Abstract
Base excision repair (BER) is a multistep process involving the sequential activity of several proteins that cope with spontaneous and environmentally induced mutagenic and cytotoxic DNA damage. Quantitative kinetic data on single proteins of BER have been used here to develop a mathematical model of the BER pathway. This model was then employed to evaluate mechanistic issues and to determine the sensitivity of pathway throughput to altered enzyme kinetics. Notably, the model predicts considerably less pathway throughput than observed in experimental in vitro assays. This finding, in combination with the effects of pathway cooperativity on model throughput, supports the hypothesis of cooperation during abasic site repair and between the apurinic/apyrimidinic (AP) endonuclease, Ape1, and the 8-oxoguanine DNA glycosylase, Ogg1. The quantitative model also predicts that for 8-oxoguanine and hydrolytic AP site damage, short-patch Polβ-mediated BER dominates, with minimal switching to the long-patch subpathway. Sensitivity analysis of the model indicates that the Polβ-catalyzed reactions have the most control over pathway throughput, although other BER reactions contribute to pathway efficiency as well. The studies within represent a first step in a developing effort to create a predictive model for BER cellular capacity.
INTRODUCTION
Past mathematical models of biological pathways include epidermal growth factor response (1), microbial metabolic pathways (2,3), bacteriophage infection (4,5), cellular signaling (6–8) and the eukaryotic cell cycle (9). These models have provided a rigorous basis to accurately evaluate alternative complex hypotheses for pathway mechanism and to predict the effect of enzyme perturbations on pathway efficiency. Using kinetic data available in the literature, we present here the first quantitative model of a human DNA repair pathway. Such cellular processes are essential for genome maintenance and protection against the development of disease and cancer (10).
DNA is subject to both spontaneous decay and chemical modification by intracellular reactive species or environmental agents (11). Events involving spontaneous hydrolytic decomposition include deamination of cytosine or cleavage of the N-glycosylic bond connecting the base with the sugar moiety of DNA, yielding products that are potentially mutagenic if unrepaired. In addition, cytotoxic and mutagenic lesions are created from attack of DNA by reactive oxygen species formed as metabolites of normal cellular respiration. Another endogenous genotoxic agent, S-adenosylmethionine can react with nucleophilic sites on DNA via transmethylation, creating alkylation lesions. Also, environmental agents, such as ultraviolet and ionizing radiation, produce hundreds of deleterious DNA products. Given the established association of defects in DNA repair with human disease, it is evident that the mutagenic and cytotoxic potential of DNA lesions depends on the level of the initial DNA damage and the cell’s capacity to execute repair (10).
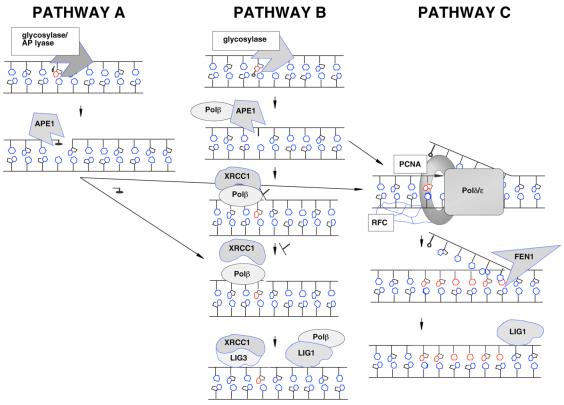
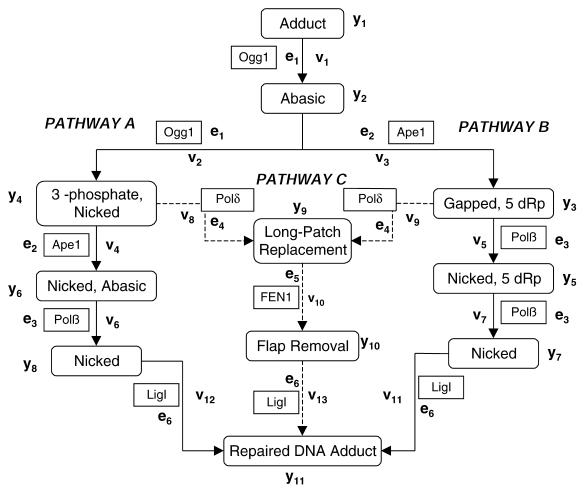
Base excision repair (BER) is a multistep process involving the sequential activity of several proteins that copes with the majority of spontaneous and endogenously produced mutagenic and cytotoxic DNA damage, e.g. base modifications, abasic sites and single-strand breaks (reviewed in 12–15). Typically, this pathway is initiated by a DNA glycosylase, a protein that recognizes and removes a damaged (e.g. 8-oxoguanine, uracil) or incorrect base (e.g. mismatch) by hydrolyzing the N-glycosidic bond (reviewed in 16–18). In some instances, DNA glycosylases exhibit not only N-glycosylase activity, but also the ability to incise at the resulting apurinic/apyrimidinic (AP) site, cleaving 3′ to the abasic residue. These ‘multifunctional’ DNA glycosylases initiate a sub-pathway of BER by generating a normal 5′-phosphate residue and a non-conventional 3′-terminus (i.e. a 3′-unsaturated aldehyde or a 3′-phosphate) (Fig. 1, pathway A). AP endonucleases, such as Ape1 in vertebrates, possess 3′-phosphodiesterase activity and operate to remove these 3′-obstructive ends, which normally prevent repair synthesis and ligation (reviewed in 19). Following removal of the 3′-blocking group, BER proceeds with gap filling, primarily by DNA polymerase β (Polβ), followed with DNA ligation by DNA ligase 1 (Lig1) or an XRCC1/Lig3 complex (reviewed in 20–22).
Figure 1.
The sub-pathways of BER. See text for details.
In most cases the DNA backbone is not incised at an AP site by a DNA glycosylase, but instead by a class II AP endonuclease, e.g. Ape1 (Fig. 1, pathway B or C). These proteins cut 5′ to the lesion (19), producing a normal 3′-hydroxyl group and a 5′-deoxyribose phosphate (abasic) fragment (a dRp group). Short-patch BER (pathway B) involves single nucleotide gap synthesis and removal of the dRp fragment by DNA Polβ (reviewed in 20). There also exists an alternative ‘long-patch’ BER pathway (pathway C) that involves the replacement of more than a single nucleotide (2–7 nt) by DNA polymerases Polδ and Polɛ (23 and references therein) or Polβ (24,25). This process utilizes PCNA and includes the excision by Fen1 of the flap-like DNA structure (which contains the 5′-dRp group) produced via polymerase strand displacement. In each BER sub-pathway, DNA Lig1 or an XRCC1/Lig3 complex completes the process by sealing the nick (Fig. 1). Using kinetic data available in the literature, we present a first step towards a complete mathematical model of the BER pathway. The model is applied towards understanding pathway mechanism and cooperativity, and we discuss its potential use as a tool for predicting individual BER capacity.
MODELS AND RESULTS
Kinetic data are available in the literature for almost all enzymatic reactions in the BER network. However, most laboratory experiments were performed with the goal of obtaining a biological understanding of the reaction mechanism and kinetics of an individual enzyme rather than a comprehensive reaction model for the entire repair pathway; thus, virtually all the data are in the form of kcat and KM (Table 1). Parameters that describe product inhibition and cooperativity have generally not been measured quantitatively for BER enzymes. Consequently, our preliminary mass action model of BER, which assumes Michaelis–Menten kinetics, necessarily lacks kinetic details. Despite this caveat, the initial model developed and tested below allows us to gain important insights into the nature of cooperativity and pathway mechanism in BER. We also assess the potential value of modeling to predict the impact of genetic variation affecting kinetic parameters or numbers of enzymes via sensitivity analysis.
Table 1. BER reaction rates.
| Enzyme | Reaction | kcat (s–1) | KM (nM) | Sources |
|---|---|---|---|---|
| Ogg1 |
8-oxoG excision |
0.00139a |
1863a |
(36,49,50,54) |
| |
AP lyase |
0.00089 |
13.7 |
(36) |
| UDG |
Uracil excision |
42 |
100 |
(55) |
| Ape1 |
AP endo |
3.08a |
32.5a |
(36,56–60) |
| |
3′-diesterase |
0.05 |
130 |
(60) |
| Polβ |
Gap filling |
0.45 |
300 |
(29,61) |
| |
dRp lyase |
0.075 |
500 |
(62) |
| Lig1 |
Ligation |
0.04 |
100 |
(56,63) |
| Fen1 |
5′-flap endo |
0.39 |
39 |
(64) |
| Polδ | Gap filling | 0.0022b | 67b | (65) |
aWhere data were found in multiple sources, the average kcat and KM are shown.
bReaction rates measured in the presence of abundant PCNA.
We note that our BER model contains fundamental assumptions of deterministic chemical kinetics and homogenous spatial distribution of BER reactions. With regard to the first assumption, the enzyme amounts used in the simulations (i.e. 104–106 molecules/cell) are far above the level at which random fluctuations in reactant concentrations predicted by stochastic kinetics become negligible (26). As for the second assumption, reactions were modeled assuming a zero average velocity of molecules, consistent with recent studies of protein movement in the nucleus using single molecule fluorescence imaging (27,28). Thus, in the case of the BER pathway, assumptions of determinism and homogeneity are likely to be consistent with in vivo conditions.
Comparing the model prediction to an in vitro pathway reconstruction: evidence for cooperativity in abasic site repair
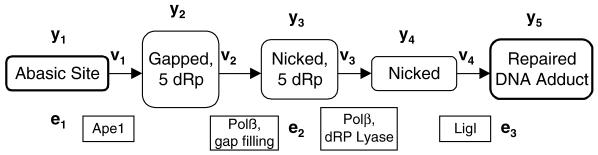
To evaluate our quantitative modeling approach against existing experimental data, we focused on the repair of abasic (AP) sites in short-patch repair (Fig. 1, pathway B), as this pathway was previously reconstructed in vitro (29). Figure 2 shows the linear reaction pathway of Srivasta et al. (29) with the definitions of substrate concentrations yi, enzyme concentrations ei and reaction velocities vi. Assuming Michaelis–Menten kinetics, the model equations are
Figure 2.
Schematic of the model for AP site repair (pathway B) showing the definition of substrate concentrations yi, enzyme concentrations ei and reaction velocities vi. Ape1 (e1) incises the abasic site generating a single nucleotide gap with a 3′-hydroxyl group and a 5′-dRp group. Subsequent to this activity, DNA Polβ (e2) fills the gap producing a nicked substrate, with a 5′-dRp group remaining. This 5′-terminus is then processed by the dRp lyase activity of Polβ prior to sealing of the nick by DNA ligase (e3).
dy1/dt = –v1 v1 = y1[k1e1/(y1 + K1)]
dy2/dt = v1 – v2 v2 = y2[k2e2/(y2 + K2)]
dy3/dt = v2 – v3 v3 = y3[k3e2/(y3 + K3)]
dy4/dt = v3 – v4 v4 = y4[k4e3/(y4 + K4)]
dy5/dt = v4
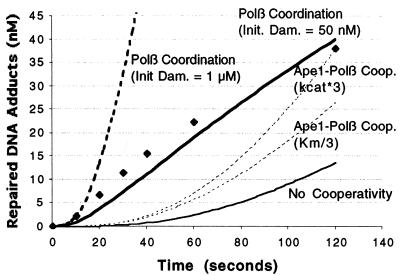
where ki is the kcat and Ki is the KM of the reaction labeled vi. The results of the simulation are shown in Figure 3. Notably, the model using average kinetic parameters obtained for the individual BER enzymes (Fig. 3, thin solid line) and reported in the literature (Table 1) underestimates the in vitro reaction rate of the ‘fully’ reconstructed assay (Fig. 3, diamonds).
Figure 3.
Model for abasic site BER pathway reconstruction experiment. The initial model using the parameters in Table 1 without cooperativity (thin solid line) underpredicts the experimental data from Srivasta et al. (29) (diamonds). The in vitro assay of Srivasta et al. (29) was performed for 120 s with an initial abasic site concentration of 1 µM and enzyme concentrations of 10 nM Ape1, 10 nM Polβ and 100 nM DNA ligase 1. We modeled a 3-fold increase in Polβ dRp lyase activity in the presence of Ape1 (dashed lines) as either a 3-fold increase in kcat or as a 3-fold decrease in KM. The model incorporating coordination between Polβ reactions (detailed in Appendix A) is simulated at initial abasic site concentrations of 1 µM (thick dashed line) and 50 nM (thick solid line). Differential equations were solved numerically using the variable-order stiff solver ‘ode15s’ of Matlab 5.3 on a Sun UltraServer workstation.
To account for this discrepancy, we investigated the effect of enzyme cooperativity suggested in the literature. The data of Bennett et al. (30) showed a 2.7-fold increase in Polβ dRp lyase activity in the presence of an Ape1 concentration of 8 nM. To simulate this effect, we increased Polβ dRp lyase catalytic efficiency (kcat/KM) 3-fold, modeled as a 3-fold increase in kcat or a 3-fold decrease in KM (Fig. 3). Neither change accurately predicts the curve of the experimentally derived data (Fig. 3, diamonds), although increasing the kcat resulted in an amount of repaired adduct at the end of the simulation that showed good agreement with the experimental outcome.
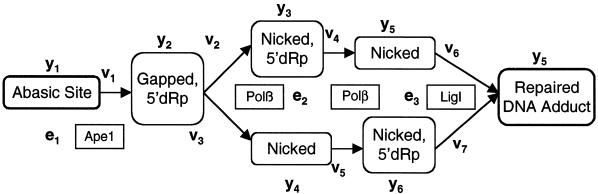
Given the inability of Ape1–Polβ cooperativity to accurately predict pathway behavior, we explored other possibilities of cooperativity not explicitly described in the literature, but which can be modeled as a change in equations requiring no additional parameter measurement. Specifically, it is known that the gap-filling and dRp lyase reactions of Polβ occur in different structural domains (31). However, it is not known whether these reactions occur coordinately. In the initial model (Fig. 2), the equations assume that these reactions are catalyzed by different Polβ molecules, where each enzyme must diffuse and locate the target site. While spatial coordination between these consecutive reactions has not been demonstrated experimentally, it is a reasonable consideration. The consequence of Polβ staying on or near the DNA was simulated by changing the expression of v3 to v3 = k3y3 and allowing for dRp lyase and gap-filling functions of Polβ to occur in any order (see Fig. 7 and equations in Appendix A). This adjusted model (Fig. 3, thick dashed line; using an initial AP site concentration of 1 µM) predicts a >10-fold increase in abasic site repair rate relative to the model incorporating Ape1–Polβ cooperativity alone (Fig. 3, dashed lines). Thus, the adjusted model of Appendix A greatly overestimates observed repair. However, the apparent saturation of the repair curve in the reconstruction assay (Fig. 3, diamonds) at abasic DNA substrate concentrations <50 nM likely suggests that only a fraction of substrate is accessible to the operational enzymes, either due to product inhibition or inadequate mixing. If we assume an initial damage concentration of 50 nM instead of 1 µM in the model, then the further adjusted model (Appendix A) agrees well (Fig. 3, thick solid line) with the experimental results of Srivasta et al. (29), including the shape of the curve. This finding suggests an alternative means of BER cooperativity not yet experimentally demonstrated.
Figure 7.
Schematic of the model described in Appendix A: the AP site repair pathway (pathway B, Fig. 2) adjusted to include coordination between gap filling and dRP lyase reactions catalyzed by Polβ.
Sensitivity of model predictions to kinetic parameters
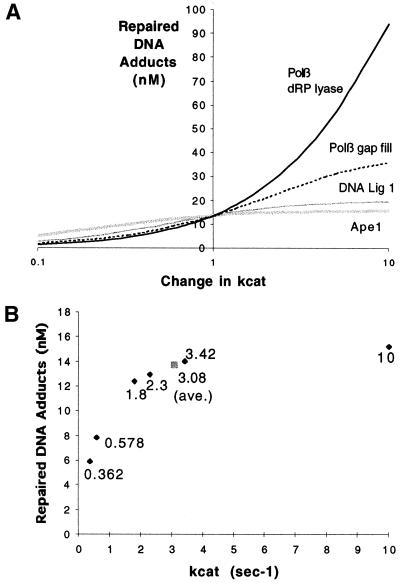
Model sensitivity analysis identifies those reactions that have the most impact on pathway throughput and thus require the most precise measurement. We studied the sensitivity of the simplest abasic site repair model (i.e. without cooperation) as described in Figure 2. We analyzed a 100-fold variation in kcat of the individual reactions, adjusting each parameter 10-fold higher or lower than the average value presented in Table 1 (Fig. 4A). The predicted repair outcome is most sensitive to the kcat of the slowest reaction, Polβ dRp lyase, yet exhibits sensitivity to the rates of other reactions, particularly Polβ gap filling.
Figure 4.
Sensitivity of the abasic site repair model (shown in Fig. 2) to changes in kcat of each reaction. (A) The final number of repaired DNA adducts (after 120 s) upon varying kcat by an order of magnitude either more or less than the average value used in the original simulation (Table 1). (B) Predicted abasic site repair (after 120 s, 1 µM initial damage) for six different experimentally measured Ape1 incision kcat parameters (Table 2) and their average (gray box). The points are labeled with the kcat value.
We also examined the differences in pathway efficiency using kinetic parameters found in the literature. In the case of Ape1 5′-abasic site incision activity (Fig. 1, pathway B), six sets of kcat and KM parameters were found (Table 2). The high degree of variability of these data (a coefficient of variation >100%) illustrates the imprecision and inconsistency of kinetic measurements, likely to be a consequence of variation in buffer choice, incubation time and quantification of chemical concentrations. As shown in Figure 4B, final repair outcome shows highest sensitivity to the slower sets of Ape1 parameters (Table 2, sets 1 and 2), where sensitivity decreases as the Ape1 reaction rate increases and the other steps in the BER pathway become lesser bottlenecks.
Table 2. Experimental data for Ape1 incision kinetics.
Comprehensive BER model: subpathway comparision
To evaluate the quantitative contribution of the subpathways of BER to overall pathway throughput, we first constructed a comprehensive model of BER using the kinetic parameters in Table 1. We selected 8-oxoguanine (8-oxoG) repair, since 8-oxoG DNA glycosylase (Ogg1) has activities that allow for pathway A (Ogg1 AP lyase activity) or pathway B (Ape1 incision), followed by either short-patch (pathway B) or long-patch repair (pathway C). The full schematic of the model and the incorporated equations are shown in Appendix B, and the kinetic parameters are those shown in Table 1, except that we have included a 3-fold increase in Polβ dRp lyase kcat, assuming Ape1 and Polβ cooperativity. No numerical kinetic data were found for DNA ligase 3, the effects of XRCC1, or Polɛ long-patch repair activity, so their reactions were excluded. For comparative purposes, we also simulated the repair of uracil, initiated by uracil DNA glycosylase (UDG), a glycosylase that lacks AP lyase activity (i.e. pathway A). There are other glycosylases for which kinetic data are present in the literature, such as endonuclease III (Nth1) (32), but we leave analysis of these glycosylases for future studies.
For these more complete BER simulations, where available, protein concentrations were based on experimental measurements of enzyme number in human cell extracts (Table 3). We found no data for the cellular abundance of Polδ or Fen1, so we assumed the same concentration as Polβ (i.e. 50 000 molecules of each per typical cell), so as not to impose bias in polymerase selection. In the absence of experimental data for Lig1, we assumed 100 000 molecules per cell, a concentration halfway between Ape1 and Polβ. Enzyme number was then converted into concentration (Table 3) using a spherical whole cell diameter of 20 µm. Because enzyme amounts were measured from whole cell extracts and there is evidence that some of these species are found throughout the cell [e.g. for Ape1 (33)], the simulation assumes enzymes are homogenously distributed in the entire cell volume.
Table 3. Determined cellular enzyme concentrations.
| Enzyme | Abundance (molecules/cell) | Source | Cellular conc. (nM) |
|---|---|---|---|
| UDG |
178 000 |
(35) |
70.6 |
| Ogg1 |
123 000 |
(35) |
48.8 |
| Ape1 |
300 000 (avg.) |
(35,66) |
119 |
| Polβ |
50 000 |
(66) |
19.8 |
| Lig1 | 100 000 | a | 39.7 |
aNot experimentally derived, see text.
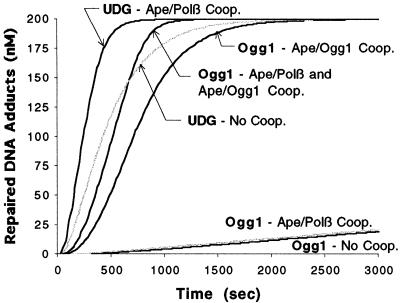
In the absence of enzyme cooperativity, the comprehensive BER model (shown in Appendix B) predicts that the rate of 8-oxoG repair is >100-fold slower than uracil repair (Fig. 5). Much of this difference arises because Ogg1 glycosylase activity is significantly slower than UDG (the kcat/KM is 105 smaller). Yet the experimentally determined rate of 8-oxoG repair using human cell extracts is only 5–7-fold slower than in vitro uracil repair (34,35). We therefore considered the recent work of Hill et al. (36) that suggests Ape1 enhances Ogg1 glycosylase activity (with a kcat of 7.55 × 10–3 s–1 and a KM of 8.9 nM when in the presence of Ape1). Since the glycosylase step is much slower than the other reactions, when modeled this increased efficiency significantly enhanced Ogg1-initiated pathway throughput (Fig. 5). Thus, with Ape1–Ogg1 cooperativity included, the difference between the modeled rates of UDG and Ogg1-initiated repair agrees qualitatively well with the biochemical experiments (35), offering support to the idea of Ape1–Ogg1 cooperativity. We note that the observed enhancement of UDG glycolysis rate by Ape1 is negligible (37) at normal physiological enzyme concentrations (about twice as much Ape1 as UDG, see Table 3), and therefore was not considered in the models here. Omitting the Ape1–Polβ dRp lyase cooperativity (30) had a similar negative impact on the kinetics of either Ogg1 or UDG-initiated repair (Fig. 5). However, unless Ape1–Ogg1 cooperativity is included, Ape1–Polβ dRp lyase has no impact on Ogg1 repair (Fig. 5), due to the dominance of Ogg1 activity in dictating pathway throughput.
Figure 5.
Results of the comprehensive BER pathway simulation. The concentration of repaired uracil sites and 8-oxoG lesions is shown for 200 nM of initial damage. Assuming a cell nucleus diameter of 5 µm, this corresponds to eight damage sites per cell. The results for models without cooperativity, with Ape1–Ogg1 and/or Ape1–Polβ dRp lyase cooperativity are shown.
To obtain further insight into potential subpathway utilization, we computed the number of 8-oxoG sites repaired by pathways A, B and C as a proportion of the total number of repaired sites. In our model (Fig. 1), pathway B (short-patch repair) uses Polβ exclusively, while pathway C (long-patch repair) uses the other polymerases, Polδ/ɛ. This is largely supported by experimental results in Fortini et al. (34), although some exceptions exist: pathway B can proceed at a low rate without Polβ (38) and pathway C can proceed with only Polβ present (24,39). With these caveats in mind, our comprehensive model (with or without cooperativity) predicts that pathway B is the dominant subpathway for 8-oxoG repair (Table 4). Even when the abundance of Polβ is reduced to 10%, pathway B is still dominant, even though pathway C does contribute more to repair. The prevalence of pathway B agrees with the results of in vitro assays using crude cell extracts (34) where the repair rate of 8-oxoG is (i) independent of PCNA, i.e. the kinetic activity of Polδ and (ii) significantly slower in the absence of Polβ. Thus, under the conditions employed, long-patch repair (pathway C) appears relatively inconsequential as a BER pathway.
Table 4. Relative contribution of BER pathwaysa.
| Pathway | With Ape1–Ogg1 cooperativity (%) | Without enzyme cooperativity (%) | With enzyme cooperativity and 90% reduction in Polβ concentration (%) |
|---|---|---|---|
| A |
0.00788 |
0.00788 |
0.00775 |
| B |
99.79 |
99.80 |
97.69 |
| C | 0.201 | 0.194 | 2.306 |
aSimulation was performed at an initial 8-oxoG site concentration of 200 nM.
Sensitivity of BER throughput to polymerase type (Polβ/δ/ɛ)
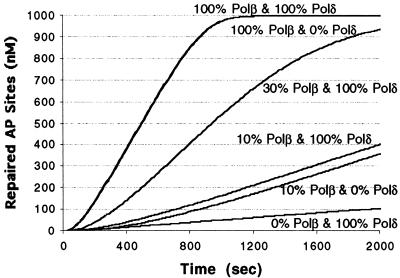
To examine in more detail the impact of the various polymerases on BER capacity, we simulated AP site repair (starting with an initial damage of 1 µM) using the model of Figure 2 with polymerase inhibition modeled as a reduction in enzyme concentration (Fig. 6). Eliminating Polδ had a minimal effect on pathway throughput, whereas reducing the Polβ abundance to 10% led to an ∼5-fold increase in repair half-life (time until 0.5 µM repaired sites appear). This latter value corresponds to an 80% reduction in repair velocity, agreeing well qualitatively with the experimental finding of Nealon et al. (40), where using Polβ or Polα/δ/ɛ inhibitors, these scientists found that repair was suppressed by 70% at 10% Polβ activity.
Figure 6.
Simulation of the effect AP site repair of inhibiting DNA polymerase activities. The initial AP site concentration was 1 µM by varying Polβ/δ concentrations. The curves at various Polβ and Polδ concentrations (with 100% representing 19.8 nM, the assumed normal cellular concentration) are indicated. Note that the curves for 100%/100% Polβ/δ and 100%/0% Polβ/δ overlap.
It has been hypothesized that the remaining BER activity under Polβ inhibition is due to switching to long-patch BER (pathway C). However, the comprehensive model (Appendix B) does not predict any significant increase in pathway C throughput at 10% Polβ concentration (Table 4). Indeed, when Polβ is at 10% and we reduce Polδ abundance to zero (thus blocking pathway C), repair is suppressed by 82%, about the same effect as inhibiting Polβ alone. The model therefore suggests that the non-proportional reduction in repair efficiency results simply from the influence of the other BER enzymes. Any reduction in Polβ activity only reduces that portion of overall pathway throughput controlled by Polβ-catalyzed reactions. Other reactions that are not necessarily ‘rate-limiting’ have some control over the pathway as well, for example DNA ligase 1 (see the sensitivity plot in Fig. 4A).
DISCUSSION
Past efforts in modeling DNA repair (41,42) have focused on systems-level aspects without including the detail of specific enzymes or reactions. Using the kinetic parameters of individual proteins in BER, we have constructed the first quantitative model of a human DNA repair process. In light of the discrepancy between the prediction of this initial model and experimental measurements of pathway kinetics, we explored the consequence of pathway cooperativity on the model prediction. These modeling studies support the existence of cooperation during AP site repair (43) and between Ape1 and Ogg1 in 8-oxoG repair (36,44,45).
In the case of abasic site repair, while incorporation of Ape1–Polβ cooperativity (30) correctly predicted the total amount of damage repaired in vitro, the adjusted model did not accurately depict the nature of the repair curve. Assessment of other potential mechanisms of cooperativity in BER revealed that coordination between the dRp lyase and gap-filling activities of Polβ more accurately reproduced both the in vitro kinetics and total product output of AP site repair. Thus, while not dismissing the possibility of Ape1–Polβ cooperativity, the model does suggest an alternative mechanism of BER coordination that will need to be tested experimentally.
Using a comprehensive model of BER, we assessed the overall contribution of the various BER subpathways to 8-oxoG and, indirectly, AP site repair. This model suggests that the AP lyase-directed pathway (pathway A) accounts for <0.01% of the total BER throughput (Table 4) and that Polδ-directed long-patch repair (pathway C) plays a minor backup role to DNA Polβ. However, it is important to point out that in some cases the contribution of long-patch BER may be underestimated in our current model. For instance, pathway C presently does not have as many established sources of quantitative cooperativity compared to pathway B. In addition, we have not considered the contribution of DNA Polɛ due to the lack of kinetic information. To fully appreciate the relative roles of the BER subpathways, we will therefore need a more complete and accurate kinetic picture. Additionally, we have not considered the repair of lesions, e.g. reduced or oxidized abasic sites, that are more likely to be processed by the alternative long-patch BER pathways. To do so would require experimental information on both the processing mechanisms and the repair kinetics for such DNA damages.
Sensitivity analysis of our model indicates that the rates of Polβ dRp lyase and gap-filling reactions are most important in determining AP site repair efficiency. This idea is supported by in vitro and in vivo data that indicate the activities of Polβ play a major role in determining BER throughput (25,46,47). However, the contribution of the other reaction steps in determining BER pathway effectiveness cannot be overlooked and will obviously be dictated by the rates and concentrations of the other enzymes in the process. Along these lines, it has been hypothesized that reduced-function, exposure-dependent susceptibility factors exist, which are potentially responsible for a large number of population cancer cases (reviewed in 48). Significant variation has been seen in DNA repair genes, and in some instances, this variation has been found to alter the primary amino acid sequence and diminish the stability or efficiency of the encoded protein [e.g. in Ogg1 (49,50) and Ape1 (51)]. A loss in BER enzyme effectiveness could reduce overall BER capacity, resulting in the inability to cope with increased DNA damage (above some threshold level) and thus accompanying genetic instability and the initiation of disease. Using our developed model, we are currently evaluating the effects of established variant protein kinetics on pathway throughput, as well as the impact of changes in enzyme and DNA damage concentrations.
The studies presented here are a first step towards being able to simulate the cell-level consequences of exposure to DNA damage, such as radiation exposure (accidental or clinical) or ingestion of a food carcinogen. A complete model will require identifying all genes and regulators relevant to DNA repair, measuring the impact of enzyme variation on reaction rates, and developing rapid and inexpensive high throughput methods for determining protein concentrations. Then, given an individual’s DNA repair genotype and a model for the DNA damage [from damage models currently under development (42,52,53)], an individual’s capacity to cope with (and potential health consequence of) a damage-induction pulse could be predicted. The studies presented here will hopefully serve as a catalyst for other investigators to develop and expand comprehensive quantitative models for DNA repair.
APPENDIX
A. Pathway B with Polβ coordination
The AP site repair model of Figure 2 has been expanded to include coordination between Polβ dRp lyase and gap filling functions. Polβ coordination is incorporated into the model by making two changes. As shown in Figure 7, the pathway is now branched so that the Polβ functions of gap filling or dRp lyase occur in either order. The greatest impact on overall pathway kinetics results from changing the expressions for the velocities of the second Polβ reaction (v4 and v5) to linear functions of the substrate concentration, modeling the persistence of the Polβ-adduct complex through both reactions (Fig. 7).
dy1/dt = –v1 v1 = y1[k1e1/(y1 + K1)]
dy2/dt = v1 – v2 – v3 v2 = y2[k2e2/(y2 + K2)]
dy3/dt = v2 – v4 v3 = y2[k3e2/(y2 + K3)]
dy4/dt = v3 – v5 v4 = k3y3
dy5/dt = v4 – v6 v5 = k2y4
dy6/dt = v5 – v7 v6 = y5[k4e3/(y5 + K4)]
dy7/dt = v6 + v7 v7 = y6[k4e3/(y6 + K4)]
B. Overall pathway model
The comprehensive model of the BER reactions, presented in Figure 8, consists of the reactions shown in Figure 1 except for those that lack published quantitative kinetic data. Polβ coordination is not considered in this model. However, there is known coordination between Ogg1 glycosylase (v1) and AP lyase (v2) functions. The model for UDG-initiated repair is not shown, but it is similar to the Ogg1 model except that pathway A (v2, v4, v6) is absent (Fig. 8).
Figure 8.
Schematic of the model described in Appendix B: the comprehensive BER reaction system (as outlined in Fig.1).
dy1/dt = –v1 v1 = y1[k1e1/(y1 + K1)]
dy2/dt = v1 – v2 – v3 v2 = k2y2
dy3/dt = v3 – v5 – v9 v3 = y2[k3e2/(y2 + K3)]
dy4/dt = v2 – v4 v4 = y4[k4e2/(y4 + K4)]
dy5/dt = v5 – v7 v5 = y3[k5e3/(y3 + K5)]
dy6/dt = v4 – v6 – v8 v6 = y6[k5e3/(y6 + K5)]
dy7/dt = v7 – v11 v7 = y5[k7e3/(y5 + K7)]
dy8/dt = v6 – v12 v8 = y6[k6e4/(y6 + K6)]
dy9/dt = v8 + v9 – v10 v9 = y3[k6e4/(y6 + K6)]
dy10/dt = v10 – v13 v10 = y9[k8e5/(y9 + K8)]
dy11/dt = v11 + v12 + v13 v11 = y7[k9e6/(y7 + K9)]
v12 = y8[k8e6/(y8 + K9)]
v13 = y10[k9e6/(y10 + K9)]
Acknowledgments
ACKNOWLEDGEMENTS
We thank our colleagues at Livermore, Drs M. E. Colvin, I. M. Jones, H. W. Mohrenweiser, B. J. Alder, K. S. Kim and B. Sutherland (Brookhaven National Laboratory) for their support during the course of these investigations and for their critical input into this manuscript. B.A.S. is supported by a LLNL Student Employee Graduate Research Fellowship. This work was performed under the auspices of the US Department of Energy by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48 and supported by ARMY (BC980514) and NIH (CA79056) grants to D.M.W. III.
REFERENCES
- 1.Knauer D.J., Wiley,H.S. and Cunningham,D.D. (1984) Relationship between epidermal growth factor receptor occupancy and mitogenic response. Quantitative analysis using a steady state system. J. Biol. Chem., 259, 5623–5631. [PubMed] [Google Scholar]
- 2.Malmberg L.H. and Hu,W.S. (1981) Kinetic analysis of cephalosporin biosynthesis in Streptomyces clavuligerus. Biotechnol. Bioeng., 38, 941–947. [DOI] [PubMed] [Google Scholar]
- 3.Leaf T.A. and Srienc,F. (1998) Metabolic modeling of polyhydroxybutyrate biosynthesis. Biotechnol. Bioeng., 57, 557–570. [DOI] [PubMed] [Google Scholar]
- 4.Arkin A., Ross,J. and McAdams,H.H. (1998) Stochastic kinetic analysis of developmental pathway bifurcation in phage infected Escherichia coli cells. Genetics, 149, 1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endy D., You,L., Yin,J. and Molineux,I.J. (2000) Computation, prediction and experimental tests of fitness for bacteriophage T7 mutants with permuted genomes. Proc. Natl Acad. Sci. USA, 97, 5375–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb T.D. and Pugh,E.N. (1992) G-protein cascades: gain and kinetics. Trends Neurosci., 15, 291–298. [DOI] [PubMed] [Google Scholar]
- 7.Bhalla U.S. and Iyengar,R. (1999) Emergent properties of networks of biological signaling pathways. Science, 283, 381–387. [DOI] [PubMed] [Google Scholar]
- 8.El-Masri H.A. and Portier,C.J. (1999) Replication potential of cells via the protein kinase C-MAPK pathway: Application of a mathematical model. Bull. Math. Biol., 61, 379–398. [DOI] [PubMed] [Google Scholar]
- 9.Yuh C.H., Bolouri,H. and Davidson,E.H. (1998) Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science, 279, 1896–1902. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 11.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 12.Wilson D.M. III and Thompson,L.H. (1997) Life without DNA repair. Proc. Natl Acad. Sci. USA, 94, 12754–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindahl T. (2000) Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res., 462, 129–135. [DOI] [PubMed] [Google Scholar]
- 14.Memislogu A. and Samson,L. (2000) Base excision repair in yeast and mammals. Mutat. Res., 451, 39–51. [DOI] [PubMed] [Google Scholar]
- 15.Nilsen H. and Krokan,H.E. (2001) Base excision repair in a network of defence and tolerance. Carcinogenesis, 22, 987–998. [DOI] [PubMed] [Google Scholar]
- 16.Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in the base excision repair of DNA. Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullough A.K., Dodson,M.L. and Lloyd,R.S. (1999) Initiation of base excision repair: glycosylase mechansms and structures. Annu. Rev. Biochem., 68, 255–285. [DOI] [PubMed] [Google Scholar]
- 18.Scharer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- 19.Wilson D.M.,III and Barsky,D. (2001) The major human abasic endonuclease Ape1: formation, consequences and repair of abasic lsions in DNA. Mutat. Res., 484, 283–307. [DOI] [PubMed] [Google Scholar]
- 20.Wilson S.H. (1998) Mammalian base excision repair and DNA polymerase beta. Mutat. Res., 407, 203–215. [DOI] [PubMed] [Google Scholar]
- 21.Tomkinson A.E. and Mackey,Z.B. (1998) Structure and function of mammalian DNA ligases. Mutat. Res., 407, 1–9. [DOI] [PubMed] [Google Scholar]
- 22.Thompson L.H. and West,M.G. (2000) XRCC1 keeps DNA from getting stranded. Mutat. Res., 459, 1–18. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto Y., Kim,K., Hurwitz,J., Gary,R., Levin,D.S., Tomkinson,A.E. and Park,M.S. (1999) Reconstitution of proliferating cell nuclear antigen-independent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem., 274, 33703–33708. [DOI] [PubMed] [Google Scholar]
- 24.Klungland A. and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podlutsky A.J., Dianova,I.I., Podust,V.N., Bohr,V.A. and Dianov,G.L. (2001) Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J., 20, 1447–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillespie D.T. (1976) A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comp. Phys., 22, 403–434. [Google Scholar]
- 27.Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- 28.Misteli T. (2001) Protein dynamics: implications for nuclear architecture and gene expression. Science, 291, 843–847. [DOI] [PubMed] [Google Scholar]
- 29.Srivasta D.K., Vande Berg,B.J., Prasad,R., Molina,J.T., Beard,W.A., Tomkinson,A.E. and Wilson,S.H. (1998) Mammalian abasic site base excision repair. J. Biol. Chem., 273, 21203–21209. [DOI] [PubMed] [Google Scholar]
- 30.Bennett R.A.O., Wilson,D.M., Wong,D. and Demple,B. (1997) Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc. Natl Acad. Sci. USA, 94, 7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beard W.A. and Wilson,S.H. (2000) Structural design of a eukaryotic DNA repair polymerase: DNA polymerase β. Mutat. Res., 460, 231–244. [DOI] [PubMed] [Google Scholar]
- 32.Dizdaroglu M., Karahalil,B., Sentürker,S., Buckley,T.J. and Roldán-Arjona,T. (1999) Excision of products of oxidative DNA base damage by human NTH1 protein. Biochemistry, 38, 243–246. [DOI] [PubMed] [Google Scholar]
- 33.Evans A.R., Limp-Foster,M. and Kelley,M.R. (2000) Going APE over ref-1. Mutat. Res., 461, 83–108. [DOI] [PubMed] [Google Scholar]
- 34.Fortini P., Parlanti,E., Sidorkina,O.M., Laval,J. and Dogliotti,E. (1999) The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem., 274, 15230–15236. [DOI] [PubMed] [Google Scholar]
- 35.Cappelli E., Degan,P. and Frosina,G. (2000) Comparative repair of the endogenous lesions 8-oxo-7,8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis, 21, 1135–1141. [PubMed] [Google Scholar]
- 36.Hill J.W., Hazra,T.K., Izumi,T. and Mitra,S. (2001) Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res., 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.E. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biade S., Sobol,R.W., Wilson,S.H. and Matsumoto,Y. (1998) Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J. Biol. Chem., 273, 898–902. [DOI] [PubMed] [Google Scholar]
- 39.Prasad R., Dianov,G.L., Bohr,V.A. and Wilson,S.H. (2000) FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem., 275, 4460–4466. [DOI] [PubMed] [Google Scholar]
- 40.Nealon K., Nicholl,I.D. and Kenny,M.K. (1996) Characterization of the DNA polymerase requirement of human base excision repair. Nucleic Acids Res., 24, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaver D.P., Jacobs,P.A., Carpenter,R.L. and Burkhart,J.G. (1997) A mathematical model for intracellular effects of toxins on DNA adduction and repair. Bull. Math. Biol., 59, 89–106. [DOI] [PubMed] [Google Scholar]
- 42.Sachs R.K., Hanfeld,P. and Brenner,D.J. (1997) The link between low-LET dose-response relations and the underlying kinetics of damage production/repair/misrepair. Int. J. Radiat. Biol., 72, 351–374. [DOI] [PubMed] [Google Scholar]
- 43.Wilson S.H. and Kunkel,T.A. (2000) Passing the baton in base excision repair. Nature Struct. Biol., 7, 176–178. [DOI] [PubMed] [Google Scholar]
- 44.Saitoh T., Shinmura,K., Yamaguchi,S., Tani,M., Seki,S., Murakami,H., Nojima,Y. and Yokota,J. (2001) Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat. Res., 486, 31–40. [DOI] [PubMed] [Google Scholar]
- 45.Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura J., La,D.K. and Swenberg,J.A. (2000) 5′-nicked apurinic/apyrimidinic sites are resitant to beta-elimination by beta-polymerase and are persistent in human cultured cells after oxidative stress. J. Biol. Chem., 275, 5323–5328. [DOI] [PubMed] [Google Scholar]
- 47.Sobol R.W., Prasad,R., Evenski,A., Baker,A., Yang,X.P., Horton,J.K. and Wilson,S.H. (2000) The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicitiy. Nature, 405, 807–810. [DOI] [PubMed] [Google Scholar]
- 48.Mohrenweiser H.W. and Jones,I.M. (1998) Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat. Res., 400, 15–24. [DOI] [PubMed] [Google Scholar]
- 49.Dherin C., Radicella,J.P., Dizdaroglu,M. and Boiteux,S. (1999) Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α-hOgg1 (Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res., 27, 4001–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Audebert M., Radicella,J.P. and Dizdaroglu,M. (2000) Effect of single mutations in the OGG1 gene found in human tumors on the substrate specificity of the Ogg1 protein. Nucleic Acids Res., 28, 2672–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadi M.Z., Coleman,M.A., Fidelis,K., Mohrenweiser,H.W. and Wilson,D.M.,III (2000) Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res., 28, 3871–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikjoo H., O’Neill,P., Goodhead,D.T. and Terrisol,M. (1997) Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol., 71, 467–483. [DOI] [PubMed] [Google Scholar]
- 53.Terrisol M. (1994) Modelling of radiation damage by 125I on a nucleosome. Int. J. Radiat. Biol., 66, 447–451. [DOI] [PubMed] [Google Scholar]
- 54.Asagoshi K., Yamada,T., Terato,H., Ohyama,Y., Monden,Y., Arai,T., Nishimura,S., Aburatani,H., Lindahl,T. and Ide,H. (2000) Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycoslylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J. Biol. Chem., 275, 4956–4964. [DOI] [PubMed] [Google Scholar]
- 55.Kavli B., Slupphaug,G., Mol.,C.D., Arvai,A.S., Peterson,S.B., Tainer,J.A. and Krokan,H.E. (1996) Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J., 15, 3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 56.Strauss P.R., Beard,W.A., Patterson,T.A. and Wilson,S.H. (1997) Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem., 272, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 57.Hang B., Rothwell,D.G., Sagi,J., Hickson,I.D. and Singer,B. (1997) Evidence for a common active site for cleavage of an AP site and the benzene-derived exocyclic adduct, 3, N4-Benzethono-dC, in the major human AP endonuclease. Biochemistry, 36, 15411–15418. [DOI] [PubMed] [Google Scholar]
- 58.Izumi T., Malecki,J., Chaudhry,M.A., Weinfeld,M., Hill,J.H., Lee,J.C. and Mitra,S. (1999) Intragenic suppression of an active site mutation in the human apurinic/apyrimidinic endonuclease. J. Mol. Biol., 287, 47–57. [DOI] [PubMed] [Google Scholar]
- 59.Rothwell D.G., Hang,B., Gorman,M.A., Freemont,P.S., Singer,B. and Hickson,I.D. (2000) Substitution of Asp-210 in HAP1 (APE/Ref-1) eliminates endonuclease activity but stabilises substrate binding. Nucleic Acids Res., 28, 2207–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suh D., Wilson,D.M.,III and Povirk,L.F. (1997) 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res., 25, 2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beard W.A., Osheroff,W.P., Prasad,R., Sawaya,M.R., Jaju,M., Wood,T.G., Kraut,J., Kunkel,T.A. and Wilson,S.H. (1996) Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase β. J. Biol. Chem., 271, 12141–12144. [DOI] [PubMed] [Google Scholar]
- 62.Prasad R., Beard,W.A., Strauss,P.R. and Wilson,S.H. (1998) Human DNA polymerase β deoxyribose phosphate lyase. J. Biol. Chem., 273, 15263–15270. [DOI] [PubMed] [Google Scholar]
- 63.Teraoka H., Sawa,M. and Tsukada,K. (1983) Kinetic studies on the reaction catalyzed by DNA ligase from calf thymus. Biochim. Biophys. Acta, 747, 117–122. [DOI] [PubMed] [Google Scholar]
- 64.Lee B.I. and Wilson,D.M.,III (1999) The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem., 274, 37763–37769. [DOI] [PubMed] [Google Scholar]
- 65.Einolf H.J. and Guengerich,F.P. (2001) Fidelity of nucleotide insertion at 8-Oxo-7,8-dihydroguanine by mammalian DNA polymerase δ. J. Biol. Chem., 276, 3764–3771. [DOI] [PubMed] [Google Scholar]
- 66.Chen D.S., Herman,T. and Demple,B. (1991) Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res., 19, 5907–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]