Abstract
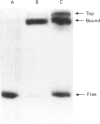
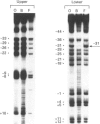
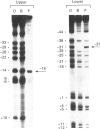
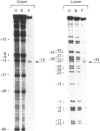
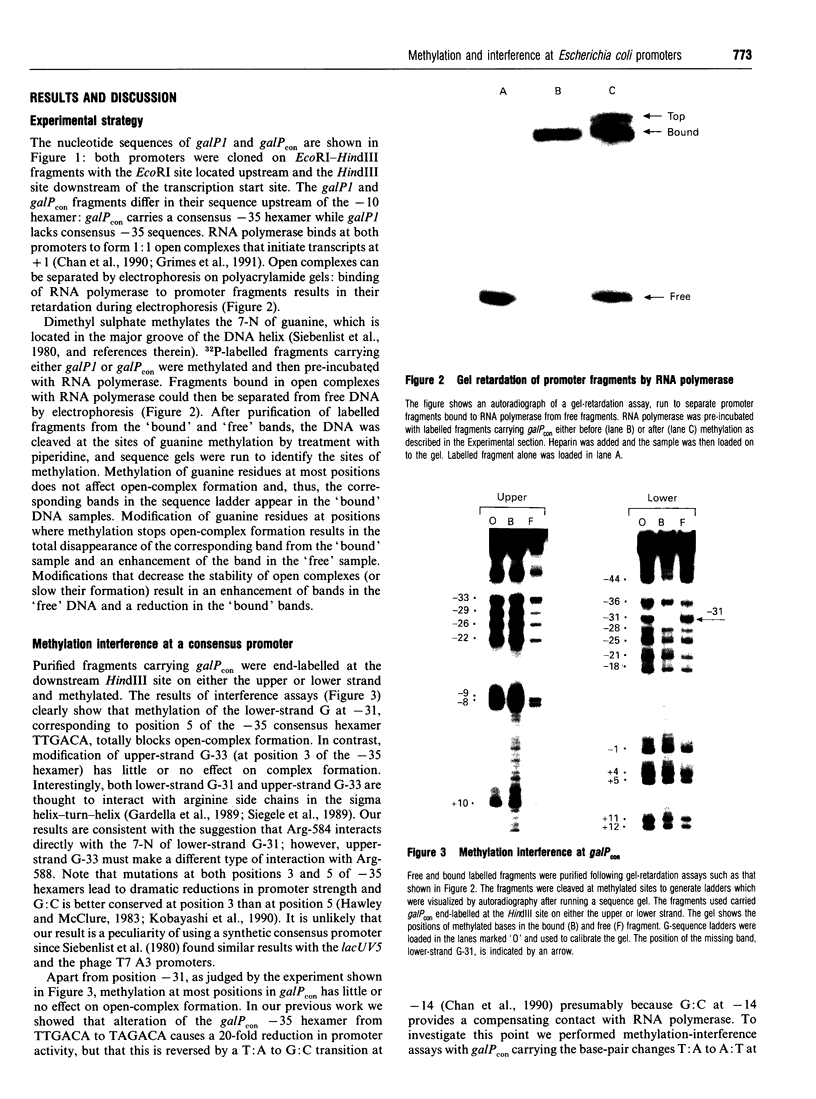
Methylation-interference assays have been used to identify guanine residues that make important contacts with RNA polymerase during open-complex formation at two related Escherichia coli promoters. Methylation of lower-strand G-31 at a gal consensus promoter completely prevents complex formation, while modification of upper-strand G-33 has no detectable effect. At galP1, which lacks a consensus -35 region, modification of lower-strand G-33 and upper-strand G-14 reduces, but does not prevent, complex formation. G-33 is the only guanine residue in the -35 region of galP1 where modification interferes with open-complex formation. Since this guanine residue is not protected in open complexes, we conclude that its modification causes alteration of, or interference with, a transient contact during the transcription initiation pathway.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bingham A. H., Ponnambalam S., Chan B., Busby S. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene. 1986;41(1):67–74. doi: 10.1016/0378-1119(86)90268-4. [DOI] [PubMed] [Google Scholar]
- Busby S., Buc H. Positive regulation of gene expression by cyclic AMP and its receptor protein in Escherichia coli. Microbiol Sci. 1987 Dec;4(12):371–375. [PubMed] [Google Scholar]
- Busby S., Dreyfus M. Segment-specific mutagenesis of the regulatory region in the Escherichia coli galactose operon: isolation of mutations reducing the initiation of transcription and translation. Gene. 1983 Jan-Feb;21(1-2):121–131. doi: 10.1016/0378-1119(83)90154-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chan B., Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989 Dec 14;84(2):227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- Chan B., Spassky A., Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus -35 region sequences. Biochem J. 1990 Aug 15;270(1):141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella T., Moyle H., Susskind M. M. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989 Apr 20;206(4):579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Gaston K., Kolb A., Busby S. Binding of the Escherichia coli cyclic AMP receptor protein to DNA fragments containing consensus nucleotide sequences. Biochem J. 1989 Jul 15;261(2):649–653. doi: 10.1042/bj2610649. [DOI] [PMC free article] [PubMed] [Google Scholar]
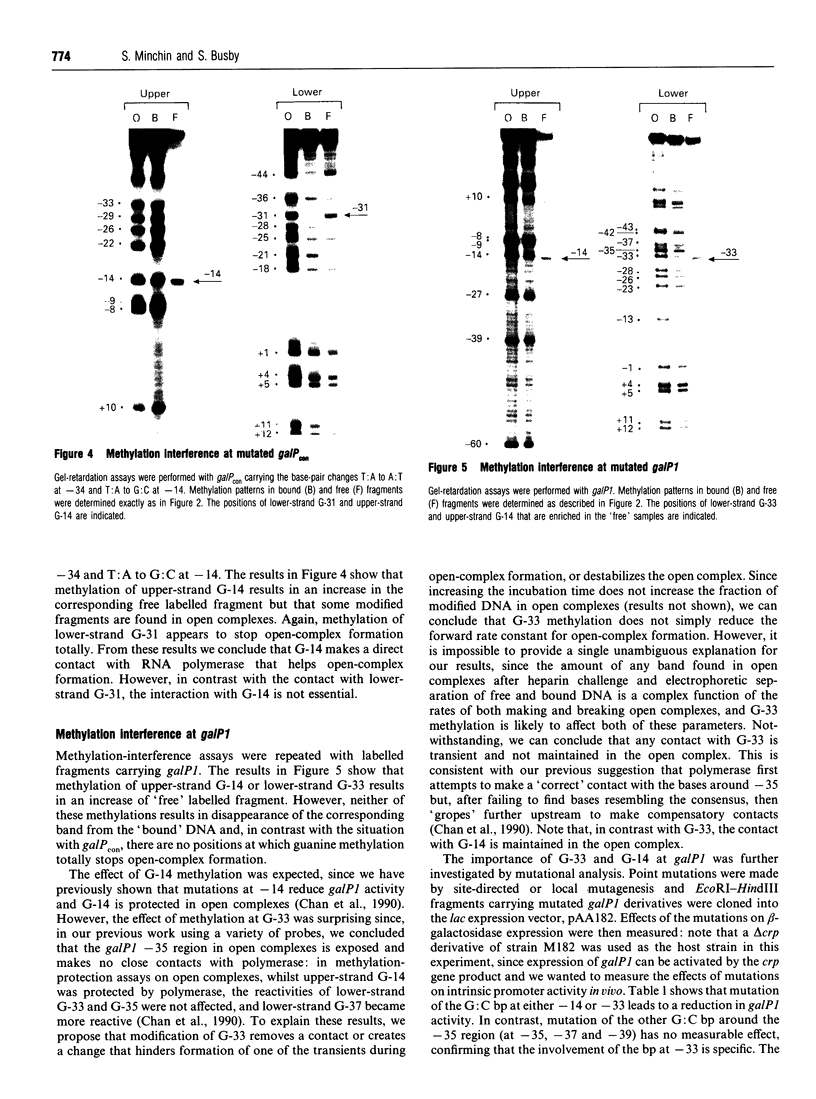
- Grimes E., Busby S., Minchin S. Different thermal energy requirement for open complex formation by Escherichia coli RNA polymerase at two related promoters. Nucleic Acids Res. 1991 Nov 25;19(22):6113–6118. doi: 10.1093/nar/19.22.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987 May 5;262(13):6389–6395. [PubMed] [Google Scholar]
- Lavigne M., Herbert M., Kolb A., Buc H. Upstream curved sequences influence the initiation of transcription at the Escherichia coli galactose operon. J Mol Biol. 1992 Mar 20;224(2):293–306. doi: 10.1016/0022-2836(92)90995-v. [DOI] [PubMed] [Google Scholar]
- Lavigne M., Kolb A., Buc H. Transcription activation by cAMP receptor protein (CRP) at the Escherichia coli gal P1 promoter. Crucial role for the spacing between the CRP binding site and the -10 region. Biochemistry. 1992 Oct 13;31(40):9647–9656. doi: 10.1021/bi00155a018. [DOI] [PubMed] [Google Scholar]
- Lodge J., Fear J., Busby S., Gunasekaran P., Kamini N. R. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol Lett. 1992 Aug 15;74(2-3):271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Peakman T., Busby S., Cole J. Transcriptional control of the cysG gene of Escherichia coli K-12 during aerobic and anaerobic growth. Eur J Biochem. 1990 Jul 31;191(2):325–331. doi: 10.1111/j.1432-1033.1990.tb19126.x. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S., Chan B., Busby S. Functional analysis of different sequence elements in the Escherichia coli galactose operon P2 promoter. Mol Microbiol. 1988 Mar;2(2):165–172. doi: 10.1111/j.1365-2958.1988.tb00018.x. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S., Webster C., Bingham A., Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal -35 region sequences. J Biol Chem. 1986 Dec 5;261(34):16043–16048. [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Siegele D. A., Hu J. C., Walter W. A., Gross C. A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989 Apr 20;206(4):591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Waldburger C., Gardella T., Wong R., Susskind M. M. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990 Sep 20;215(2):267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991 May 20;219(2):201–215. doi: 10.1016/0022-2836(91)90562-k. [DOI] [PubMed] [Google Scholar]