Abstract
Background
Metagenomic binning, the clustering of assembled contigs that belong to the same genome, is a crucial step for recovering metagenome-assembled genomes (MAGs). Contigs are linked by exploiting consistent signatures along a genome, such as read coverage patterns. Using coverage from multiple samples leads to higher-quality MAGs; however, standard pipelines require all-to-all read alignments for multiple samples to compute coverage, becoming a key computational bottleneck.
Results
We present fairy (https://github.com/bluenote-1577/fairy), an approximate coverage calculation method for metagenomic binning. Fairy is a fast k-mer-based alignment-free method. For multi-sample binning, fairy can be faster than read alignment and accurate enough for binning. Fairy is compatible with several existing binners on host and non-host-associated datasets. Using MetaBAT2, fairy recovers of MAGs with completeness and contamination relative to alignment with BWA. Notably, multi-sample binning with fairy is always better than single-sample binning using BWA ( more complete MAGs on average) while still being faster. For a public sediment metagenome project, we demonstrate that multi-sample binning recovers higher quality Asgard archaea MAGs than single-sample binning and that fairy’s results are indistinguishable from read alignment.
Conclusions
Fairy is a new tool for approximately and quickly calculating multi-sample coverage for binning, resolving a computational bottleneck for metagenomics.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01861-6.
Keywords: Metagenomic binning, Coverage calculation, K-mers, Alignment-free
Background
Direct shotgun sequencing of microbiomes has allowed for the recovery of metagenome-assembled genomes (MAGs), unlocking unprecedented insights into the ecology of even unculturable organisms [1]. The computational process of generating MAGs first requires assembling the sequenced reads into contiguous sequences called contigs. After assembly, contigs are grouped into MAGs by binning through either automated algorithms [2–5] or manual curation [6].
Binning is done by leveraging consistent information across an entire genome. Such genomic signatures [7] include k-mer frequencies (e.g., tetranucleotide frequencies) and sequencing coverage, which are commonly used in binning algorithms. Combining coverage information from multiple samples is an effective way of increasing the resolving power of binning algorithms [8]. It has been shown that multi-sample coverage is vastly superior to single-sample coverage for binning and produces better MAGs that even quality-control software such as CheckM [9] may not be able to detect [10].
Coverage calculation is usually done by aligning reads back to contigs (e.g., using BWA [11] or BowTie2 [12]). For a project with n samples and n resulting assemblies, computing multi-sample coverage naively requires aligning each sample to each assembly, resulting in read-alignment runs. This quadratic scaling becomes prohibitive when the number of samples is large. Co-assembly, where all reads are assembled to give one set of contigs, is a potential solution, but co-assembly can be memory intensive and collapse similar strains [13]. Another method is split-binning [4], where all contigs from a set of assemblies are concatenated together and then aligned to. This is faster but still relatively time and memory-intensive. Thus, many large-scale studies still do single-sample binning [14].
If only coverage is needed, read alignment is computationally wasteful because the exact base alignments are not needed. Alignment-free methods are faster and provide an intriguing alternative. For example, pseudoalignment [15, 16] has been used for coverage calculation [17, 18]. Additionally, direct k-mer counts have also been used to separate strains in de Bruijn graphs [19] and applied to multi-sample coverage for binning [20].
Our contributions
In this paper, we present a much faster, alignment-free method of computing multi-sample coverage for metagenomic binning. Our method, fairy, is built on top of our metagenomic profiler sylph [21], but fairy is specifically adapted for metagenomic binning of contigs.
Fairy’s algorithm is related to previously developed k-mer sketching algorithms [22, 23], but we are not aware of detailed investigations justifying their effectiveness for the specific task of MAG recovery. Thus, we investigate the effectiveness of fairy on a diverse set of environments and methods. We confirm that fairy recovers multi-sample bins of similar quality relative to alignment at a fraction of the runtime, justifying the usage of k-mer sketching techniques and paving the way for more computationally efficient pipelines.
Methods
The key computational insight of fairy is that indexing metagenomic reads with a sparse set of k-mers is much more efficient than indexing genomes for coverage calculation. This is due to the redundancy of k-mers within the reads, allowing fairy to be 2–3 orders of magnitude faster than alignment. A variant of this technique was previously employed for compressively accelerated all-mapping, but that task still required base-level alignment, whereas fairy eschews that step [24].
Repurposing a fast metagenomic profiler for approximate coverage computation
Fairy’s codebase was forked and independently developed from sylph, a metagenomic profiler we developed [21]. However, fairy is extended and repurposed for metagenomic binning. Fairy and sylph are algorithmically similar, but they differ in three ways:
Default parameter choices. We will make the differences explicit below.
User interface. This includes command line options, inputs, and output formats―fairy is a coverage calculator rather than a metagenomic profiler.
A coverage variance computation step. This is useful for some binners such as MetaBAT2 [2].
Fairy’s output is the same as the commonly used jgi_summarize_bam_contig_depths script from MetaBAT2 and can be toggled to be compatible with binners such as MaxBin2 and SemiBin2 as well. We recapitulate fairy’s key algorithmic steps below, also shown in Fig. 1A. We outline technical details pertaining to sylph’s algorithm in Additional file 1: Supplementary Methods.
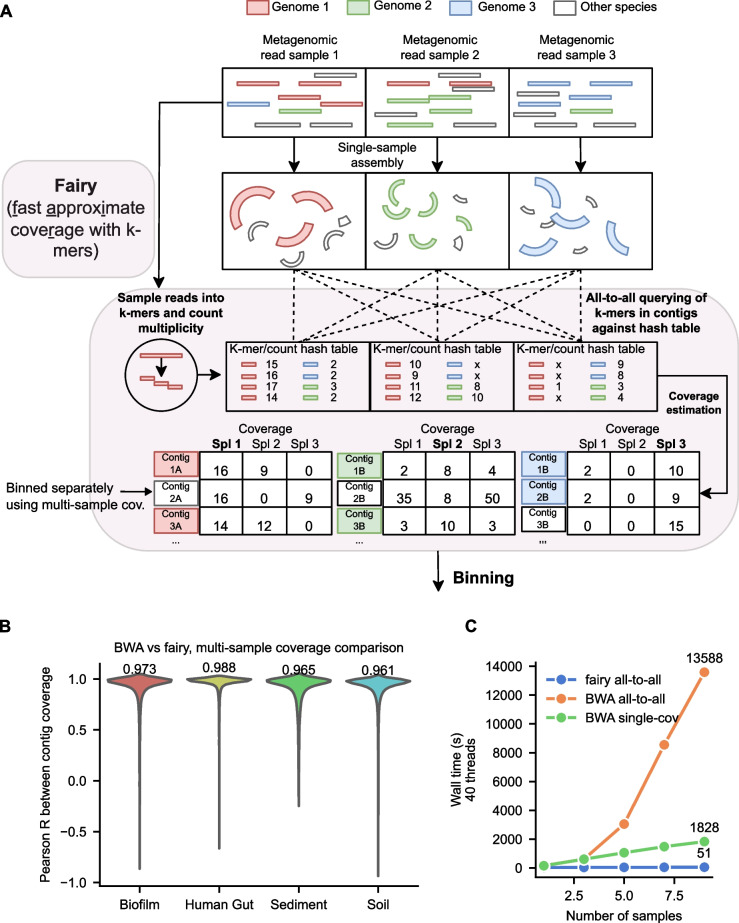
Fig. 1.
Fast approximate k-mer coverage estimates for multi-sample metagenomic binning. A Outline of fairy’s k-mer-based algorithm. Fairy’s processing steps are outlined in light red. Fairy indexes (or sketches) the reads into subsampled k-mer-to-count hash tables. K-mers from contigs are then queried against the hash tables to estimate coverage. Finally, fairy’s output is used for binning and is compatible with several binners (e.g., MetaBAT2, MaxBin2). Notice that white contigs (e.g., Contig 2A) have similar coverage to the colored contigs in the same sample, but additional samples help clarify that the white contigs should be binned separately. B Pearson R values between fairy and BWA’s multi-sample coverages for contigs in an arbitrary assembly from the dataset. Median values are shown above the plots. C Wall time with 40 threads for fairy vs BWA on 1, 3, 5, 7, and 9 gut samples in all-to-all mode (multi-sample) and single-sample mode
Sparse sketching of k-mers
Fairy sparsely samples k-mers from reads and assemblies using the FracMinHash [25] method to sample approximately 1/50 k-mers (whereas sylph defaults to a 1/200 sampling rate). For each metagenomic sample, the k-mers in the reads and their associated multiplicities within the sample are stored in a hash table. The hash table indices (one for each sample) are written to disk and loaded as needed, whereas the assemblies’ k-mers are kept in memory.
Contig querying and containment
For an assembly, every contig’s sampled k-mers are queried against every metagenomic sample’s hash table. For a contig, fairy requires 8 k-mers at minimum to be contained in the sample (whereas sylph requires 50) to proceed with coverage calculation. In addition to the minimum k-mer threshold, fairy calculates a containment ANI in the same manner as sylph (Additional file 1: Supplementary Methods).
Containment ANI measures the average nucleotide identity of the contig to all sequences in the metagenome. Intuitively, the containment ANI is a continuous absence-presence measure for the contig in the reads. For example, if only one of a contig’s 1000 k-mers is present in the reads’ k-mers, it will have low containment ANI and is likely not present in the sample. If the containment ANI is , fairy assumes the contig is not present in the sample at species-level, and thus assigns a coverage of 0.
We choose 95% as a presence-absence threshold by default, corresponding to species-level ANI thresholds used in practice [26]. Strain-level MAG binning from strain-resolved assemblies (e.g., with PacBio HiFi reads [27]) may require higher ANI thresholds, but higher thresholds would reduce sensitivity for multi-sample binning. In the “Results” section, we discuss some issues with the 95% threshold for PacBio HiFi MAG binning.
Coverage calculation from k-mers
For a contig passing the containment ANI threshold, the coverage is estimated in three different ways. Let M be the median k-mer multiplicity of the contig’s k-mers within the sample. We calculate the coverage exactly as is done with sylph’s “effective coverage” estimator [21], briefly restated below (see Additional file 1: Supplementary Methods for more information). Fairy’s output coverage is:
If : a statistical estimator using Poisson coverage assumptions.
If : a robust mean of k-mer counts, trimming off large k-mer counts.
If : median of the k-mer counts.
Coverage variance calculation
The variance is calculated as the sample variance of the contig’s non-zero k-mer multiplicities in the sample with the 10 and 90 percentile k-mer multiplicities trimmed to remove long-tailed k-mer coverage outliers (e.g., due to mobile elements).
Why fairy is fast for multi-sample computation
Fairy’s speed comes from the fact that each sample’s reads are only processed once. Processing reads into hash tables turns out to be far more computationally costly than querying a contig’s k-mers against the hash table. Thus, for n samples, fairy only requires n costly read-processing steps and fast query procedures. This makes fairy scale well for multi-sample binning compared to read alignment.
Benchmarking procedure
We benchmark on real datasets with multiple samples [28–34] and subsetted the samples when the number was , the size of the largest dataset, to avoid extremely long computations; exact accessions are available in Additional file 3: Supplementary Table 2. We opted to benchmark on real data using CheckM2 as opposed to synthetic data to show that fairy has good performance on realistic datasets. It has been found that CheckM’s relative performance is strongly correlated with ground-truth performance on simulated data [35]. In particular, we primarily care about relative results rather than true contamination/completeness.
Assembly and MAG generation
For generating assemblies and MAGs using read alignment for binning, our procedure was as follows:
Short-read metagenomes: we generated assemblies using ATLAS [36] v2.18.1 with default settings. We mapped short-reads using BWA and used CoverM [37] with the -m metabat option, which is identical to using the jgi_summarize_bam_contig_depths script from MetaBAT2 with default settings.
Nanopore long-read metagenomes: we generated assemblies using metaFlye [38] with default settings. We mapped reads using minimap2 [39] and used the jgi_summarize_bam_contig_depths script with minimum mapping quality 5 and alignment identity (as opposed to default 97%) due to lower accuracy of nanopore reads.
PacBio HiFi metagenomes: we used MetaMDBG [40] for assembly and minimap2 with jgi_summarize_bam_contig_depths (default settings) for coverage calculation.
For all above data types, fairy v0.5.1 was then used with default parameters on the resulting assemblies and reads to generate another set of coverage profiles. Fairy’s coverage profiles were used to generate another set of MAGs that we compared to the MAGs generated from alignment-based coverages.
Binning and evaluation
For short-read assemblies, binning was conducted with MetaBAT2 v2.15 [2], MaxBin2 v2.2.7 [3], MetaBinner v1.4.4 [5], VAMB v3.0.9 [4], and SemiBin v2.1 [41] (with the -a option). Notably, we used an older version of VAMB because the newer version required BAM inputs, which fairy can not generate. For long-read assemblies, we only used MetaBAT2, as it seemed to be more commonly used for existing long-read pipelines [27, 33, 40]. All binners were run with standard settings, except using a minimum contig length of 1500 when possible to emulate existing pipelines [10]. Some binners can only be run with a BAM file as opposed to a custom coverage profile, so we could not benchmark them [42]. Finally, we use CheckM2 [9] to evaluate the contamination and completeness of the resulting bins.
Results
Fairy is concordant with read-alignment coverage while being times faster
For four short-read datasets (Fig. 1B), we took one arbitrary assembly and examined all present contigs. For n samples, each contig has n coverage values. We calculated the Pearson correlation between the n coverages that fairy output and BWA’s n coverages. The median Pearson R value was for all datasets, indicating good concordance. Sediment and soil metagenomes are often more complex (i.e., more low-abundance organisms and overall diversity) than gut metagenomes, likely explaining the lower concordance between BWA and fairy on these datasets (0.988 for gut vs 0.965 for sediment and 0.961 for soil).
Fairy is more than faster than BWA for multi-sample coverage (Fig. 1C) when using just 9 of the short-read human gut metagenomes. While fairy technically requires a quadratic number of coverage calculations, much of the processing is in the linear-time indexing step. For read-alignment, the quadratic time cost of all-to-all alignment is clear for even a small number of samples.
Multi-sample coverage calculation quickly becomes a bottleneck, but not for fairy
For the soil dataset (10 samples) with 40 cores, assembly with SPAdes took approximately 15 h, while read alignment took more than 40 h. Theoretically, if we used 100 samples instead of 10, assembly would take h and finish within a week, whereas all-to-all alignment would take h (around 167 days), which is not feasible. On this dataset, fairy took 9 minutes for indexing and 7 minutes for querying; most of the querying time was spent on disk I/O, and we did not even use an SSD. In the worst case, 100 samples would still take less than a day for fairy.
For memory, fairy scales with the size and complexity of the metagenome. Processing the 10 aforementioned soil samples in parallel took 40 GB of RAM (4 GB per sample), which is still relatively small compared to assembly, and if memory is a constraint, fewer samples can be processed in parallel.
Multi-sample binning with fairy is better than single-sample binning with alignment
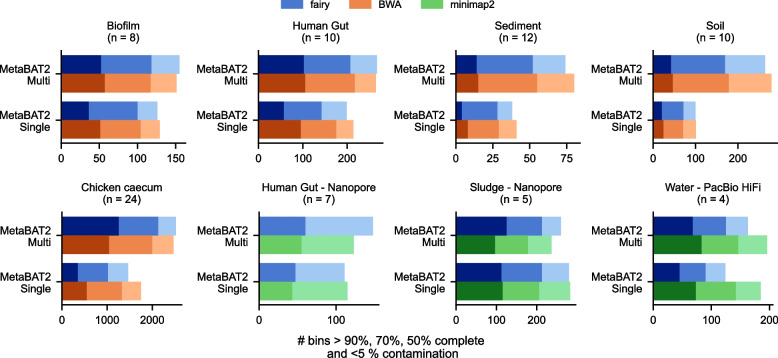
We compared fairy’s binning results with BWA and minimap2 using MetaBAT2 as the binner (Fig. 2). For every short-read dataset, fairy’s multi-sample binning outperforms single-sample binning, even with read alignment. This is especially prominent for complex sediment or soil metagenomes. We also note that single-sample binning contains contamination which may not be detectable by CheckM [10]. The long-read sludge metagenome is an exception, with minimap2 performing worse for multi-sample binning. We speculate that this is due to parameters (i.e., for coverage and binning) not being tuned as carefully for long reads. Compared to short-read binning, optimal parameter choices have not been explored as much for long-read binning―we do not claim our choice of alignment-based coverage parameters is optimal.
Fig. 2.
Binning results over multiple datasets for MetaBAT2. Fairy’s concordance with BWA and minimap2 for multi-sample and single-sample coverage binning using MetaBAT2. Darker shades indicate higher completeness (90%, 70%, and 50% completeness thresholds with < 5% contamination). minimap2 was used with long-read datasets (PacBio HiFi or nanopore), whereas BWA was used with short-read datasets. Sample accessions are available in Additional file 3: Supplementary Table 2
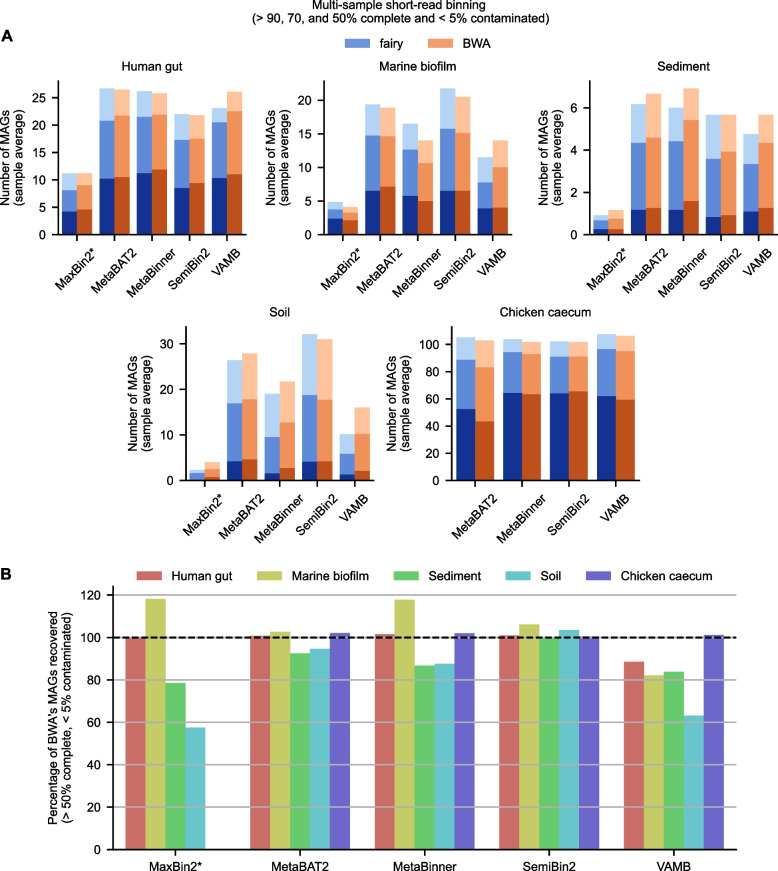
Fairy recovers comparable quality bins to alignment for multi-sample short-read metagenomes
Next, we analyzed fairy versus BWA across different binning algorithms (Fig. 3 and Additional file 1: Supplementary Figure 1). We focus on multi-sample, short-read binning. Overall, no binning method or coverage method was consistently the best (Fig. 3A). The mean percentage of recovered bins (> 50% complete; < 5% contaminated) as a percentage of BWA’s # of bins is 88.6%, 98.5%, 99.1%, 83.8%, and 102.2% for MaxBin2, MetaBAT2, MetaBinner, VAMB, and SemiBin2 respectively (Fig. 3B).
Fig. 3.
Short-read, multi-sample binning comparison across binning algorithms. A Number of bins recovered on average per sample with or completeness and contamination. Darker indicates higher completeness. *MaxBin2 did not complete on the chicken caecum dataset due to a software error. B Percentage of bins in (A) with completeness and contamination obtained with fairy relative to BWA over four different binners. This was calculated as . Higher than 100% indicates superior performance to BWA. Complete results are available in Additional file 2: Supplementary Table 1
MaxBin2 produced very few bins on the soil and sediment datasets, so fairy’s large relative recovery dropoff represents a small absolute dropoff. We also note that for VAMB, our single-sample assembly with multi-sample coverage approach differs from the “multisplit” approach that VAMB suggests using, perhaps impacting performance. MaxBin2 failed to complete for the chicken caecum dataset where fairy outperformed BWA on all other binners (102%, 102%, 101%, and 100% recovered compared to BWA for MetaBAT2, MetaBinner, VAMB, and SemiBin2 respectively).
On our host-associated datasets (human gut and chicken caecum), fairy produces more complete bins than BWA for all binners except VAMB. Fairy particularly excelled for high-quality bins ( complete and contaminated) on the caecum dataset; fairy recovered 120% of BWA’s bins on the caecum dataset using MetaBAT2, a 20% increase in recovery. In Additional file 1: Supplementary Figure 2, we show an example where MetaBAT2 recovered a 98% complete bin with fairy, but the bin was split into two using BWA’s coverages.
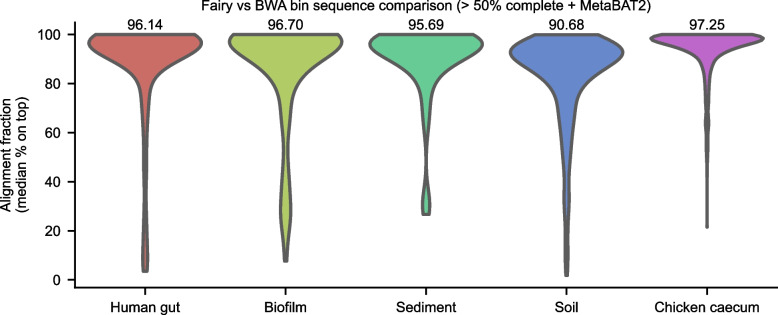
Fairy’s bins are genomically similar to bins recovered by alignment
We also verified that fairy’s short-read, multi-sample bins have similar genomic content to BWA’s bins, and not just similar completeness and contamination statistics (Fig. 4). We compared MetaBAT2 + fairy’s bins against MetaBAT2 + BWA’s bins across the same sample using skani [43]. For each of MetaBAT2 + fairy’s bins, we considered the highest aligned fraction match with ANI as the best hit. Over all of fairy’s complete bins, we found that the median aligned fraction against the best hit BWA bin was on all datasets except soil, which still had median aligned fraction. Thus more than half of fairy’s bins shared 90% of the same genomic content as one of BWA’s bins.
Fig. 4.
Alignment fraction of fairy’s bins versus BWA’s bins (multi-sample, short-read bins with MetaBAT2). For each sample, fairy’s bins with completeness were compared to all of BWA’s bins using skani [43]. For each bin, the highest alignment fraction match with ANI was taken. Median alignment fraction values are shown above each violin plot
When to use fairy versus alignment
Our results suggest that fairy performs well for multi-sample binning and especially for host-associated metagenomes. We outline two situations below where our results suggest that fairy can not yet replace read alignment.
Caveat 1: fairy is not as good as alignment for single-sample binning
For single-sample coverage with short reads, the performance of fairy is noticeably worse than BWA (Fig. 2). Using MetaBAT2, fairy recovers only 60% of BWA’s high-quality ( completeness and contamination) human gut bins for single-sample binning, but 97% of BWA’s high-quality bins for multi-sample binning. We do not recommend fairy for single-sample binning―single-sample coverage calculation is not a bottleneck anyway.
Caveat 2: fairy is usable with nanopore long-reads, but not high-fidelity reads
For nanopore long-reads, our MetaBAT2 results show that fairy is competitive with minimap2’s alignment coverage (Fig. 2). However, fairy’s results are markedly worse than minimap2 when using PacBio HiFi reads. We found that this is because HiFi assemblers assemble similar strains instead of collapsing them, and our k-mer-based approach is not suitable for calculating coverage for extremely similar strains.
It may however be possible to tune fairy’s ANI thresholds for strain-level HiFi binning. Although PacBio HiFi metagenomic samples are still uncommon relative to short reads, we believe that strain-level binning is an interesting avenue to explore for future work. Nevertheless, we do not recommend fairy in its current version for PacBio HiFi MAG recovery.
Case study: fairy recovers comparable Asgard archaea genomes in sediment
The study of Asgard archaea (or Asgardarchaeota) has served an important role in the study of eukaryogenesis [44]. Asgard archaea genomes were first recovered through metagenomics [45], and metagenomics continues to be important in their study [30]. In Fig. 5, we investigate the quality of Asgard archaea MAGs recovered from the sediment dataset (also used in Figs. 2 and 3). Sediment metagenomes are known to be complex [46], so this presents an interesting challenge for fairy.
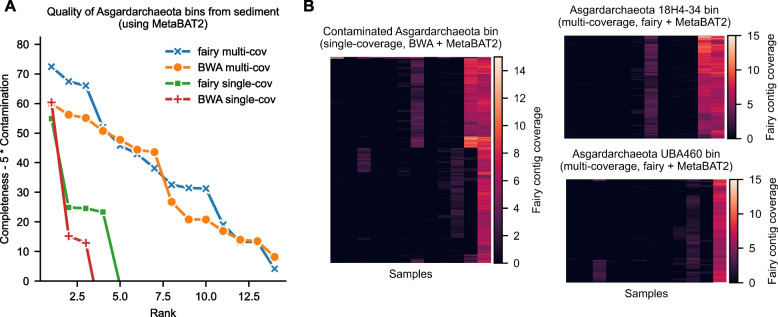
Fig. 5.
Investigating Asgard archaea from sediment metagenomes using MetaBAT2 with fairy and BWA. A Multi-sample binning, whether with fairy or BWA, recovers much higher quality Asgard archaea bins. B Coverage-contig heatmaps for specific bins. Left: single-sample binning produces a contaminated bin with two Asgard archaea (BWA’s bin pictured). Coverage patterns (fairy’s coverage) show clear contamination. Right: multi-sample binning disentangle the two genomes and correctly generates two bins (fairy’s bins pictured)
For MetaBAT2 bins from the sediment metagenomes, we compared single-sample versus multi-sample binning and found that multi-sample binning produces much higher quality Asgard bins (Fig. 5A). Fairy is even slightly better than BWA when ranking the bins under the Completeness Contamination metric.
We visualized an explicit example of where single-sample coverage binning failed in Fig. 5B. In the sample DRR310882, both fairy and BWA (with MetaBAT2) produced a contaminated bin with two Asgard archaea. BWA’s bin (visualized in Fig. 5B, left) had a 55% contamination estimate. The two erroneously binned Asgard genomes have similar coverage in this sample and could not be separated with k-mer frequencies. However, multi-sample coverage information disentangles the two genomes, and both fairy and BWA do so successfully. Fairy’s resulting multi-sample bins (Fig. 5B, right), with genus assignments 18H4-34 and UBA460 using GTDB-Tk [47], had only 2.66% and 4.25% contamination respectively.
Discussion
Fairy’s role in MAG recovery pipelines
Fairy is orders of magnitude faster than read alignment and solves a key computational bottleneck. The speed increase is not controversial. In some cases, fairy may give better results (e.g., the biofilm and chicken caecum datasets in Fig. 2), but there may be a slight sensitivity loss compared to read alignment on complex metagenomes such as sediment or soil. However, we show that when all-to-all alignment is not feasible, multi-sample binning with fairy should always be preferred over single-sample read alignment, thus filling an important niche for large-sample projects.
Furthermore, we envision fairy as complementary to all-to-all read alignment. Users can first use fairy to obtain a set of good-quality multi-sample bins and immediately analyze their data while they wait days/weeks for their all-to-all read alignments finish.
For default parameters, fairy’s accuracy is encouraging
Binning accuracy depends on both the binner and the coverage calculation method. We used standard pipelines for obtaining read-alignment coverage and default parameters for all binners so that our results represent a realistic situation where fairy is used instead of, say, BWA. We have seen that fairy’s coverages have specific patterns not seen in read-alignment coverages (e.g., 0-coverage dropouts due to ANI thresholds; see Additional file 1: Supplementary Figure 2). It would be interesting to understand how binning parameters work in conjunction with coverages obtained by k-mer sketching techniques like fairy, and if alternative parameterizations could lead to better results.
Conclusion
In this paper, we provide a new k-mer-based coverage calculation method for metagenomic binning called fairy. Fairy is magnitudes faster than read alignment and enables multi-sample binning on much larger datasets than before. In particular, we show that recovered bins are of competitive completeness and contamination to read-mapping approaches and sometimes even better.
Interesting future avenues to explore include investigating other approaches to calculate coverage for binning as well as understanding theoretically what coverage characteristics lead to good MAG recovery. We do not yet understand the exact tradeoffs, pitfalls, and issues for how coverage calculation affects binning. While we expect an inevitable speed-accuracy tradeoff across different methods, a deeper understanding would guide practitioners, allowing optimal MAG recovery pipeline design for their needs.
Supplementary Information
Additional file 1: Supplementary Methods. Supplementary Figures.
Additional file 2: Supplementary Table 1.
Additional file 3: Supplementary Table 2.
Acknowledgements
Not applicable.
Authors’ contributions
J.S. devised the study, ran the experiments, and wrote the software. Y.W.Y supervised the study. Both authors wrote and edited the manuscript.
Funding
J.S. was supported by an NSERC CGS-D. This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) grant RGPIN-2022-03074 and DND/NSERC Supplement DGDND-2022-03074.
Availability of data and materials
No new datasets were generated or analyzed during the current study. All analyzed samples are publicly available and accessions can be found in Additional file 3: Supplementary Table 2. Scripts for reproducing figures can be found at https://github.com/bluenote-1577/fairy-test/.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jim Shaw, Email: jshaw@math.toronto.edu.
Yun William Yu, Email: ywyu@cmu.edu.
References
- 1.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35(9):833–44. 10.1038/nbt.3935. 10.1038/nbt.3935 [DOI] [PubMed] [Google Scholar]
- 2.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7. 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed]
- 3.Wu YW, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32(4):605–7. 10.1093/bioinformatics/btv638. 10.1093/bioinformatics/btv638 [DOI] [PubMed] [Google Scholar]
- 4.Nissen JN, Johansen J, Allesøe RL, Sønderby CK, Armenteros JJA, Grønbech CH, et al. Improved metagenome binning and assembly using deep variational autoencoders. Nat Biotechnol. 2021;39(5):555–60. 10.1038/s41587-020-00777-4. 10.1038/s41587-020-00777-4 [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Huang P, You R, Sun F, Zhu S. MetaBinner: a high-performance and stand-alone ensemble binning method to recover individual genomes from complex microbial communities. Genome Biol. 2023;24(1):1. 10.1186/s13059-022-02832-6. 10.1186/s13059-022-02832-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavia MJ, Chede A, Wu Z, Cadillo-Quiroz H, Zhu Q. BinaRena: a dedicated interactive platform for human-guided exploration and binning of metagenomes. Microbiome. 2023;11(1):186. 10.1186/s40168-023-01625-8. 10.1186/s40168-023-01625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10(8):R85. 10.1186/gb-2009-10-8-r85. 10.1186/gb-2009-10-8-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11(11):1144–6. 10.1038/nmeth.3103. 10.1038/nmeth.3103 [DOI] [PubMed] [Google Scholar]
- 9.Chklovski A, Parks DH, Woodcroft BJ, Tyson GW. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat Methods. 2023;20(8):1203–12. 10.1038/s41592-023-01940-w. 10.1038/s41592-023-01940-w [DOI] [PubMed] [Google Scholar]
- 10.Mattock J, Watson M. A comparison of single-coverage and multi-coverage metagenomic binning reveals extensive hidden contamination. Nat Methods. 2023;1–4. 10.1038/s41592-023-01934-8. [DOI] [PubMed]
- 11.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013;(arXiv:1303.3997). 10.48550/arXiv.1303.3997.
- 12.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11(12):2864–8. 10.1038/ismej.2017.126. 10.1038/ismej.2017.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(3):649-662.e20. 10.1016/j.cell.2019.01.001. 10.1016/j.cell.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–7. 10.1038/nbt.3519. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 16.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–9. 10.1038/nmeth.4197. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alneberg J, Bennke C, Beier S, Bunse C, Quince C, Ininbergs K, et al. Ecosystem-wide metagenomic binning enables prediction of ecological niches from genomes. Commun Biol. 2020;3(1):1–10. 10.1038/s42003-020-0856-x. 10.1038/s42003-020-0856-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zorrilla F, Buric F, Patil KR, Zelezniak A. metaGEM: reconstruction of genome scale metabolic models directly from metagenomes. Nucleic Acids Res. 2021;49(21):e126. 10.1093/nar/gkab815. 10.1093/nar/gkab815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quince C, Nurk S, Raguideau S, James R, Soyer OS, Summers JK, et al. STRONG: metagenomics strain resolution on assembly graphs. Genome Biol. 2021;22(1):214. 10.1186/s13059-021-02419-7. 10.1186/s13059-021-02419-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krivonosova K, Gorshkov Y, Nurk S. Estimating differential abundance profiles for metagenomic series binning. https://bioinformaticsinstitute.ru/sites/default/files/4krivonosova_150918.pdf. Accessed 23 Apr 2024.
- 21.Shaw J, Yu YW. Metagenome profiling and containment estimation through abundance-corrected k-Mer Sketching with sylph. bioRxiv. 2023;2023.11.20.567879. 10.1101/2023.11.20.567879.
- 22.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17(1):132. 10.1186/s13059-016-0997-x. 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce NT, Irber L, Reiter T, Brooks P, Brown CT. Large-scale sequence comparisons with sourmash. F1000Research. 2019;8:1006. 10.12688/f1000research.19675.1. [DOI] [PMC free article] [PubMed]
- 24.Yorukoglu D, Yu YW, Peng J, Berger B. Compressive mapping for next-generation sequencing. Nat Biotechnol. 2016;34(4):374–6. 10.1038/nbt.3511. 10.1038/nbt.3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irber L, Brooks PT, Reiter T, Pierce-Ward NT, Hera MR, Koslicki D, et al. Lightweight compositional analysis of metagenomes with FracMinHash and minimum metagenome covers. 2022;2022.01.11.475838. 10.1101/2022.01.11.475838.
- 26.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114. 10.1038/s41467-018-07641-9. 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng X, Cheng H, Portik D, Li H. Metagenome assembly of high-fidelity long reads with Hifiasm-Meta. Nat Methods. 2022;19(6):671–4. 10.1038/s41592-022-01478-3. 10.1038/s41592-022-01478-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glendinning L, Stewart RD, Pallen MJ, Watson KA, Watson M. Assembly of hundreds of novel bacterial genomes from the chicken caecum. Genome Biol. 2020;21(1):34. 10.1186/s13059-020-1947-1. 10.1186/s13059-020-1947-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olm MR, Butterfield CN, Copeland A, Boles TC, Thomas BC, Banfield JF. The source and evolutionary history of a microbial contaminant identified through soil metagenomic analysis. mBio. 2017;8(1). 10.1128/mbio.01969-16. [DOI] [PMC free article] [PubMed]
- 30.Medvedeva S, Sun J, Yutin N, Koonin EV, Nunoura T, Rinke C, et al. Three families of asgard archaeal viruses identified in metagenome-assembled genomes. Nat Microbiol. 2022;7(7):962–73. 10.1038/s41564-022-01144-6. 10.1038/s41564-022-01144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Ding W, Li YX, Tam C, Bougouffa S, Wang R, et al. Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat Commun. 2019;10:517. 10.1038/s41467-019-08463-z. 10.1038/s41467-019-08463-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sereika M, Petriglieri F, Jensen TBN, Sannikov A, Hoppe M, Nielsen PH, et al. Closed genomes uncover a saltwater species of Candidatus Electronema and shed new light on the boundary between marine and freshwater cable bacteria. ISME J. 2023;17(4):561–9. 10.1038/s41396-023-01372-6. 10.1038/s41396-023-01372-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gounot JS, Chia M, Bertrand D, Saw WY, Ravikrishnan A, Low A, et al. Genome-centric analysis of short and long read metagenomes reveals uncharacterized microbiome diversity in Southeast Asians. Nat Commun. 2022;13(1):6044. 10.1038/s41467-022-33782-z. 10.1038/s41467-022-33782-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidhu C, Kirstein IV, Meunier CL, Rick J, Fofonova V, Wiltshire KH, et al. Dissolved storage glycans shaped the community composition of abundant bacterioplankton clades during a North Sea spring phytoplankton bloom. Microbiome. 2023;11(1):77. 10.1186/s40168-023-01517-x. 10.1186/s40168-023-01517-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP–a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6(1):158. 10.1186/s40168-018-0541-1. 10.1186/s40168-018-0541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieser S, Brown J, Zdobnov EM, Trajkovski M, McCue LA. ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinformatics. 2020;21(1):257. 10.1186/s12859-020-03585-4. 10.1186/s12859-020-03585-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aroney STN, Newell RJP, Nissen J, Camargo AP, Tyson GW, Woodcroft BJ. CoverM: read coverage calculator for metagenomics. 2024. Zenodo. 10.5281/zenodo.10531254.
- 38.Kolmogorov M, Bickhart DM, Behsaz B, Gurevich A, Rayko M, Shin SB, et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods. 2020;17(11):1103–10. 10.1038/s41592-020-00971-x. 10.1038/s41592-020-00971-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100. 10.1093/bioinformatics/bty191. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benoit G, Raguideau S, James R, Phillippy AM, Chikhi R, Quince C. High-quality metagenome assembly from long accurate reads with metaMDBG. Nat Biotechnol. 2024;1–6. 10.1038/s41587-023-01983-6. [DOI] [PMC free article] [PubMed]
- 41.Pan S, Zhao XM, Coelho LP. SemiBin2: self-supervised contrastive learning leads to better MAGs for short- and long-read sequencing. Bioinformatics. 2023;39(Supplement-1):i21–9. 10.1093/bioinformatics/btad209. 10.1093/bioinformatics/btad209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu CC, Dong SS, Chen JB, Wang C, Ning P, Guo Y, et al. MetaDecoder: a novel method for clustering metagenomic contigs. Microbiome. 2022;10(1):46. 10.1186/s40168-022-01237-8. 10.1186/s40168-022-01237-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw J, Yu YW. Fast and robust metagenomic sequence comparison through sparse chaining with skani. Nat Methods. 2023;1–5. 10.1038/s41592-023-02018-3. [DOI] [PMC free article] [PubMed]
- 44.Eme L, Tamarit D, Caceres EF, Stairs CW, De Anda V, Schön ME, et al. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature. 2023;618(7967):992–9. 10.1038/s41586-023-06186-2. 10.1038/s41586-023-06186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521(7551):173–9. 10.1038/nature14447. 10.1038/nature14447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chevrette MG, Bratburd JR, Currie CR, Stubbendieck RM. Experimental microbiomes: models not to scale. mSystems. 2019;4(4). 10.1128/msystems.00175-19. [DOI] [PMC free article] [PubMed]
- 47.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36(6):1925–7. 10.1093/bioinformatics/btz848. 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Methods. Supplementary Figures.
Additional file 2: Supplementary Table 1.
Additional file 3: Supplementary Table 2.
Data Availability Statement
No new datasets were generated or analyzed during the current study. All analyzed samples are publicly available and accessions can be found in Additional file 3: Supplementary Table 2. Scripts for reproducing figures can be found at https://github.com/bluenote-1577/fairy-test/.