Abstract
Quiescent adult neural stem cells (NSCs) in the mammalian brain arise from proliferating NSCs during development. Beyond acquisition of quiescence, an adult NSC hallmark, little is known about the process, milestones, and mechanisms underlying the transition of developmental NSCs to an adult NSC state. Here, we performed targeted single-cell RNA-seq analysis to reveal the molecular cascade underlying NSC development in the early postnatal mouse dentate gyrus. We identified two sequential steps, first a transition to quiescence followed by further maturation, each of which involved distinct changes in metabolic gene expression. Direct metabolic analysis uncovered distinct milestones, including an autophagy burst before NSC quiescence acquisition and cellular reactive oxygen species level elevation along NSC maturation. Functionally, autophagy is important for the NSC transition to quiescence during early postnatal development. Together, our study reveals a multi-step process with defined milestones underlying establishment of the adult NSC pool in the mammalian brain.
In brief
Jimenez-Cyrus et al. report the molecular cascade underlying the neural stem cell transition to the quiescent adult state in the early postnatal mouse dentate gyrus. Neural stem cells sequentially progress through multiple states accompanied by transcriptional and metabolic changes. Autophagy functionally promotes the neural stem cell transition to quiescence.
Graphical Abstract
INTRODUCTION
Neural stem cells (NSCs) supply the mammalian brain with new neurons and glia.1 NSCs persist in adulthood in a quiescent state in two niches, the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) in the dentate gyrus (DG) of the hippocampus.2,3 Quiescent NSCs in the adult SGZ can reactivate to generate DG granule neurons that integrate into the existing hippocampal circuitry, contributing to learning and memory and affective cognition.4–7 Conversely, dysregulated adult hippocampal neurogenesis is associated with neurological disorders, such as depression, epilepsy, and Alzheimer’s disease.7–10 The capacity for adult neurogenesis-associated plasticity is ultimately dependent on the source of new neurons, the quiescent adult NSC pool. However, the adult NSC pool is finite and depletes as adulthood progresses, resulting in an age-dependent decline in neurogenesis and cognitive function.11–14 Therefore, the capacity for lifelong neurogenesis-associated plasticity is determined by the initial establishment of the quiescent adult NSC pool during early postnatal development.15,16 Very little is known about how the adult NSC pool is established,17,18 information that will be key to understanding how NSCs persist beyond development to contribute to adult neuroplasticity.
Quiescence, a non-dividing state from which cells can reenter cell cycle, is a defining feature of adult stem cells in many somatic tissues,19,20 and the transition to quiescence is the only known major milestone that marks the shift from a developmental to an adult NSC state in both the SGZ and SVZ.15,16,21,22 Adult SGZ NSCs originate from a Hopx-expressing lineage of NSCs that shift to quiescence during early postnatal development.16 Other properties, such as morphology and localization,16,23–25 cell division and self-renewal properties,26–29 and molecular signatures,16,30,31 also distinguish developmental NSCs from adult NSCs, but the timing of these changes and their contribution to the adult NSC state remain unknown. A detailed analysis during critical developmental stages is required to fill these gaps and reveal mechanisms that drive NSC development into an adult state and establishment of the adult NSC pool. Given that somatic stem cells in many tissues undergo a developmental shift from a proliferating to a quiescent state, knowledge gained from DG NSC development may provide insight into the establishment of adult somatic stem cells in other tissues.19
Distinct metabolic states are associated with stem cell function.32 For example, adult NSCs are generally characterized by low levels of transcription, translation, and ribosome biogenesis and favor glycolysis to oxidative phosphorylation as their primary energy source.19,20,33 In contrast, as adult NSCs differentiate, they exhibit higher levels of transcription, translation, and ribosome biogenesis and favor oxidative phosphorylation.34 Recent studies suggest that certain aspects of cellular metabolism, like the lysosomal pathway and lipogenesis, regulate NSC activation from quiescence.35,36 In addition, fatty acid oxidation and mitochondrial pyruvate metabolism are required to maintain quiescence.37,38 Despite what we have begun to learn about the metabolic control of adult NSCs, little is known about how cellular metabolism may regulate NSC development and the transition into a quiescent adult state.
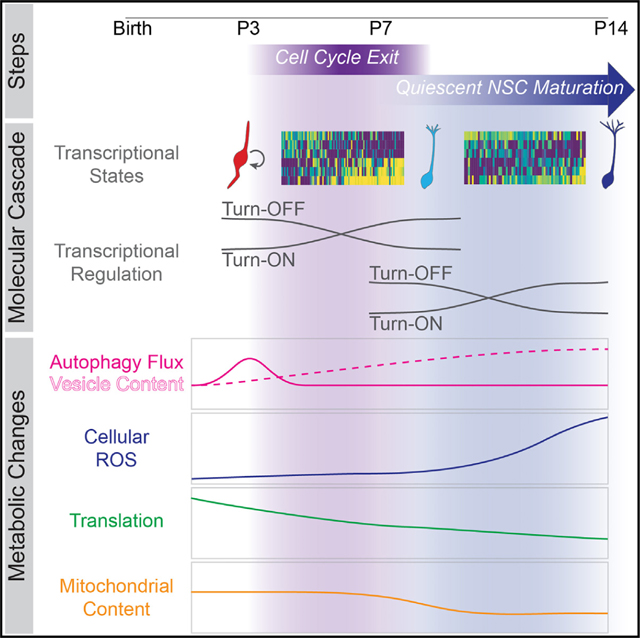
Here we took advantage of the genetic marking of Hopx-expressing DG NSCs in mice16 and applied single-cell RNA-sequencing to construct a molecular cascade of gene expression changes underlying NSC development into an adult state. We found that NSC development into an adult state is a protracted, multi-step process, rather than an abrupt switch that occurs as NSCs enter quiescence. We further identified a sequence of molecular, cellular, and metabolic milestones that mark each developmental step and showed that autophagy promotes the NSC transition to quiescence during early postnatal development. Collectively, our results delineate a previously unstudied developmental process that is critical to establishing the adult NSC pool in the mammalian hippocampus and support the notion that NSCs in the DG undergo continuous development across the lifespan.
RESULTS
NSCs exist in multiple proliferative and quiescent states during early postnatal development
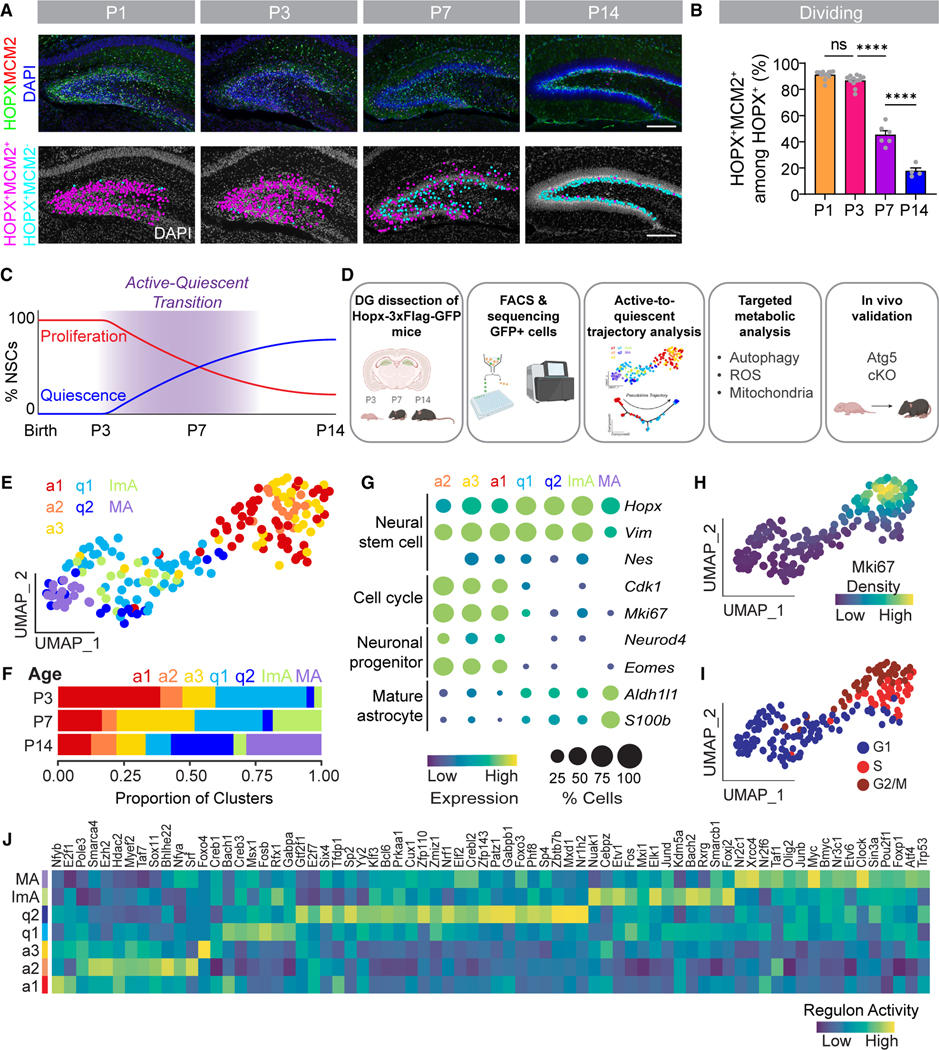
We investigated changes in DG NSC properties at multiple time points across early postnatal development (postnatal day 1 [P1], P3, P7, P14) that encompass the transition to quiescence using the Hopx-3xFlag-GFP mouse line,39 which labels DG NSCs, including a lineage that acquires quiescence to establish the adult SGZ NSC pool.16 First, we performed immunostaining of MCM2, a cell cycle marker expressed during all cell cycle phases (G1–S–G2–M) but rapidly downregulated during cell cycle exit (G0),40,41 to determine when DG NSCs exit the cell cycle during the early postnatal period. We found that the majority of the HOPX+ NSC population in the DG was MCM2+ at P1 and P3, and it was not until P7 that a significant proportion of NSCs was MCM2− and had exited the cell cycle (Figures 1A–1C), confirming our previous finding.16 The majority of NSCs were quiescent and located in the SGZ by P14 (Figure 1A), a stage when the adult neurogenic niche is clearly delineated.25 We wondered if changes in cell cycle phase occupancy between P1 and P3 might precede the NSC shift to quiescence. Using GEMININ to mark the S/G2/M phases of cell cycle,42 we found that the proportion of HOPX+MCM2+ NSCs in the S/G2/M phase increased from P1 to P3, and this increase was sustained at P7 (Figures S1A and S1B). Then, we used histone H3 phosphorylated on serine 10 (PH3) to identify dividing NSCs that were in the G2/M phases of cell cycle. We found that the proportion of HOPX+MCM2+ NSCs in the G2/M phases also increased from P1 to P3, and this increase was sustained at P7 (Figures S1C and S1D). These results showed that DG NSCs increased their occupancy in the S/G2/M phases before exiting cell cycle, suggesting that some changes in NSC properties may not be coupled to the shift to quiescence.
Figure 1. Single-cell RNA-sequencing reveals transcriptomic states of NSCs during the transition to a quiescent adult state.
(A) Sample confocal images of HOPX+MCM2+ dentate gyrus (DG) NSCs at P1, P3, P7, and P14 from Hopx-3xFlag-GFP mice (top panels) and corresponding diagrams of dividing HOPX+MCM2+ NSCs (pink dots) and non-dividing HOPX+ MCM2− NSCs (blue dots). Scale bars, 200 μm.
(B) Quantification of the percentage of MCM2+ cells among all HOPX+ NSCs in the DG. Each dot represents data from one mouse. Values represent mean ± SEM (n = 4–12 mice; ****p < 0.0001; ns: p > 0.05; one-way ANOVA with Tukey’s post hoc test).
(C) Timeline of the DG NSC transition from an active to quiescent state across early postnatal development (postnatal day 3 [P3], P7, and P14).
(D) GFP+ cells were isolated from the DG of Hopx-3xFlag-GFP mice at P3, P7, and P14 for single-cell RNA-sequencing (scRNA-seq; Figures 1, 2, 3, and 4), which informed a targeted metabolic analysis (Figure 5) and an in vivo functional assay to validate autophagy as a regulator of NSC development (Figure 6).
(E) UMAP of all cells (excluding ependymal cells) analyzed by scRNA-seq in (D) separated into 7 clusters (a1, active-1; a2, active-2; a3, active-3; q1, quiescent-1; q2, quiescent-2; ImA, immature astrocyte; MA, mature astrocyte) based on unsupervised Monocle2 clustering and supervised identification of a mature astrocyte cluster.
(F) Bar graph of the distribution of cells in each cluster at different ages.
(G) Bubble plot of sample cell-type-specific marker gene expression in each cluster.
(H) Kernel density estimation plot showing the distribution of Mki67 in each cell on the UMAP (E).
(I) Predicted cell cycle phase (G1, S, and G2/M) in each cell on the UMAP (E).
(J) Heatmap of regulon activity in each cluster calculated by gene regulatory network (GRN) analysis.
See also Figure S1.
Next, we used single-cell RNA-sequencing (scRNA-seq) as a global approach to identify a transcriptome-wide molecular cascade in DG NSCs as they transition into a quiescent state during early postnatal development. We dissected DGs from Hopx-3xFlag-GFP mice at P3, P7, and P14, time points that span the transition into quiescence16 (Figure 1C) and isolated HOPX-GFP+ cells via fluorescence-activated cell sorting (FACS) for scRNA-seq analysis using SMART-seq2 to have better coverage of transcripts43 (Figure 1D; Table S1). After quality control and removal of a small number of ependymal cells, our dataset formed six clusters via unsupervised clustering (Figures S1E and S1F; Table S1). Because Hopx is also expressed in a limited number of DG astrocytes,44 we used Abhd3, Gjb6, and Gabrg1 co-expression31,45 as the criteria to delineate a seventh cluster containing mature astrocytes (Figures 1E, S1G, and S1H). The mature astrocyte cluster (MA) only included cells from P14 (Figure 1F), had high expression of astrocyte marker genes (Figures 1G and S1I), and its transcriptome correlated with mature astrocytes from a previous hippocampal scRNA-seq dataset31 (Figure S1J). Among the seven clusters, we also identified an immature astrocyte cluster (ImA), which was largely composed of cells from P7 (Figure 1F), had moderate expression of astrocyte marker genes (Figure S1I), and its transcriptome also correlated with immature astrocytes from a previous hippocampal scRNA-seq dataset31 (Figure S1J). The remaining five clusters represented different NSC states based on marker expression: three clusters (a1, a2, and a3) represented actively dividing NSC states, and two clusters (q1 and q2) represented quiescent NSC states (Figure 1G). Cells from actively dividing states (Mki67high) grouped together in one corner of the Uniform Manifold Approximation and Projection (UMAP), and cells from quiescent states (Mki67low) grouped together in the opposing corner (Figure 1H). Each dividing cluster and age included cells from multiple phases of the cell cycle (Figures S1K and S1L), and cells in each cell cycle phase grouped together on the UMAP (Figure 1I). Each NSC state had a unique age distribution (Figures 1F and S1M) and distinct gene regulatory network (GRN) activity (Figure 1J). Together, this analysis provided a transcriptional atlas of multiple NSC states that exist during the transition to quiescence.
NSCs continue to mature after quiescence acquisition
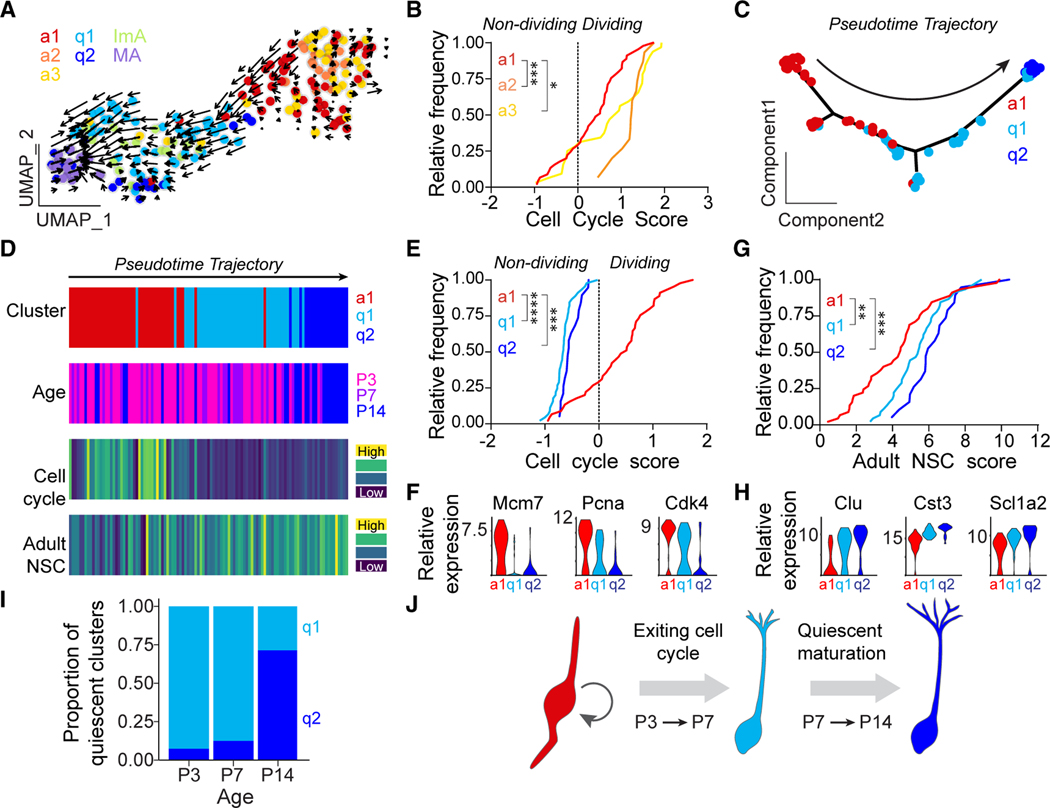
Next, we used RNA velocity,46 a high-dimensional vector that represents the predicted future state of individual cells,46 to determine which cluster(s) of actively dividing NSCs were most likely transitioning into quiescence. We found that large RNA velocities in the a1 group pointed toward the q1 cluster, whereas only small, randomly directed velocities were found in the a2 and a3 dividing NSC clusters, suggesting that a2 and a3 were either in a steady state or their future state was not present in the cells analyzed (Figure 2A). Removing the astroglial clusters (ImA and MA) resulted in similar RNA velocities in the a1 group pointed toward the q1 cluster (Figure S2A). In addition, we found that the a2 and a3 groups had higher cell cycle scores than the a1 group, indicating lower cell cycle gene expression in a1 (Figure 2B). Together, these data suggested that the a1 dividing NSC cluster was most likely transitioning into a quiescent state.
Figure 2. NSCs exit cell cycle and undergo additional maturation before reaching an adult state.
(A) RNA velocity vectors overlaid on the cluster UMAP (Figure 1E).
(B) Distribution curve of cell cycle scores for individual cells of all three active clusters (a1, a2, and a3; *p < 0.05, ***p < 0.001; Kruskal-Wallis rank-sum test with Dunn’s post hoc test).
(C) Principal component analysis (PCA) showing the pseudotime trajectory of cells from clusters a1, q1, and q2 that represents the NSC transition to a quiescent adult state. Arrow indicates direction of pseudotime trajectory.
(D) Heatmaps of cell cycle scores and adult NSC scores of individual cells in pseudotime order with the corresponding cluster and age identification. Cell cycle score was calculated as in (B).
(E) Distribution curve of cell cycle scores for individual cells from clusters a1, q1, and q2 (***p < 0.001, ****p < 0.0001; Kruskal-Wallis rank-sum test with Dunn’s post hoc test).
(F) Violin plots of sample cell cycle gene expression for clusters a1, q1, and q2.
(G) Distribution curve of adult NSC scores for individual cells from clusters a1, q1, and q2 (**p < 0.01, ***p < 0.001; Kruskal-Wallis rank-sum test with Dunn’s post hoc test).
(H) Violin plots of sample adult NSC gene expression for clusters a1, q1, and q2.
(I) Bar plot of the proportion of cells in quiescent NSC clusters (q1 and q2) at each age.
(J) Graphical summary showing that DG NSCs first exit cell cycle between P3 to P7 and then acquire a more mature transcriptomic signature from P7 to P14.
We then focused our analysis on a1, q1, and q2. Large RNA velocities in the q1 cluster pointed toward the q2 cluster, suggesting that cells dynamically moved through q1 into q2 (Figure 2A). We generated an inferred pseudotime trajectory that ordered the cells into sequential transcriptional states beginning with a1, followed by q1 and then q2 (Figures 2C, 2D, and S2B). Following the real-time developmental time course, the first half of the pseudotime was dominated by cells from P3 and P7, while the second half of the pseudotime contained many cells from P14 (Figures 2D and S2C). We found that only cells in a1 had a positive cell cycle score, indicating that they were dividing, while cells in q1 and q2 expressed very low levels of cell cycle genes (Figures 2E and 2F). In contrast, we found that an Adult NSC score associated with quiescent adult NSC gene expression34 gradually increased along the pseudotime (Figure 2D). The Adult NSC score increased from a1 to q1 and from q1 to q2, suggesting that adult NSC gene expression continues to increase after cell cycle exit (Figures 2G and 2H). The quiescent clusters followed developmental age, with q1 dominant at P3/P7 and q2 dominant at P14 (Figure 2I). We also observed progressive acquisition of the adult NSC signature with age (Figure S2D). We wondered if the adult NSC signature strengthens beyond P14, so we integrated our early postnatal dataset with our previously published adult NSC dataset,34 which included 5 clusters that spanned the beginning of the neurogenic lineage from quiescent NSC (S1, S2, and S3) to active NSC/early neuronal progenitor (S4 and S5). We observed overlap between datasets but segregation of dividing clusters and quiescent clusters (Figures S2E and S2F). Our q1 and q2 quiescent clusters were most similar to the S2 cluster in the adult NSC dataset but less similar to the S1 cluster, which anchors the quiescent end of the pseudotime34 (Figure S2G), suggesting that quiescent NSCs continue to mature beyond P14.
Together, these data suggest that acquisition of an adult NSC state is a multi-step developmental process: NSCs first exit the cell cycle and enter an immature quiescent state between P3 and P7, and then they undergo further maturation between P7 and P14 toward an adult NSC state (Figure 2J).
Distinct molecular landscapes characterize each step of NSC development
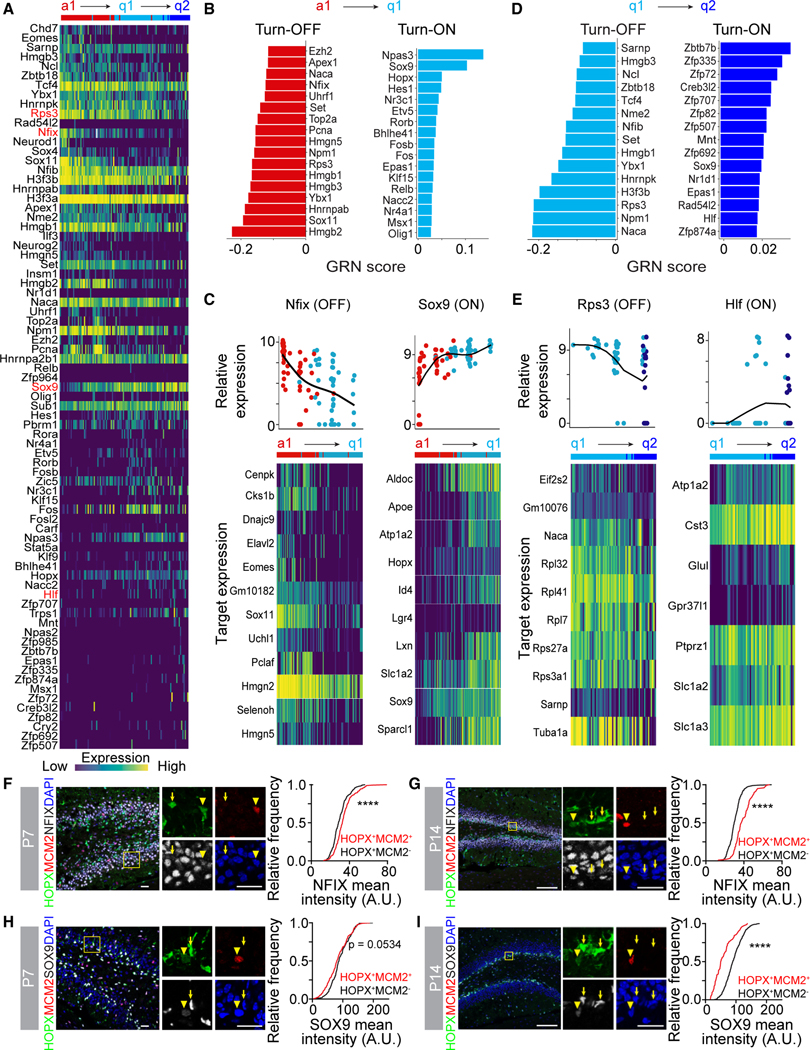
We conducted GRN analysis on each step of the pseudotime (a1 to q1 and q1 to q2) to identify distinct gene networks that characterize each step of NSC development. The GRN score considers changes in expression of the transcriptional regulator and its predicted target genes, collectively called a regulon. We found that transcriptional regulator expression cascaded across the pseudotime (Figure 3A), and we identified regulons that were either increased or decreased across the a1 to q1 step (Figures 3B and 3C) and the q1 to q2 step (Figures 3D and 3E). Interestingly, some regulons only changed across one step, like Nfix and Hlf, while others continuously changed across the entire pseudotime, like Rps3 and Sox9. Immunostaining confirmed that NFIX protein expression was lower in quiescent NSCs (HOPX+MCM2−) than proliferating NSCs (HOPX+MCM2+) at P7 and P14 (Figures 3F and 3G), and that SOX9 protein expression was higher in quiescent NSCs than proliferating NSCs at P7 and P14 (Figures 3H and 3I). Notably, regulons that changed across both steps often had a different composition of target genes across each step, and a target gene’s contribution to the regulon could be different across each step (Figures S3A and S3B). Some of the identified regulons, like Sox11 and Nfix, are known to be required for proper DG morphogenesis during development.47–49 Others, like Npas3, a transcription factor implicated in schizophrenia and intellectual disability,50–52 are known to be required for normal proliferation and neurogenesis in the adult mouse DG.53 We also identified regulons that have no known role in regulating NSCs, like Hlf, which maintains hematopoietic stem cell quiescence.54–56 Collectively, these results revealed distinct gene networks underlying each step of NSC development into an adult state.
Figure 3. Distinct gene regulatory network activity at each step of NSC development.
(A) Heatmap of transcriptional regulator gene expression across the pseudotime trajectory (Figure 2D) identified by GRN analysis. Transcriptional regulators highlighted in red are plotted in (C) and (E), along with their target gene expression.
(B) Bar plot of regulon GRN scores whose activity turned off (left panel) or turned on (right panel) across the a1 to q1 step of the pseudotime trajectory.
(C) Dot plot of transcriptional regulator gene expression across the a1 to q1 step of the pseudotime (Nfix and Sox9, upper panels) and heatmap of corresponding target gene expression (lower panels).
(D) Bar plot of regulon GRN scores whose activity turned off (left panel) or turned on (right panel) across the q1 to q2 step of the pseudotime trajectory.
(E) Dot plot of transcriptional regulator gene expression across the q1 to q2 step of the pseudotime (Rps3 and Hlf, upper panels) and heatmap of corresponding target gene expression (lower panels).
(F–I) Sample confocal images of NFIX (F and G) or SOX9 (H and I) expression in HOPX+ NSCs by immunostaining at P7 (F and H) or P14 (G and I) in the DG (left panels). Boxed area is shown at higher magnification (middle panels; arrowheads = dividing [HOPX+MCM2+] NSCs, arrows = quiescent [HOPX+MCM2−] NSCs). Scale bars: 20 μm. Cumulative frequency distribution plots of NFIX or SOX9 mean intensity (right panels) in dividing NSCs (red line) and quiescent NSCs (black line). (n > 300 cells from 3 mice; ****p < 0.0001; Kolmogorov-Smirnov test).
Metabolism-related gene expression changes with each step of NSC development
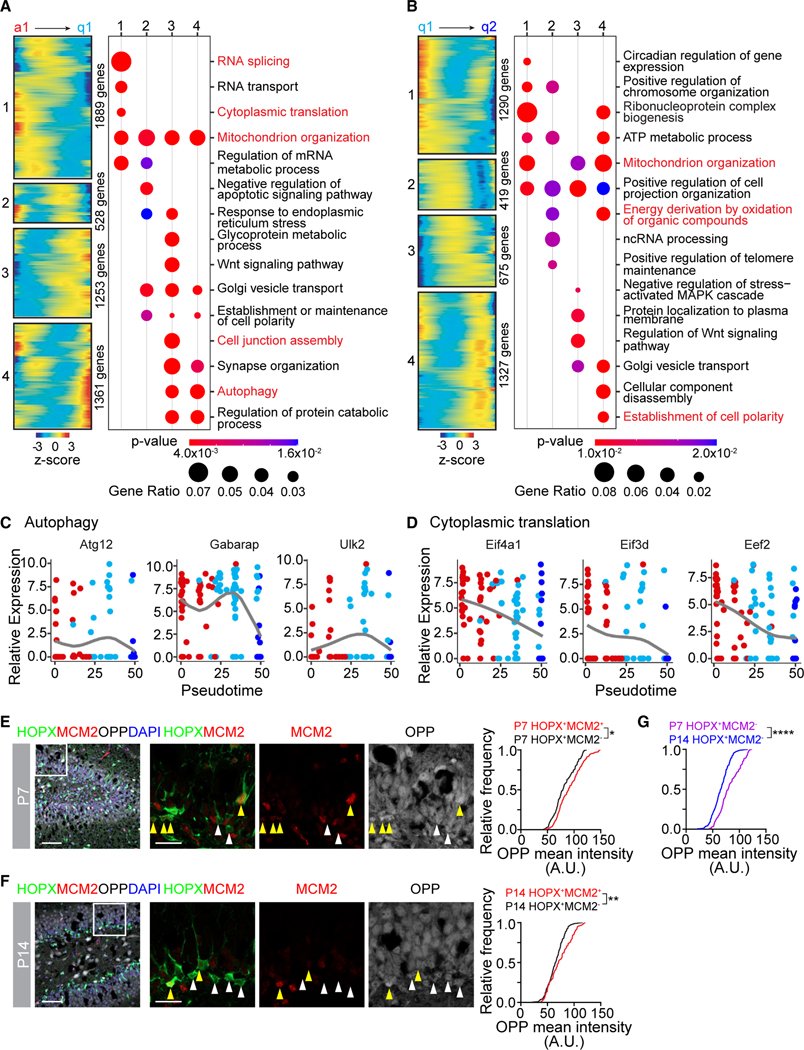
Next, we elucidated the impact of gene expression changes on cell biology in each step of NSC development. We identified kinetic patterns of gene expression across the a1 to q1 and q1 to q2 steps of the pseudotime using CellRouter57 (Figures 1A and 4B; Table S2) and performed Gene Ontology (GO) analysis on each kinetic pattern (Figures 1A and 1B; Table S3). Genes related to transcription and RNA splicing decreased in expression across the a1 to q1 step of quiescence acquisition (Figures 1A and S4A). Interestingly, we found increased expression of genes involved in cell junction assembly, protein catabolism, and autophagy along the a1 to q1 trajectory (Figures 1A and S4A). Genes involved in autophagy were specifically upregulated in the q1 state (Figure 1C). Genes involved in the establishment of cell polarity and metabolic processes, such as ATP metabolic process and energy derivation by oxidation of organic compounds, increased in expression across the q1 to q2 step of NSC maturation (Figures 1B and S4B). Some biological processes, like those related to mitochondrion organization and translation, exhibited gene expression changes across both steps of the trajectory (Figures 1A, 1B, S4A, and S4B). Decreased expression of genes related to translation indicated downregulation of protein synthesis at the cell cycle exit step and the maturation step (Figure 1D), which we confirmed in an independent assay that measured global protein translation using OPP (O-propargyl-puromycin) incorporation (Figures 1E–1G). Together, these results revealed a timeline of cell biology changes during NSC development into an adult state and highlighted a significant metabolic transformation.
Figure 4. Cell biology changes at each step of NSC development.
(A) Heatmaps of gene expression patterns (#1, 2, 3, and 4) across the a1 to q1 step of the pseudotime (left panel) and bubble plots of Gene Ontology (GO) analysis of biological processes analysis of each pattern (right panel). Genes from GO terms in red are plotted in (C) and (D) and Figure S4A.
(B) Heatmaps of gene expression patterns (#1, 2, 3, and 4) across the q1 to q2 step of the pseudotime (left panel) and bubble plots of GO analysis of biological processes analysis of each pattern (right panel). Genes from GO terms in red are plotted in Figure S4B.
(C) Dot plot of autophagy-related gene expression across the pseudotime.
(D) Dot plot of cytoplasmic translation-related gene expression across the pseudotime.
(E and F) Sample confocal images of OPP (O-propargyl-puromycin) staining in dividing (HOPX+MCM2+) and quiescent (HOPX+MCM2−) NSCs at P7 (E) or P14 (F) (left panel). Boxed area is shown at higher magnification (middle panels; yellow arrowheads = dividing [HOPX+MCM2+] NSCs, white arrowheads = quiescent [HOPX+MCM2−] NSCs). Scale bars: 50 μm (left panels) and 20 μm (middle panels). Cumulative frequency distribution plots of OPP mean intensity (right panel) in dividing NSCs (red line) and quiescent NSCs (black line). (n > 300 cells; *p < 0.05; **p < 0.01; Kolmogorov-Smirnov test).
(G) Cumulative frequency distribution plot of OPP mean intensity in quiescent (HOPX+MCM2−) NSCs at P7 (purple line) and P14 (blue line). Same data in (E) and (F) are replotted (****p < 0.0001; Kolmogorov-Smirnov test).
Sequential metabolic milestones mark each step of NSC development
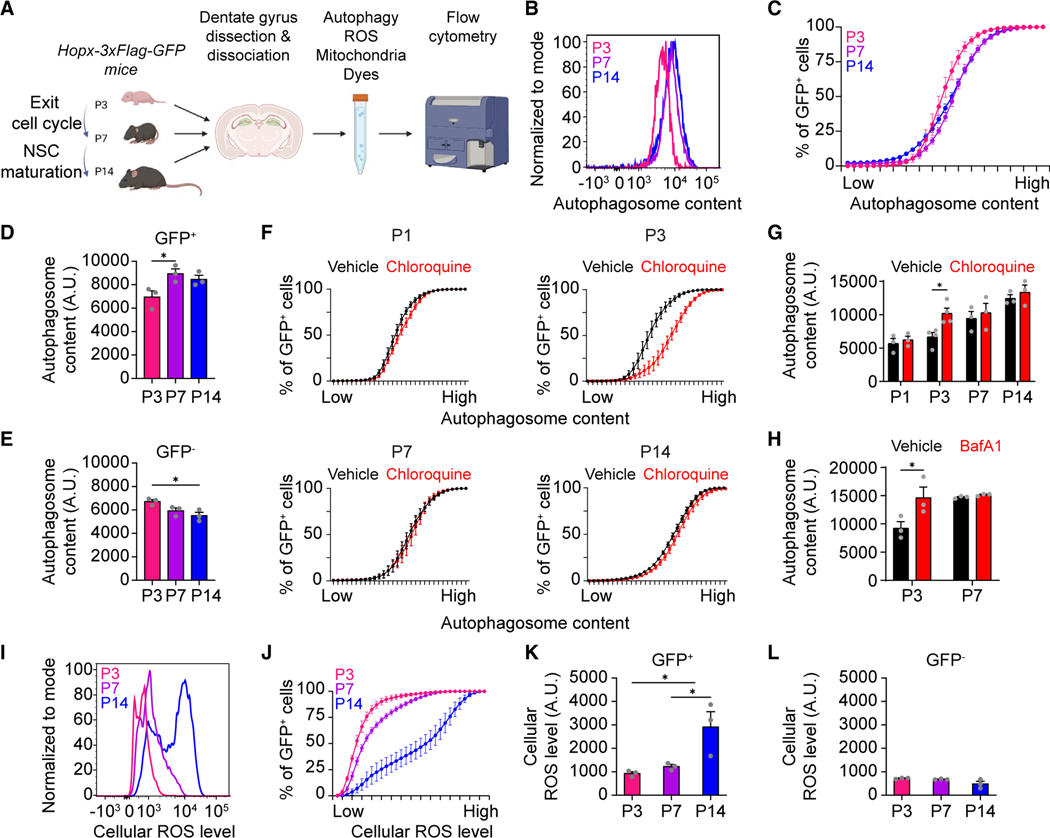
To test whether distinct metabolic changes indeed underlie each step of NSC development at the cellular level, we used cell permeable dyes and flow cytometry to experimentally assess multiple metabolic parameters in individual DG NSCs isolated from Hopx-3xFlag-GFP mice at P3, P7, and P14 (Figures 5A and S5A).
Figure 5. Targeted metabolic analysis reveals distinct changes in autophagy and ROS in NSCs during the transition to a quiescent adult state.
(A) Diagram of metabolic analysis workflow. DG cells were isolated from Hopx-3xFlag-GFP mice at P3, P7, and P14 and loaded with dyes to measure autophagic vesicle content, cellular reactive oxygen species (ROS), and mitochondrial mass in via flow cytometry.
(B–E) Flow cytometry analysis of autophagic vesicle content in GFP+ cells (B–D) and GFP− cells (E) using Autophagy Assay Kit (Red) shown as a histogram plot of individual samples (B), a cumulative distribution plot of all samples (C), and a bar plot of the average of all samples (D and E; individual dots represent data from each experiment). Values represent mean ± SEM (n = 3 experiments; 3 mice were pooled for each experimental sample; *p < 0.05; one-way ANOVA with Tukey’s post hoc test).
(F and G) Flow cytometry analysis of autophagic vesicle content in GFP+ cells after vehicle or chloroquine (10 μM) treatment shown as a cumulative distribution plot of all samples (F) and as a bar plot of the average of all samples (G). Values represent mean ± SEM (n = 3 experiments; 3 mice were pooled for each experimental sample; *p < 0.05; two-way ANOVA with Šídák’s multiple comparisons test).
(H) Bar plot of flow cytometry analysis of autophagic vesicle content in GFP+ cells after vehicle or bafilomycin A (BafA1; 10 nM) treatment. Values represent mean ± SEM (n = 3 experiments; 3 mice were pooled for each experimental sample; *p < 0.05; two-way ANOVA with Šídák’s multiple comparisons test).
(I–L) Flow cytometry analysis of cellular ROS levels in GFP+ cells (I–K) and GFP− cells (L) using dihydroethidium (DHE). Similar as in (B)–(E). Values represent mean ± SEM (n = 3 experiments; 3 mice were pooled for each experimental sample; *p < 0.05; one-way ANOVA with Tukey’s post hoc test).
Also see Figure S5.
Autophagy is a conserved catabolic process that degrades and recycles intracellular components for turnover of cytoplasmic content, but autophagy can also be induced in response to stress to eliminate toxic or damaged components and to serve as a source of energy.58,59 Previous studies suggested that autophagy may be important for DG development,60 as well as for proliferation and survival of adult NSCs and their progeny.61,62 Given our observation that autophagy-related genes were upregulated as NSCs exited the cell cycle (Figures 1A and 1C), we examined whether changes in autophagy might occur in sync with cell cycle exit between P3 and P7. We used the commercial Autophagy Assay Kit (Red),63 which labels autophagosomes and autolysosomes (Figure S5B), and we confirmed the specificity of the dye using cells derived from transgenic mice expressing GFP-LC364–66 (Figure S5C). We found that autophagic vesicle content increased in NSCs (GFP+) from P3 to P7, and this higher level was maintained at P14 and 6 weeks (Figures 5B–5D and S5E). An increase in autophagic vesicle content can correspond to enhanced autophagy, but it can also occur if lysosomal degradation is reduced.67 We measured autophagic flux in NSCs using treatment with chloroquine to distinguish between these two possibilities.68 We found that the NSC autophagic flux was highest at P3 and decreased to low basal levels at P7 and P14 (Figures 5F and 5G). Increased autophagic flux at P3 was confirmed using bafilomycin A167,68 (Figure 5H). To examine whether high autophagic flux might simply be correlated with NSC proliferation, we also measured autophagic flux at P1, another age when the majority of NSCs are dividing (Figures 1A and 1B). To our surprise, we found that NSCs had a low basal level of autophagic flux at P1, similar to P7 and P14 (Figures 5F and 5G). Together, these results demonstrated that NSCs undergo a burst of autophagic flux at P3, right as they prepare to exit cell cycle. Importantly, NSCs exhibited a distinct developmental trajectory of autophagic vesicle content and flux compared to the surrounding cellular environment (GFP−), suggesting that temporal changes in autophagy are specific to NSC development, rather than a tissue-wide phenomenon (Figures 5E, S5D, and S5F).
Redox balance and buildup of cellular reactive oxygen species (ROS) contributes to oxidative stress that can be toxic for the cell, but ROS can also function as a second messenger signal.69–71 High levels of cellular ROS are associated with the adult quiescent NSC state in the DG,72 but cellular ROS levels in developmental NSCs have not been studied. We used the superoxide indicator dihydroethidium (DHE)72 to measure cellular ROS levels in NSCs across early postnatal development (Figure 5A). We found that cellular ROS was relatively low in NSCs (GFP+) at P3 and P7, similar to the surrounding cellular environment (GFP− cells; Figures 5I–5L). However, levels of cellular ROS markedly increased in NSCs at P14 and furthermore at 6 weeks, distinguishing them from the surrounding cellular environment in the DG (Figures 5I–5L, S5G, and S5H). These results demonstrated that the high ROS level associated with the adult quiescent NSC state in the DG is acquired after P7 and is associated with NSC maturation after quiescence.
Mitochondrial function is critical for NSC self-renewal, proliferation, and differentiation,73 but mitochondrial respiration is also a major source of cellular ROS.74 We wondered if the increased cellular ROS associated with the quiescent NSC state was due to changes in mitochondria. We used MitoTracker Deep Red staining75 to show that mitochondrial mass did not significantly differ in DG NSCs between P3 and P7, but it was reduced after P7 at P14 and 6 weeks (Figures S5I–S5K). The changes in mitochondrial mass were anti-correlated to the changes in cellular ROS during this period, suggesting that increased cellular ROS in NSCs was not due to increased mitochondrial respiration. The changes in NSC mitochondrial mass and cellular ROS levels both occurred after P7, suggesting that these metabolic transformations were associated with NSC maturation after quiescence acquisition.
Together, these data demonstrate that distinct metabolic milestones occur sequentially during DG NSC development: first, a burst of autophagy flux corresponds to the initiation of the shift to quiescence, and then increased ROS accompanies NSC maturation into an adult state.
Autophagy promotes the NSC transition to quiescence
Timing is key to cellular mechanisms that promote the developmental transition to quiescence. The transient autophagic burst at P3, when DG NSCs are on the cusp of transitioning to quiescence, suggested that autophagy may be involved in this developmental transition. To test this hypothesis, we deleted autophagy-related gene 5 (Atg5),76 a critical component of the autophagy machinery that decreased in expression across the pseudotime trajectory (Figure 6A). Hopx-CreERT2::mTmG Atg5wt/wt and Hopx-CreERT2::mTmG Atg5Flx/Flx mice were administered tamoxifen at P1, and NSC proliferation was assayed at P7 and P14 (Figure 6B). Atg5 deletion increased the proportion of DG NSCs that were proliferating (HOPX+MCM2+) but decreased the total number of GFP+ NSCs at both P7 and P14 (Figures 6C–6H). In addition, Atg5 deletion increased the proportion of GFP+ cells that were neurons (PROX1+) and the total number of GFP+ neurons at both P7 (Figures S6A–S6C) and P14 (Figures S6D–S6F). Together, these results suggest that autophagy promotes quiescence, and in its absence, NSCs continue to divide and generate neurons at the expense of preserving the NSC pool. Thus, autophagy is an important metabolic regulator of DG NSC quiescence acquisition during early postnatal development.
Figure 6. Blocking autophagy attenuates the NSC transition to quiescence.
(A) Dot plot of Atg5 gene expression across the pseudotime (Figure 2D).
(B) Experimental paradigm to knock out Atg5 in DG NSCs in vivo. Hopx-CreERT2::mTmG Atg5wt/wt and Hopx-CreERT2::mTmG Atg5Flx/Flx mice were administered tamoxifen at P1 and analyzed at P7 and P14.
(C) Sample confocal images of GFP+HOPX+ cells in the DG at P7 in Hopx-CreERT2::mTmG Atg5wt/wt and Hopx-CreERT2::mTmG Atg5Flx/Flx mice (left panels). Boxed area is shown at higher magnification (right panels) to view GFP+HOPX+MCM2+ cells (arrows) and GFP+HOPX+MCM2− cells (arrowheads). Scale bars, 50 μm (left panels) and 25 μm (right panels).
(D) Quantification of the percentage of MCM2+ cells among all GFP+HOPX+ cells in the DG at P7. Each dot represents data from one mouse. Values represent mean ± SEM (n = 4 mice; **p < 0.01; unpaired t test).
(E) Quantification of the number of GFP+HOPX+ cells in the DG at P7. Each dot represents data from one mouse. Values represent mean ± SEM (n = 4 mice).
(F) Sample confocal images of GFP+HOPX+ cells in the DG at P14 in Hopx-CreERT2::mTmG Atg5wt/wt and Hopx-CreERT2::mTmG Atg5Flx/Flx mice (left panels). Boxed area is shown at higher magnification (right panels) to view GFP+HOPX+MCM2+ cells (arrows) and GFP+HOPX+MCM2− cells (arrowheads). Scale bars, 50 μm (left panels) and 25 μm (right panels).
(G) Quantification of the percentage of MCM2+ cells among all GFP+HOPX+ cells in the DG at P14. Each dot represents data from one mouse. Values represent mean ± SEM (n = 4 mice; *p < 0.05; unpaired t test).
(H) Quantification of the number of GFP+HOPX+ cells in the DG at P14. Each dot represents data from one mouse. Values represent mean ± SEM (n = 4 mice; *p < 0.05; unpaired t test).
Also see Figure S6.
DISCUSSION
Our study reveals the stepwise nature of DG NSC development into an adult state and supports the concept of progressive NSC development. The cellular, molecular, and metabolic changes that collectively underlie development into an adult state occur sequentially, before or after quiescence acquisition, rather than a synchronous switch at cell cycle exit. Using our timeline combined with an in vivo functional assay, we identified autophagy as an early mechanism promoting NSC quiescence, consistent with a recent independent study demonstrating a role for Atg7 in the NSC quiescence transition.18 Insights into the changes identified in this study that underlie NSC development will be critical to revealing the brain’s endogenous capacity for neurogenesis-associated plasticity across the lifespan.
Our study attained a high-resolution picture of DG NSC development by analyzing multiple parameters in individual NSCs along a developmentally relevant time course. We analyzed transcriptomic and metabolic changes in a defined population of Hopx+ DG NSCs during the critical period of quiescence acquisition and revealed a timeline of defined milestones that underlie NSC development into an adult state. Previous studies have used unbiased scRNA-seq to show that the molecular profile of NSCs changes across embryonic and postnatal development in the neocortex and hippocampus,31,77 but these studies focused on a single parameter (gene expression) and did not functionally test candidate mechanisms driving development. In contrast, our multimodal approach revealed autophagy as a functional regulator of NSC development into a quiescent state. First, we used our scRNA-seq data to identify two quiescent NSC states that were age segregated to early (P3/7) and late (P14) postnatal development and constructed an inferred developmental trajectory that showed NSCs sequentially progressing from a dividing state to an early quiescent state to a late quiescent state. We identified distinct regulatory landscapes that characterized each step of NSC development, including known transcriptional regulators of DG development47–49 or adult neurogenesis,53,78 as well as others that have no known function in DG NSCs, like Hlf and Npm1, but play a role in hematopoietic stem cell function and leukemia.54,56,79,80 Next, we demonstrated that distinct metabolic milestones characterize each step of NSC development. A burst of autophagy occurs as NSCs prepare to exit the cell cycle, followed by decreased levels of global translation that occur as NSCs exit cell cycle and continue as quiescent NSCs mature. In addition, after quiescence acquisition, mitochondrial content decreases to a plateau, and levels of cellular ROS significantly increase. Finally, we tested the causal role of autophagy in NSC development and showed that blocking autophagy in vivo attenuated the NSC transition into a quiescent adult state.
Our data support a concept of protracted DG NSC development that may extend across the lifespan. The size of the adult DG NSC pool decreases across adulthood,13,14 and DG NSCs become more dormant with advancing age,16,81 both of which contribute to reduced levels of neurogenesis in aging.11,12 More recent studies have identified additional changes in adult NSC properties, such as the capacity for self-renewal, as early as 6 months old in mice, only about 25% of the way through the lifespan.14,82,83 Together with our findings, these results suggest that the properties of NSCs continue to change well after quiescence acquisition and are likely to undergo a lifelong progression.
Though there is clear evidence that hippocampal neurogenesis occurs throughout adulthood in rodents27,84–86 and non-human primates,87–92 there is conflicting evidence for the existence of active neurogenesis in the adult human hippocampus.93–101 Neurogenesis in adulthood is fundamentally dependent on the presence of a long-lived, quiescent NSC population. Accumulating evidence demonstrates that the protracted nature of DG development and the order of developmental milestones is conserved between species, despite differences in the timing.16,84,85,97,102–111 Thus, the relative timing of milestones in rodents could be used as a guide for studying human DG development. For example, the establishment of a long-lived, quiescent NSC population in the mouse DG occurs immediately following peak neurogenesis,107 and peak neurogenesis occurs during mid-gestation in the human DG.97,104,105 Future work investigating human DG NSC development during the fetal period will be essential to understanding the potential for lifelong human neurogenesis.
While this study focused on stem cells in the brain, many organs in the body harbor a population of adult stem cells, and the principles identified here may apply broadly to the developmental establishment of other somatic stem cell populations. Many studies have identified the developmental origins of hematopoietic stem cells,112 muscle stem cells (satellite cells),113–115 and intestinal stem cells,116 but the process(es) leading to the establishment of the adult stem cell state remains largely unclear. Our study presents a methodological outline to approach the study of stem cell development in other systems. An unrealized feature of many somatic stem cell populations is that quiescence acquisition is not coupled to the establishment of the adult stem cell state. For example, SVZ NSCs become quiescent embryonically21,22 but do not acquire an adult molecular signature or localization to the center of a pinwheel structure until after birth.77,117 Similarly, a significant proportion of muscle stem cells become quiescent before they acquire satellite cell characteristics and locate to their adult niche under the basal lamina.118 Hematopoietic stem cells colonize the bone marrow niche as early as E16.5 in mice119 but do not acquire an adult molecular signature, long-term repopulating activity, or quiescence until after birth.120,121 What drives stem cell maturation into an adult state remains largely unknown, but our work suggests that it may be driven by changes in stem cell metabolism.18 Distinct metabolic changes occur as quiescent adult stem cells reactivate, and many of these changes are common across systems.19,33 For example, quiescent adult stem cells have low levels of transcription and translation that get upregulated with activation.122–125 Additionally, quiescent adult stem cells have increased heterochromatin and lower chromatin accessibility, which is reversed upon activation.126–128 Many previous studies have suggested that adult quiescent stem cells rely on glycolysis as their primary energy source but switch to mitochondrial respiration upon activation.129–133 However, recent studies demonstrate a more prominent role for mitochondrial activity during quiescence,37,134 suggesting that there is still a lot to learn about the relationship between metabolism and stem cell function. Future work will benefit from comparisons between systems, given the potential for common mechanisms to establish and maintain adult stem cell pools. Detailed time course analyses across embryonic and postnatal development will be required to better delineate different phases of stem cell development and to uncover both common and tissue-specific mechanisms regulating the establishment of adult stem cell populations.
Limitations of the study
First, this study focused on the intrinsic properties of DG NSCs and did not investigate extrinsic changes in the environment that may influence the establishment of the adult NSC pool. There are undoubtedly changes in the signaling environment during this dynamic period of early postnatal development,30,107 and future studies should investigate how these changes impact NSC development. Second, our study was designed based on previous knowledge that by the end of the second postnatal week the majority (>80%) of NSCs have shifted to quiescence,16 and the SGZ niche resembles its adult form.25 Previous studies have demonstrated changes in NSC behavior across adulthood14,82; therefore, it is likely that additional changes occur to the NSCs after P14. Third, our scRNA-seq data demonstrate the heterogeneity of the Hopx+ population, which could not be controlled for in the rest of our experiments. Future experiments could use index sorting to link transcriptomic and metabolic data at the single-cell level. Finally, though we tested the causality of autophagy in regulating the establishment of the quiescent adult NSC state, we did not test the causality of other milestones, like cellular ROS. Future studies could determine whether other milestones identified here are drivers of NSC development.
In summary, our deliberate and comprehensive investigation of the establishment of the quiescent adult NSC pool in the DG demonstrates that acquisition of the adult NSC state is not simply a switch from proliferation to quiescence but is instead a progressive process. Many somatic stem cell populations undergo a similar developmental process to establish their quiescent adult stem cell pools, and our study identifies a framework that may be universal across systems. Further detailed study of the developmental process that establishes adult stem cell pools will be critical to understanding the body’s capacity for lifelong plasticity and regeneration.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hongjun Song (shongjun@pennmedicine.upenn.edu)
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA-seq data have been deposited at the NCBI GEO: GSE232833. All original code has been deposited at GitHub: https://github.com/dvjim/AtoQ. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
All animal procedures used in this study were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Animals were housed in a 14 h light/10 h dark cycle with food and water ad libitum. Both male and female mice from the same litters were used for all experiments and data were pooled together. Hopx3FlagGFP knock-in mice (Jackson #029271; RRID #IMSR_JAX:029271) harboring a 3xFLAG tagged HOPX and EGFP under the Hopx promoter were used to visualize Hopx expression in the brain and to identify and isolate Hopx-expressing cells from the DG.39 Hopx3FlagGFP transgenic mice used in this study were back-crossed for at least five generations and maintained on a C57BL/6 background. Hopx3FlagGFP/+ genotyped mice were used in all experiments. GFP-LC3 mice (RIKEN BioResource Research Center #RBRC00806; strain name B6.Cg-Tg(CAG-EGFP/LC3)53Nmi/NmiRbrc; GFP-LC3#53; RRID # IMSR_RBRC00806) expressing a fusion cDNA encoding enhan ced GFP jointed at its C terminus to rat microtubule-associated protein 1 light chain 3 beta under the control of the CAG promoter were used to visualize the occurrence of autophagy.65 The GFP-LC3 transgenic mouse line was maintained as heterozygotes bred to C57BL/6. Triple transgenic Hopx-CreERT2::mTmG; Atg5Flx/Flx mice were generated by mating the following three transgenic mouse lines: (1) Hopx-CreERT2 knock-in mice (Jackson #017606; RRID # IMSR_JAX:017606) that harbor a tamoxifen-inducible CreERT2 fusion gene,149 (2) Rosa26flox-mT-stop-flox-mG mice (Jackson #007676; RRID # IMSR_JAX:007676), which harbor loxP sites on either side of a membrane-targeted tdTomato (mT) cassette followed by an STOP sequence, upstream of a membrane-target EGFP (mG) cassette, such that in the presence of Cre the mT cassette is deleted, replaced by mG expression,150 and (3) Atg5Flx/Flx mice (RIKEN BioResource Research Center # RBRC02975; strain name B6.129S-Atg5<tm1Myok>; RRID # IMSR_RBRC02975), in which Exon 3 of the Atg5 gene is flanked by loxP sites.76 Hopx-CreERT2::mTmG; Atg5w/w and Hopx-CreERT2::mTmG; Atg5Flx/Flx mice were generated as littermates and used for experimentation. Transgenic mice were genotyped using primer sets provided by The Jackson Laboratory or RIKEN BioResource Research Center. Genomic DNA was isolated in a solution of 25 mM NaOH and 0.2 mM EDTA at 95°C for 1 h, followed by vortexing and centrifugation.
METHOD DETAILS
Tissue processing and immunohistology
Animals were transcardially perfused with ice-cold DPBS, followed by ice-cold 4% paraformaldehyde (PFA). Brains were fixed for 2 h in 4% PFA at 4°C, and then cryoprotected in 30% sucrose solution in 1x PBS overnight at 4°C. Brains were frozen in Tissue Freezing Medium (GeneralData) and stored at −80°C until coronal brain sections (25 μm) were collected onto Fisherbrand Superfrost Plus Microscope Slides (Fisher Scientific, 12–550-15) using a cryostat (Leica, CM3050S). Slides with brain sections were stored at −20°C until immunohistochemistry was performed.
All immunostaining was performed in sets that included sections from each age being analyzed to prevent the potentially confounding batch effects of doing multiple sets of immunostaining. Antibodies used in this study can be found in the key resources table. Mouse anti-Flag (Sigma, Cat# F1804) was used to identify Hopx3FlagGFP+ cells, denoted in images simply as “HOPX”. Slides with brain sections were washed in TBS with 0.05% Triton X-100 and then underwent antigen retrieval: sections were incubated in 1x Target Retrieval Solution (Agilent Dako) at ~95°C in a steamer for 30 min and cooled to room temperature for 45 min. Slides were then washed in TBS with 0.05% Triton X-100 and incubated in primary antibody solution (3.33% donkey serum and 0.05% Triton X-100 in TBS) overnight at 4°C. Slides were washed in PBS and then incubated in secondary antibody solution (3.33% donkey serum and 0.05% Triton X-100 in TBS) and DAPI nuclear stain (Roche) for 1–2 h at room temperature. Alexa Fluor 488, 555, 647 (Invitrogen) secondary antibodies were used at 1:250 dilutions and DAPI was used at a 1:1000 dilution. After a second set of PBS washes, sections were mounted with ProLong Gold Antifade Mountant (Thermo Fisher Scientific). If GFP immunostaining was performed in conjunction with other antibodies, then GFP primary and secondary antibody steps were completed prior to antigen retrieval. All brain sections from Hopx-CreERT2::mTmG mice underwent antigen retrieval to quench the endogenous mTomato signal.
Confocal microscopy and image processing
Brain sections were imaged using a Zeiss LSM 810 confocal microscope (Carl Zeiss). Either 40X or 63X objectives were used for imaging. For each imaging experiment, the exact same imaging settings (laser power and gain) were used to image sections from each staining set to reduce variability. Images were analyzed using ImageJ software.
Single-cell RNA-sequencing
Dentate gyrus dissection and dissociation
DG was rapidly dissected from fresh Hopx3FlagGFP/+ mouse brain in ice-cold DPBS under a dissecting microscope. Tissue was then dissociated using a MACS Neural Tissue Dissociation Kit with Papain (Miltenyi Biotec, 130–092-628) according to the manufacturer’s guidelines. The single cell suspension was then resuspended in 22% Percoll in Hibernate A (Brain Bits) and spun for 10 min at 700 g at 4°C without brakes. The myelin/debris supernatant was removed and the cell pellet was resuspended in Hibernate A Low Fluorescence (Brain Bits). Cells were incubated with a 1:50,000 dilution of MitoTracker Deep Red FM (Thermo Fisher Scientific) for 2 min at room temperature to label viable cells, and then were washed and put through a cell-strainer. GFP+/− cells were sorted using a BD FACS Aria II SORP cell sorter. Cells were collected at P3 (8 pups pooled), P7 (8 pups pooled), or P14 (8 pups pooled) and sorted into individual wells of a 96-well plate containing 2.85 μL SMART-Seq lysis buffer.
Sequencing library preparation
Library preparation was performed as previously described with minor adjustments for pg concentrations.16 Briefly, mRNA was isolated from single cells using the SMART-seq protocol151 and incubated at 72°C for 3 min in lysis buffer. A custom-designed primer with a 30-deoxythymidine anchor (1 μL of 12 μM) was added and incubated at 72°C for 3 min to anneal the poly-adenylated RNA. First-strand synthesis was performed using a custom-designed TSO oligo by adding 6.05 μL of RT buffer (1 μL SMARTScribe Transcriptase RT, 0.3 μL 200 mM MgCl2, 0.5 μL RNase inhibitor, 1 μL dNTP, 0.25 μL 100 mM DTT, 1 μL of 12 μM TSO) to the lysed cell sample. Cells were incubated at 42°C for 90 min, followed by enzyme inactivation at 70°C for 10 min. RT product was amplified for 20 cycles using the Advantage 2 Polymerase system (2 μL of Advantage 2 Polymerase, 2 μL of dNTPs, 2 μmL 12 μM custom-designed amplifying PCR primer, 29 μL of water) and PCR thermocycling with following parameters: 95°C 1 min, 95°C 15 s, 65°C 30 s, 68°C 6 min, 72°C 10 min with a final extension at 72°C 10 min. PCR products were purified using a 1:1 ratio of SPRI Beads and cDNA concentration was measured by Qubit.
Tagmentation was performed using a homemade Tn5 enzyme-buffer system, which included a Tn5 enzyme at 1:32 dilution and 2XTD buffer (20 mM TAPS-NaOH, 10 mM MgCl2 and 8%PEG-8000).43,152 Each 5 μL tagmentation reaction was incubated at 55°C for 10 min and immediately quenched using 1.25 μL 0.2% SDS. After quenching, Universal Illumina barcodes and 2x KAPA HiFi HotStart ReadyMix were used to amplify the library, bringing the reaction to 50 μL. PCR thermocycling with the following parameters was used: 72°C 5 min, 95°C 1 min, 95°C 20 s, 55°C 30 s, 72°C 30 s with a final extension at 72°C 1 min for X15 cycles. PCR products were purified using a 1:1 ratio of SPRI Beads and cDNA concentration was measured by Qubit.
Sequencing, reads alignment and pre-processing
Samples were then multiplexed at equal nM concentration to the lowest library concentration, and all libraries from each age were pooled 1:1, then diluted to a final concentration of 2 nM and sequenced on an Illumina NextSeq 550 instrument using Illumina 75-cycle High Output Kit v2.5 (20024906). The raw data was converted to “.fastq” files using bcl2fastq (v2.17.1) software. Single-end sequencing reads were pre-processed trimmed of sequencing adaptors and poly-A sequence contaminants using Cutadapt.136 To digitally count at gene-level, the RSEM (v.1.2.28) package135 where fastq files were imported, which outputs STAR (v.2.7.1a)145 summarized estimates of gene-level counts using mouse reference genome assembly (GRCm38 Mouse Gencode release V19). Tximport package (v. 1.18.0)142 was then used to concatenate individualized cell output as a matrix using gene-level RSEM settings. Raw digital expression matrices were generated, and we observed a range of gene expressions in different samples due to variability of sequencing depth.
Quality control, cell clustering and visualization
A raw expression matrix for cells was loaded as log transformed TPM value Seurat objects (v 4.0.2)153 in R (v4.0.3) using the function “CreateSeuratObject”. Summed gene expression count across all cells <200 TPM were discarded, cells with <500 or >7000 genes expressed were discarded, cells with >10% genes mapped to mitochondrial genes were discarded, and the retained cells underwent library size normalization with scuttle (v1.0.4) and SCT transformation with Seurat.147,148,153 Due to the nature of our samples, we did not regress out cycling genes for downstream analysis. To cluster cells, we performed density peak clustering from Monocle (v.2.18.0)138 using the ‘clusterCells’ function with ‘dpFeautres’ default settings. To visualize cells we used principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) with the ‘RunPCA’ and ‘RunUMAP’ functions in Seurat. Three methods were used to determine the optimal range of clusters and number of principal components for downstream analysis: (1) the WSS and ‘elbow’; (2) the ‘Silhouette’ method using ‘approxSilhouette’ from bluster (v1.0.0)143,154; and (3) hierarchical clustering from clustree (v0.5.0).139 The elbow method assessed the total intra-cluster variation by way of within-cluster-sum of squared error as a function of the number of clusters and assessed the percentage of variance explained by successive PCs to select the optimal PC to linearize cell-cell relationships.34 The Silhouette method uses the silhouette coefficients of over a range of k clusters, 2–8, to measure the consistency of similarity of each cell in one cluster to cells in the neighboring clusters and identify the peak as the optimum, k.154 Clustree allows for visual interrogation of over clustering as cluster resolution increases.139 Final evaluation of cluster separation by graph-based clusters was done using pairwise modularity between communities, which observed the total weights of edges between nodes of the same cluster using ‘pairwiseModularity’ function, and cluster purity quantifying the degree to which cells from multiple clusters intermingled in expression space via the ‘neighborPurity’ function from bluster. The resulting cell clusters were projected to the UMAP previously computed for visualizations, while PCA cell embeddings were used for RNA velocity analysis. The stability of cluster identity across P3, P7, and P14 was assessed using Pearson’s correlation via pseudo-bulk analysis, which showed high correlation of cluster signatures across age samples. To remove ependymal cell contamination, we selected out cells with expression of Foxj1. Mature astrocytes were identified in this dataset by using Monocle’s ‘newCellTypeHierarchy’ function and gene expression of Abhd3 > 3, Gjb6 ≥ 1, and Gabrg1 >0. These genes were used to distinguish astrocytes from NSCs because they have previously been identified to be highly expressed in mature astrocytes and lowly expressed in adult DG NSCs.27,34 To benchmark our assigned cell type identities, we conducted a comprehensive analysis that involved a Pearson’s correlation test of our astroglial lineage clusters (ImA and MA). These clusters were compared with astroglial lineage and NSC clusters (Astro-adult, Astro-juv, Immature-Astro, RGL, RGL_young) obtained from the Hochgerner et al. dataset,27 a single-cell RNA-seq dataset of the developing mouse hippocampus. In addition to this developmental dataset, we performed a comparative analysis by juxtaposing our data with an adult DG cell dataset.34 This comparison aimed to evaluate and validate the activity and quiescence of our cells, while also anticipating the future (adult) distribution of q2 and MA clusters. To achieve these objectives, we integrated normalized matrices, using the Hochgerner et al.31 clustering as an anchoring criterion through Seurat’s canonical correlation analysis based integration workflow (‘FindIntegrationAnchors’ and ‘IntegrateData’ functions).155 The resulting manifold, featuring 7 clusters, was employed for downstream analysis. Subsequently, we directly compared this validated manifold with the Shin et al. dataset40 using the same integration methods, ensuring a consistent and reliable comparative assessment across each query dataset. We then conducted a Pearson’s correlation across all groups and visualized the integrated cell distributions using UMAP. This confirmed our q1 and q2 quiescent clusters were most similar to the S2 cluster from Shin et al., but less similar to the S1 cluster which anchors the quiescent end of the pseudotime.40
Single-cell RNA-sequencing analyses
RNA velocity
RNA Velocity (velocyto.R.0.6)46 was used to forecast the future expression state of individual cells. The vector flows, representing the direction and magnitude of changes in gene expression for each cell, were utilized within a manifold constricted from the gene expression data. The velocity magnitude and directions within this manifold were then used to identify clusters serving as either the source or sink of the trajectory. Count estimates of unspliced and spliced mRNA were computed with the following parameters: velocyto run_smartseq2-d $Bamlocation $mm10_rmsk.gtf.gz –o $output, where the repeat masker files were generated from the UCSC browser. The resulting matrix of unspliced, spliced, and spanning estimates were filtered using the default setting and imported into the ‘gene.relative.velocity.estimate’ function using PCA embeddings to calculate cell distance. Default settings were used except for the cell kNN pooling, for which we used a k of 10 and a fit quantile of 0.2. Top genes with >0.6 gamma fit were subsetted to confirm the directionality of trajectory. This generated vectors for each cell, identifying a change in expression in the original cell state and the predicted cell state at future time t identified across the pseudotime trajectory. The rate of transcription was designated by the vector magnitude. Velocity vectors were visualized using UMAP embedding.
Cell scores
A Cell Cycle Score was calculated for each cell by taking the average of the S score and G2/M score assigned using the Seurat ‘Cell-CycleScoring’ function. A higher score indicated higher expression of S or G2/M phase markers, while a lower score indicated that cells were likely not cycling and/or in G1 phase. An Adult NSC Score was calculated for each cell using expression of quiescent adult hippocampal NSC enriched genes identified in a previous single-cell RNA-seq dataset.34 In the previous dataset, adult hippocampal quiescent NSCs and their immediate progeny were aligned in a pseudotime trajectory, and the top 1000 downregulated genes across the pseudotime were identified as quiescent adult hippocampal NSC enriched genes.34 The average expression of these 1000 genes was used to calculate an Adult NSC Score for each cell.
Pseudotime trajectory
To construct the active to quiescent NSC trajectory, we loaded relevant clusters (a1, q1, and q2) of P3, P7, and P14 cells into Monocle using the default ‘dpFeatures’ and ICA DDRTree method along with a census method using uniform option processing for pseudotime projection. The root state was identified as a cluster with steady-state velocities and the state contained the majority of cells with the earliest developmental age for each individual analysis. A single main active to quiescent trajectory was used and divided into two separate trajectory paths (a1-q1 and q1-q2) as no major branching or deviation off the core cellular trajectory from active to quiescence was expected.
Gene regulatory network (GRN) analysis: SCENIC
To identify steady-state regulons within a particular cluster, we used the SCENIC R package (v.1.2.4)146 algorithm, which was run on the RNA slotted Seurat matrix, which included library size normalized data. Default filtering parameters and settings were used to subset the log-transformed expression matrix and build a GRN by first using GENIE3156 to identify potential targets for each transcriptional regulator based on inferred co-expression, which identifies non-linear relationships of genes, formatting the identified targets into co-expression modules. Then, using RcisTarget, over-represented and putative regulons were identified via DNA motifs and pruned using cis-regulatory motif analyses from co-expression modules mm10_refseq-r80_10kb_up_and_down_tss.mc9nr.-feather. Finally, to match cell states and cell types with regulators, network activity in each cell was identified by scoring each regulon in the cell by calculating area under the curve using AUCell. We set seed for GENIE3 for reproducibility as it is based on a Random Forest approach.
CellRouter
For dynamic transcriptional regulator analysis across pseudotime, we applied the CellRouter57 GRN analysis feature using Monocle-imported pseudotime applied to normalized data. We considered genes above the 80% quantile as positively correlated with the trajectory progression and genes below the 10% quantile as anti-correlated with the trajectory progression. These selected genes were then used to create a global GRN by sub-setting identified transcriptional regulators and using a Z score threshold to identify regulatory interactions via the CLR algorithm.157,158 GRN scores ranked transcriptional regulators based on correlation with progression, the correlation of transcriptional regulator predicted targets, the number of targets and the Spearman’s rank correlation.
Kinetic patterns of gene expression
To identify genes with similar expression patterns across each step of the pseudotime, we used CellRouter’s57 ‘smoothDynamics’, which uses loess regression to predict gene expression, and ‘clusterGenesPseudotime’, which uses partitioning around medoid (PAM) clustering. Gene patterns were extracted and visualized with Monocle using ‘plot_pseudotime_heatmap’ function.
Gene ontology (GO) analysis
To map enrichment patterns of gene ontology (GO) terms of biological processes onto the kinetic patterns of gene expression, we applied the “compareCluster” function from clusterProfiler (v. 3.19.1)144,159 with default parameters to the full gene list outputted from the kinetic clustering identified by CellRouter. A full list of GO terms is summarized in Table S3. GO terms with <4 counts were removed and semantic simplification of GO terms was conducted at a 0.7 redundancy rate to identify globally significant terms. A p-value threshold of 0.05 was initially used to identify potentially significant results. Subsequently, the false discovery rate (FDR) was controlled at a q-value threshold of <0.2 to refine the selection of significantly enriched Gene Ontology (GO) terms.
Measuring global translation levels with OPP incorporation
OPP incorporation into ex vivo hippocampal slices was used to measure acute global translation levels in DG NSCs in Hopx3FlagGFP mice at P7 and P14. Ex vivo acute slices were used because OPP did not cross the blood-brain barrier very well, despite reaching peripheral organs, like the liver, when OPP was injected intraperitoneally in vivo (data not shown). All samples (P7 and P14) were processed (dissections, OPP incorporation, staining, imaging) in parallel on the same day to reduce variability. Brains were rapidly dissected from Hopx3FlagGFP/+ mice and sliced coronally into 400 μm thick slices in ice-cold, oxygenated cutting solution (206 mM Sucrose, 2.8 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3 26 mM, 1 mM CaCl2 2H2O, 2 mM MgSO47H2O, 1 mM MgCl26H2O, 0.4 mM Sodium Ascorbate, 10 mM D-Glucose) on a vibratome (Leica VT 1200S) at 0.30 mm/s feed speed and 1.35 mm amplitude adjustment. Brain slices were cut within 20 min of dissection and were transferred into room temperature, oxygenated aCSF (124 mM NaCl, 2.8 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 2 mM CaCl22H2O, 2 mM MgSO47H2O, 0.4 mM Sodium Ascorbate, 10 mM D-Glucose) where they were equilibrated for 1 h. After the initial recovery period, brain slices were transferred to a new holding experimental chamber where they were incubated in 20 μM OPP in oxygenated aCSF for 1 h. Brain slices were immediately fixed in 4% PFA for 15 min and then cryoprotected in 30% sucrose solution in 1x PBS overnight at 4°C. Brain slices were frozen in TFM Tissue Freezing Medium (GeneralData) and stored at −80°C until they were re-sliced and coronal brain sections (10 μm) were collected onto Fisherbrand Superfrost Plus Microscope Slides (Fisher Scientific, 12–550-15) using a cryostat (Leica, CM3050S). Slides with re-sliced brain sections were stored at −20°C until staining was performed.
OPP was detected using the Click-iT Plus OPP Alexa Fluor 647 Protein Synthesis Assay Kit (Invitrogen, C10458) per the manufacturer’s guidelines. Immunohistochemistry was performed immediately following OPP detection as described above, and all steps were performed in the dark to preserve the OPP fluorescent signal. Stained brain sections were imaged using a 40X objective on a Zeiss LSM 810 confocal microscope (Carl Zeiss). For each 400 μm thick brain slice that was re-sliced, 3 single-plane images of the DG were collected from 3 different brain sections. The exact same imaging settings (laser power and gain) were used to image sections stained in parallel to reduce variability.
Primary cortical astrocyte culture
Primary cortical astrocyte cultures were prepared as previously described.160 Cerebral cortices were dissected from brains of GFP-LC3 transgenic66,160 or non-transgenic neonatal mice at P0-P1. Cortical tissue was digested using 0.25% trypsin for 10 min at 37°C, then triturated with a 5-mL pipet to break up large pieces of tissue, and then triturated with a P1000 pipet until a homogeneous suspension was achieved. Cells were passed through a 40-μm pore cell strainer (Falcon Brand Products, 352,340) and plated at a density of 2,000,000–3,000,000 cells per 10-cm dish. Cells were plated and grown in glial media (DMEM [Thermo Fisher Scientific/Gibco, 11965084] supplemented with 10% heat inactivated fetal bovine serum [Hyclone, SH30071.03], 2 mM Glutamax [Thermo Fisher Scientific/Gibco, 35,050–061], 100 U/ml penicillin and 100 μg/mL streptomycin [Thermo Fisher Scientific/Gibco, 15140122]) at 37°C in a 5% CO2 incubator. The day after plating, and every 3–4 days following, glia were fed by replacing the glial media. After reaching 75–90% confluence (~8–10 days after plating), astrocytes were trypsinized and plated for experiments. Prior to media changes, or splitting, the culture dishes were tapped to dislodge any microglia which were then discarded with the media change. Glia were plated at a density of 225,000 cells per 35-mm fluorodish (World Precision Instruments, FD35–100) coated with 500 μg/mL poly-L-lysine (Sigma, P2636) for live-cell imaging. Astrocytes were grown in glial media for 5 DIV at 37°C in a 5% CO2 incubator.
Primary cortical astrocyte cultures were incubated with Autophagy Assay Kit (Red) (1x concentration per manufacturer’s guidelines, Abcam) for 1 h at 37°C before live imaging to detect co-localization of GFP-LC3 and Autophagy Assay Kit (Red) dye. Live cell imaging was performed on a BioVision spinning disk confocal microscope system with a Leica DMi8 inverted widefield microscope, a Yokagawa W1 spinning disk confocal, and a Photometrics Prime 95B scientific complementary metal-oxide-semiconductor camera, and an environmental chamber to maintain temperature at 37°C. Images were acquired with the VisiView software using a 63x/1.4 NA plan apochromat oil-immersion objective and solid-state 488 nm and 561 nm lasers for excitation. Z-stacks spanning the entire depth of the cell were acquired at 0.2 μm sections. Primary cortical astrocyte cultures were imaged immediately following treatment in HibE (Brain Bits, HE-Lf) imaging solution supplemented with 2% B-27, 2 mM Glutamax, 100 U/mL penicillin and 100 μg/mL streptomycin.
Flow cytometry and analysis
Dentate gyrus dissection and dissociation
DG was rapidly dissected from fresh Hopx3FlagGFP/+ mouse brain in ice-cold DPBS under a dissecting microscope. Tissue was then dissociated using a MACS Neural Tissue Dissociation Kit with Papain (Miltenyi Biotec, 130–092-628) according to the manufacturer’s guidelines. Dissociated cells were washed in Hank’s balanced salt solution (HBSS) (PAA; GE Healthcare), the cell pellet was resuspended in 1 mL of growth medium (neurobasal medium + B27 + 1x Glutmax + 1x PenStrep) or HBSS, and the single cell suspension was filtered through a 40 μm cell sieve (Falcon; BD Biosciences).
Metabolic dye labeling
After DG dissection and dissociation, cell suspensions were incubated with Autophagy Assay Kit (Red) (1x concentration per manufacturer’s guidelines, Abcam), MitoTracker Deep Red FM (25 nM, Thermo Fisher Scientific) and dihydroethidium (DHE; 5 μM, Thermo Fisher Scientific), along with the live/dead vitality dye, DAPI (1:2500 dilution, BD Biosciences), for 30 min at 37°C. Post incubation, the cells were centrifuged to remove excess dye, resuspended in PBS and immediately subjected to flow cytometry.
Autophagic flux measurement
Acute treatment with autophagy inhibitors, chloroquine or bafilomycin A167,68, was used to assess autophagic flux ex vivo because chloroquine did not reliably cross the blood-brain barrier (data not shown). After DG dissection and dissociation, cell suspensions were incubated in media (without growth factors) with either (1) saline or chloroquine (10 μM, Sigma-Aldrich), or (2) DMSO or bafilomycin A1(10 nM, Sigma-Aldrich) for 3 h at 37°C. During the last 30 min of incubation, the cells were additionally incubated with the metabolic dyes and DAPI as described above. Post incubation, the cells were centrifuged to remove excess dye, resuspended in PBS and immediately subjected to flow cytometry.
Flow cytometry and data analysis
Flow cytometry72 was conducted on a BD FACSymphony A3 Cell Analyzer (BD Biosciences). Compensation experiments were extensively performed with single-dye staining of each dye and FMO (fluorescence minus one) controls, at all specific ages. Gating was performed to exclude dead cells and doublets (Figure S5A). The levels of the metabolic dyes within GFP+ and GFP− cells were both evaluated. FlowJo (V10.7.1) was used to perform all cytometric analyses.
Tamoxifen injection
A stock solution of 66.67 mg/mL tamoxifen (Sigma, T5648) was prepared in a 5:1 solution of corn oil:ethanol at 37°C with occasional vortexing until dissolved. Tamoxifen was injected intraperitoneally into Hopx-CreERT2::mTmG; Atg5w/w and Hopx-CreERT2::mTmG; Atg5Flx/Flx mice at 333 mg/kg on P1.
QUANTIFICATION AND STATISTICAL ANALYSIS
For quantification of cell cycle, ≥3 tiled images of the entire DG region were imaged per mouse from 4 to 12 mice at P1, P3, P7 and P14. Brain sections of equivalent dorsal-ventral location were quantified for each immunostaining set to control for any differences along the dorsal-ventral axis of the hippocampus. ≥300 GFP+ cells from the entire DG (P1, P3, P7) or from the entire SGZ (P14) were quantified using the Point Tool in ImageJ.
For quantification of NFIX and SOX9, ≥2 images of the entire DG region were imaged per mouse from 3 mice at P7 and P14. Brain sections of equivalent dorsal-ventral locations were quantified for each immunostaining set to control for any differences along the dorsal-ventral axis of the hippocampus. The mean intensity of NFIX or SOX9 was measured in ≥300 GFP+ cells throughout the entire DG region (P7) or throughout the entire the SGZ region (P14). A GFP+ cell was identified, a region of interest was drawn around the nucleus of the GFP+ cell, and the Measure function in ImageJ was used to quantify the mean intensity of staining. Then, MCM2 status was noted for each cell.
For quantification of acute global translation levels, the mean intensity of OPP was measured in ≥300 GFP+ cells throughout the entire DG region (P7) or throughout the entire the SGZ region (P14). A GFP+ cell was identified, a region of interest was drawn around the cell body of the GFP+ cell, and the Measure function in ImageJ was used to quantify the mean intensity of OPP. Then, MCM2 status was noted for each cell.
For quantification of GFP+ cells in Hopx-CreERT2::mTmG; Atg5Flx/Flx and Hopx-CreERT2::mTmG; Atg5w/w DG, samples were blinded to genotype and 3 tiled mages of the entire DG region were imaged per mouse from 4 mice per genotype at P7 and P14. GFP+ cells from the entire DG region (P7) or from the entire SGZ and granule cell regions (P14) were quantified using the Point Tool in ImageJ. GFP+ cells were then assessed for colocalization with other markers, such as HOPX, MCM2 or PROX1. Volumetric analyses were performed by measuring the volume of the entire DG region (P7) or the granule cell layer (P14).
Statistical analysis was performed using GraphPad Prism version 9.00 (GraphPad Software Inc.) or R. Detailed tests are listed in each figure legend.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse anti-Flag | Sigma-Aldrich | Cat# F1804; RRID: AB_262044 |
| Goat anti-Mcm2 | R&D Systems | Cat# AF5778; RRID: AB_2141963 |
| Rabbit anti-Geminin | Proteintech | Cat# 10802–1-AP; RRID: AB_2110945 |
| Rabbit anti-phospho-histone H3 (SER10) | Cell Signaling Technology | Cat# 53348S; RRID: AB_2799431 |
| Rabbit anti-Sox9 | Millipore | Cat# AB5535; RRID: AB_2239761 |
| Rabbit anti-Nfix | Thermo Fisher Scientific | Cat# PA5–30897; RRID: AB_2548371 |
| Goat anti-GFP | Rockland Inc. | Cat#600–101-215 RRID: AB_218182 |
| Mouse anti-Mcm2 (BM28) | BD Biosciences | Cat#610701 RRID: AB_398024 |
| Rabbit anti-Hopx | Proteintech | Cat# 11419–1-AP; RRID: AB_10693525 |
| Rabbit anti-Prox1 | Abcam | Cat# ab199359; RRID: AB_2868427 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| DAPI | Roche | 10236276001 |
| DAPI | BD Biosciences | 564907 |
| Paraformaldehyde | Electron Microscopy Sciences | 100504–162 |
| Triton X-100 | Sigma-Aldrich | T9284 |
| Donkey serum | Millipore-Sigma | S30–100ML |
| TFM Tissue Freezing Medium | GeneralData | TFM-5 |
| ProLong™ Gold Antifade Mountant | Thermo Fisher Scientific | P36930 |
| Percoll | GE Healthcare | 17–0891-02 |
| Hibernate A | Brain Bits | HA |
| Hibernate A Low Fluorescence | Brain Bits | HALF |
| Hibernate E Low Fluorescence | Brain Bits | HELF |
| MitoTracker™ Deep Red FM - Special Packaging | Thermo Fisher Scientific | M22426 |
| Dihydroethidium | Thermo Fisher Scientific | D1168 |
| Autophagy Assay Kit (Red) | Abcam | ab270790 |
| Target Retrieval Solution, Concentrated x 10 | Agilent Dako | S169984–2 |
| Chloroquine diphosphate salt | Sigma-Aldrich | C6628–25G |
| Bafilomycin A1 | Sigma-Aldrich | 19–148 |
| OPP (O-propargyl-puromycin) | Click Chemistry Tools | 1407–5 |
| DMEM | Gibco | 11965084 |
| Fetal Bovine Serum | Hyclone | SH30071.03 |
| Glutamax | Gibco | 35050061 |
| Penicillin-Streptomycin | Gibco | 15140122 |
| Poly-L-lysine | Sigma-Aldrich | P2636 |
| B-27 | Gibco | 17504044 |
|
| ||
| Critical commercial assays | ||
|
| ||
| MACS Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | 130–092-628 |
| Click-iT™ Plus OPP Alexa Fluor™ 647 Protein Synthesis Assay Kit | Thermo Fisher Scientific | C10458 |
| Agencourt RNAClean XP | Beckman Coulter | A63987 |
| SMARTScribe Reverse Transcriptase | Clontech | 639538 |
| Advantage 2 PCR Kit | Clontech | 639206 |
| KAPA HiFi PCR Kit with dNTPs | KAPA | KK2102 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32854 |
| EZ-Tn5 Custom Transposome Construction Kit with pMOD-2 Transposon Construction Vector | Epicenter | TNP10622 |
| 75-cycle High Output Kit v2.5 | Illumina | 20024906 |
| Click-iT™ Plus EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit | Thermo Fisher Scientific | C10634 |
|
| ||
| Deposited data | ||
|
| ||
| scRNA-seq data | This Paper | NCBI GEO: GSE232833 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Hopxtm3.1Joe/J | The Jackson Laboratory | Cat# 029271; RRID: IMSR_JAX:029271 |
| Mouse: GFP-LC3 | RIKEN BioResource Research Center |
Cat# RBRC00806; RRID: IMSR_RBRC00806 |
| Mouse: C57BL/6NCrl | Charles River Laboratory | Cat# CRL:27; RRID: IMSR_CRL:27 |
| Mouse: Hopxtm2.1(cre/ERT2)Joe/J | The Jackson Laboratory | Cat# 017606; RRID: IMSR_JAX:017606 |
| Mouse: B6.129(Cg)-Gt(ROSA) 26Sortm4(ACTB-tdTomato,EGFP)Luo/J |
The Jackson Laboratory | Cat# 007676; RRID: IMSR_JAX:007676 |
| Mouse: B6.129S-Atg5<tm1Myok> | RIKEN BioResource Research Center | Cat# RBRC02975; RRID: IMSR_RBRC02975 |
|
| ||
| Oligonucleotides | ||
|
| ||
| SMARTSeq_CDS_Primer | AACGAGTGGAATCACTG CTGAGTACTTTTTTTTTTT TTTTTTTTTTTTTTTTTTTVN |
|
| SMARTSeq_Amp | AACGAGTGGAATCACTGCTGAGT | |
| SMARTSeq_TSO_RNAOligo | AACGAGTGGAATCACTGCTG AGTACATrGrG+G |
|
|
| ||
| Software and algorithms | ||
|
| ||
| Code | This Paper | GitHub: https://github.com/dvjim/AtoQ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism | GraphPad Software Inc | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | FlowJO, LLC | https://www.flowjo.com/ |
| BD FACSDiva™ | BD Biosciences | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| R | Open source | https://www.r-project.org/ |
| RSEM | Li and Dewey135 | https://deweylab.github.io/RSEM/ |
| Cutadapt | Martin136 | https://cutadapt.readthedocs.io/en/stable/ |
| Samtools | Li et al., 2009137 | http://www.htslib.org/ |
| Monocle | Qiu et al.138 | http://cole-trapnell-lab.github.io/monocle-release/ |
| Clustree | Zappia and Oshlack139 | http://lazappi.github.io/clustree. |
| Seurat | Satija and Hafemeister, 2019140; Hao et al., 2020141 | https://satijalab.org/seurat/ |
| Tximport | Soneson et al.142 | https://github.com/mikelove/tximport |
| CellRouter | Lummertz da Rocha et al.57 | https://github.com/edroaldo/cellrouter |
| bluster | Lun and Hicks143 | https://bioconductor.org/packages/release/bioc/html/bluster.html |
| ClusterProfiler | Yu et al.144 | https://yulab-smu.top/software/ |
| VelocytoR | La Manno et al.46 | http://velocyto.org/ |
| STAR | Dobin et al.145 | https://github.com/alexdobin/STAR/releases |
| SCENIC | Aibar et al.146 | https://scenic.aertslab.org/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html; RRID: SCR_010279 |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html; RRID:SCR_014199 |
| Zen 2 | Carl Zeiss | https://www.zeiss.com; RRID: SCR_013672 |
| scuttle | Lun et al.147; McCarthy et al.148 | https://bioconductor.org/packages/release/bioc/html/scuttle.html |
|
| ||
| Other | ||
|
| ||
| Fluorodish | World Precision Instruments | FD35–100 |
Highlights.
NSC development into an adult state is a multi-step process with defined milestones
Molecular cascade underlying NSC quiescence acquisition and subsequent maturation
Sequential metabolic changes in autophagy and ROS during NSC development
Autophagy promotes NSC quiescence during early postnatal development
ACKNOWLEDGMENTS
We thank members of the Ming and Song laboratories for comments and suggestions and B. Temsamrit, E. LaNoce, G. Alepa, and A. Angelucci for laboratory support. This work was supported by grants from the National Institutes of Health (R01NS110716 to S.M., R35NS097370 to G.-l.M., R35NS116843 to H.S., and K01MH125144 to A.M.B.) and from Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to G.-l.M.). Some figures were created with BioRender.com.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114339.
DECLARATION OF INTERESTS
The authors declare no other competing interests.
REFERENCES
- 1.Gage FH (2019). Adult neurogenesis in mammals. Science 364, 827–828. 10.1126/science.aav6885. [DOI] [PubMed] [Google Scholar]
- 2.Bond AM, Ming GL, and Song H. (2015). Adult Mammalian Neural Stem Cells and Neurogenesis: Five Decades Later. Cell Stem Cell 17, 385–395. 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ming GL, and Song H. (2011). Adult Neurogenesis in the Mammalian Brain: Significant Answers and Significant Questions. Neuron 70, 687–702. 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christian KM, Song H, and Ming G.l. (2014). Functions and Dysfunctions of Adult Hippocampal Neurogenesis. Annu. Rev. Neurosci. 37, 243–262. 10.1146/annurev-neuro-071013-014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anacker C, and Hen R. (2017). Adult hippocampal neurogenesis and cognitive flexibility − linking memory and mood. Nat. Rev. Neurosci. 18, 335–346. 10.1038/nrn.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller SM, and Sahay A. (2019). Functions of adult-born neurons in hippocampal memory interference and indexing. Nat. Neurosci. 22, 1565–1575. 10.1038/s41593-019-0484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toda T, Parylak SL, Linker SB, and Gage FH (2019). The role of adult hippocampal neurogenesis in brain health and disease. Mol. Psychiatr. 24, 67–87. 10.1038/s41380-018-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger T, Lee H, Young AH, Aarsland D, and Thuret S. (2020). Adult Hippocampal Neurogenesis in Major Depressive Disorder and Alzheimer’s Disease. Trends Mol. Med. 26, 803–818. 10.1016/j.molmed.2020.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Bielefeld P, Durá I, Danielewicz J, Lucassen PJ, Baekelandt V, Abrous DN, Encinas JM, and Fitzsimons CP (2019). Insult-induced aberrant hippocampal neurogenesis: Functional consequences and possible therapeutic strategies. Behav. Brain Res. 372, 112032. 10.1016/j.bbr.2019.112032. [DOI] [PubMed] [Google Scholar]
- 10.Jessberger S, and Parent JM (2015). Epilepsy and Adult Neurogenesis. Cold Spring Harbor Perspect. Biol. 7, a020677. 10.1101/cshperspect.a020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn HG, Dickinson-Anson H, and Gage FH (1996). Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J. Neurosci. 16, 2027–2033. 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben Abdallah NM-B, Slomianka L, Vyssotski AL, and Lipp H-P (2010). Early age-related changes in adult hippocampal neurogenesis in C57 mice. Neurobiol. Aging 31, 151–161. 10.1016/j.neurobiolaging.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Encinas JM, Michurina TV, Peunova N, Park J-H, Tordo J, Peterson DA, Fishell G, Koulakov A, and Enikolopov G. (2011). Division-Coupled Astrocytic Differentiation and Age-Related Depletion of Neural Stem Cells in the Adult Hippocampus. Cell Stem Cell 8, 566–579. 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrayeva A, Bay M, Pu E, Jörg DJ, Peng L, Jun H, Zhang N, Aaron D, Lin C, Resler G, et al. (2021). Early stem cell aging in the mature brain. Cell Stem Cell 28, 955–966.e7. 10.1016/j.stem.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, Fang L, Fernández G, and Pleasure SJ (2013). The Ventral Hippocampus Is the Embryonic Origin for Adult Neural Stem Cells in the Dentate Gyrus. Neuron 78, 658–672. 10.1016/j.neuron.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg DA, Su Y, Jimenez-Cyrus D, Patel A, Huang N, Morizet D, Lee S, Shah R, Ringeling FR, Jain R, et al. (2019). A Common Embryonic Origin of Stem Cells Drives Developmental and Adult Neurogenesis. Cell 177, 654–668.e15. 10.1016/j.cell.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Yoon K, Kim N-S, Ming GL, and Song H. (2023). Cell-autonomous and non-cell-autonomous roles of NKCC1 in regulating neural stem cell quiescence in the hippocampal dentate gyrus. Stem Cell Rep. 18, 1468–1481. 10.1016/j.stemcr.2023.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calatayud-Baselga I, Casares-Crespo L, Franch-Ibáñez C, Guijarro-Nuez J, Sanz P, and Mira H. (2023). Autophagy drives the conversion of developmental neural stem cells to the adult quiescent state. Nat. Commun. 14, 7541. 10.1038/s41467-023-43222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Velthoven CTJ, and Rando TA (2019). Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell 24, 213–225. 10.1016/j.stem.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbán N, Blomfield IM, and Guillemot F. (2019). Quiescence of Adult Mammalian Neural Stem Cells: A Highly Regulated Rest. Neuron 104, 834–848. 10.1016/j.neuron.2019.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, and Alvarez-Buylla A. (2015). Embryonic Origin of Postnatal Neural Stem Cells. Cell 161, 1644–1655. 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furutachi S, Miya H, Watanabe T, Kawai H, Yamasaki N, Harada Y, Imayoshi I, Nelson M, Nakayama KI, Hirabayashi Y, and Gotoh Y. (2015). Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 18, 657–665. 10.1038/nn.3989. [DOI] [PubMed] [Google Scholar]
- 23.Brunne B, Zhao S, Derouiche A, Herz J, May P, Frotscher M, and Bock HH (2010). Origin, maturation, and astroglial transformation of secondary radial glial cells in the developing dentate gyrus. Glia 58, 1553–1569. 10.1002/glia.21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seki T, Sato T, Toda K, Osumi N, Imura T, and Shioda S. (2014). Distinctive population of Gfap-expressing neural progenitors arising around the dentate notch migrate and form the granule cell layer in the developing hippocampus. J. Comp. Neurol. 522, 261–283. 10.1002/cne.23460. [DOI] [PubMed] [Google Scholar]
- 25.Nicola Z, Fabel K, and Kempermann G. (2015). Development of the adult neurogenic niche in the hippocampus of mice. Front. Neuroanat. 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilley JA, Yang C-P, and Kernie SG (2011). Developmental profiling of postnatal dentate gyrus progenitors provides evidence for dynamic cell-autonomous regulation. Hippocampus 21, 33–47. 10.1002/hipo.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming G.l., and Song H. (2011). In Vivo Clonal Analysis Reveals Self-Renewing and Multipotent Adult Neural Stem Cell Characteristics. Cell 145, 1142–1155. 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namba T, Mochizuki H, Suzuki R, Onodera M, Yamaguchi M, Namiki H, Shioda S, and Seki T. (2011). Time-Lapse Imaging Reveals Symmetric Neurogenic Cell Division of GFAP-Expressing Progenitors for Expansion of Postnatal Dentate Granule Neurons. PLoS One 6, e25303. 10.1371/journal.pone.0025303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilz G-A, Bottes S, Betizeau M, Jörg DJ, Carta S, April S, Simons BD, Helmchen F, and Jessberger S. (2018). Live imaging of neurogenesis in the adult mouse hippocampus. Science 359, 658–662. 10.1126/science.aao5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond AM, Ming G, and Song H. (2021). Chapter Two - Ontogeny of adult neural stem cells in the mammalian brain. In Current Topics in Developmental Biology Molecular Mechanisms of Neural Development and Insights into Disease, Bashaw GJ, ed. (Academic Press; ), pp. 67–98. 10.1016/bs.ctdb.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochgerner H, Zeisel A, Lönnerberg P, and Linnarsson S. (2018). Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci. 21, 290–299. 10.1038/s41593-017-0056-2. [DOI] [PubMed] [Google Scholar]
- 32.Ly CH, Lynch GS, and Ryall JG (2020). A Metabolic Roadmap for Somatic Stem Cell Fate. Cell Metabol. 31, 1052–1067. 10.1016/j.cmet.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 33.de Morree A, and Rando TA (2023). Regulation of adult stem cell quiescence and its functions in the maintenance of tissue integrity. Nat. Rev. Mol. Cell Biol. 24, 334–354. 10.1038/s41580-022-00568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming G.l., and Song H. (2015). Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–372. 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leeman DS, Hebestreit K, Ruetz T, Webb AE, McKay A, Pollina EA, Dulken BW, Zhao X, Yeo RW, Ho TT, et al. (2018). Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283. 10.1126/science.aag3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knobloch M, Braun SMG, Zurkirchen L, von Schoultz C, Zamboni N, Araúzo-Bravo MJ, Kovacs WJ, Karalay O, Suter U, Machado RAC, et al. (2013). Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 493, 226–230. 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrelli F, Scandella V, Montessuit S, Zamboni N, Martinou J-C, and Knobloch M. (2023). Mitochondrial pyruvate metabolism regulates the activation of quiescent adult neural stem cells. Sci. Adv. 9, eadd5220. 10.1126/sciadv.add5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knobloch M, Pilz G-A, Ghesquière B, Kovacs WJ, Wegleiter T, Moore DL, Hruzova M, Zamboni N, Carmeliet P, and Jessberger S. (2017). A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates Adult Neural Stem Cell Activity. Cell Rep. 20, 2144–2155. 10.1016/j.celrep.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeda N, Jain R, LeBoeuf MR, Padmanabhan A, Wang Q, Li L, Lu MM, Millar SE, and Epstein JA (2013). Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development 140, 1655–1664. 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams GH, and Stoeber K. (2007). Cell cycle markers in clinical oncology. Curr. Opin. Cell Biol. 19, 672–679. 10.1016/j.ceb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, and Williams GH (2001). DNA replication licensing and human cell proliferation. J. Cell Sci. 114, 2027–2041. 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- 42.McGarry TJ, and Kirschner MW (1998). Geminin, an Inhibitor of DNA Replication, Is Degraded during Mitosis. Cell 93, 1043–1053. 10.1016/S0092-8674(00)81209-X. [DOI] [PubMed] [Google Scholar]
- 43.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 44.Li D, Takeda N, Jain R, Manderfield LJ, Liu F, Li L, Anderson SA, and Epstein JA (2015). Hopx distinguishes hippocampal from lateral ventricle neural stem cells. Stem Cell Res. 15, 522–529. 10.1016/j.scr.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batiuk MY, Martirosyan A, Wahis J, de Vin F, Marneffe C, Kusserow C, Koeppen J, Viana JF, Oliveira JF, Voet T, et al. (2020). Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220. 10.1038/s41467-019-14198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Lin L, Lai H, Parada LF, and Lei L. (2013). Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev. Dynam. 242, 638–653. 10.1002/dvdy.23962. [DOI] [PubMed] [Google Scholar]
- 48.Miller JA, Nathanson J, Franjic D, Shim S, Dalley RA, Shapouri S, Smith KA, Sunkin SM, Bernard A, Bennett JL, et al. (2013). Conserved molecular signatures of neurogenesis in the hippocampal subgranular zone of rodents and primates. Development 140, 4633–4644. 10.1242/dev.097212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heng YHE, McLeay RC, Harvey TJ, Smith AG, Barry G, Cato K, Plachez C, Little E, Mason S, Dixon C, et al. (2014). NFIX Regulates Neural Progenitor Cell Differentiation During Hippocampal Morphogenesis. Cerebr. Cortex 24, 261–279. 10.1093/cercor/bhs307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamnasaran D, Muir WJ, Ferguson-Smith MA, and Cox DW (2003). Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J. Med. Genet. 40, 325–332. 10.1136/jmg.40.5.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pickard BS, Malloy M.p., Porteous D.j., Blackwood Dh. r., and Muir, W.j. (2005). Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am. J. Med. Genet. B Neuropsychiatr. Genet. 136B, 26–32. 10.1002/ajmg.b.30204. [DOI] [PubMed] [Google Scholar]
- 52.Rossi JJ, Rosenfeld JA, Chan KM, Streff H, Nankivell V, Peet DJ, Whitelaw ML, and Bersten DC (2021). Molecular characterisation of rare loss-of-function NPAS3 and NPAS4 variants identified in individuals with neurodevelopmental disorders. Sci. Rep. 11, 6602. 10.1038/s41598-021-86041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pieper AA, Wu X, Han TW, Estill SJ, Dang Q, Wu LC, Reece-Fincanon S, Dudley CA, Richardson JA, Brat DJ, and McKnight SL (2005). The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc. Natl. Acad. Sci. USA 102, 14052–14057. 10.1073/pnas.0506713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lehnertz B, Chagraoui J, MacRae T, Tomellini E, Corneau S, Mayotte N, Boivin I, Durand A, Gracias D, and Sauvageau G. (2021). HLF expression defines the human hematopoietic stem cell state. Blood 138, 2642–2654. 10.1182/blood.2021010745. [DOI] [PubMed] [Google Scholar]
- 55.Komorowska K, Doyle A, Wahlestedt M, Subramaniam A, Debnath S, Chen J, Soneji S, Van Handel B, Mikkola HKA, Miharada K, et al. (2017). Hepatic Leukemia Factor Maintains Quiescence of Hematopoietic Stem Cells and Protects the Stem Cell Pool during Regeneration. Cell Rep. 21, 3514–3523. 10.1016/j.celrep.2017.11.084. [DOI] [PubMed] [Google Scholar]
- 56.Tang W, He J, Huang T, Bai Z, Wang C, Wang H, Yang R, Ni Y, Hou J, Wang J, et al. (2021). Hlf Expression Marks Early Emergence of Hematopoietic Stem Cell Precursors With Adult Repopulating Potential and Fate. Front. Cell Dev. Biol. 9, 728057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lummertz da Rocha E, Rowe RG, Lundin V, Malleshaiah M, Jha DK, Rambo CR, Li H, North TE, Collins JJ, and Daley GQ (2018). Reconstruction of complex single-cell trajectories using Cell-Router. Nat. Commun. 9, 892. 10.1038/s41467-018-03214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang NC (2020). Autophagy and Stem Cells: Self-Eating for Self-Renewal. Front. Cell Dev. Biol. 8, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murley A, and Dillin A. (2023). Macroautophagy in quiescent and senescent cells: a pathway to longevity? Trends Cell Biol. 33, 495–504. 10.1016/j.tcb.2022.10.004. [DOI] [PubMed] [Google Scholar]
- 60.Wang C, Liang C-C, Bian ZC, Zhu Y, and Guan J-L (2013). FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat. Neurosci. 16, 532–542. 10.1038/nn.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yazdankhah M, Farioli-Vecchioli S, Tonchev AB, Stoykova A, and Cecconi F. (2014). The autophagy regulators Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis. 5, e1403. 10.1038/cddis.2014.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xi Y, Dhaliwal JS, Ceizar M, Vaculik M, Kumar KL, and Lagace DC (2016). Knockout of Atg5 delays the maturation and reduces the survival of adult-generated neurons in the hippocampus. Cell Death Dis. 7, e2127. 10.1038/cddis.2015.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams PA, Parham LR, Acheampong KK, Danan CH, Ma X, Simon LA, Naughton KE, Karakasheva T, Whelan KA, Brady DC, et al. (2022). IGF2BP1/IMP1 contributes to autophagy modulation directly via MAP1LC3B. Preprint at bioRxiv. 10.1101/2022.02.28.482365. [DOI] [Google Scholar]
- 64.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, and Ohsumi Y. (2004). In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111. 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, and Mizushima N. (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 66.Dong A, Kulkarni VV, and Maday S. (2019). Methods for Imaging Autophagosome Dynamics in Primary Neurons. In Autophagy: Methods and Protocols Methods in Molecular Biology N. Ktistakis and O. Florey, eds. (Springer; ), pp. 243–256. 10.1007/978-1-4939-8873-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moulis M, and Vindis C. (2017). Methods for Measuring Autophagy in Mice. Cells 6, 14. 10.3390/cells6020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.du Toit A, Hofmeyr J-HS, Gniadek TJ, and Loos B. (2018). Measuring autophagosome flux. Autophagy 14, 1060–1071. 10.1080/15548627.2018.1469590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sinenko SA, Starkova TY, Kuzmin AA, and Tomilin AN (2021). Physiological Signaling Functions of Reactive Oxygen Species in Stem Cells: From Flies to Man. Front. Cell Dev. Biol. 9, 714370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye Z-W, Zhang J, Townsend DM, and Tew KD (2015). Oxidative stress, redox regulation and diseases of cellular differentiation. Biochim. Biophys. Acta 1850, 1607–1621. 10.1016/j.bbagen.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holmström KM, and Finkel T. (2014). Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411–421. 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 72.Adusumilli VS, Walker TL, Overall RW, Klatt GM, Zeidan SA, Zocher S, Kirova DG, Ntitsias K, Fischer TJ, Sykes AM, et al. (2021). ROS Dynamics Delineate Functional States of Hippocampal Neural Stem Cells and Link to Their Activity-Dependent Exit from Quiescence. Cell Stem Cell 28, 300–314.e6. 10.1016/j.stem.2020.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khacho M, Harris R, and Slack RS (2019). Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat. Rev. Neurosci. 20, 34–48. 10.1038/s41583-018-0091-3. [DOI] [PubMed] [Google Scholar]
- 74.Zorov DB, Juhaszova M, and Sollott SJ (2014). Mitochondrial Reactive Oxygen Species (ROS) and ROS-Induced ROS Release. Physiol. Rev. 94, 909–950. 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lugli E, Troiano L, Ferraresi R, Roat E, Prada N, Nasi M, Pinti M, Cooper EL, and Cossarizza A. (2005). Characterization of cells with different mitochondrial membrane potential during apoptosis. Cytometry A. 68, 28–35. 10.1002/cyto.a.20188. [DOI] [PubMed] [Google Scholar]
- 76.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, and Mizushima N. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889. 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 77.Borrett MJ, Innes BT, Jeong D, Tahmasian N, Storer MA, Bader GD, Kaplan DR, and Miller FD (2020). Single-Cell Profiling Shows Murine Forebrain Neural Stem Cells Reacquire a Developmental State when Activated for Adult Neurogenesis. Cell Rep. 32, 108022. 10.1016/j.celrep.2020.108022. [DOI] [PubMed] [Google Scholar]
- 78.Guo N, McDermott KD, Shih Y-T, Zanga H, Ghosh D, Herber C, Meara WR, Coleman J, Zagouras A, Wong LP, et al. (2022). Transcriptional regulation of neural stem cell expansion in the adult hippocampus. Elife 11, e72195. 10.7554/eLife.72195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Falini B, Brunetti L, Sportoletti P, and Martelli MP (2020). NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood 136, 1707–1721. 10.1182/blood.2019004226. [DOI] [PubMed] [Google Scholar]
- 80.Morganti C, Ito K, Yanase C, Verma A, Teruya-Feldstein J, and Ito K. (2022). NPM1 ablation induces HSC aging and inflammation to develop myelodysplastic syndrome exacerbated by p53 loss. EMBO Rep. 23, e54262. 10.15252/embr.202154262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kalamakis G, Brüne D, Ravichandran S, Bolz J, Fan W, Ziebell F, Stiehl T, Catalá -Martinez F, Kupke J, Zhao S, et al. (2019). Quiescence Modulates Stem Cell Maintenance and Regenerative Capacity in the Aging Brain. Cell 176, 1407–1419.e14. 10.1016/j.cell.2019.01.040. [DOI] [PubMed] [Google Scholar]
- 82.Harris L, Rigo P, Stiehl T, Gaber ZB, Austin SHL, Masdeu MDM, Edwards A, Urbán N, Marciniak-Czochra A, and Guillemot F. (2021). Coordinated changes in cellular behavior ensure the lifelong maintenance of the hippocampal stem cell population. Cell Stem Cell 28, 863–876.e6. 10.1016/j.stem.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Y, Bottes S, Fisch R, Zehnder C, Cole JD, Pilz G-A, Helmchen F, Simons BD, and Jessberger S. (2023). Chronic in vivo imaging defines age-dependent alterations of neurogenesis in the mouse hippocampus. Nat. Aging 3, 380–390. 10.1038/s43587-023-00370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altman J, and Das GD (1965). Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124, 319–335. 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 85.Angevine. (1965). Time of neuron origin in the hippocampal region: An autoradiographic study in the mouse. Exp. Neurol. 11, 1–39. 10.1016/0014-4886(65)90121-4. [DOI] [PubMed] [Google Scholar]
- 86.Seri B, García-Verdugo JM, McEwen BS, and Alvarez-Buylla A. (2001). Astrocytes Give Rise to New Neurons in the Adult Mammalian Hippocampus. J. Neurosci. 21, 7153–7160. 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gould E, Tanapat P, McEwen BS, Flügge G, and Fuchs E. (1998). Proliferation of granule cell precursors in the dentate gyrus of adult monkeys is diminished by stress. Proc. Natl. Acad. Sci. USA 95, 3168–3171. 10.1073/pnas.95.6.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kornack DR, and Rakic P. (1999). Continuation of neurogenesis in the hippocampus of the adult macaque monkey. Proc. Natl. Acad. Sci. USA 96, 5768–5773. 10.1073/pnas.96.10.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ngwenya LB, Peters A, and Rosene DL (2006). Maturational sequence of newly generated neurons in the dentate gyrus of the young adult rhesus monkey. J. Comp. Neurol. 498, 204–216. 10.1002/cne.21045. [DOI] [PubMed] [Google Scholar]
- 90.Jabès A, Lavenex PB, Amaral DG, and Lavenex P. (2010). Quantitative analysis of postnatal neurogenesis and neuron number in the macaque monkey dentate gyrus. Eur. J. Neurosci. 31, 273–285. 10.1111/j.1460-9568.2009.07061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kohler SJ, Williams NI, Stanton GB, Cameron JL, and Greenough WT (2011). Maturation time of new granule cells in the dentate gyrus of adult macaque monkeys exceeds six months. Proc. Natl. Acad. Sci. USA 108, 10326–10331. 10.1073/pnas.1017099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yuan T-F, Li J, Ding F, and Arias-Carrion O. (2014). Evidence of adult neurogenesis in non-human primates and human. Cell Tissue Res. 358, 17–23. 10.1007/s00441-014-1980-z. [DOI] [PubMed] [Google Scholar]
- 93.Eriksson PS, Perfilieva E, Björk-Eriksson T, Alborn A-M, Nordborg C, Peterson DA, and Gage FH (1998). Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317. 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 94.Knoth R, Singec I, Ditter M, Pantazis G, Capetian P, Meyer RP, Horvat V, Volk B, and Kempermann G. (2010). Murine Features of Neurogenesis in the Human Hippocampus across the Lifespan from 0 to 100 Years. PLoS One 5, e8809. 10.1371/journal.pone.0008809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, Boström E, Westerlund I, Vial C, Buchholz BA, et al. (2013). Dynamics of Hippocampal Neurogenesis in Adult Humans. Cell 153, 1219–1227. 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V, Rosoklija GB, Stankov A, Arango V, Dwork AJ, et al. (2018). Human Hippocampal Neurogenesis Persists throughout Aging. Cell Stem Cell 22, 589–599.e5. 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381. 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moreno-Jiménez EP, Flor-García M, Terreros-Roncal J, Rábano A, Cafini F, Pallas-Bazarra N, Ávila J, and Llorens-Martín M. (2019). Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560. 10.1038/s41591-019-0375-9. [DOI] [PubMed] [Google Scholar]
- 99.Tobin MK, Musaraca K, Disouky A, Shetti A, Bheri A, Honer WG, Kim N, Dawe RJ, Bennett DA, Arfanakis K, and Lazarov O. (2019). Human Hippocampal Neurogenesis Persists in Aged Adults and Alzheimer’s Disease Patients. Cell Stem Cell 24, 974–982.e3. 10.1016/j.stem.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Franjic D, Skarica M, Ma S, Arellano JI, Tebbenkamp ATN, Choi J, Xu C, Li Q, Morozov YM, Andrijevic D, et al. (2022). Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron 110, 452–469.e14. 10.1016/j.neuron.2021.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Su Y, Li S, Kennedy BC, Zhang DY, Bond AM, Sun Y, Jacob F, Lu L, Hu P, et al. (2022). Molecular landscapes of human hippocampal immature neurons across lifespan. Nature 607, 527–533. 10.1038/s41586-022-04912-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bayer SA (1980). Development of the hippocampal region in the rat I. Neurogenesis examined with 3H-thymidine autoradiography. J. Comp. Neurol. 190, 87–114. 10.1002/cne.901900107. [DOI] [PubMed] [Google Scholar]
- 103.Bayer SA (1980). Development of the hippocampal region in the rat II. Morphogenesis during embryonic and early postnatal life. J. Comp. Neurol. 190, 115–134. 10.1002/cne.901900108. [DOI] [PubMed] [Google Scholar]
- 104.Yang P, Zhang J, Shi H, Zhang J, Xu X, Xiao X, and Liu Y. (2014). Developmental profile of neurogenesis in prenatal human hippocampus: An immunohistochemical study. Int. J. Dev. Neurosci. 38, 1–9. 10.1016/j.ijdevneu.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 105.Cipriani S, Journiac N, Nardelli J, Verney C, Delezoide A-L, Guimiot F, Gressens P, and Adle-Biassette H. (2017). Dynamic Expression Patterns of Progenitor and Neuron Layer Markers in the Developing Human Dentate Gyrus and Fimbria. Cerebr. Cortex 27, 358–372. 10.1093/cercor/bhv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cipriani S, Ferrer I, Aronica E, Kovacs GG, Verney C, Nardelli J, Khung S, Delezoide A-L, Milenkovic I, Rasika S, et al. (2018). Hippocampal Radial Glial Subtypes and Their Neurogenic Potential in Human Fetuses and Healthy and Alzheimer’s Disease Adults. Cerebr. Cortex 28, 2458–2478. 10.1093/cercor/bhy096. [DOI] [PubMed] [Google Scholar]
- 107.Bond AM, Berg DA, Lee S, Garcia-Epelboim AS, Adusumilli VS, Ming GL, and Song H. (2020). Differential Timing and Coordination of Neurogenesis and Astrogenesis in Developing Mouse Hippocampal Subregions. Brain Sci. 10, 909. 10.3390/brainsci10120909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bond AM, Ming GL, and Song H. (2022). What Is the Relationship Between Hippocampal Neurogenesis Across Different Stages of the Lifespan? Front. Neurosci. 16, 891713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Amrein I, Isler K, and Lipp H-P (2011). Comparing adult hippocampal neurogenesis in mammalian species and orders: influence of chronological age and life history stage. Eur. J. Neurosci. 34, 978–987. 10.1111/j.1460-9568.2011.07804.x. [DOI] [PubMed] [Google Scholar]
- 110.Charvet CJ, and Finlay BL (2018). Comparing Adult Hippocampal Neurogenesis Across Species: Translating Time to Predict the Tempo in Humans. Front. Neurosci. 12, 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Snyder JS (2019). Recalibrating the Relevance of Adult Neurogenesis. Trends Neurosci. 42, 164–178. 10.1016/j.tins.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 112.Gao X, Xu C, Asada N, and Frenette PS (2018). The hematopoietic stem cell niche: from embryo to adult. Development 145, dev139691. 10.1242/dev.139691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gros J, Manceau M, Thomé V, and Marcelle C. (2005). A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435, 954–958. 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 114.Relaix F, Rocancourt D, Mansouri A, and Buckingham M. (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948–953. 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 115.Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomès D, and Tajbakhsh S. (2005). Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 19, 1426–1431. 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guiu J, Hannezo E, Yui S, Demharter S, Ulyanchenko S, Maimets M, Jørgensen A, Perlman S, Lundvall L, Mamsen LS, et al. (2019). Tracing the origin of adult intestinal stem cells. Nature 570, 107–111. 10.1038/s41586-019-1212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Redmond SA, Figueres-Oñate M, Obernier K, Nascimento MA, Parraguez JI, López-Mascaraque L, Fuentealba LC, and Alvarez-Buylla A. (2019). Development of Ependymal and Postnatal Neural Stem Cells and Their Origin from a Common Embryonic Progenitor. Cell Rep. 27, 429–441.e3. 10.1016/j.celrep.2019.01.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Picard CA, and Marcelle C. (2013). Two distinct muscle progenitor populations coexist throughout amniote development. Dev. Biol. 373, 141–148. 10.1016/j.ydbio.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 119.Christensen JL, Wright DE, Wagers AJ, and Weissman IL (2004). Circulation and Chemotaxis of Fetal Hematopoietic Stem Cells. PLoS Biol. 2, e75. 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hall TD, Kim H, Dabbah M, Myers JA, Crawford JC, Morales-Hernandez A, Caprio CE, Sriram P, Kooienga E, Derecka M, et al. (2022). Murine fetal bone marrow does not support functional hematopoietic stem and progenitor cells until birth. Nat. Commun. 13, 5403. 10.1038/s41467-022-33092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, and Eaves CJ (2006). Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J. Clin. Invest. 116, 2808–2816. 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Baser A, Skabkin M, Kleber S, Dang Y, Gülcüler Balta GS, Kalamakis G, Göpferich M, Ibañez DC, Schefzik R, Lopez AS, et al. (2019). Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104. 10.1038/s41586-019-0888-x. [DOI] [PubMed] [Google Scholar]
- 123.Wang R, and Amoyel M. (2022). mRNA Translation Is Dynamically Regulated to Instruct Stem Cell Fate. Front. Mol. Biosci. 9, 863885. 10.3389/fmolb.2022.863885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Corté s-Garrido R, Gkatza N, et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340. 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Signer RAJ, Magee JA, Salic A, and Morrison SJ (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54. 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Machado L, Esteves de Lima J, Fabre O, Proux C, Legendre R, Szegedi A, Varet H, Ingerslev LR, Barrès R, Relaix F, and Mourikis P. (2017). In Situ Fixation Redefines Quiescence and Early Activation of Skeletal Muscle Stem Cells. Cell Rep. 21, 1982–1993. 10.1016/j.celrep.2017.10.080. [DOI] [PubMed] [Google Scholar]
- 127.Boonsanay V, Zhang T, Georgieva A, Kostin S, Qi H, Yuan X, Zhou Y, and Braun T. (2016). Regulation of Skeletal Muscle Stem Cell Quiescence by Suv4–20h1-Dependent Facultative Heterochromatin Formation. Cell Stem Cell 18, 229–242. 10.1016/j.stem.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 128.Cebrián-Silla A, Alfaro-Cervelló C, Herranz-Pérez V, Kaneko N, Park DH, Sawamoto K, Alvarez-Buylla A, Lim DA, and García-Verdugo JM (2017). Unique Organization of the Nuclear Envelope in the Post-natal Quiescent Neural Stem Cells. Stem Cell Rep. 9, 203–216. 10.1016/j.stemcr.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T, et al. (2013). Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61. 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang Y-H, Israelsen WJ, Lee D, Yu VWC, Jeanson NT, Clish CB, Cantley LC, Vander Heiden MG, and Scadden DT (2014). Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 158, 1309–1323. 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flores A, Schell J, Krall AS, Jelinek D, Miranda M, Grigorian M, Braas D, White AC, Zhou JL, Graham NA, et al. (2017). Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat. Cell Biol. 19, 1017–1026. 10.1038/ncb3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Beckervordersandforth R, Ebert B, Schäffner I, Moss J, Fiebig C, Shin J, Moore DL, Ghosh L, Trinchero MF, Stockburger C, et al. (2017). Role of Mitochondrial Metabolism in the Control of Early Lineage Progression and Aging Phenotypes in Adult Hippocampal Neurogenesis. Neuron 93, 560–573.e6. 10.1016/j.neuron.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Simsek T, Kocabas F, Zheng J, DeBerardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, and Sadek HA (2010). The Distinct Metabolic Profile of Hematopoietic Stem Cells Reflects Their Location in a Hypoxic Niche. Cell Stem Cell 7, 380–390. 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liang R, Arif T, Kalmykova S, Kasianov A, Lin M, Menon V, Qiu J, Bernitz JM, Moore K, Lin F, et al. (2020). Restraining Lysosomal Activity Preserves Hematopoietic Stem Cell Quiescence and Potency. Cell Stem Cell 26, 359–376.e7. 10.1016/j.stem.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 17, 10–12. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 137.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Subgroup, 1000 Genome Project Data Processing (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zappia L, and Oshlack A. (2018). Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience 7, giy083. 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hafemeister C, and Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology 20, 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soneson C, Matthes KL, Nowicka M, Law CW, and Robinson MD (2016). Isoform prefiltering improves performance of count-based methods for analysis of differential transcript usage. Genome Biol. 17, 12. 10.1186/s13059-015-0862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lun A, and Hicks S. (2022). Bluster: Clustering Algorithm for Bioconductor. [Google Scholar]
- 144.Yu G, Wang L-G, Han Y, and He Q-Y (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS 16, 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lun ATL, McCarthy DJ, and Marioni JC (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122. 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, and Epstein JA (2011). Interconversion Between Intestinal Stem Cell Populations in Distinct Niches. Science 334, 1420–1424. 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L. (2007). A global double-fluorescent Cre reporter mouse. genesis 45, 593–605. 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 151.Ramsköld D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782. 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, and Sandberg R. (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098. 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 153.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kaufman L, and Rousseeuw PJ (1990). Finding Groups in Data: An Introduction to Cluster Analysis. [Google Scholar]
- 155.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huynh-Thu VA, Irrthum A, Wehenkel L, and Geurts P. (2010). Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLoS One 5, e12776. 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, and Collins JJ (2014). CellNet: network biology applied to stem cell engineering. Cell 158, 903–915. 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, and Gardner TS (2007). Large-Scale Mapping and Validation of Escherichia coli Transcriptional Regulation from a Compendium of Expression Profiles. PLoS Biol. 5, e8. 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yuan R, Hahn Y, Stempel MH, Sidibe DK, Laxton O, Chen J, Kulkarni A, and Maday S. (2023). Proteasomal inhibition preferentially stimulates lysosome activity relative to autophagic flux in primary astrocytes. Autophagy 19, 570–596. 10.1080/15548627.2022.2084884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at the NCBI GEO: GSE232833. All original code has been deposited at GitHub: https://github.com/dvjim/AtoQ. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.