ABSTRACT
The interaction between the gut microbiota and invariant Natural Killer T (iNKT) cells plays a pivotal role in colorectal cancer (CRC). The pathobiont Fusobacterium nucleatum influences the anti-tumor functions of CRC-infiltrating iNKT cells. However, the impact of other bacteria associated with CRC, like Porphyromonas gingivalis, on their activation status remains unexplored. In this study, we demonstrate that mucosa-associated P. gingivalis induces a protumour phenotype in iNKT cells, subsequently influencing the composition of mononuclear-phagocyte cells within the tumor microenvironment. Mechanistically, in vivo and in vitro experiments showed that P. gingivalis reduces the cytotoxic functions of iNKT cells, hampering the iNKT cell lytic machinery through increased expression of chitinase 3-like-1 protein (CHI3L1). Neutralization of CHI3L1 effectively restores iNKT cell cytotoxic functions suggesting a therapeutic potential to reactivate iNKT cell-mediated antitumour immunity. In conclusion, our data demonstrate how P. gingivalis accelerates CRC progression by inducing the upregulation of CHI3L1 in iNKT cells, thus impairing their cytotoxic functions and promoting host tumor immune evasion.
KEYWORDS: iNKT cells, CRC, Porphyromonas gingivalis, CHI3L1
Introduction
Colorectal cancer (CRC) is the third most prevalent cancer worldwide and the second leading cause of cancer-related death.1 The mutational landscape and the mechanisms of tumor initiation in CRC have been widely described, but colon carcinogenesis also depends on the interaction between cancer cells and the tumor microenvironment (TME).2 Indeed, the polarization and activation profiles of immune cells within the TME are highly informative to predict CRC patient survival or their response to therapy, highlighting the importance of the inflammatory microenvironment for CRC tumorigenesis.2 Microbiota-elicited inflammation is an important contributor to CRC pathogenesis regardless of pre-cancer inflammatory history.3 Pro-carcinogenic bacteria are able to initiate and promote colon cancer, partly through mechanisms that are not fully understood.4 Porphyromonas gingivalis is an opportunistic oral pathogen associated with different inflammatory diseases and cancers5,6 and specifically enriched in CRC patients.7,8 P. gingivalis accelerates epithelial cell proliferation through the MAPK/ERK signaling pathway9 and upregulates the expression of senescence and proinflammatory genes through the local production of butyrate.10 Moreover, P. gingivalis promotes CRC immune subversion through activation of the hematopoietic NOD-like receptor protein 3 inflammasome in tumor-infiltrating myeloid cells.11 Recently, we demonstrated that tumor-infiltrating invariant Natural Killer T (iNKT) cells favor a proinflammatory yet immunosuppressive TME in CRC by sensing Fusobacterium nucleatum, an opportunistic pathogen in the oral cavity.12 Indeed, F. nucleatum induces the iNKT cell-mediated recruitment of immunosuppressive PMN-MDSCs-like neutrophils, supporting CRC progression.12
iNKT cells are lipid-specific, evolutionary conserved, T lymphocytes13 expressing a semi-invariant αβ T cell receptor (Vα24-Jα18/Vβ11 in humans and Vα14-Jα18 in mice) together with NK surface receptors and manifest both adaptive and innate/cytotoxic functional properties.14 iNKT cells are specialized in the recognition of glycolipid antigens presented by the non-polymorphic MHC-Ib molecule CD1d. The α-galactosylceramide (αGalCer) is the prototypical iNKT cell antigen,15 although other related microbial and endogenous glycolipid antigens have been identified.16–19 iNKT cells are active players in cancer immune surveillance20 showing either direct or indirect anti-tumor effects on cancer cells.21 Moreover, iNKT cells are able to kill CRC cells through the perforin–granzyme pathway.22 Nevertheless, the role of iNKT cells in cancer biology is still controversial, which may reflect the negative impact of the TME on their functions. We and others demonstrated that in CRC, the TME impairs the killing ability of tumor infiltrating iNKT cells promoting immunosuppressive responses in myeloid cells12,23 and favoring liver metastasis.24 The suppression of iNKT cell anti-tumor functions by the TME is associated with poor overall survival in solid and hematologic tumors, but the environmental factors modulating their functionality are incompletely elucidated.20 Nonetheless, the functional impairment of iNKT cells is reversible upon modulation of the iNKT cell activation status.12,25 Here, we address the contribution of P. gingivalis to the activation status and functions of tumor-infiltrating iNKT cells in CRC. We demonstrate that P. gingivalis imprints a protumour phenotype on iNKT cells which in turn affects the intratumour mononuclear-phagocyte cell landscape. Mechanistically, P. gingivalis impairs iNKT cell cytotoxicity by interfering with the iNKT cell lytic machinery through the upregulation of chitinase 3-like-1 protein (CHI3L1) while promoting recruitment of neutrophils within the TME.
Results
Intestinal colonization by Porphyromonas gingivalis promotes CRC through iNKT cells
We have recently demonstrated that tumor-infiltrating iNKT cells contribute to CRC tumorigenesis through interaction with F. nucleatum.12 Given the common pathogenic features of P. gingivalis (Pg) and F. nucleatum,5,26 we hypothesized that Pg might similarly induce a protumour phenotype in iNKT cells. Thus, we classified our previously described cohort of CRC patients based on the presence of Pg in their mucosal-associated microbiota, which had been characterized in our prior study.12 We observed a significant enrichment of tumor-infiltrating iNKT cells and neutrophils in Pgpositive as compared to Pgnegative CRC patients (Figure 1(a,b)). The enrichment of both neutrophils and iNKT cells is consistent with our previous observations which underscore the iNKT cell-mediated recruitment of tumor-associated neutrophils (TANs) in CRC lesions.12
Figure 1.
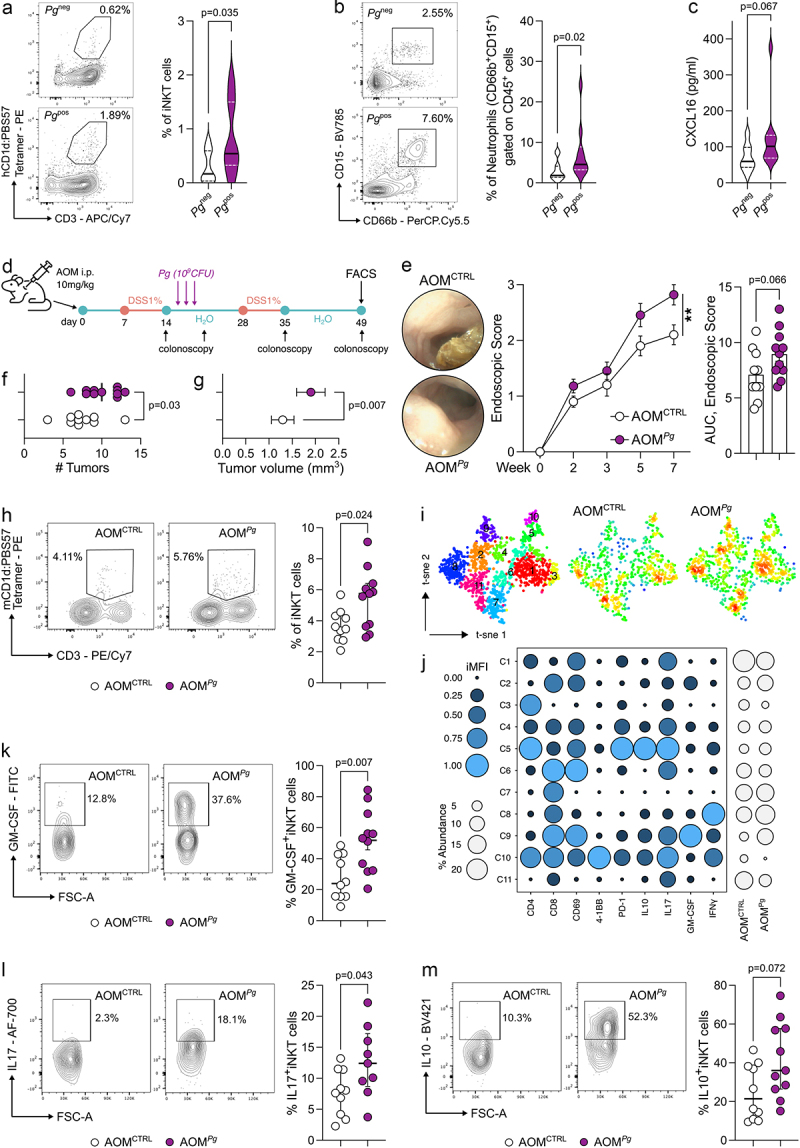
P. gingivalis promotes colorectal tumorigenesis by modulating iNKT cell functions. A) frequency of tumor-infiltrating iNKT cells and B) tumor-associated neutrophils in CRC patients (n = 31) positive (Pgpos; n = 16) or negative (Pgneg; n = 15) for P. gingivalis in their mucosa-associated microbiota with representative dot plots. C) CXCL16 concentration from tissue lysates (200 µg of total protein). D) schematic representation of the AOM-DSS experimental plan. E) tumour endoscopic score, AUC and representative endoscopic pictures, F) number and G) volume of tumors from AOM-DSS treated C57BL/6 animals orally gavaged with PBS (AOMCTRL) or 109 CFUs of P. gingivalis (AOMPg). H) frequency of tumor-infiltrating iNKT cells in AOMCTRL and AOMPg C57BL/6 mice with representative plots. I) t-sne map of iNKT cells based on phenograph metaclustering analysis of AOMCTRL and AOMPg tumor samples. J) balloon plot of the scaled integrated mean fluorescent intensity (iMFI) of phenograph clusters generated in panel J. K-M) frequency of tumor-infiltrating K) GM-CSF+ L) IL17+ and M) IL10+ iNKT cells in AOMCTRL and AOMPg C57BL/6 mice with representative plots. Data (n = 10, AOMCTRL; n = 11, AOMPg) from two pooled independent experiments representative of at least three.
The upregulation of IL17A, CD274 (PD-L1), and CCL20 in Pgpositive patients suggested that iNKT cells might accumulate within a TME favoring a Th17-like protumour response (Supplementary Figure S1A), where the cytotoxic functions akin to Th1-like responses are dampened. This hypothesis is further suggested by the diminished expression of TBX21, encoding for the Th1 transcription factor T-bet (Suplementary Figure S1A). Moreover, we found a trend increase (p = 0.067) in the concentration of intratumour CXCL16, a key chemokine responsible for recruiting iNKT cells,27,28 in Pgpositive patients (Figure 1c).
Next, we evaluated the contribution of Pg to CRC pathogenesis using the chemical azoxymethane–dextran sodium sulfate (AOM–DSS) model of colitis-associated CRC (Figure 1d). Pg-treated C57BL/6 (B6) mice (AOMPg) showed an increased tumor burden compared to control tumor-bearing mice (Figure 1e-g). A multidimensional immunophenotyping of T cells by Phenograph unsupervised clustering showed a different distribution of CD3+T cell density between AOMPg vs AOMCTRL B6 mice (Supplementary Figure S1B-C) although the frequencies of tumor-infiltrating CD4+ and CD8+ T cells as well as of NK cells did not differ by manual gating FACS analysis (Supplementary Figure S1D). In contrast, iNKT cells were significantly enriched in AOMPg samples (Figure 1h). The metaclustering analysis of the iNKT cell cluster C2 (Supplementary Figure S1C) showed that tumor-infiltrating iNKT cells in AOMPg mice are characterized by an overall increased expression of IL17, GM-CSF, and IL10 (C9; p = 0.08, Mann–Whitney U test) (Figure 1i-j). We further confirmed the overall higher expression of GM-CSF, IL17, and IL10 in iNKT cells by manual gating FACS analysis (Figure 1k-m and Supplementary Figure S1E). Notably, different iNKT cell clusters showed positivity for CD8, a quite rare feature among iNKT cells in mice29 whose existence may be dictated by defects in mechanisms of negative selection on developing iNKT cells.30 As expected, CD8+iNKT cells were almost absent in the thymus and barely present in the spleen of C57BL/6 mice at steady-state (Supplementary Figure S2A). However, a relevant fraction of colonic iNKT cells (~13%) expressed the CD8 as single-positive or double-positive (DP) cells in colon and CRC samples (Supplementary Figure S2B-C) suggesting unique tissue-specific features of colonic iNKT cells.
To demonstrate that Pg promotes CRC by modulating the iNKT cell functions, we induced tumorigenesis in iNKT cell deficient Cd1d-/- and Traj18−/− mice (Jα18−/−). The results obtained underscore the essential role of iNKT cells in Pg-mediated CRC tumorigenesis. Indeed, tumor-bearing iNKT knockout mice treated with Pg exhibited a comparable tumor burden to untreated AOM-DSS Cd1d-/- and Jα18−/− animals (Figure 2a-c). Intriguingly, frequencies and phenotypes of NK cells and conventional T cells remained largely unchanged in Jα18−/− mice irrespective of Pg treatment (Supplementary Figure S1F), further supporting the notion of a specific interplay between Pg and iNKT cells.
Figure 2.
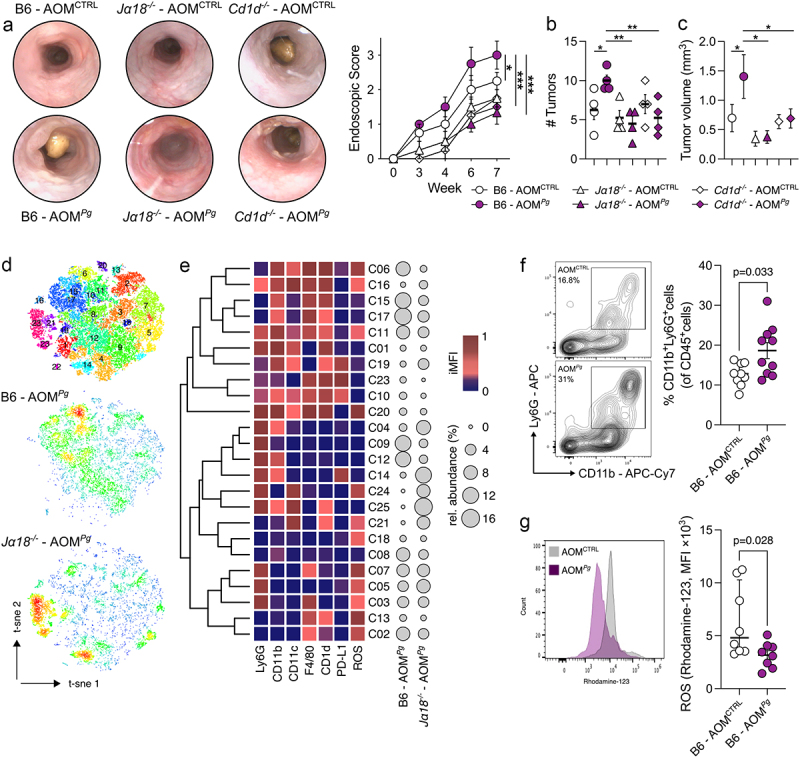
iNKT cells are essential to promote P. gingivalis-elicited colorectal tumorigenesis. A) Tumour endoscopic score and representative endoscopic pictures, B) number and C) volume of tumors from AOM-DSS treated B6, Traj18−/− (Jα18−/−) and Cd1d−/− mice orally gavaged with PBS (AOMCTRL) or 109 CFUs of P. gingivalis (AOMPg); data (n = 4) from one representative experiment. D) t-sne map of intratumour myeloid cells based on phenograph clustering from AOM-DSS, P. gingivalis treated C57BL/6 (B6 - AOMPg) and Traj18−/− mice (Jα18−/− - AOMPg). E) heatmap of scaled integrated MFI data from phenograph clustering analysis; relative abundance of the identified clusters is also shown. F) frequency of CD11b+Ly6G+ and G) respiratory burst quantification of tumor-associated neutrophils from AOM-DSS treated C57BL/6 mice orally gavaged with PBS (AOMCTRL) or 109 CFUs of P. gingivalis (AOMPg), with representative dot plots. Data (n = 8-10) from two pooled independent experiments representative of at least three.
Then, we performed a multidimensional Phenograph analysis of tumor-infiltrating myeloid cells in tumor-bearing B6 and Jα18−/− mice treated with Pg (Figure 2d-e) to investigate how Pg may condition the composition of myeloid cell populations through iNKT cells. Pg specifically induced the iNKT cell-dependent enrichment of CD11b+monocytic cells (C08; p = 0.03, One-way ANOVA), F4/80+CD1d+ activated (ROS+) macrophages (C02; p = 0.06, One-way ANOVA) and Ly6G+CD11b+neutrophils (C12; p = 0.08, One-way ANOVA) (Figure 2e). In agreement with our previous study,12 we observed the enrichment of TANs in Pg-treated B6 mice compared to untreated AOM-DSS animals (Figure 2f). This enrichment is accompanied by TANs reduced respiratory burst capacity (Figure 2g), suggesting a diminished cytotoxic potential.31
In summary, these data show that Pg promotes CRC by modulating the iNKT cell phenotype and the mononuclear-phagocyte cell landscape.
Porphyromonas gingivalis impairs iNKT cell cytotoxicity while promoting iNKT cell-mediated recruitment of TANs
To unravel the mechanisms governing the interaction between Pg and iNKT cells, we performed a series of experiments involving the priming of intestinal and circulating human iNKT cell lines22,32 with monocyte-derived dendritic cells (moDC) loaded with Pg. Subsequently, we conducted a range of in vitro functional assays and RNA sequencing on iNKT cells (Figure 3a). iNKT cells primed with Pg display a pronounced enrichment in genes associated with neutrophil chemotaxis, marked by the upregulation of key chemokines such as CXCL1, CXCL2, CXCL5, CXCL8, CCL2, and CCL4 (Figure 3b-c, Supplementary Figure S3A-B, Supplementary Table S1) as compared to iNKT cells activated by αGalCer, the prototype agonist of these cells.33 This finding strongly suggests that Pg can foster the iNKT cell-mediated recruitment of neutrophils. To validate this, we performed a migration assay that confirmed the capability of Pg-primed iNKT cells to induce neutrophil chemotaxis (Figure 3d). Furthermore, iNKT cell activation induced by Pg led to a reduction in the respiratory burst capability of neutrophils (Figure 3e) coupled with an elevated expression of the immunosuppressive marker PD-L1 (Figure 3f).
Figure 3.
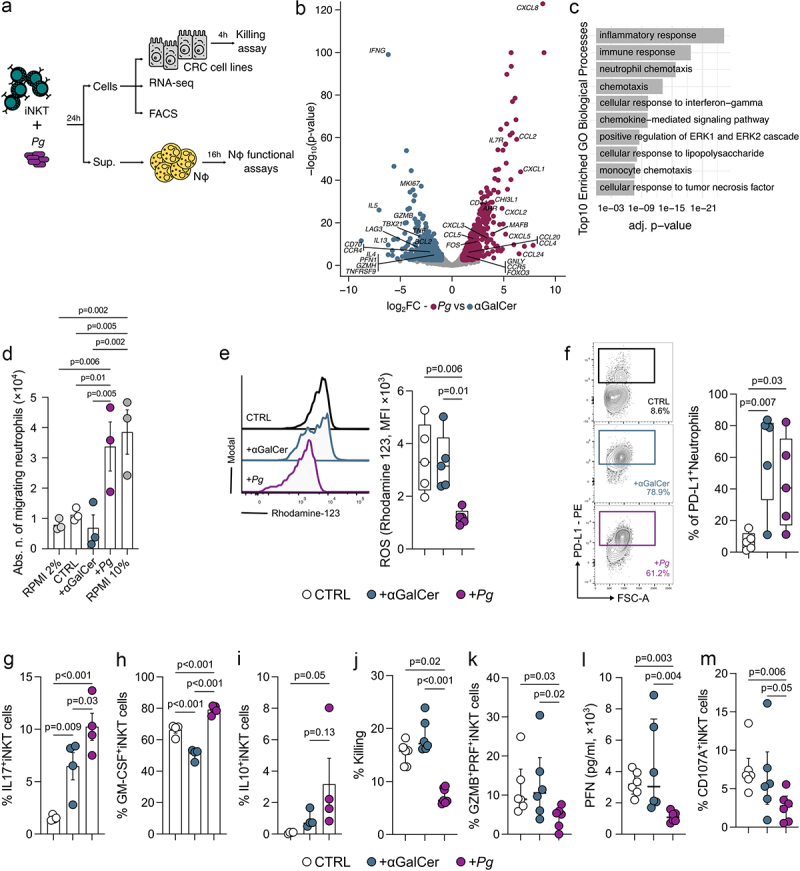
P. gingivalis promotes the iNKT cell-mediated recruitment of TANs while impairing iNKT cell cytotoxicity. A) schematic representation of the experimental plan. B) volcano plot representing the differentially expressed genes (DEGs) in Pg- vs αGalCer-primed iNKT cells; the volcano plot shows for each gene (dots) the differential expression [log2fold-change (log2FC)] and its associated statistical significance (log10p-value). Dots indicate those genes with an fdr-corrected p < 0.05 and log2FC > |1|. C) gene ontology (GO) analysis of differentially expressed genes (Bonferroni-corrected p < 0.05 and log2FC >2). D) absolute numbers of migrating neutrophils upon exposure to RPMI + 2%FBS (negative control), RPMI + 10%FBS (positive control), unloaded moDC (CTRL), αGalCer- and pg-primed (+Pg) iNKT cell supernatants. Results are representative of three (n = 3) independent experiments. p < 0.05 (*), p < 0.01 (**), one-way ANOVA. E) respiratory burst assay quantification and F) frequency of PD-L1+ cells from neutrophils exposed to the culture supernatants of unloaded moDC (CTRL), αGalCer- and Pg-primed (+Pg) iNKT cells with representative plots. G-I) frequency of G) IL17+ H) GM-CSF+ and I) IL10+ iNKT cells upon stimulation with P. gingivalis. J) percentage of killed tumor cells by unloaded moDC (CTRL), αGalCer- and Pg-primed (+Pg) iNKT cells. K) frequency of GZMB+PFN+iNKT cells. L) perforin concentration in the culture supernatant of unloaded moDC (CTRL), αGalCer- and Pg-primed (+Pg) iNKT cells. M) frequency of CD107a+iNKT cells. Data are representative of at least three independent experiments.
From a phenotypic perspective, Pg induced a Th17-like profile in iNKT cells, characterized by the expression of IL17, GM-CSF, and IL10 (Figure 3g-i). Functionally, Pg abrogated the cytotoxic capabilities of iNKT cells against colon adenocarcinoma cell lines (Figure 3j) because of the reduced expression of granzyme B (GMZB) and perforin (PFN) (Figure 3k-l) and impaired lytic degranulation as shown by decreased CD107a expression (Figure 3m). Nevertheless, this mechanism appears to be independent of antigen presentation, as inhibition of CD1d-mediated signaling neither reduce the expression of IL-17, GM-CSF, and IL-10 in Pg-primed iNKT cells, nor does its rescue iNKT cell cytotoxicity (Supplementary Figure S3C-F). This suggests that CD1d may not be central to this response. Nevertheless, some lines of evidence have shown that the upregulation of Th17 genes on CD4+T cells by P. gingivalis is mediated by TLR-4 signaling,34 whereas IL-10 expression is mediated by TLR-2.35 Thus, we primed iNKT cells with Pg in the presence of neutralizing antibodies for TLR-2 and TLR-4. We observed that the inhibition of both TLRs provoked a reduction in the frequencies of IL17, GM-CSF, and IL-10 positive iNKT cells (Supplementary Figure S3G-I). iNKT cell cytotoxic functions were not affected by TLR blockade (Supplementary Figure S3J) suggesting that the activation of iNKT cells by Pg might partly involve TLR signaling.
Collectively, these data suggest that Pg has the potential to foster CRC tumorigenesis by compromising the cytotoxic functions of iNKT cells.
Porphyromonas gingivalis impairs iNKT cell cytotoxicity through CHI3L1
The RNA-seq analysis of Pg-primed iNKT cells revealed the upregulation of CHI3L1 (Figure 3b). CHI3L1 is known to be a proinflammatory protein36 that exerts its influence on the cytotoxic machinery of NK cells, resulting in the inhibition of their killing functions.37 Thus, we hypothesized that Pg might undermine iNKT cell cytotoxicity through a similar mechanism. In agreement with its mRNA expression, we measured a higher concentration of CHI3L1 in the culture supernatant of Pg-primed iNKT cells (Figure 4a).
Figure 4.
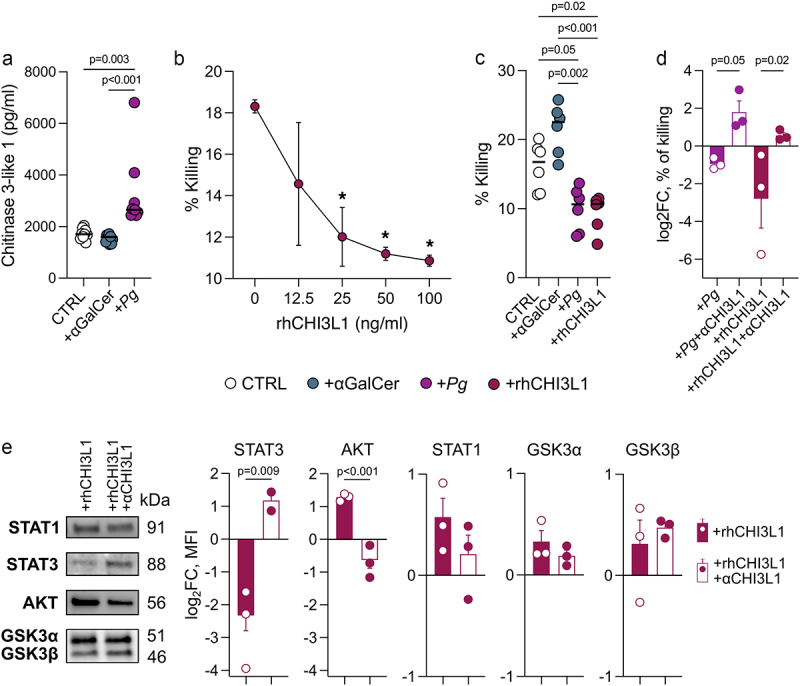
P. gingivalis exerts its effect on iNKT cells through CHI3L1. A) CHI3L1 concentration in the culture supernatant of Pg-primed (+Pg) iNKT cells and unloaded moDC (CTRL). B) percentage of killed tumor cells by iNKT cells treated with increasing doses of rhCHI3L1. C) percentage of killed tumor cells by iNKT cells basal activated (CTRL), primed with 109 CFUs of P. gingivalis (+Pg) or treated with rhCHI3L1 (100 ng/ml). D) Log2 fold-change of the percentage of killed tumor cells by iNKT cells primed with 109 CFUs of P. gingivalis (+Pg) or treated with rhCHI3L1 (100 ng/ml) upon neutralization with anti-CHI3L1 antibody (full bars); data have been normalized vs iNKT cells primed with unloaded moDC. E) western blot analysis; MFI is expressed as log2 fold-change normalized vs iNKT cells primed with unloaded moDC. Data are representative of at least three independent experiments.
In order to assess whether CHI3L1 directly affects iNKT cell cytotoxicity, we pre-treated human iNKT cell lines with varying concentrations of CHI3L1 before their co-incubation with target cells. Our findings indicate that CHI3L1 indeed impairs the iNKT cell cytotoxicity in a dose-dependent manner (Figure 4b), mirroring the effects observed with Pg treatment (Figure 4c). To further investigate this aspect, we neutralized CHI3L1 using an anti-human CHI3L1 antibody on iNKT cells treated with both Pg and CHI3L1. This intervention successfully restored iNKT cell functions (Figure 4d) and STAT3 signaling (Figure 4e), a transcription factor critical to promote tumor-specific cytotoxic T cell development and effector functions in cancer.38 Collectively, these results highlight the potential of targeting CHI3L1 as a therapeutic strategy to restore iNKT cell activity.
Discussion
Numerous studies have unveiled the capacity of the gut microbiome to regulate iNKT cell functions in health and disease.18,28,32,39–48 Nonetheless, the mechanisms by which gut microbes exert their influence on iNKT cells are not fully elucidated, and warrant further investigations for their safe use for adoptive cell therapies.20,25 The identification of such mechanisms assumes paramount importance given our recent findings in which we demonstrated that tumor-infiltrating iNKT cells bear unfavorable prognostic implications in human CRC because of the functional interaction with the periodontal pathogen F. nucleatum.12 Although iNKT cells are recognized as crucial components of anti-tumor immunity and their infiltration within tumor lesions is regarded as a positive prognostic factor,49,50 in the presence of F. nucleatum, iNKT cells acquire a protumour Th17-like phenotype that facilitates the recruitment of immune suppressive TANs and fosters CRC progression.12
P. gingivalis is a keystone periodontal pathogen that is enriched in CRC and associated with poor overall and relapse-free survival.10,11 Here, we show that P. gingivalis increases the intratumour abundance of pro-inflammatory, yet immune suppressive iNKT cells in CRC patients and in in vivo models of CRC. Prior studies have indeed emphasized the elicitation of IL17- and IL10-mediated responses by P. gingivalis.35,51–54 P. gingivalis has the potential to enhance Th17 responses by upregulating the expression of key cytokines such as IL-6, IL-23, and IL-1β on myeloid cells,51–53 while simultaneously exerting a suppressive effect on IL-12 production.53 iNKT cell activation and differentiation into NKT17 relies on the presence of these cytokines,55–57 suggesting that P. gingivalis may similarly induce a Th17-like phenotype in iNKT cells. Remarkably, the involvement of gingipains and FimA, two significant P. gingivalis virulence factors54 involved in bacterial adhesion to tissues,58,59 emerge as pivotal determinants of IL17 and IL10 production and the promotion of pathogenic responses in chronic inflammatory conditions.35,51,52 In this context, the suppression of anti-tumor immunity by P. gingivalis might resemble the mechanism used by F. nucleatum, where adhesion of the Fap2 virulence factor to TIGIT receptor inhibits NK cell cytotoxicity.60 This mechanism could partly explain why modulation of the iNKT pro-tumor phenotype by P. gingivalis seems to be independent of antigen presentation by the CD1d antigen-presenting molecule. Moreover, P. gingivalis fosters an inflammatory response in CRC achieved through the recruitment of tumor-infiltrating CD11b+ myeloid cells and activation of the NLPR3 inflammasome.11 However, evidence regarding the mechanism by which P. gingivalis mediates the active recruitment of these myeloid cells within the TME is still lacking. Our study suggests that the Pg-mediated recruitment of myeloid cells in tumor lesions might be predominantly orchestrated by iNKT cells, as we previously demonstrated for F. nucleatum.12 Indeed, upon exposure to F. nucleatum, iNKT cells actively recruit neutrophils through an IL-8 mediated mechanism,12 a mechanism described also in CRC and PDAC cells.61,62 Similarly, Pg-primed iNKT cells upregulated the expression of several neutrophil chemotaxis genes such as CXCL8, CXCL1, CXCL3, CXCL5, and CCL4 promoting the active recruitment of these cells in vitro and the enrichment of myeloid cells within tumor lesions in vivo. iNKT cells elicited remarkable changes in the intratumour mononuclear-phagocyte cell landscape in response to P. gingivalis, while the composition of conventional CD4+ and CD8+ T cells within the TME remained unaltered, as also shown in previous studies.11 Furthermore, P. gingivalis showed a unique and distinguishing pathogenic trait compared to F. nucleatum, i.e., the impairment of iNKT cell cytotoxic functions. The immune-mediated elimination of cancer cells depends on the lytic granule machinery of cytotoxic lymphocytes, including iNKT cells, CD8+ cytotoxic T lymphocytes, and NK cells. Cancer cells can use a variety of evasion mechanisms to prevent these cells from killing them. P. gingivalis was shown to interfere with the iNKT cell lytic granule machinery, since we observed a decreased secretion of perforin and reduced expression of the degranulation marker CD107a upon treatment with Pg. Our data suggest that P. gingivalis impairs iNKT cell cytotoxicity by inducing the production of CHI3L1 by iNKT cells, thus diminishing their ability to eliminate CRC cells.
CHI3L1 protein is a mammalian member of the evolutionarily conserved chitinase protein family. Although its precise physiological function remains incompletely understood, aberrant expression of CHI3L1 is linked to the development of various human diseases, including cancer.36 CHI3L1 expression by T cells promotes lung metastasis by dampening antitumour Th1 responses.63 When acting on NK cells, a population that shares phenotypic and functional features with iNKT cells, CHI3L1 disrupts the proper alignment of the microtubule-organizing center and lytic granules at the immunological synapse.37 Remarkably, we show that neutralization of CHI3L1 is sufficient to restore iNKT cell activity, even in the presence of exogenous CHI3L1 supplementation. CHI3L1 can signal through CD44, one of its physiological receptors,64–66 which is upregulated in Pg-primed iNKT cells. This signaling may act as an immune checkpoint to suppress iNKT cell activation via the PI3K/AKT pathway, a known repressor of STAT3 transcription38,67–69 Indeed, although STAT3 is a cell-intrinsic regulator of human unconventional T cell numbers and NKT17 function,57,70 we observed that CHI3L1 downregulated STAT3 while inducing AKT signaling. This suggests that STAT3 could be mainly involved in the modulation of iNKT cell cytotoxic functions rather than in the polarization of an NKT17 response, as demonstrated for conventional CD8+T cells.38 However, upon neutralization of CHI3L1, we did not find differences in the expression of GSK3, which is important for the induction of Th17 responses by leading to the activation of STAT3,71 nor in STAT1, which is important for the polarization of Th1 responses in opposition to STAT372 and is an important regulator of NK cell cytotoxicity.73 This warrants further investigations to dissect the molecular mechanisms induced by Pg modulating iNKT cell functions. Moreover, we acknowledge that a direct demonstration of iNKT cells impacting CRC development due to the exacerbation of their pro-tumoral effect by Pg in vivo is still lacking. Further investigations are therefore required, including adoptive transfer experiments of iNKT cells from Pg-treated and non-treated mice into iNKT cell-deficient mice, as well as the use of littermate controls to account for microbiota and uncontrolled genetic differences.
In conclusion, our data demonstrate that P. gingivalis fuels CRC progression by inducing iNKT cells to express CHI3L1, an immune checkpoint that can suppress iNKT cell cytotoxicity favoring host tumor immune evasion.
Materials & methods
Human samples
Samples were collected with informed consent from patients (n = 31) diagnosed with colorectal adenocarcinoma between January 2017 and July 2022 undergoing surgical resection at IRCCS Policlinico Ospedale Maggiore, Milan, Italy, as approved by the Institutional Review Board (Milan, Area B) with permission number 566_2015.
Tumour-associated microbiota isolation
The tumor-associated microbiota was obtained at the moment of surgery from tumor tissue by scraping. Handling, DNA extraction, sequencing, and analysis were performed as previously described.12
Isolation of tumour-infiltrating cells
Tumor samples were taken transversally to collect both marginal and core tumor zones. Human lamina propria mononuclear cells (LPMCs) were isolated as previously described.74 Briefly, the dissected intestinal mucosa was freed of mucus and epithelial cells in sequential steps with DTT (0.1 mmol/l) and EDTA (1 mmol/l) (Sigma-Aldrich) and then digested with collagenase D (400 U/ml) (Worthington Biochemical Corporation) for 5 h at 37°C in agitation. LPMCs were then separated with a Percoll gradient.
Mice
C57BL/6, B6.129S6-Del(3Cd1d2-Cd1d1)1Sbp/J (Cd1d-/-), and B6(Cg)-Traj18tm1.1Kro/J (Traj18−/−) mice75 (provided by G. Casorati and P. Dellabona, San Raffaele Scientific Institute) were housed and bred at the European Institute of Oncology (IEO) animal facility (Milan, Italy) in SPF conditions. Sample size was chosen based on previous experience. No sample exclusion criteria were applied. No littermate controls were used for experiments. No method of randomization was used during group allocation, and investigators were not blinded. Age-matched male and female mice were used for experiments. Animal experimentation was approved by the Italian Ministry of Health (Auth. 10/21 and Auth. 1217/20) and by the animal welfare committee (OPBA) of the European Institute of Oncology (IEO), Italy.
Porphyromonas gingivalis culture condition
P. gingivalis strain DSM 20709 (ATCC 33277) was maintained on Columbia agar supplemented with 5% sheep blood or in Columbia broth (Difco, Detroit, MI, USA) under anaerobic conditions at 37°C. Columbia broth was supplemented with hemin at 5 μg·mL −1 and menadione at 1 μg·mL −1. Bacterial cell density was adjusted to 1 × 1010 CFU·mL−1 and heat-killed at 95°C for 15 min before being stored at −80°C until use in downstream experimentation.
Animal experiments
Adult mice (7–8 weeks old) were injected intraperitoneally with azoxymethane (AOM, Merck) dissolved in isotonic saline solution at a concentration of 10 mg/kg body weight. After 7 d, mice were given 1% (w/v) dextran sodium sulfate (DSS MW 40 kD; TdB Consultancy) in their drinking water for 7 d followed by 14 days of recovery. The cycles were repeated for a total of two DSS cycles, and mice sacrificed at day 49. After the first cycle of DSS treatment, during the recovery phase, mice were orally gavaged for 3 d with 200 μl of PBS (control) or 109 CFUs (colony forming unit) suspension of P. gingivalis DSM 20,709. The treatment schedule is shown in Figure 1d.
Murine colonoscopy
Colonoscopy was performed weekly for tumor monitoring using the Coloview system (TP100 Karl Storz, Germany). Tumor endoscopic score has been quantified as previously described.76 During the endoscopic procedure mice were anesthetized with 3% isoflurane.
Murine cells isolation
Single-cell suspensions were prepared from the colon of C57BL/6 and Traj18−/− mice as previously described.48 Briefly, cells were isolated via incubation with 5 mM EDTA at 37°C for 30 min, followed by mechanical disruption with GentleMACS (Miltenyi Biotec). After filtration with 100-µm and 70-µm nylon strainers (BD), the LPMCs were counted and stained for immunophenotyping.
Cell lines
The different cell lines used in this work are listed in Supplementary Table S2. Human iNKT cell lines were generated from sorted CD45+CD3+CD1d:PBS57Tet+ cells from total LPMCs isolated from intestinal surgical specimens and PBMCs from healthy donor buffy coats, as previously described.32 Sorted iNKT cells were stimulated with phytohemagglutinin (PHA, 1 µg·mL−1, Sigma–Aldrich) and irradiated peripheral blood feeders. PBMCs used as feeders were irradiated at 12.5 Gy. Stimulated cells were then expanded for 15 days by subculturing them every 2–3 days and maintained in RPMI-1640 medium with stable glutamine, 5% v/v human serum, and 100 IU·mL−1 IL-2 (Proleukin). All cells were maintained in a humidified incubator with 95% air, 5% CO2 at 37°C.
Pg-priming of iNKT cell
1 × 105 monocyte-derived dendritic cells (moDCs) were pulsed with αGalCer (100 ng/ml) or heat-inactivated P. gingivalis (Pg) (4 × 105 CFU) and co-cultured with iNKT cells (2 × 105 cells) in RPMI-1640 supplemented with 10% FBS, Pen/Strep. After 24 h, iNKT cell activation status was estimated by extracellular or intracellular staining. iNKT cells co-cultured with unloaded moDCs were used as control (CTRL).
iNKT cell cytotoxicity assay
iNKT cell cytotoxicity toward the human CRC cell lines Colo205 and RKO (American Type Culture Collection, ATCC) was performed as previously described.22 In neutralization experiments, anti-CD1d (10 µg/ml, clone CD1d42, BD Biosciences) or anti-CHI3L1 (10 µg/ml, clone mYA, Millipore) were pre-incubated with rhCHI3L1 (R&D Systems) or Pg-primed iNKT cells for 24 h at 37°C before performing the assay.
Neutrophil isolation
Neutrophils were isolated from whole blood samples by dextran sedimentation (4% diluted in HBSS). Red blood cells were lysed using ACK lysis buffer (Life Technologies) and neutrophils separated with Percoll gradient.
iNKT cell–neutrophil co-culture assay
P. gingivalis primed-iNKT cells (2 × 105 cells) were co-cultured with freshly isolated neutrophils in a 1:1 ratio, in RPMI-1640 supplemented with 10% FBS. After 24 h cells were stained for the expression of extracellular marker and ROS production.
Neutrophil migration assay
Freshly isolated neutrophils were seeded on top of a 3 μm-pore transwell (SARSTEDT) in 200 μl of RPMI-1640 + 2% FBS. Five hundred microliter of chemoattracting medium was added to the bottom of transwells and neutrophil migration was allowed for 4 h at 37°C. RPMI-1640 + 10% FBS was used as positive control. The total number of cells at the bottom of the plate was stained and counted using the FACSCelesta flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) with plate-acquisition mode and defined volumes.
Neutrophil survival assay
Freshly isolated neutrophils were cultured with RPMI-1640 + 10% FBS supplemented with the culture supernatants (10%) from Pg-primed iNKT cells for 16 h at 37°C. Cells were then stained with FITC Annexin V Apoptosis Detection Kit with 7-AAD (Biolegend) following manufacturer’s instruction and acquired at a FACS Celesta flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Respiratory burst assay
ROS production was quantified using the neutrophil/monocyte respiratory burst assay (Cayman) following manufacturer’s instructions.
ELISA assay
Detection of Perforin and CHI3L1 in iNKT cell culture supernatants was performed using the Human Perforin Elisa Flex (Mabtech) and the Human CHI3L1 DuoSet ELISA (R&D systems) according to manufacturers’ instructions.
Flow cytometry
Cells were washed and stained with the combination of mAbs purchased from different vendors, as listed in Supplementary Table S2. iNKT cells were stained and identified using human or mouse CD1d:PBS57 Tetramer (NIH Tetramer Core Facility) diluted in PBS with 1% heat-inactivated FBS for 30 min at 4°C. Unloaded CD1d tetramers served as negative controls (Supplementary Figure S4). For intracellular cytokine labeling, cells were incubated for 3 h at 37°C in RPMI-1640 + 10% FBS with PMA (50 ng/ml, Merck), Ionomycin (1 μg/ml, Merck), and Brefeldin A (10 μg/ml, Merck). Before, intracellular staining cells were fixed and permeabilized using Cytofix/Cytoperm (BD). Samples were analyzed with a FACSCelesta flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and the FlowJo software (Version 10.8, TreeStar, Ashland, OR, USA). For the multidimensional analysis using t-SNE visualization and Phenograph clustering77 of T cells, samples were pregated as live lymphocytes CD45+ (CD45.2 BV510-conjugated, Biolegend) and CD3+ (CD3 PE-Cy7-conjugated, Biolegend) and stained with the following mAbs: CD279 (PD1) APC-conjugated, Biolegend; CD4 APCCy7-conjugated, Biolegend; CD69 PerCPcy5.5-conjugated, BD Biosciences; CD8a BV650-conjugated, BD Biosciences; GM-CSF FITC-conjugated, Biolegend; IFNγ BV785-conjugated, BD Biosciences; IL10 BV421-conjugated, Biolegend; IL17A Alexa Fluor-700-conjugated, Biolegend; CD137 (4-1BB) BV605-conjugated, BD Biosciences; PBS57-CD1d Tetramer PE-conjugated. For the analysis of myeloid cells, samples were pregated as live lymphocytes CD45+ (CD45.2 BV510-conjugated Biolegend) and stained with the following mAbs: CD11b APC-Cy7-conjugated, TONBO; CD1d PerCPCy5.5-conjugated, BD Biosciences; CD19 BV421-conjugated, BD Biosciences; CD11c BV605-conjugated, BD Biosciences; CD274 (PDL1) BV786-conjugated, BD Biosciences; F4/80 PE-conjugated, TONBO; Ly6G APC-conjugated, TONBO. ROS expression was detected in FITC with the Neutrophil/Monocyte Respiratory Burst Assay Kit Cayman chemical. FCS files were quality checked for live, singlets and antibody agglomerates and normalized to avoid batch effects. Data were cleaned for antibody aggregates by checking each parameter in a bimodal plot. The different cell populations were down-sampled to 3000 events per sample using the DownSample plugin (Version 3.3.1) of FlowJo to create uniform population sizes. Down-sampled populations were exported as FCS files with applied compensation correction. Files were then uploaded to RStudio environment (Version 3.5.3) using the flowCore package (Version 1.38.2). Data were transformed using logicleTransform() function present in the flowCore package. To equalize the contribution of each marker they were interrogated for their density distribution using the densityplot() function of the flowViz package (Version 1.36.2). Each marker was normalized using the Per-channel normalization based on landmark registration using the gaussNorm() function present in the package flowStats (Version 3.30.0). Peak.density, peak.distance, and number of peaks were chosen according to each marker expression. Normalized files were analyzed using the cytofkit package through the cytofkit_GUI interface. For data visualization, we used the t-Distributed Stochastic Neighbor Embedding (t-SNE) method, while for clustering we used the phenograph algorithm. t-SNE plots were visualized on the cytofkitShinyAPP with the following parameters: perplexity = 50, iterations = 1000, k = 50. FCS for each cluster were generated and re-imported in FlowJo to be manually analyzed for the determination of the integrated MFI. The iMFI of different markers was scaled from 0 to 1 and used to identify phenograph clusters.77
Bulk RNA sequencing of human iNKT cells
Total RNA (from 1 × 106 cells) was isolated with the RNeasy kit (Qiagen), and RNA quality was checked with the Agilent 2100 Bioanalyzer (Agilent Technologies). 0.5–1 μg were used to prepare libraries for RNA-seq with the Illumina TruSeq RNA Library Prep Kit v2 following the manufacturer’s instructions. RNA-seq libraries were then run on the Agilent 2100 Bioanalyzer (Agilent Technologies) for quantification and quality control and pair-end sequenced on the Illumina NovaSeq platform.
RNA sequencing data analysis
RNA-seq reads were preprocessed using the FASTX-Toolkit tools. Quality control was performed using FastQC. Pipelines for primary analysis (filtering and alignment of the reference genome of the raw reads) and secondary analysis (expression quantification, differential gene expression) have been integrated and run in the HTS-flow system.78 Differentially expressed genes were identified using the Bioconductor Deseq2 package.79 P-values were False Discovery Rate corrected using the Benjamini–Hochberg procedure implemented in DESeq2. Functional enrichment analyses to determine Gene Ontology categories and KEGG pathways were performed using the DAVID Bioinformatics Resources (DAVID Knowledgebase v2022q2) (https://david.ncifcrf.gov).80
Western blot
Total protein extracts from iNKT cells were prepared as previously described,81 and separated on Mini-PROTEAN TGX Stain-Free Precast Gels (4–15%, Bio-Rad Laboratories, Hercules, California, USA) to enhance transfer efficiency and detection of proteins with stain-free enabled imagers. Samples were then transferred on nitrocellulose membranes (Bio-Rad Laboratories, Hercules, California, USA) and incubated overnight at 4°C with the following primary antibodies: STAT-1 (1:600, E-Ab -32,977, Elabscience, Houston, Texas, USA); STAT-3 (1:600, E-Ab -40,131, Elabscience, Houston, Texas, USA); GSK-3ab (1:500, sc-7291, Santa Cruz, Dallas, Texas, USA); Akt 1/2/3 (1:500, Ab -179,463, Abcam Cambridge, United Kingdom). Membranes were detected with peroxidase conjugated secondary antibodies (Agilent Technologies, California, USA) and developed by ECL (Amersham Biosciences, United Kingdom). Image Lab Software from Bio-Rad was used to analyze band intensity.
Supplementary Material
Acknowledgments
We thank the IEO Animal Facility for the excellent animal husbandry and the NIH Tetramer Facility for providing human and murine CD1d:PBS57 tetramers. We are grateful to the équipe of the General and Emergency Surgery Unit, Ospedale Maggiore Policlinico, Milano for their tireless work. We thank Prof Paolo Dellabona and Prof Giulia Casorati for providing the Traj18−/− mice. We thank Maria Rita Giuffrè, Valentina Pasquale, Elisa Cirrincione, Luca Iachini, Luana Tripodi, and Monica Molinaro for their assistance with the experiments. Panels 1D and 3A were created using icons from the Noun Project (https://thenounproject.com/).
Funding Statement
This work was made possible thanks to the financial support of Associazione Italiana per la Ricerca sul Cancro [Start-Up 2013 14378, Investigator Grant - IG 2019 22923 to FF] and of Italy’s Ministry of Health [GR-2016-0236174 to FF and FC]. This work has received funding from the European Union - NextGenerationEU through the Italian Ministry of University and Research under the PNRR - M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to FF. This work has been and partially supported by the Italian Ministry of Health with Ricerca Corrente and 5 × 1000fund.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2388801.
Data availability statement
RNA sequencing data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-12281 and E-MTAB-14021. 16S rRNA gene sequencing data are available in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession number PRJEB56178.
Authors contribution
FF and FS conceived the study. FF, FS, ADB, and GL designed the experiments. GL, ADB, FP, CA, AB, and AF performed the experiments. FF and FS supervised the experiments. FC, MV, LB, YT, GP, and MR contributed with reagents and resources. GL performed multidimensional FACS data analysis. FS performed RNAseq data analyses. FS wrote the first draft of the manuscript. All authors reviewed and critically edited the manuscript. Both GL and ADB contributed equally and have the right to list their names first on their CVs. All authors contributed to the article and approved the submitted version.
References
- 1.Deo SVS, Sharma J, Kumar SG.. Report on global cancer burden: challenges and opportunities for surgical oncologists. Ann Surg Oncol. 2022;2020;29(11):6497–17. doi: 10.1245/s10434-022-12151-6. [DOI] [PubMed] [Google Scholar]
- 2.Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 2021;21(10):653–667. doi: 10.1038/s41577-021-00534-x. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Pitmon E, Wang K. Microbiome, inflammation and colorectal cancer. Semin Immunol. 2017;32:43–53. doi: 10.1016/j.smim.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, Letellier E. Microbiome in colorectal cancer: how to get from meta-omics to mechanism? Trends Microbiol. 2020;28(5):401–423. doi: 10.1016/j.tim.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15(1):30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333(1):1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 7.Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, O’Riordain M, Shanahan F, O’Toole PW. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66(4):633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7(1):11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu W, Jia Y, Chen X, Li H, Wang Z, Cheng B. Intracellular Porphyromonas gingivalis promotes the proliferation of colorectal cancer cells via the MAPK/ERK signaling pathway. Front Cell Infect Microbiol. 2020;10:584798. doi: 10.3389/fcimb.2020.584798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, Sato S, Yoshida Y, Tsuji S, Uemura K, et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun. 2021;12(1):5674. doi: 10.1038/s41467-021-25965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Jia Y, Wen L, Mu W, Wu X, Liu T, Liu X, Fang J, Luan Y, Chen P, et al. Porphyromonas gingivalis promotes colorectal carcinoma by activating the hematopoietic NLRP3 Inflammasome. Cancer Res. 2021;81(10):2745–2759. doi: 10.1158/0008-5472.CAN-20-3827. [DOI] [PubMed] [Google Scholar]
- 12.Lattanzi G, Strati F, Diaz-Basabe A, Perillo F, Amoroso C, Protti G, Rita Giuffrè M, Iachini L, Baeri A, Baldari L, et al. iNKT cell-neutrophil crosstalk promotes colorectal cancer pathogenesis. Mucosal Immunol. 2023;16(3):326–340. doi: 10.1016/j.mucimm.2023.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Crosby CM, Kronenberg M. Tissue-specific functions of invariant natural killer T cells. Nat Rev Immunol. 2018;18(9):559–574. doi: 10.1038/s41577-018-0034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25(1):297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 15.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al. CD1d-restricted and TCR-Mediated activation of V α 14 NKT cells by Glycosylceramides. Science. 1997;278(5343):1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 16.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong C-H, Kronenberg M, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434(7032):520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 17.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7(9):978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 18.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg R, Kasper D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156(1–2):123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kain L, Webb B, Anderson BL, Deng S, Holt M, Costanzo A, Zhao M, Self K, Teyton A, Everett C, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian α-linked glycosylceramides. Immunity. 2014;41(4):543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delfanti G, Dellabona P, Casorati G, Fedeli M. Adoptive immunotherapy with engineered iNKT cells to target cancer cells and the suppressive microenvironment. Front Med (Lausanne). 2022;9:897750. doi: 10.3389/fmed.2022.897750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Basabe A, Strati F, Facciotti F. License to kill: when iNKT cells are granted the use of lethal cytotoxicity. Int J Mol Sci. 2020;21(11):3909. doi: 10.3390/ijms21113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz-Basabe A, Burrello C, Lattanzi G, Botti F, Carrara A, Cassinotti E, Caprioli F, Facciotti F. Human intestinal and circulating invariant natural killer T cells are cytotoxic against colorectal cancer cells via the perforin–granzyme pathway. Mol Oncol. 2021;15(12):3385–3403. doi: 10.1002/1878-0261.13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Sedimbi S, Lofbom L, Singh AK, Porcelli SA, Cardell SL. Unique invariant natural killer T cells promote intestinal polyps by suppressing TH1 immunity and promoting regulatory T cells. Mucosal Immunol. 2018;11(1):131–143. doi: 10.1038/mi.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannou AD, Kempski J, Shiri AM, Lucke J, Zhang T, Zhao L, Zazara DE, Cortesi F, Riecken K, Amezcua Vesely MC, et al. Tissue resident iNKT17 cells facilitate cancer cell extravasation in liver metastasis via interleukin-22. Immunity. 2023;56(1):125–42 e12. doi: 10.1016/j.immuni.2022.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delfanti G, Cortesi F, Perini A, Antonini G, Azzimonti L, de Lalla C, Garavaglia C, Squadrito ML, Fedeli M, Consonni M, et al. Tcr-engineered iNKT cells induce robust antitumor response by dual targeting cancer and suppressive myeloid cells. Sci Immunol. 2022;7(74):eabn6563. doi: 10.1126/sciimmunol.abn6563. [DOI] [PubMed] [Google Scholar]
- 26.Metzger Z, Lin YY, Dimeo F, Ambrose WW, Trope M, Arnold RR. Synergistic pathogenicity of Porphyromonas gingivalis and Fusobacterium nucleatum in the mouse subcutaneous chamber model. J Endod. 2009;35(1):86–94. doi: 10.1016/j.joen.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Germanov E, Veinotte L, Cullen R, Chamberlain E, Butcher EC, Johnston B. Critical role for the chemokine receptor CXCR6 in homeostasis and activation of CD1d-restricted NKT cells. J Immunol. 2008;181(1):81–91. doi: 10.4049/jimmunol.181.1.81. [DOI] [PubMed] [Google Scholar]
- 28.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360(6391):360(6391. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H, Hong C, Shin J, Oh S, Jung S, Park YK, Hong S, Lee GR, Park S-H. The presence of CD8+ invariant NKT cells in mice. Exp Mol Med. 2009;41(12):866–872. doi: 10.3858/emm.2009.41.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh S, Lee H, Shin JH, Hong C, Park SH. Murine CD8(+) invariant natural killer T cells are negatively selected by CD1d expressed on thymic epithelial cells and dendritic cells. Immunol Invest. 2018;47(1):89–100. doi: 10.1080/08820139.2017.1385621. [DOI] [PubMed] [Google Scholar]
- 31.Gershkovitz M, Caspi Y, Fainsod-Levi T, Katz B, Michaeli J, Khawaled S, Lev S, Polyansky L, Shaul ME, Sionov RV, et al. TRPM2 mediates neutrophil killing of disseminated tumor cells. Cancer Res. 2018;78(10):2680–2690. doi: 10.1158/0008-5472.CAN-17-3614. [DOI] [PubMed] [Google Scholar]
- 32.Burrello C, Pellegrino G, Giuffre MR, Lovati G, Magagna I, Bertocchi A, Cribiù FM, Boggio F, Botti F, Trombetta E, et al. Mucosa-associated microbiota drives pathogenic functions in ibd-derived intestinal iNKT cells. Life Sci Alliance. 2019;2(1):e201800229. doi: 10.26508/lsa.201800229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. CD1d-mediated recognition of an α-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188(8):1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia L, Wu R, Han N, Fu J, Luo Z, Guo L, Su Y, Du J, Liu Y. Porphyromonas gingivalis and lactobacillus rhamnosus GG regulate the Th17/Treg balance in colitis via TLR4 and TLR2. Clin & Trans Imm. 2020;9(11):e1213. doi: 10.1002/cti2.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaddis DE, Maynard CL, Weaver CT, Michalek SM, Katz J. Role of TLR2-dependent IL-10 production in the inhibition of the initial ifn-gamma T cell response to Porphyromonas gingivalis. J Leukoc Biol. 2013;93(1):21–31. doi: 10.1189/jlb.0512220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao T, Su Z, Li Y, Zhang X, You Q. Chitinase-3 like-protein-1 function and its role in diseases. Signal Transduct Target Ther. 2020;5(1):201. doi: 10.1038/s41392-020-00303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darwich A, Silvestri A, Benmebarek MR, Mouries J, Cadilha B, Melacarne A, Morelli L, Supino D, Taleb A, Obeck H, et al. Paralysis of the cytotoxic granule machinery is a new cancer immune evasion mechanism mediated by chitinase 3-like-1. J Immunother Cancer. 2021;9(11):e003224. doi: 10.1136/jitc-2021-003224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Q, Zhao X, Li R, Liu D, Pan B, Xie B, Chi X, Cai D, Wei P, Xu W, et al. STAT3 regulates CD8+ T cell differentiation and functions in cancer and acute infection. J Exp Med. 2023;220(4). doi: 10.1084/jem.20220686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, Brugman S, Yamaguchi K, Ishikawa H, Aiba Y, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest. 2009;119(5):1241–1250. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, Braun, J., Mazmanian, S.K. and Kronenberg, M. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143(2):418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maricic I, Marrero I, Eguchi A, Nakamura R, Johnson CD, Dasgupta S, Hernandez CD, Nguyen PS, Swafford AD, Knight R, et al. Differential activation of hepatic invariant NKT cell subsets plays a key role in progression of nonalcoholic steatohepatitis. J Immunol. 2018;201(10):3017–3035. doi: 10.4049/jimmunol.1800614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Guinoa J S, Jimeno R, Gaya M, Kipling D, Garzon MJ, Dunn-Walters D, Ubeda C, Barral P. CD 1d-mediated lipid presentation by CD 11c + cells regulates intestinal homeostasis. Embo J. 2018;37(5). doi: 10.15252/embj.201797537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma C, McCallen J, McVey JC, Trehan R, Bauer K, Zhang Q, Ruf B, Wang S, Lai CW, Trinchieri G, et al. CSF-1R+ macrophages control the gut microbiome–enhanced liver invariant NKT function through IL-18. J Immunol. 2023;211(7):1099–1107. doi: 10.4049/jimmunol.2200854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Q, Kuypers M, Philpott DJ, Mallevaey T. The dialogue between unconventional T cells and the microbiota. Mucosal Immunol. 2020;13(6):867–876. doi: 10.1038/s41385-020-0326-2. [DOI] [PubMed] [Google Scholar]
- 45.Lin Q, Kuypers M, Liu Z, Copeland JK, Chan D, Robertson SJ, Kontogiannis J, Guttman DS, Banks EK, Philpott DJ, et al. Invariant natural killer T cells minimally influence gut microbiota composition in mice. Gut Microbes. 2022;14(1):2104087. doi: 10.1080/19490976.2022.2104087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Q, Kuypers M, Baglaenko Y, Cao E, Hezaveh K, Despot T, de Amat Herbozo C, Cruz Tleugabulova M, Umaña JM, McGaha TL, et al. The intestinal microbiota modulates the transcriptional landscape of iNKT cells at steady-state and following antigen exposure. Mucosal Immunol. 2024;17(2):226–237. doi: 10.1016/j.mucimm.2024.02.002. [DOI] [PubMed] [Google Scholar]
- 47.de Aguiar CF, Castoldi A, Amano MT, Ignacio A, Terra FF, Cruz M, Felizardo RJF, Braga TT, Davanzo GG, Gambarini V, et al. Fecal IgA levels and gut microbiota composition are regulated by invariant natural killer T cells. Inflamm Bowel Dis. 2020;26(5):697–708. doi: 10.1093/ibd/izz300. [DOI] [PubMed] [Google Scholar]
- 48.Burrello C, Strati F, Lattanzi G, Diaz-Basabe A, Mileti E, Giuffre MR, Lopez G, Cribiù FM, Trombetta E, Kallikourdis M, et al. IL10 secretion endows intestinal human iNKT cells with regulatory functions towards pathogenic T lymphocytes. J Crohns Colitis. 2022;16(9):1461–1474. doi: 10.1093/ecco-jcc/jjac049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tachibana T, Onodera H, Tsuruyama T, Mori A, Nagayama S, Hiai H, Imamura M. Increased intratumor Vα24-positive natural killer T cells: a prognostic factor for primary colorectal carcinomas. Clin Cancer Res. 2005;11(20):7322–7327. doi: 10.1158/1078-0432.CCR-05-0877. [DOI] [PubMed] [Google Scholar]
- 50.Metelitsa LS, Wu HW, Wang H, Yang Y, Warsi Z, Asgharzadeh S, Groshen S, Wilson SB, Seeger RC. Natural killer T cells infiltrate neuroblastomas expressing the chemokine CCL2. J Exp Med. 2004;199(9):1213–1221. doi: 10.1084/jem.20031462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moutsopoulos NM, Kling HM, Angelov N, Jin W, Palmer RJ, Nares S, Osorio M, Wahl SM. Porphyromonas gingivalis promotes Th17 inducing pathways in chronic periodontitis. J Autoimmun. 2012;39(4):294–303. doi: 10.1016/j.jaut.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Y, Kobayashi R, Hashizume-Takizawa T, Kurita-Ochiai T. Porphyromonas gingivalis infection enhances Th17 responses for development of atherosclerosis. Arch Oral Biol. 2014;59(11):1183–1191. doi: 10.1016/j.archoralbio.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Glowczyk I, Wong A, Potempa B, Babyak O, Lech M, Lamont RJ, Potempa J, Koziel J. Inactive gingipains from P. gingivalis selectively skews T cells toward a Th17 phenotype in an IL-6 dependent manner. Front Cell Infect Microbiol. 2017;7:140. doi: 10.3389/fcimb.2017.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zenobia C, Hajishengallis G. Porphyromonas gingivalis virulence factors involved in subversion of leukocytes and microbial dysbiosis. Virulence. 2015;6(3):236–243. doi: 10.1080/21505594.2014.999567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clancy-Thompson E, Chen GZ, Tyler PM, Servos MM, Barisa M, Brennan PJ, Ploegh HL, Dougan SK. Monoclonal invariant NKT (iNKT) cell mice reveal a role for both tissue of origin and the TCR in development of iNKT functional subsets. J Immunol. 2017;199(1):159–171. doi: 10.4049/jimmunol.1700214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith E, Croca S, Waddington KE, Sofat R, Griffin M, Nicolaides A, Isenberg DA, Torra IP, Rahman A, Jury EC, et al. Cross-talk between iNKT cells and monocytes triggers an atheroprotective immune response in SLE patients with asymptomatic plaque. Sci Immunol. 2016;1(6). doi: 10.1126/sciimmunol.aah4081. [DOI] [PubMed] [Google Scholar]
- 57.Bortoluzzi S, Dashtsoodol N, Engleitner T, Drees C, Helmrath S, Mir J, Toska A, Flossdorf M, Öllinger R, Solovey M, et al. Brief homogeneous TCR signals instruct common iNKT progenitors whose effector diversification is characterized by subsequent cytokine signaling. Immunity. 2021;54(11):2497–513 e9. doi: 10.1016/j.immuni.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 58.Pathirana RD, Nm O-S, Visvanathan K, Hamilton JA, Reynolds EC. Flow cytometric analysis of adherence of porphyromonas gingivalis to oral epithelial cells. Infect Immun. 2007;75(5):2484–2492. doi: 10.1128/IAI.02004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinberg A, Belton CM, Park Y, Lamont RJ. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65(1):313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umana A, Zhang Y, Peng H, Duncan AJ, Wang Y, Li L, et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal. 2020;13(641). doi: 10.1126/scisignal.aba9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Udayasuryan B, Ahmad RN, Nguyen TTD, Umana A, Monet Roberts L, Sobol P, Jones SD, Munson JM, Slade DJ, Verbridge SS, et al. Fusobacterium nucleatum induces proliferation and migration in pancreatic cancer cells through host autocrine and paracrine signaling. Sci Signal. 2022;15(756):eabn4948. doi: 10.1126/scisignal.abn4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim DH, Park HJ, Lim S, Koo JH, Lee HG, Choi JO, Oh JH, Ha S-J, Kang M-J, Lee C-M, et al. Regulation of chitinase-3-like-1 in T cell elicits Th1 and cytotoxic responses to inhibit lung metastasis. Nat Commun. 2018;9(1):503. doi: 10.1038/s41467-017-02731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geng B, Pan J, Zhao T, Ji J, Zhang C, Che Y, Yang J, Shi H, Li J, Zhou H, et al. Chitinase 3-like 1-CD44 interaction promotes metastasis and epithelial-to-mesenchymal transition through β-catenin/erk/akt signaling in gastric cancer. J Exp Clin Cancer Res. 2018;37(1):208. doi: 10.1186/s13046-018-0876-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guetta-Terrier C, Karambizi D, Akosman B, Zepecki JP, Chen JS, Kamle S, Fajardo JE, Fiser A, Singh R, Toms SA, et al. Chi3l1 is a modulator of glioma stem cell states and a Therapeutic target in Glioblastoma. Cancer Res. 2023;83(12):1984–1999. doi: 10.1158/0008-5472.CAN-21-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shan Z, Li L, Atkins CL, Wang M, Wen Y, Jeong J, Moreno NF, Feng D, Gui X, Zhang N, et al. Chitinase 3-like-1 contributes to acetaminophen-induced liver injury by promoting hepatic platelet recruitment. Elife. 2021;10:10. doi: 10.7554/eLife.68571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klement JD, Paschall AV, Redd PS, Ibrahim ML, Lu C, Yang D, Celis E, Abrams SI, Ozato K, Liu K, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Invest. 2018;128(12):5549–5560. doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krasilnikov M, Ivanov VN, Dong J, Ronai Z. ERK and PI3K negatively regulate stat-transcriptional activities in human melanoma cells: implications towards sensitization to apoptosis. Oncogene. 2003;22(26):4092–4101. doi: 10.1038/sj.onc.1206598. [DOI] [PubMed] [Google Scholar]
- 69.Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in cancer immunotherapy. Mol Cancer. 2020;19(1):145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson RP, Ives ML, Rao G, Lau A, Payne K, Kobayashi M, Arkwright PD, Peake J, Wong M, Adelstein S, et al. STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med. 2015;212(6):855–864. doi: 10.1084/jem.20141992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beurel E, Yeh WI, Michalek SM, Harrington LE, Jope RS. Glycogen synthase kinase-3 is an early determinant in the differentiation of pathogenic Th17 cells. J Immunol. 2011;186(3):1391–1398. doi: 10.4049/jimmunol.1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Villarino AV, Gallo E, Abbas AK. STAT1-activating cytokines limit Th17 responses through both T-bet-dependent and -independent mechanisms. J Immunol. 2010;185(11):6461–6471. doi: 10.4049/jimmunol.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Putz EM, Majoros A, Gotthardt D, Prchal-Murphy M, Zebedin-Brandl EM, Fux DA, Schlattl A, Schreiber RD, Carotta S, Müller M, et al. Novel non-canonical role of STAT1 in natural killer cell cytotoxicity. Oncoimmunology. 2016;5(9):e1186314. doi: 10.1080/2162402X.2016.1186314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caprioli F, Sarra M, Caruso R, Stolfi C, Fina D, Sica G, MacDonald TT, Pallone F, Monteleone G. Autocrine regulation of IL-21 production in human T lymphocytes. J Immunol. 2008;180(3):1800–1807. doi: 10.4049/jimmunol.180.3.1800. [DOI] [PubMed] [Google Scholar]
- 75.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M, et al. Requirement for V α 14 NKT cells in IL-12-Mediated rejection of tumors. Science. 1997;278(5343):1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 76.Becker C, Fantini MC, Neurath MF. High resolution colonoscopy in live mice. Nat Protoc. 2006;1(6):2900–2904. doi: 10.1038/nprot.2006.446. [DOI] [PubMed] [Google Scholar]
- 77.Brummelman J, Haftmann C, Nunez NG, Alvisi G, Mazza EMC, Becher B, Lugli E. Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nat Protoc. 2019;14(7):1946–1969. doi: 10.1038/s41596-019-0166-2. [DOI] [PubMed] [Google Scholar]
- 78.Bianchi V, Ceol A, Ogier AG, de Pretis S, Galeota E, Kishore K, Bora P, Croci O, Campaner S, Amati B, et al. Integrated systems for NGS data management and analysis: open issues and available solutions. Front Genet. 2016;7:75. doi: 10.3389/fgene.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bella P, Farini A, Banfi S, Parolini D, Tonna N, Meregalli M, Belicchi M, Erratico S, D’Ursi P, Bianco F, et al. Blockade of IGF2R improves muscle regeneration and ameliorates Duchenne muscular dystrophy. EMBO Mol Med. 2020;12(1):e11019. doi: 10.15252/emmm.201911019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-12281 and E-MTAB-14021. 16S rRNA gene sequencing data are available in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession number PRJEB56178.


