli and purified by Ni-NTA affinity chromatography as described (Wu-Baer et al. 2003). In vitro ubiquitination assays were carried out using purified recombinant proteins and purified RNAP IIA and RNAP IIO. Ubiquitination reaction mixtures contained 50 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 5 μg of His-HA-Ub, 30 ng of rabbit E1 (Affinity Research Products), 0.2 μg of His-UbcH5c, and purified BRCA1/BARD1 heterodimer (an amount containing 20 ng of the BRCA1 subunit) and 100 ng of either RNAP IIO or RNAP IIA as indicated. After incubation for 40 min at 37°C, reactions were terminated with 5× SDS buffer, separated by SDS-PAGE, and analyzed by immunoblotting with a Ub-specific monoclonal antibody (P4D1, Santa Cruz) and a RNAP II-specific polyclonal antibody (N20, Santa Cruz).
siRNA knckdown of BRCA1 and BARD1 expression in HeLa cells
The siRNA specific for human BRCA1 (NNGGAACCUGUC UCCACAAAGUU) (Bruun et al. 2003) and human BARD1 (NNCAGUAACAUGUCCGAUGAAUU) (Choudhury et al. 2004) were synthesized by Dharmacon RNA Technologies. The control RNA duplex used as nonsilencing siRNA was also obtained from Dharmacon. HeLa cells were gown in a 10-cm plate in complete DMEM. At 60% confluence, the cells were transfected with 240 pmol of siRNA and 60 μL of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After culturing the cells for additional 18 h, the transfection procedure was repeated, and the cells were harvested for analysis 48 h after the initial transfection. Before harvesting, a fraction of the cells was exposed to UV and incubated for 2 h with the indicated drugs. NEs were prepared as described above and analyzed by Western blot and used in 3′ cleavage reactions. For the double knockdown cells, the first transfection was with BRCA1 RNAi and the second one with BARD1 RNAi.
Acknowledgments
We thank A. Norris for technical assistance, C. Shin for help with splicing reactions, and I. Boluk for help preparing the manuscript. This work was supported by NIH grants GM28983 (J.L.M.) and CA88545 (R.B.), and USAMRAA award DAMD170210727 (F.K.).
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1309505.
References
- Abbott D.W., Thompson, M.E., Robinson-Benion, C., Tomlinson, G., Jensen, R.A., and Holt, J.T. 1999. BRCA1 expression restores radiation resistance in BRCA1-deficient cancer cells through enhancement of transcription-coupled DNA repair. J. Biol. Chem. 274: 18808–18812. [DOI] [PubMed] [Google Scholar]
- Baer R. and Ludwig, T. 2002. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12: 86–91. [DOI] [PubMed] [Google Scholar]
- Balajee A.S., May, A., Dianova, I., and Bohr, V.A. 1998. Efficient PCNA complex formation is dependent upon both transcription coupled repair and genome overall repair. Mutat. Res. 409: 135–146. [DOI] [PubMed] [Google Scholar]
- Beaudenon S.L., Huacani, M.R., Wang, G., McDonnell, D.P., and Huibregtse, J.M. 1999. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell Biol. 19: 6972–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman D.B., Du, L., van der Zee, S., and Warren, S.L. 1995. Transcription-dependent redistribution of the largest subunit of RNA polymerase II to discrete nuclear domains. J. Cell. Biol. 129: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman D.B., Halaban, R., van Gool, A.J., Henning, K.A., Friedberg, E.C., and Warren, S.L. 1996. UV-induced ubiquitination of RNA polymerase II: A novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. 93: 11586–11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun D., Folias, A., Akkari, Y., Cox, Y., Olson, S., and Moses, R. 2003. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair 2: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Chen A., Kleiman, F.E., Manley, J.L., Toru, O., and Pan, Z.Q. 2002. Auto-ubiquitination of the BRCA1/BARD1 RING ubiquitin ligase. J. Biol. Chem. 277: 22085–22092. [DOI] [PubMed] [Google Scholar]
- Chiba N. and Parvin, J.D. 2001. Redistribution of BRCA1 among four different protein complexes following replication blockage. J. Biol. Chem. 276: 38549–38554. [DOI] [PubMed] [Google Scholar]
- ____. 2002. The BRCA1 and BARD1 association with the RNA polymerase II holoenzyme. Cancer Res. 62: 4222–4228. [PubMed] [Google Scholar]
- Choudhury A.D., Xu, H., and Baer, R. 2004. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J. Biol. Chem. 279: 33909–33918. [DOI] [PubMed] [Google Scholar]
- Chuvpilo S., Zimmer, M., Kerstan, A., Glockner, J., Avots, A., Escher, C., Fischer, C., Inashkina, I., Jankevics, E., Berberich-Siebelt, F., et al. 1999. Alternative polyadenylation events contribute to the induction of NF-Atc in effector T cells. Immunity 10: 261–269. [DOI] [PubMed] [Google Scholar]
- Dong Y., Hakimi, M.A., Chen, X., Kumaraswamy, E., Cooch, N.S., Godwin, A.K., and Shiekhattar, R. 2003. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 12: 1087–1099. [DOI] [PubMed] [Google Scholar]
- Dornan D., Wertz, I., Shimizu, H., Arnott, D., Frantz, G.D., Dowd, P., O'Rourke, K., Koeppen, H., and Dixit, V.M. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429: 86–92. [DOI] [PubMed] [Google Scholar]
- Hashizume R., Fukuda, M., Maeda, I., Nishikawa, H., Oyake, D., Yabuki, Y., Ogata, H., and Ohta, T. 2001. The RING heterodimer BRCA1–BARD1 is an ubiquitin ligase inactivated by a cancer-derived mutation. J. Biol. Chem. 276: 14537–14540. [DOI] [PubMed] [Google Scholar]
- Hirose Y., and Manley, J.L. 1997. Creatine phosphate, not ATP, is required for 3′ end cleavage of mammalian pre-mRNA in vitro. J. Biol. Chem. 272: 29636–29642. [DOI] [PubMed] [Google Scholar]
- ____. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96. [DOI] [PubMed] [Google Scholar]
- ____. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Huibregtse J.M., Yang, J.C., and Beaudenon, S.L. 1997. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. 94: 3656–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. 2002. Homologous repair of DNA damage and tumorigenesis: The BRCA connection. Oncogene 21: 8981–8993. [DOI] [PubMed] [Google Scholar]
- Jin Y., Xu, X.L., Yang, M.C., Wei, F., Ayi, T.C., Bowcock, A.M., and Baer, R. 1997. Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc. Natl. Acad. Sci. 94: 12075–12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman F.E., and Manley, J.L. 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285: 1576–1579. [DOI] [PubMed] [Google Scholar]
- ____. 2001. The BARD1–CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104: 743–753. [DOI] [PubMed] [Google Scholar]
- Krum S.A., Miranda, G.A., Lin, C., and Lane, T.F. 2003. BRCA1 associates with processive RNA polymerase II. J. Biol. Chem. 278: 52012–52020. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., and Spector, D.L. 2003. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4: 605–612. [DOI] [PubMed] [Google Scholar]
- Le Page F., Randrianarison, V., Marot, D., Cabannes, J., Perricaudet, M., Feunteun, J., and Sarasin, A. 2000. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesions in human cells. Cancer Res. 60: 5548–5552. [PubMed] [Google Scholar]
- Lee K.B., Wang, D., Lippard, S.J., and Sharp, P.A. 2002. Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl. Acad. Sci. 99: 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel L., Gregory, S.M., Becker, K.I., and Sweder, K.S. 2000. Transcription-coupled DNA repair in yeast transcription factor IIE (TFIIE) mutants. Nucleic Acids Res. 28: 835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Zheng, J., Lu, Y., and Bregman, D.B. 2001. Ultraviolet radiation alters the phosphorylation of RNA polymerase II large subunit and accelerates its proteasome-dependent degradation. Mutat. Res. 486: 259–274. [DOI] [PubMed] [Google Scholar]
- Mallery D.L., Vandenberg, C.J., and Hiom, K. 2002. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 21: 6755–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne L.V. and Lehmann, A.R. 1982. Failure of RNA synthesis to recover after UV irradiation: An early defect in cells from individuals with Cockayne's syndrome and Xxeroderma Pigmentosum. Cancer Res. 42: 1473–1478. [PubMed] [Google Scholar]
- McCracken S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M., and Bentley, D.L. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- McKay B.C., Chen, F., Clarke, S.T., Wiggin, H.E., Harley, L.M., and Ljungman, M. 2001. UV light-induced degradation of RNA polymerase II is dependent of Cockayne's syndrome A and B proteins but not p53 or MLH1. Mutat. Res. 485: 93–105. [DOI] [PubMed] [Google Scholar]
- Mitsui A. and Sharp, P.A. 1999. Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc. Natl. Acad. Sci. 96: 6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M. and Tansey, W. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4: 192–201. [DOI] [PubMed] [Google Scholar]
- Mullenders L.H.F. 1998. Transcription response and nucleotide excision repair. Mutat. Res. 409: 59–64. [DOI] [PubMed] [Google Scholar]
- Neumann S., Petfalski, E., Brugger, B., Grobhans, H., Wieland, F., Tollervey, D., and Hurt, E. 2003. Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO J. 4: 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H., Ooka, S., Sato, K., Arima, K., Okamoto, J., Klevit, R.E., Fukuda, M., and Ohta, T. 2004. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1–BARD1 ubiquitin ligase. J. Biol. Chem. 279: 3916–3924. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Cortez, D., Bowers, B., Elledge, S.J., and Gellert, M. 2001. Direct DNA binding by Brca1. Proc. Natl. Acad. Sci. 98: 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner J.N., Balasubramanian, B., Corden, J., Warren, S.L., and Bregman, D.B. 1998. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II: Implications for transcription-coupled DNA repair. J. Biol. Chem. 273: 5184–5189. [DOI] [PubMed] [Google Scholar]
- Sato K., Hayami, R., Wu, W., Nishikawa, T., Nishikawa, H., Okuda, Y., Ogata, H., Fukuda, M., and Ohta, T. 2004. Nucleophosmin/B23 is a candidate substrate for the BRCA1–BARD1 ubiquitin ligase. J. Biol. Chem. 279: 30919–30922. [DOI] [PubMed] [Google Scholar]
- Schul W., Groenhout, B., Koberna, K., Takagaki, Y., Jenny, A., Manders, E.M.M., Raska, I., van Driel, R., and de Jong, L. 1996. The RNA 3′ cleavage factors CstF-64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 15: 2883–2892. [PMC free article] [PubMed] [Google Scholar]
- Scorilas A. 2002. Polyadenylate polymerase (PAP) and 3′ end pre-mRNA processing: Function, assays, and association with disease. Crit. Rev. Clin. Lab. Sci. 39: 193–224. [DOI] [PubMed] [Google Scholar]
- Scully R., Anderson, S.F., Chao, D.M., Wei, W., Ye, L., Young, R.A., Livingston, D.M., and Parvin, J.D. 1997a. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. 94: 5605–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Chen, J., Ochs, R.L., Keegan, K., Hoekstra, M., Feunteun, J., and Livingston, D.M. 1997b. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435. [DOI] [PubMed] [Google Scholar]
- Shatkin A.J. and Manley, J.L. 2000. The ends of the affair: Capping and polyadenylation. Nat. Struct. Biol. 7: 838–842. [DOI] [PubMed] [Google Scholar]
- Starita L.M. and Parvin, J.D. 2003. The multiple nuclear functions of BRCA1: Transcription, ubiquitination and DNA repair. Curr. Opin. Cell. Biol. 15: 345–350. [DOI] [PubMed] [Google Scholar]
- Starita L.M., Machida, Y., Sankaran, S., Elias, J.E., Griffin, K., Schlegel, B.P., Gygi, S.P., and Parvin, J.D. 2004. BRCA1-dependent ubiquitination of γ-tubulin regulates centrosome number. Mol. Cell. Biol. 24: 8457–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup J.Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3: 21–29. [DOI] [PubMed] [Google Scholar]
- Takagaki Y. and Manley, J.L. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2: 761–771. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Seipelt, R.L., Peterson, M.L., and Manley, J.L. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952. [DOI] [PubMed] [Google Scholar]
- Tornaletti S., Reines, D., and Hanawalt, P.C. 1999. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J. Biol. Chem. 274: 24124–24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom V., Jaspers, N.G.J., and Vermeulen, W. 2002. When machines get stuck-obstructed RNA polymerase II: Displacement, degradation or suicide. Bioessays 24: 780–784. [DOI] [PubMed] [Google Scholar]
- Venkitaraman A.R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108: 171–182. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S.J., and Qin, J. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Dev. 14: 927–939. [PMC free article] [PubMed] [Google Scholar]
- Woudstra E.C., Gilbert, C., Fellows, J., Jansen, L., Brouwer, J., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. 2002. A Rad26–Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415: 929–933. [DOI] [PubMed] [Google Scholar]
- Wu-Baer F., Lagrazon, K., Yuan, W., and Baer, R. 2003. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278: 34743–34746. [DOI] [PubMed] [Google Scholar]
- Xia Y., Pao, G.M., Chen, H.W., Verma, I.M., and Hunter, T. 2003. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 278: 5255–5263. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]