Summary
Despite extensive global research into genetic predisposition for severe COVID-19, knowledge on the role of rare host genetic variants and their relation to other risk factors remains limited. Here, 52 genes with prior etiological evidence were sequenced in 1,772 severe COVID-19 cases and 5,347 population-based controls from Spain/Italy. Rare deleterious TLR7 variants were present in 2.4% of young (<60 years) cases with no reported clinical risk factors (n = 378), compared to 0.24% of controls (odds ratio [OR] = 12.3, p = 1.27 × 10−10). Incorporation of the results of either functional assays or protein modeling led to a pronounced increase in effect size (ORmax = 46.5, p = 1.74 × 10−15). Association signals for the X-chromosomal gene TLR7 were also detected in the female-only subgroup, suggesting the existence of additional mechanisms beyond X-linked recessive inheritance in males. Additionally, supporting evidence was generated for a contribution to severe COVID-19 of the previously implicated genes IFNAR2, IFIH1, and TBK1. Our results refine the genetic contribution of rare TLR7 variants to severe COVID-19 and strengthen evidence for the etiological relevance of genes in the interferon signaling pathway.
Keywords: SARS-CoV-2, host genetics, toll-like receptor 7, targeted sequencing, rare variants, variant collapsing analysis, burden analysis, innate immunity, immune deficiency, infection
This study explores genetic factors in severe COVID-19 in a Spanish/Italian cohort. Rare deleterious TLR7 variants were found in 2.4% of young cases. Incorporation of functional assay or protein modeling data yielded increased effect sizes. Further, TLR7 signals in females suggest additional mechanisms beyond X chromosome recessive inheritance.
Introduction
The SARS-CoV-2 pandemic has posed major challenges to societies and health care systems around the world. Clinically, SARS-CoV-2 infection results in a broad spectrum of outcomes, ranging from the complete absence of symptoms to severe illness and even death secondary to the associated lung disease (severe COVID-19). Extensive research has been conducted to elucidate the causes of these inter-individual differences, with the aim of informing drug development programs and designing strategies for individual risk prediction in future viral pandemics. This has demonstrated that the observed variability is explained in part by demographic and clinical risk factors. Specifically, increased age; male sex; and comorbidities like diabetes, coronary artery disease (CAD), high body weight, and hypertension,1,2,3 as well as the presence of auto-antibodies4 have been suggested to be associated with severe COVID-19. In addition, research has shown robust associations between severe COVID-19 and common genetic variants in the host, which are typically characterized by a minor allele frequency (MAF) of >1% and modest effect sizes.5,6,7,8,9,10
Monogenic causes have been suggested in individuals with severe COVID-19, as based on the identification of highly penetrant pathogenic variants in TLR7 [OMIM: 300365], TBK1 [OMIM: 604834], and IFNAR1 [OMIM: 107450] in individual families.11,12,13,14 To date, only a limited number of studies have performed systematic investigations of the role of rare genetic variants in large severe COVID-19 cohorts.9,15,16 At the population level, the most compelling evidence for this to date has been reported for rare variants in the X chromosome gene TLR7.17,18,19,20,21,22 The corresponding protein TLR7 (toll-like receptor 7) is a receptor for single-stranded RNA and is central to SARS-CoV-2 host defense.23 The suggested pathomechanism of TLR7 rare variants in males with severe COVID-19 is X-linked recessive loss of function.19 Since TLR7 escapes X-inactivation,24 this hypothesis does not explain recent findings of rare deleterious TLR7 variants in females with severe COVID-19.15
Given prior epidemiological evidence for a contribution of age, sex, and additional clinical risk factors to the risk for severe COVID-19, the aim of the present study was to empower the search for rare variant associations by performing stratified analyses in two ethnically homogeneous cohorts. For this purpose, 52 candidate genes for severe COVID-19, including TLR7, were sequenced in 1,772 individuals from Spain and Italy who had been hospitalized for COVID-19 and had required respiratory support, and 5,347 individuals from the general Spanish/Italian populations. Notably, the severe COVID-19 cases were recruited prior to vaccine availability, thus allowing analysis of the virus-naive host reaction to SARS-CoV-2 infection. All individuals had undergone previous array-based genotyping as part of prior genome-wide association studies (GWASs).25,26 The candidate gene sequencing approach was based on the cohort’s informed consent on targeted follow-up sequencing. Together with available clinical information, sequencing data were then analyzed for single-variant associations and gene burden using different stratified approaches, including distinct phenotype definitions and variant pathogenicity levels.
Subjects and methods
Candidate gene selection
The available informed consent documentation allowed follow-up sequencing only and precluded systematic approaches such as exome sequencing (ES). Therefore, 55 genes were selected in August 2020, based on evidence available at that time. These comprised 14 genes from early GWAS loci5,25; five genes from diagnostic ES11,13; and 36 genes with functional evidence, which have been implicated previously in viral defense or pathogen immunity (Figure 1A). For each gene, the evidence for selection is presented in Table S1. Three genes (CCL3, CXCL1, CFD) were subsequently excluded from the analysis, since the size of the respective covered region post quality control (QC) was less than 50% of the originally targeted region. Detailed information on the coverage of these genes, and the number of variant sites per gene, is provided in Table S1.
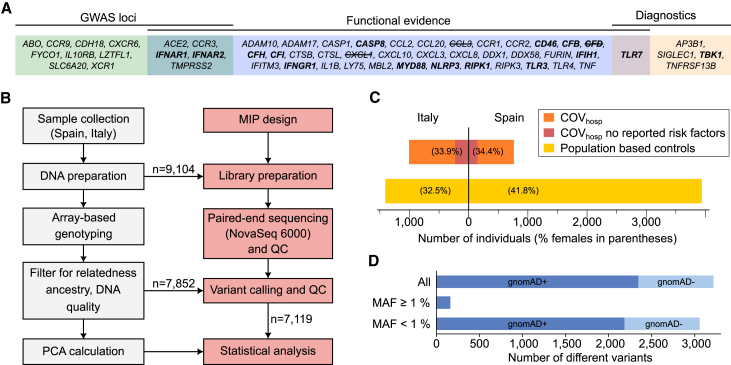
Figure 1.
Study design and cohort characteristics
(A) Candidate genes included in targeted sequencing, grouped according to source of evidence (details in Table S1). Genes known to cause human inborn errors of immunity27 are highlighted in bold, and genes excluded during quality control due to low sequencing coverage are crossed out.
(B) Workflow describing the main steps of sample preparation, genotyping, sequencing, and computational processing. Boxes colored in gray indicate steps that were performed in previous studies.25,26 MIP, molecular inversion probe; PCA, principal-component analysis; QC, quality control.
(C) Number of individuals in the Italian (left) and Spanish (right) subcohorts. The number of COVhosp individuals with no reported risk factors (as described in Table 1) is highlighted in red. The proportion of females is shown in parentheses.
(D) Number of variants observed in the cohort in relation to their minor allele frequency (MAF). In the present study, variants with MAF <1% were denoted as rare variants, while all others were considered common. Intensity of color shading indicates whether (dark) or not (light) variants have been reported with allele frequency in gnomAD r2.1 exomes.
Study design and phenotype definition
Coding regions were sequenced using single-molecule molecular inversion probes (MIPs),28 in 9,104 Spanish/Italian individuals from the Severe COVID-19 GWAS cohort25,26 (see supplemental methods). Following post-sequencing QC, which included the use of array-based genotype data for population inference and relatedness filtering (Figure 1B), a total of 7,119 individuals remained for analysis. Data analysis included (1) single-variant association analysis and (2) rare variant collapsing analysis. Both analyses were performed using four case-control definitions (Table 1) that involved one main analysis comprising the entire cohort, and three stratified analyses. The stratified analyses were performed in order to investigate the contribution of rare variants in individuals with otherwise low epidemiological risk (POPlowrisk, COVhosp by risk factors) and the potential contribution of rare variants to the level of disease severity (COVhosp by respiratory support). Each of the four analyses was repeated separately for males and females, in view of prior reports of sex differences in etiological risk.3 Notably, some COVhosp individuals (66 of 1,772) did not have sufficient information on comorbidities and were therefore excluded from the risk factor-based stratifications (POPlowrisk, COVhosp by risk factors).
Table 1.
Case-control definitions used in the present study
| Analysis | Cases | n cases (females/males) | Controls | n controls (females/males) |
|---|---|---|---|---|
| Case-control definitions for analyses involving population-based controls (POP) | ||||
| (1) POPall | Individuals hospitalized for COVID-19 who required respiratory support (COVhosp) | 1,772 (605/1,167) | Individuals from the general population with unknown SARS-CoV-2/COVID-19 status (population controls) | 5,347 (2,102/3,245) |
| (2) POPlowrisk | COVhosp with no reported risk factorsa | 378 (126/252) | Same as above | 5,347 (2,102/3,245) |
| Case-control definitions for analyses involving COVID-19 hospitalized individuals (COVhosp) only | ||||
| (3) COVhosp by risk factors | COVhosp with no reported risk factorsa | 378 (126/252) | COVhosp with two or more of the reported risk factorsa | 726 (244/482) |
| (4) COVhosp by respiratory support | COVhosp requiring respiratory support level 3 (intubation) or 4 (ECMO, highest level) | 478 (115/363) | COVhosp requiring respiratory support level 1 (oxygen mask only, lowest level) | 661 (284/377) |
Risk factors for which phenotype data were broadly available: age ≥60 years, diabetes, hypertension, coronary artery disease. Notably, 66 of 1,772 COVhosp individuals did not have sufficient information on comorbidities and were therefore excluded from the risk factor-based stratification (POPlowrisk, COVhosp by risk factors). ECMO, extracorporeal membrane oxygenation.
Cohort characteristics
The recruitment procedure, sample collection, and DNA extraction were conducted by the Severe COVID-19 GWAS group (Figure 1B) and are described elsewhere.26 Approvals were obtained from the relevant ethics committees (listed in supplemental methods) and informed consent was obtained. Individuals hospitalized for severe COVID-19 (COVhosp) were collected at several centers in Spain and Italy in 2020 as part of the first outbreaks of the pandemic in Europe. Severe COVID-19 was defined as requiring respiratory support, i.e., the necessity for oxygen supplementation. While other definitions exist, this approach was chosen to ensure feasibility.26 Following QC (see next paragraph) the cohort comprised (1) 1,772 COVhosp individuals (n = 1,008 from Italy, n = 764 from Spain; Figure 1C; Table 1); and (2) 5,347 population-based controls (n = 1,408 from Italy, n = 3,939 from Spain). In total, 38% of all individuals were female. Respiratory support for COVhosp individuals was documented as the maximum support required during hospitalization: oxygen mask only (level 1, lowest), non-invasive ventilation (level 2), invasive ventilation (level 3), or extracorporeal membrane oxygenation (ECMO) (level 4, highest). For the majority of the COVhosp individuals (1,706 of 1,772) data were available on comorbid CAD, diabetes, and hypertension (see Figure S1 for further information including subcohort [Italy/Spain]-specific distribution of risk factors).
QC and data processing
After library preparation and sequencing using MIPs28 (2×150 base pairs [bp], paired-end, see supplemental methods), data were processed using an MIP-specific pipeline that included several filter and QC steps (supplemental methods) and various tools.29,30,31,32,33,34,35 DNA QC, population inference, and relatedness filtering had been performed previously by the Severe COVID-19 GWAS group25,26 using their array-based genotype data.
Two patients in the Asano et al.19 study had phenotypes, age, sex, and rare TLR7 variants that were identical to those in the present data, suggesting a sample overlap. After recontacting the groups responsible for the recruitment of these two individuals, a total of 82 individuals who may have been common to other research groups were identified. Rare TLR7 variants of previously reported individuals are labeled accordingly (Table S2).
Single-variant analysis
Analysis of the present cohort
An additive non-singleton single-variant association test was performed using logistic regression with plink36 v2.0 and Firth correction, as well as age, sex, age2, age∗sex, and the first 10 principal components (PCs) as covariates. The number of PCs was chosen in accordance with Degenhardt et al.26 and the COVID-HGI exome-wide association study.15 As the target region spans only about 0.003% of the human genome, the PCs were calculated using the respective array-based genome-wide genotype data (Degenhardt et al.26) to maximize the capture of population structure. As case-control ratios and other sample characteristics were substantially different between both populations, logistic regression was performed separately for the Italian and the Spanish cohorts, and the results were then meta-analyzed using METAL.37 We applied two thresholds for multiple testing: The “strict” threshold was established using the Bonferroni method, which involved correction for the number of analyses (four case-control definitions, three sex-based stratifications) and the number of tested variants (strict, α = 6.7 × 10−6). To take the potential correlation of the different analyses into account, a “lenient” significance threshold was applied, involving correction for the number of tested variants only (lenient, α = 4.1 × 10−5).
Replication cohorts
Whenever COVID-HGI release 7 analysis A2 summary statistics7 were used as the replication cohort, this refers to the leave-one-out-HOSTAGE dataset (which excludes all individuals who were common to the present cohort and the COVID-HGI). For comparison and meta-analysis of the present single-variant association results with those of the Regeneron dataset,16 the results of the POPall analysis and the POPlowrisk analysis (without sex stratification) were followed up for all variants with OR >5 and p < 0.05 in the present cohort. When associations for these variants were reported in the Regeneron browser (see web resources), the respective results were filtered for (1) the use of exome data (instead of imputed data); (2) a phenotype corresponding to that used in the present study (“COVID-19 positive severe vs. COVID-19 negative or COVID-19 status unknown” or “COVID-19 positive hospitalized vs. COVID-19 negative or COVID-19 status unknown,” as defined in Kosmicki et al.16); (3) “European” or “pan-ancestry” ancestry; and (4) the analysis type “meta-analysis.” For each variant, the results of the analysis that included the maximal number of cases were selected.
Gene-based rare variant collapsing analysis
Variant collapsing (or burden testing) is a widely used approach that is applied to increase statistical power for the testing of rare variants. Here, variants from distinct genetic regions (e.g., in the present study, genes or gene groups) are combined, and testing is performed for these variant groups rather than for single variants.
Definition of variant classes
The present analyses considered two allele frequency groups: MAF <1% and MAF <0.1% (defined as maximal MAF in this cohort or in gnomAD r2.1 non-Finnish European [NFE] exomes). Cohort allele frequencies were calculated using plink v2.0. Deleteriousness classes SYN, M1, M3, M4, and C10+M1 were used. M1, M3, and M4 are similar to those described in Kosmicki et al.16 The M1 class is restricted to pLoF variants that are defined as having an Ensembl variant effect predictor (VEP)34 impact of “HIGH.” M3 contains all M1 variants, plus variants with a VEP impact of “moderate” but not missense and missense variants for which five of five prediction algorithms (SIFT, PolyPhen2-HDIV database, PolyPhen2-HVAR database, LRT, MutationTaster) predict deleteriousness. M4 contains all M3 variants plus missense variants for which at least one of the five algorithms predicts a deleterious effect. SYN contains synonymous variants only, and functions as a control class. C10+M1 contains all pLoF (M1) variants and all variants with a CADD v1.638 (combined annotation dependent depletion) score greater than 10, as used by Kousathanas et al.9
TLR7-specific variant definitions
For TLR7, two additional gene-specific deleteriousness classes were created. The first one comprised biochemically loss-of-function (bLoF) variants, i.e., all variants reported as being loss of function on the basis of biochemical tests in previous research.18,19,20 Synonymous TLR7 variants were inspected for potential cryptic splicing effects using spliceAI.39 The second class (3D-P) comprised variants that were deemed pathogenic or likely pathogenic based on protein structural analyses. Herefore, each of the mutation sites was analyzed in the context of its structural environment and with regard to changes in protein folding stability. The latter analyses aimed to infer pathogenicity from the extent of mutation-induced changes in the structural integrity of the TLR7 dimer (see supplemental methods40,41,42,43).
Statistical analysis
For the statistical analysis of the collapsed variants, the Cochran-Mantel-Haenszel (CMH) test (plink v1.9 implementation, dominant model) was used, as previously described.44 While other methods exist, the CMH test was chosen as it was developed for case-control studies with subgroups of different characteristics by performing internal stratification while still generating overall test statistics for the entire cohort.45 Moreover, the CMH test can handle rare events,46 which is especially useful for rare variant collapsing analysis. The stratification categories used for the CMH test were subcohort (Italy, Spain) and sex (male, female). Similar to the single-variant association analyses, two thresholds for statistical correction were applied: The “strict” definition was performed according to Bonferroni, and accounted for all performed tests (tested genes, variant categories, case-control definitions, α = 8.7 × 10−6). The “lenient” threshold considered that the case-control definitions and the different variant categories are correlated and therefore corrected for the number of tested genes only (α = 9.6 × 10−4). Data from the GenOMICC-study9 were used for a replication attempt, details for which are provided in the supplemental methods.
Results
Single-variant analyses identify etiological variant in TBK1
Within the 52 genes, 3,218 high-confidence variants were identified across the entire cohort, 95% of which were rare (n = 3,059; MAF <1%). Of these rare variants, 28.6% had no reported frequency in gnomAD r2.1 exomes (n = 874, Figures 1D and S2). More specifically, 2,007 singletons (i.e., variants that occur in only one individual) were observed, including 111 putative loss-of-function (pLoF) variants. These were present in 31 COVhosp individuals, and 77 population-based controls (1.75% vs. 1.44%; three individuals carried two variants, respectively). Within the subset of COVhosp individuals with no reported risk factors, eight singleton pLoFs were observed in seven individuals (1.85%), all of which were heterozygous and two of which were found in one individual (Table S3). For these seven individuals, the distribution of age and level of respiratory support did not differ significantly from those of the remaining COVhosp individuals with no reported risk factors (Welch’s p > 0.39).
Next, formal association testing for the 1,211 non-singleton variants was performed using Firth’s logistic regression and the covariates age, sex, age2, age∗sex, and 10 PCs obtained from prior array-based genotyping (see subjects and methods). This was performed separately for the Spanish and Italian cohorts, and the results were meta-analyzed using inverse variance based meta-analysis (Figures 2 and S3). Overall, seven variants had p values below the strict significance threshold (see subjects and methods). All of these seven variants were associated at genome-wide significance (and with the same direction of effect) in the independent data freeze of the global COVID-19 Host Genetics Initiative (HGI)7 (release 7, see subjects and methods). Variants associated with nominal significance (p < 0.05) and gnomAD r2.1 NFE exomes-AF > 0.01% are reported in Table S4.
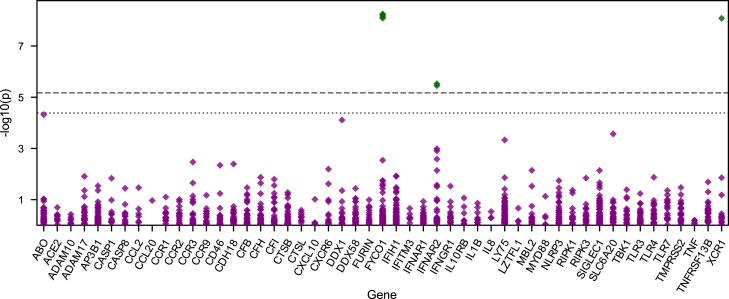
Figure 2.
Association analysis for individual variants
The p values (y axis, negative log10) obtained in the association analysis of 1,211 non-singleton variants from the POPall analysis. Variants are grouped according to the genes (x axis, sorted alphabetically) in which they are located. Results for case-control definitions other than POPall are provided in Figure S3. Dotted line: Lenient significance threshold, correcting for the number of variants tested (α = 4.1 × 10−5). Dashed line: Strict significance threshold, also taking into account multiple testing due to additional case-control definitions (α = 6.7 × 10−6). Variants with p values below the lenient significance threshold are marked in green and were only found in genes selected based on prior GWAS evidence, i.e., FYCO1 and XCR1 at 3p21.31, IFNAR2 at 21q22.11.
Given the limited statistical power for single-variant analyses, candidate variants (defined as high effect size estimates [OR > 5] and nominal significance [p < 0.05]) from the POPall and POPlowrisk analyses (non-sex-stratified) were followed up in the Regeneron dataset (see subjects and methods). A total of 62 variants, all of which had an MAF <0.2% and were absent from the COVID-19 HGI data, met these criteria. Of those, 38 variants were also present in the Regeneron dataset (Table S5). The most significant variant was a missense variant in TBK1 (p.Arg358His, chr12:64878163:G:A (hg19), CADD = 23.3, REVEL = 0.259), which showed effect sizes of >20 in both cohorts (Regeneron: OR = 24.2, confidence interval = [3.64, 160.47], p = 0.00097; present study: OR = 30.0 [2.71, 332.6], p = 0.0056). In a meta-analysis of both cohorts, this variant showed strong association with severe COVID-19 (p = 1.67 × 10−5, OR = 26.3 [5.93, 116.2]).
Gene-based rare variant collapsing analysis confirms TLR7 association
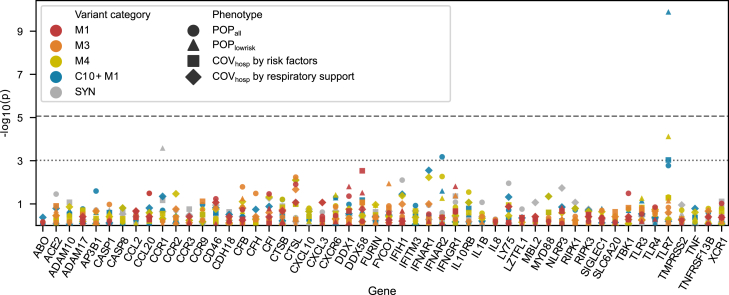
To increase statistical power, gene-based collapsing analyses were performed. For this purpose, variants were assigned to (1) two allele frequency groups (MAF <0.1% and MAF <1%); and (2) five classes of deleteriousness (M1, M3, M4, C10+M1, SYN; see subjects and methods). Variant counts per class are provided in Figure S2. For each combination of MAF, deleteriousness, and gene, statistical association analyses were performed using the CMH test. The results are reported in Figure 3 for MAF <0.1% and in Figure S4 for both MAF <1% and sex-stratified analyses, respectively. At strict threshold definition (subjects and methods), significant associations were obtained for TLR7 in (1) the POPlowrisk analysis overall (C10+M1, MAF <0.1%; carriers: 9/378 cases vs. 13/5,347 controls; p = 1.27 × 10−10, OR = 12.3 [4.7, 32.2]; Figure 3) and (2) the female-only subgroup (C10+M1, MAF <0.1%; 4/126 vs. 5/2102; p = 1.75 × 10−9, OR = 24.8 [5.9, 105.2]; Figure S4). Suggestive evidence (at lenient threshold, see subjects and methods) was obtained for two additional genes: (1) IFNAR2 [OMIM: 602376] (POPall, C10+M1, MAF<1%; 60/1772 vs. 73/5347; p = 2.61 ×10−4, OR = 1.9 [1.3, 2.7]; Figure S4) and (2) IFIH1 [OMIM: 606951] (COVhosp by respiratory support, C10+M1, MAF <1%; 54/478 vs. 36/661; p = 3.60 × 10−4, OR = 2.2 [1.4, 3.4]; Figure S4). All associations with nominal significance (p < 0.05) are listed in Table S6.
Figure 3.
Results of the gene-based collapsing analysis for rare variants with MAF <0.1%
The p values (y axis, negative log10) are plotted for 52 genes (x axis, sorted alphabetically). The various case-control definitions (see Table 1; excluding sex-stratified analyses) are depicted as symbols, while variant deleteriousness classes are coded according to color (M1: pLoF, M3 and M4: pLoF and moderate effect variants including missense in two graduations, C10+M1: CADD >10 or pLoF, SYN: synonymous, see subjects and methods). Dashed line: Strict significance threshold, correcting for all tests conducted: (α = 8.7 × 10−6). Dotted line: Lenient significance threshold, correcting for the number of genes tested (α = 9.6 × 10−4). Results for sex-stratified analyses and variants with MAF <1% are provided in Figure S4.
To investigate whether genes with related functions were enriched for rare variants, eight gene sets were defined (Table S7) and a collapsing analysis based on each gene set was conducted. No significant results were obtained after strict correction for multiple testing (Figure S5). Nevertheless, the most significant associations were observed for the set of immunodeficiency genes (n = 15), and this remained nominally significant even after the exclusion of TLR7.
Identification of a low-frequency TLR7 risk variant in the Spanish population
In view of both the highly significant results presented above and robust prior evidence for the involvement of TLR7 in severe COVID-19,11,12,18,19,20,21 more detailed investigations were performed to characterize the rare TLR7 variant associations in the present cohort. Overall, the identified TLR7 variants comprised 26 missense, one 3′UTR, and 16 synonymous (maximum spliceAI score: 0.02) variants, but no pLoF variants (see Table S2). Two COVhosp individuals (one male case, one female case; none of the population-based controls) carried two distinct rare variants in TLR7 respectively. The male individual (p.M854I, p.L988S) was previously reported in an independent study by Asano et al.19 (see subjects and methods). In the female individual, biallelic occurrence of the two deleterious variants (p.A448V, p.R920K) could cause X-linked recessive disease. While no direct assessment of compound heterozygosity based on MIP sequencing data was possible, in silico haplotype assessment using the variant co-occurrence tool of gnomAD v2.1.1 (see web resources)47 suggested that the two variants map to different haplotypes.
The analyses also identified a missense variant exclusive to the Spanish subcohort (rs202129610, p.D332G). This was present in two population-based controls (MAF = 0.038%, one female, one male), and three COVhosp individuals (MAF = 0.33%, one female, two males), The frequency further increased in COVhosp individuals with no reported risk factors (MAF = 1.0%). The variant was nominally significant in the single-variant logistic regression analysis (POPlowrisk, OR = 5.77 [1.49, 22.3], p = 0.011), but was absent from the Regeneron dataset and the in silico pathogenicity prediction of this variant was ambiguous (CADD = 18.45, REVEL = 0.078). However, a previous study reported that this variant was hypomorphic, as based on in vitro experiments (7% NF-κB activity19). This variant is absent from European individuals in gnomAD v3.1.2 and has only been reported to date in Latino/Admixed Americans (population-specific MAF of 0.019%).
Incorporation of functional and protein data increases TLR7 rare variant effect sizes
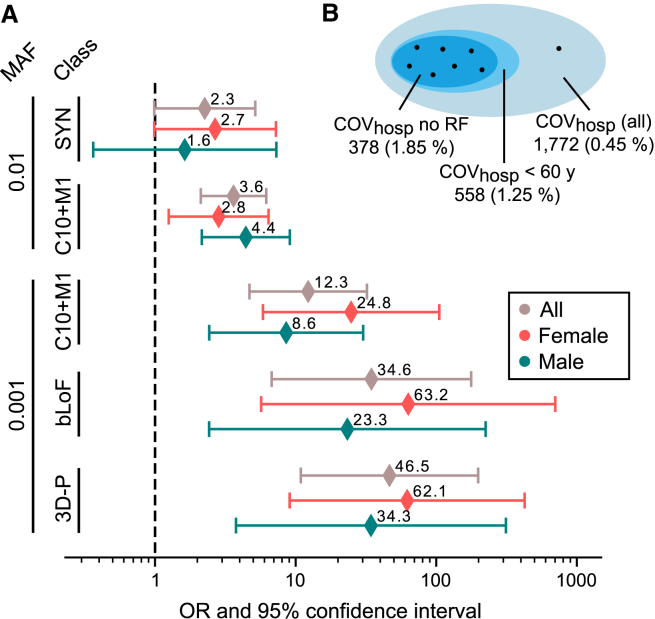
Seventeen of the 26 TLR7 missense variants have previously been analyzed in vitro. In these experiments, seven variants were reported to decrease or even abolish the function of TLR7.18,19,20 These seven variants were combined to a new deleteriousness class (bLoF, biochemically loss of function, as proposed in Matuozzo et al.21) for the rare variant collapsing analysis. The resulting OR (POPlowrisk, bLoF, MAF<0.1%; 4/378 vs. 3/5347; p = 1.73 × 10−10, OR = 34.6 [6.8,177.2]; Figure 4A) was substantially higher than effect sizes based on in silico prediction alone (POPlowrisk, C10+M1, MAF <0.1%; OR = 12.3; see above).
Figure 4.
Forest plot for TLR7 rare variant gene burden according to variant classification
(A) Odds ratios (ORs) of collapsed variants in TLR7 are shown for POPlowrisk at different minor allele frequency groups (MAF) and deleteriousness predictions (class). Within each group, results are presented for all individuals and for sex-stratified analyses. Error bars indicate 95% confidence intervals. SYN, synonymous; C10+M1, CADD>10 or pLoF; bLoF, biochemical evidence for a loss-of-function effect; 3D-P, variant class based on 3D protein structure, see subjects and methods. SYN variants with MAF <0.001 were only present in controls (OR = 0.0, no confidence interval calculable).
(B) Presence of 3D-P TLR7 (MAF<0.1%) variant carriers (black dots) in all COVhosp individuals (gray blue), COVhosp with age <60 y (light blue) and COVhosp with no reported risk factors (“no RF,” dark blue). The number of individuals within each set is indicated by area and is specified in the outer legend. Percentages in brackets represent carrier ratios.
To create a structure-based variant class, protein structure data for TLR7 were used for 3D modeling and protein energy calculation (subjects and methods, supplemental methods). Based on this approach, eight of the 26 rare missense variants were classified as either damaging (n = 4) or probably damaging (n = 4) to the protein structure, and were aggregated into a new variant class (3D-P). Statistical analysis of this 3D-P class yielded even higher ORs (POPlowrisk, 3D-P, MAF <0.1%; 7/378 vs. 4/5,347; p = 1.74 × 10−15, OR = 46.5 [10.9, 198.7]) than the aforementioned variant classifications (see Figures 4A and S6). In alignment with prior studies that identified TLR7 associations in younger individuals,11,12,18,19,20,21 the analysis was repeated by defining cases as individuals with severe COVID-19 aged <60 years, with no consideration of other risk factors, and comparing these individuals with all population controls. Using the 3D-P TLR7 (MAF <0.1%) class, the proportion of carriers increased across the following three subgroups: all COVhosp individuals (0.45%); younger COVhosp individuals (age <60 years, 1.25%); COVhosp individuals with no reported risk factors (1.85%; Figure 4B).
Investigation of domain- and sex-specific variant effects in TLR7
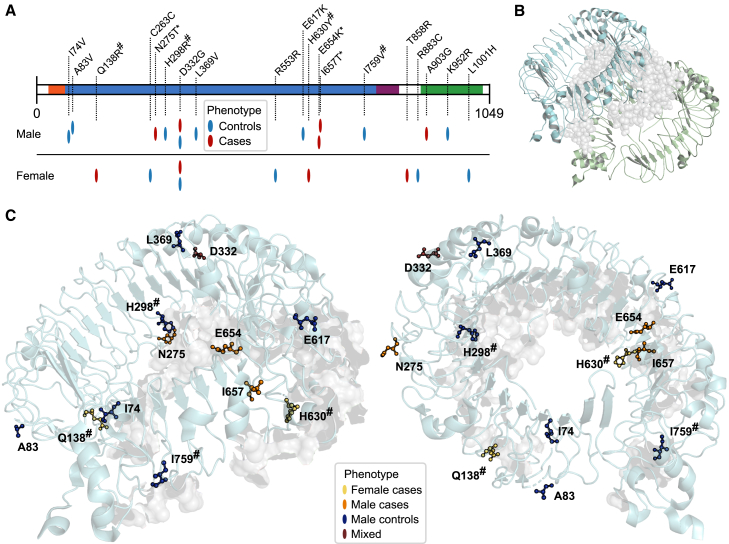
To date, X-linked TLR7 deficiency, as mediated by raredeleterious variants, has mainly been reported in males,11,12,18,19,20 and a classical X-linked recessive mode of inheritance has been suggested.11,12,18,19 However, two recent association studies also reported an enrichment of rare variants in females.15,21 Given the present finding of an enrichment of rare heterozygous TLR7 variants in females, and previous observations of TLR7 escaping X-inactivation in immune cells,24 analyses were performed to explore other potential mutational mechanisms. First, the distribution of deleterious rare variants across the TLR7 protein was studied in females with no reported risk factors (i.e., POPlowrisk; C10+M1, MAF <0.1%). In female cases, an overrepresentation of these variants was observed in the leucine-rich-repeat (LRR) domain (see Figure 5A). Since the LRR domain is involved in the dimerization of TLR7 monomers, which is essential for the activation of downstream signaling pathways,48 we hypothesized that missense variants located in this domain could potentially confer a dominant-negative effect by affecting protein dimerization. We approached this by using the TLR7 protein structure, and observed that four non-synonymous variants (Q138R, H298R, H630Y, I759V; all singleton, all missense) in the entire cohort were located within 5 Ångström of the dimerization interface (I5AN; hashed residue labels in Figure 5). Two of these I5AN variants (Q138R, H630Y) were present in female COVhosp individuals with no reported risk factors, and were among the 3D-P variants (indicating a damaging structural effect, see above). No I5AN variant was observed in female controls (POPlowrisk females, I5AN, MAF <1%; 2/126 vs. 0/2101; p = 2.1 × 10−6; Figure 5). The two other variants (H298R, I759V) were observed in male controls (POPlowrisk males, I5AN, MAF<1%; 0/252 vs. 2/3245; p = 0.65).
Figure 5.
Location of rare TLR7 variants within TLR7 protein domains
(A) Rare, deleterious TLR7 variants (POPlowrisk, C10+M1, MAF <0.1%) are mapped on the protein domains of TLR7 (x axis: amino acid position). Phenotype, according to the POPlowrisk case-control definition, and the sex of variant carriers is indicated by color or caption. Variants of carriers previously reported in Asano et al.19 (see subjects and methods and Table S2) are indicated by asterisks (∗). TLR7 domains: LRR-NT (leucine-rich repeat, N terminal, aa 27–65) orange; LRR regions 1–26 (aa 66–786) blue; LRR-CT (leucine-rich repeat, C terminal, aa 787–839), violet; TIR (Toll/interleukin-1 receptor) domain (aa 889–1033), green.
(B) TLR7 dimer overview, interface highlighted as gray surface (also in C).
(C) Non-synonymous variants from (A) are highlighted in the 3D conformation of one TLR7 subunit (PDB ID: 5GMH) and are presented from two angles. Phenotype (POPlowrisk, see A) and sex of the variant carriers are indicated by color coding. Variants within 5 Ångström of the subunit interface are highlighted by a hash (#, also in A). Variants located downstream of position T858 could not be plotted due to absence of the respective residues from the structure. Visualized using PyMOL Molecular Graphics System (Version 2.5.5 Schrödinger, LLC).
To replicate the domain- and sex-specific TLR7 findings, analyses were performed in the cohort of the GenOMICC study, which has generated one of the largest collections of genome sequencing (GS) data from individuals with severe COVID-19 to date.9 Overall, only very few numbers of TLR7 variants were observed in females, and no I5AN variant was observed in either female cases or controls. Detailed results are shown in Table S8 and methodical information is presented in the supplemental methods.
Discussion
The present study investigated the contribution of rare genetic variants within 52 candidate genes to the etiology of severe COVID-19 and their relation to clinical risk factors, via the performance of joint and stratified analyses in two large, ethnically homogeneous cohorts recruited in the pre-vaccine era of the SARS-CoV-2 pandemic. The present findings reinforce prior genetic evidence for an etiological role of the X-chromosomal gene TLR7 in severe COVID-19 through the identification of a robust enrichment of deleterious rare variants. Notably, this enrichment was particularly pronounced in young individuals with severe COVID-19 with no reported demographic or clinical risk factors, and was also present in the female-only subgroup. Together with results from protein structural modeling, this suggests the existence of more complex pathomechanisms of TLR7 variants, beyond X-linked recessive loss of function. The analyses also generated statistical evidence that rare variants in three additional genes of the interferon signaling pathway, specifically IFNAR2, IFIH1, and TBK1, contribute to severe COVID-19, though these findings require further follow-up.
TLR7 is a cytosolic receptor that recognizes single-stranded RNA, and is a central component of the interferon signaling pathway during SARS-CoV-2 host defense.23 Multiple lines of evidence suggest that deleterious variants within TLR7 play a causal role in severe COVID-19,11,12,18,19,20,21 and this eventually resulted in recognition of TLR7 deficiency as an inborn error of immunity27 [OMIM: 301051]. Research suggests that TLR7 deficiency is more frequent in younger (<60 years) patients with severe COVID-19,21 which is consistent with the hypothesis that the contribution of host genetic factors is larger in young individuals,49 as has been demonstrated for other risk loci for severe COVID-19, e.g., at the key GWAS locus 3p21.31.50 To refine the subgroup in which severe COVID-19 secondary to TLR7 deficiency is prevalent, the present analyses extended the list of non-genetic risk factors beyond age by including available data on diabetes, hypertension, and CAD. The largest effect size for the association of rare deleterious TLR7 variants with severe COVID-19 was observed in young individuals with none of the aforementioned risk factors. Specifically, in these cases, an approximately 10-fold increase in the proportion of individuals carrying variants that were predicted to be deleterious was observed (2.4% vs. 0.24% in population-based controls, C10+M1, MAF <0.1%). Variant classification via 3D protein structural analysis (3D-P, MAF <0.1%) further refined this overrepresentation to 1.85% in young individuals with severe COVID-19 and none of the listed risk factors, compared with 0.07% in population-based controls.
In the female-only subgroup, the present analyses identified a strong enrichment of rare TLR7 variants that were predicted to be damaging. While such an enrichment has been observed in previous independent cohorts,15,21 the underlying mechanisms were not explored. The proposed X-linked recessive model19 suggests that TLR7 deficiency would be restricted to females with biallelic deleterious mutations. While we identified one female with presumed compound heterozygosity, this individual was not among the cases of the POPlowrisk analysis and did not contribute to the observed burden. We therefore suggest the existence of an additional pathomechanism in heterozygous females, which may be dominant-negative in nature. We hypothesized that an affected TLR7 monomer would interfere with dimerization, thereby reducing TLR7 function by >50%. In support of this, an overrepresentation of TLR7 missense variants that surrounded the dimerization interface in 3D space was observed in female cases. This observation adds to accumulating evidence for an allelic series underlying TLR7 dosage and its relevance to human immune disorders. The most recent support for this was provided by reports of hypermorphic or gain-of-function mutations in TLR7, which underlie monogenic forms of systemic lupus erythematosus51 [OMIM: 301080]. However, we were unable to obtain additional confirmation from the GenOMICC cohort due to power limitations, such as the very low number of variant observations and the differing cohort characteristics, including recruitment criteria. Future functional in vitro investigation of the pathogenic variants that were found in the present female cases are required to confirm our hypothesis.
The present analyses also identified a missense TLR7 variant (rs202129610, p.D332G) that was specific to the Spanish subcohort. This variant, which has in vitro evidence for deleteriousness,19 was observed in three of 764 severe COVID-19 cases from Spain (MAF = 0.33%), including two out of 147 young hospitalized individuals with no additional risk factors (MAF = 1.0%). This is substantially higher than the allele frequency observed in the present Spanish controls (MAF = 0.038%), as well as estimates from the Latin American population groups from the gnomAD data v3.1.2 (0.019%).
Besides the results for TLR7, the present analyses generated several other interesting findings that require replication in larger cohorts. Specifically, associations with severe COVID-19 were found for IFNAR2 and IFIH1 in the rare variant collapsing analysis and for a rare missense TBK1 variant in the single-variant analysis. All of the three genes are involved in the interferon signaling pathway,23 and prior evidence for involvement in severe COVID-19 has been presented.13,52,53 The observed rare TBK1 missense variant (p.Arg358His) was found in two of 378 young cases with no reported risk factors and only one of 5,347 controls. Although statistical evidence for this variant was not robust to multiple testing in our study alone, its independent replication in the Regeneron dataset adds to the prior finding of a rare deleterious TBK1 variant in a child with severe COVID-19.13 Furthermore, our observation of an enrichment of rare variants in the broader group of immunodeficiency genes, even after the exclusion of TLR7, suggests that this set of genes is likely to harbor a substantial proportion of the rare variant risk for severe COVID-19.
While our results contribute to ongoing work into the role of rare variants within the overall host genetic architecture of severe COVID-19, the present study had some inherent limitations. First, the candidate gene approach, which was selected due to a lack of informed consent for more systematic ES/GS analyses, limited the number of analyzed genes to 52. This prevented identification of additional risk genes, and also poses challenges regarding population substructure that might cause confounding in rare variant studies.54 To address the latter, we took advantage of the availability of prior array-based genotypes,25,26 which decreased the risk of false-positive findings due to population stratification. Second, gene selection was performed in August 2020, and thus subsequently reported risk genes were not examined, e.g., those located at loci that have been reported in recent global GWAS.7,10 Third, comorbidity data were limited, and did not include the now well-established risk factor increased weight—usually measured as body mass index (BMI)—which is one of the strongest clinical predictors of severe COVID-19.3 However, CAD, diabetes, and hypertension are all correlated with BMI, which suggests that the present analyses captured this effect at least in part. Of note, following initial evidence on hypertension being an independent risk factor for severe COVID-19,2 subsequent studies have reported ambiguous results.55 Given that individual array-based genotypes are available for the individuals included in the present study, future refinement analyses might include the evaluation of genetically mediated obesity via the integration of polygenic risk scores. Finally, in the present analysis, the selection of variants with a deleterious effect on protein function was mainly based on computational prediction tools, since (with the exception of some variants within TLR7) experimental data on genetic variants are limited. Particularly for missense variants, computational prediction tools are imperfect, and misclassification probably decreased the power of the gene-based collapsing analyses. However, a tailored, molecular modeling approach for missense variants within TLR7 was used in order to fine-tune the statistical analyses and led to increased effect size estimates. In the future, new approaches, such as novel computational prediction tools that build more strongly on protein structural information,56,57,58 and data from deep mutational scanning experiments, could improve statistical power, and enhance the information content of the present data.
Despite the residual open questions, our stratified analysis approach refined the association between rare deleterious TLR7 variants and severe COVID-19. We suggest a candidate pathomechanism in females, which was identified on the basis of the integration of cohort-level sequencing data and information on protein structure.
Data and code availability
Individual-level data, including raw sequencing data and genotypes, are unavailable for sharing due to consent restrictions. Single-variant summary statistics (MAF >0.01%) and the results of the burden analyses are made available at Zenodo (https://doi.org/10.5281/zenodo.11148109). Code used for the analyses in the study is openly available and referenced throughout the paper.
Acknowledgments
We thank Julia Fazaal, Anna Carreras, Alessio Aghemo, Antonio Voza, and Maurizio Cecconi (for laboratory and clinical support); Beatriz Cortés (for data transfer support); and Alberto Mantovani and Stefano Duga (for scientific input). We thank the staff of the Basque Biobank in Spain, members of the COVICAT study group, the staff of GCAT|Genomes for Life, and the Baillie Gifford/Baillie Gifford Science Pandemic Hub (University of Edinburgh). This research received support from the Solve-RD project (to A.H.; funded from the European Union’s Horizon 2020 research and innovation program (no. 779257)), the BONFOR program of the Medical Faculty, University of Bonn (Gerok stipend to A.S., O-149.0134), the Fondazione IRCCS Ca' Granda "FoGS 2021" genomic study (to L.V.C.V.) and the Banca Intesa San Paolo. G.R. was supported by the Europees Fonds voor Regionale Ontwikkeling (to R.A., EFRO, R0005582). J.K.B. gratefully acknowledges funding support from a Wellcome Trust Senior Research Fellowship (223164/Z/21/Z), UKRI grants MC_PC_20004, MC_PC_19025, MC_PC_1905, MRNO2995X/1, and MC_PC_20029, Sepsis Research (Fiona Elizabeth Agnew Trust), and a BBSRC Institute Strategic Program Grant to the Roslin Institute (BB/P013732/1, BB/P013759/1). The study was partially funded by the Cariplo Foundation in Milan. It received support from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through 286/2020B01 (428994620), LU1944-3/1, and infrastructure support from the DFG Clusters of Excellence 2167 “PrecisionMedicine in Chronic Inflammation (PMI)” (EXC 2167-390884018) and 2151 “ImmunoSensation” (390873048). Sequencing was performed at the West German Genome Center (WGGC; INST 216/981-1) and the NGS Core Facility Bonn.
Author contributions
Study conceptualisation and design: J.Bo., C.I.v.d.M., A.F., A.H., A.S., K.U.L.; Sample and data acquisition: R.A., B.-S.L., L.V.C.V., R.d.C., L.B., A.J., J.K.B., S.May, A.A., J.M.B., J.Ba., N.B., P.B., M.B., J.F., S.Mar., D.P., L.R., N.S., A.F., D.E., A.S., K.U.L.; Analysis and Interpretation: J.Bo., C.I.v.d.M., G.R., E.C., E.P.-C., B.Z., J.H., K.R., A.H., A.S., K.U.L.; Manuscript writing: J.Bo., A.S., K.U.L., with contributions from C.I.v.d.M., G.R., R.A., A.H.,; Coordination and funding acquisition: J.Bo., R.A., J.L.S., O.R., K.U.L.; All authors reviewed the final manuscript.
Declaration of interests
K.U.L. is a co-founder of LAMPseq Diagnostics GmbH.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100323.
Contributor Information
Kerstin U. Ludwig, Email: kerstin.ludwig@uni-bonn.de.
The Spanish/Italian Severe COVID-19 Sequencing group:
Valeria Rimoldi, Elvezia M. Paraboschi, Alessandra Bandera, Flora Peyvandi, Giacomo Grasselli, Francesco Blasi, Francesco Malvestiti, Serena Pelusi, Cristiana Bianco, Lorenzo Miano, Angela Lombardi, Pietro Invernizzi, Alessio Gerussi, Giuseppe Citerio, Andrea Biondi, Maria Grazia Valsecchi, Marina Elena Cazzaniga, Giuseppe Foti, Ilaria Beretta, Mariella D'Angiò, Laura Rachele Bettini, Xavier Farré, Susana Iraola-Guzmán, Manolis Kogevinas, Gemma Castaño-Vinyals, Koldo Garcia-Etxebarria, Beatriz Nafria, Mauro D'Amato, and Adriana Palom
GenOMICC Investigators:
Colin Begg, Sara Clohisey, Charles Hinds, Peter Horby, Julian Knight, Lowell Ling, David Maslove, Danny McAuley, Johnny Millar, Hugh Montgomery, Alistair Nichol, Peter J.M. Openshaw, Alexandre C. Pereira, Chris P. Ponting, Kathy Rowan, Malcolm G. Semple, Manu Shankar-Hari, Charlotte Summers, Timothy Walsh, J. Kenneth Baillie, Latha Aravindan, Ruth Armstrong, Heather Biggs, Ceilia Boz, Adam Brown, Richard Clark, Sara Clohisey, Audrey Coutts, Judy Coyle, Louise Cullum, Sukamal Das, Nicky Day, Lorna Donnelly, Esther Duncan, Angie Fawkes, Paul Fineran, Max Head Fourman, Anita Furlong, James Furniss, Bernadette Gallagher, Tammy Gilchrist, Ailsa Golightly, Fiona Griffiths, Katarzyna Hafezi, Debbie Hamilton, Ross Hendry, Andy Law, Dawn Law, Rachel Law, Sarah Law, Rebecca Lidstone-Scott, Louise Macgillivray, Alan Maclean, Hanning Mal, Sarah McCafferty, Ellie Mcmaster, Jen Meikle, Shona C. Moore, Kirstie Morrice, Lee Murphy, Sheena Murphy, Mybaya Hellen, Wilna Oosthuyzen, Chenqing Zheng, Jiantao Chen, Nick Parkinson, Trevor Paterson, Katherine Schon, Andrew Stenhouse, Mihaela Das, Maaike Swets, Helen Szoor-McElhinney, Filip Taneski, Lance Turtle, Tony Wackett, Mairi Ward, Jane Weaver, Nicola Wrobel, Marie Zechner, Mybaya Hellen, Gill Arbane, Aneta Bociek, Sara Campos, Neus Grau, Tim Owen Jones, Rosario Lim, Martina Marotti, Marlies Ostermann, Manu Shankar-Hari, Christopher Whitton, Zoe Alldis, Raine Astin-Chamberlain, Fatima Bibi, Jack Biddle, Sarah Blow, Matthew Bolton, Catherine Borra, Ruth Bowles, Maudrian Burton, Yasmin Choudhury, David Collier, Amber Cox, Amy Easthope, Patrizia Ebano, Stavros Fotiadis, Jana Gurasashvili, Rosslyn Halls, Pippa Hartridge, Delordson Kallon, Jamila Kassam, Ivone Lancoma-Malcolm, Maninderpal Matharu, Peter May, Oliver Mitchelmore, Tabitha Newman, Mital Patel, Jane Pheby, Irene Pinzuti, Zoe Prime, Oleksandra Prysyazhna, Julian Shiel, Melanie Taylor, Carey Tierney, Suzanne Wood, Anne Zak, Olivier Zongo, Stephen Bonner, Keith Hugill, Jessica Jones, Steven Liggett, Evie Headlam, Nageswar Bandla, Minnie Gellamucho, Michelle Davies, Christopher Thompson, Marwa Abdelrazik, Dhanalakshmi Bakthavatsalam, Munzir Elhassan, Arunkumar Ganesan, Anne Haldeos, Jeronimo Moreno-Cuesta, Dharam Purohit, Rachel Vincent, Kugan Xavier, Kumar Rohit, Frater Alasdair, Malik Saleem, Carter David, Jenkins Samuel, Zoe Lamond, Wall Alanna, Jaime Fernandez-Roman, David O. Hamilton, Emily Johnson, Brian Johnston, Maria Lopez Martinez, Suleman Mulla, David Shaw, Alicia A.C. Waite, Victoria Waugh, Ingeborg D. Welters, Karen Williams, Anna Cavazza, Maeve Cockrell, Eleanor Corcoran, Maria Depante, Clare Finney, Ellen Jerome, Mark McPhail, Monalisa Nayak, Harriet Noble, Kevin O'Reilly, Evita Pappa, Rohit Saha, Sian Saha, John Smith, Abigail Knighton, David Antcliffe, Dorota Banach, Stephen Brett, Phoebe Coghlan, Ziortza Fernandez, Anthony Gordon, Roceld Rojo, Sonia Sousa Arias, Maie Templeton, Megan Meredith, Lucy Morris, Lucy Ryan, Amy Clark, Julia Sampson, Cecilia Peters, Martin Dent, Margaret Langley, Saima Ashraf, Shuying Wei, Angela Andrew, Archana Bashyal, Neil Davidson, Paula Hutton, Stuart McKechnie, Jean Wilson, David Baptista, Rebecca Crowe, Rita Fernandes, Rosaleen Herdman-Grant, Anna Joseph, Denise O'Connor, Meryem Allen, Adam Loveridge, India McKenley, Eriko Morino, Andres Naranjo, Richard Simms, Kathryn Sollesta, Andrew Swain, Harish Venkatesh, Jacyntha Khera, Jonathan Fox, Gillian Andrew, J. Kenneth Baillie, Lucy Barclay, Marie Callaghan, Rachael Campbell, Sarah Clark, Dave Hope, Lucy Marshall, Corrienne McCulloch, Kate Briton, Jo Singleton, Sohphie Birch, Lutece Brimfield, Zoe Daly, David Pogson, Steve Rose, Ceri Battle, Elaine Brinkworth, Rachel Harford, Carl Murphy, Luke Newey, Tabitha Rees, Marie Williams, Sophie Arnold, Petra Polgarova, Katerina Stroud, Charlotte Summers, Eoghan Meaney, Megan Jones, Anthony Ng, Shruti Agrawal, Nazima Pathan, Deborah White, Esther Daubney, Kay Elston, Lina Grauslyte, Musarat Hussain, Mandeep Phull, Tatiana Pogreban, Lace Rosaroso, Erika Salciute, George Franke, Joanna Wong, Aparna George, Laura Ortiz-Ruiz de Gordoa, Emily Peasgood, Claire Phillips, Laura Ortiz-Ruiz de Gordoa, Emily Peasgood, Claire Phillips, Michelle Bates, Jo Dasgin, Jaspret Gill, Annette Nilsson, James Scriven, Carlos Castro Delgado, Deborah Dawson, Lijun Ding, Georgia Durrant, Obiageri Ezeobu, Sarah Farnell-Ward, Abiola Harrison, Rebecca Kanu, Susannah Leaver, Elena Maccacari, Soumendu Manna, Romina Pepermans Saluzzio, Joana Queiroz, Tinashe Samakomva, Christine Sicat, Joana Texeira, Edna Fernandes Da Gloria, Ana Lisboa, John Rawlins, Jisha Mathew, Ashley Kinch, William James Hurt, Nirav Shah, Victoria Clark, Maria Thanasi, Nikki Yun, Kamal Patel, Sara Bennett, Emma Goodwin, Matthew Jackson, Alissa Kent, Clare Tibke, Wiesia Woodyatt, Ahmed Zaki, Azmerelda Abraheem, Peter Bamford, Kathryn Cawley, Charlie Dunmore, Maria Faulkner, Rumanah Girach, Helen Jeffrey, Rhianna Jones, Emily London, Imrun Nagra, Farah Nasir, Hannah Sainsbury, Clare Smedley, Tahera Patel, Matthew Smith, Srikanth Chukkambotla, Aayesha Kazi, Janice Hartley, Joseph Dykes, Muhammad Hijazi, Sarah Keith, Meherunnisa Khan, Janet Ryan-Smith, Philippa Springle, Jacqueline Thomas, Nick Truman, Samuel Saad, Dabheoc Coleman, Christopher Fine, Roseanna Matt, Bethan Gay, Jack Dalziel, Syamlan Ali, Drew Goodchild, Rhiannan Harling, Ravi Bhatterjee, Wendy Goddard, Chloe Davison, Stephen Duberly, Jeanette Hargreaves, Rachel Bolton, Miriam Davey, David Golden, Rebecca Seaman, Shiney Cherian, Sean Cutler, Anne Emma Heron, Anna Roynon-Reed, Tamas Szakmany, Gemma Williams, Owen Richards, Yusuf Cheema, Hollie Brooke, Sarah Buckley, Jose Cebrian Suarez, Ruth Charlesworth, Karen Hansson, John Norris, Alice Poole, Alastair Rose, Rajdeep Sandhu, Brendan Sloan, Elizabeth Smithson, Muthu Thirumaran, Veronica Wagstaff, Alexandra Metcalfe, Mark Brunton, Jess Caterson, Holly Coles, Matthew Frise, Sabi Gurung Rai, Nicola Jacques, Liza Keating, Emma Tilney, Shauna Bartley, Parminder Bhuie, Sian Gibson, Amanda Lyle, Fiona McNeela, Jayachandran Radhakrishnan, Alistair Hughes, Bryan Yates, Jessica Reynolds, Helen Campbell, Maria Thompsom, Steve Dodds, Stacey Duffy, Sandra Greer, Karen Shuker, Ascanio Tridente, Reena Khade, Ashok Sundar, George Tsinaslanidis, Isobel Birkinshaw, Joseph Carter, Kate Howard, Joanne Ingham, Rosie Joy, Harriet Pearson, Samantha Roche, Zoe Scott, Hollie Bancroft, Mary Bellamy, Margaret Carmody, Jacqueline Daglish, Faye Moore, Joanne Rhodes, Mirriam Sangombe, Salma Kadiri, James Scriven, Maria Croft, Ian White, Victoria Frost, Maia Aquino, Rajeev Jha, Vinodh Krishnamurthy, Lai Lim, Rajeev Jha, Vinodh Krishnamurthy, Li Lim, Edward Combes, Teishel Joefield, Sonja Monnery, Valerie Beech, Sallyanne Trotman, Christine Almaden-Boyle, Pauline Austin, Louise Cabrelli, Stephen Cole, Matt Casey, Susan Chapman, Stephen Cole, Clare Whyte, Yolanda Baird, Aaron Butler, Indra Chadbourn, Linda Folkes, Heather Fox, Amy Gardner, Raquel Gomez, Gillian Hobden, Luke Hodgson, Kirsten King, Michael Margarson, Tim Martindale, Emma Meadows, Dana Raynard, Yvette Thirlwall, David Helm, Jordi Margalef, Kristine Criste, Rebecca Cusack, Kim Golder, Hannah Golding, Oliver Jones, Samantha Leggett, Michelle Male, Martyna Marani, Kirsty Prager, Toran Williams, Belinda Roberts, Karen Salmon, Peter Anderson, Katie Archer, Karen Austin, Caroline Davis, Alison Durie, Olivia Kelsall, Jessica Thrush, Charlie Vigurs, Laura Wild, Hannah-Louise Wood, Helen Tranter, Alison Harrison, Nicholas Cowley, Michael McAlindon, Andrew Burtenshaw, Stephen Digby, Emma Low, Aled Morgan, Naiara Cother, Tobias Rankin, Sarah Clayton, Alex McCurdy, Cecilia Ahmed, Balvinder Baines, Sarah Clamp, Julie Colley, Risna Haq, Anne Hayes, Jonathan Hulme, Samia Hussain, Sibet Joseph, Rita Kumar, Zahira Maqsood, Manjit Purewal, Leonie Benham, Zena Bradshaw, Joanna Brown, Melanie Caswell, Jason Cupitt, Sarah Melling, Stephen Preston, Nicola Slawson, Emma Stoddard, Scott Warden, Bethan Deacon, Ceri Lynch, Carla Pothecary, Lisa Roche, Gwenllian Sera Howe, Jayaprakash Singh, Keri Turner, Hannah Ellis, Natalie Stroud, Jodie Hunt, Joy Dearden, Emma Dobson, Andy Drummond, Michelle Mulcahy, Sheila Munt, Grainne O'Connor, Jennifer Philbin, Chloe Rishton, Redmond Tully, Sarah Winnard, Susanne Cathcart, Katharine Duffy, Alex Puxty, Kathryn Puxty, Lynne Turner, Jane Ireland, Gary Semple, Kate Long, Simon Whiteley, Elizabeth Wilby, Bethan Ogg, Amanda Cowton, Andrea Kay, Melanie Kent, Kathryn Potts, Ami Wilkinson, Suzanne Campbell, Ellen Brown, Julie Melville, Jay Naisbitt, Rosane Joseph, Maria Lazo, Olivia Walton, Alan Neal, Peter Alexander, Schvearn Allen, Joanne Bradley-Potts, Craig Brantwood, Jasmine Egan, Timothy Felton, Grace Padden, Luke Ward, Stuart Moss, Susannah Glasgow, Lynn Abel, Michael Brett, Brian Digby, Lisa Gemmell, James Hornsby, Patrick MacGoey, Pauline O'Neil, Richard Price, Natalie Rodden, Kevin Rooney, Radha Sundaram, Nicola Thomson, Bridget Hopkins, James Scriven, Laura Thrasyvoulou, Heather Willis, Martyn Clark, Martina Coulding, Edward Jude, Jacqueline McCormick, Oliver Mercer, Darsh Potla, Hafiz Rehman, Heather Savill, Victoria Turner, Charlotte Downes, Kathleen Holding, Katie Riches, Mary Hilton, Mel Hayman, Deepak Subramanian, Priya Daniel, Oluronke Adanini, Nikhil Bhatia, Maines Msiska, Rebecca Collins, Ian Clement, Bijal Patel, A. Gulati, Carole Hays, K. Webster, Anne Hudson, Andrea Webster, Elaine Stephenson, Louise McCormack, Victoria Slater, Rachel Nixon, Helen Hanson, Maggie Fearby, Sinead Kelly, Victoria Bridgett, Philip Robinson, Julie Camsooksai, Charlotte Humphrey, Sarah Jenkins, Henrik Reschreiter, Beverley Wadams, Yasmin Death, Victoria Bastion, Daphene Clarke, Beena David, Harriet Kent, Rachel Lorusso, Gamu Lubimbi, Sophie Murdoch, Melchizedek Penacerrada, Alastair Thomas, Jennifer Valentine, Ana Vochin, Retno Wulandari, Brice Djeugam, Gillian Bell, Katy English, Amro Katary, Louise Wilcox, Michelle Bruce, Karen Connolly, Tracy Duncan, Helen T-Michael, Gabriella Lindergard, Samuel Hey, Claire Fox, Jordan Alfonso, Laura Jayne Durrans, Jacinta Guerin, Bethan Blackledge, Jade Harris, Martin Hruska, Ayaa Eltayeb, Thomas Lamb, Tracey Hodgkiss, Lisa Cooper, Joanne Rothwell, Angela Allan, Felicity Anderson, Callum Kaye, Jade Liew, Jasmine Medhora, Teresa Scott, Erin Trumper, Adriana Botello, Liana Lankester, Nikitas Nikitas, Colin Wells, Bethan Stowe, Kayleigh Spencer, Craig Brandwood, Lara Smith, Richard Clark, Katie Birchall, Laurel Kolakaluri, Deborah Baines, Anila Sukumaran, Elena Apetri, Cathrine Basikolo, Bethan Blackledge, Laura Catlow, Bethan Charles, Paul Dark, Reece Doonan, Jade Harris, Alice Harvey, Daniel Horner, Karen Knowles, Stephanie Lee, Diane Lomas, Chloe Lyons, Tracy Marsden, Danielle McLaughlan, Liam McMorrow, Jessica Pendlebury, Jane Perez, Maria Poulaka, Nicola Proudfoot, Melanie Slaughter, Kathryn Slevin, Melanie Taylor, Vicky Thomas, Danielle Walker, Angiy Michael, Matthew Collis, Tracey Cosier, Gemma Millen, Neil Richardson, Natasha Schumacher, Heather Weston, James Rand, Nicola Baxter, Steven Henderson, Sophie Kennedy-Hay, Christopher McParland, Laura Rooney, Malcolm Sim, Gordan McCreath, Louise Akeroyd, Shereen Bano, Matt Bromley, Lucy Gurr, Tom Lawton, James Morgan, Kirsten Sellick, Deborah Warren, Brian Wilkinson, Janet McGowan, Camilla Ledgard, Amelia Stacey, Kate Pye, Ruth Bellwood, Michael Bentley, Jeremy Bewley, Zoe Garland, Lisa Grimmer, Bethany Gumbrill, Rebekah Johnson, Katie Sweet, Denise Webster, Georgia Efford, Karen Convery, Deirdre Fottrell-Gould, Lisa Hudig, Jocelyn Keshet-Price, Georgina Randell, Katie Stammers, Maria Bokhari, Vanessa Linnett, Rachael Lucas, Wendy McCormick, Jenny Ritzema, Amanda Sanderson, Helen Wild, Anthony Rostron, Alistair Roy, Lindsey Woods, Sarah Cornell, Fiona Wakinshaw, Kimberley Rogerson, Jordan Jarmain, Robert Parker, Amie Reddy, Ian Turner-Bone, Laura Wilding, Peter Harding, Caroline Abernathy, Louise Foster, Andrew Gratrix, Vicky Martinson, Priyai Parkinson, Elizabeth Stones, Llucia Carbral-Ortega, Georgia Bercades, David Brealey, Ingrid Hass, Niall MacCallum, Gladys Martir, Eamon Raith, Anna Reyes, Deborah Smyth, Letizia Zitter, Sarah Benyon, Suzie Marriott, Linda Park, Samantha Keenan, Elizabeth Gordon, Helen Quinn, Kizzy Baines, Lenka Cagova, Adama Fofano, Lucie Garner, Helen Holcombe, Sue Mepham, Alice Michael Mitchell, Lucy Mwaura, Krithivasan Praman, Alain Vuylsteke, Julie Zamikula, Bally Purewal, Vanessa Rivers, Stephanie Bell, Hayley Blakemore, Borislava Borislavova, Beverley Faulkner, Emma Gendall, Elizabeth Goff, Kati Hayes, Matt Thomas, Ruth Worner, Kerry Smith, Deanna Stephens, Louise Mew, Esther Mwaura, Richard Stewart, Felicity Williams, Lynn Wren, Sara-Beth Sutherland, Emily Bevan, Jane Martin, Dawn Trodd, Geoff Watson, Caroline Wrey Brown, Amy Collins, Waqas Khaliq, Estefania Treus Gude, Olugbenga Akinkugbe, Alasdair Bamford, Emily Beech, Holly Belfield, Michael Bell, Charlene Davies, Gareth A.L. Jones, Tara McHugh, Hamza Meghari, Lauran O'Neill, Mark J. Peters, Samiran Ray, Ana Luisa Tomas, Iona Burn, Geraldine Hambrook, Katarina Manso, Ruth Penn, Pradeep Shanmugasundaram, Julie Tebbutt, Danielle Thornton, Jade Cole, Michelle Davies, Rhys Davies, Donna Duffin, Helen Hill, Ben Player, Emma Thomas, Angharad Williams, Denise Griffin, Nycola Muchenje, Mcdonald Mupudzi, Richard Partridge, Jo-Anna Conyngham, Rachel Thomas, Mary Wright, Maria Alvarez Corral, Reni Jacob, Cathy Jones, Craig Denmade, Sarah Beavis, Katie Dale, Rachel Gascoyne, Joanne Hawes, Kelly Pritchard, Lesley Stevenson, Amanda Whileman, Patricia Doble, Joanne Hutter, Corinne Pawley, Charmaine Shovelton, Marius Vaida, Deborah Butcher, Susie O'Sullivan, Nicola Butterworth-Cowin, Norfaizan Ahmad, Joann Barker, Kris Bauchmuller, Sarah Bird, Kay Cawthron, Kate Harrington, Yvonne Jackson, Faith Kibutu, Becky Lenagh, Shamiso Masuko, Gary H. Mills, Ajay Raithatha, Matthew Wiles, Jayne Willson, Helen Newell, Alison Lye, Lorenza Nwafor, Claire Jarman, Sarah Rowland-Jones, David Foote, Joby Cole, Roger Thompson, James Watson, Lisa Hesseldon, Irene Macharia, Luke Chetam, Jacqui Smith, Amber Ford, Samantha Anderson, Kathryn Birchall, Kay Housley, Sara Walker, Leanne Milner, Helena Hanratty, Helen Trower, Patrick Phillips, Simon Oxspring, Ben Donne, Catherine Jardine, Dewi Williams, Alasdair Hay, Rebecca Flanagan, Gareth Hughes, Scott Latham, Emma McKenna, Jennifer Anderson, Robert Hull, Kat Rhead, Carina Cruz, Natalie Pattison, Rob Charnock, Denise McFarland, Denise Cosgrove, Ashar Ahmed, Anna Morris, Srinivas Jakkula, Asifa Ali, Megan Brady, Sam Dale, Annalisa Dance, Lisa Gledhill, Jill Greig, Kathryn Hanson, Kelly Holdroyd, Marie Home, Diane Kelly, Ross Kitson, Lear Matapure, Deborah Melia, Samantha Mellor, Tonicha Nortcliffe, Jez Pinnell, Matthew Robinson, Lisa Shaw, Ryan Shaw, Lesley Thomis, Alison Wilson, Tracy Wood, Lee-Ann Bayo, Ekta Merwaha, Tahira Ishaq, Sarah Hanley, Bethan Deacon, Meg Hibbert, Carla Pothecary, Dariusz Tetla, Chrsitopher Woodford, Latha Durga, Gareth Kennard-Holden, Debbie Branney, Jordan Frankham, Sally Pitts, Nigel White, Shondipon Laha, Mark Verlander, Alexandra Williams, Abdelhakim Altabaibeh, Ana Alvaro, Kayleigh Gilbert, Louise Ma, Loreta Mostoles, Chetan Parmar, Kathryn Simpson, Champa Jetha, Lauren Booker, Anezka Pratley, Colene Adams, Anita Agasou, Tracie Arden, Amy Bowes, Pauline Boyle, Mandy Beekes, Heather Button, Nigel Capps, Mandy Carnahan, Anne Carter, Danielle Childs, Denise Donaldson, Kelly Hard, Fran Hurford, Yasmin Hussain, Ayesha Javaid, James Jones, Sanal Jose, Michael Leigh, Terry Martin, Helen Millward, Nichola Motherwell, Rachel Rikunenko, Jo Stickley, Julie Summers, Louise Ting, Helen Tivenan, Louise Tonks, Rebecca Wilcox, Maureen Holland, Natalie Keenan, Marc Lyons, Helen Wassall, Chris Marsh, Mervin Mahenthran, Emma Carter, Thomas Kong, Helen Blackman, Ben Creagh-Brown, Sinead Donlon, Natalia Michalak-Glinska, Sheila Mtuwa, Veronika Pristopan, Armorel Salberg, Eleanor Smith, Sarah Stone, Charles Piercy, Jerik Verula, Dorota Burda, Rugia Montaser, Lesley Harden, Irving Mayangao, Cheryl Marriott, Paul Bradley, Celia Harris, Susan Anderson, Eleanor Andrews, Janine Birch, Emma Collins, Kate Hammerton, Ryan O'Leary, Michele Clark, Sarah Purvis, Russell Barber, Claire Hewitt, Annette Hilldrith, Karen Jackson-Lawrence, Sarah Shepardson, Maryanne Wills, Susan Butler, Silvia Tavares, Amy Cunningham, Julia Hindale, Sarwat Arif, Sarah Bean, Karen Burt, Michael Spivey, Carrie Demetriou, Charlotte Eckbad, Sarah Hierons, Lucy Howie, Sarah Mitchard, Lidia Ramos, Alfredo Serrano-Ruiz, Katie White, Fiona Kelly, Daniele Cristiano, Natalie Dormand, Zohreh Farzad, Mahitha Gummadi, Kamal Liyanage, Brijesh Patel, Sara Salmi, Geraldine Sloane, Vicky Thwaites, Mathew Varghese, Anelise C. Zborowski, John Allan, Tim Geary, Gordon Houston, Alistair Meikle, Peter O'Brien, Miranda Forsey, Agilan Kaliappan, Anne Nicholson, Joanne Riches, Mark Vertue, Miranda Forsey, Agilan Kaliappan, Anne Nicholson, Joanne Riches, Mark Vertue, Elizabeth Allan, Kate Darlington, Ffyon Davies, Jack Easton, Sumit Kumar, Richard Lean, Daniel Menzies, Richard Pugh, Xinyi Qiu, Llinos Davies, Hannah Williams, Jeremy Scanlon, Gwyneth Davies, Callum Mackay, Joannne Lewis, Stephanie Rees, Metod Oblak, Monica Popescu, Mini Thankachen, Andrew Higham, Kerry Simpson, Jayne Craig, Rosie Baruah, Sheila Morris, Susie Ferguson, Amy Shepherd, Luke Stephen Prockter Moore, Marcela Paola Vizcaychipi, Laura Gomes de Almeida Martins, Jaime Carungcong, Inthakab Ali Mohamed Ali, Karen Beaumont, Mark Blunt, Zoe Coton, Hollie Curgenven, Mohamed Elsaadany, Kay Fernandes, Sameena Mohamed Ally, Harini Rangarajan, Varun Sarathy, Sivarupan Selvanayagam, Dave Vedage, Matthew White, Mandy Gill, Paul Paul, Valli Ratnam, Sarah Shelton, Inez Wynter, Siobhain Carmody, Valerie Joan Page, Claire Marie Beith, Karen Black, Suzanne Clements, Alan Morrison, Dominic Strachan, Margaret Taylor, Michelle Clarkson, Stuart D'Sylva, Kathryn Norman, Fiona Auld, Joanne Donnachie, Ian Edmond, Lynn Prentice, Nikole Runciman, Dario Salutous, Lesley Symon, Anne Todd, Patricia Turner, Abigail Short, Laura Sweeney, Euan Murdoch, Dhaneesha Senaratne, Michaela Hill, Thogulava Kannan, Wild Laura, Rikki Crawley, Abigail Crew, Mishell Cunningham, Allison Daniels, Laura Harrison, Susan Hope, Ken Inweregbu, Sian Jones, Nicola Lancaster, Jamie Matthews, Alice Nicholson, Gemma Wray, Helen Langton, Rachel Prout, Malcolm Watters, Catherine Novis, Anthony Barron, Ciara Collins, Sundeep Kaul, Heather Passmore, Claire Prendergast, Anna Reed, Paula Rogers, Rajvinder Shokkar, Meriel Woodruff, Hayley Middleton, Oliver Polgar, Claire Nolan, Vicky Thwaites, Kanta Mahay, Dawn Collier, Anil Hormis, Victoria Maynard, Cheryl Graham, Rachel Walker, Victoria Maynard, Ellen Knights, Alicia Price, Alice Thomas, Chris Thorpe, Teresa Behan, Caroline Burnett, Jonathan Hatton, Elaine Heeney, Atideb Mitra, Maria Newton, Rachel Pollard, Rachael Stead, Vishal Amin, Elena Anastasescu, Vikram Anumakonda, Komala Karthik, Rizwana Kausar, Karen Reid, Jacqueline Smith, Janet Imeson-Wood, Denise Skinner, Jane Gaylard, Dee Mullan, Julie Newman, Denise Skinner, Jane Gaylard, Dee Mullan, Julie Newman, Alison Brown, Vikki Crickmore, Gabor Debreceni, Joy Wilkins, Liz Nicol, Waqas Khaliq, Rosie Reece-Anthony, Mark Birt, Alison Ghosh, Emma Williams, Louise Allen, Eva Beranova, Nikki Crisp, Joanne Deery, Tracy Hazelton, Alicia Knight, Carly Price, Sorrell Tilbey, Salah Turki, Sharon Turney, Joshua Cooper, Cheryl Finch, Sarah Liderth, Alison Quinn, Natalia Waddington, Tina Coventry, Susan Fowler, Michael MacMahon, Amanda McGregor, Anne Cowley, Judith Highgate, Anne Cowley, Judith Highgate, Alison Brown, Jane Gregory, Susan O'Connell, Tim Smith, Luigi Barberis, Shameer Gopal, Nichola Harris, Victoria Lake, Stella Metherell, Elizabeth Radford, Amelia Daniel, Joanne Finn, Rajnish Saha, Nikki White, Amy Easthope, Phil Donnison, Fiona Trim, Beena Eapen, Jenny Birch, Laura Bough, Josie Goodsell, Rebecca Tutton, Patricia Williams, Sarah Williams, Barbara Winter-Goodwin, Ailstair Nichol, Kathy Brickell, Michelle Smyth, Lorna Murphy, Samantha Coetzee, Alistair Gales, Igor Otahal, Meena Raj, Craig Sell, Paula Hilltout, Jayne Evitts, Amanda Tyler, Joanne Waldron, Kate Beesley, Sarah Board, Agnieszka Kubisz-Pudelko, Alison Lewis, Jess Perry, Lucy Pippard, Di Wood, Clare Buckley, Peter Barry, Neil Flint, Patel Rekha, Dawn Hales, Lara Bunni, Claire Jennings, Monica Latif, Rebecca Marshall, Gayathri Subramanian, Peter J. McGuigan, Christopher Wasson, Stephanie Finn, Jackie Green, Erin Collins, Bernadette King, Andy Campbell, Sara Smuts, Joseph Duffield, Oliver Smith, Lewis Mallon, Watkins Claire, Liam Botfield, Joanna Butler, Catherine Dexter, Jo Fletcher, Atul Garg, Aditya Kuravi, Poonam Ranga, Emma Virgilio, Zakaula Belagodu, Bridget Fuller, Anca Gherman, Olumide Olufuwa, Remi Paramsothy, Carmel Stuart, Naomi Oakley, Charlotte Kamundi, David Tyl, Katy Collins, Pedro Silva, June Taylor, Laura King, Charlotte Coates, Maria Crowley, Phillipa Wakefield, Jane Beadle, Laura Johnson, Janet Sargeant, Madeleine Anderson, Ailbhe Brady, Rebekah Chan, Jeff Little, Shane McIvor, Helena Prady, Helen Whittle, Bijoy Mathew, Ben Attwood, Penny Parsons, Geraldine Ward, Pamela Bremmer, West Joe, Baird Tracy, Ruddy Jim, Ellie Davies, Lisa Roche, Sonia Sathe, Catherine Dennis, Alastair McGregor, Victoria Parris, Sinduya Srikaran, Anisha Sukha, Rachael Campbell, Noreen Clarke, Jonathan Whiteside, Mairi Mascarenhas, Avril Donaldson, Joanna Matheson, Fiona Barrett, Marianne O'Hara, Laura Okeefe, Clare Bradley, Christine Eastgate-Jackson, Helder Filipe, Daniel Martin, Amitaa Maharajh, Sara Mingo Garcia, Glykeria Pakou, Mark De Neef, Kathy Dent, Elizabeth Horsley, Muhmmad Nauman Akhtar, Sandra Pearson, Dorota Potoczna, Sue Spencer, Melanie Clapham, Rosemary Harper, Una Poultney, Polly Rice, Tim Smith, Rachel Mutch, Luigi Barberis, Lisa Armstrong, Hayley Bates, Emma Dooks, Fiona Farquhar, Brigid Hairsine, Chantal McParland, Sophie Packham, Rehana Bi, Barney Scholefield, Lydia Ashton, Linsha George, Sophie Twiss, David Wright, Manish Chablani, Amy Kirkby, Kimberley Netherton, Kim Davies, Linda O'Brien, Zohra Omar, Igor Otahal, Emma Perkins, Tracy Lewis, Isobel Sutherland, Karen Burns, Andrew Higham, Dr Ben Chandler, Kerry Elliott, Janine Mallinson, Alison Turnbull, Prisca Gondo, Bernard Hadebe, Abdul Kayani, Bridgett Masunda, Taya Anderson, Dan Hawcutt, Laura O'Malley, Laura Rad, Naomi Rogers, Paula Saunderson, Kathryn Sian Allison, Deborah Afolabi, Jennifer Whitbread, Dawn Jones, Rachael Dore, Matthew Halkes, Pauline Mercer, Lorraine Thornton, Joy Dawson, Sweyn Garrioch, Melanie Tolson, Jonathan Aldridge, Ritoo Kapoor, David Loader, Karen Castle, Sally Humphreys, Ruth Tampsett, Katherine Mackintosh, Amanda Ayers, Wendy Harrison, Julie North, Suzanne Allibone, Roman Genetu, Vidya Kasipandian, Amit Patel, Ainhi Mac, Anthony Murphy, Parisa Mahjoob, Roonak Nazari, Lucy Worsley, Andrew Fagan, Thomas Bemand, Ethel Black, Arnold Dela Rosa, Ryan Howle, Shaman Jhanji, Ravishankar Rao Baikady, Kate Colette Tatham, Benjamin Thomas, Dina Bell, Rosalind Boyle, Katie Douglas, Lynn Glass, Emma Lee, Liz Lennon, Austin Rattray, Abigail Taylor, Rachel Anne Hughes, Helen Thomas, Alun Rees, Michaela Duskova, Janet Phipps, Suzanne Brooks, Michelle Edwards, Victoria Parris, Sheena Quaid, Ekaterina Watson, Adam Brayne, Emma Fisher, Jane Hunt, Peter Jackson, Duncan Kaye, Nicholas Love, Juliet Parkin, Victoria Tuckey, Lynne Van Koutrik, Sasha Carter, Benedict Andrew, Louise Findlay, Katie Adams, Jen Service, Alison Williams, Claire Cheyne, Anne Saunderson, Sam Moultrie, Miranda Odam, Kathryn Hall, Isheunesu Mapfunde, Charlotte Willis, Alex Lyon, Chunda Sri-Chandana, Joslan Scherewode, Lorraine Stephenson, Sarah Marsh, David Brealey, John Hardy, Henry Houlden, Eleanor Moncur, Eamon Raith, Ambreen Tariq, Arianna Tucci, Maria Hobrok, Ronda Loosley, Heather McGuinness, Helen Tench, Rebecca Wolf-Roberts, Val Irvine, Benjamin Shelley, Amy Easthope, Claire Gorman, Abhinav Gupta, Elizabeth Timlick, Rebecca Brady, Colin Begg, Barry Milligan, Arianna Bellini, Jade Bryant, Anton Mayer, Amy Pickard, Nicholas Roe, Jason Sowter, Alex Howlett, Katy Fidler, Emma Tagliavini, and Kevin Donnelly
Web resources
Supplemental information
References
- 1.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet Lond. Engl. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P., et al. OpenSAFELY: factors associated with COVID-19 death in 17 million patients. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H.-H., Eto S., Garcia-Prat M., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID-19 Host Genetics Initiative Mapping the human genetic architecture of COVID-19. Nature. 2021;600:472–477. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.COVID-19 Host Genetics Initiative A first update on mapping the human genetic architecture of COVID-19. Nature. 2022;608:E1–E10. doi: 10.1038/s41586-022-04826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanai M., Andrews S.J., Cordioli M., Stevens C., Neale B.M., Daly M., Ganna A., Pathak G.A., Iwasaki A., Karjalainen J., et al. A second update on mapping the human genetic architecture of COVID-19. Nature. 2023;621:E7–E26. doi: 10.1038/s41586-023-06355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 9.Kousathanas A., Pairo-Castineira E., Rawlik K., Stuckey A., Odhams C.A., Walker S., Russell C.D., Malinauskas T., Wu Y., Millar J., et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. 2022;607:97–103. doi: 10.1038/s41586-022-04576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pairo-Castineira E., Rawlik K., Bretherick A.D., Qi T., Wu Y., Nassiri I., McConkey G.A., Zechner M., Klaric L., Griffiths F., et al. GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19. Nature. 2023;617:764–768. doi: 10.1038/s41586-023-06034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M., et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solanich X., Vargas-Parra G., van der Made C.I., Simons A., Schuurs-Hoeijmakers J., Antolí A., Del Valle J., Rocamora-Blanch G., Setién F., Esteller M., et al. Genetic Screening for TLR7 Variants in Young and Previously Healthy Men With Severe COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.719115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt A., Peters S., Knaus A., Sabir H., Hamsen F., Maj C., Fazaal J., Sivalingam S., Savchenko O., Mantri A., et al. TBK1 and TNFRSF13B mutations and an autoinflammatory disease in a child with lethal COVID-19. NPJ Genom. Med. 2021;6:55. doi: 10.1038/s41525-021-00220-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abolhassani H., Landegren N., Bastard P., Materna M., Modaresi M., Du L., Aranda-Guillén M., Sardh F., Zuo F., Zhang P., et al. Inherited IFNAR1 Deficiency in a Child with Both Critical COVID-19 Pneumonia and Multisystem Inflammatory Syndrome. J. Clin. Immunol. 2022;42:471–483. doi: 10.1007/s10875-022-01215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler-Laporte G., Povysil G., Kosmicki J.A., Cirulli E.T., Drivas T., Furini S., Saad C., Schmidt A., Olszewski P., Korotko U., et al. Exome-wide association study to identify rare variants influencing COVID-19 outcomes: Results from the Host Genetics Initiative. PLoS Genet. 2022;18 doi: 10.1371/journal.pgen.1010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosmicki J.A., Horowitz J.E., Banerjee N., Lanche R., Marcketta A., Maxwell E., Bai X., Sun D., Backman J.D., Sharma D., et al. Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am. J. Hum. Genet. 2021;108:1350–1355. doi: 10.1016/j.ajhg.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fallerini C., Daga S., Mantovani S., Benetti E., Picchiotti N., Francisci D., Paciosi F., Schiaroli E., Baldassarri M., Fava F., et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. Elife. 2021;10 doi: 10.7554/eLife.67569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., Zhang P., Meertens L., Bolze A., Materna M., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantovani S., Daga S., Fallerini C., Baldassarri M., Benetti E., Picchiotti N., Fava F., Gallì A., Zibellini S., Bruttini M., et al. Rare variants in Toll-like receptor 7 results in functional impairment and downregulation of cytokine-mediated signaling in COVID-19 patients. Genes Immun. 2022;23:51–56. doi: 10.1038/s41435-021-00157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matuozzo D., Talouarn E., Marchal A., Zhang P., Manry J., Seeleuthner Y., Zhang Y., Bolze A., Chaldebas M., Milisavljevic B., et al. Rare predicted loss-of-function variants of type I IFN immunity genes are associated with life-threatening COVID-19. Genome Med. 2023;15:22. doi: 10.1186/s13073-023-01173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q., Matuozzo D., Le Pen J., Lee D., Moens L., Asano T., Bohlen J., Liu Z., Moncada-Velez M., Kendir-Demirkol Y., et al. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J. Exp. Med. 2022;219 doi: 10.1084/jem.20220131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Made C.I., Netea M.G., van der Veerdonk F.L., Hoischen A. Clinical implications of host genetic variation and susceptibility to severe or critical COVID-19. Genome Med. 2022;14:96. doi: 10.1186/s13073-022-01100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Souyris M., Cenac C., Azar P., Daviaud D., Canivet A., Grunenwald S., Pienkowski C., Chaumeil J., Mejía J.E., Guéry J.-C. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
- 25.Severe Covid-19 GWAS Group. Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degenhardt F., Ellinghaus D., Juzenas S., Lerga-Jaso J., Wendorff M., Maya-Miles D., Uellendahl-Werth F., ElAbd H., Rühlemann M.C., Arora J., et al. Detailed stratified GWAS analysis for severe COVID-19 in four European populations. Hum. Mol. Genet. 2022;31:3945–3966. doi: 10.1093/hmg/ddac158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tangye S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2022;42:1473–1507. doi: 10.1007/s10875-022-01289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiatt J.B., Pritchard C.C., Salipante S.J., O’Roak B.J., Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle E.A., O’Roak B.J., Martin B.K., Kumar A., Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinforma. Oxf. Engl. 2014;30:2670–2672. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis M.P.A., van Dongen S., Abreu-Goodger C., Bartonicek N., Enright A.J. Kraken: A set of tools for quality control and analysis of high-throughput sequence data. Methods San Diego Calif. 2013;63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasimuddin M., Misra S., Li H., Aluru S. 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS) 2019. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems; pp. 314–324. [DOI] [Google Scholar]
- 32.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mbatchou J., Barnard L., Backman J., Marcketta A., Kosmicki J.A., Ziyatdinov A., Benner C., O’Dushlaine C., Barber M., Boutkov B., et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 36.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. Oxf. Engl. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 40.Buß O., Rudat J., Ochsenreither K. FoldX as Protein Engineering Tool: Better Than Random Based Approaches? Comput. Struct. Biotechnol. J. 2018;16:25–33. doi: 10.1016/j.csbj.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parra R.G., Schafer N.P., Radusky L.G., Tsai M.-Y., Guzovsky A.B., Wolynes P.G., Ferreiro D.U. Protein Frustratometer 2: a tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Res. 2016;44:W356–W360. doi: 10.1093/nar/gkw304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai M.-Y., Zheng W., Balamurugan D., Schafer N.P., Kim B.L., Cheung M.S., Wolynes P.G. Electrostatics, structure prediction, and the energy landscapes for protein folding and binding. Protein Sci. 2016;25:255–269. doi: 10.1002/pro.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirulli E.T., Lasseigne B.N., Petrovski S., Sapp P.C., Dion P.A., Leblond C.S., Couthouis J., Lu Y.-F., Wang Q., Krueger B.J., et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantel N., Haenszel W. Statistical Aspects of the Analysis of Data From Retrospective Studies of Disease. J. Natl. Cancer Inst. 1959;22:719–748. doi: 10.1093/jnci/22.4.719. [DOI] [PubMed] [Google Scholar]
- 46.Efthimiou O. Practical guide to the meta-analysis of rare events. Health. 2018;21:72–76. doi: 10.1136/eb-2018-102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo M.H., Francioli L.C., Stenton S.L., Goodrich J.K., Watts N.A., Singer-Berk M., Groopman E., Darnowsky P.W., Solomonson M., Baxter S., et al. Inferring compound heterozygosity from large-scale exome sequencing data. Nat. Genet. 2024;56:152–161. doi: 10.1038/s41588-023-01608-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z., Ohto U., Shibata T., Krayukhina E., Taoka M., Yamauchi Y., Tanji H., Isobe T., Uchiyama S., Miyake K., Shimizu T. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity. 2016;45:737–748. doi: 10.1016/j.immuni.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Cruz R., Diz-de Almeida S., López de Heredia M., Quintela I., Ceballos F.C., Pita G., Lorenzo-Salazar J.M., González-Montelongo R., Gago-Domínguez M., Sevilla Porras M., et al. Novel genes and sex differences in COVID-19 severity. Hum. Mol. Genet. 2022;31:3789–3806. doi: 10.1093/hmg/ddac132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakanishi T., Pigazzini S., Degenhardt F., Cordioli M., Butler-Laporte G., Maya-Miles D., Bujanda L., Bouysran Y., Niemi M.E.K., Palom A., et al. Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. J. Clin. Invest. 2021;131 doi: 10.1172/JCI152386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown G.J., Cañete P.F., Wang H., Medhavy A., Bones J., Roco J.A., He Y., Qin Y., Cappello J., Ellyard J.I., et al. TLR7 gain-of-function genetic variation causes human lupus. Nature. 2022;605:349–356. doi: 10.1038/s41586-022-04642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fricke-Galindo I., Martínez-Morales A., Chávez-Galán L., Ocaña-Guzmán R., Buendía-Roldán I., Pérez-Rubio G., Hernández-Zenteno R.J., Verónica-Aguilar A., Alarcón-Dionet A., Aguilar-Duran H., et al. IFNAR2 relevance in the clinical outcome of individuals with severe COVID-19. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.949413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muñiz-Banciella M.G., Albaiceta G.M., Amado-Rodríguez L., Del Riego E.S., Alonso I.L., López-Martínez C., Martín-Vicente P., García-Clemente M., Hermida-Valverde T., Enríquez-Rodriguez A.I., et al. Age-dependent effect of the IFIH1/MDA5 gene variants on the risk of critical COVID-19. Immunogenetics. 2023;75:91–98. doi: 10.1007/s00251-022-01281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Connor T.D., Kiezun A., Bamshad M., Rich S.S., Smith J.D., Turner E., NHLBIGO Exome Sequencing Project, ESP Population Genetics, Statistical Analysis Working Group. Leal S.M., Akey J.M. Fine-scale patterns of population stratification confound rare variant association tests. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gallo G., Calvez V., Savoia C. Hypertension and COVID-19: Current Evidence and Perspectives. High Blood Press. Cardiovasc. Prev. 2022;29:115–123. doi: 10.1007/s40292-022-00506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt A., Röner S., Mai K., Klinkhammer H., Kircher M., Ludwig K.U. Predicting the pathogenicity of missense variants using features derived from AlphaFold2. Bioinforma. Oxf. Engl. 2023;39:btad280. doi: 10.1093/bioinformatics/btad280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao H., Hamp T., Ede J., Schraiber J.G., McRae J., Singer-Berk M., Yang Y., Dietrich A.S.D., Fiziev P.P., Kuderna L.F.K., et al. The landscape of tolerated genetic variation in humans and primates. Science. 2023;380 doi: 10.1126/science.abn8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng J., Novati G., Pan J., Bycroft C., Žemgulytė A., Applebaum T., Pritzel A., Wong L.H., Zielinski M., Sargeant T., et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science. 2023;381 doi: 10.1126/science.adg7492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual-level data, including raw sequencing data and genotypes, are unavailable for sharing due to consent restrictions. Single-variant summary statistics (MAF >0.01%) and the results of the burden analyses are made available at Zenodo (https://doi.org/10.5281/zenodo.11148109). Code used for the analyses in the study is openly available and referenced throughout the paper.