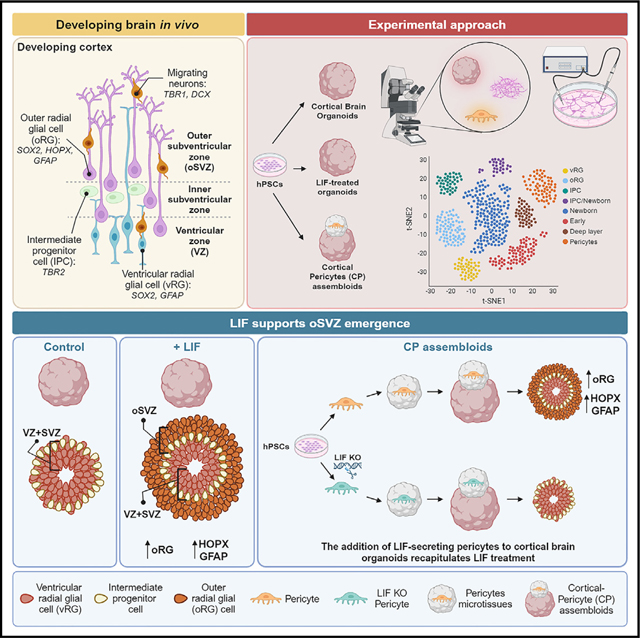
SUMMARY
Outer radial glia (oRG) emerge as cortical progenitor cells that support the development of an enlarged outer subventricular zone (oSVZ) and the expansion of the neocortex. The in vitro generation of oRG is essential to investigate the underlying mechanisms of human neocortical development and expansion. By activating the STAT3 signaling pathway using leukemia inhibitory factor (LIF), which is not expressed in guided cortical organoids, we define a cortical organoid differentiation method from human pluripotent stem cells (hPSCs) that recapitulates the expansion of a progenitor pool into the oSVZ. The oSVZ comprises progenitor cells expressing specific oRG markers such as GFAP, LIFR, and HOPX, closely matching human fetal oRG. Finally, incorporating neural crest-derived LIF-producing cortical pericytes into cortical organoids recapitulates the effects of LIF treatment. These data indicate that increasing the cellular complexity of the organoid microenvironment promotes the emergence of oRG and supports a platform to study oRG in hPSC-derived brain organoids routinely.
In brief
Walsh et al. show that exogenous LIF treatment promotes the emergence of outer radial glia (oRG). LIF-treated organoids recapitulate the formation of an outer subventricular zone (oSVZ). A similar effect is recapitulated by increasing the microenvironment complexity and incorporating neural crest-derived LIF-secreting pericytes.
Graphical Abstract
INTRODUCTION
During brain development, the progenitors of the human neocortex are organized into distinct proliferative compartments. These are the ventricular zone (VZ) and the subventricular zone (SVZ), which give rise to the outer neuronal layers in the cortical plate. The VZ and SVZ contain different classes of neural progenitors: apical radial glia (aRG) in the VZ, basal radial glia, also known as outer radial glia (oRG), intermediate progenitors, and transient amplifying cells in the SVZ.1 Like aRG, oRG self-renew and give rise to cortical neurons via asymmetric cell divisions.2 Unlike aRG, oRG do not possess an apical process or undergo dynamic interkinetic nuclear migration within the VZ.3,4 In gyrencephalic species (e.g., human, macaque, ferret), the SVZ is vastly expanded and subdivided into the inner SVZ (iSVZ) and outer SVZ (oSVZ). oRG primarily populate the oSVZ, and oRG are mainly absent in the developing cerebral cortex of lissencephalic species (e.g., mouse, rat, rabbit), linking the expansion of this compartment to neocortical expansion and folding.5–7
In current guided cerebral organoid differentiation protocols in which hPSCs are directed toward cerebral cortical identity via small molecule treatment,8–10 oRG are less prominent and often lack the expression of Glial Fibrillary Acid Protein (GFAP) (Figures S1A–S1E). GFAP is a comprehensive marker for glial lineage cell types, including oRG11–13; however, GFAP expression appears only very late in hPSC-based 3D culture (>90 days).9,10,14–17 Minimally guided cerebral organoid differentiation protocols from hPSCs eventually lead to the spontaneous production of oRG18 (Figure S1E). However, the factors that promote the emergence of oRG and the contribution of this population to the brain microenvironment still need to be better understood. The guided generation of the oRG population is thus essential to recapitulate proper human-specific neocortex development and to study the mechanisms underlying human neocortical expansion in vitro using cerebral organoid models.
Transcriptional profiling studies for oRG suggest that the elevation of leukemia inhibitory factor receptor (LIFR)/STAT3 signaling may contribute to oRG generation or expansion in the human cortex.19 LIF promotes neural stem cell (NSC) self-renewal in the human brain by promoting the proliferation or formation of glial progenitors in the SVZ.20 Moreover, neural progenitor cell appearance was enhanced by LIF treatment in mouse embryoid bodies,21 and the addition of LIF to in vitro cerebral organoids elevated pSTAT3 staining and led to a >3-fold increase in oRG-like cells and thickening of the SVZ.22 However, it remains unclear how transcriptionally similar LIF-induced oRG are to the fetal human counterpart, their impact on organoid development, and which cells within the brain microenvironment regulate LIF expression and signaling.
Here, we have developed an efficient, robust, and reproducible cortical organoid differentiation method in which an oSVZ is present, and abundant oRG arise upon LIF treatment. Transcriptional analysis shows that hPSC-derived oRG closely match fetal oRG. Finally, incorporating LIF-producing cortical pericytes into cerebral organoids partially mimics the effects observed upon LIF treatment, suggesting that non-neural cell types might contribute to normal oSVZ and oRG development in humans. This study provides a reliable in vitro cerebral organoid system for studying human-specific neocortex development and investigating the mechanisms involved in human neocortical expansion.
RESULTS
LIF promotes the formation of oRG-like cells in hPSC-derived cortical organoids
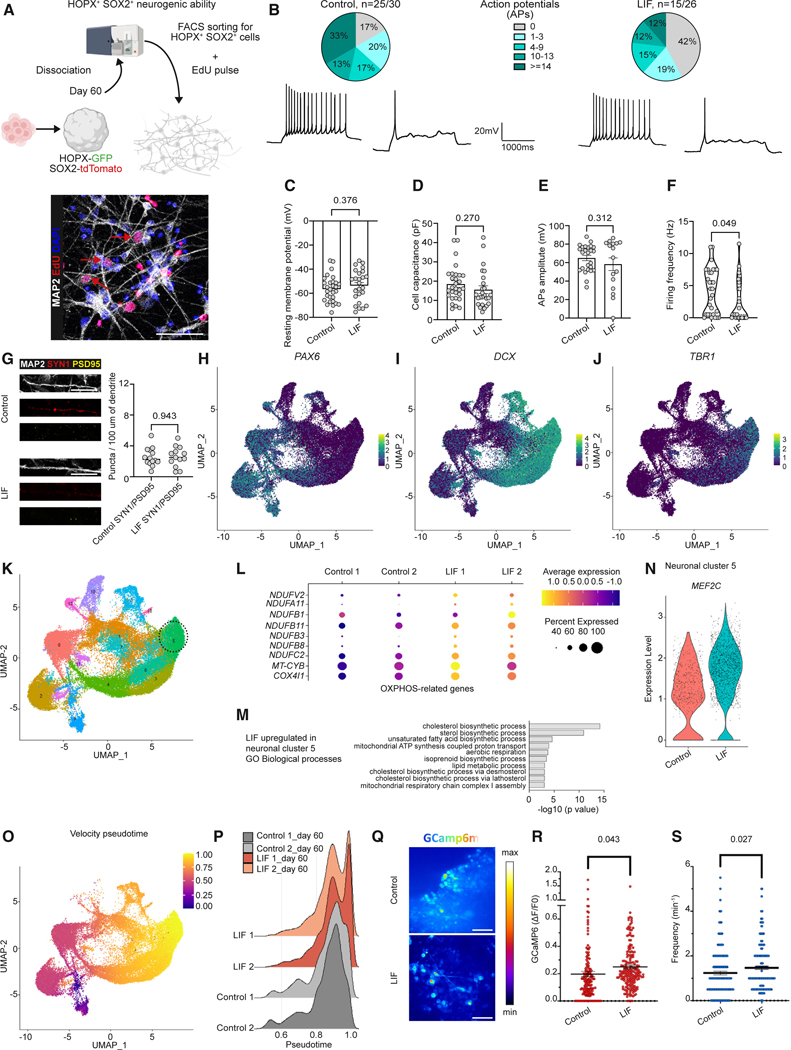
To develop an organoid model that incorporates oRG more efficiently without interfering with neuronal differentiation, we tested the impact of LIF treatment on a cerebral cortical differentiation protocol that we previously developed.23 Exogenous LIF treatment was selected as a candidate strategy for oRG generation, given the robust expression of LIFR in oRG19 and lack of the LIF ligand expression in cortical patterned organoids (Figures S1A–S1I). Neural induction was performed using dual SMAD inhibition in combination with WNT inhibition, as described by Cederquist et al.23 and Rosebrock et al.,15 on days 0–4, followed by dual SMAD inhibition alone until day 8 (Figure 1A). On day 8, two different conditions were compared: (1) standard differentiation toward neural and neuronal cell fate (control condition) and (2) treatment with human LIF from day 8 onward to promote oRG fate (Figure 1A). To test whether LIF promotes the appearance of an oSVZ under our culture conditions, we performed immunocytochemistry at an intermediate time point within the differentiation period (day 60) and used antibodies against SOX2, TBR1, and EOMES/TBR2 to visualize the neural rosette morphology and size in control and LIF-treated organoids (Figures 1B and S2A–S2D). In line with what was reported by Watanabe et al.,22 LIF treatment leads to a thickening of the progenitor zones and the emergence of an oSVZ in cerebral organoids treated with LIF (Figures 1B, 1C, and S2A–S2D), whereas VZ and SVZ size and organization were comparable between conditions.
Figure 1. LIF promotes oRG emergence in hPSC-derived guided cortical organoids.
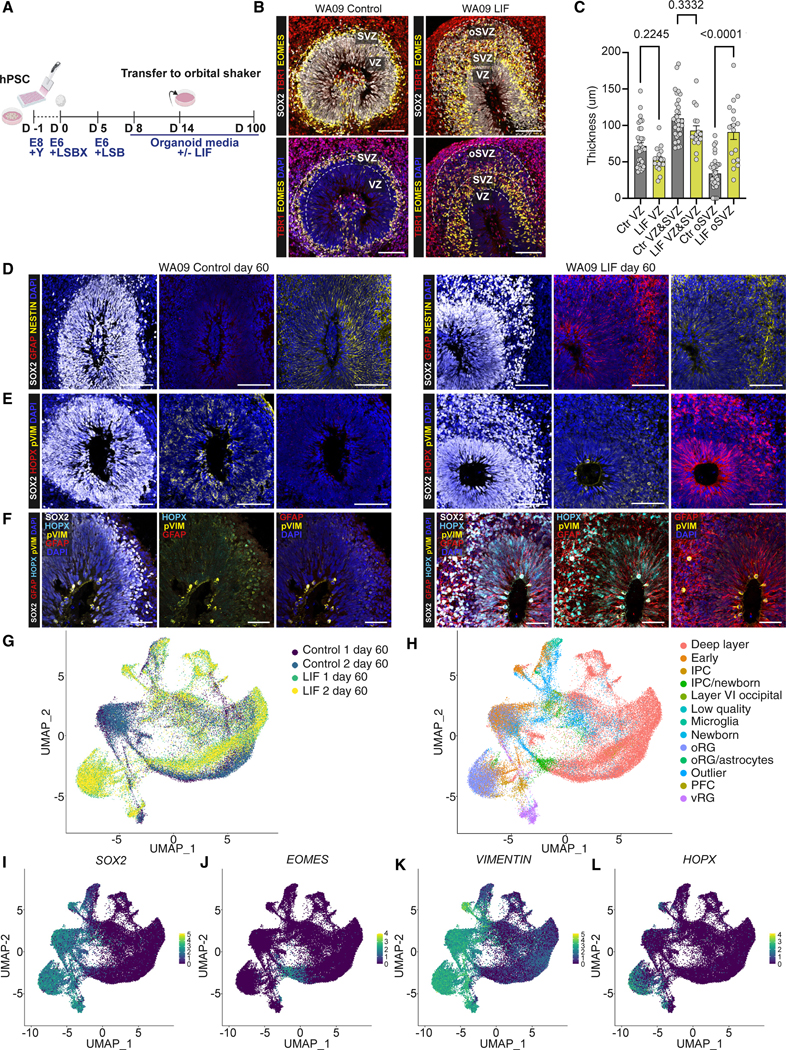
(A) Schematic outline of the cortical organoid differentiation protocol with or without LIF treatment. Y corresponds to ROCK inhibitor (Y-27632, 10 μM); LSBX corresponds to LDN193189 (100 nM), SB431542 (10 μM), and XAV939 (5 μM); LSB corresponds to LDN193189 (100 nM) and SB431542 (10 μM); for organoid media composition, see the relative section in STAR Methods. Human LIF was added at 10 ng/mL.
(B) SOX2 (gray), TBR1 (red), and EOMES (yellow) staining in control and LIF-treated WA09 cortical organoids, sectioned and stained at day 60. The dotted lines highlight the regions of the ventricular zone (VZ), subventricular zone (SVZ), and outer subventricular zone (oSVZ). Nuclei are stained in blue with DAPI. Scale bars, 100 μm.
(C) Rosette quantifications of WA09 control and LIF-treated cortical organoids at day 60, based on the separation of the regions as shown in (B). One-way ANOVA with Tukey’s test. Adjusted p values are shown on the graph.
(D) SOX2 (gray), GFAP (red), and NESTIN (yellow) staining in day 60 control and LIF-treated WA09 brain organoids. Scale bars, 100 μm.
(E) SOX2 (gray), HOPX (red), and phospho-VIMENTIN (pVIM, yellow) staining in day 60 control and LIF-treated WA09 brain organoids. Scale bars, 100 μm.
(F) Quadruple staining for HOPX (cyan), GFAP (red), pVIM (yellow), and SOX2 (gray), in day 60 control and LIF-treated WA09 brain organoids. Scale bars, 50 μm.
(G and H) Single-cell RNA-seq (scRNA-seq) experiments at day 60 showing plots for original identity (G) and label transfer, cell type annotation, and data integration with in vivo dataset from Bhaduri et al.24 (H). Two independent batches of 10 organoids each were processed and analyzed by scRNA-seq for both control and LIF conditions.
(I–L) Feature plots depicting the distribution of the expression of SOX2 (I), EOMES (J), VIMENTIN (K), and HOPX (L) in control and LIF-treated cortical organoids on day 60.
To further characterize the cells that comprise this oSVZ expansion, we performed immunofluorescent labeling for the RG markers SOX2, NESTIN, and GFAP.19 The VZ and oSVZ of LIF-treated organoids were positive for SOX2 and NESTIN. In contrast, SOX2 in the control organoids was primarily restricted to the VZ, where most NESTIN+ fibers were found to originate (Figures 1D and S2E–S2G). We also noticed a striking upregulation of GFAP in both the VZ and oSVZ of LIF-treated organoids, which, unlike SOX2 and NESTIN, is typically not expressed in organoids until very late in their development9,10,14–17,25 (Figures S1A–S1E). After observing the presence of RG markers within the oSVZ, we assessed the expression of the human oRG marker HOPX19 and phospho-VIMENTIN (pVIM), which marks dividing RG. Here, we observed a striking increase in HOPX+ cells and, importantly, phospho-VIMENTIN+/HOPX+ cells within the oSVZ (Figure 1E). Quadruple staining of HOPX, GFAP, SOX2, and pVIM confirmed our observations (Figure 1F). The presence of pVIM+/HOPX+ within the oSVZ (Figure 1F) suggests that, unlike aRG, which divide against the apical surface, these HOPX+ cells can divide within the oSVZ, which is a property of in vivo oRG. These observations were consistent across multiple human embryonic stem cell (hESC) and induced pluripotent stem cell (hiPSC) lines, demonstrating the reproducibility of the protocol (Figures S2A–S2G). Together, these data indicate that the expanded oSVZ within LIF-treated organoids appears to comprise a population of oRG-like cells. The LIF effect could be recapitulated in several published differentiation protocols, including a minimally guided differentiation protocol (Figure S1J)18,26 and another published differentiation protocol guided toward the cortical forebrain region (Figure S1K).27
Next, we investigated the transcriptional profile at day 60 of control and LIF-treated organoids using single-cell RNA-sequencing (scRNA-seq) (Figures 1G and 1H). Using canonical markers, we identified SOX2+ VIMENTIN+ progenitor and EOMES+ intermediate progenitor populations in control and LIF-treated organoids (Figures 1I–1K). However, we observed a cellular population expressing the oRG marker HOPX, specifically in LIF-treated organoids (Figure 1L). To determine whether this population may represent bona fide oRG, we performed a cell-type annotation by label transfer with the in vivo dataset from Bhaduri et al.24 (Figure 1H). This analysis allowed us to identify an LIF-specific oRG cell population in our dataset that globally matches fetal oRG (Figure 1H). Moreover, while these cells were positive for SOX2 (Figure 1I), they were negative for EOMES/TBR2 (Figure 1J), which is consistent with an oRG identity as defined by Matsumoto and colleagues.28 Our data revealed an LIF-induced expansion of HOPX+ cells, suggesting the formation of progenitor cells with possible oRG-like identity. This HOPX+ population was absent on day 40 (Figures S3A–S3D), implying that oRG emerged only after day 40 but before day 60 under these culture conditions.
LIF-induced oRG-like cells are transcriptionally similar to fetal oRG
To probe the molecular identity of the LIF-induced oRG population further, we next characterized the transcriptional profile of the GFAP+ HOPX+ cells within the scRNA-seq data. We identified cluster 2 (Figure 2A) as the LIF-induced oRG cluster (Figure 1H). Interestingly, we observed a population of SOX2+ VIMENTIN+ progenitor cells within the control organoids, the cluster 0 (Figure 2A), that appeared molecularly similar to the LIF-induced oRG cluster but was lacking the expression of crucial oRG marker genes (Figure 2B). We postulated that this may be a population of immature or pre-oRG cells that still require an inductive signal from LIF to become oRG molecularly and functionally. This was supported by the label transfer experiment we had previously performed, which identified cells of cluster 0 as primarily “early” RG (Figure 1H).24
Figure 2. In vitro, LIF-induced oRG-like cells are transcriptionally similar to fetal oRG.
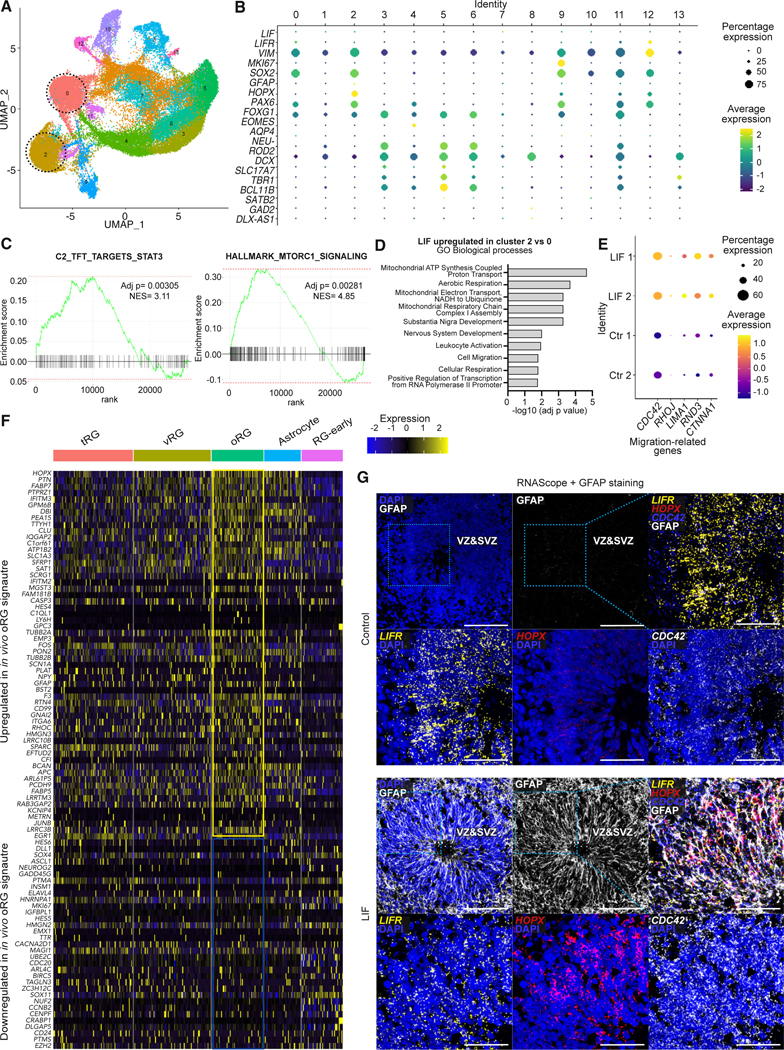
(A) scRNA-seq Seurat clusters in control and LIF-treated organoids on day 60. The progenitor cluster 0 from control cortical organoids and the oRG cluster 2 from LIF-treated cortical organoids are highlighted.
(B) Genes from analysis in (A) to mark selected populations of interest (neuronal cells, neuronal progenitors, astrocytes, and progenitor cells, including oRG) in control and LIF-treated organoids.
(C) Enrichment analysis showing upregulation of STAT3 (left) and mTORC1 signaling (right) pathways in LIF-enriched cluster 2 compared to control cluster 0.
(D) Gene Ontology (GO) Biological Process terms enriched in the LIF-specific cluster 2 compared to control cluster 0.
(E) Migration-related genes upregulated in the LIF-specific cluster 2 compared to control cluster 0.
(F) scRNA-seq signature enriched in LIF-treated organoids at day 60 (cluster 2) compared to control organoids (cluster 0) and mapped to fetal brain populations.29 The transcriptional profile of hPSC-derived oRG cells is enriched in fetal oRG compared to other radial glia populations and fetal astrocytes.
(G) RNAscope for the LIFR, HOPX, and CDC42 transcripts, combined with a GFAP staining in control and LIF-treated WA09 cortical organoids at day 60. Nuclei are stained with DAPI and depicted either in blue. Scale bars, 50 μm.
We then compared control-specific cluster 0 (GFAP− HOPX−) with LIF-specific cluster 2 (GFAP+ HOPX+) on day 60 (Figures 2A, 2B, and S3E). All the cells expressing HOPX and GFAP belonged to LIF-treated organoids (Figure 1G). We then compared LIF-specific cluster 2 to control-specific cluster 0 and found a significant enrichment of STAT3 and mTORC1 signaling pathways, consistent with the data derived from fetal oRG19,30 (Figure 2C). Similarly, we observed a specific upregulation of pathways related to mitochondrial respiration and cell migration (Figures 2D and 2E) as previously observed in fetal oRG.19 The genes related to cell migration included members of the Rho family of GTPases (Figure 2E), whose activity has been shown to regulate oRG migration and morphology.30,31 We then asked whether LIF-induced oRG cells had a molecular identity similar to that of in vivo oRG (Figure 2F). To address this, we compared the transcriptional profile of LIF-induced oRG and the single-cell transcriptomics datasets of developing fetal brain samples.29 The fetal transcriptomics data include various populations of radial glia (tRG, vRG, and oRG), fetal astrocytes, and an early RG population. The transcriptomic profile of hPSC-derived oRG nicely mapped into the transcriptional signature specific to fetal oRG29 (Figure 2F), further corroborating that LIF-induced oRG cells closely match the transcriptional profile of their in vivo counterparts. To compare their transcriptional profile to hPSC-derived astrocytes, we analyzed published transcriptomics datasets derived from scRNA-seq of hPSC-derived brain organoids after 590 days of culture.10 Those data include signatures of VZ/fetal astrocytes, oRG cells, and astrocytes (Figures S2H–S2L). The control-specific cluster 0 and the LIF-specific cluster 2 both correlated with the previously published oRG-related signature in organoids (Figure S2K), which highlights that the fetal datasets are more helpful in detecting fine differences between different radial glia populations.24,29 Cluster 9, equally shared by control and LIF-treated organoids, correlates with a VZ/fetal astrocyte-related signature (Figure S2J). None of the identities are specifically enriched in the astrocyte-related signature at day 60 (Figure S2L).
Next, we performed RNAscope to gain spatial information regarding the expression of the oRG-related transcripts HOPX, LIFR, and CDC42, a small GTPase of the Rho family related to cell migration (Figure 2G)32. We combined the RNAScope with immunofluorescence staining against the GFAP protein (Figure 2G) and further validated its expression upon LIF treatment. Consistent with the scRNA-seq data, LIFR and CDC42 were expressed in both control and LIF-treated organoids in the VZ and SVZ, but CDC42 was upregulated in LIF organoids. Strikingly, HOPX was strongly expressed only upon LIF treatment, and its expression spread to the outer areas of the neural rosette (Figure 2G). This reflects the observations made in fetal tissues, where HOPX+ cells are enriched in the oSVZ but also span through multiple layers.24,33
In later organoids at day 100, scRNA-seq revealed that a few HOPX+ cells also started appearing in control organoids, but these cells remained largely negative for GFAP (Figures S4A–S4D). These findings suggest that the LIF-specific cluster 2 represents bona fide oRG. At the same time, the control condition generates a putative pre-oRG population of progenitors expressing LIFR that, without the inductive signal from LIF, remain transcriptionally distinct from HOPX+ GFAP+ oRG cells.
Neurogenic competence of LIF-induced oRG
To investigate the neurogenic capacity of oRG in LIF-treated organoids, we developed a cell-sorting strategy to isolate those cells and directly assess their differentiation potential in monolayer culture (Figure 3A). We generated a SOX2-tdTomato HOPX-GFP dual reporter line, which contained a heterozygous P2A-H2B-tdTomato transgene knocked into the SOX2 locus, and a heterozygous P2A-GFP-PGK-Puro cassette knocked into the HOPX locus in the WA09 hESC line. LIF-treated organoids from the dual reporter line were dissociated at day 60 of differentiation, and cells were fluorescence-activated cell sorting (FACS) sorted for SOX2 (tdTomato) and HOPX (GFP) double positivity. SOX2+ HOPX+ cells were then replated as a monolayer in organoid media. 2 weeks after replating, cells were treated with EdU for 24 h and cultured in organoid media for 2 additional weeks (Figure 3A). This experiment revealed that some EdU+ cells gave rise to MAP2+ neurons, demonstrating that LIF-induced SOX2+ HOPX+ cells possess neurogenic abilities (Figure 3A).
Figure 3. Neurogenic competence of LIF-induced oRG.
(A) FACS sorting strategy to isolate SOX2+ HOPX+ progenitor cells from LIF-treated brain organoids and test their neurogenic differentiation potential in a monolayer culture. MAP2 (gray) and EdU (red) staining on dissociated monolayers from LIF-treated organoids. Nuclei are stained with DAPI in blue. Scale bar, 50 μm.
(B–F) Manual patch-clamp results for the total number of recorded action potentials (APs) on dissociated monolayers from control and LIF-treated brain organoids (B). Patch-clamp results for the resting membrane potential (C), cell capacitance (D), AP amplitude (E), and firing frequency (F); Welch’s t test.
(G) MAP2 (gray), SYN1 (red), and PSD95 (yellow) staining and quantifications of monolayer cultures derived upon dissociation of control and LIF-treated brain organoids. Welch’s t test. Scale bars, 25 μm.
(H–J) Feature plots depicting the distribution of the expression of PAX6 (H), DCX (I), and TBR1 (J) in control and LIF-treated organoids at day 60.
(K) Seurat clusters in control and LIF-treated organoids at day 60. The neuronal cluster 5 is highlighted.
(L) OXPHOS-related genes enriched in cluster 5 at day 60.
(M) GO Biological Process terms involved in metabolism enriched in cluster 5 and upregulated in LIF-treated organoids.
(N) Violin plot showing expression levels of MEF2C in control and LIF-treated organoids at day 60 in the neuronal cell cluster 5.
(O) Velocity pseudotime plots showing all neural cells undergoing maturation.
(P) Ridge plots showing expression levels of neural cells that undergo maturation in LIF-treated organoids compared to control organoids at day 60.
(Q–S) Calcium imaging (Q) and analysis showing quantification of Ca2+ intensity (Welch’s t test) (R) and frequency (Welch’s t test) (S) measured in control and LIF-treated organoids at day 60. Scale bars, 50 μm.
We then focused on the neuronal cells generated in control and LIF-treated organoids. To better characterize their electrophysiological profile, we dissociated control and LIF-treated organoids at day 60, replated the cells into a monolayer for 3 weeks, and performed a manual patch-clamp procedure (Figures 3B–3F). We did not observe any significant electrophysiological difference in resting membrane potential (Figure 3C), cell capacitance (Figure 3D), and action potential (AP) amplitude (Figure 3E). However, we did observe a higher firing frequency (Figure 3F) in control neurons. The increased firing frequency was modest but significant, and we could not explain this difference by an apparent maturation phenotype since the other parameters were not significantly changed, including the expression of the synaptic proteins SYN1 and PSD95 on MAP2+ neurites (Figure 3G).
Brain organoids originate from several waves of neurogenesis, which give rise to neurons born at different time points. Upon organoid dissociation, neuronal networks are destroyed, and no specific neuronal population is selected for replating. The advantage of our scRNA-seq datasets is that we can specifically analyze any neuronal populations of interest in the organoids. We identified cluster 5 as the most mature deep-layer neuronal cluster based on the absence of PAX6 and the highest expression of DCX and TBR1 (Figures 3H–3K). Gene Ontology (GO) analysis of cluster 5 showed enrichment in mitochondrial and aerobic respiration in LIF-treated organoids (Figures 3L and 3M), which has been linked before to enhanced neuronal maturation.34 In line with these observations, we noticed an increase of MEF2C expression, specifically in neurons from LIF-treated organoids in cluster 5 (Figure 3N), a transcription factor shown to be upregulated in in vivo transplanted organoids24 and involved in synapse refinement.34,35 To sort cells along their developmental trajectory and decipher the relationships between progenitor and neuronal clusters, we performed RNA velocity analysis and defined a velocity pseudotime.36 Pseudotime analysis revealed that neurons from LIF-treated organoids were farther along their pseudotime trajectory than those of control organoids (Figures 3O and 3P), and cluster 5 was confirmed to be the most mature neuronal cluster based on the pseudotime analysis.
Finally, we investigated Ca2+ activity in intact control and LIF-treated organoids. We found that LIF-treatment promoted an increased Ca2+ amplitude and frequency (Figures 3Q–3S and Videos S1 and S2), a sign that excitability and neuronal activity might be enhanced in the superficial layers of LIF-treated organoids.
These data confirm the neurogenic ability of LIF-induced oRG and suggest that their presence does not impact the overall neuronal activity and maturation of the organoid. However, the data also indicate that there might be some neuronal subpopulations, e.g., deep-layer neurons, that show different properties in an organoid that also originated from oRG, and it will be exciting in future studies to define maturation speed depending on the progenitor of origin.
Brain pericytes secrete LIF and promote oRG emergence in human cortical organoids
We asked how LIF ligand is produced in the brain to understand whether a cell type that secretes LIF could be added to cortical organoids to obviate the need for exogenous LIF. In other organs, such as the heart, vascular pericytes are known to secrete LIF.37 In the brain, pericytes that originate either from mesodermal progenitors or the neural crest (NC) play a prominent role in numerous vascular functions, including regulation of cerebral blood flow, maintenance of the blood-brain barrier (BBB) and the BBB-cerebrospinal fluid (CSF) barrier, control of vascular development, and angiogenesis.38,39 NC-derived pericytes facilitate neuroinflammatory processes, possess stem-like properties, and form part of the neurovascular unit, a group of cells that control interactions between neurons and the cerebral vasculature to meet the demands of the brain,38 and they are greatly enriched in the developing choroid plexus.40
To investigate LIF expression in brain pericytes, we analyzed human fetal brain sections at gestational week (GW) 8 and 11 and stained for PDGFRβ and LIF expression. From these samples, we found strong LIF positivity in PDGFRβ+ cells of the choroid plexus (Figure 4A), a region that is one of the main producers of CSF.39 In the developing human choroid plexus, PDGFRβ is also expressed by mesenchymal cell types, including pericytes, suggesting that fetal pericytes are some of the cells secreting LIF. This is particularly interesting considering that LIF is expressed in minimally guided brain organoids that contain choroid plexus, and those organoids also show the emergence of a GFAP+ HOPX+ oRG population (Figures S1E and S1I).18
Figure 4. Pericytes secrete LIF and promote oRG emergence.
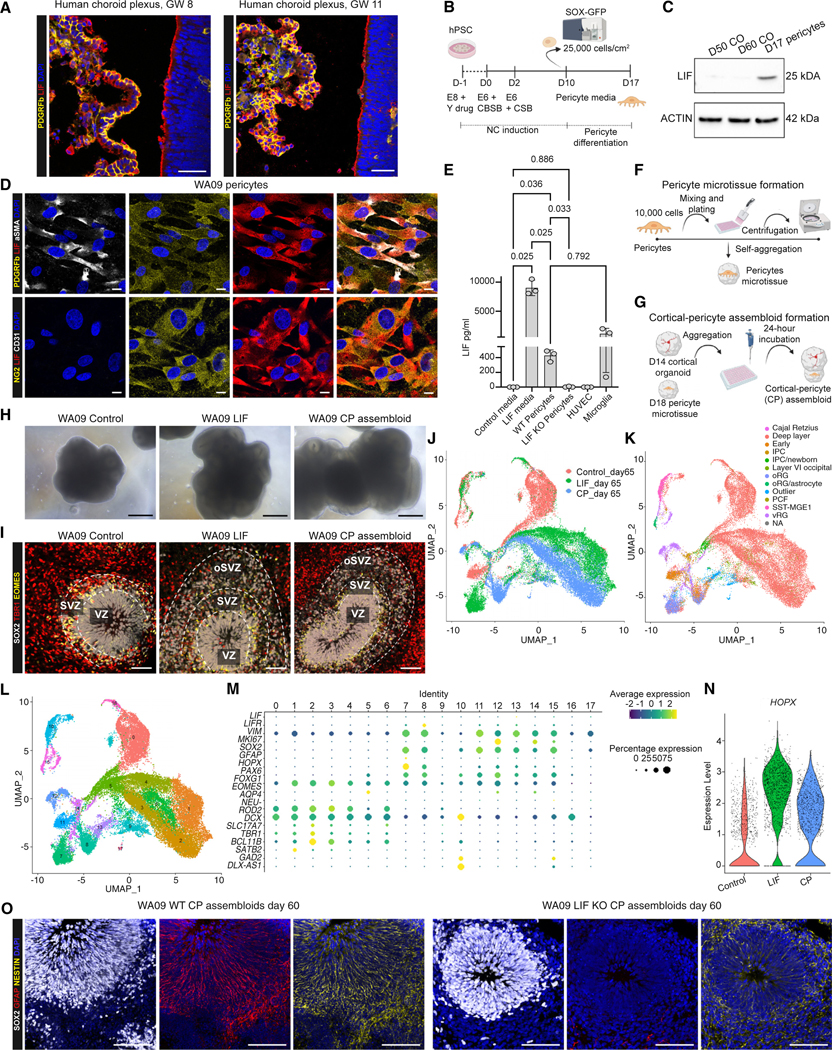
(A) PDGFRβ (yellow) and LIF (red) staining in human fetal choroid plexus at gestational week (GW) 8 and 11. Nuclei are stained in blue with Hoechst. Scale bars, 50 μm.
(B) Schematic representation of the pericyte differentiation protocol from WA09-SOX10-GFP hPSC-derived neural crest (NC) cells. CBSB corresponds to CHIR (600 nM), BMP4 (1 ng/mL), and SB (10 μM). CSB corresponds to CHIR (1.5 μM) and SB (10 μM). On day 10, hPSC-derived NC cells were sorted based on GFP expression to derive SOX10+ NC cells. SOX10+ cells were maintained in pericyte medium to generate NC-derived pericytes.
(C) Western blot data showing LIF expression in day 17 NC-derived pericytes but not in control cortical organoids (COs) at day 50 and 60 of differentiation.
(D) Immunofluorescence images of day 17 NC-derived pericytes for pericyte markers PDGFRβ (yellow), NG2 (yellow), and αSMA (white), endothelial marker CD31 (white), and LIF (red). Cell nuclei are shown by DAPI. Scale bars, 10 μm.
(E) ELISA assay for LIF in control NC-derived pericytes, LIF KO NC-derived pericytes, HUVEC cells, and hPSC-derived microglia. Positive and negative controls are cortical organoid media with or without LIF addition, one-way ANOVA with Tukey’s test.
(F and G) Schematic outline showing the protocol to form pericyte microtissues (F) and cortical-pericyte (CP) assembloids (G).
(H) Representative bright-field images of the morphological appearance of WA09 control organoids, LIF-treated organoids, and CP assembloids at day 60. Scale bars, 100 μm.
(I) SOX2 (gray), TBR1 (red), and EOMES (green) staining showing rosette areas in control, LIF-treated organoids, and CP assembloids. Cell nuclei are stained by DAPI and shown in blue. Scale bars, 100 μm.
(J–L) scRNA-seq experiments showing plots for original identity (J), label transfer, cell type annotation, and data integration with in vivo dataset from Bhaduri et al.24 (K), and Seurat clusters (L) in control organoids, LIF-treated organoids, and CP assembloids at day 65.
(M) Genes selected from (J) to mark selected populations of interest (dividing cells, neuronal cell types, and neuronal progenitors, astrocytes, postmitotic neurons, and radial glia) in control organoids, LIF-treated organoids, and CP assembloids at day 65.
(N) Violin plots showing expression levels of HOPX in control organoids, LIF-treated organoids, and CP assembloids at day 65.
(O) SOX2 (gray), GFAP (red), and NESTIN (yellow) staining in day 60 WT CP assembloids and LIF KO CP assembloids. Cell nuclei were stained by DAPI. Scale bars, 100 μm.
We wondered whether incorporating NC-derived pericytes in our control organoids could rescue the limited oSVZ and oRG development, thus mimicking exogenous LIF treatment. Firstly, we developed a two-step protocol to differentiate hPSCs into NC-derived pericytes using an NC induction protocol previously established41,42 followed by a pericyte differentiation method based on the work of Faal et al.43 (Figure 4B). We used the SOX10eGFP/w hWA09 hESC line in which enhanced green fluorescent protein (eGFP) is targeted to the genomic locus of the NC transcription factor SOX10.44 This allows the monitoring and sorting of NC cells based on eGFP expression. The NC cells are then differentiated in NC-derived pericytes that express LIF (Figure 4C), the typical pericyte markers (aSMA, PDGFRβ, and NG2) (Figure 4D), and are negative for the endothelial cell marker CD31. Finally, LIF secretion was confirmed by ELISA (Figure 4E).
Next, NC-derived pericytes were aggregated into spheroid microtissues in V-bottom 96-well microplates, as depicted in Figure 4F, similar to what has been previously described for microtissues containing cardiac myocytes.45 Pericyte microtissues were then fused to control cerebral organoids to generate cortical-pericyte (CP) assembloids as depicted in Figure 4G. Control and LIF-treated organoids were differentiated in parallel as negative and positive control, respectively. Visual assessment of rosette areas (Figure 4H) and immunocytochemistry (Figure 4I) revealed rosette expansion and oSVZ appearance in CP assembloids compared to control organoids, similar to LIF-treated organoids.
We then compared the transcriptional profile of CP assembloids, control, and LIF-treated organoids by scRNA-seq (Figures 4J–4N) and detected the appearance of an oRG cluster in CP assembloids (Figure 4K). In those CP assembloids, pericyte markers were enriched in cluster 17 (Figures 4L, S4E, and S4F). We then compared the transcriptional signature of cluster 17 to the published scRNA-seq data of human fetal pericytes, endothelial cells, and smooth muscle cells (SMCs) from a recent fetal neocortex atlas.46 We first calculated the top 2000 differentially expressed genes for both the pericytes in CP assembloids and the human fetal dataset using a Wilcoxon rank-sum test. The p values were then adjusted for multiple testing using the Benjamini-Hochberg method. Fetal cells and hPSC-derived pericytes were then compared using the Jaccard index to quantify the degree to which genes are either expressed or not expressed within the same cell type.47 This comparison confirmed that hPSC-derived pericytes transcriptionally resemble fetal pericytes rather than endothelial cells or SMCs (Figure S4G). Overall, the morphological and transcriptional results indicated that including NC-derived pericytes in cerebral organoids could mimic the exogenous LIF treatment and promote oSVZ and oRG emergence.
Finally, to prove that pericytes promote the emergence of an oRG population mainly through LIF secretion, we generated a doxycycline (dox)-inducible LIF knockout (KO) W09 hESC line, and we repeated the CP assembloids experiment with either control (no dox) LIF-expressing pericytes or with dox-induced LIF KO pericytes. LIF KO pericytes did not produce LIF, as shown by ELISA (Figure 4E), and remarkably, they did not promote the typical emergence of GFAP+ SOX2+ NESTIN+ cells (Figure 4O) and the expansion of oSVZ (Figure S4H), as conversely done by the no dox control pericytes.
Overall, our data indicate that the inclusion of LIF-secreting non-neural-derived lineages, such as cortical pericytes, into cerebral organoids significantly impacts the recapitulation of the physiological development of human cortical lineages.
DISCUSSION
In this study, we have established an hPSC-based protocol for the induction and expansion of the oRG population in guided cortical organoids, which matches the human fetal counterpart. This was achieved by adding human recombinant LIF or incorporating NC-derived LIF-secreting pericytes. Interestingly, several guided cortical organoid differentiation protocols lack or show a delayed appearance of GFAP+ HOPX+ oRG (Figure S1).9,10,14 In contrast, some minimally guided differentiation protocols, commonly associated with the formation of choroid plexus structures, a region that produces LIF both in vitro18 and in vivo (Figure 4A), contain populations of GFAP+ HOPX + oRG (Figures S1E and S1I).
Two distinct types of oRG were previously identified based on HOPX expression in the gyrencephalic cerebral cortex of ferrets: HOPX + and HOPX − oRG. Compared to HOPX − counterpart, HOPX + oRG have a higher self-renewal activity and preferentially produce upper-layer neurons in prospective gyral regions, resulting in the formation of cortical folds in gyrencephalic brains.28 Within the SOX2 + VIMENTIN + progenitor cells, we also identified HOPX− and HOPX + cells in LIF-treated organoids. In the future, it will be interesting to isolate these two cell populations directly from LIF-treated organoids and compare their self-renewal properties and differentiation capabilities.
The fact that LIF-induced oRG simultaneously express GFAP and HOPX, in combination with the transcriptional similarity to fetal oRG from Bhaduri et al.24,29 and Nowakowski et al.,29 indicates that we have obtained and expanded bona fide oRG. Our work highlights the importance of the microenvironment in differentiating NSCs into oRG. The population of SOX2+ VIMENTIN+ cells in control organoids also expresses LIFR (Figure S3E), though they fail to express oRG-specific markers and do not expand into the oSVZ. Our results indicate that the emergence of oRG requires extrinsic signals and highlights LIF as a critical factor in this process. Indeed, in the majority of the current guided differentiation protocols we have analyzed, we could not find any cell type producing LIF (Figures S1F–S1H), which prevents a sustained activation of the STAT3 pathway known to be required for oRG maintenance.19
Interestingly, the addition of pericytes, a cell type that contributes to the formation of the choroidal BBB-CSF barrier and produces LIF (Figure 4A), partially rescues the lack of oRG in our protocol (Figure 4), while dox-induced LIF KO pericytes do not (Figure 4O). These data underline the potential implication of the microenvironment not only in sustaining brain-specific cells but also in regulating their differentiation and the resulting tissue architecture. Beyond pericytes, other cell types are known to secrete LIF in the human brain, and it would be interesting to investigate whether they also might play a similar role in oRG induction and expansion. One example is the microglia,48 whose LIF secretion was confirmed in hPSC-derived microglia by ELISA assays (Figure 4E). Overall, the CP assembloids offer a proof-of-principle demonstration that by increasing the cellular complexity of the organoid microenvironment and adding a cell type secreting LIF, it is possible to activate the STAT3 pathway and promote oRG emergence. In line with this finding, a previously published protocol of minimally guided differentiation18 generated a subset of non-telencephalon-specific cells expressing LIF (Figure S1I), and those organoids also contain a GFAP+ HOPX+ population (Figure S1E).
Our data show that oRG cells derived from LIF-treated cortical organoids possess neurogenic abilities (Figure 4A). When we compared neurons derived from control and LIF-treated organoids upon dissociation into a monolayer, we did not observe any significant electrical difference by manual patch clamping (Figures 3B–3F) or synaptic protein quantifications (Figure 3G). However, when we looked at specific neuronal clusters by scRNA-seq, such as the deep neuronal cluster 5, we observed a transcriptional profile related to neuronal maturation (Figures 3K–3P).49 On this line, Ca2+ imaging performed on intact control and LIF-treated organoids showed boosted neuronal activity (Figures 3Q–3S). Overall, these data suggest that global neuronal activity is not impacted by oRG but that there might be some differences depending on specific neuronal subpopulations. Future lineage tracing studies will be required to address the neuronal properties and maturation speed depending on the progenitor of origin and to define to which extent cortical expansion can be recapitulated by oRG cells in brain organoids.
Of note, the scRNA-seq data also showed a trend for an increased generation of cortical-derived interneurons (Figures 2B and S4A–S4D),50,51 but this population was also sporadically observed in some control organoids, and additional work will be required to analyze oRG-derived cell types in this protocol.
Overall, our study describes a cortical organoid system that incorporates an expanded germinal zone (oSVZ) and progenitor cells (oRG), two features characteristic of primate cortical development. These findings open the possibility of using this system for future mechanistic studies comparing neocortical development and expansion across different species. Finally, we demonstrate that the microenvironment, including LIF-secreting cortical pericytes, is essential for recapitulating aspects of neocortical development such as oSVZ and oRG emergence. This finding points to the importance of developing more complex organoid models incorporating non-neural cells to study their contribution to neocortex development and cortical neuronal physiology.
Limitations of the study
To characterize neuronal activity in control and LIF-treated cortical organoids, we performed manual patch clamping. The results showed no significant increase in neuronal activity and other maturation parameters in either of the two conditions. However, the electrophysiology analysis was performed on 2D neuronal cultures following organoid dissociation and replating. The dissociation likely limited the possibility of detecting differences in the neuronal network, and we cannot exclude that some cells, such as immature neurons, might be more likely to survive upon dissociation. Our Ca2+ activity analysis partially circumvents this limitation by analyzing intact organoids, where we observed increased Ca2+ activity for LIF-treated samples. This is further supported by our scRNA-seq pseudo-time analysis, which suggests that deep-layer neurons (cluster 5 in Figure 3K) from LIF-treated organoids are developmentally more advanced than their counterparts in control organoids. However, electrophysiology in specific neuronal populations should be performed in the future to assess any possible changes in electrical activity in LIF-treated organoids. Future work should also more formally address whether neuronal activity and maturation speed depend on the subtype of radial glia from which neurons originate.
Our study proposes that adding LIF-secreting non-neural cell types, such as cortical pericytes, to a guided cortical organoid protocol bypasses the need for exogenous LIF addition. These data suggest that a complex brain microenvironment supports the emergence of a broader set of radial glia populations. Pericytes are likely not the only cell type in the brain that secretes LIF. The analysis of fetal tissues at GW8 and GW11 (Figure 4A) indeed suggests that PDGFRβ− cells also express LIF, and we already documented this for microglia (Figure 4E).48 The population of GFAP+ HOPX+ cells in CP assembloids remains reduced compared to that in LIF-treated organoids. It is possible that increasing pericyte numbers or adding other LIF-secreting cell types of the brain microenvironment would enable an equally efficient generation of oRG. Future studies will be essential to elucidate the role of LIF-secreting cell types in brain development and their broader function beyond LIF secretion.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact: arianna.baggioli@ior.usi.ch.
Materials availability
This study did not generate new unique reagents. The hESC and iPSC lines used in this study will be made available upon request from the Studer laboratory at Memorial Sloan Kettering Cancer Center or the Baggiolini lab at the Institute of Oncology Research under a materials transfer agreement with the institute.
Data and code availability
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table. Datasets generated in this study have been deposited at Gene Expression Omnibus (GEO): GSE224346.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| SOX2 Monoclonal antibody (Btjce), eBioscience | Thermo Fisher scientific | Cat#14-9811-82; RRID:AB_11219471 |
| EOMES Monoclonal Antibody (WD1928), eBioscience | Thermo Fisher Scientific | Cat#14-4877-82; RRID:AB_2572882 |
| Recombinant Anti-TBR1 antibody [EPR8138(2)] | Abcam | Cat#ab183032; RRID:AB_2936859 |
| phospho-vimentin (Ser55) | MBL International | Cat#D076-3; RRID:AB_592963 |
| Anti-HOPX polyclonal antibody | Sigma-Aldrich | Cat#HPA030180; RRID:AB_10603770 |
| Glial Fibrillary Acidic Protein | Agilent | Cat#Z033429-2; RRID:AB_10013382 |
| Purified Mouse Anti-Nestin | BD Biosciences | Cat#611658; RRID:AB_399176 |
| CD31, Endothelial Cell Antibody (Concentrate) | Agilent | Cat#M0823; RRID:AB_2114471 |
| Monoclonal Anti-Actin, α-Smooth Muscle | Sigma-Aldrich | Cat#A2547; RRID:AB_476701 |
| Anti-LIF antibody [IMG39N7D10] | Abcam | Cat#ab138002; RRID:AB_3083551 |
| PDGFRB Monoclonal Antibody (G.290.3) | Thermo Fisher Scientific | Cat#MA5-15143; RRID:AB_10985851 |
| Anti-NG2 antibody | Abcam | Cat#ab129051; RRID:AB_2877152 |
| MAP2 antibody - Neuronal Marker | Abcam | Cat#ab5392; RRID:AB_2138153 |
| Rabbit Anti-Synapsin I Antibody, Unconjugated | Sigma-Aldrich | Cat#S193; RRID:AB_261457 |
| PSD-95 Monoclonal Antibody (7E3-1B8) | Thermo Fisher Scientific | Cat#MA1-046; RRID:AB_2092361 |
| GFAP Antibody | Novus Bio | Cat#NBP1-05198; RRID:AB_1556315 |
| CD271 (NGF Receptor) Monoclonal Antibody (ME20.4), PE, eBioscience | Thermo Fisher Scientific | Cat#12-9400-42; RRID:AB_2572710 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 |
Thermo Fisher Scientific | Cat#A32790; RRID:AB_2762833 |
| Donkey anti-Rat IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 555 |
Thermo Fisher Scientific | Cat#A48270; RRID:AB_2896336 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 |
Thermo Fisher Scientific | Cat#A31571; RRID:AB_162542 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 594 |
Thermo Fisher Scientific | Cat#A32754; RRID:AB_2762827 |
| Donkey anti-Chicken IgY (H + L) Highly Cross Adsorbed Secondary Antibody, Alexa Fluor 488 |
Thermo Fisher Scientific | Cat#A78948; RRID:AB_2921070 |
| HRP Anti-beta Actin antibody [AC-15] | Abcam | Cat#ab49900; RRID:AB_867494 |
| Anti-rat IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7077; RRID:AB_10694715 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| One Shot Stbl3 Chemically Competent E. coli | Thermo Fisher Scientific | Cat#C737303 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Essential 8 medium | Thermo Fisher Scientific | Cat#A1517001 |
| Vitronectin (VTN-N) Recombinant Human Protein, Truncated | Thermo Fisher Scientific | Cat#A14700 |
| UltraPure 0.5M EDTA, pH 8.0 | Thermo Fisher Scientific | Cat#15575020 |
| ReLeSR | StemCell Technologies | Cat#100-0483 |
| Accutase solution | Sigma-Aldrich | Cat#A6964 |
| ROCK inhibitor Y-27632 | Bio-Techne (Tocris) | Cat#1254 |
| Essential 6 Medium | Thermo Fisher Scientific | Cat#A1516401 |
| LDN-193189 | Reprocell (Stemgent) | Cat#04-0074 |
| SB 431542 | Bio-Techne (Tocris) | Cat#1614 |
| XAV 939 | Bio-Techne (Tocris) | Cat#3748 |
| Neurobasal Medium | Thermo Fisher Scientific | Cat#21103049 |
| DMEM/F-12, HEPES | Thermo Fisher Scientific | Cat#31330038 |
| B-27 Supplement (50X), minus vitamin A | Thermo Fisher Scientific | Cat#12587010 |
| N-2 Supplement (100X) | Thermo Fisher Scientific | Cat#17502048 |
| GlutaMAX Supplement | Thermo Fisher Scientific | Cat#35050038 |
| 2-Mercaptoethanol | Thermo Fisher Scientific | Cat#21985023 |
| Normocin - Antimicrobial Reagent | Invivogen | Cat#ant-nr-2 |
| Insulin solution human | Sigma-Aldrich | Cat#I9278 |
| Recombinant Human LIF | Peprotech | Cat#300-05 |
| STEMdiff Cerebral Organoid Kit | StemCell Technologies | Cat#8570 |
| Matrigel Basement Membrane Matrix, LDEV-free, 10 mL | Corning | Cat#354234 |
| KnockOut Serum Replacement | Thermo Fisher Scientific | Cat#10828028 |
| MEM Non-Essential Amino Acids Solution (100X) | Thermo Fisher Scientific | Cat#11140050 |
| Penicillin-Streptomycin | Sigma-Aldrich | Cat#P4333 |
| Dorsomorphin | MedChemExpress | Cat#HY-13418A |
| Recombinant Human FGF-basic (154 a.a.) | Peprotech | Cat#100-18B |
| Animal-Free Recombinant Human EGF | Peprotech | Cat#AF-100-15 |
| Recombinant Human/Murine/Rat BDNF | Peprotech | Cat#450-02 |
| Recombinant Human NT-3 | Peprotech | Cat#450-03 |
| Recombinant Human BMP-4 Protein | Bio-Techne (R&D Systems) | Cat#314-BP |
| CHIR 99021 | Bio-Techne (Tocris) | Cat#4423 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Pericyte Medium | Sciencell Research Laboratories |
Cat#1201 |
| Trypsin-EDTA (0.05%) in DPBS (1x) | Capricorn Scientific | Cat#TRY-1B |
| Puromycin dihydrochloride from Streptomyces alboniger | Sigma-Aldrich | Cat#P8833 |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D5207 |
| RiboLock RNase Inhibitor (40 U/μL) | Thermo Fisher Scientific | Cat#EO0381 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat#A9418 |
| Poly-L-ornithine hydrobromide | Sigma-Aldrich | Cat#P3655 |
| Cultrex Mouse Laminin I, Pathclear | Bio-Techne (R&D Systems) | Cat#3400-010-02 |
| Fibronectin Human Protein, Plasma | Thermo Fisher Scientific | Cat#33016015 |
| Click-iT™ Plus EdU Cell Proliferation Kit for Imaging | Thermo Fisher Scientific | Cat #C10640 |
| BrainPhys™ Neuronal Medium and SM1 Kit | StemCell Technologies | Cat #05792 |
| Halt Protease and Phosphatase Inhibitor Cocktail (100X) | Thermo Fisher Scientific | Cat#78440 |
| NuPAGE LDS Sample Buffer (4X) | Thermo Fisher Scientific | Cat#NP0007 |
| NuPAGE™ 4 to 12%, Bis-Tris, 1.0-1.5 mm, Mini Protein Gels | Thermo Fisher Scientific | Cat#NP0335BOX |
| NuPAGE™ MES SDS Running Buffer (20X) | Thermo Fisher Scientific | Cat#NP0002 |
| Nitrocellulose Membranes, 0.45 μm | Thermo Fisher Scientific | Cat#88018 |
| NuPAGE™ Transfer Buffer (20X) | Thermo Fisher Scientific | Cat#NP0006 |
| Nonfat Dry Milk | Cell Signaling Technology | Cat#9999 |
| OCT EMBEDDING MATRIX - 125mL | CellPath | Cat#KMA-0100-00A |
| Normal Donkey Serum | Abcam | Cat#ab7475 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| DPBS, calcium, magnesium | Thermo Fisher Scientific | Cat#14040091 |
| TWEEN 20 | Sigma-Aldrich | Cat#P9416 |
| Normal Goat Serum Blocking Solution | Vector Laboratories | Cat#S-1000-20 |
| Hoechst 33258, Pentahydrate (bis-Benzimide) - 10 mg/mL Solution in Water |
Thermo Fisher Scientific | Cat#H3569 |
| Glycergel Mounting Medium, Aqueous | Agilent | Cat#C056330-2 |
| BrainPhys Imaging Optimized Medium | StemCell Technologies | Cat#05796 |
| Fluoromount-G Mounting Medium | Thermo Fisher Scientific | Cat#00-4958-02 |
| Neutral Buffered Formalin | Epredia | Cat#5701 |
| DPBS, no calcium, no magnesium | Thermo Fisher Scientific | Cat#14190094 |
| EGM-2 Endothelial Cell Growth Medium-BulletKit | Lonza | Cat#CC-3162 |
| IMDM | Thermo Fisher Scientific | Cat# 21980032 |
| Fetal Bovine Serum Advanced (FBS Advanced), Collected in South America |
Capricorn Scientific | Cat# FBS-11A |
| RNAscope H202 and Protease Reagents | ACD Bio | Cat#322381 |
| RNAscope Probe- Hs-HOPX | ACD Bio | Cat#423001 |
| RNAscope Probe- Hs-LIFR | ACD Bio | Cat#441021 |
| RNAscope Probe- Hs-CDC42-O1-C3 | ACD Bio | Cat#502651-C3 |
| TSA Vivid Fluorophore 520 | ACD Bio | Cat#323271 |
| TSA Vivid Fluorophore 570 | ACD Bio | Cat#323272 |
| TSA Vivid Fluorophore 650 | ACD Bio | Cat#323273 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Human LIF ELISA kit | RayBiotech | Cat#ELH-LIF1 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| Chromium Single Cell 3ʹ GEM, Library & Gel Bead Kit v3 | 10x Genomics | Cat# PN-1000075 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | ACD Bio | Cat#323110 |
| Papain Dissociation System | Worthington Biochemical | Cat#LK003150 |
|
| ||
| Deposited data | ||
|
| ||
| Single Cell RNA sequencing dataset Generation of human cerebral organoids with a structured outer subventricular zone” |
This manuscript | GEO Accession #: GSE224346 |
| Single Cell RNA sequencing dataset “Comparison of Cortical Organoids to Human Cortex Identifies Cellular Identity and Maturation Differences” | Bhaduri et al.24 | GEO Accession: GSE132672 |
| Single Cell RNA sequencing dataset “ | Nowakowski et al.29 | dbGaP Accession: phs000989.v3.p1 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| WA09 | WiCell | RRID:CVCL_9773 |
| WA01 | WiCell | RRID:CVCL_9771 |
| HUVEC-C | ATCC | Cat # CRL-1730 RRID:CVCL_2959 |
| HEK293T | ATCC | Cat # CRL-3216 RRID:CVCL_0063 |
|
| ||
| Oligonucleotides | ||
|
| ||
| TCCGTCACAGACTAAGGAGA | IDT | sgRNA targeting HOPX locus |
| GCGGGAAGTCCGTCACGTTG | IDT | sgRNA targeting LIF locus |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pX330-U6-Chimeric_BB-CBh-hSpCas9 | Cong et al.52 | Addgene #42230 |
| Plasmid: lenti sgRNA(MS2)_puro optimized backbone | Joung et al.53 | Addgene #73797 |
| Plasmid: psPAX2 | psPAX2 was a gift from Didier Trono | Addgene #12260 |
| Plasmid: pMD2.G | pMD2.G was a gift from Didier Trono | Addgene #12259 |
| Plasmid: pGP-CMV-GCaMP6m | Chen et al.54 | Addgene #40754 |
|
| ||
| Software and algorithms | ||
|
| ||
| 10x Genomics Cellranger DNA | 10x Genomics | RRID:SCR_023221 |
| Seurat | Butler et al.55 | RRID:SCR_016341 |
| fgsea | Bioconductor | RRID:SCR_020938 |
| scVelo | La Manno et al.56 | RRID:SCR_018168 |
| LoomPy | Linnarsson Lab |
https://loompy.org RRID:SCR_016666 |
| scanpy | Wolf et al.57 | RRID:SCR_018139 |
| Celltypist | Xu et al.58 Dominguez Conde59 |
www.celltypist.org/RRID:SCR_024893 |
| Biorender | Biorender | www.biorender.com |
| LASX Office 1.4.5 27713 | Leica Microsystems | www.leica-microsystems.com |
| GraphPad Prism | GraphPad Software | http://www.graphpad.com/scientific-software/prism/ RRID: SCR_002798 |
|
| ||
| Other | ||
|
| ||
| PrimeSurface® 3D culture: Ultra-low Attachment Plates: 96 well, V bottom, Clear plates |
S-Bio | Cat#MS-9096VZ |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
hPSC lines
Human pluripotent stem cells (H9 (WA09, female); H9-S0X10-GFP; H9-SOX2-TdTomato-HOPX-GFP; H9-iCas960; H1 (WA01, male), MSK-SRF00161 (female) and CS5DZLiCTR-n5 (male) from https://data.answerals.org/home) were maintained in Essential 8 medium (Thermo, #A1517001) in feeder-free conditions on vitronectin (rhVTN-N) substrate (Thermo, #A14700). hPSCs were passaged as clumps with an EDTA (Thermo, #15575020) dissociation solution (0.5 mM EDTA/PBS). Cells were maintained at 37°C and 5% CO2 and were routinely tested for mycoplasma and periodically assessed for genomic integrity by karyotyping.
Cell lines
HEK293-T cells were maintained in IMDM (Thermo, #21980032) supplemented with 10% of Heat Inactivated FBS (Capricorn, # FBS-11A) and utilized to produce the lentivirus used for generating the LIF-KO line.
HUVEC cells were cultured in EGM-2 Endothelial Cell Growth Medium (Lonza, #CC-3162) and used for the ELISA test. Both cell lines were passaged using Trypsin-EDTA (0.05%) in DPBS (Capricorn, #TRY-1B) and replated.
METHOD DETAILS
Experimental design
To ensure the reproducibility of the differentiation protocol provided, several members of Studer and Baggiolini laboratories independently performed aspects of the cerebral organoid differentiation method (with and without human LIF supplementation), and the protocol is consistently used in the two groups across multiple hPSC lines. No specific methods were used for randomization, and investigators were not blinded to the methods of differentiation. No statistical methods were utilized to determine sample size.
Generation of H9-SOX10-GFP reporter line
H9 SOX10:GFP (H9-SOX10-GFP) reporter lines were generated using CRISPR/Cas9-based HDR.62 Briefly, a sgRNA was designed to target a sequence close to the stop codon of the SOX10 gene, and the target was cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene plasmid #42230) to make the gene targeting constructs. A donor plasmid containing a 450 bp left homology arm, followed by a P2A-H2B-GFP cassette, a floxed puromycin selection cassette, and a 450 bp right homology arm was used as the donor template for knock-in. The sgRNA and the donor plasmid were electroporated into H9 cells using a Lonza 4D-Nucleofector instrument with Solution “Primary Cell P3” and Pulse Code “CB-150”. 0.5 μg/ml Puromycin was added to the 3 days post-electroporation for 4 days. Single-cell clones were generated, PCR and sanger-sequencing were used to correctly identify knock-in clones.
Generation of H9-SOX2-TdTomato-HOPX-GFP dual reporter line
H9 SOX2:tdTomato/HOPX::GFP (H9-SOX2-TdTomato-HOPX-GFP) dual reporter line was generated using CRISPR/Cas9 based HDR method.62 To generate the dual reporter, the HOPX::GFP were knock-in on top of the validated H9 SOX2:tdTomato cells.42 Briefly, the sgRNA was designed targeting a sequence close to the stop codon of HOPX gene (sgRNA target: TCCGTCACAGAC TAAGGAGA). The sgRNA target sequence was cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene plasmid #42230). A donor plasmid containing a 400 bp left homology arm, followed by a P2A-H2B-GFP cassette, a floxed puromycin selection cassette (loxP-PGK-puro-loxP), and a 400 bp right homology arm was used as the template for HDR. The HOPX sgRNA and the donor plasmids were electroporated into H9 SOX2:tdTomato cells using Lonza 4D-Nucleofector instrument with Solution “Primary Cell P3” and Pulse Code “CB-150”. 0.5 μg/ml Puromycin was added to the 3 days post-electroporation cells for 4 days. Single-cell clones were generated, PCR and sanger-sequencing were used to correctly identify knock-in clones.
Cortical brain organoid differentiation protocol and LIF treatment
On day −1, hPSCs were dissociated with Accutase (Sigma, #A6964) for 5 min or PBS/EDTA for 10 min at 37°C and allowed to aggregate into spheroids of 10 000 cells each in V-bottom 96 well microplates (S-Bio, #MS-9096VZ) in E8 medium with 10 μM Y-27632 ROCK inhibitor (Bio-Techne, 1254/50). The next day (day 0), medium was changed to perform double SMAD and WNT inhibition by using E6 media (Thermo, A1516401) supplemented with 3 small molecules (100 nM LDN193189, Stemgent #04–0074; 10 μM SB431542, R&D #1614; 5 μM XAV939, R&D #3748) and media change was performed every day until day 4. On day 5, we interrupted WNT inhibition, and the medium was switched to Essential (E6) supplemented only with 100 nM LDN193189 and 10 μM SB431542, with media change every day until day 7. On day 8, the medium was changed to an N2/B27 based organoid medium composed of Neurobasal (Gibco, #21103049), B-27 supplement w/o vitamin A 1:100 (Gibco, #12587010), N2 supplement 1:200 (Gibco, # 17502048), GlutaMax 1:100 (Gibco, #35050038), 0.055 mM cell culture grade 2-mercaptoethanol solution (Gibco, #21985023), 1:1000 Normocin (Invivogen, #ant-nr-2), and 2.5 μg/ml Insulin (Sigma, #I9278–5mL),+/− 10 ng/mL Human LIF (Peprotech, #300–05). Medium change was performed every other day and on day 14 organoids were moved to an orbital shaker on 6 or 10-cm dishes. Medium (+/− LIF treatment) was then changed to a Monday-Wednesday-Friday schedule.
Minimally guided brain organoid differentiation protocol with LIF treatment
For the minimally guided differentiation protocol we used the STEMdiff Cerebral Organoid Kit (StemCell Technologies, #8570) that is based on the four-stage protocol published by Lancaster et al.26,63 following manufacturer instructions. Briefly, 9000 hPSCs were plated in each well of a V-bottom 96 well microplates to induce embryoid body formation (Day 0–5) and the germ layer differentiation. Then, the media was changed to prompt the induction of neural ectoderm (Day 5–7) and subsequently organoids were included in Matrigel (Corning, #354234) droplets to induce the expansion of neuroepithelial buds after switching to an expansion media. From Day 10, the plate with Matrigel droplets containing organoids was moved on a shaking platform and organoids were exposed to a maturation media +/− LIF treatment. Media was changed 2 times per week.
Additional guided differentiation protocol with LIF treatment
To test the effect of LIF on another cortical brain organoid differentiation protocol, we selected the one published by Caporale et al.,27 based on the protocol established by Pasca et al.64
To recapitulate the protocol, on day −2 hPSCs were resuspended at a concentration of 2×105 cell/ml in E8 + 5μM of ROCKi and 100 μL/well of cell suspension were seeded into a V-bottom 96 well microplates and the plates were centrifuged at 850 rpm for 3 min to promote the formation of embryoid bodies. On day 0 the medium is switched to a neural induction medium containing 80% DMEM/F12 medium (Gibco, #11330057), 20% Knockout serum (Gibco, #10828028), 1:100 Non-essential amino acids (Gibco, #11140′050), 0.1 mM cell culture grade 2-mercaptoethanol solution, 1:100 GlutaMax, penicillin 100 U/mL and streptomycin 100 μg/mL (Sigma, #P4333), 7 μM Dorsomorphin (MedChem express, #HY-13418A) and 10 μM SB431542 (R&D #1614).
From day 0 to day 4 medium change was performed every day with the very same medium while on day 5 of differentiation the neural induction medium was substituted with a complete neurobasal medium, composed of Neurobasal medium, 1:50 B-27 supplement w/o vitamin A, 1:100 GlutaMax, penicillin 100 U/mL and streptomycin 100 μg/mL and 0.1 mM cell culture-grade 2-mercaptoethanol solution supplemented with 20 ng/mL FGF2 (Peprotech, #100–18B) and 20 ng/mL EGF (Peprotech, #AF-100–15). Neurobasal medium is added until day 24 with daily medium change for the first 7 days. The LIF treatment was started at day 7 of differentiation. On day 12, organoids were transferred to 10 cm dishes and placed on a standard orbital shaker with medium change every other day. On day 23, FGF and EGF were replaced with 20 ng/mL BDNF (Peprotech, #450–02) and 20 ng/mL neurotrophin-3 (Peprotech, #450–03) to promote differentiation of neural progenitors. From day 42 onwards, complete neurobasal medium without BDNF and NT3 was used, performing media changes every other day.
Differentiation of hPSCs into neural crest-derived pericytes
Neural crest induction was performed using previously established protocols ((Fan et al.42; Tchieu et al; 41) based on an initial SMAD inhibition and a WNT activation followed by increased levels of WNT activation from days 2–10 (Figure 4B).
Prior to starting the differentiation, on day −1, H9-SOX10-GFP were plated as a high-density monolayer (150 000 cells per cm2) on Matrigel in E8 medium with 10μM ROCKi. On day 0, the medium was changed to E6 supplemented with 1 ng/mL BMP4 (R&D #314-BP) + 10μM SB431542 + 600nM CHIR99021 (R&D #4421) to inhibits TGFβ and BMP activating WNT via glycogen synthase kinase-3 β (GSK3-β) inhibition. On day 2, the medium was changed to E6 supplemented with 10μM SB431542 + 1.5μM CHIR99021. Medium was refreshed every day throughout the differentiation protocol up to day 10. On day 10, hPSCs-derived neural crest cells were sorted using a BD-FACS Aria6 cell sorter by the Flow Cytometry Core Facilities of MSKCC and IOR to derive GFP+ (SOX10+ cells of H9-SOX10-GFP) neural crest (NC) cells. DAPI (Sigma, #D9542) was used to exclude dead cells. NC cells were differentiated into pericyte-like cells as described previously (Faal et al.43) by seeding 25 000 cells/cm2 single cells onto Matrigel-coated plates. Cells were maintained in pericyte medium (ScienCell, #1201) to generate neural crest-derived pericytes and medium was refreshed every other day. When confluent, cells were passaged using Trypsin-EDTA (0.05%) in DPBS (Capricorn#TRY-1B) and replated at a concentration of 25000 cells/cm2 in pericyte medium.
Pericyte microtissue formation
Pericyte microtissues were generated similarly to what was done previously for MT-CM microtissues.45 Briefly, hPSC-pericytes among day 17–25 were detached using Trypsin for 5 min at 37°C, centrifuged for 3 min at 1100 rpm and resuspended in pericyte medium with 10μM ROCK inhibitor. Pericytes were diluted to 10 000 cells per 100 μL of medium and cell suspensions were seeded on V-bottom 96 well microplates (S-Bio) and centrifuged for 10 min at 1100 rpm. Pericyte microtissues were incubated at 37°C, 5% CO2 for 24 h prior to cortical-pericyte assembloid formation.
Cortical-pericyte assembloid formation
To generate cortical-pericyte assembloids, day 14-cortical organoids (without LIF treatment) were moved on top of pericyte microtissues to induce their fusion by close proximity in V-bottom 96 well microplates. Plates were centrifuged for 1 min at 1100 rpm and placed in the incubator for 24 h at 37°C, 5% CO2. After 24 h, tissues were fused and assembloids were moved to an orbital shaker on 10 cm dishes. Medium (organoid medium without LIF treatment) was changed on a Monday-Wednesday-Friday schedule.
Dox-induced LIF KO pericytes
To obtain LIF KO pericytes we used a doxycycline (dox)-inducible Cas9 system53 to knockout LIF in hPSCs-derived pericytes.
To target the LIF locus we designed a guide RNA (5′-GCGGGAAGTCCGTCACGTTG-3′) using a CRISPR design tool (http://crispor.tefor.net/).65 The gRNA was cloned into the Lentiguide Puro vector (Addgene, #73797), following Zhang lab indications.53 Then, the LentiGuide Puro vector was transfected together with psPAX2 (Addgene, #12260) and pMD2.G (Addgene, #12259) into HEK293T cells and lentivirus was collected after 48 h and concentrated using the Amicon Ultra-15 Centrifugal Filter Units (Millipore, #UFC910024).
Then, the H9-iCas9 line,60,66 engineered to express Cas9 upon dox induction, was infected with the lentivirus carrying the Lentiguide Puro vector and cells expressing the gRNA were selected upon a 72hrs of 1ug/ml Puromycin (Sigma, #P8833) treatment.
After positive selection, H9-iCas9 cells were differentiated into pericytes as described above but sorted at day 10 for NGFR positivity (CD271 PE, Invitrogen #12–9400-42). At day 17 of differentiation, pericytes were re-selected with Puromycin for 48hrs and then treated for 72hrs with 1ug/ml Doxycycline (Sigma, #D5807) to induce LIF knock-out. Knockout pericyte assembloids were generated as described above.
Organoid and assembloid dissociation for downstream applications
10 organoids/assembloids for each condition and for each time point were dissociated for single-cell RNA-sequencing, FACS sorting of SOX2+ HOPX+ cells, and patch clamp electrophysiology using a papain dissociation method following manufacturer instructions (http://www.worthington-biochem.com/PDS/). Briefly, samples were chopped using a scalpel into smaller pieces and moved into papain solution. The samples were then incubated at 37°C with constant agitation on a shaking platform for 1 h. After incubation, the mixtures were pipetted up and down with a P1000 until tissues were completely dissolved. The samples were then transferred to 15 mL tubes and centrifuged at 300g for 5 min at room temperature. Supernatants were discarded and cell pellets were immediately resuspended in DNase dilute albumin-inhibitor solution. Then, 1 mL of albumin-inhibitor solution was added drop-by-drop to each centrifuged tube (sample) to create a phase separation and, finally, tubes were centrifuged at 70g for 6 min at room temperature. For single-cell RNA-sequencing, cells were resuspended in FACS buffer solution (4mL of FACS buffer +20 μL of RiboLock RNase inhibitor) at a final concentration of 200 000 of cells/200 μL of FACS buffer solution for each sample.
For FACS sorting and replating of SOX2+ HOPX+ cells, cells were resuspended in MACS buffer (1L PBS + 4mL EDTA + 5g BSA) and sorted using a BD-FACS Aria6 cell sorter at the Flow Cytometry Core Facility of MSKCC to derive TdTomato+ GFP+ cells (SOX2+ HOPX+ cells). 4, 6-diamidino-2-phenylindole (DAPI) was used to exclude dead cells. SOX2+ HOPX+ cells were replated in organoid medium (without LIF) by seeding 100’000 cells/cm2 onto 1:1000 Poly-Ornithine (Sigma, #P3655), 1:500 Laminin (Bio-Techne, #3400–010-02), 1:500 Fibronectin (Gibco, # 33016015) coated plates with 10 μM Y-27632 ROCK inhibitor. Medium was refreshed every 3 days. After 2 weeks, cells were treated with EdU (3uM) for 24 h and kept in culture for additional 2 weeks in organoid medium before fixation.
2D neurons replating for patch-clamp and synaptic protein analysis: Neurons derived from dissociated organoids were plated on PO/LAM/FN-coated plate in brain organoid media with 10 μM Y-27632 ROCK inhibitor. After 24 h, the media was removed to get rid of dead and unattached cells and replaced with regular brain organoid media. After 5 days, half of the media was replaced with a 1:1 mixture of brain organoids/BrainPhys medium (StemCell Technologies. #5790). From then onwards, every 5 days half of the medium was replaced with BrainPhys medium67 and after 21 days neurons were fixed for immunofluorescence analysis or subjected to electrophysiology experiments.
Western blot
hPSC-derived pericytes and organoids without LIF treatment were lysed with RIPA buffer +1:1000 Halt Protease and Phosphatase Inhibitor Cocktail (Thermo, #78440) and then sonicated with a Bioruptor Plus sonication device for 8 cycles with High Power (sonication cycle: 15 s ON, 60 s OFF). Supernatant was collected upon 15 min of centrifugation at >13000 rpm at 4°C and quantified using the Pierce BCA Protein Assay Kit (Thermo, # 23227) following manufacturer instructions. Equal amounts of protein were boiled in NuPAGE LDS sample buffer (Invitrogen, #NP0007) at 95°C for 5 min and separated using NuPAGE 4%–12% Bis-Tris Protein Gel (Invitrogen, #NP0335BOX) in NuPAGE MES SDS Running Buffer (Invitrogen, #NP0002). Proteins were electrophoretically transferred to a nitrocellulose membrane (Thermo, #88018) with NuPAGE Transfer Buffer (Invitrogen, #NP0006). Blots were blocked for 60 min at RT in TBS-T + 5% nonfat milk (Cell Signaling, #9999) and incubated with the respective primary antibody at 4°C. The following primary antibodies were used: mouse HRP anti-beta actin (abcam, ab49900, 1:10000), rat anti-LIF (abcam, ab138002, 1:500). Primary antibodies were detected using the secondary anti-rat IgG HRP-linked (Cell Signaling, #7077S, 1:1000) together with the SuperSignal West Femto Chemiluminescent Substrate (Thermo, #34095).
Histology and immunofluorescence (IF)
Organoids and assembloids were washed in PBS and then fixed in 4% PFA for 1–2 h at 37°C. After fixation, organoids were washed three to four times with PBS and then cryoprotected in 30% sucrose/PBS and included in OCT (CellPath, #KMA-0100–00A). Included frozen organoids were cut with an Epredia CryoStar NX50 Cryostat in 20μm slices and sections were mounted on SuperFrost Plus slides (Epredia, #J1810AMNZ).
To perform immunofluorescence staining, sections were treated for 1hr with a 2% BSA (Sigma, #A2153), 5% donkey serum (Abcam, #ab7475), 0.3% Triton X-100 (Sigma, #T8787) in 1X PBS (Gibco, #14080055) blocking solution and then incubated with primary antibodies diluted in blocking solution overnight at 4°C or for 1h 30 min at RT. Afterward, sections were washed three/four times with PBS-T (0.05% Tween 20 (Sigma, #P9416) and incubated with secondary antibodies diluted in a 2%BSA, 0.05% Tween 20 in PBS
solution for 1 h at RT in dark. After 3 washes in PBS-T, DAPI stain was used to mark cell nuclei. Images were captured using a Leica Stellaris 5 or a Leica Stellaris 8 confocal microscope.
Antibodies used: 1:300 rat anti-SOX2 (Invitrogen, #14–9811-82), 1:300 mouse anti EOMES/TBR2 (Invitrogen, #14–4877-82), 1:400 rabbit anti-TBR1 (Abcam, ab183032), 1:200 mouse anti-Phosphorylated Vimentin (pVIM) (MBL, #d076–3), 1:200 rabbit anti-HOPX (Sigma, #HPA030180), 1:400 rabbit anti-GFAP (Dako, #Z033429–2); 1:300 mouse anti-Nestin (Abcam, #611658).
The hPSCs-derived pericytes and organoid-derived neurons were cultured on 8-Well Glass Bottom μ-Slide (iBidì, #80827) and fixed with 4% PFA for 20 min at RT, washed three times with PBS and blocked with a 2% BSA, 5% donkey serum, 0.05% Tween 20 in PBS blocking solution for 1 h. Primary antibodies were diluted in blocking solution and incubated with the samples for 1 h and 30 min at RT. Then, samples were washed 3 times with PBS-T and secondary antibodies were diluted in PBS-T 2% BSA and incubated with the samples in dark at RT for 1 h. Samples were washed 3 times with PBS-T and stained with DAPI to identify cell nuclei. Microscopy was performed using a Leica Stellaris 8 confocal microscope.
Primary antibodies used: 1:400 Anti-Human CD31 Clone JC70A (Dako, M0823); 1:500 mouse anti-α-Smooth Muscle Actin (Sigma Aldrich, #A2547); 1:200 rat anti LIF (abcam, #ab138002), 1:100 rabbit anti-PDGRβ (Invitrogen, #MA5–15143), 1:100 rabbit anti-NG2 (abcam, #ab129051); 1:2000 chicken anti-MAP2 (abcam, #ab5392); 1:100 rabbit anti-Synapsin I (Sigma, #S193); 1:100 mouse anti PSD-95 (Thermo, #MA1–146).
Secondary antibody used: 1:500 donkey anti-rabbit Alexa Fluor Plus 488 (Invitrogen, #A32790); 1:500 donkey anti-rat Alexa Fluor Plus 555 (Invitrogen, #A48270); 1:500 donkey anti-mouse Alexa Fluor 647 (Invitrogen, #A31571).
ELISA assay
The ELISA assays were performed using the Human LIF ELISA kit from RayBiotech (#ELH-LIF-1) following manufacturer protocol. Briefly, cell culture conditioned media were collected from hPSCs-derived pericytes, LIF-KO pericytes, HUVEC cells and hPSCs-derived microglia cells and were concentrated using the Amicon Ultra-15 Centrifugal Filter Unit (Sigma, #UFC910024) following manufacturer instructions. Brain organoids media with or without LIF were used as negative and positive controls, respectively. The ELISA 96-well plate was incubated with the various conditioned media for 2.5 h and then washed to remove unbound molecules. Then, biotinylated antibodies and streptavidin solution bound to a luminescent dye were added and incubated to detect the LIF molecules for 1 h and 45 min respectively. Finally, the substrate to induce the colorimetric chemical reaction was incubated for 30 min after which a stop solution was added to interrupt the reaction. The 450 nm absorbance signal was read using a Cytation 3 instrument. All the samples were analyzed as a biological triplicate.
Human fetal brain sections and immunohistochemistry
Brains were dissected out, fixed (24 h, 4°C) in 4% paraformaldehyde (PFA) and incubated in sucrose 30% for 24 h at 4°C. Brains were then frozen in optimal cutting temperature (OCT) medium on dry ice. Frozen sections (15 μm) were obtained using a cryostat (Leica, Germany) and stored at − 80°C. Sections were thawed just prior to staining and fixed with 4% PFA for 10 min followed by rinsing in PBS. Sections were then dehydrated in 5 min dehydration steps in 70% and 100% ethanol, respectively, washed with PBS, permeabilized with 0.5% Triton X-100 (Euroclone, #BK39487S) in PBS for 10 min, washed in PBS and retrieved with Sodium Citrate 10 mM at 90°C for 30 min. After antigen retrieval, the sections were washed with PBS and blocked with 10% NGS (Vector) and 0.2% Triton X-100 in PBS at room temperature for 1 h. Primary antibodies were diluted in solution containing 3% NGS and 0.1% Triton X-100 in PBS at 4°C overnight. The following day, sections were washed three times in PBS at RT. Secondary antibodies conjugated to Alexa fluorophores 488, 568 or 647 (Molecular Probe, Life Technologies) were used 1:500 in solution containing 3% NGS and 0.1% Triton X-100 in PBS at RT for 1 h mixed with 5 μg/mL Hoechst 33258 (Thermo, #H3569) to visualize nuclei. The sections were then washed once in PBS with 0.1% Triton X-100 and twice in PBS and finally mounted with Dako Glycergel (Aqueous Mounting Medium, Agilent #C056330–2) at RT overnight. The following day the sections were dry enough to be visualized under the microscope and then stored at 4°C. Images were acquired with a widefield and a confocal microscope (Leica SP5).
Single-cell RNA-sequencing data processing and analysis
Single cell suspensions were diluted to 1000 cells/μl in 1X PBS with 0.04% BSA and 0.2U/μl Ribolock RNAse inhibitor (Thermo #EO0382) for sequencing. Single-cell RNA-sequencing (scRNA-seq) was performed for a target recovery of 10 000 cells/sample using 10X Genomics Chromium Single Cell 3′ Kit, Version 3 according to the manufacturer’s protocol. Libraries were sequenced on an Illumina NovaSeq. The CellRanger pipeline (Version 6.1.2) was used to demultiplex and align reads to the GRCh38 reference genome to generate a cell-by-gene count matrix. Data analysis was performed with R v4.1 using Seurat v4.2.0.68 Cells expressing between 200 and 5000 genes and less than 10% counts in mitochondrial genes were kept for analysis. Gene counts were normalized by total counts per cell and ScaleData was used to regress out cell cycle gene expression variance as determined by the CellCycleScoring function. PCA was performed on scaled data for the top 2000 highly variable genes and a JackStraw significance test and ElbowPlot were used to determine the number of PCs for use in downstream analysis. A Uniform Manifold Approximation and Projection (UMAP) on the top 35 PCs was used for dimensional reduction and data visualization. FindNeighbors on the top 35 PCs and FindClusters with a resolution of 2.0 were used to identify clusters. Differential expression analysis was performed by identifying either oRG-like clusters or neuronal clusters based on marker expression and performing FindMarkers within these groups between LIF-treated and control cells using a Wilcoxon Rank-Sum test. GSEA analysis and plots were performed with in R with the fgsea package v1.2.0, for the analysis, a Wilcoxon rank-sum test was performed with presto v1.0.0 using wilcoxonauc() and gene sets for each cluster were ranked based on the area under the receiver operator curve (auc) value. Annotated gene sets for enrichment analysis were obtained from the GSEA MSigDB (gsea-msigdb.org 69,70; and enrichment scores were calculated with the fgsea() function and plotted with the plotEnrichment() function of the fgsea package. For trajectory and pseudotime analyses, Velocity v0.17.1756 was used to define spliced vs. unspliced reads, and loompy.combine (Loompy v2.0.17; http://loompy.org) was used to merge samples into a single loom file. Downstream analysis was carried out in Python v3.9.12 with Scanpy v1.9.157 and scVelo v0.2.436 scvelo.pp.filter_and_normalize(min_shared_counts = 20, n_top_genes = 5000, flavor = “seurat”) was used for normalization and log transformation and moments and nearest neighbors were then calculated with scvelo.pp.moments. Velocities were estimated using the stochastic model with scvelo.tl.veloctiy(mode = ”stochastic”) and pseudotimes obtained with scv.tl.velocity_pseudotime.
Label transfer with CellTypist
Cortical human fetal data from Bhaduri et al.24 were downloaded from the UCSC cell browser. Data processing was performed Scanpy v1.9.157 and CellTypist v0.1.9 was used for analysis. Data was normalized and log transformed. Ribosomal genes were removed from the analysis and CellTypist was used to train a model based on the “Subtype” metadata annotation. Gene selection was performed by generating a quick model using celltypist.train(max_iter = 5, use_SDG = True) and selecting the top 400 genes for each Subtype based on absolute regression coefficients (4590 genes total). These selected genes were used to train a more accurate model with celltypist.train (max_iter = 100). Label transfer to our combined LIF and Control samples was performed with celltypist.annoate (majority_voting = True).
Comparison of fetal scRNAseq datasets
For comparison of radial glia and astrocyte signatures in previously published fetal data, single cell datasets from Bhaduri et al.24 and Nowakowski et al.29 were processed in R v4.1 using Seurat v4.2.0.68 Each dataset was normalized and scaled with a cell cycle regression as described above. For the Nowakowski et al.29 dataset, the subset of the “WGCNAcluster” containing the labels: “tRG”, “oRG”, “vRG, “RG-early” or “Astrocyte” identities was extracted for subsequent heatmap construction and analysis. The same was done for the dataset from Bhaduri et al.24 using the “Subset” labels: “early”, “vRG”, “oRG”, “late”, “org/Astrocyte”, and “tRG”.
Jaccard index
To compare the pericyte signature in cluster 17 to the published scRNA-seq datasets that include human pericytes, endothelial cells, and SMC from the fetal neocortex,46 we first calculated the top 2000 differentially expressed genes in the hPSC-derived pericytes and the human fetal data using a Wilcoxon rank-sum test. The p values were then adjusted for multiple testing using the Benjamini-Hochberg method. The ex vivo and in vitro data were then compared using the Jaccard index to quantify the degree to which genes are either expressed or not expressed within the same cell type.47
Calcium imaging and analysis
hPSC-derived cortical brain organoids were infected with lentiviruses encoding GCamp6m at day 50 of differentiation and cultured in BrainPhys Imaging Optimized Medium (Stem Cell Technologies, # 05796) for a week before the imaging. On the day of the imaging, organoids were equilibrated in imaging buffer for 30 min (25 mM HEPES, 140 mM NaCl, 8 mM KCl, 1 mM MgCl2, 10 mM glucose, 4 mM CaCl2, 10 μM glycine, 0.1% BSA pH 7.4, pre-warmed to 37°C) and transferred into imaging cuvettes. GCamp6m fluorescence on intact organoids was recorded by light-sheet microscopy on TruLive3D Imager (Bruker) under environmental control (37°C; 95% O2 – 5% CO2). Multiple fields of view from 3 to 4 organoids per condition were imaged for ~2 min at a frame rate of 10 frames/second (1200 frames/time lapse) and at 31.3x effective magnification. Analysis was performed as previously described.49,71 Briefly, the live-imaging image stack was converted to TIFF format and loaded into optimized scripts in MATLAB. Neurons were identified based on GCaMP6m expression and regions of Interest (ROIs) were placed on the neuron somas to calculate the raw GCaMP6m intensity of each neuron over time. The signal intensity of each trace was normalized to the baseline (ΔF/F0) for spike detection. The amplitude of Ca2+ spikes was calculated from the normalized GCaMp6m intensity for all the detected spikes in each trace (mean ΔF/F0 of detected spikes for each neuron). The frequency of Ca2+ spikes was calculated as the number of detected spikes in each trace per minute of recording.
Patch clamp electrophysiology
Neurons derived from dissociated MSK-SRF001 organoids were plated on 35mm dishes at a density of 250 000 cells/cm2. Neurons were cultured for 20 days before performing the patch clamp experiments. Cells were initially cultured in organoid media and slowly switch to BrainPhys.67 Whole cell patch clamp recordings using HEKA EPC10 USB amplifier were achieved by applying negative pressure upon giga-seal formation. Extracellular solution contained (in mM) 140 NaCl, 2 MgCl2, 2 CaCl2, 10 HEPES, 3 KCl, 10 D-Glucose, pH 7.4. Pipette solution contained (in mM) 4 NaCl, 120 K-gluconate, 10 HEPES, 10 EGTA, 3 Mg-ATP, 0.5 CaCl2, 1 MgCl2 pH 7.2. Patch pipettes ~7–9 MOhm were pulled from borosilicate glass capillaries (Harvard Apparatus, ì #30–0038) using a two-steps vertical pipette puller PC 100 (Narishige). Liquid junction correction of 10 mV was used in all experiments. For current clamp experiments cells were hold at 0 pA (resting membrane voltage) for 500 ms. Afterward, incrementally increasing currents (5 pA every sweep) were injected starting from −20 pA to +60 pA during 2000 ms. At the end of the stimulation period, cells were allowed to recover to resting conditions by clamping at 0 pA for 500 ms. Data were filtered at 10 kHz and digitized with 20–100 kHz. For voltage clamp experiments, cells were kept 300 ms at −80 mV and stimulated with stepwise increments of +10 mV during 1000 ms (−80 mV to +60 mV). Cells were allowed to recover by clamping to −80 mV during 300 ms at the end of the stimulation period. Peak Na+ currents were normalized to whole cell capacitance assessed after gaining access to the cell.
RNAscope
RNAscope for HOPX, LIFR and CDC42 was achieved according to the manufacturer’s protocol (RNAscope Multiplex Fluorescent Reagent Kit v2 Assay) with minor adaptations followed by an IF staining for GFAP. After the preparation of the OCT samples, as mentioned before, several serial sections of 10 um thickness were cut and subsequently air dried. Dried sections were washed for 5 min in PBS 1x to remove the OCT excess, baked for 30 min at 60°C and post-fixed in pre-chilled 10% NBF (Neutral Buffered Formalin) for 15 min at 4°C. The sections were then dehydrated in ethanol (50%,70% and 100% for 5 min each) and air dried. For the pretreatment steps, the sections were covered for 10 min at RT with RNAscope Hydrogen Peroxide, and then incubated 30 min at 40°C with RNAscope Protease III (RNAscope H2O2 and protease reagents ACDBio #322381).
A mix of the probes for HOPX-C1 (RTU, ACDBio #423001), LIFR-C2 (1:50, ACDBio #441021-C2) and CDC42-C3 (1:50, ACDBio #502651-C3) was prepared according to the manufacturer’s protocol, and used to incubate the samples for 2h at 40°C. The following steps of Amplification and Development of the signal were also performed following exactly the RNAscope Multiplex Fluorescent Reagent Kit v2 Assay protocol (ACDBio, #323110). The fluorophores we used were TSA Vivid 520 for HOPX (1:1000, ACDBio #323271), TSA Vivid 570 for LIFR (1:1500, ACDBio #323272) and TSA Vivid 650 for CDC42 (1:1500, ACDBio #323273).
Once completed the RNAscope protocol, but prior to DAPI staining, we performed an IF staining against GFAP. All the steps of the IF stainings were done at RT and avoiding the direct light exposure as following: after 30 min in Protein blocking buffer (DAKO #X0909) the samples were incubated for 1 h with 1:400 rabbit anti-GFAP (Dako. #Z033429–2) and, after two wash steps in PBS-T, with 1:400 donkey anti-rabbit AF594 (Invitrogen, #A11012) for 30 min. Then, sections were exposed to DAPI (RTU RNAscope Multiplex FL v2 DAPI, ACDBio,#323110) and the slides were mounted with Fluoromount-G (Invitrogen, #00–4958-02) and air dried for 30 min at RT.
QUANTIFICATION AND STATISTICAL ANALYSIS
The data presented in this study are representative of at least three independent batches of differentiation.
Rosettes were quantified with the LasX software. The length of the VZ was measured for the region marked by SOX2+ cells only. The length of the VZ/SVZ was measured, including the regions encompassing both SOX2+ cells (VZ) and the region marked by both SOX2+ EOMES+/TBR2+ cells until the denser layer of TBR2+ neural stem cells (SVZ). The oSVZ was defined as the region that contained SOX2+ TBR2+ cells outside the denser EOMES+/TBR2+ layer.
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications. Data distribution was assumed normal, but this was not formally tested. Data are presented as the mean ± sem. All statistical analyses were performed using GraphPad Prism 9: one-way ANOVA with Tukey’s tests to compare multiple groups and t test to compare two groups. Statistical differences were considered significant with p < 0.05 as indicated in figure legends. All reported measurements are from distinct samples. For scRNA-seq experiments, two independent batches of 10 organoids each were processed and analyzed by scRNA-seq for both control and LIF conditions on day 40, 60 and 100, whereas one batch of 10 organoids were processed and analyzed for control, LIF condition and CP assembloids on day 65.
Supplementary Material
Highlights.
LIF treatment induces the emergence of outer radial glia (oRG)
LIF-treated cortical organoids show an expansion of an outer subventricular zone (oSVZ)
Neural crest-derived LIF-producing pericytes partially recapitulate exogenous LIF treatment
The cellular microenvironment regulates neural progenitor diversity
ACKNOWLEDGMENTS
We thank all the Studer and Baggiolini lab members for their insightful comments and feedback on this manuscript. We would also like to thank the Flow Cytometry core, the Molecular Cytology core, the Integrated Genomic Operation at Memorial Sloan Kettering Cancer Center (MSKCC); the Imaging core, the Flow Cytometry core, and the Histology core at the Institute of Oncology Research (IOR); and Dr. David Penton and the Electrophysiology core facility at the University of Zurich (UZH) for outstanding technical and scientific support. Figures contain images created with BioRender. We acknowledge using the Integrated Genomics Operation Core, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. This work was supported in part through the NIH grants 1R01 MH135403, R01AG054720, and R21NS116545 (L.S.), the grant from the Department of Defense (DOD) AL200169 - W81XWH2110140 (L.S.), and the support from the Tri-institutional stem cell initiative. This work was also partially supported by the SNSF grant 211664 (A.B). A.B. was supported by the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center (GMTEC) Scholars Fellowship Program and by the Foundation for the Institute of Oncology Research. G.C. was supported by the European Molecular Biology Organization (EMBO) long-term fellowship (ALTF 311-2015) and the New York State Stem Cell Science (NYSTEM) postdoctoral training award (C32559GG); E.G. by the Rubicon (2020/30766/ZONMW); and R.W. by the NIH F32 (5F32MH116590-03). Processing of the human fetal brain specimen was funded by the European Union’s Horizon 2020 Research and Innovation Program under grant agreement no. 874758 - Consortium Nsc-Reconstruct: Novel Strategies for Cell based Neural Reconstruction (2020–23) awarded to E.C. and R.A.B. R.A.B. was supported by the NIHR Cambridge Biomedical Research Centre (NIHR203312). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. This research was funded in part by the Wellcome Trust 203151/Z/16/Z. For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) license to any Author Accepted Manuscript version arising from this submission. Ethics number is 096/085.
Footnotes
DECLARATION OF INTERESTS
L.S. is a scientific cofounder and paid consultant of BlueRock Therapeutics Inc. L.S. is a scientific cofounder of DaCapo Brainscience.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114031.
REFERENCES
- 1.Florio M, and Huttner WB (2014). Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194. 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 2.Shitamukai A, and Matsuzaki F. (2012). Control of asymmetric cell division of mammalian neural progenitors. Dev. Growth Differ 54, 277–286. 10.1111/j.1440-169X.2012.01345.x. [DOI] [PubMed] [Google Scholar]
- 3.Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, and Huttner WB (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci 13, 690–699. 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 4.Hansen DV, Lui JH, Parker PRL, and Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 5.Lewitus E, Kelava I, and Huttner WB (2013). Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Front. Hum. Neurosci 7, 424. 10.3389/fnhum.2013.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reillo I, de Juan Romero C, García-Cabezas MÁ, and Borrell V. (2011). A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex 21, 1674–1694. 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 7.Sun T, and Hevner RF (2014). Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci 15, 217–232. 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pașca SP, Arlotta P, Bateup HS, Camp JG, Cappello S, Gage FH, Knoblich JA, Kriegstein AR, Lancaster MA, Ming GL, et al. (2022). A nomenclature consensus for nervous system organoids and assembloids. Nature 609, 907–910. 10.1038/s41586-022-05219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, et al. (2019). Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell 176, 743–756.e17. 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, and Paşca SP (2017). Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 95, 779–790.e6. 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bignami A, and Dahl D. (1977). Specificity of the glial fibrillary acidic protein for astroglia. J. Histochem. Cytochem 25, 466–469. 10.1177/25.6.69656. [DOI] [PubMed] [Google Scholar]
- 12.Johnson K, Barragan J, Bashiruddin S, Smith CJ, Tyrrell C, Parsons MJ, Doris R, Kucenas S, Downes GB, Velez CM, et al. (2016). Gfap-positive radial glial cells are an essential progenitor population for later-born neurons and glia in the zebrafish spinal cord. Glia 64, 1170–1189. 10.1002/glia.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang SC (2001). Defining glial cells during CNS development. Nat. Rev. Neurosci 2, 840–843. 10.1038/35097593. [DOI] [PubMed] [Google Scholar]
- 14.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 112, 15672–15677. 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosebrock D, Arora S, Mutukula N, Volkman R, Gralinska E, Balaskas A, Aragonés Hernández A, Buschow R, Brändl B, Müller FJ, et al. (2022). Enhanced cortical neural stem cell identity through short SMAD and WNT inhibition in human cerebral organoids facilitates emergence of outer radial glial cells. Nat. Cell Biol 24, 981–995. 10.1038/s41556-022-00929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uzquiano A, Kedaigle AJ, Pigoni M, Paulsen B, Adiconis X, Kim K, Faits T, Nagaraja S, Antón-Bolaños N, Gerhardinger C, et al. (2022). Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell 185, 3770–3788.e27. 10.1016/j.cell.2022.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527. 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrini L, Bonfio C, Chadwick J, Begum F, Skehel M, and Lancaster MA (2020). Human CNS barrier-forming organoids with cerebrospinal fluid production. Science 369, eaaz5626. 10.1126/science.aaz5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer S, and Patterson PH (2006). Leukemia inhibitory factor promotes neural stem cell self-renewal in the adult brain. J. Neurosci 26, 12089–12099. 10.1523/JNEUROSCI.3047-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z, Li JJ, Zhen CH, Feng LY, and Ding XY (2006). Effect of leukemia inhibitory factor on embryonic stem cell differentiation: implications for supporting neuronal differentiation. Acta Pharmacol. Sin 27, 80–90. 10.1111/j.1745-7254.2006.00254.x. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, Coppola G, Pearson CA, Yamauchi K, Gong D, et al. (2017). Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 21, 517–532. 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, and Studer L. (2019). Specification of positional identity in forebrain organoids. Nat. Biotechnol 37, 436–444. 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, Jung D, Schmunk G, Haeussler M, Salma J, et al. (2020). Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148. 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verkerke M, Berdenis van Berlekom A, Donega V, Vonk D, Sluijs JA, Butt NF, Kistemaker L, de Witte LD, Pasterkamp RJ, Middeldorp J, and Hol EM (2024). Transcriptomic and morphological maturation of human astrocytes in cerebral organoids. Glia 72, 362–374. 10.1002/glia.24479. [DOI] [PubMed] [Google Scholar]
- 26.Lancaster MA, and Knoblich JA (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc 9, 2329–2340. 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporale N, Leemans M, Birgersson L, Germain PL, Cheroni C, Borbély G, Engdahl E, Lindh C, Bressan RB, Cavallo F, et al. (2022). From cohorts to molecules: Adverse impacts of endocrine disrupting mixtures. Science 375, eabe8244. 10.1126/science.abe8244. [DOI] [PubMed] [Google Scholar]
- 28.Matsumoto N, Tanaka S, Horiike T, Shinmyo Y, and Kawasaki H. (2020). A discrete subtype of neural progenitor crucial for cortical folding in the gyrencephalic mammalian brain. Elife 9, e54873. 10.7554/eLife.54873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews MG, Subramanian L, and Kriegstein AR (2020). mTOR signaling regulates the morphology and migration of outer radial glia in developing human cortex. Elife 9, e58737. 10.7554/eLife.58737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ostrem BEL, Lui JH, Gertz CC, and Kriegstein AR (2014). Control of outer radial glial stem cell mitosis in the human brain. Cell Rep. 8, 656–664. 10.1016/j.celrep.2014.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govek EE, Hatten ME, and Van Aelst L. (2011). The role of Rho GTPase proteins in CNS neuronal migration. Dev. Neurobiol 71, 528–553. 10.1002/dneu.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Li Z, Liu G, Li X, and Yang Z. (2022). Developmental Origins of Human Cortical Oligodendrocytes and Astrocytes. Neurosci. Bull 38, 47–68. 10.1007/s12264-021-00759-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwata R, Casimir P, Erkol E, Boubakar L, Planque M, Gallego López IM, Ditkowska M, Gaspariunaite V, Beckers S, Remans D, et al. (2023). Mitochondria metabolism sets the species-specific tempo of neuronal development. Science 379, eabn4705. 10.1126/science.abn4705. [DOI] [PubMed] [Google Scholar]
- 35.Lyons GE, Micales BK, Schwarz J, Martin JF, and Olson EN (1995). Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci 15, 5727–5738. 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 37.Su H, Cantrell AC, Zeng H, Zhu SH, and Chen JX (2021). Emerging Role of Pericytes and Their Secretome in the Heart. Cells 10, 548. 10.3390/cells10030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown LS, Foster CG, Courtney JM, King NE, Howells DW, and Sutherland BA (2019). Pericytes and Neurovascular Function in the Healthy and Diseased Brain. Front. Cell. Neurosci 13, 282. 10.3389/fncel.2019.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liddelow SA (2015). Development of the choroid plexus and blood-CSF barrier. Front. Neurosci 9, 32. 10.3389/fnins.2015.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dani N, Herbst RH, McCabe C, Green GS, Kaiser K, Head JP, Cui J, Shipley FB, Jang A, Dionne D, et al. (2021). A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell 184, 3056–3074.e21. 10.1016/j.cell.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tchieu J, Zimmer B, Fattahi F, Amin S, Zeltner N, Chen S, and Studer L. (2017). A Modular Platform for Differentiation of Human PSCs into All Major Ectodermal Lineages. Cell Stem Cell 21, 399–410.e7. 10.1016/j.stem.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan Y, Hackland J, Baggiolini A, Hung LY, Zhao H, Zumbo P, Oberst P, Minotti AP, Hergenreder E, Najjar S, et al. (2023). hPSC-derived sacral neural crest enables rescue in a severe model of Hirschsprung’s disease. Cell Stem Cell 30, 264–282.e9. 10.1016/j.stem.2023.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faal T, Phan DTT, Davtyan H, Scarfone VM, Varady E, Blurton-Jones M, Hughes CCW, and Inlay MA (2019). Induction of Mesoderm and Neural Crest-Derived Pericytes from Human Pluripotent Stem Cells to Study Blood-Brain Barrier Interactions. Stem Cell Rep. 12, 451–460. 10.1016/j.stemcr.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Lo L, Dormand E, and Anderson DJ (2003). SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 38, 17–31. 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 45.Giacomelli E, Bellin M, Sala L, van Meer BJ, Tertoolen LGJ, Orlova VV, and Mummery CL (2017). Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008–1017. 10.1242/dev.143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu K, Bendl J, Rahman S, Vicari JM, Coleman C, Clarence T, Latouche O, Tsankova NM, Li A, Brennand KJ, et al. (2023). Multi-omic profiling of the developing human cerebral cortex at the single-cell level. Sci. Adv 9, eadg3754. 10.1126/sciadv.adg3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skinnider MA, Squair JW, and Foster LJ (2019). Evaluating measures of association for single-cell transcriptomics. Nat. Methods 16, 381–386. 10.1038/s41592-019-0372-4. [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi M, Niidome T, Matsuda S, Akaike A, Kihara T, and Sugimoto H. (2007). Microglia-derived interleukin-6 and leukaemia inhibitory factor promote astrocytic differentiation of neural stem/progenitor cells. Eur. J. Neurosci 25, 649–658. 10.1111/j.1460-9568.2007.05309.x. [DOI] [PubMed] [Google Scholar]
- 49.Ciceri G, Baggiolini A, Cho HS, Kshirsagar M, Benito-Kwiecinski S, Walsh RM, Aromolaran KA, Gonzalez-Hernandez AJ, Munguba H, Koo SY, et al. (2024). Nature 626, 881–890. 10.1038/s41586-023-06984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delgado RN, Allen DE, Keefe MG, Mancia Leon WR, Ziffra RS, Crouch EE, Alvarez-Buylla A, and Nowakowski TJ (2022). Individual human cortical progenitors can produce excitatory and inhibitory neurons. Nature 601, 397–403. 10.1038/s41586-021-04230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews MG, Siebert C, Wang L, White ML, Ross J, Morales R, Donnay M, Bamfonga G, Mukhtar T, McKinney AA, et al. (2023). LIF signaling regulates outer radial glial to interneuron fate during human cortical development. Cell Stem Cell 30, 1382–1391.e5. 10.1016/j.stem.2023.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, and Zhang F. (2017). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc 12, 828–863. 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu C, Prete M, Webb S, Jardine L, Stewart BJ, Hoo R, He P, Meyer KB, and Teichmann SA (2023). Automatic cell-type harmonization and integration across Human Cell Atlas datasets. Cell 186, 5876–5891.e20. 10.1016/j.cell.2023.11.026. [DOI] [PubMed] [Google Scholar]
- 59.Domínguez Conde C, Xu C, Jarvis LB, Rainbow DB, Wells SB, Gomes T, Howlett SK, Suchanek O, Polanski K, King HW, et al. (2022). Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197. 10.1126/science.abl5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.González F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, and Huangfu D. (2014). An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15, 215–226. 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M, Zhong A, Wu Y, Sidharta M, Beaury M, Zhao X, Studer L, and Zhou T. (2022). Transient inhibition of p53 enhances prime editing and cytosine base-editing efficiencies in human pluripotent stem cells. Nat. Commun 13, 6354. 10.1038/s41467-022-34045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong A, Li M, and Zhou T. (2020). Protocol for the Generation of Human Pluripotent Reporter Cell Lines Using CRISPR/Cas9. STAR Protoc. 1, 100052. 10.1016/j.xpro.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678. 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baggiolini A, Callahan SJ, Montal E, Weiss JM, Trieu T, Tagore MM, Tischfield SE, Walsh RM, Suresh S, Fan Y, et al. (2021). Developmental chromatin programs determine oncogenic competence in melanoma. Science 373, eabc1048. 10.1126/science.abc1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, Gorris M, Galet B, Palomares V, Brown J, et al. (2015). Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. USA 112, E2725–E2734. 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Z, and Südhof TC (2021). A simple Ca(2+)-imaging approach to neural network analyses in cultured neurons. J. Neurosci. Methods 349, 109041. 10.1016/j.jneumeth.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table. Datasets generated in this study have been deposited at Gene Expression Omnibus (GEO): GSE224346.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| SOX2 Monoclonal antibody (Btjce), eBioscience | Thermo Fisher scientific | Cat#14-9811-82; RRID:AB_11219471 |
| EOMES Monoclonal Antibody (WD1928), eBioscience | Thermo Fisher Scientific | Cat#14-4877-82; RRID:AB_2572882 |
| Recombinant Anti-TBR1 antibody [EPR8138(2)] | Abcam | Cat#ab183032; RRID:AB_2936859 |
| phospho-vimentin (Ser55) | MBL International | Cat#D076-3; RRID:AB_592963 |
| Anti-HOPX polyclonal antibody | Sigma-Aldrich | Cat#HPA030180; RRID:AB_10603770 |
| Glial Fibrillary Acidic Protein | Agilent | Cat#Z033429-2; RRID:AB_10013382 |
| Purified Mouse Anti-Nestin | BD Biosciences | Cat#611658; RRID:AB_399176 |
| CD31, Endothelial Cell Antibody (Concentrate) | Agilent | Cat#M0823; RRID:AB_2114471 |
| Monoclonal Anti-Actin, α-Smooth Muscle | Sigma-Aldrich | Cat#A2547; RRID:AB_476701 |
| Anti-LIF antibody [IMG39N7D10] | Abcam | Cat#ab138002; RRID:AB_3083551 |
| PDGFRB Monoclonal Antibody (G.290.3) | Thermo Fisher Scientific | Cat#MA5-15143; RRID:AB_10985851 |
| Anti-NG2 antibody | Abcam | Cat#ab129051; RRID:AB_2877152 |
| MAP2 antibody - Neuronal Marker | Abcam | Cat#ab5392; RRID:AB_2138153 |
| Rabbit Anti-Synapsin I Antibody, Unconjugated | Sigma-Aldrich | Cat#S193; RRID:AB_261457 |
| PSD-95 Monoclonal Antibody (7E3-1B8) | Thermo Fisher Scientific | Cat#MA1-046; RRID:AB_2092361 |
| GFAP Antibody | Novus Bio | Cat#NBP1-05198; RRID:AB_1556315 |
| CD271 (NGF Receptor) Monoclonal Antibody (ME20.4), PE, eBioscience | Thermo Fisher Scientific | Cat#12-9400-42; RRID:AB_2572710 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 |
Thermo Fisher Scientific | Cat#A32790; RRID:AB_2762833 |
| Donkey anti-Rat IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 555 |
Thermo Fisher Scientific | Cat#A48270; RRID:AB_2896336 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 |
Thermo Fisher Scientific | Cat#A31571; RRID:AB_162542 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 594 |
Thermo Fisher Scientific | Cat#A32754; RRID:AB_2762827 |
| Donkey anti-Chicken IgY (H + L) Highly Cross Adsorbed Secondary Antibody, Alexa Fluor 488 |
Thermo Fisher Scientific | Cat#A78948; RRID:AB_2921070 |
| HRP Anti-beta Actin antibody [AC-15] | Abcam | Cat#ab49900; RRID:AB_867494 |
| Anti-rat IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7077; RRID:AB_10694715 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| One Shot Stbl3 Chemically Competent E. coli | Thermo Fisher Scientific | Cat#C737303 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Essential 8 medium | Thermo Fisher Scientific | Cat#A1517001 |
| Vitronectin (VTN-N) Recombinant Human Protein, Truncated | Thermo Fisher Scientific | Cat#A14700 |
| UltraPure 0.5M EDTA, pH 8.0 | Thermo Fisher Scientific | Cat#15575020 |
| ReLeSR | StemCell Technologies | Cat#100-0483 |
| Accutase solution | Sigma-Aldrich | Cat#A6964 |
| ROCK inhibitor Y-27632 | Bio-Techne (Tocris) | Cat#1254 |
| Essential 6 Medium | Thermo Fisher Scientific | Cat#A1516401 |
| LDN-193189 | Reprocell (Stemgent) | Cat#04-0074 |
| SB 431542 | Bio-Techne (Tocris) | Cat#1614 |
| XAV 939 | Bio-Techne (Tocris) | Cat#3748 |
| Neurobasal Medium | Thermo Fisher Scientific | Cat#21103049 |
| DMEM/F-12, HEPES | Thermo Fisher Scientific | Cat#31330038 |
| B-27 Supplement (50X), minus vitamin A | Thermo Fisher Scientific | Cat#12587010 |
| N-2 Supplement (100X) | Thermo Fisher Scientific | Cat#17502048 |
| GlutaMAX Supplement | Thermo Fisher Scientific | Cat#35050038 |
| 2-Mercaptoethanol | Thermo Fisher Scientific | Cat#21985023 |
| Normocin - Antimicrobial Reagent | Invivogen | Cat#ant-nr-2 |
| Insulin solution human | Sigma-Aldrich | Cat#I9278 |
| Recombinant Human LIF | Peprotech | Cat#300-05 |
| STEMdiff Cerebral Organoid Kit | StemCell Technologies | Cat#8570 |
| Matrigel Basement Membrane Matrix, LDEV-free, 10 mL | Corning | Cat#354234 |
| KnockOut Serum Replacement | Thermo Fisher Scientific | Cat#10828028 |
| MEM Non-Essential Amino Acids Solution (100X) | Thermo Fisher Scientific | Cat#11140050 |
| Penicillin-Streptomycin | Sigma-Aldrich | Cat#P4333 |
| Dorsomorphin | MedChemExpress | Cat#HY-13418A |
| Recombinant Human FGF-basic (154 a.a.) | Peprotech | Cat#100-18B |
| Animal-Free Recombinant Human EGF | Peprotech | Cat#AF-100-15 |
| Recombinant Human/Murine/Rat BDNF | Peprotech | Cat#450-02 |
| Recombinant Human NT-3 | Peprotech | Cat#450-03 |
| Recombinant Human BMP-4 Protein | Bio-Techne (R&D Systems) | Cat#314-BP |
| CHIR 99021 | Bio-Techne (Tocris) | Cat#4423 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Pericyte Medium | Sciencell Research Laboratories |
Cat#1201 |
| Trypsin-EDTA (0.05%) in DPBS (1x) | Capricorn Scientific | Cat#TRY-1B |
| Puromycin dihydrochloride from Streptomyces alboniger | Sigma-Aldrich | Cat#P8833 |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D5207 |
| RiboLock RNase Inhibitor (40 U/μL) | Thermo Fisher Scientific | Cat#EO0381 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat#A9418 |
| Poly-L-ornithine hydrobromide | Sigma-Aldrich | Cat#P3655 |
| Cultrex Mouse Laminin I, Pathclear | Bio-Techne (R&D Systems) | Cat#3400-010-02 |
| Fibronectin Human Protein, Plasma | Thermo Fisher Scientific | Cat#33016015 |
| Click-iT™ Plus EdU Cell Proliferation Kit for Imaging | Thermo Fisher Scientific | Cat #C10640 |
| BrainPhys™ Neuronal Medium and SM1 Kit | StemCell Technologies | Cat #05792 |
| Halt Protease and Phosphatase Inhibitor Cocktail (100X) | Thermo Fisher Scientific | Cat#78440 |
| NuPAGE LDS Sample Buffer (4X) | Thermo Fisher Scientific | Cat#NP0007 |
| NuPAGE™ 4 to 12%, Bis-Tris, 1.0-1.5 mm, Mini Protein Gels | Thermo Fisher Scientific | Cat#NP0335BOX |
| NuPAGE™ MES SDS Running Buffer (20X) | Thermo Fisher Scientific | Cat#NP0002 |
| Nitrocellulose Membranes, 0.45 μm | Thermo Fisher Scientific | Cat#88018 |
| NuPAGE™ Transfer Buffer (20X) | Thermo Fisher Scientific | Cat#NP0006 |
| Nonfat Dry Milk | Cell Signaling Technology | Cat#9999 |
| OCT EMBEDDING MATRIX - 125mL | CellPath | Cat#KMA-0100-00A |
| Normal Donkey Serum | Abcam | Cat#ab7475 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| DPBS, calcium, magnesium | Thermo Fisher Scientific | Cat#14040091 |
| TWEEN 20 | Sigma-Aldrich | Cat#P9416 |
| Normal Goat Serum Blocking Solution | Vector Laboratories | Cat#S-1000-20 |
| Hoechst 33258, Pentahydrate (bis-Benzimide) - 10 mg/mL Solution in Water |
Thermo Fisher Scientific | Cat#H3569 |
| Glycergel Mounting Medium, Aqueous | Agilent | Cat#C056330-2 |
| BrainPhys Imaging Optimized Medium | StemCell Technologies | Cat#05796 |
| Fluoromount-G Mounting Medium | Thermo Fisher Scientific | Cat#00-4958-02 |
| Neutral Buffered Formalin | Epredia | Cat#5701 |
| DPBS, no calcium, no magnesium | Thermo Fisher Scientific | Cat#14190094 |
| EGM-2 Endothelial Cell Growth Medium-BulletKit | Lonza | Cat#CC-3162 |
| IMDM | Thermo Fisher Scientific | Cat# 21980032 |
| Fetal Bovine Serum Advanced (FBS Advanced), Collected in South America |
Capricorn Scientific | Cat# FBS-11A |
| RNAscope H202 and Protease Reagents | ACD Bio | Cat#322381 |
| RNAscope Probe- Hs-HOPX | ACD Bio | Cat#423001 |
| RNAscope Probe- Hs-LIFR | ACD Bio | Cat#441021 |
| RNAscope Probe- Hs-CDC42-O1-C3 | ACD Bio | Cat#502651-C3 |
| TSA Vivid Fluorophore 520 | ACD Bio | Cat#323271 |
| TSA Vivid Fluorophore 570 | ACD Bio | Cat#323272 |
| TSA Vivid Fluorophore 650 | ACD Bio | Cat#323273 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Human LIF ELISA kit | RayBiotech | Cat#ELH-LIF1 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| Chromium Single Cell 3ʹ GEM, Library & Gel Bead Kit v3 | 10x Genomics | Cat# PN-1000075 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | ACD Bio | Cat#323110 |
| Papain Dissociation System | Worthington Biochemical | Cat#LK003150 |
|
| ||
| Deposited data | ||
|
| ||
| Single Cell RNA sequencing dataset Generation of human cerebral organoids with a structured outer subventricular zone” |
This manuscript | GEO Accession #: GSE224346 |
| Single Cell RNA sequencing dataset “Comparison of Cortical Organoids to Human Cortex Identifies Cellular Identity and Maturation Differences” | Bhaduri et al.24 | GEO Accession: GSE132672 |
| Single Cell RNA sequencing dataset “ | Nowakowski et al.29 | dbGaP Accession: phs000989.v3.p1 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| WA09 | WiCell | RRID:CVCL_9773 |
| WA01 | WiCell | RRID:CVCL_9771 |
| HUVEC-C | ATCC | Cat # CRL-1730 RRID:CVCL_2959 |
| HEK293T | ATCC | Cat # CRL-3216 RRID:CVCL_0063 |
|
| ||
| Oligonucleotides | ||
|
| ||
| TCCGTCACAGACTAAGGAGA | IDT | sgRNA targeting HOPX locus |
| GCGGGAAGTCCGTCACGTTG | IDT | sgRNA targeting LIF locus |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pX330-U6-Chimeric_BB-CBh-hSpCas9 | Cong et al.52 | Addgene #42230 |
| Plasmid: lenti sgRNA(MS2)_puro optimized backbone | Joung et al.53 | Addgene #73797 |
| Plasmid: psPAX2 | psPAX2 was a gift from Didier Trono | Addgene #12260 |
| Plasmid: pMD2.G | pMD2.G was a gift from Didier Trono | Addgene #12259 |
| Plasmid: pGP-CMV-GCaMP6m | Chen et al.54 | Addgene #40754 |
|
| ||
| Software and algorithms | ||
|
| ||
| 10x Genomics Cellranger DNA | 10x Genomics | RRID:SCR_023221 |
| Seurat | Butler et al.55 | RRID:SCR_016341 |
| fgsea | Bioconductor | RRID:SCR_020938 |
| scVelo | La Manno et al.56 | RRID:SCR_018168 |
| LoomPy | Linnarsson Lab |
https://loompy.org RRID:SCR_016666 |
| scanpy | Wolf et al.57 | RRID:SCR_018139 |
| Celltypist | Xu et al.58 Dominguez Conde59 |
www.celltypist.org/RRID:SCR_024893 |
| Biorender | Biorender | www.biorender.com |
| LASX Office 1.4.5 27713 | Leica Microsystems | www.leica-microsystems.com |
| GraphPad Prism | GraphPad Software | http://www.graphpad.com/scientific-software/prism/ RRID: SCR_002798 |
|
| ||
| Other | ||
|
| ||
| PrimeSurface® 3D culture: Ultra-low Attachment Plates: 96 well, V bottom, Clear plates |
S-Bio | Cat#MS-9096VZ |