Abstract
Pseudomonas putida M10 was originally isolated from factory waste liquors by selection for growth on morphine. The NADP(+)-dependent morphine dehydrogenase that initiates morphine catabolism is encoded by a large plasmid of 165 kb. Treatment of P. putida M10 with ethidium bromide led to the isolation of a putative plasmid-free strain that was incapable of growth on morphine. The structural gene for morphine dehydrogenase, morA, has been located on the plasmid by oligonucleotide hybridization, by coupled transcription-translation of cloned restriction fragments and by nucleotide sequence analysis and is contained within a 1.7 kb SphI fragment that has been cloned into Escherichia coli. The cloned dehydrogenase enzyme is expressed at high levels in E. coli resulting in a 65-fold increase in morphine dehydrogenase activity in cell-free extracts compared with P. putida M10. Morphine dehydrogenase was rapidly purified to homogeneity, as judged by SDS/PAGE, by a one-step affinity chromatography procedure on Mimetic Orange 3 A6XL. The properties of the purified enzyme were identical with those previously reported for P. putida M10 morphine dehydrogenase. The morA gene was sequenced and the deduced amino acid sequence confirmed by N-terminal amino acid sequencing of the over-expressed protein. The predicted amino acid sequence of morA, deduced from the nucleotide sequence, indicated that morphine dehydrogenase did not belong to the non-metal-requiring short-chain class of dehydrogenases, but was more closely related to the aldo-ketoreductases.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Marks C. B., Lazarus R., Miller J., Stafford K., Seymour J., Light D., Rastetter W., Estell D. Production of 2-Keto-L-Gulonate, an Intermediate in L-Ascorbate Synthesis, by a Genetically Modified Erwinia herbicola. Science. 1985 Oct 11;230(4722):144–149. doi: 10.1126/science.230.4722.144. [DOI] [PubMed] [Google Scholar]
- Bohren K. M., Bullock B., Wermuth B., Gabbay K. H. The aldo-keto reductase superfamily. cDNAs and deduced amino acid sequences of human aldehyde and aldose reductases. J Biol Chem. 1989 Jun 5;264(16):9547–9551. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
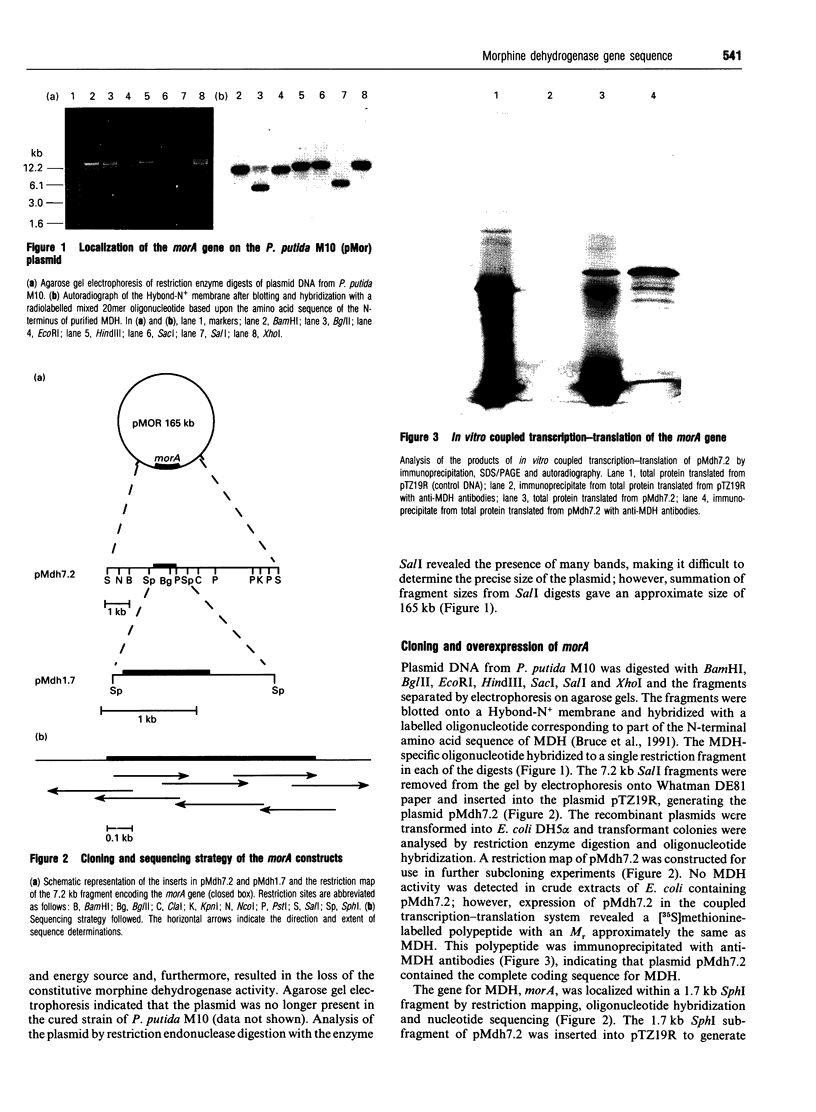
- Bruce N. C., Wilmot C. J., Jordan K. N., Stephens L. D., Lowe C. R. Microbial degradation of the morphine alkaloids. Purification and characterization of morphine dehydrogenase from Pseudomonas putida M10. Biochem J. 1991 Mar 15;274(Pt 3):875–880. doi: 10.1042/bj2740875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce N. C., Wilmot C. J., Jordan K. N., Trebilcock A. E., Gray Stephens L. D., Lowe C. R. Microbial degradation of the morphine alkaloids: identification of morphine as an intermediate in the metabolism of morphine by Pseudomonas putida M10. Arch Microbiol. 1990;154(5):465–470. doi: 10.1007/BF00245229. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Flinta C., Persson B., Jörnvall H., von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem. 1986 Jan 2;154(1):193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez A., Martin B., Murphy H. R., Uchida S., Murer H., Cowley B. D., Jr, Handler J. S., Burg M. B. Molecular cloning of cDNA coding for kidney aldose reductase. Regulation of specific mRNA accumulation by NaCl-mediated osmotic stress. J Biol Chem. 1989 Oct 5;264(28):16815–16821. [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., von Bahr-Lindström H., Jany K. D., Ulmer W., Fröschle M. Extended superfamily of short alcohol-polyol-sugar dehydrogenases: structural similarities between glucose and ribitol dehydrogenases. FEBS Lett. 1984 Jan 9;165(2):190–196. doi: 10.1016/0014-5793(84)80167-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Morris H. R., Williams D. H., Midwinter G. G., Hartley B. S. A mass-spectrometric sequence study of the enzyme ribitol dehydrogenase from Klebsiella aerogenes. Biochem J. 1974 Sep;141(3):701–713. doi: 10.1042/bj1410701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C., Wistow G., Carper D. Rat lens aldose reductase: characterization of its primary structure, mRNA, and gene complexity. Prog Clin Biol Res. 1989;290:211–220. [PubMed] [Google Scholar]
- Oechsner U., Magdolen V., Bandlow W. A nuclear yeast gene (GCY) encodes a polypeptide with high homology to a vertebrate eye lens protein. FEBS Lett. 1988 Sep 26;238(1):123–128. doi: 10.1016/0014-5793(88)80240-0. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondeau J. M., Tête-Favier F., Podjarny A., Reymann J. M., Barth P., Biellmann J. F., Moras D. Novel NADPH-binding domain revealed by the crystal structure of aldose reductase. Nature. 1992 Jan 30;355(6359):469–472. doi: 10.1038/355469a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade S. Z., Early S. L., Williams T. R., Kézdy F. J., Heinrikson R. L., Grimshaw C. E., Doughty C. C. Sequence analysis of bovine lens aldose reductase. J Biol Chem. 1990 Mar 5;265(7):3628–3635. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Winters C. J., Molowa D. T., Guzelian P. S. Isolation and characterization of cloned cDNAs encoding human liver chlordecone reductase. Biochemistry. 1990 Jan 30;29(4):1080–1087. doi: 10.1021/bi00456a034. [DOI] [PubMed] [Google Scholar]