Abstract
Auxiliary factors likely play an important role in determining the polyadenylation efficiency of mammalian pre-mRNAs. We previously identified an auxiliary factor, hnRNP H/H′, which stimulates 3′-end processing through an interaction with sequences downstream of the core elements of the SV40 late polyadenylation signal. Using in vitro reconstitution assays we have demonstrated that hnRNP H/H′ can stimulate processing of two additional model polyadenylation signals by binding at similar relative downstream locations but with significantly different affinities. A short tract of G residues was determined to be a common property of all three hnRNP H/H′ binding sites. A survey of mammalian polyadenylation signals identified potential G-rich hnRNP H/H′ binding sites at similar downstream locations in ∼34% of these signals. All of the novel G-rich elements tested were found to bind hnRNP H/H′ protein and the processing of selected signals identified in the survey was stimulated by the protein both in vivo and in vitro. Downstream G-rich tracts, therefore, are a common auxiliary element in mammalian polyadenylation signals. Sequences capable of binding hnRNP H protein with varying affinities may play a role in determining the processing efficiency of a significant number of mammalian polyadenylation signals.
INTRODUCTION
The 3′-end of most mammalian mRNAs is generated by a site-specific cleavage event that is tightly coupled to the addition of 150–200 adenylate residues (reviewed in 1,2). Two core elements that signal this event lie in the pre-mRNA on either side of the cleavage site. A highly conserved AAUAAA element is located ∼10–25 bases upstream of the cleavage site and a U-rich element is positioned ∼15–30 bases downstream (3–5). This bipartite signal provides a cooperative binding site for two general polyadenylation factors, CPSF and CstF, which serve as a scaffold for assembly of the other general factors involved in the enzymatic reactions of 3′-end processing (6,7).
Polyadenylation is closely coordinated with other aspects of gene expression in the nucleus. Transcription termination is linked to polyadenylation, and several general polyadenylation factors can be found in association with the CTD of RNA polymerase II (8–10). The splicing efficiency of adjacent introns is also influenced by polyadenylation (11,12). Polyadenylation itself can be a regulated event. Several cellular and viral transcription units contain alternative polyadenylation signals that are used in a development-, growth- or tissue-specific manner (reviewed in 13). A key question is how polyadenylation signals are regulated, especially when many contain highly conserved core elements.
The relative level of CstF has been proposed to be involved in regulated polyadenylation site usage (14), particularly because the downstream U-rich element that serves as the binding site for the factor contains a less conserved consensus sequence than the canonical upstream AAUAAA element (3,4,15). Changes in CstF concentration, however, cannot account for all of the regulation of 3′-end processing that has been observed (16–18). It is likely that the sequence/structural context of the core polyadenylation signal elements (19,20), as well as auxiliary sequence elements that lie upstream (21,22) and/or downstream (23) of the core elements, may also play a role in determining 3′-end processing efficiency.
We have previously shown that auxiliary sequences which lie downstream of the core elements of the polyadenylation signal can influence 3′-end processing efficiency (23). The best-characterized auxiliary downstream element to date is a 14 base G-rich sequence (GRS) (GGGGGAGGUGUGGG) located at position +32 relative to the cleavage site in the SV40 late (SVL) polyadenylation signal (24). Deleting, making point mutations in or moving the GRS further downstream all significantly decreased the processing efficiency of the SVL polyadenylation signal (25). The GRS serves as a binding site for a nuclear protein originally called DSEF-1. The results of peptide sequencing of purified DSEF-1 protein are consistent with it being either hnRNP H or hnRNP H′ (which has 96% identity with hnRNP H) (26,27). These two proteins, along with the closely related hnRNP F protein, represent a family of RNA binding proteins that show preference for binding G-rich RNAs. Addition of recombinant hnRNP H′ protein to in vitro polyadenylation reactions stimulates 3′-end processing efficiency in a sequence-specific fashion, consistent with a role for hnRNP H′ as an auxiliary 3′-end processing factor (26). Several hnRNP proteins, including A1, H and the related F protein, have previously been shown to influence splicing efficiency (28–30). It is therefore possible that several hnRNP proteins may play a role in differential mRNA processing.
In this study we have addressed three hypotheses regarding the possible mechanistic role of hnRNP H protein in 3′-end processing. First, hnRNP H may regulate 3′-end processing efficiency through differential binding to the downstream region of polyadenylation signals. Using in vitro reconstitution assays, we have demonstrated that hnRNP H binds to several model polyadenylation signals with significantly different affinities. The relative binding affinity of hnRNP H proteins for the polyadenylation signal directly affects their ability to stimulate processing of a polyadenylation signal. Second, we hypothesize that properly localized hnRNP H binding sites might be common in polyadenylation signals. Mapping of novel hnRNP H binding sites in two model polyadenylation signals suggested that a common motif in hnRNP H binding is a short tract of G residues. Using this streamlined consensus sequence we identified hnRNP H binding sites in the downstream region of ∼34% of signals in a database. Finally, we have demonstrated that these newly identified G-rich tracts are indeed hnRNP H binding sites and are capable of stimulating polyadenylation both in vitro and in vivo. These data suggest that hnRNP H is a positive regulator of numerous mammalian polyadenylation signals.
MATERIALS AND METHODS
Plasmids, RNAs and recombinant proteins
pSVL has a 241 bp BamHI–BclI fragment containing the SV40 late polyadenylation signal inserted into the BamHI site of pSP65 (31). Transcription of DraI-linearized templates yields a 224 base RNA (SVL). The transcription template to generate SVL-GEM RNA, in which positions 202–224 of the transcript (including the GRS element) are substituted by polylinker-derived sequences, was prepared by PCR. PCR reactions containing pSVL, an SP6 promoter-specific primer (5′-CATACGATTTAGGTGACACT) and a mutagenic downstream primer (5′-ACCGAGCTCGAATTCCGTGTATTCTGAACCTGAAACAT) yielded a 247 bp DNA fragment. Direct transcription of the PCR product gave a 224 base RNA (SVL-GEM).
pGEM2µMPA has a 199 bp RsaI fragment that contains the polyadenylation signal of the membrane-bound immunoglobulin µ heavy chain inserted at the SmaI site of pGEM2 (32). Transcription of BamHI-linearized templates gave a 232 base RNA (µM). Transcription of BsrI-linearized templates gave a 198 base RNA (µM198). Transcription templates for the µM33nt and µM26nt RNAs were made by hybridizing the following DNA oligonucleotides and their appropriate complementary strands: µM33nt, 5′-CATACGATTTAGGTGACACTATAGAATACAGAAATGGAACAGTG-CAGTCTGATGGTGGGGATC; µM28nt, 5′-CATACGATTTAGGTGACACTATAGAATACAGAAATGGAACAGT-GCAGTCTGATGGT.
pIVA2 contains the 155 bp BamHI–PvuII fragment of the adenovirus plasmid pAd5E1B cloned into the HincII–BamHI sites of pGEM4 (31). Transcription of BglI-cut template yielded a 158 base RNA (IVA2). Transcription of MnlI-cut template gave a 130 base RNA (IVA2130). The transcription template to generate IVA2-GEM RNA, in which positions 134–158 of the transcript (the region 3′ of the core downstream U-rich element) are substituted by polylinker-derived sequences, was prepared by PCR. PCR reactions containing pIVA2, an SP6 promoter-specific and a mutagenic downstream primer (5′-GAGCTCGAATTCCGTGTATTCCGGTTTAAAAC) yielded a 181 bp DNA fragment. Direct transcription of the PCR product gave a 158 base RNA (IVA2-GEM). The template for IVA228nt RNA was derived by hybridizing the oligonucleotide 5′-CATACGATTTAGGTGACACTATAGAATACACCATTGGGAGGGGAGGAAGCCTTCAGGGC and its appropriate complement.
Templates to generate short transcripts containing novel G-rich elements found in a survey of polyadenylation signals were generated by hybridizing the following synthetic DNA oligonucleotides and their appropriate complementary strands: HUMMK, 5′-CATACGATTTAGGTGACACTATAGAATACAGGGAGTGGGAAGGTGGGGAGAAAG; OCACE3, 5′-CATACGATTTAGGTGACACTATAGAATACAGGGCAGGGATGGGGGAGGGACCAC; HUMTNP1, 5′-CATACGATTTAGGTGACACTATAGAATACAGGTGGGTTTGG-GGAAGAA; MUSPKCD10, 5′-CATACGATTTAGGTGACACTATAGAATACAGGGGTGTTGGAAAGGGAATTC; RATCRP2A, 5′-CATACGATTTAGGTGACACTATAGAATACAGGGGGCAGTAGTTGGCTTTTG; RNLAPG, 5′-CATACGATTTAGGTGACACTATAGAATACAGGGGGCAGTA-GTTGGCTTTTG. Note that the first 23 bases of each oligonucleotide represent the upstream portion of an SP6 promoter.
In order to construct the core elements and downstream region of the RATCRP2A polyadenylation signal, the oligonucleotide 5′-GAGTGAATTCAATAAACTTTTTTGGAGAATGTTTAAAAAGCCTTTTGGGGGCAGTAGTTGGAT-CCGAGC and its appropriate complement were hybridized and inserted in pGEM4 that was cut with BamHI and EcoRI. The resulting plasmid was linearized with XbaI and transcribed to produce RATCRP2A RNA.
Capped RNAs were prepared by in vitro transcription reactions using SP6 RNA polymerase in the presence of [32P]UTP. Transcripts were purified from 5% acrylamide–7 M urea gels prior to use (31).
A 1.35 kb DNA fragment, containing the entire open reading frame of hnRNP H′, was subcloned into pGEX2TZQ (a pGEX2T derivative; 33) to form phnRNP H′. Recombinant GST fusion proteins were produced by IPTG induction of Escherichia coli transformed with the indicated plasmids and purified on glutathione–Sepharose columns. Proteins were stored in 50 mM Tris, pH 8.0, 10 mM reduced glutathione, 10% glycerol at –80°C.
In vitro cleavage and polyadenylation assays
Cleavage and polyadenylation reactions were performed in the in vitro system derived from HeLa nuclear extracts (34) as previously described (31) using equimolar amounts (∼15 fmol) of capped RNAs that were radiolabeled to the same specific activity. The α,β-methylene analog of ATP and EDTA was substituted for ATP and phosphocreatine in reaction mixtures to allow cleavage to occur but prevent addition of adenosines to the 3′ end of the 5′ cleavage product. Reaction products were analyzed on 5% acrylamide gels containing 7 M urea. In reactions where endogenous hnRNP H protein in the extract was sequestered by GRS competitor RNA, the indicated amounts of synthetic RNA oligomer were added directly to reaction mixtures.
Gel shift analyses
Gel shift assays were performed as described previously (26) using heparin to reduce non-specific protein–RNA interactions and 5% acrylamide gels in a 50 mM Tris–borate–EDTA running buffer. Quantitative assays to determine binding constants were performed using a fixed amount of radiolabeled SVL RNA (16.8 fmol) and increasing amounts of protein. The relative amounts of protein-bound and free RNA were determined using phosphorimaging and ImageQuant software.
In vivo analysis of transfected cells
The mouse AxJ plasma cell line was transfected with the Sigma FLAG tag vector carrying either no insert, the hnRNP H′ coding sequence in-frame or hnRNP F coding sequences. Transfection and analyses of recombinant hnRNP:FLAG epitope-tagged proteins was performed as previously described (35). Poly(A)+ RNA was isolated from the transfected cells and run on denaturing formaldehyde gels, blotted to Nitran+ membranes, and exposed to UV light to crosslink the RNA to the membrane. The blots were probed with [32P]G-labeled riboprobes made from oligonucleotides containing a T7 promoter and nucleotides 4571–4620 of psmb5 and 1261–1310 of Gpi1 or from cDNA clones for CstF-64 (35) and MUSPKCD10 (oligonucleotide 5′-GATCCGGGAGTGAGGAGGGTGTTTGGGGAGGGTGGGACAATTTG-GAGGGTGGGAGGCACG and its appropriate complement were inserted into the EcoRI and BamHI sites of pGem4).
RESULTS
Polyadenylation signals have different affinities for hnRNP H protein
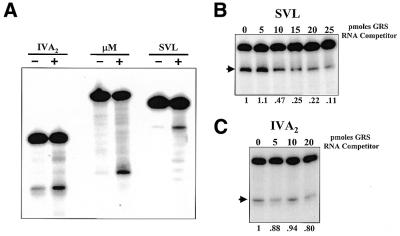
A complex of general polyadenylation factors assembles on the core elements of the polyadenylation signal and mediates 3′-end processing of pre-mRNAs (1,2,7). Since polyadenylation signals can be differentially used in a tissue- or development-specific fashion (13), proteins in addition to the core polyadenylation factors may play an important role in influencing the efficiency of usage of polyadenylation signals. We have recently reported that recombinant DSEF-1 protein can interact with several polyadenylation signals and stimulate 3′-end processing (26). DSEF-1 is identical to either hnRNP H or the very closely related H′ protein. For simplicity, we will refer to DSEF-1 protein here as hnRNP H. As seen in Figure 1A, when recombinant hnRNP H protein was added to an in vitro cleavage/polyadenylation system that was programmed with sub-optimal levels of HeLa nuclear extracts, cleavage efficiency of the SVL, IVA2 and µM polyadenylation signals was stimulated ∼4-fold. While a single time point is shown for simplicity, similar levels of stimulation were observed in time-course studies (data not shown). Control RNA-binding proteins or GST alone had no effect on 3′-end processing efficiency (26; data not shown). Several independent HeLa nuclear extracts gave similar results as hnRNP H protein stimulated processing 3–7-fold depending on the extract (Figs 2 and 4 and data not shown). Finally, similar levels of stimulation by hnRNP H protein were also observed in polyadenylation assays (26; data not shown). These data suggest that hnRNP H protein is a candidate factor that may help regulate the usage of polyadenylation signals.
Figure 1.
HnRNP H protein shows signal-specific effects on 3′-end processing. (A) The IVA2, µM and SVL polyadenylation substrate RNAs were incubated in the in vitro 3′-end cleavage system in the absence (– lanes) or presence (+ lanes) of 500 ng recombinant hnRNP H protein. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The presence of hnRNP H increased the processing efficiency of the IVA2, µM and SVL polyadenylation signals by 3.7 ± 0.8-, 3.9 ± 0.8- and 3.6 ± 0.5-fold, respectively. (B) The SVL polyadenylation substrate RNA was incubated in the in vitro 3′-end cleavage system in the presence of the indicated amounts of a synthetic competitor RNA, GRS, that can bind and sequester hnRNP H protein. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The processing efficiency relative to 0 pmol GRS RNA competitor is shown below each lane. (C) The IVA2 polyadenylation substrate RNA was incubated in the in vitro 3′-end cleavage system in the presence of the indicated amounts of synthetic GRS competitor RNA. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The processing efficiency relative to 0 pmol GRS RNA competitor is shown below each lane. The arrows on the left of (B) and (C) indicate the positions of the 5′ cleavage product.
Figure 2.
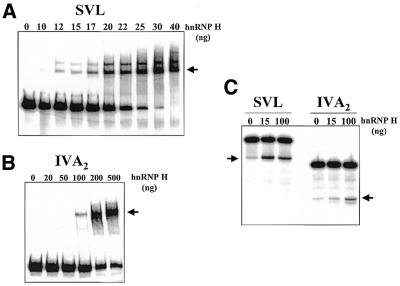
Differential binding affinity of hnRNP H affects its ability to stimulate 3′-end processing of the SVL and IVA2 polyadenylation signals. SVL (A) and IVA2 (B) RNAs were incubated with the indicated amount of recombinant hnRNP H protein. Heparin-resistant RNA–protein complexes were analyzed on 5% native acrylamide gels. The arrows on the right of each panel indicate the positions of hnRNP H–RNA complexes. (C) The indicated amounts of hnRNP H protein were added to in vitro cleavage reactions containing either the SVL or IVA2 polyadenylation signal RNAs. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. hnRNP H at 15 ng had a 4.2-fold greater effect on the SVL versus the IVA2 polyadenylation signal. hnRNP H at 100 ng stimulated both signals to a similar extent (6.8-fold).
Figure 4.
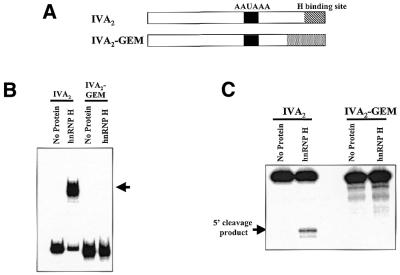
The downstream hnRNP H binding site is required for efficient processing and stimulation of the IVA2 polyadenylation signal. (A) Diagrammatic representation of the RNAs used in this study. (B) The IVA2 polyadenylation substrate RNA or IVA2-GEM, a derivative that contains a substitution in the hnRNP H binding region, were incubated with 500 ng recombinant hnRNP H protein. Heparin-resistant RNA–protein complexes were analyzed on a 5% native acrylamide gel. The arrow on the right indicates the position of hnRNP H–RNA complexes. (C) IVA2 or IVA2-GEM RNAs were incubated in the in vitro 3′-end cleavage system in the absence (no protein lanes) or presence (hnRNP H lanes) of 1 µg recombinant hnRNP H protein. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The arrow on the left indicates the position of the 5′ cleavage product. The addition of hnRNP H to the IVA2 polyadenylation signal resulted in a 6.8-fold increase in processing efficiency in this assay, while the protein had no effect on processing of the IVA2-GEM RNA substrate.
While the SVL, IVA2 and µM polyadenylation signals are all stimulated by exogenous hnRNP H protein to a similar extent, individual polyadenylation signals surprisingly reacted differently to changes in the availability of endogenous hnRNP H proteins present in the HeLa nuclear extract. The level of available hnRNP H proteins was altered in extracts by the addition of a synthetic RNA oligonucleotide, GRS (5′-GGGGGAGGUGUGGG), which contains the G-rich binding site for hnRNP H protein that was originally identified in the SVL polyadenylation signal (24). The GRS RNA efficiently sequestered endogenous hnRNP H protein in extracts, as seen by an inhibition of UV crosslinking of hnRNP H protein to the SVL RNA substrate (data not shown). The effect of sequestration of endogenous hnRNP H protein on the processing efficiency of the SVL polyadenylation signal was evaluated first. As expected, based on previous observations (24–26), addition of increasing amounts of GRS RNA to in vitro cleavage reactions decreased processing of the SVL polyadenylation signal to ∼10–20% of untreated levels (Fig. 1B). An RNA oligomer containing an unrelated sequence had no effect on in vitro processing efficiency and all of the effects observed with the GRS oligomer could be reversed by the addition of recombinant hnRNP H protein (25; data not shown). The observed decrease in 3′-end processing correlated with the ability of the GRS RNA to inhibit UV crosslinking of hnRNP H protein to the SVL polyadenylation signal (data not shown). These data demonstrate that hnRNP H protein influences the basal rate of processing of the SVL polyadenylation signal in HeLa extracts. Similar assays were next carried out using the IVA2 polyadenylation signal. Surprisingly, sequestration of hnRNP H protein by increasing amounts of the GRS RNA did not affect the basal processing efficiency of the IVA2 polyadenylation signal (Fig. 1C). In addition, although the IVA2 polyadenylation signal contains a binding site for hnRNP H (26), it was not efficiently crosslinked to endogenous hnRNP H protein in HeLa extracts (data not shown). Reduction in the endogenous level of hnRNP H protein in extracts also failed to influence the processing efficiency of the µM polyadenylation signal (data not shown). These data suggest that, while all three polyadenylation signals can interact with and be stimulated by recombinant hnRNP H protein (Fig. 1A), the SVL polyadenylation signal interacts in a distinct fashion with the auxiliary factor relative to the other two signals.
We hypothesized that the specific behavior of the SVL polyadenylation signal in response to hnRNP H protein in these assays could be due to two possibilities. First, there may be a difference between endogenous and recombinant hnRNP H proteins (i.e. post-translational modifications) that influences their ability to stimulate 3′-end processing. Alternatively, the binding affinity for hnRNP H may vary among polyadenylation signals. The low levels of endogenous hnRNP H protein relative to the amount of recombinant hnRNP H protein used in Figure 1 may only be sufficient to stimulate polyadenylation signals that have a high affinity for the protein. In order to directly test this latter hypothesis, the Kd for hnRNP H binding was measured for the three polyadenylation signals noted above by titrations and gel shift analyses. As seen in Figure 2, the SVL polyadenylation signal has a much higher affinity for hnRNP H protein (Kd = 2.2 × 10–8; Fig. 2A) than does the IVA2 polyadenylation signal (Kd = 1.3 × 10–7; Fig. 2B) or the µM signal (Kd = 1.1 × 10–7; data not shown). In order to assess the functional significance of the differential affinity of hnRNP H for various polyadenylation signals, the ability of recombinant hnRNP H to stimulate 3′-end processing was measured in titration studies. As seen in Figure 2C, low levels of hnRNP H efficiently stimulated processing of the SVL polyadenylation signal, but had no effect on processing of the IVA2 signal, which contains a lower affinity protein binding site. High levels (100 ng) of hnRNP H effectively stimulated both polyadenylation signals.
We conclude that the SVL polyadenylation signal has a relatively high affinity for hnRNP H protein and can be stimulated by low amounts of protein. The IVA2 and µM polyadenylation signals bind hnRNP H protein with ∼20-fold lower affinity. The level of hnRNP H protein in extracts, therefore, is not sufficient to bind or stimulate these polyadenylation signals to any significant extent. These observations suggest that variations in the local concentration of hnRNP H protein in the cell nucleus may differentially affect the processing of a polyadenylation signal depending on the nature of its hnRNP H binding site. This property is what would be expected of an auxiliary factor involved in the regulation of 3′-end processing.
Functional hnRNP H binding sites are relatively conserved with respect to sequence and location 3′ to the core downstream U-rich element
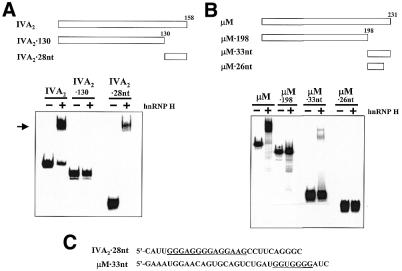
If hnRNP H is an auxiliary factor for polyadenylation, we would predict that the location of its binding site would be relatively conserved in the downstream regions of the polyadenylation signals that it stimulates. We previously mapped the hnRNP H binding site in the SVL polyadenylation signal to a 14 base G-rich element located 32 bases downstream of the cleavage site (24). In order to localize the hnRNP H binding sites on the IVA2 and µM polyadenylation signals, a series of 3′ deletion derivatives was prepared and tested for protein binding by gel shift analysis. As seen in Figure 3A, deletion of 28 bases from the 3′-end of the IVA2 polyadenylation signal eliminates hnRNP H binding (compare lanes IVA2 and IVA2130). A short RNA representing the deleted region was prepared (IVA228nt) and used in gel shift assays in order to verify that the 28 base region was sufficient for hnRNP H binding. As seen in Figure 3A, lanes IVA228nt, the 28mer efficiently bound to hnRNP H protein. We conclude that the hnRNP H binding site of the IVA2 polyadenylation signal lies within this 28 base segment which is located 36 nt downstream of the cleavage site and 3′ of the core downstream U-rich element. This is a similar location to the hnRNP H binding site previously mapped at +32 in the SVL polyadenylation signal (24). In addition, a 9 of 11 base G-rich region lies within the 28 base element (Fig. 3C). This G-rich region in the IVA2 hnRNP H binding site is also similar to the 11 of 14 base G-rich element of the hnRNP H-binding site of the SVL signal.
Figure 3.
hnRNP H protein binds to G-rich sequences located at a conserved downstream position in the SVL, IVA2 and µM polyadenylation signals. (A) The IVA2 polyadenylation substrate RNA or the indicated derivatives diagrammed at the top were incubated with 500 ng recombinant hnRNP H protein. Heparin-resistant RNA–protein complexes were analyzed on a 5% native acrylamide gel. The arrow on the left indicates the position of hnRNP H–RNA complexes. (B) The µM polyadenylation substrate RNA or the indicated derivatives diagrammed at the top were incubated with 500 ng recombinant hnRNP H protein. Heparin-resistant RNA–protein complexes were analyzed on a 5% native acrylamide gel. (C) Sequences required for hnRNP H binding by the IVA2 and µM polyadenylation signals. Conserved G-rich regions that may be required for binding are underlined.
A similar hnRNP H gel shift analysis was performed with deletion derivatives of the µM polyadenylation signal. As seen in Figure 3B, deletion of 33 bases from the 3′-end of the µM RNA (µM198) abolished hnRNP H binding (compare lanes µM and µM198). A 33 base RNA representing the deleted region (µM33nt) was able to bind hnRNP H protein, demonstrating that this downstream region is sufficient for binding. Inspection of the 33 base region from the µM polyadenylation signal also revealed a G-rich region (6 of 7 bases) located near the 3′-end of the binding region (Fig. 3C) that may be involved in hnRNP H binding. A 7 base deletion from the 3′-end of the µM33nt RNA that removes a significant portion of this G-rich region abolishes hnRNP H binding (Fig. 3B, lanes µM26nt). These data provide additional support for the important contribution of G-rich tracts to hnRNP H–RNA interactions (25,36). The 3′ portion of this G-rich tract in the µM RNA substrate, however, is not part of the natural polyadenylation signal. It was created by fusion of a RsaI site with a SmaI site in the vector DNA (32). Therefore, the hnRNP H binding site of the µM polyadenylation substrate RNA provides additional evidence for the important role of G residues and downstream location in hnRNP H protein binding and function in 3′-end processing efficiency. The µM RNA was not used for further study, however, because of the synthetic nature of its hnRNP H binding site.
We wished to determine whether the 28 nt G-rich region of the IVA2 polyadenylation signal that was sufficient for hnRNP H binding was also necessary for stimulation of 3′-end processing. We created a variant of IVA2 RNA that had the region +40 to +63 3′ of the cleavage site substituted by polylinker sequences (IVA2-GEM; Fig. 4A). IVA2-GEM RNA lost its ability to interact with hnRNP H protein by gel shift analysis (Fig. 4B) as well as was no longer stimulated by the addition of recombinant hnRNP H protein in in vitro cleavage assays (Fig. 4C, compare lanes IVA2 with IVA2-GEM). A similar substitution derivative of the SVL polyadenylation signal that lacked the GRS element also failed to both bind hnRNP H protein as well as have its 3′ cleavage efficiency stimulated by the addition of recombinant protein (25; data not shown). We conclude that the G-rich hnRNP H binding site located downstream of the core downstream element of the IVA2 polyadenylation signal is required for efficient 3′-end processing.
The identification of a relatively conserved hnRNP H binding site with respect to location and G-richness on the three polyadenylation signals we have studied allowed us to more accurately predict additional polyadenylation signals that may be capable of interacting with hnRNP H protein. We surveyed four databases of randomly compiled mammalian polyadenylation signals for G-rich regions located within 100 bases downstream of the predicted U-rich CstF-binding site. One database was developed by Dr Clint MacDonald (6); the others were compiled by searching human genomic entries, chromosome-specific entries, or mouse genomic entries for poly(A) and refined by identification of a properly located AAUAAA or AUUAAA core element. Interestingly, 76 of the 219 polyadenylation signals surveyed contained significant G-rich regions at this dowsntream location (Table 1). A similar percentage of mouse and human polyadenylation signals contained downstream G-rich regions, suggesting that the relative presence of this element is conserved among species. In addition, a bias for G-rich sequences downstream of the poly(A) addition site was also observed previously in an analysis of the distribution of random quartets and pentamers (37). In order to test if these G-rich regions were indeed hnRNP H binding sites, short RNAs containing these sites were prepared and tested for hnRNP H interaction by band shift analysis. As seen in Figure 5, all six RNAs effectively bound to hnRNP H protein with a binding affinity similar to the IVA2 polyadenylation signal (data not shown). We conclude that these G-rich regions contain bona fide hnRNP H-binding sites.
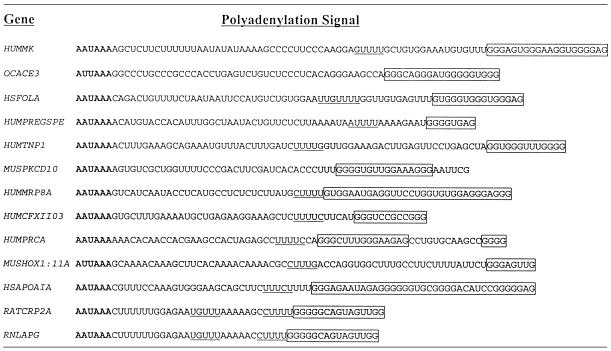
Table 1. Sequences of a subset of the 76 polyadenylation signals identified in a database search that contain G-rich sequences downstream of the core elements.
The core AAUAAA elements are in bold, core U-rich elements are underlined and the G-rich sequences are boxed.
Figure 5.
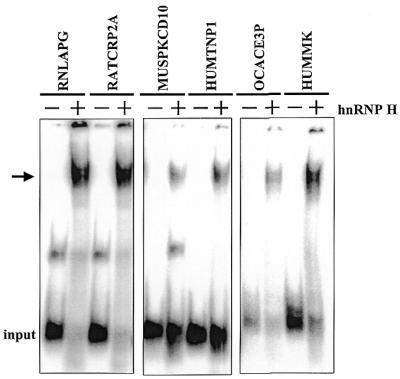
G-rich regions located in the downstream region of ∼34% of polyadenylation signals surveyed are capable of directing the binding of hnRNP H protein. Small RNAs containing the G-rich downstream region from the indicated polyadenylation signal (Table 1) were incubated with 1 µg recombinant hnRNP H protein. Heparin-resistant RNA–protein complexes were analyzed on a native 5% acrylamide gel.
Processing of a newly identified cellular polyadenylation signal that contains a downstream G-rich tract is stimulated by hnRNP H protein
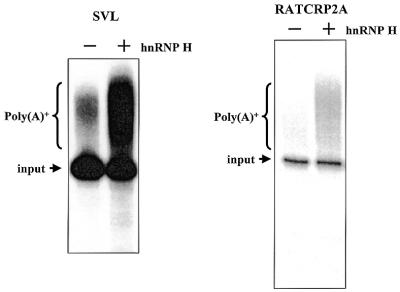
The data described above suggest that ∼34% of mammalian polyadenylation signals contain binding sites for hnRNP H protein that are in an appropriate location for the protein to influence 3′-end processing. In order to test this hypothesis, we prepared an RNA substrate that contains the core elements and downstream G-rich tract of the RATCRP2A gene. Since this signal was processed rather inefficiently in vitro, polyadenylation assays were performed instead of cleavage assays to obtain more quantitative data. As we have previously shown, hnRNP H protein stimulates 3′-end processing in both cleavage and polyadenylation assays to a similar extent (26). The RATCRP2A RNA substrate was very inefficiently processed in in vitro polyadenylation assays (Fig. 6, lane –). The addition of 250 ng hnRNP H protein, however, stimulated the processing of this weak polyadenylation signal ∼4-fold (Fig. 6, lane +). We conclude that the representative RATCRP2A polyadenylation signal responds in a similar fashion to the three other polyadenylation signals we have tested that contain downstream G-rich motifs in that it can both bind and be stimulated by hnRNP H protein. These data suggest that hnRNP H is an auxiliary factor that influences the processing of a significant proportion of cellular polyadenylation signals through an interaction with a conserved G-rich auxiliary downstream element.
Figure 6.
Processing of the cellular RATCRP2A polyadenylation signal is stimulated by hnRNP H protein. Substrate RNA containing the SVL or RATCRP2A polyadenylation signal was incubated in the in vitro polyadenylation system in the absence (– lane) or presence (+ lane) of 250 ng recombinant hnRNP H protein . Reaction products were identified on a 5% acrylamide gel containing 7 M urea. The positions of input and polyadenylated RNA products of the reaction are indicated on the left. The addition of hnRNP H protein to the reaction stimulated processing by 4.1 ± 0.8-fold.
Overexpression of hnRNP H′ in vivo enhances expression of genes with G-rich auxiliary downstream elements
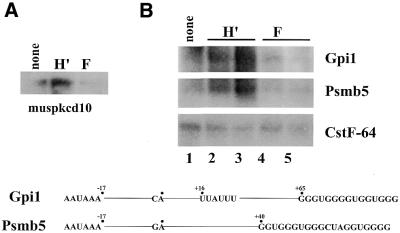
In order to investigate the effect of overexpression of hnRNP H on polyadenylation efficiency in vivo, we have recently established several independent clones of the mouse AxJ myeloma cell line that express a FLAG-tagged version of hnRNP H′. These clones showed a 10–30% increase in hnRNP H protein expression over endogenous levels. Cell lines were also obtained that contained empty vector as well as hnRNP F protein as controls. First, expression of the mouse cAMP-dependent protein kinase α-subunit (MUSPKCD10) was examined. The MUSPKCD10 polyadenylation signal contains a G-rich sequence in its downstream region as outlined in Table 1. As seen in Figure 7A, expression of the MUSPKCD10 transcript was stimulated in cell lines that overexpressed H′ protein, but not in control cell lines or one that expressed hnRNP F. In order to confirm the ability of hnRNP H protein to stimulate expression from genes that contain G-rich elements in their polyadenylation signals, two additional mouse genes identified in our survey were examined. As diagrammed at the bottom of Figure 7, the mouse Gpi1 and Psmb5 genes both contain G-rich elements located downstream of the polyadenylation/cleavage site. As seen in Figure 7B, expression of both Gpi1 and Psmb5 mRNAs was specifically stimulated in two independent cell lines that overexpressed hnRNP H′ protein. Expression of CstF-64 mRNA, which lacks a G-rich element in its polyadenylation signal, was unaffected in these lines. Furthermore, we observed no change in Ig sec mRNA levels (which also lacks a G-rich element) or in the splicing of CD45 mRNAs in these studies (data not shown). The overall stimulation of expression by hnRNP H′ protein of the Gpi1, Psmb5 and MUSPKCD10 mRNAs in these studies was 3–4-fold. These data demonstrate that relative levels of hnRNP H′ protein can selectively influence gene expression and 3′-end processing efficiency both in vivo and in vitro.
Figure 7.
Expression of transcripts containing G-rich sequences downstream of their polyadenylation/cleavage sites is stimulated in hnRNP H′-transfected cells. AxJ plasma cells were transfected with an expresssion vector carrying either no insert (none lanes), hnRNP H′ coding sequence (lanes H′) or hnRNP F coding sequences (lanes F). Poly(A)+ RNA (1 µg) was run on a denaturing formaldehyde gel, blotted to membrane and probed for the indicated endogenous mouse mRNAs. (A) MUSPKCD10 gene (organization of the polyadenylation signal is outlined in Table 1). (B) Lane 1, cells transfected with vector containing no insert; lanes 2 and 3, vector plus hnRNP H′ coding region; lanes 4 and 5, vector plus hnRNP F coding region. Blots were probed for the mRNA indicated on the right. (Bottom) Organization of the AAUAAA motif, U-rich CstF-binding site and G-rich elements in the mouse Gpi1 and Psmb5 polyadenylation signals. Numbers indicate the position relative to the CA or GA cleavage site. A consensus CstF-binding site could not be identified in the downstream region of the Psmb5 signal.
DISCUSSION
The studies reported above advance our understanding of several aspects of hnRNP H protein, a candidate auxiliary factor for 3′-end processing. First, we have demonstrated that polyadenylation signals show differences in their affinity for hnRNP H protein. This observation raises the possibility that changes in localized concentrations of hnRNP H protein could have a dramatic, signal-specific effect on polyadenylation efficiency in vivo. Second, we have identified several new hnRNP H-binding sites in polyadenylation signals. These data suggest that downstream G-rich tracts are a conserved feature of a significant percentage of mammalian polyadenylation signals. Finally, we have demonstrated that a polyadenylation signal that contains a downstream G-rich motif can be stimulated by hnRNP H protein. This allows for a better understanding of the sequence requirements for a polyadenylation signal to be efficiently stimulated by hnRNP H and suggests an important role for hnRNP H in transcript-specific regulation of polyadenylation site usage.
The identification of a relatively conserved hnRNP H-binding site with respect to location and G-richness in approximately one-third of mammalian polyadenylation signals surveyed, suggests that the protein may play a rather widespread role in efficient 3′-end processing. One particularly interesting example of a new hnRNP H binding site is in the rabbit angiotension converting enzyme pre-mRNA (OCACE3P). This pre-mRNA is alternatively polyadenylated in a tissue-specific fashion. The regulated upstream polyadenylation signal contains non-consensus core elements and a G-rich hnRNP H binding site in an appropriate downstream location. Perhaps hnRNP H plays a direct role in regulating this 3′-end processing event. It will be interesting to determine the levels of hnRNP H protein in various tissues and see if alterations in protein levels correlate with polyadenylation site usage. Previous data by Dreyfuss and co-workers, in fact, have suggested significant variation in the tissue distribution of hnRNP H protein using a monoclonal antibody that detects both hnRNP H and the closely related hnRNP F protein (38). Future studies will involve assessing in vivo expression patterns of hnRNP H protein using specific anti-peptide antibodies that we have recently developed (39).
This study establishes downstream G-rich auxiliary elements as a common feature of numerous polyadenylation signals that likely play an important role in determining the efficiency of poly(A) site selection. We have previously reported that hnRNP H protein can stimulate assembly of the CPSF/CstF scaffold onto the core elements of a polyadenylation signal (26). Recent data by Hans and Alwine (20) suggest that the downstream G-rich hnRNP H binding site of the SVL polyadenylation signal has a significant influence on the overall secondary structure of the core elements. We suggest that as the SVL polyadenylation signal (or any polyadenylation signal that contains a downstream G-rich element) is transcribed, it may fold into a sub-optimal conformation that does not form an efficient binding site for the CPSF/CstF factors that are carried to the nascent transcript by the CTD of the large subunit of RNA polymerase II (8,9). Local concentrations of hnRNP H protein may determine whether the auxiliary factor will bind to the G-rich element of the nascent transcript, alter its structure and present the core elements to the general polyadenylation factors in a productive fashion. Alternatively, hnRNP H can bind to the nascent polyadenylation signal and directly interact with CstF to stimulate complex assembly onto the AAUAAA and U-rich elements. In total, these models would provide a mechanistic explanation for regulated usage of alternative polyadenylation sites in a transcription unit in the absence of significant changes in the level of CstF-64 protein (17,18,40).
Two other proteins that have been implicated in regulated RNA processing are members of the hnRNP H family of RNA binding proteins. The hnRNP F protein binds avidly to poly(G), can influence splicing in vitro (41) and has been implicated in the regulation of splice site choice in neurons (29,30,36). While the RNA binding domains of hnRNP H and F are rather conserved, significant differences exist, however, in the C-terminal region of these two proteins. Recent data has in fact suggested that the binding of hnRNP F to a polyadenylation signal may inhibit rather than stimulate 3′-end processing (39). This suggests that hnRNP H and hnRNP F may not simply play redundant roles in 3′-end processing. H29, a small 29 kDa relative of the hnRNP H/F family of proteins, contains a large degree of similarity to the central domain and RRM3 of hnRNP H, but differs significantly in the C-terminal domain (42). H29 is also differentially expressed during heat shock and has been suggested to stimulate RNA splicing. In summary, this network of RNA binding proteins with different combinations of RNA binding and effector domains may play an important role in regulating gene expression at the level of splicing and 3′-end processing.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank members of the Wilusz laboratory for their assistance throughout this study, Kathleen Martincic for excellent technical assistance and Dr Bob Donnelly of the NJMS Molecular Resource Facility for oligonucleotide synthesis. This work was supported by grants from the National Institutes of Health, GM56434 and CA80062 to J.W. and CA86433 to C.M.
REFERENCES
- 1.Wahle E. and Ruegsegger,U. (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMSMicrobiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F., MacDonald,C.C. and Wilusz,J. (1995) Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res., 23, 2614–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA, 96, 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheets M.D., Ogg,S.C. and Wickens,M.P. (1990) Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res., 18, 5799–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacDonald C.C., Wilusz,J. and Shenk,T. (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol., 14, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murthy K.G. and Manley,J.L. (1995) The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- 8.Hirose Y. and Manley,J.L. (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- 9.McCracken S., Fong,N., Yankulov,K., Ballantyne,S., Pan,G., Greenblatt,J., Patterson,S.D., Wickens,M. and Bentley,D.L. (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- 10.Yonaha M. and Proudfoot,N.J. (1999) Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell, 3, 593–600. [DOI] [PubMed] [Google Scholar]
- 11.Nesic D. and Maquat,L.E. (1994) Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev., 8, 363–375. [DOI] [PubMed] [Google Scholar]
- 12. Niwa,M., Rose,S.D. and Berget,S.M. (1990) In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev., 4, 1552–1559. [DOI] [PubMed] [Google Scholar]
- 13.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagaki Y., Seipelt,R.L., Peterson,M.L. and Manley,J.L. (1996) The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell, 87, 941–952. [DOI] [PubMed] [Google Scholar]
- 15.Beyer K., Dandekar,T. and Keller,W. (1997) RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem., 272, 26769–26779. [DOI] [PubMed] [Google Scholar]
- 16.Phillips C., Schimpl,A., Dietrich-Goetz,W., Clements,J. and Virtanen,A. (1996) Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory poly(A) site. Eur. J. Immunol., 26, 3144–3152. [DOI] [PubMed] [Google Scholar]
- 17.Martincic K., Campbell,R., Edwalds-Gilbert,G., Souan,L., Lotze,M. and Milcarek,C. (1998) Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl Acad. Sci. USA, 95, 11095–11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takami Y., Kikuchi,H. and Nakayama,T. (1999) Chicken histone deacetylase-2 controls the amount of the IgM H-chain at the steps of both transcription of its gene and alternative processing of its pre-mRNA in the DT40 cell line. J. Biol. Chem., 274, 23977–23990. [DOI] [PubMed] [Google Scholar]
- 19.Graveley B.R., Fleming,E.S. and Gilmartin,G.M. (1996) RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol. Cell. Biol., 16, 4942–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hans H. and Alwine,J.C. (2000) RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol. Cell. Biol., 20, 2926–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carswell S. and Alwine,J.C. (1989) Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol. Cell. Biol., 9, 4248–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreira A., Takagaki,Y., Brackenridge,S., Wollerton,M., Manley,J.L. and Proudfoot,N.J. (1998) The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev., 12, 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F. and Wilusz,J. (1998) Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res., 26, 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian Z. and Wilusz,J. (1991) An RNA-binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol., 11, 5312–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagga P.S., Ford,L.P., Chen,F. and Wilusz,J. (1995) The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′-end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res., 23, 1625–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagga P.S., Arhin,G.K. and Wilusz,J. (1998) DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res., 26, 5343–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honore B., Rasmussen,H.H., Vorum,H., Dejgaard,K., Liu,X., Gromov,P., Madsen,P., Gesser,B., Tommerup,N. and Celis,J.E. (1995) Heterogeneous nuclear ribonucleoproteins H, H′ and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem., 270, 28780–28789. [DOI] [PubMed] [Google Scholar]
- 28.Bai Y., Lee,D., Yu,T. and Chasin,L.A. (1999) Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res., 27, 1126–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou M.Y., Rooke,N., Turck,C.W. and Black,D.L. (1999) hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol., 19, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min H., Chan,R.C. and Black,D.L. (1995) The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev., 9, 2659–2671. [DOI] [PubMed] [Google Scholar]
- 31.Wilusz J. and Shenk,T. (1988) A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell, 52, 221–228. [DOI] [PubMed] [Google Scholar]
- 32.Peterson M.L. and Perry,R.P. (1989) The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol. Cell. Biol., 9, 729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian Z. and Wilusz,J. (1994) GRSF-1: a poly(A)+ mRNA binding protein which interacts with a conserved G-rich element. Nucleic Acids Res., 22, 2334–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore C.L. and Sharp,P.A. (1985) Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell, 41, 845–855. [DOI] [PubMed] [Google Scholar]
- 35.Veraldi K.L., Arhin,G.K., Martincic,K., Chung-Ganster,L.H., Wilusz,J. and Milcarek,C. (2001) hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol., 21, 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson M.S. and Dreyfuss,G. (1988) RNA binding specificity of hnRNP proteins: a subset bind to the 3′-end of introns. EMBO J., 7, 3519–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nussinov R. (1986) Sequence signals which may be required for efficient formation of mRNA 3′-termini. Nucleic Acids Res., 14, 3557–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamma H., Portman,D.S. and Dreyfuss,G. (1995) Cell type-specific expression of hnRNP proteins. Exp. Cell Res., 221, 187–196. [DOI] [PubMed] [Google Scholar]
- 39.Veraldi K.L., Arhin,G.K., Martincic,K., Chung-Ganster,L.H., Wilusz,J. and Milcarek,C. (2001) hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol., 21, 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwalds-Gilbert G. and Milcarek,C. (1995) Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol., 15, 6420–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gamberi C., Izaurralde,E., Beisel,C. and Mattaj,I. (1997) Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol. Cell. Biol., 17, 2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahe D., Mahl,P., Gattoni,R., Fischer,N., Mattei,M.G., Stevenin,J. and Fuchs,J.P. (1997) Cloning of human 2H9 heterogeneous nuclear ribonucleoproteins. Relation with splicing and early heat shock-induced splicing arrest. J. Biol. Chem., 272, 1827–1836. [DOI] [PubMed] [Google Scholar]