Abstract
We defined the minimal core promoter sequences responsible for efficient and accurate initiation of cucumber mosaic virus (CMV) subgenomic RNA4. The necessary sequence maps to positions −28 to +15 relative to the initiation cytidylate used to initiate RNA synthesis in vivo. Positions −28 to −5 contain a 9-bp stem and a 6-nucleotide purine-rich loop. Considerable changes in the stem and the loop are tolerated for RNA synthesis, including replacement with a different stem-loop. In a template competition assay, the stem-loop and the initiation cytidylate are sufficient to interact with the CMV replicase. Thus, the mechanism of core promoter recognition by the CMV replicase appears to be less specific in comparison to the minimal subgenomic core promoter of the closely related brome mosaic virus.
Viruses that encode multiple proteins in an RNA have evolved different strategies to translate all of the coding sequences. The Bromoviridae family of plant viruses, with genomes of capped plus-stranded RNAs, transcribes subgenomic-length RNAs by initiating synthesis from an internal sequence within the minus-strand RNA (for a review, see reference 19). The subgenomic promoter is defined as a sequence that interacts with the viral replicase to direct the accurate initiation of RNA synthesis.
In Brome mosaic virus (BMV), genus Bromovirus, family Bromoviridae, several sequence motifs appear to contribute to efficient subgenomic RNA synthesis in protoplasts. These include an upstream A-U-rich sequence, a polyuridylate tract, a 20-nucleotide (nt) core promoter, and a downstream A-U-rich sequence (9, 18). The polyuridylate sequence is of variable length; it is required for BMV infection in vivo (31) and binds to the replicase in vitro (1). The BMV core promoter directs the BMV replicase to recognize the initiation nucleotide. A mutation in the core promoter can compensate for the lack of a polyuridylate sequence (31). Using a reductionist approach, our laboratory determined that the core promoter alone is required and sufficient for accurate and efficient in vitro initiation of transcripts less than 15 nt in length (1). A systematic mutational analysis of the subgenomic core promoter revealed that four essential nucleotides, at positions −17, −14, −13, and −11 relative to the initiation cytidylate (+1C), are recognized by the RNA-dependent RNA polymerase (RdRp) in a sequence-specific manner (25). The base and some ribose moieties of these key nucleotides responsible for RdRp recognition were identified using RNAs containing chemically synthesized nucleotide analogs (26).
Jaspars (14) used nucleotide sequence alignment to demonstrate that a structure upstream of the subgenomic RNA initiation cytidylate is found in alfalfa mosaic virus (AMV) and several other members of the Bromoviridae family. Haasnoot et al. (13) demonstrated that a stem-loop structure is correlated to AMV subgenomic RNA synthesis. We biochemically examined the cucumber mosaic virus (CMV) subgenomic promoter to expand the information on the structure and sequence required for transcription by viral replicases.
CMV, genus Cucumovirus, family Bromoviridae, is an economically important plant pathogen and can be classified into two major serological subgroups, I and II. Nucleotide sequence alignment of RNA3 indicated that subgroup I can be divided into two other subgroups, IA and IB (24). The CMV RNA genome consists of RNA1 (∼3.4 kb), RNA2 (∼3 kb), and RNA3 (∼2.2 kb). As in BMV and AMV, the capsid is translated from subgenomic RNA4 (∼1 kb) (22). A second, low-abundance subgenomic transcript, RNA4a, is produced from the minus strand of RNA2 (8). The cis-acting sequences for CMV RNA4 synthesis have been characterized (4). Using a construct containing two subgenomic promoters for RNA4, Boccard and Baucombe (4) demonstrated that the sequence from 70 nt upstream to 30 nt downstream of the initiation cytidylate (+1C; complement of nt 1167 in the Kin strain of CMV) was required to direct RNA synthesis in transfected protoplasts. A truncated promoter beginning 30 nt upstream of the initiation cytidylate also directed subgenomic RNA4 synthesis but at a reduced level (4). Here we further define the sequence needed to direct CMV subgenomic RNA synthesis using an enriched CMV replicase that can accurately direct the synthesis of genomic minus-strand, genomic plus-strand, and subgenomic RNA in vitro (29). Deletion analysis indicates that a stem-loop at nt −28 to −5 upstream of the +1C is responsible for the efficient and accurate initiation of subgenomic RNA synthesis. This stem-loop structure is conserved in all subgroups of CMV.
MATERIALS AND METHODS
RNA synthesis and purification.
Transcription conditions were as previously described (20). Briefly, the DNA strands were purified via denaturing polyacrylamide gel electrophoresis (PAGE) and then adjusted to a final concentration of 8 μM. One microliter of each DNA was used in a 20-μl transcription reaction mixture containing 40 mM Tris (pH 8.1), 1 mM spermidine, 0.01% Triton X-100, 80 mg of polyethylene glycol 8000, and 4 mM each nucleoside triphosphate. The T7 RNA polymerase used was purified using the protocol of Grodberg and Dunn (10). RNAs of the correct size were purified by preparative denaturing gel electrophoresis and excised from the gel after UV shadowing. The gel slices were crushed and ground to small pieces, and the RNA was eluted from the polyacrylamide with 0.4 M ammonium acetate at 30°C overnight. Following precipitation with ethanol, the RNA concentration was determined by spectrophotometry and checked by staining with 0.25% toluidine blue on an analytical gel.
RdRp activity assay and product analysis.
CMV replicase was enriched from CMV-infected tobacco by a method described previously (29), modeled after previously published protocols (11, 36). A standard assay consisted of a 20-μl reaction mixture containing 0.25 pmol of template (unless stated otherwise), 5 μl of replicase preparation, 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM dithiothreitol, 0.5% (vol/vol) Triton X-100, 1 mM MnCl2, 200 μM ATP and UTP, 500 μM GTP, and 250 nM [α-P32]CTP (Amersham). Reaction mixtures were incubated at 25°C for 60 min, and reactions were stopped by phenol-chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen and 0.4 M ammonium acetate. Products were separated by electrophoresis on 10 to 20% denaturing (8 M urea) polyacrylamide gels. Gels were wrapped in plastic and exposed to film at −80°C. Product bands were quantified using a PhosphorImager (Molecular Dynamics). All values presented are the mean of at least three independent assays, all of which varied by less than 20%. Percent synthesis is calculated based on the sum of the intensities from 15- and 16-nt products relative to the wild-type proscript (see below).
Native gel analysis.
RNA conformation native gel analysis was performed in 10% polyacrylamide gels (38% acrylamide and 2% bisacrylamide) containing 0.5× Tris-borate-EDTA (30). After preelectrophoresis for 20 min, a 100-pmol sample in RNA renaturing buffer (50 mM Tris-HCl [pH 7.4], 100 mM KCl, 0.1 mM EDTA) was heated to 90°C for 2 min before the samples were placed on ice for at least 10 min. The sample was then adjusted to contain 0.5× Tris-borate-EDTA, 5% glycerol, and bromophenol blue. Electrophoresis was performed at a constant 150 V at 5°C until bromophenol blue migrated to within 3 to 5 cm from the bottom of the gel (17 by 15 by 0.08 cm). The gel was then stained with toluidine blue to visualize the RNAs.
RESULTS
Sequence required for initiation of CMV subgenomic RNA in vitro.
Boccard and Baulcombe (4) found that the minimal necessary sequence for CMV subgenomic RNA4 synthesis maps to nt −30 to +30 relative to the initiation cytidylate. While the results from protoplast analysis are biologically relevant, it can be difficult to determine the contributions from multiple effects, such as changes in stability of the RNA. Biochemical analysis for RNA synthesis was done to complement and extend the protoplast results.
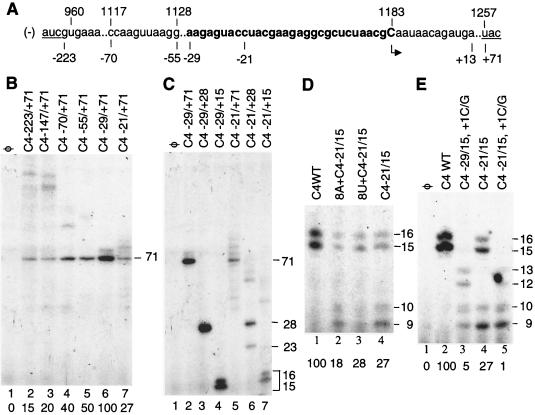
To examine the promoter for subgenomic RNA synthesis, we used CMV replicase enriched from tobacco plants infected with CMV strain Fny (29) and in vitro-transcribed proscripts. The latter were so named because these RNAs contain both promoter and template sequence. The first proscript is a 294-nt RNA containing the complement to the entire intercistronic region of CMV RNA3 (Fig. 1A). The 3′ end of RNA C4−223/+71 is 1 nt downstream of the complement of the 3a translational termination codon, and the 5′ end is 5 nt upstream of the translation initiation codon of the capsid protein-coding sequence. C4−223/+71 directed the synthesis of several RNAs, including a major product of ∼71 nt, the length expected from initiation at the site used predominantly in vivo (the complement of nt 1183 in RNA3) (Fig. 1B, lane 2). Faint higher-molecular-weight bands may be due to either initiation at alternative sites or RNA recombination by the CMV replicase after correct initiation. Similar recombination events have been observed and characterized with the BMV and cowpea chlorotic mottle virus replicases (M.-J. Kim and C. C. Kao, submitted for publication).
FIG. 1.
Sequences required to direct the initiation of CMV subgenomic RNA synthesis. (A) Partial sequence containing the complement to the intercistronic region of minus-strand CMV RNA3. Numbers correspond to the nucleotide positions of CMV strain Fny RNA3. The underlined sequences at the left and right correspond to the complement of the termination and initiation codons of the CMV 3a and capsid proteins, respectively. The sequence in bold contains the 29 nt 3′ of the initiation cytidylate. The sequence between positions 1117 and 1128 corresponds to the complement of the B-box sequence, a motif demonstrated to be important in BMV RNA replication (9, 23). The arrow identifies the initiation cytidylate for the initiation of CMV subgenomic RNA synthesis used in vivo (4). (B) Effects of deletion of the sequence 3′ of the initiation cytidylate. Proscripts used in the reactions are indicated at the top. “φ” indicates that no template was in the reaction. The expected 71-nt product is denoted to the right of the autoradiograph from a 7 M urea–10% polyacrylamide gel. (C) Effects of deletion of the sequence 5′ of the initiation cytidylate. Since the deletions decrease the template lengths, the products of RNA synthesis are of different lengths. The gel was of two densities; the bottom portion contained 20% acrylamide, and the top contained 10% acrylamide. The border of the two gel densities lies slightly above the 71-nt band. (D) Examination of the length 3′ of the initiation cytidylate required for accurate and efficient RNA synthesis in a 7 M urea–20% polyacrylamide gel. The expected products should be 15- and 16-nt. The 9- and 10-nt products were incorrectly initiated. (E) The +1C used in the initiation of RNA synthesis, and the effects of the 3′ sequence.
Deletions of the 3′ sequence of C4−223/+71 were tested to identify the minimal promoter sequence required for correct initiation of RNA synthesis. The following proscripts contain identical template sequences that end at nt +71. Truncation to make the 3′ ends at positions −147, −70, −55, and −29 all retained correct initiation of the ca. 71-nt product. An increase in the abundance of the correctly initiated product was observed with this series of deletions (Fig. 1B, lanes 3 to 6). However, having the 3′ end at −21 resulted in significant decrease in RNA synthesis and the appearance of some lower-molecular-weight bands (Fig. 1B, lane 7; Fig. 1C, lane 5). Thus, the 3′ end of the CMV subgenomic core promoter maps to between nt −29 and −21, in good agreement with the results of Boccard and Baulcombe (4).
To map the 5′ end of the functional proscript, the constructs contained the same 3′ end at nt −29 and different 5′ ends at positions +71, +28, and +15 nt. All three proscripts directed synthesis (Fig. 1C, lanes 2 to 4) at comparable levels after normalizing for radiolabeled CMP incorporation, indicating that the template sequence from positions +15 to +71 does not contain a signal that regulates RNA synthesis. With proscript CA−29/+15, the expected 15-nt product and a 16-nt product were resolved by PAGE. The latter was likely generated by the nontemplated addition of one nucleotide after the replicase reaches the 5′ end of the template (25). Nontemplated nucleotide addition is a common property of several RdRps and has been observed with CMV-associated RNAs (21, 25, 36, 38).
Three proscripts that have identical 3′ ends at position −21 and template lengths of 71, 28, and 15 nt were examined to determine if there is an effect on efficient and accurate initiation. Consistent with the above observation made with C4−21/+71 (Fig. 1B, lane 7), a proscript containing only 21 nt 3′ of the initiation cytidylate resulted in not only lower levels of the correct-size products but also an increase in the abundance of incorrectly initiated products (Fig. 1C, lanes 5 to 7). However, truncations at the 5′ end from nt +71 to +15 did not affect the amount of the correctly initiated products, confirming that a 15-nt template sequence is sufficient for efficient RNA synthesis. Since this template length allows high-resolution analysis of the replicase products, it was routinely used in subsequent experiments.
The products from −29/+15 and −21/+15 were examined by high-resolution PAGE. The 15- and 16-nt products from both proscripts were well separated in a 20% denaturing gel, and the difference in their production from the two proscripts was reproducible (Fig. 1D, lanes 1 and 4). Due to the use of CTP as the radiolabeled nucleotide, products that initiated from the +1C but were aborted before position +9 would not be radiolabeled. The reaction with −29/+15 produced only the 15- and 16-nt RNAs. In contrast, proscript −21/+15 directed the synthesis of 9- and 10-nt RNAs that could have initiated from the cytidylate at the +7 position (Fig. 1A; Fig. 1D, lane 4). Mutation of the +7C to a guanylate abolished synthesis of the 9- and 10-nt products from −21/+15, demonstrating that misinitiation occurred when the core promoter was only 21 nt (M.-H. Chen, unpublished data).
To determine whether positions −29 to −22 contain a specific sequence that contributes to replicase recognition, or simply serves as a required length, we added either eight adenylates or uridylates to the 3′ end of −21/+15 to result in proscripts of the same length as −29/+15. These RNAs not only were less efficient in directing the synthesis of the 15- and 16-nt products in comparison to −29/+15 but also produced the 9- and 10-nt misinitiated products (Fig. 1D, lanes 2 and 3). Therefore, the sequence upstream from −29 to +15 is required for efficient and accurate initiation of CMV subgenomic RNA synthesis.
The use of the +1C in the initiation of the 15- and 16-nt RNAs was examined by changing the +1C to a G in the context of both −29/+15 and −21/+15 (Fig. 1E, lanes 3 and 5). These changes abolished the synthesis of the 15- and 16-nt products from proscripts −29/+15, +1C/G and −21/+15,+1C/G (Fig. 1E, lanes 3 and 5), indicating that the 15-nt product is initiated from +1C and terminated at the end of the template. Also, this result supports our hypothesis that the 16-nt band initiated correctly and likely has the addition of a nontemplated nucleotide at the 3′ end of the RNA. Thus, the initiation of RNA synthesis from proscript −29/+15 takes place at the cytidylate used in vivo (4). The 9- and 10-nt products from proscript −21/+15,+1C/G were unaffected in comparison to −21/+15, confirming that they resulted from incorrect initiation instead of premature termination. With both −29/+15,+1C/G and −21/+15,+1C/G, prominent 12- and 13-nt products were observed. Based on their sizes, these RNAs likely initiated from the +4 uridylate. In addition, proscript 29/+15, +1C/G also produced the 9- and 10-nt RNAs. These results suggest that the fully functional core promoter will dictate the site of initiation and that +1C is the preferred initiation site. However, initiation can take place at an alternative cytidylate and/or uridylate when the RNA contains changes in the core promoter or the authentic initiation cytidylate.
Strain variation of the putative CMV core promoter sequence.
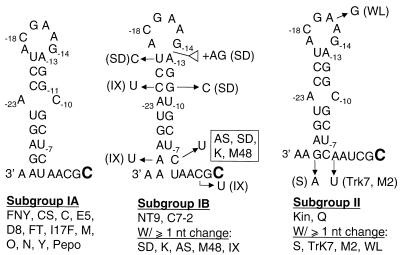
The biochemical analysis defined a sequence that could be analyzed for the features (structure or sequence) that direct RNA synthesis. The mfold RNA secondary structure prediction program (13) predicted that nt −28 to −5 upstream of the +1C in CMV strain Fny should form a stem of 9 bp, with a central A-C mismatch and a 6-nt purine-rich loop (Fig. 2A). The predicted RNA secondary structure of RNA C4−70/+15 also contained the same stem-loop at positions −28 to −5 (6). The stability of this secondary structure, as predicted by mfold, is −7.6 kcal/mol. The sequence from −4 to the end of the template was predicted to be unstructured.
FIG. 2.
Comparison of predicted RNA secondary structures for the sequence 3′ of the initiation cytidylate in three CMV subgroups. The initiation cytidylate used in vivo in strains Fny (subgroup I) and Kin (subgroup II) and the corresponding cytidylates in other CMV strains are in bold letters (4, 22). The CMV isolates used to generate the prototype structure are indicated directly under the viral subgroups, and strains that vary from the prototype are listed under the heading “W/≥ 1 nt change.” Nucleotides that diverged from the prototype in each subgroup are indicated with an arrow. The white triangle in the middle structure denotes the insertion of two nucleotides in strain SD.
CMV isolates can be divided into three phylogenetic subgroups: IA, IB, and II (24). To examine the possible relevance of features in the stem-loop, the intercistronic (minus-strand RNA3s from 25 different CMV isolates were aligned. The high degree of similarity in the sequence encompassing the core promoter makes the alignments unambiguous. For example, the template sequence of 3′-GCAA-5′ is found in all 25 strains at positions −1 to +3 (Fig. 2 and data not shown), helping to align the sequences. The sequences examined varied according to the three phylogenetic subgroups. Subgroup IA, which includes Fny, has the same sequence in all 12 strains examined. The seven subgroup IB isolates examined had several sequence changes relative to the IA motif. First, the predicted stem is shorter than those in the IA subgroup, indicating that some flexibility in the stem length is acceptable for CMV subgenomic RNA transcription. Also, the central A-C mismatch in the IA stem was not conserved in the IB isolates, indicating that it may not be an essential feature in replicase interaction. Strain SD in subgroup IB was particularly divergent relative to the others. In addition to numerous substitutions in the stem sequence (including a change of nt −11 from a G to a C that could destabilize the stem), strain SD also has two additional purines in the already purine-rich loop.
Six subgroup II isolates were examined, including the Kin isolate characterized by Boccard and Baulcombe (4). The subgroup II stem-loop was similar to the one in subgroup IA, with a 8-bp stem with a central A-C mispair. Subgroup II isolate S has a change in the terminal GC base pair that shortens the stem by one base pair. Strain WL has a transition of the −16 A to a G. The minor variations in the stem-loop sequence of the three CMV subgroups indicate some flexibility in the requirements for this sequence to act as a core promoter element.
Effects of sequence changes on RNA conformations.
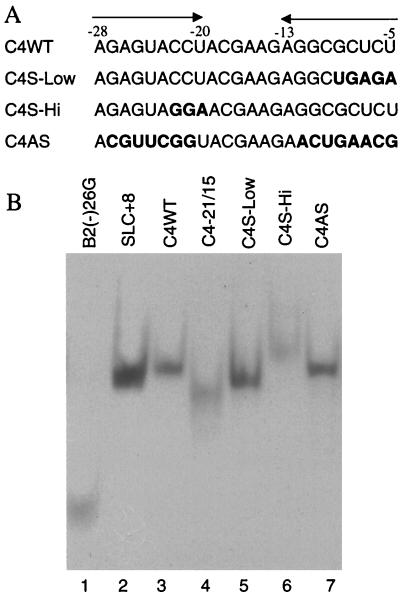
Previously we found that nondenaturing gel electrophoresis was able to distinguish different conformations of RNAs of the same length (30). Five CMV proscripts were tested: C4−29/+15 (henceforth referred to as C4WT), C4−21/+15, C4S-Hi, which has a mutation designed to destabilize the upper portion of the stem, C4S-Low, which should be destabilized in the lower stem, and C4AS, which contains a completely different 8-bp stem but maintains the loop sequence of C4WT (Fig. 3A). The stem in C4-AS comes from the core promoter for the BMV tRNA-like structure, SLC, whose structure was solved using multidimensional nuclear magnetic resonance spectroscopy and is known to exist in an A-form helix with a 3-nt loop (16). In a denaturing gel, all of the RNAs tested migrate according to their expected lengths (Chen, unpublished). In a nondenaturing gel, proscript C4WT migrated as a discrete band, indicating that any RNA structure that is present is likely due to intramolecular interactions and not due to the existence of a duplex of two molecules of C4WT. The observed mobility of C4WT was similar to that of SLC+8, a 45-nt RNA that forms a stable stem-loop and an 8-nt single-stranded sequence (Fig. 3, lane 2) (6, 16). Proscript C4−21/+15 migrated to a lower position in the gel, as expected for its lower molecular mass, and was more smeared, suggesting that the RNA exists as a mixture of molecular conformations (Fig. 3B, lane 4). C4S-Low migrated at a position reproducibly lower than that of C4WT, perhaps due to a longer single-stranded region (Fig. 3, lane 5). C4-Hi migrated as a higher-molecular-weight band in the nondenaturing gel, consistent with a larger loop that should make the RNA less compact or with an alternative structure (Fig. 3, lane 6). C4AS has mobility similar to that of C4WT, suggesting that both RNAs fold into a stem-loop structure. The altered mobility from the changes in the putative stem was also consistent with the existence of a stable stem-loop C4WT.
FIG. 3.
Analysis of RNA structure using native gel electrophoresis. (A) Sequences of the RNAs tested. Each of the RNAs tested is 44 nt in length, spanning from positions −29 to +15. However, only the portions relevant to formation of the stem-loop are shown, with the nucleotides that form the stem indicated by arrows. Nucleotide changes that affect the stem are shown in bold letters. C4AS contains a sequence from the stem of the BMV SLC, whose structure has been solved by nuclear magnetic resonance spectroscopy (Kim and Kao, submitted). (B) RNAs stained with toluidine blue after PAGE in a 10% nondenaturing gel. B2(−)26G is a 27-nt RNA whose secondary structure was reported by Sivakumaran et al. (30). SLC+8 is a 45-nt RNA whose structure was reported by Kim and Kao (submitted).
Effects of stem changes on RNA synthesis in vitro.
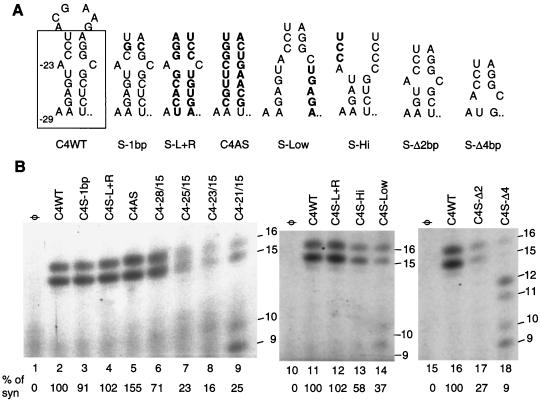
The role of the stem-loop in C4WT in directing RNA synthesis was examined systematically. The change of one base pair in the stem in proscript S-1bp did not affect RNA synthesis (Fig. 4B, lane 3). No effects on synthesis were observed when both sides of the stem were switched to their complementary sequences (lane 4). A change that should form a stem of a different sequence, C4AS, reproducibly directed about 1.5-fold of the synthesis from C4WT (Fig. 4A; Fig. 4B, lane 5). Furthermore, the absence of the 9- and 10-nt products indicates that these changes did not result in misinitiation. These results indicate that the sequence of the stem is not important for efficient and accurate initiation or the level of RNA synthesis, which is in agreement with the analysis of the sequences of CMV strains that showed some variations in the stem (Fig. 2). Also, the A-C mispair present in subgroups IA and II, but absent in subgroup IB isolates, appears unimportant in vitro.
FIG. 4.
Effects of changes in the stem in the CMV subgenomic core promoter on the level and accuracy of RNA synthesis. (A) Summary of most of the RNA constructs tested for the ability to direct RNA synthesis. Nucleotides changed from the prototype C4WT are indicated in bold letters. The predicted RNA secondary structures that resulted from the changes are also shown. (B) Autoradiograph of the results from RNA synthesis assays from several proscripts. The proscripts tested are indicated above the lanes, and sizes of the products are indicated in nucleotides at the left. “% of syn” indicates the percentage of the CMV replicase products made from the specified template relative to C4WT tested in the same set of reactions; “φ” indicates that no template was added to the reaction. A 7 M urea–20% polyacrylamide gel was used for analysis of the 15- and 16-nt products shown in this and subsequent figures.
Since C4−21/+15 directed a significant amount of misinitiation (Fig. 1D), we wanted to determine what changes in C4WT would result in misinitiation. Mutants that retained 28, 25, or 23 nt upstream of the initiation cytidylate were examined. Proscripts C4−25/+15 and C4−23/+15 resulted in reduced synthesis (Fig. 4B, lanes 7 and 8). However, the 9- and 10-nt misinitiated products were less abundant in reactions with C4−23/+15, while they were easily observed C4WT (Fig. 1E, lane 4; Fig. 4B, lane 9). This result suggests that a truncated stem of ∼3 bp near the loop might be sufficient to prevent misinitiation.
To confirm the importance of the stem, changes that should destabilize the stem were also tested. Proscript C4S-Hi has nt −22 to −20 changed to a sequence no longer complementary to −11 to −13. Proscript C4S-Low contains changes of −5 to −9 and is predicted by the mfold program to form an alternative structure. Both changes affected RNA structure, as determined by changes in the mobility of the RNA in a nondenaturing gel in comparison to C4WT (Fig. 3), and both resulted in the reduced ability to direct RNA synthesis (Fig. 4B, lanes 13 and 14). With C4S-Low, small amounts of the 9- and 10-nt products were observed (Fig. 4B, lane 14). Finally, proscripts that lack 2 (C4S-Δ2) or 4 (C4S-Δ4) bp of the stem were reduced in RNA synthesis (Fig. 4B, lanes 17 and 18). C4S-Δ4 had not only a more severe reduction of the expected 15- and 16-nt products in comparison to C4S-Δ2 but also an increase in the amount of misinitiated 9- and 10-nt products (Fig. 4B, lane 18). Taken together, these results confirm our previous hypothesis based on the comparison of CMV RNA sequences that a stem of ca. 7 bp in length is more than sufficient for efficient RNA synthesis. However, changes in the sequence of the stem can be tolerated as long as a stem is formed.
Effect of loop nucleotide changes.
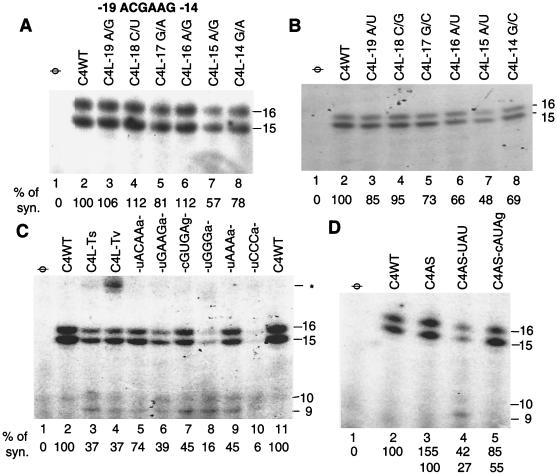
The loop nucleotides in the different CMV isolates were rich in purines, but the SD isolate had two additional purines inserted in the loop (Fig. 2). To examine the features in the loop required for RNA synthesis, a relatively conservative change of each of the six nucleotides (3′ ACGAAG 5′) to its Watson-Crick (W-C) transition was made separately and examined for RNA synthesis by the CMV replicase (Fig. 5A). Most of the nucleotides could be individually changed without a significant effect on the efficiency and specificity of initiation, except for C4L−15A/G, whose change decreased synthesis to 57% relative to C4WT (Fig. 5A, lane 7). Next, each of the six nucleotides was individually changed to their W-C transversions. Changes at positions −19, −18, and −17 did not affect RNA synthesis, but slight reductions were observed with changes of the individual purines at positions −16 and −14 (lanes 3 to 8). Reduced synthesis was also observed with W-C transitional mutant C4L−14G/A (78%) (lane 8) but not with C4L−16A/G (112%) (lane 6), although the reduction was not as significant as that from transversions. More significant reduction was observed with a change at position −15 (57%) (Fig. 5B, lane 7). The accuracy of initiation was not affected by these mutations. These results indicate that a specific adenylate at position −15 and probably accompanied with purine bases are favored even though the specific nucleotide requirements at other positions in the loop for RNA synthesis in vitro are relatively lax.
FIG. 5.
Effects of changes in the hexanucleotide loop within the CMV core promoter. (A) Effects of single-base W-C transversions of the six loop nucleotides. The sequence of the wild-type loop from positions −19 to −14 is in bold. The constructs used are named so as to indicate the identity of the original base before the slash and the nucleotide of the substitution after the slash. Positions of the 15- and 16-nt replicase products are shown at the right. “% of syn” indicates the percentage of the CMV replicase products made from the specified template relative to C4WT tested in the same set of reactions; “φ” indicates that no template was added to the RNA synthesis reaction. (B) Effects of single-base W-C transversions of the six loop nucleotides. (C) Effects of multiple nucleotide substitutions and deletions in the loop. C4-Ts has all of the loop nucleotides changed to 3′ GUAGGA 5′; C4-Tv has all of the loop nucleotides changed to 3′ UGUUUC 5′. Changes in the other constructs are indicated according to the final sequence of the loop. The putative closing base pairs are in lowercase letters. Products of the RNA synthesis reaction are indicated at the left. The asterisk denotes a misinitiation product of ca. 25 nt. (D) Effects of changes in both the stem and the loop nucleotides. C4AS contains an alternative stem sequence shown in Fig. 4. The nucleotides in the loop are identified in the names of the proscripts. Where changed, the identities of the closing nucleotides are indicated in lowercase letters.
More drastic changes to the loop nucleotides were examined. W-C transitions of all six nucleotides in the loop were made in a proscript named C4L-Ts. C4L-Ts directed RNA synthesis at 37% relative to C4WT and resulted in small amounts of misinitiated products (Fig. 5C, lane 2). Changing the loop sequence to their W-C transversions (3′UGCUUC5′) in proscript C4L-Tv also resulted in approximately 37% of the level of synthesis in comparison to C4WT. Minor amounts of 9- and 10-nt misinitiated products were observed with both C4L-Ts and C4L-Tv. However, a prominent longer misinitiated product was observed with C4L-Tv (lane 4). We decided to next alter the number of nucleotides in the loop. Two different proscripts, with U-A closing base pairs and four loop nucleotides of ACAA and GAAG (same as those in C4WT), were found to have only a minor detrimental effect on the efficiency and accuracy of RNA synthesis (lanes 5 and 6, respectively). However, since a U-A closing base pair is relatively unstable and could result in a 6-nt loop (with one fewer base pair in the stem), we changed the loop to a tetraloop of the GNRA class (34) with a CG closing base pair. The latter proscript directed 45% synthesis of the 15- and 16-nt products in comparison to C4WT, with a noticeable increase in the abundance of the 9- and 10-nt misinitiated products. Next, a 3-nt loop of the sequences of GGG, AAA, and CCC were tested in the context of a UA closing base pair. All three constructs had an increased amount of the 9- and 10-nt misinitiated products relative to C4WT (lanes 8 to 10). However, the amount of the 15- and 16-nt products differed more significantly. A loop of AAA was able to direct 45% synthesis of C4WT (lane 9), while loops of GGG or CCC resulted in RNA synthesis of only 16 or 6%, respectively, relative to C4WT (lanes 8 and 10). All of these changes indicate that one or more adenylates are preferred for efficient and accurate synthesis by the CMV replicase. The only RNA that directed synthesis without an adenylate in the loop was C4-Tv (lane 4). Whether this is due to some compensatory effects of the neighboring nucleotides is not clear.
We next changed both the stem and the loop (Fig. 5D). Consistent with previous results, C4AS, containing an altered stem sequence and the normal 6-nt loop, directed 1.5-fold more synthesis than C4WT without incorrectly initiated products (Fig. 4B, lane 5; Fig. 5D, lane 3). However, an altered stem with a triloop of UAU and a UA closing base pair reduced RNA synthesis to 27% relative to synthesis from C4AS and also produced the 9- and 10-nt misinitiated RNAs (Fig. 5D, lane 4). A triloop with two adenylates (AUA) and a CG closing base pair was able to accurately direct synthesis of only the 15- and 16-nt products, but at 55% relative to C4AS (Fig. 5D, lane 5). These results are in agreement with our previous observation that there is a preference for one or more adenylates at the 5′ portion of the loop.
Proscript sequence needed to interact with replicase.
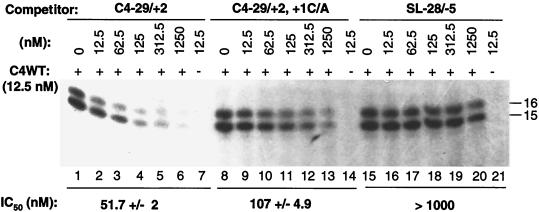
The observation that some changes in the stem-loop will affect the use of an initiation site indicates that both the stem-loop and the initiation cytidylate may interact with the replicase. To determine if the sequence interacted directly with the CMV replicase, we used a template competition assay. In this assay, increasing concentrations of a competitor RNA that does not produce visible products were added to reaction mixtures containing a constant amount of C4WT. RNA containing nt −29 to +2, with the −2C changed to U to remove a potential alternative initiation cytidylate, decreased the synthesis from C4WT significantly. C4−29/+2 was able to reduce synthesis from C4WT to 50% when present at 52 nM. This concentration, the IC50, could be used to compare the relative inhibitory activities of different competitors. The IC50 for −29/+2, with the +1C changed to an adenylate, was 107 nM (Fig. 6), demonstrating that the initiation cytidylate does contribute to the interaction with the replicase. An RNA containing the sequence from −28 to −5, with only the stem-loop necessary for efficient and accurate initiation, was a poor competitor, with an IC50 of >1 μM (Fig. 6). Therefore, both the stem within the CMV core subgenomic promoter and the initiation cytidylate are needed to stably interact with the CMV replicase.
FIG. 6.
Minimal proscript sequence required to interact with the CMV replicase, as identified by a template competition assay. The reaction measures the synthesis from C4WT as affected by the three competitor RNAs listed at the top. The IC50s were calculated from three independent experiments, and 1 standard deviation from the mean is listed after the mean. For SL-28/+5, reduction of synthesis to 50% was never obtained in three independent experiments.
Spatial requirements between the stem-loop and the initiation cytidylate.
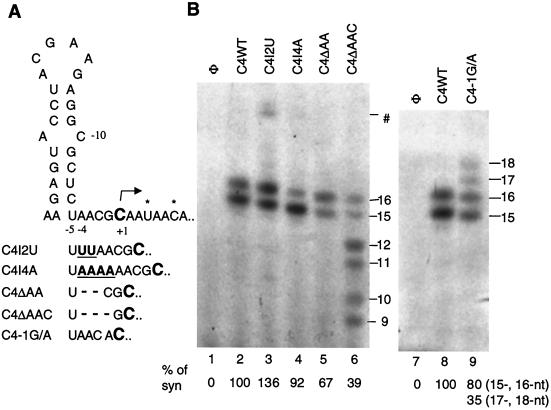
Since both the stem-loop and the initiation cytidylate are required to interact with the CMV replicase, we examined the effect of altering the number of nucleotides between the two elements. The single-stranded sequence between −5 and −1 was targeted for changes because it lies between the stem and the +1C. Two uridylates or four adenylates inserted between positions −5 and −4 in proscripts C4I2U and C4I4A, respectively, had little effect on the efficiency of RNA synthesis, except for a minor amount of a longer product from C4I2U, which likely initiated from −6C, based on the length of the product (Fig. 7A; Fig. 7B, lanes 3 and 4). Deletion of the two adenylates at −4 and −3 in proscript C4ΔAA resulted in decreased synthesis, and some misinitiated products were produced (Fig. 7B; lane 5). Removal of the three nucleotides from −4 to −2 in proscript C4ΔAAC ignificantly reduced accurate initiation, and increased amounts of RNAs of 9, 10, 11, and 12 nt were observed, probably due to misinitiation at +6C and +3C (Fig. 7B, lane 6). These results indicate that there is some flexibility in the spatial requirements between the stem-loop and the initiation site, and insertions are better tolerated than deletions.
FIG. 7.
Effects of changing the spacing between the stem-loop and the initiation cytidylate. (A) Schematic of the region affected by insertions and deletions. The initiation cytidylate is in bold and a larger font. Asterisks identify potential alternative initiation sites. Underlined bold letters indicate insertions; nucleotides deleted are indicated by dashes. (B) RNA synthesis from the mutant proscripts identified in panel A. Sizes of the initiation products are indicated in nucleotides at the right. The symbol ⋕ identifies a longer product in the reaction with C4I2U.
Some spatial flexibility in the position of the initiation template nucleotide is tolerated by the CMV replicase in vitro, which raises the question of why the −2C is not used as an initiation site, especially in the presence of 2-nt insertions between −5 and −4. Two previous observations may be relevant to the role of the −2C. First, Adkins et al. (2) previously showed that an adenylate is strongly preferred at the +2 position in the initiation of genomic and subgenomic plus-strand synthesis of alpha-like viruses. Second, Sivakumaran et al. (30) established that guanylates and cytidylates are generally not found in the first four positions after the initiation of genomic plus-strand RNA synthesis in alphaviruses. These observations suggest that the −2C is not a suitable initiation site because the −1 position is a G. Therefore, C4−1G/A was made, changing the −1G to an A. C4−1G/A initiated RNA synthesis from the −2C, as evidenced by the products that are 17 and 18 nt in length, at 35% in comparison to the correctly initiated products of 15 and 16 nt (Fig. 7B, lane 9). The −1G is conserved in all of the CMV strains examined (Fig. 2) and is also found in the templates for subgenomic RNAs of BMV and CMV. It may be required to lend specificity to the recognition of the +1C.
DISCUSSION
Due to its importance as a plant pathogen, CMV has been well characterized in terms of host range, symptom determinants, and other biological properties. However, the biochemical characterization of the CMV RNA replication signals has lagged behind that of other viruses. We seek to take a reductionist's approach to define the sequences responsible for directing CMV RNA synthesis. The results can be compared and contrasted to those for other members of the Bromoviridae. For the CMV subgenomic RNA synthesis, we have defined a stem-loop sequence from −28 to −5 relative to the initiation cytidylate that contributes to efficient and accurate initiation of subgenomic RNA synthesis.
Our results are in general agreement with and extend the analysis of CMV replication in protoplasts by Boccard and Baulcombe (4). In protoplasts, nt −30 to +67 containing the CMV subgenomic promoter could direct synthesis of the native subgenomic RNA4 and of a second copy of the subgenomic promoter placed within the capsid-coding sequence, although synthesis is lower than an RNA with 70 nt upstream of the initiation cytidylate (4). We found that in vitro, the presence of sequence 3′ of position −28 did not improve RNA synthesis. The differences observed by Boccard and Baulcombe (4) may have been due to some factor(s) that enhances RNA synthesis which is absent in our replicase preparation or to the fact that the sequence from −70 to −30 has some effects in vivo other than on RNA synthesis. Nonetheless, the finding that only 28 nt upstream of the initiation site is sufficient to direct basal levels of RNA synthesis indicates that the core subgenomic promoter is contained within this sequence. We have demonstrated that the stem-loop and the initiation cytidylate are minimal replicase recognition elements. Both contribute to the specific selection of the initiation template nucleotide.
While the stem-loop structure upstream of the CMV subgenomic RNA4 initiation site contributes to the efficiency and accuracy of initiation, it is somewhat surprising that many of the changes made in the sequence allowed RNA synthesis. While the presence of a stem-loop is required, complete replacement of the stem and loop nucleotides with at least some alternative sequences retained efficient and accurate RNA synthesis (Fig. 5D). The sequences found in various CMV strains support the claim that some variations in the stem and the loop are tolerated.
Yoshinari et al. (37) and Deiman et al. (7) have previously proposed that a nonspecific structure upstream of an initiation site may be sufficient for the specific recognition of the initiation site. Despite the fact that some mutations in the stem-loop were still recognizable and able to direct RNA synthesis by CMV replicase, we note that not all nonspecific secondary structures can direct efficient and accurate initiation (e.g., structures in which the loops were changed to CCC or GGG [Fig. 5C]). In fact, several changes in the stem and the loop caused initiation to occur at both +1 and other positions. Therefore, while the sequences in the stem and loop can be changed and retain significant levels of RNA synthesis, the wild-type sequence has already apparently been optimized for efficient and accurate initiation of RNA synthesis.
CMV, BMV, and AMV subgenomic promoters.
RNA structures that appear to direct RNA synthesis by viral replicases have been demonstrated in several species (for review, see references 5 and 19). In several of these cases, the sequence that directs correct initiation of RNA synthesis includes a stem-loop upstream of the initiation site (e.g., reference 32). Also, for subgenomic RNA synthesis of barley yellow dwarf virus and AMV, stem-loops are apparently required (12, 17). The identification of a stem-loop directing efficient and accurate CMV subgenomic RNA synthesis contributes to this paradigm.
The requirement for AMV subgenomic RNA synthesis in vitro differs from the situation with the CMV subgenomic core promoter, where the stem sequence can be totally changed with no adverse effects on the efficiency and accuracy of RNA synthesis. Exchanging the two sides of the AMV stem, especially in the lower part of the stem, caused significant reduction in RNA synthesis. Also, the structure of the AMV stem-loop was insufficient for the initiation of AMV subgenomic RNA synthesis, since a single-stranded region from positions −37 to −31 was required (11). It is possible that the specific nucleotides in the AMV stem and single-stranded regions, or a structure other than the one proposed, contribute to replicase recognition.
With the BMV subgenomic core promoter, several specific nucleotides at positions −17, −14, −13, and −11 that exist in an apparently unstructured region of the core promoter direct replicase interaction and accuracy of initiation (25). Nucleotide moieties at these four positions that can affect replicase recognition have been identified (26). Within positions −17 to −11, stringent spatial requirements exist with respect to the BMV subgenomic core promoter. One or two nucleotides inserted or deleted between positions −17 and −14 and between positions −14 and −13 resulted in severe decreases in RNA synthesis (33). The situation is quite different with the CMV core subgenomic promoter, where removal of several loop nucleotides and even replacement of the endogenous stem-loop with a different stem-loop still retained more than 50% of the synthesis of the wild-type proscript (Fig. 5). We conclude that the CMV replicase has more relaxed recognition requirements compared to the BMV replicase. Thus, the previous hypothesis of a common recognition mode for the subgenomic promoters proposed by our lab (25) was incorrect.
Haasnoot et al. (11) suggested that the recognition nucleotides in the BMV subgenomic core promoter also exist in a stem-loop secondary structure. The stem was proposed to be formed by the base pairing of nt −22 to −17 and −14 to −9, partly because transversions of both the −17 and −13 mutations resulted in 17% of wild-type core promoter activity in comparison to the single changes at either position (11). This claim is in contrast to our analysis of the BMV subgenomic core promoter (1, 25–27, 33), and such low RNA synthesis by the double mutation indicates a specific requirement for the bases at those positions, not the formation of a base pair. Furthermore, mutations affecting both sides of the putative stem could retain significant levels of RNA synthesis in comparison to a wild-type RNA (1, 28). Finally, UV spectrometry performed continuously from 0 to 100°C revealed that a functional BMV subgenomic proscript, −20/−13, does not possess a stable structure in solution, even at 10°C (our RNA synthesis assays can be performed at up to 40°C (C. C. Kao, unpublished data). A stable structure in the BMV subgenomic promoter does not appear to exist a priori in solution to direct recognition by the BMV replicase. This is not to say that RNA structure does not play a role in BMV subgenomic RNA initiation. We have reported evidence consistent with the model that intramolecular RNA interactions may form after replicase binding in an induced-fit mechanism (3, 33). A prominent difference between the CMV and BMV subgenomic core promoters is the presence of a polyuridylate tract 3′ of the BMV core promoter. The polyuridylate tract may keep the core sequence accessible for replicase interaction and obviate the need to evolve a specific structure that then dictates subgenomic RNA synthesis (1). In CMV, the presence of eight uridylates was unable to compensate for the truncation of the stem-loop (Fig. 1D). Thus, even closely related viruses could adapt different strategies for the initiation of RNA synthesis.
Despite the differences in the initial recognition of the CMV and BMV subgenomic core promoters by the appropriate viral polymerases, several features are common to both promoters. First, they both have relatively short sequences that interact with the replicase and direct accurate initiation of RNA synthesis in vitro. Second, there is a flexible spacer sequence between the key recognition elements and the initiation cytidylate. Third, the initiation cytidylates are required by both replicases to increase the stability of the interaction between the proscripts and the replicases (Fig. 7 and reference 26). Fourth, the cytidylate used in vivo is apparently the optimal one for initiation due to the spacing between the recognition element and the +1C. For example, the −1G renders the −2C an ineffective initiation nucleotide, and the A-U-rich sequence immediately after initiation seems to play a role in efficient RNA synthesis. Given these similarities, it is quite likely that an induced-fit model will best describe the interaction between the CMV replicase and its core promoter. Finally, motifs other than the core promoter may be capable of directing viral subgenomic RNA synthesis. Long-range interactions between RNA elements have been reported to regulate subgenomic RNA synthesis in several viruses (15, 19, 28).
ACKNOWLEDGMENTS
Helpful discussions and encouragement from K. Sivakumaran and editing by L. Kao are much appreciated.
The Kao laboratory is supported by the NSF (MCB9507344) and USDA (9702126) and by a fellowship from the Samuel Noble Foundation. M.J.R. is supported by funding from the Samuel Robert Noble Foundation. C.C.K. acknowledges a Linda and Jack Gill fellowship.
REFERENCES
- 1.Adkins S, Siegel R, Sun J H, Kao C C. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA. 1997;3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins S, Stawicki S S, Faurote G, Siegel R, Kao C C. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 3.Adkins S, Kao C C. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology. 1998;252:1–8. doi: 10.1006/viro.1998.9449. [DOI] [PubMed] [Google Scholar]
- 4.Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 5.Buck K W. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman M R, Kao C C. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J Mol Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 7.Deiman B A L M, Konen A K, Verlaan P W G, Pleij C W A. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998;72:3965–3972. doi: 10.1128/jvi.72.5.3965-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding S W, Anderson B J, Haase H R, Symon R H. New overlapping gene encoded by the cucumber mosaic virus genome. Virology. 1994;198:593–601. doi: 10.1006/viro.1994.1071. [DOI] [PubMed] [Google Scholar]
- 9.French R, Ahlquist P. Characterization and engineering of subgenomic RNA. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grodberg J, Dunn J J. Purification of T7 RNA polymerase. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haasnoot P C J, Brederode F T, Olsthoorn R C L, Bol J F. A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA. 2000;6:708–716. doi: 10.1017/s1355838200992471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes R J, Buck K W. Infectious cucumber mosaic virus RNA transcribed in vitro from clones obtained from cDNA amplified using the polymerase chain reaction. J Gen Virol. 1990;71:2503–2508. doi: 10.1099/0022-1317-71-11-2503. [DOI] [PubMed] [Google Scholar]
- 13.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaspars E M S. A core promoter hairpin is essential for subgenomic RNA synthesis in alfalfa mosaic alfamovirus and is conserved in other Bromoviridae. Virus Genes. 1999;17:233–242. doi: 10.1023/a:1008065704102. [DOI] [PubMed] [Google Scholar]
- 15.Kim K-H, Hemenway C L. The 5′ nontranslated region of potato virus X affects both genomic and subgenomic RNA synthesis. J Virol. 1996;70:5533–5540. doi: 10.1128/jvi.70.8.5533-5540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C H, Kao C, Tinoco I. RNA motifs that determine specificity between a viral RNA replicase and its promoter. Nat Struct Biol. 2000;7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 17.Koev G, Mohan B R, Miller W A. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J Virol. 1999;73:2876–2885. doi: 10.1128/jvi.73.4.2876-2885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsh L E, Dreher T W, Hall T C. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 1988;16:981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller W A, Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- 20.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neufeld K L, Galarza J M, Richards O C, Summers D F, Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palukaitis P, Roossinck M J, Dietgen R G, Francki R I B. Cucumber mosaic virus. Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 23.Pogue G, Hall T C. The requirement for a 5′ stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992;66:674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roossinck M J, Zhang L, Hellwald K H. Rearrangements in the 5′ nontranslated region and phlogenetic analyses of cucumber mosaic virus RNA3 indicate radial evolution of three subgroups. J Virol. 1999;73:6752–6758. doi: 10.1128/jvi.73.8.6752-6758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel R W, Bellon L, Beigelman L, Kao C C. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1998;95:11613–11618. doi: 10.1073/pnas.95.20.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel R W, Bellon L, Beigelman L, Kao C C. Use of DNA, RNA and chimeric templates by a viral RNA-dependent RNA polymerase. J Virol. 1999;73:6424–6429. doi: 10.1128/jvi.73.8.6424-6429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sit T L, Vaewhongs, Lommel S A. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 29.Sivakumaran K, Bao Y, Roossinck M J, Kao C C. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and Cucumber mosaic virus. J Virol. 2000;74:10323–10331. doi: 10.1128/jvi.74.22.10323-10331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sivakumaran K, Kim C H, Tayon R, Jr, Kao C C. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J Mol Biol. 1999;294:667–682. doi: 10.1006/jmbi.1999.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smirnyagina E, Lin N, Ahlquist P. The polymerase-like core of brome mosaic virus 2a protein, lacking a region interacting with viral 1a protein in vitro, maintains activity and 1a selectivity in RNA replication. J Virol. 1996;70:4729–4736. doi: 10.1128/jvi.70.7.4729-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song C, Simon A E. Requirement of a 3′-terminal stem-loop in in vivo transcription by an RNA dependent RNA polymerase. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 33.Stawicki S S, Kao C C. Spatial requirements for promoter recognition by a viral RNA-dependent RNA polymerase. J Virol. 1999;73:198–204. doi: 10.1128/jvi.73.1.198-204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese C R, Winker S, Gutell R R. Architecture of ribosomal RNA: constraints on the sequence of “tetraloops.”. Proc Natl Acad Sci USA. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G, Kaper J M. Competition of viral and satellite RNAs of cucumber mosaic virus for replication in vitro and by viral RNA-dependent RNA polymerase. Res Virol. 1995;146:61–67. doi: 10.1016/0923-2516(96)80590-5. [DOI] [PubMed] [Google Scholar]
- 36.Wu G, Kaper J M. Requirement of 3′-terminal guanosine in (−)-strand RNA for in vitro replication of cucumber mosaic virus satellite RNA by viral RNA-dependent RNA polymerase. J Mol Biol. 1994;238:655–667. doi: 10.1006/jmbi.1994.1326. [DOI] [PubMed] [Google Scholar]
- 37.Yoshinari S, Nagy P D, Simon A E, Dreher T W. CCA initiation boxes without unique promoter elements support in vitro transcription by three RNA-dependent RNA polymerases. RNA. 2000;6:698–707. doi: 10.1017/s1355838200992410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]