Abstract
BUD31, a splicing factor, is linked to various cancers. This study examines BUD31's expression, prognostic value, mutation profile, genomic instability, tumor immune environment, and role in clear cell renal cell carcinoma (ccRCC), focusing on cell cycle regulation via alternative splicing. BUD31 expression was analyzed using TCGA and GTEx databases across 33 cancers. Techniques included IHC staining, survival analysis, Cox regression, and nomogram construction. Mutation landscape, genomic instability, and tumor immune microenvironment were evaluated. Functional assays on ccRCC cell lines involved BUD31 knockdown, RNA sequencing, and alternative splicing analysis. BUD31 was upregulated in multiple tumors, including ccRCC. High BUD31 expression correlated with worse survival outcomes and was identified as an independent predictor of poor prognosis in ccRCC. High BUD31 expression also correlated with increased genomic instability and a less active immune microenvironment. BUD31 knockdown inhibited cell proliferation, migration, and invasion in vitro and reduced tumor growth in vivo. RNA sequencing identified 390 alternative splicing events regulated by BUD31, including 17 cell cycle-related genes. KEGG analysis highlighted pathways involved in cell cycle regulation, indicating BUD31's role in promoting cell cycle progression through alternative splicing. BUD31 is upregulated in various tumors and is associated with poor outcomes, increased genomic instability, and a suppressed immune microenvironment in ccRCC. BUD31 promotes cell cycle progression via alternative splicing, suggesting it as a prognostic biomarker and potential therapeutic target in ccRCC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-024-01451-8.
Keywords: BUD31, Clear cell renal cell carcinoma, Prognosis, Cell cycle, Alternative splicing, Cancer biomarker
Introduction
Renal cell carcinoma (RCC) is a prevalent and highly aggressive malignancy, with the clear cell variant (ccRCC) being the most common subtype. ccRCC is notorious for its resistance to conventional chemotherapeutic and radiotherapeutic treatments, presenting significant challenges in oncology [1]. The discovery and characterization of biomarkers are crucial for understanding cancer pathophysiology and developing targeted therapeutic strategies. In this context, BUD31 has recently emerged as a gene of significant interest due to its involvement in various cellular processes and its potential role in cancer progression [2–4].
BUD31, a spliceosome-associated protein, plays a critical role in alternative splicing, a post-transcriptional process that generates multiple mRNA variants from a single gene, thereby contributing to proteomic diversity and cellular complexity [4–8]. Dysregulation of alternative splicing has been implicated in various cancers [9–12], including ccRCC, where it may drive tumor progression by affecting the expression of genes involved in key pathways such as cell cycle regulation [13–15]. Despite previous studies indicating that BUD31 is upregulated in several tumor types and suggesting its role in oncogenesis [2, 4, 16]. Its specific contributions to ccRCC remain incompletely understood.
Our study aims to investigate the role of BUD31 in ccRCC by examining its expression levels, functional impact on cell proliferation, migration, and invasion, and its influence on alternative splicing and cell cycle pathways. We utilized comprehensive bioinformatics analyses to identify significant correlations between BUD31 expression and key clinical outcomes, genomic instability, and immune microenvironment parameters in ccRCC. Previous research has highlighted the activation of the PI3K pathway as a promoter of cell cycle progression, which we explored further in the context of BUD31 knockdown and its regulation of PIK3AP1, a gene implicated in PI3K signaling [17–21].
Our findings indicate that BUD31 is upregulated in ccRCC and correlates with poor survival outcomes, increased genomic instability, and a less active immune microenvironment. Functional assays revealed that BUD31 knockdown inhibited cell proliferation, migration, and invasion in vitro and reduced tumor growth in vivo. RNA sequencing identified numerous alternative splicing events regulated by BUD31, particularly involving cell cycle-related genes. KEGG pathway analysis further highlighted BUD31's role in promoting cell cycle progression through alternative splicing.
By integrating genomic, transcriptomic, and functional data, this study provides new insights into the molecular mechanisms by which BUD31 promotes ccRCC progression. These findings underscore BUD31's potential as a prognostic biomarker and a therapeutic target, offering significant implications for personalized treatment strategies in ccRCC.
Materials and methods
Data collection and processing
From UCSC Xena database (https://xena.ucsc.edu/), we obtained a unified and standardized general cancer dataset TCGA Pan Cancer (PANCAN) and its clinical data, excluding cancers with less than three samples in a single cancer, and finally obtained the expression data of 37 cancers. We downloaded the KIRC-related datasets of GSE53757, GSE66272, and GSE36895 from the geo database (https://www.ncbi.nlm.nih.gov/geo/). In addition, the mutation data of GDC TCGA-KIRC were obtained from UCSC Xena database and processed by MuTect2 software. We examined the expression of Bud31 protein in renal cell carcinoma by searching the Human Protein Map Database (HPA, http://www.ProteinAtlas.org/).
Study of Bud31 and clinical data
Single-factor Cox regression was performed using the survival package, the optimal blockade of Bud31 RNA expression was found by survminer package, and then the Kaplan–Meier (KM) method was used to study the survival rate and prognostic value of Bud31 in different cancer types. In TCGA-KIRC, we performed multifactorial Cox regression to examine the prognostic value of Bud31 expression and related clinical parameters such as TNM stage, stage, class, age, gender on OS, DSS, and PFI survival data, and nomogram of OS was constructed using the rms package. characteristic (ROC) curve analysis using the time ROC package to determine OS, DSS, and PFI at 1, 3, and 5 years. The sensitivity and specificity of Bud31 were analyzed using the area under the curve (AUC) results, and the constructed comparison was used as a prognostic indicator. p < 0.05 was the significance threshold.
Genetic mutation landscape and genome heterogeneity analysis
The tmb function of the R software package maftools [22] (version 2.8.05) was used to calculate TMB (tumor mutation burden) for each KIRC sample, and the infer heterogeneity function was used to calculate MATH (mutant allelic tumor heterogeneity) for each sample. At the same time, we also obtained the purity (tumor purity) data, ploidy (tumor ploidy) data, HRD (homologous recombination deficiency) data, and LOH (Loss of heterozygosity) data of each TCGA-KIRC tumor sample from a previous study [23]. We divided TCGA-KIRC tumor samples into high and low expression groups based on the median expression of Bud31 in KIRC and explored the distribution differences of the above indicators in different groups. p < 0.05 was the threshold of significance.
Immune infiltration and RNA modification analysis
The R ESTIMATE package was used to calculate stromal, immune, and ESTIMATE scores for each TCGA-KIRC tumor sample based on the observed gene expression profile. Furthermore, the correlation between Bud31 and the proportion of different immune cells within KIRC was analyzed using Cibersort and TIMER. In addition, five types of immune pathways (chemokines (41), receptors (18), MHC (21), immune inhibitors (24), immune stimulants (46)), and RNA modification-related gene sets (m6A, m1A, m5C) and investigated the Pearson's correlation with Bud31 expression in KIRC.
Alternative splicing analysis using rMATS software
Alternative splicing events were analyzed using the rMATS (replicate Multivariate Analysis of Transcript Splicing) software. Total RNA was extracted from renal clear cell carcinoma (ccRCC) cells with knockdown of the BUD31 gene and from control cells. RNA sequencing (RNAseq) was performed to obtain high-quality sequencing data. The sequencing reads were then aligned to the human reference genome using the STAR aligner. In post-alignment, the rMATS software was employed to identify and quantify differential alternative splicing events between BUD31 knockdown and control samples. rMATS utilizes a statistical model to analyze splicing events from RNAseq data, accounting for biological replicates and identifying five types of alternative splicing events: skipped exons (SE), retained introns (RI), alternative 5' splice sites (A5SS), alternative 3' splice sites (A3SS), and mutually exclusive exons (MXE). The resulting splicing events were filtered for significance using a false discovery rate (FDR) threshold of < 0.05. Additionally, the inclusion levels of each splicing event were calculated to determine the extent of alternative splicing. Differentially spliced genes were further subjected to KEGG pathway enrichment analysis to identify the biological pathways affected by BUD31-regulated splicing events, with a particular focus on pathways related to cell cycle regulation.
GO, KEGG and GSEA analysis
The clusterProfiler package was utilized to perform GO, KEGG, GSEA analysis [24]. Base on the “c5.go.bp.v2023.2.Hs.symbols” gene set, the "c2.cp.kegg.v7.4.symbols" gene set, the "c2.cp.wikipathways.v2023.2.Hs.symbols" gene set, the “c2.cp.reactome.v2023.2.Hs.symbols” gene set and the HALLMARK gene set from the MSigDB database (https://www.gsea-msigdb.org/gsea/index.jsp), different pathways and molecular mechanisms in the WT and shBud31 groups were evaluated. Adjust p < 0.05 was the threshold of significance.
Cell lines and culture
The human renal cell carcinoma (RCC) cell lines 786O, OS-RC-2, ACHN, CAKI-1, A498, and 769P, as well as the human renal proximal tubular epithelial cell line HK2, were purchased from the American Type Culture Collection (ATCC). The culture media used for the 786O, 769P, and OS-RC-2 cell lines was RPMI-1640 (Invitrogen-Gibco), which was incubated at 37 °C in a humidified atmosphere containing 5% CO2. CAKI-1 cells were cultured in McCoy's 5A (Gibco), while HK2 and HEK293T cells were maintained in DMEM (Gibco). The ACHN and A498 cell lines were cultured in Minimum Essential Medium (MEM) from Gibco. The cell culture media were supplemented with 10% concentration of Fetal Bovine Serum (FBS) from Thermo Fisher Scientific and 1% penicillin–streptomycin from Gibco.
Establishment of plasmids and stable transfected cells, total RNA isolation, reverse transcription, and quantitative real-time polymerase chain reaction
The plausibility of all plasmids was verified through DNA sequencing. Subsequently, recombinant lentiviral vectors were transfected into HEK293T cells, in conjunction with pMD2.G and psPAX2 plasmids, to generate recombinant lentivirus. The lentivirus was used to infect the target cells, which were then exposed to a 14-day puromycin treatment. After confirming the efficacy of knockdown plasmids, the surviving cells were utilized for subsequent experiments. The TRIzol Reagent (Invitrogen, USA) was utilized for the purpose of extracting total RNA from tissues or cells. Following this, the synthesis of cDNA was conducted utilizing the Prime Script RT Reagent Kit. (Takara, Shiga, Japan). The ChamQ Universal SYBR qPCR Master Mix (Vazyme #Q711) was employed for qRT-PCR, the oligonucleotide primers used for quantitative PCR are shown in Supplementary Table S2.
Colony formation, EdU incorporation, and CCK-8 assay
In colony formation assays, approximately 1000 cells were initially introduced into six-well plates and subsequently cultivated for a period of 2 weeks. The enumeration of cell colonies was conducted subsequent to the application of crystal violet staining. The Cell-Light EdU Apollo488 in vitro Kit (RiboBio, Guangzhou, China) was utilized to stain the nucleus of proliferating cells with red fluorescence in EdU incorporation assays, while all nuclei were stained with blue fluorescent light. In the CCK-8 assay, the cells were subjected to the corresponding kit and then seeded into 96-well microplates at a concentration of 1000 cells per well. The absorbance at 450 nm was measured for each well at different time intervals.
Wound healing and transwell experiments
Wound healing assay: According to the density of 2.5 × 105 cells per well, 786O, OS-RC2 cells transfected with the control plasmids or shBud31 plasmids were inoculated in the 6-well plate. After 24 h, 200-μL pipetting head was used to scratch the cells in the plate and the serum-free medium was replaced to collect images under an inverted microscope (IX81, Olympus Company, Japan) at 0 h, 24 h. Transwell assay: The upper chamber of Transwell assay was coated with a layer of Matrigel matrix glue (Corning Company, USA) (matrix glue: Serum-free medium = 1:8). Then, the cells were resuspended and counted using serum-free medium. The cells were seeded into the upper chamber at a density of 5 × 104, and the lower chamber was added 600 μL full medium. After 24 h, the cells were fixated with 4% paraformaldehyde for 15 min; 1% crystal violet was used to staining cells for 10 min. The remaining cells in the upper chamber were slightly wiped off, and images were collected under a positive microscope (BX53, Olympus Company, Japan). In addition, the number of cells passing through the chamber was counted in four random fields under the microscope.
Western blot analysis
Protein extracts were obtained from ccRCC cells using RIPA buffer with protease inhibitors. Protein concentrations were measured using a BCA assay. Samples containing 30 µg of protein were separated by 10% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with 5% milk in TBST for 1 h, then incubated overnight at 4 °C with primary antibody. After washing, membranes were exposed to HRP-conjugated secondary antibody (1:5000) for 1 h at room temperature. Protein bands were visualized using ECL and imaged on X-ray film. Band intensities were analyzed using ImageJ. The antibody brand and dilution ratio are as follows: Bud31,Proteintech,1:1000; β-actin, Proteintech,1:1000; CDK2, Proteintech,1:1000; CDK4, Proteintech,1:1000; CDK6, Proteintech,1:1000; P21, Proteintech,1:1000; P27, Proteintech,1:1000.
Subcutaneous xenograft experiment
All mouse experiments were conducted in accordance with protocols approved by The Animal Care Committees of Wuhan University Medical Research Institute and followed guidelines for animal welfare. Four-week-old BALB/c female nude mice were purchased from GemPharmatech LLC. The mice were subjected to subcutaneous administration of either OS-RC-2 cells with stable transfected shBUD31 or control cells, with a quantity of 5 × 106 cells per mouse, specifically targeting the left axillary region.
Tumor dimensions were assessed on a weekly basis. After 30 days, mice were euthanized using CO2 gas, subcutaneous tumors were removed and images were collected. The tumor volume was calculated with the formula volume = (length × width2)/2.
Statistical analysis
The data are presented as means ± standard deviation (SD), and all experiments were repeated in triplicate. All statistical analyses were performed by GraphPad Prism 9.0 or R software (version 4.3.2). Data were normalized and preprocessed using the “limma” package [25] and log2 transformed, and differentially expressed genes were identified using the following criteria: log2 Fold Change (FC) ≥ 1 and adj p value < 0.05. Differences between two groups were compared using unpaired Student’s t-test or Wilcoxon rank sum test. Correlations between genes expression or biological pathways were calculated using Pearson’s correlation analysis. p < 0.05 was the threshold of significance. Statistical significance is described as follows ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
Results
Bud31 expression is upregulated in multiple tumors including ccRCC.
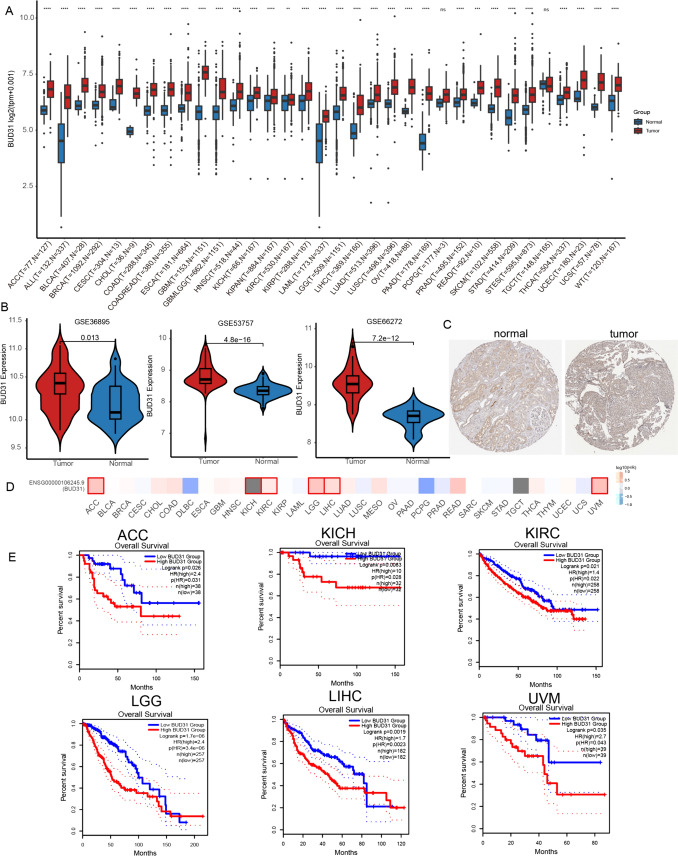
TCGA and GTEx database analyses of tumor tissues from 33 cancer types showed that Bud31 mRNA expression levels were tumor-specific and showed that the Bud31 mRNA levels were upregulated in the adrenocortical carcinoma (ACC), bladder urothelial carcinoma(BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head-and-neck cancer (HNSC), kidney renal clear cell carcinoma (KIRC), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), and uterine carcinosarcoma (UCS) tumor tissues compared to the corresponding normal tissues, while decreased in testicular germ cell tumors (TGCTs) (Fig. 1A). In addition, the expression results of Bud31 in the KIRC datasets GSE36895, GSE53757, and GSE66272 were the same as those in the TCGA dataset (Fig. 1B). IHC staining in the HPA database further confirmed the upregulation of Bud31 at the protein level in KIRC (Fig. 1C). Kaplan–Meier curves indicated that patients expressing higher levels of Bud31 exhibited a shorter OS relative to patients expressing lower levels of this gene. (Fig. 1D, E). Together, these data indicate that the consistent pattern of higher Bud31 expression in tumor samples across most cancer types could imply that Bud31 plays an important role in tumorigenesis or tumor maintenance. It may also suggest that Bud31 has the potential to be a biomarker for the presence of cancer or possibly a target for therapeutic intervention.
Figure 1.
Differential expression and prognostic value of Bud31 in various malignancies. A Expression profile of Bud31 across multiple cancer types, depicted as box plots. Data are segregated into normal (blue) and tumor (red) groups for each study identifier. B Violin plots illustrating the distribution of Bud31 expression in normal versus tumor samples from three independent datasets, with p-values indicating statistical significance. C Comparative immunohistochemical analysis of Bud31 protein levels in normal (left) and neoplastic (right) tissue sections. D Heatmap correlation matrix of Bud31 expression levels across different cancer subtypes, with intensity gradients representing expression magnitude. E Kaplan–Meier survival plots evaluating the correlation between Bud31 expression and overall survival in six distinct cancer cohorts, stratified by high (red) and low (blue) expression levels, accompanied by p-values and hazard ratios. * P < 0.05, **P < 0.01, ***P < 0.001
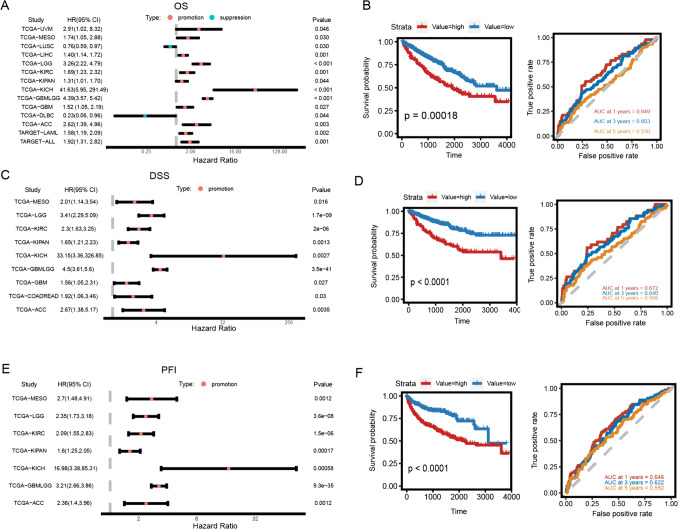
Bud31 expression correlates with prognostic outcomes in diverse tumors, including clear cell renal cell carcinoma (ccRCC)
The relationship between BUD31 expression and clinical outcomes, including overall survival (OS), disease-specific survival (DSS), and progression-free survival (PFS), was evaluated across various cancer types. High BUD31 expression was found to be associated with worse OS in several cancers, including UVM, MESO, LIHC, KIRC, KICH, GBMLGG, and ACC, as indicated by the hazard ratios (HR) in Fig. 2A. For example, the HR for UVM was 2.91 (p = 0.030), for MESO was 7.41 (p = 0.030), and for KIRC was 1.90 (p < 0.001). Kaplan–Meier survival curves in Fig. 2B show that patients with high BUD31 expression had significantly lower OS compared to those with low BUD31 expression in ccRCC (p =0.00018). Similarly, high BUD31 expression correlated with worse DSS in cancers such as MESO, LGG, KIRC, KICH, GBMLGG, COADREAD, and ACC, as shown in Fig. 2C. For instance, the HR for LGG was 3.41 (p < 0.0001), for KIRC was 2.31 (p =0.000002), and for KICH was 33.16 (p = 0.0027). Kaplan–Meier curves in Fig. 2D confirm that high BUD31 expression is significantly associated with reduced DSS in ccRCC (p < 0.0001). For PFS, high BUD31 expression predicted poorer outcomes in cancers including MESO, LGG, KIRC, COADREAD, KICH, GBMLGG, and ACC, as depicted in Figure E. For example, the HR for MESO was 2.70 (p = 0.0012), for LGG was 2.35 (p = 0.0000036), and for KIRC was 2.09 (p = 0.0000015). Kaplan–Meier survival curves in Figure F demonstrate that high BUD31 expression is significantly linked to poorer PFS in ccRCC (p < 0.0001). Overall, these results highlight that high BUD31 expression is consistently associated with worse clinical outcomes across multiple cancer types, including ccRCC, suggesting its potential as a prognostic marker for poor survival in cancer patients.
Figure 2.
Analysis of Bud31's influence on cancer survival. A Forest plot showing hazard ratios for overall survival across different cancers. B Kaplan–Meier curve for overall survival in KIRC, comparing high and low Bud31 levels. The ROC curve shows the Bud31's accuracy in predicting survival. C Forest plot for disease-specific survival, with hazard ratios indicating risk across various cancers. D Kaplan–Meier curve for disease-specific survival in KIRC, with the ROC curve showing predictive accuracy based on Bud31 levels. E Forest plot for progression-free survival, with hazard ratios for different cancers. F Kaplan–Meier curve for progression-free survival in KIRC, with the ROC curve indicating the Bud31's predictive accuracy
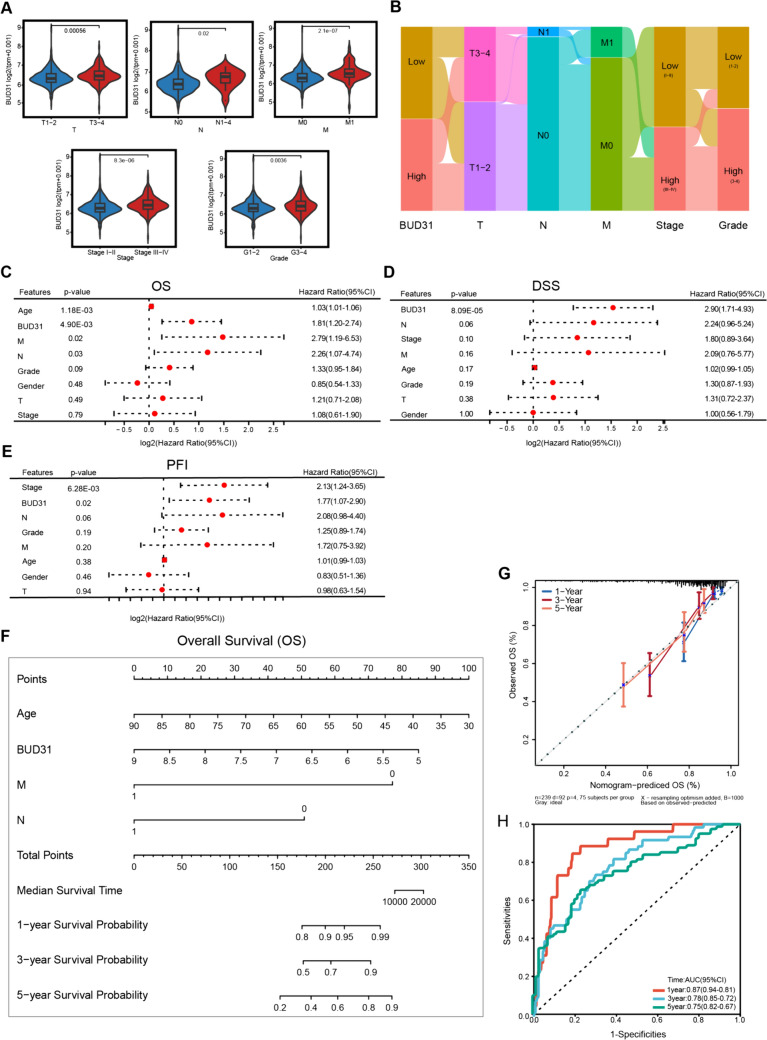
The correlation between Bud31 expression and clinicopathological parameters in tumor patients includes kidney renal clear cell carcinoma (KIRC).
The relationship between BUD31 gene expression and clinicopathological characteristics of clear cell renal cell carcinoma (ccRCC) patients was analyzed. Higher BUD31 expression is significantly associated with advanced tumor stages (T3-4 vs. T1-2, p = 0.00056), lymph node metastasis (N1-4 vs. N0, p = 0.02), distant metastasis (M1 vs. M0, p = 2.1e-07), advanced clinical stages (Stage III-IV vs. Stage I-II, p = 8.5e-07), and higher tumor grades (G3-4 vs. G1-2, p = 0.0093), as shown in Fig. 3A. The Sankey diagram in Fig. 3B further illustrates the distribution of BUD31 expression across various stages and grades, emphasizing its higher expression in more advanced and aggressive ccRCC subtypes. Multivariate Cox regression analyses for overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) (Figures C, D, and E) identify high BUD31 expression as a significant independent predictor of worse OS (HR = 1.81, p = 4.90E-03), DSS (HR = 2.90, p = 8.09E-05), and PFI (HR = 1.77, p = 0.02). Other significant predictors include age and metastasis status. A nomogram (Fig. 3F) constructed to predict 1-, 3-, and 5-year OS probabilities based on BUD31 expression, age, and metastasis status shows good predictive performance. The calibration plot (Fig. 3G) demonstrates good agreement between nomogram-predicted and observed OS probabilities. Time-dependent ROC curves (Fig. 3H) for predicting OS at 1, 3, and 5 years yield AUC values of 0.70, 0.75, and 0.72, respectively, indicating that BUD31 expression has good predictive accuracy for OS in ccRCC patients. Overall, these findings indicate that high BUD31 expression is significantly associated with advanced disease and poor prognosis in ccRCC patients, underscoring its potential as a prognostic biomarker.
Figure 3.
BUB31's prognostic significance in KIRC. A Bud31 expression is plotted against tumor characteristics, showing variations in early versus advanced stages, node involvement, metastasis, and grade. B The Sankey diagram traces Bud31 levels across stages and grades in KIRC. C A forest plot reveals how Bud31 and other clinical factors are related to overall survival in KIRC patients. D Another forest plot assesses the impact of BUB31 on disease-specific survival. E A forest plot examines Bud31's association with the time KIRC patients remain disease-free after treatment. F A nomogram scores patient characteristics to predict overall survival. G A calibration curve compares the nomogram's survival predictions to actual outcomes. H ROC curves display the nomogram's effectiveness in predicting survival over time
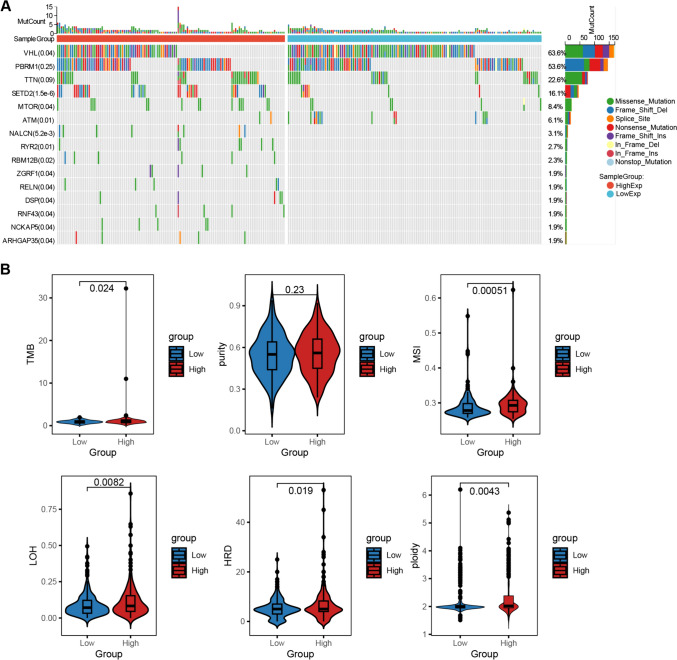
Implications of Bud31 expression on mutation landscape and genomic instability in ccRCC
The impact of BUD31 expression on the mutation landscape and genomic instability in clear cell renal cell carcinoma (ccRCC) was analyzed. As shown in Fig. 4A, the mutation profile of ccRCC samples stratified by high and low BUD31 expression reveals that high BUD31 expression is associated with a higher overall mutation count compared to low BUD31 expression. The most frequently mutated genes include VHL, PBRM1, TTN, SETD2, and MTOR, among others, with various types of mutations such as missense, frame shift, and non-sense mutations observed. Figure 4B presents violin plots comparing various genomic instability metrics between high and low BUD31 expression groups. Tumor mutation burden (TMB) is significantly higher in the high BUD31 expression group (p = 0.024). Similarly, microsatellite instability (MSI) is higher in the high BUD31 expression group (p = 0.00051). Metrics such as loss of heterozygosity (LOH) and homologous recombination deficiency (HRD) are also elevated in the high BUD31 expression group, with p-values of 0.0082 and 0.019, respectively. Additionally, ploidy is higher in the high BUD31 expression group (p = 0.0043). However, no significant difference in tumor purity is observed between the groups (p = 0.23). These results indicate that high BUD31 expression in ccRCC is associated with increased genomic instability, as evidenced by higher TMB, MSI, LOH, HRD, and ploidy. This suggests that BUD31 may play a role in promoting genetic alterations and instability in ccRCC, potentially contributing to tumor progression and aggressiveness.
Figure 4.
The impact of BUD31 expression on the mutation landscape and genomic instability in KIRC. A The waterfall plot delineates the correlation between Bud31 expression levels and prevalent oncogenic mutations in KIRC. B The association between Bud31 expression levels and tumor genomic heterogeneity in KIRC
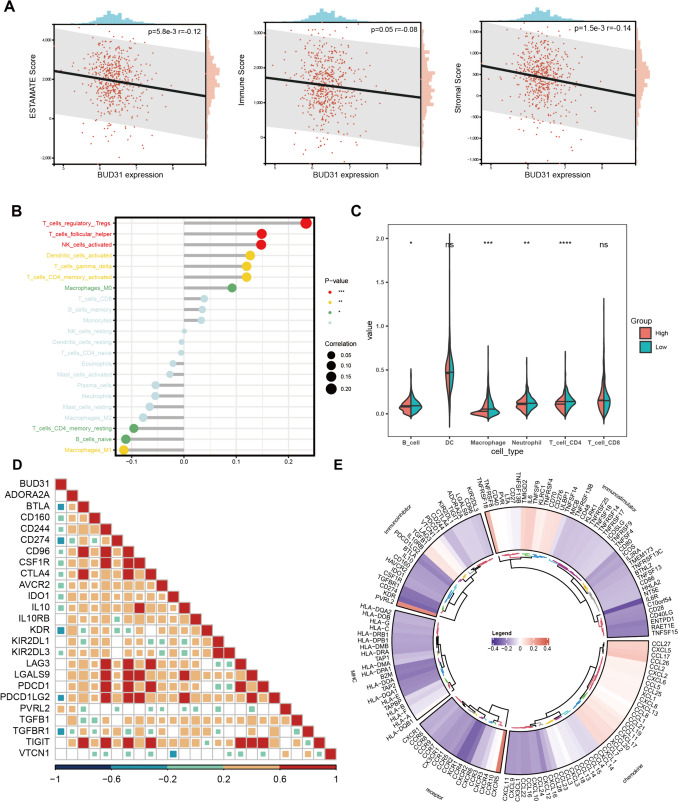
The relationship between BUD31 expression and the tumor immune microenvironment in KIRC
Our study focused on the role of BUD31 in clear cell renal cell carcinoma (ccRCC) and revealed significant correlations between BUD31 expression and various immune-related metrics. BUD31 expression exhibited a significant negative correlation with the ESTIMATE score (p = 5.8e-3, r = − 0.12), immune score (p = 0.05, r = − 0.08), and stromal score (p = 1.5e-3, r = − 0.14), indicating that higher BUD31 expression is associated with lower overall immune, stromal, and composite scores in ccRCC (Fig. 5A). Further analysis using the CIBERSORT algorithm (Fig. 5B) showed that BUD31 expression negatively correlated with regulatory T cells, follicular helper T cells, and activated NK cells, while a slight positive correlation was observed with macrophages. The TIMER algorithm (Fig. 5C) revealed significant differences in dendritic cells (DCs), macrophages, and CD4 + T cells between high and low BUD31 expression groups, but not in B cells and CD8 + T cells. A heatmap of the correlation matrix between BUD31 and various immune checkpoint and regulatory genes indicated notable negative correlations with genes such as PDCD1, LAG3, and TGFB1, suggesting BUD31's role in modulating immune checkpoint pathways (Fig. 5D). Additionally, a circular heatmap showed that most immunoinhibitory and MHC genes exhibited a negative correlation with BUD31, while chemokines and some receptors showed positive correlations (Fig. 5E). In summary, the data suggest that in ccRCC, BUD31 expression is inversely correlated with immune, stromal, and ESTIMATE scores, implicating its role in immune suppression within the tumor microenvironment. The correlation with specific immune cell types and key immune checkpoint genes highlights BUD31's potential influence on immune evasion mechanisms in ccRCC, suggesting its relevance as a target for therapeutic interventions in cancer immunotherapy.
Figure 5.
The relationship between BUD31 expression and the tumor immune microenvironment in KIRC. A The relationship between Bud31 expression levels and tumor immune infiltration in KIRC. B The dot plot illustrates the correlation between BUD31 expression and different immune cell types using CIBERSORT. C Violin plots display the distribution of immune cell types between high and low BUD31 expression groups using TIMER. D The correlation matrix of BUD31 and various immune checkpoint and regulatory genes. E The correlation between BUD31 expression and the expression of various immune-related genes
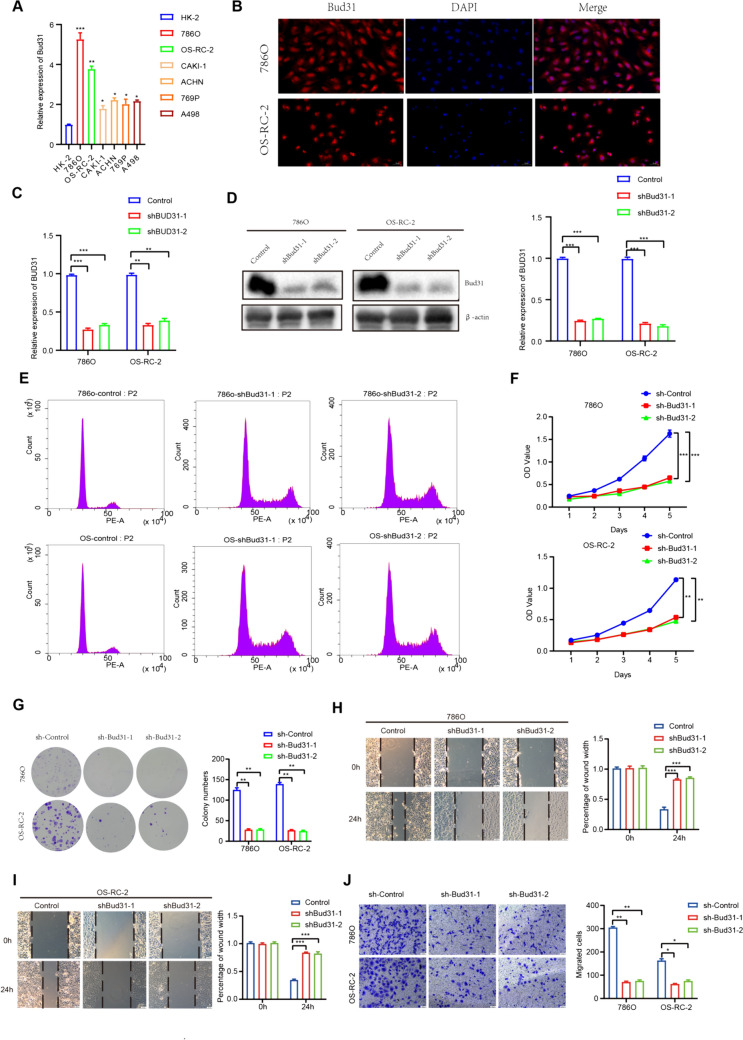
BUD31 promotes proliferation, migration, and invasion of ccrcc cells both in vitro and in vivo.
Next, we analyzed Bud31 expression levels in various ccRCC cell lines by qRT-PCR. Bud31 mRNA levels were significantly higher in the 786O and OS-RC-2 cell lines compared to the other ccRCC cell lines (Fig. 6A). The immunofluorescence colocalization assays reveal a predominant nuclear localization of the Bud31 protein within the cells (Fig. 6B). We then used specific Bud31-targeting shRNAs to knockdown the expression levels of Bud31 in the 786O and OS-RC-2 cells (Fig. 6C, D). Histograms derived from flow cytometry cell cycle analysis indicate that the downregulation of sh-Bud31 may exert an influence on the progression of the cell cycle (Fig. 6E). CCK-8 assay, flat plate colony formation, wound healing assay, and transwell assay results showed that Bud31 knockdown suppressed the cell proliferation, migration, and invasion of 786O and OS-RC-2 (Fig. 6F–J).
Figure 6.
Consequences of Bud31 downregulation in renal carcinoma cell lines. A Bar chart indicating Bud31 transcript levels, with marked expression in renal carcinoma cells. B Immunofluorescence depicting nuclear localization of Bud31 (red), counterstained with DAPI (blue). C Graph showing significant diminution in Bud31 mRNA post-targeted shRNA interference. D Western blot confirming downregulation of Bud31 protein following shRNA application. E Flow cytometric analysis reflecting cell cycle disruption subsequent to Bud31 reduction. F CCK-8 assay illustrating suppressed proliferation in Bud31-deficient cells. G Clonogenic assay outcomes demonstrating reduced colony formation capability upon Bud31 knockdown. H–I Wound healing assays evidencing decreased cellular motility following Bud31 attenuation. J Transwell migration assays substantiate the inhibitory effect on cell motility post-Bud31 knockdown
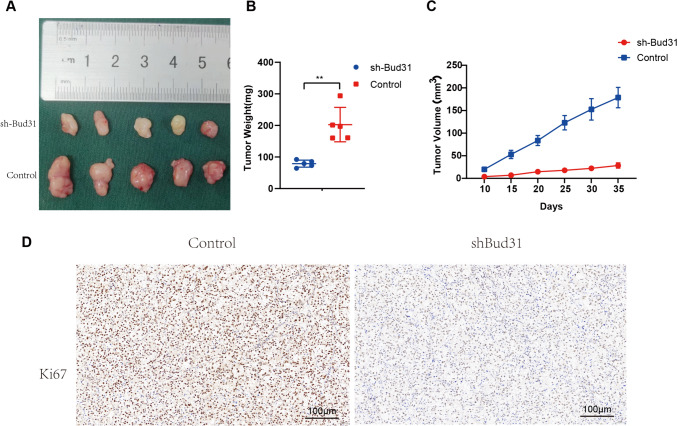
The effect of BUD31 knockdown on tumor growth was assessed in vivo using a xenograft model. Representative images of tumors excised from mice (Fig. 7A) show that tumors from the shBUD31 group were visibly smaller than those from the control group. Quantitative analysis of tumor weight (Fig. 7B) revealed a significant reduction in the weight of tumors from the shBUD31 group compared to the control group (p < 0.01). Tumor volume measurements over time (Fig. 7C) further demonstrated that tumors in the shBUD31 group grew significantly slower than those in the control group. Immunohistochemical staining for Ki67, a marker of cell proliferation (Fig. 7D), showed markedly reduced staining intensity in tumors from the shBUD31 group compared to the control group, indicating decreased proliferation in BUD31 knockdown tumors. Overall, these results indicate that BUD31 knockdown significantly inhibits tumor growth and proliferation in vivo, suggesting that BUD31 plays a critical role in the progression of renal clear cell carcinoma.
Figure 7.
The effect of BUD31 knockdown on tumor growth was assessed in vivo using a xenograft model. A representative images of tumors excised from mice injected with control and shBUD31-transfected cells. B Plot of tumor weights indicating significantly lighter tumors in the sh-Bud31 group than in controls (**P < 0.01). C Graph of tumor volumes over 35 days, with a clear trend of reduced growth in the sh-Bud31 group. D Ki67 staining of tumor sections reveals less cell proliferation in the sh-Bud31 group compared to the control group
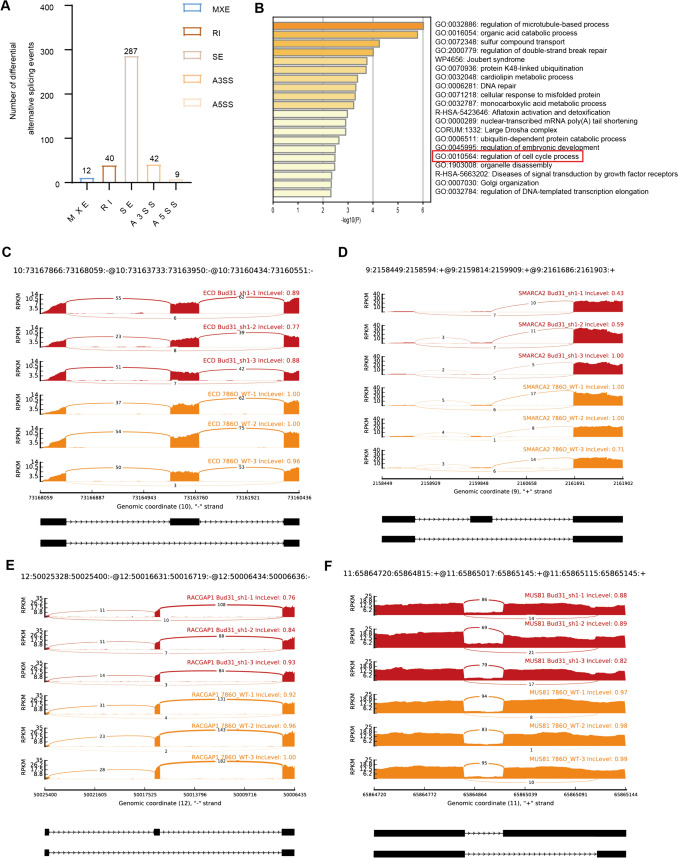
BUD31 promotes cell cycle progression by regulating alternative splicing.
As an important splicing factor, BUD31 plays a crucial role in the alternative splicing of cells. Using RNA sequencing (RNAseq) on renal clear cell carcinoma cells with knockdown of the BUD31 gene. Further analysis of alternative splicing using rMATS software identified a total of 390 differential alternative splicing events (Fig. 8A). Among these, there were 287 skipped exons (SE), 40 retained introns (RI), 42 alternative 3' splice sites (A3SS), 12 mutually exclusive exons (MXE), and nine alternative 5' splice sites (A5SS). Using KEGG enrichment analysis of differentially spliced genes, it was found that the main enrichment pathways are regulation of microtubule-based process, organic acid catabolic process, and regulation of cell cycle process. Among them, 17 genes related to the cell cycle were identified, such as ECD, SMARCA2, RACGAP1, MUS81, PBRM1, and CCNL1 (Fig. 8B–F).
Figure 8.
The extensive impact of BUD31 knockdown on alternative splicing in renal clear cell carcinoma cells. A The distribution of differentially spliced events identified by rMATS analysis in renal clear cell carcinoma cells with BUD31 knockdown. B The results of KEGG enrichment analysis for differentially spliced genes. C–F RNA sequencing read coverage across specific genomic regions, comparing BUD31 knockdown samples
Bud31 promotes the proliferation of ccRCC cells by influencing the cell cycle pathway.
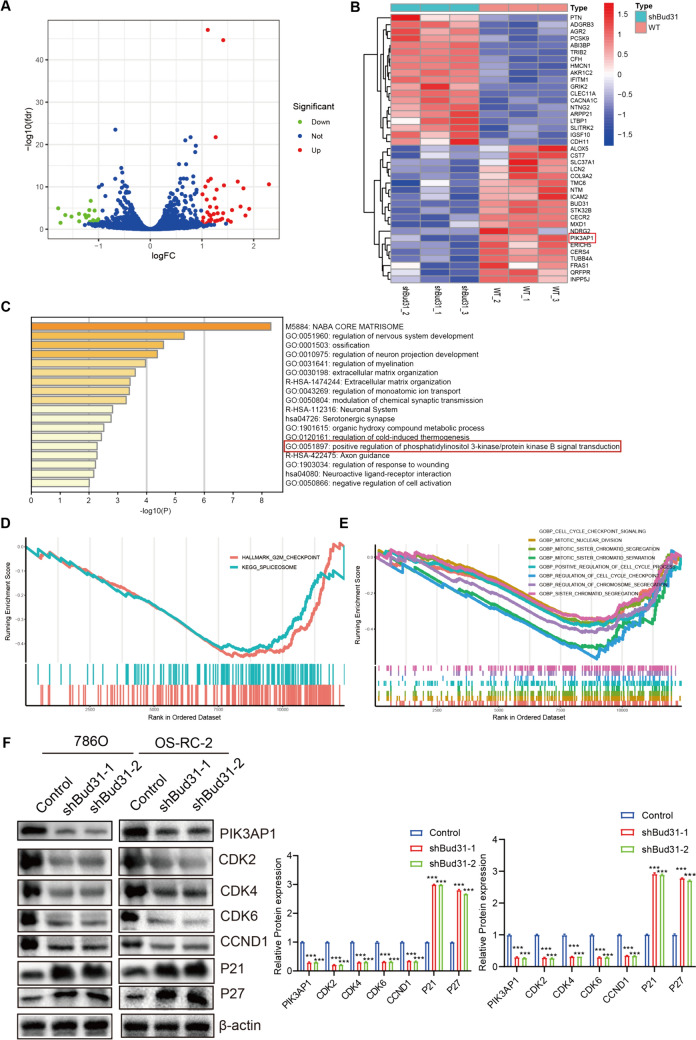
Knockdown of BUD31 resulted in significant changes in gene expression. As shown in Fig. 9A, the volcano plot highlights the genes with significant differential expression, with BUD31 prominently downregulated. Figure 9B presents a heatmap of differentially expressed genes, revealing distinct expression patterns between BUD31 knockdown (Bud31_sh1) and wild-type (WT) samples. Key genes such as ALOX5, DTNB, ADGRB3, and PTPRB exhibited notable changes in expression. Further KEGG enrichment analysis (Fig. 9C) identified significant pathways associated with the differentially expressed genes, including regulation of nervous system development (GO:0051960), ossification (GO:0001503), regulation of neuron projection development (GO:0010975), myelination (GO:0031641), and extracellular matrix organization (GO:0030198). Notably, the positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction (GO:0014068) was also significantly enriched. Gene set enrichment analysis (GSEA) (Fig. 9D, E) highlighted the enrichment of gene sets related to cell cycle regulation, such as "HALLMARK_G2M_CHECKPOINT" and "KEGG_SPLICEOSOME." Additional GSEA results indicated significant enrichment scores for pathways involved in cell cycle checkpoints, mitotic spindle assembly, and chromosome segregation, including gene sets like GOBP_CELL_CYCLE_CHECKPOINT_SIGNALING, GOBP_MITOTIC_SPINDLE_ASSEMBLY, and GOBP_MITOTIC_SISTER_CHROMATID_SEGREGATION. Western blot analysis (Fig. 9F) demonstrated that BUD31 knockdown led to decreased levels of cell cycle-related proteins CDK2, CDK4, CDK6, and CCND1, and increased levels of the cell cycle inhibitors P21 and P27 in both 786O and OS-RC-2 cells (p < 0.001). These findings collectively underscore the critical role of BUD31 in regulating gene expression and alternative splicing, with significant implications for cell cycle regulation and other essential cellular processes.
Figure 9.
The comprehensive analysis reveals that BUD31 knockdown significantly impacts gene expression. A The volcano plot of gene expression changes upon BUD31 knockdown compared to control (WT) samples. B The heatmap of differentially expressed genes between BUD31 knockdown and WT samples. C A bar chart of the KEGG enrichment analysis, identifying significant pathways associated with differentially expressed genes. D–E The results of gene set enrichment analysis (GSEA). F The expression of cell cycle-related proteins through Western blot analysis
BUD31 regulates the cell cycle pathway by affecting PIK3AP1 expression.
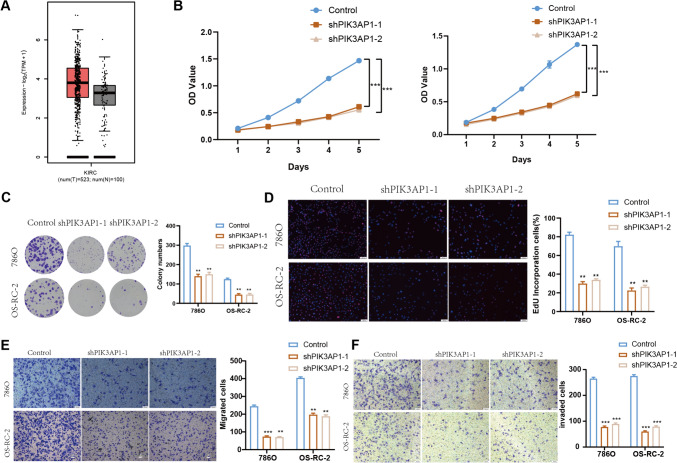
Through differential gene enrichment analysis of BUD31, we identified its enrichment in the PI3K activation pathway. Further differential gene analysis indicated that PIK3AP1 primarily functions within this pathway. Consequently, we hypothesize that BUD31 regulates cell cycle progression by modulating PIK3AP1 expression and subsequently activating the PI3K pathway. As shown in Fig. 10A, PIK3AP1 expression is significantly higher in kidney renal clear cell carcinoma (KIRC) tissues compared to normal tissues. Cell proliferation assays (Fig. 10B) demonstrate that PIK3AP1 knockdown significantly reduces cell proliferation compared to control cells over a 5-day period (p < 0.001). Colony formation assays (Fig. 10C) reveal that PIK3AP1 knockdown markedly decreases the number of colonies formed by 786O and OS-RC-2 cells (p < 0.01). EdU incorporation assays (Fig. 10D) show a substantial reduction in DNA synthesis in PIK3AP1 knockdown cells, with significantly fewer EdU-positive cells in both 786O and OS-RC-2 cell lines compared to controls (p < 0.01). Migration assays (Fig. 10E) demonstrate that PIK3AP1 knockdown significantly impairs the migratory ability of both cell lines (p < 0.01). Invasion assays (Fig. 10F) highlight a significant reduction in the invasive capacity of PIK3AP1 knockdown cells compared to control cells in both 786O and OS-RC-2 lines (p < 0.001). Overall, these results indicate that PIK3AP1 knockdown inhibits cell proliferation, colony formation, migration, and invasion in renal clear cell carcinoma cells.
Figure 10.
The impact of PIK3AP1 knockdown on renal clear cell carcinoma cells. A PIK3AP1 expression in kidney renal clear cell carcinoma (KIRC) tissues compared to normal tissues. B The results of CCK-8 assays. C Colony formation assays, where PIK3AP1 knockdown markedly decreased the number of colonies formed by 786O and OS-RC-2 cells (p < 0.01). D EdU incorporation assays, indicating a substantial reduction in DNA synthesis in PIK3AP1 knockdown cells. E migration assays, demonstrating that PIK3AP1 knockdown significantly impairs the migratory ability of both cell lines (p < 0.01). F invasion assays, highlighting a significant reduction in the invasive capacity of PIK3AP1 knockdown cells (p < 0.001)
Discussion
Our study presents a comprehensive analysis of Bud31 expression in various cancer types, with a focus on clear cell renal cell carcinoma (ccRCC). The significant upregulation of Bud31 in multiple tumor tissues, as revealed by The Human Protein Atlas and TCGA database analyses, underscores its potential role in tumorigenesis and progression [2, 16]. This observation aligns with recent findings indicating the critical role of gene expression alterations in cancer development [26, 27].
The strong association between elevated Bud31 expression and poor prognosis across a range of cancers, particularly in ccRCC, implicates Bud31 as a key player in cancer progression [2, 4, 16, 28]. This is consistent with recent studies highlighting the prognostic value of gene expression profiles in cancer [29, 30]. Furthermore, the observed correlation between high Bud31 expression and advanced clinical features such as increased tumor size, nodal involvement, distant metastasis, and higher histological grade in KIRC, resonates with findings from similar studies on other oncogenes [31–35]. Our findings suggest that Bud31 could serve as a valuable prognostic biomarker in ccRCC, as supported by recent research emphasizing the importance of biomarkers in personalized medicine [2, 16].
Furthermore, BUD31 appears to play a critical role in modulating the tumor immune microenvironment. Higher BUD31 expression is negatively correlated with immune, stromal, and ESTIMATE scores, indicating a less active immune environment. Detailed analysis using CIBERSORT and TIMER algorithms showed significant correlations between BUD31 expression and various immune cell types. Notably, regulatory T cells, follicular helper T cells, and activated NK cells negatively correlated with BUD31 expression, while macrophages showed a slight positive correlation. These findings suggest that BUD31 may promote immune evasion mechanisms in ccRCC by altering the immune microenvironment [36–39].
Functionally, BUD31 promotes cell proliferation, migration, and invasion both in vitro and in vivo. Knockdown of BUD31 significantly inhibited these malignant behaviors in ccRCC cell lines, further confirming its oncogenic role. RNA sequencing (RNAseq) and alternative splicing analysis identified 390 differential splicing events upon BUD31 knockdown, affecting key pathways related to cell cycle regulation [40–42]. Notably, BUD31 knockdown led to decreased levels of cell cycle-related proteins CDK2, CDK4, CDK6, and CCND1, while increasing the levels of cell cycle inhibitors P21 and P27.
Additionally, our differential gene enrichment analysis revealed that BUD31 is enriched in the PI3K activation pathway. Specifically, PIK3AP1, a gene primarily functioning in the PI3K pathway, was significantly regulated by BUD31. We hypothesize that BUD31 regulates cell cycle progression by modulating PIK3AP1 expression and subsequently activating the PI3K pathway [43–47]. Functional assays confirmed that PIK3AP1 knockdown inhibited cell proliferation, colony formation, migration, and invasion in ccRCC cells, supporting the role of the PI3K pathway in mediating the oncogenic effects of BUD31.
While our study provides significant insights into the role of BUD31 in ccRCC, it has certain limitations. Firstly, the use of cell lines and animal models may not fully replicate the complexity of human ccRCC. Future studies should involve larger, more diverse patient cohorts, and clinical samples to validate our findings. Secondly, although we identified numerous alternative splicing events regulated by BUD31, the exact molecular mechanisms through which BUD31 influences these splicing events remain unclear. Further research is needed to elucidate the detailed splicing mechanisms and their functional consequences. Additionally, exploring the potential interactions between BUD31 and other spliceosomal components could provide deeper insights into its role in splicing regulation. Finally, while our study highlights the role of BUD31 in immune evasion, the precise pathways through which BUD31 modulates the tumor immune microenvironment require further investigation. Understanding these pathways could open new avenues for developing immune-based therapies targeting BUD31. Moreover, the potential therapeutic benefits of targeting BUD31, either alone or in combination with other treatments, should be explored in clinical trials to assess its efficacy and safety in cancer therapy.
In conclusion, BUD31 is a critical player in the progression of ccRCC, influencing tumor growth, genomic stability, immune evasion, and cell cycle regulation. Its role in alternative splicing and activation of the PI3K pathway highlights its potential as a therapeutic target. Future research should focus on further elucidating the molecular mechanisms of BUD31 and exploring targeted therapies that could mitigate its oncogenic effects in ccRCC and other cancers.
Conclusion
This study highlights the critical role of BUD31 in cancer, particularly in clear cell renal cell carcinoma (ccRCC). BUD31 is significantly upregulated in various tumors and associated with poor clinical outcomes. High BUD31 expression correlates with advanced stages, metastasis, higher tumor grades, and worse overall survival, indicating its potential as a prognostic biomarker. BUD31 is linked to increased genomic instability, evidenced by higher tumor mutation burden (TMB), microsatellite instability (MSI), and other genomic alterations. Additionally, high BUD31 expression is associated with reduced immune cell infiltration, suggesting a role in immune evasion.
Functionally, BUD31 promotes cell proliferation, migration, and invasion in vitro and tumor growth in vivo. It regulates key cell cycle-related genes and pathways and significantly impacts alternative splicing, affecting critical genes involved in cell cycle regulation. In summary, BUD31 is a key player in tumor progression and a potential target for prognosis and therapy in ccRCC.
List of abbreviations: The information is shown in Supplementary Table 1.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Thanks to the Xu Hua laboratory for providing technical support.
Author contributions
Xiaoliang Wu was contributed conceptualization, data curation, formal analysis, investigation, methodology, project administration, validation, visualization, writing–original draft, and writing–review and editing. Ruixin Fan was involved in data curation, formal analysis, methodology, investigation, methodology, resources, software, validation, visualization, and writing–original draft. Yangjun Zhang and Xiangyang Yao was performed investigation and methodology. Chen Duan was done methodology and resources. Kai Liu was responsible for Data curation and Formal analysis. Dongxu Lin was attributed methodology and software. Zhong Chen did conceptualization, funding acquisition, methodology, supervision, and writing–review and editing.
Funding
This work was supported by National Natural Science Foundation of China, grant number (82270812).
Data availability
No datasets were generated or analyzed during the current study.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
The animal study was reviewed and approved by Animal Care Committees of Wuhan University Medical Research Institute.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoliang Wu and Ruixin Fan have contributed equally to this study.
References
- 1.Jonasch E, Walker CL, Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat Rev Nephrol. 2021;17:245–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Wang S, Qin J, et al. Splicing factor BUD31 promotes ovarian cancer progression through sustaining the expression of anti-apoptotic BCL2L12. Nat Commun. 2022;13:6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu CL, Liu JS, Wu PL, et al. Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis. Mol Oncol. 2014;8:1575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu TY, Simon LM, Neill NJ, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanjuan-Sanjuan A, Alors-Perez E, Sanchez-Frias M, et al. Splicing machinery is impaired in oral squamous cell carcinomas and linked to key pathophysiological features. Int J Mol Sci. 2024;25(13):2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang F, Dai Z, Yu J, et al. RBM7 deficiency promotes breast cancer metastasis by coordinating MFGE8 splicing switch and NF-kB pathway. Elife. 2024. 10.7554/eLife.95318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan D, Long L, Li C, et al. Splicing factor hnRNPA1 regulates alternative splicing of LOXL2 to enhance the production of LOXL2Delta13. J Biol Chem. 2024;300: 107414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee S, Nagasawa CK, Widen SG, Garcia-Blanco MA. Parsing the roles of DExD-box proteins DDX39A and DDX39B in alternative RNA splicing. Nucleic Acids Res. 2024. 10.1038/s41467-024-49874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamoski D, Reis LMD, Mafra AC, et al. HuR controls glutaminase RNA metabolism. Nat Commun. 2024;15:5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Xiong B, Shuai X, et al. Downregulation of HNRNPA1 induced neoantigen generation via regulating alternative splicing. Mol Med. 2024;30:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Liao R, Miao C, et al. Oncofetal SNRPE promotes HCC tumorigenesis by regulating the FGFR4 expression through alternative splicing. Br J Cancer. 2024;131:77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Zhang S, Li Y, et al. The regulatory network of hnRNPs underlying regulating PKM alternative splicing in tumor progression. Biomolecules. 2024;14(5):566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W, Zhai ZY, Huang ZY, et al. Dual RNA sequencing of Helicobacter pylori and host cell transcriptomes reveals ontologically distinct host-pathogen interaction. mSystems. 2024;9: e0020624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin M, Mi Y, Li F, et al. ZNF131 facilitates the growth of hepatocellular carcinoma by acting as a transcriptional activator of SMC4 expression. Biochem Biophys Res Commun. 2024;696: 149515. [DOI] [PubMed] [Google Scholar]
- 15.Saha D, Banerjee S, Bashir S, Vijayraghavan U. Context dependent splicing functions of Bud31/Ycr063w define its role in budding and cell cycle progression. Biochem Biophys Res Commun. 2012;424:579–85. [DOI] [PubMed] [Google Scholar]
- 16.Koedoot E, Fokkelman M, Rogkoti VM, et al. Uncovering the signaling landscape controlling breast cancer cell migration identifies novel metastasis driver genes. Nat Commun. 2019;10:2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan ZW, Liu Y, Zhang PP, et al. Da-Chai-Hu-Tang formula inhibits the progression and metastasis in HepG2 cells through modulation of the PI3K/AKT/STAT3-induced cell cycle arrest and apoptosis. J Ethnopharmacol. 2024;331: 118293. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Wang H, Chen S, et al. Cystatin SA attenuates gastric cancer cells growth and increases sensitivity to oxaliplatin via PI3K/AKT signaling pathway. J Cancer Res Clin Oncol. 2024;150:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao T, Ye W, Zhang R, et al. Dual-regulated oncolytic adenovirus carrying ERCC1-siRNA gene possesses potent antitumor effect on ovarian cancer cells. Mol Med Rep. 2024. 10.3892/mmr.2024.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Liu L, Xu X, Zeng H. KIF15 promotes the development and progression of chordoma via activating PI3K-AKT signalling pathway. Heliyon. 2024;10: e29386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XQ, Cheng XJ, Wu J, et al. Targeted inhibition of the PI3K/AKT/mTOR pathway by (+)-anthrabenzoxocinone induces cell cycle arrest, apoptosis, and autophagy in non-small cell lung cancer. Cell Mol Biol Lett. 2024;29:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity. 2018;48(812–830): e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu T, Hu E, Xu S, et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2: 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren J, Huang B, Li W, et al. RNA-binding protein IGF2BP2 suppresses metastasis of clear cell renal cell carcinoma by enhancing CKB mRNA stability and expression. Transl Oncol. 2024;42: 101904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen SY, Zhang FL, Zhang YL, et al. Spermatid perinuclear RNA-binding protein promotes UBR5-mediated proteolysis of dicer to accelerate triple-negative breast cancer progression. Cancer Lett. 2024;586: 216672. [DOI] [PubMed] [Google Scholar]
- 28.Choudhry M, Gamallat Y, Khosh Kish E, et al. Downregulation of BUD31 promotes prostate cancer cell proliferation and migration via activation of p-akt and vimentin in vitro. Int J Mol Sci. 2023;24(7):6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv H, Mu Y, Zhang C, et al. Comparative analysis of single-cell transcriptome reveals heterogeneity and commonality in the immune microenvironment of colorectal cancer and inflammatory bowel disease. Front Immunol. 2024;15:1356075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Chiaro P, Nacci L, Arco F, et al. Mapping functional to morphological variation reveals the basis of regional extracellular matrix subversion and nerve invasion in pancreatic cancer. Cancer Cell. 2024;42(4):662-681.e10. [DOI] [PubMed] [Google Scholar]
- 31.Peak T, Tian Y, Patel A, et al. Pathogenic roles for RNASET2 in clear cell renal cell carcinoma. Lab Invest. 2024;104(5):102041. [DOI] [PubMed] [Google Scholar]
- 32.Sainero-Alcolado L, Garde-Lapido E, Snaebjornsson MT, et al. Targeting MYC induces lipid droplet accumulation by upregulation of HILPDA in clear cell renal cell carcinoma. Proc Natl Acad Sci USA. 2024;121: e2310479121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu L, Zhang Y, Guo L, et al. Kinome-wide siRNA screen identifies a DCLK2-TBK1 oncogenic signaling axis in clear cell renal cell carcinoma. Mol Cell. 2024;84(776–790): e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Lin Y, Zheng S, et al. CBX3 promotes clear cell renal carcinoma through PI3K/AKT activation and aberrant immunity. J Transl Med. 2023;21:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Miao C, Wang Y, et al. SPTBN1 abrogates renal clear cell carcinoma progression via glycolysis reprogramming in a GPT2-dependent manner. J Transl Med. 2022;20:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Modugno F, Di Carlo A, Spada S, et al. Tumoral and stromal hMENA isoforms impact tertiary lymphoid structure localization in lung cancer and predict immune checkpoint blockade response in patients with cancer. EBioMedicine. 2024;101: 105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu D, Mihoayi M, Ablikim Y, Arikin A. RNA splicing regulator EIF3D regulates the tumor microenvironment through immunogene-related alternative splicing in head and neck squamous cell carcinoma. Aging (Albany NY). 2024;16:5929–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An W, Yang Q, Xi Y, et al. Identification of SRSF10 as a promising prognostic biomarker with functional significance among SRSFs for glioma. Life Sci. 2024;338: 122392. [DOI] [PubMed] [Google Scholar]
- 39.Wahid M, Pratoomthai B, Egbuniwe IU, et al. Targeting alternative splicing as a new cancer immunotherapy-phosphorylation of serine arginine-rich splicing factor (SRSF1) by SR protein kinase 1 (SRPK1) regulates alternative splicing of PD1 to generate a soluble antagonistic isoform that prevents T cell exhaustion. Cancer Immunol Immunother. 2023;72:4001–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salib A, Jayatilleke N, Seneviratne JA, et al. MYCN and SNRPD3 cooperate to maintain a balance of alternative splicing events that drives neuroblastoma progression. Oncogene. 2024;43:363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu N, Ren Y, Bao Y, et al. PUF60 promotes cell cycle and lung cancer progression by regulating alternative splicing of CDC25C. Cell Rep. 2023;42: 113041. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, Yang HT, Zhang B, et al. The RNA-binding proteins hnRNP H and F regulate splicing of a MYC-dependent HRAS exon in prostate cancer cells. Proc Natl Acad Sci USA. 2023;120: e2220190120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang F, Li K, Yao X, et al. Corrigendum to A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer [EBioMedicine 44 (2019) 311–321]. EBioMedicine. 2021;69: 103469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie C, Zhou H, Qin D, et al. Bcl-2 inhibition combined with PPARalpha activation synergistically targets leukemic stem cell-like cells in acute myeloid leukemia. Cell Death Dis. 2023;14:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J, Tang T, Li J, et al. Overexpression of MicroRNA 142–5p suppresses the progression of cervical cancer through targeting phosphoinositol-3-kinase adaptor protein 1 expression. Mol Cell Biol. 2021;41: e0036320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu G, Sun BY, Sun J, et al. BRG1 regulates lipid metabolism in hepatocellular carcinoma through the PIK3AP1/PI3K/AKT pathway by mediating GLMP expression. Dig Liver Dis. 2022;54:692–700. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Zhang Z, Hu J, et al. MiR-1246 regulates the PI3K/AKT signaling pathway by targeting PIK3AP1 and inhibits thyroid cancer cell proliferation and tumor growth. Mol Cell Biochem. 2022;477:649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study.