Abstract
Quinoprotein (2,7,9-tricarboxy-1H-pyrrolo-[2,3-f]quinoline-4,5-dione quinone form (PQQ)-containing) ethanol dehydrogenase (EDH) from Pseudomonas aeruginosa ATCC 17933 was purified to homogeneity. EDH has an alpha 2 beta 2 configuration and subunits comparable in size to those of methanol dehydrogenase (MDH). Compared with other PQQ-containing dehydrogenases, Ca2+ is rather loosely bound and it seems necessary for PQQ binding and stability of EDH. Two soluble cytochromes c were detected in extracts from ethanol-grown cells and both were purified. One of these has an alpha-band at 551 nm for its reduced form, the oxidized form being an excellent electron acceptor for the semiquinone form of EDH. Since this cytochrome is quite different from the already known cytochrome c551 (operating in nitrate respiration) of this organism, it is indicated here as cytochrome cEDH. Comparison of the N-terminal amino acid sequence of cytochrome cEDH with the complete sequence of cytochrome cL (the electron acceptor of MDH), cytochrome cH (the electron acceptor of cytochrome cL) and cytochrome c551 revealed some similarity only to internal stretches of amino acids of the last two. The other soluble cytochrome appeared to be the already-known cytochrome c556. Since it was not an electron acceptor for cytochrome cEDH (neither for EDH), cytochrome cH is lacking in the quinoprotein-EDH-ethanol oxidation system of P. aeruginosa. It seems, therefore, that the respiratory chains for MDH and EDH are different.
Full text
PDF

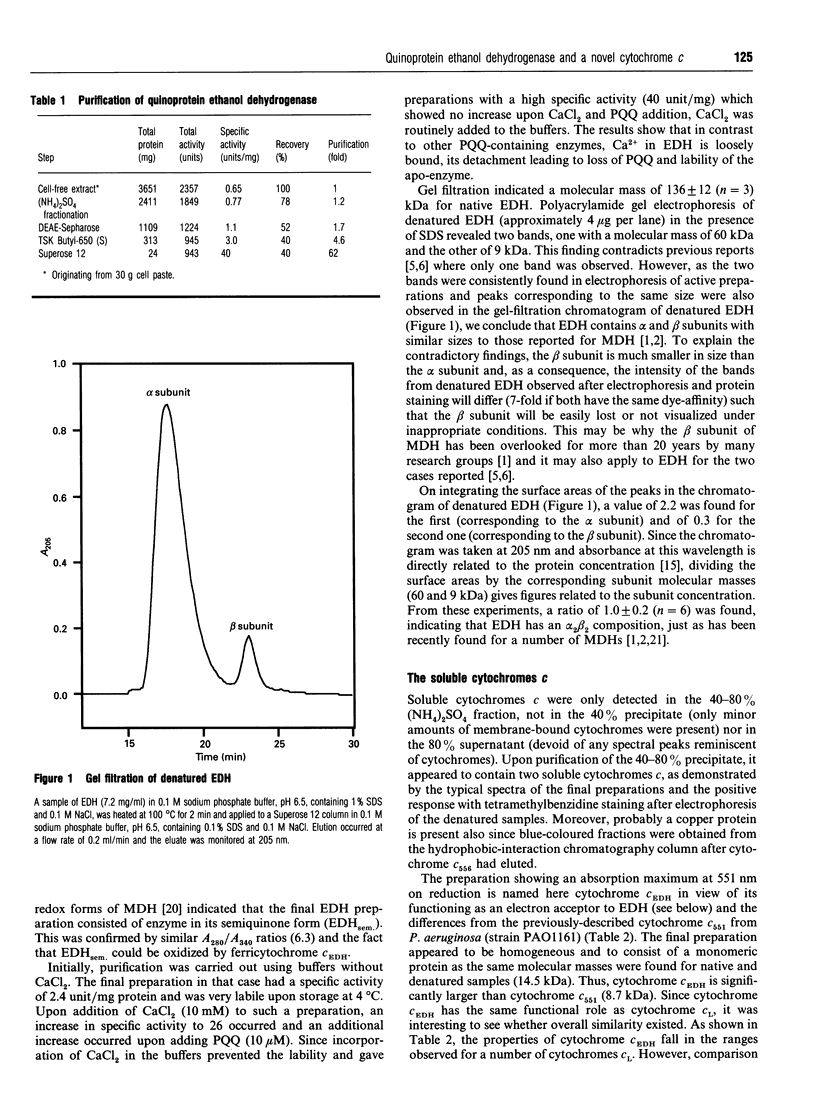

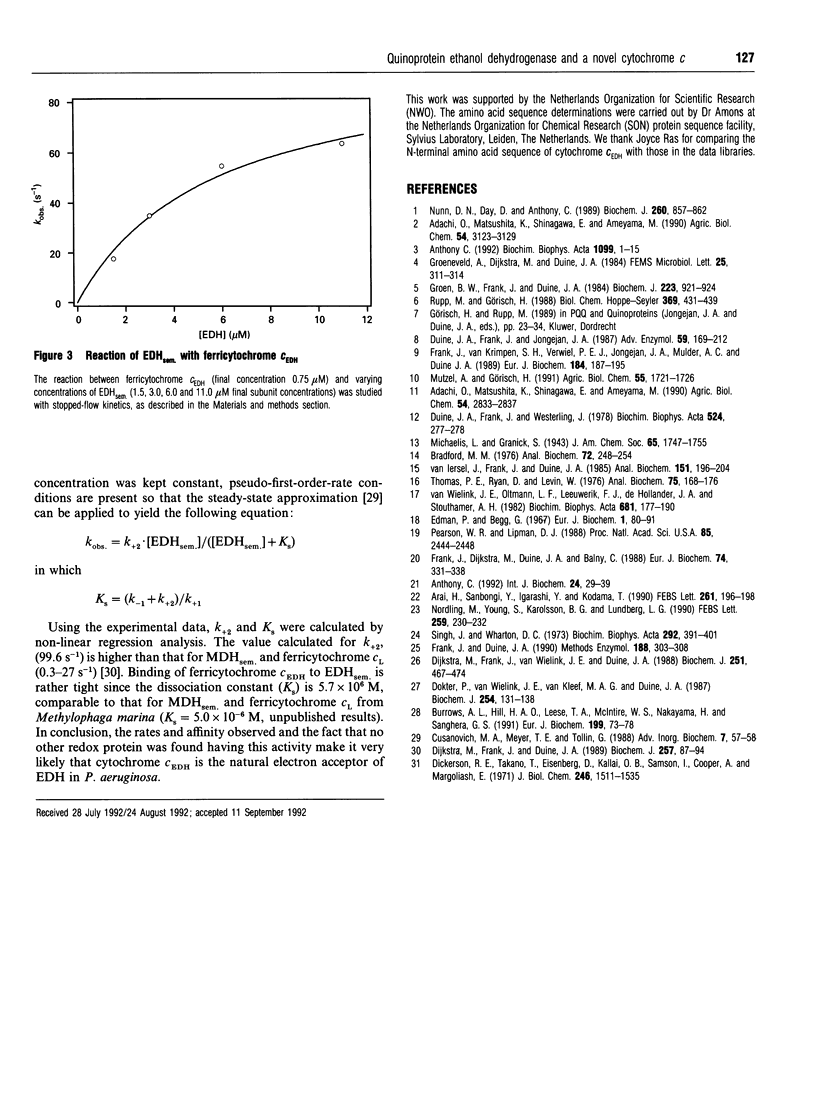
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony C. The c-type cytochromes of methylotrophic bacteria. Biochim Biophys Acta. 1992 Jan 30;1099(1):1–15. [PubMed] [Google Scholar]
- Anthony C. The structure of bacterial quinoprotein dehydrogenases. Int J Biochem. 1992;24(1):29–39. doi: 10.1016/0020-711x(92)90226-q. [DOI] [PubMed] [Google Scholar]
- Arai H., Sanbongi Y., Igarashi Y., Kodama T. Cloning and sequencing of the gene encoding cytochrome c-551 from Pseudomonas aeruginosa. FEBS Lett. 1990 Feb 12;261(1):196–198. doi: 10.1016/0014-5793(90)80669-a. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burrows A. L., Hill H. A., Leese T. A., Mcintire W. S., Nakayama H., Sanghera G. S. Direct electrochemistry of the enzyme, methylamine dehydrogenase, from bacterium W3A1. Eur J Biochem. 1991 Jul 1;199(1):73–78. doi: 10.1111/j.1432-1033.1991.tb16093.x. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Takano T., Eisenberg D., Kallai O. B., Samson L., Cooper A., Margoliash E. Ferricytochrome c. I. General features of the horse and bonito proteins at 2.8 A resolution. J Biol Chem. 1971 Mar 10;246(5):1511–1535. [PubMed] [Google Scholar]
- Dijkstra M., Frank J., Jr, Duine J. A. Studies on electron transfer from methanol dehydrogenase to cytochrome cL, both purified from Hyphomicrobium X. Biochem J. 1989 Jan 1;257(1):87–94. doi: 10.1042/bj2570087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra M., Frank J., Jr, van Wielink J. E., Duine J. A. The soluble cytochromes c of methanol-grown Hyphomicrobium X. Evidence against the involvement of autoreduction in electron-acceptor functioning of cytochrome cL. Biochem J. 1988 Apr 15;251(2):467–474. doi: 10.1042/bj2510467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokter P., van Wielink J. E., van Kleef M. A., Duine J. A. Cytochrome b-562 from Acinetobacter calcoaceticus L.M.D. 79.41. Its characteristics and role as electron acceptor for quinoprotein glucose dehydrogenase. Biochem J. 1988 Aug 15;254(1):131–138. doi: 10.1042/bj2540131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Jongejan J. A. Enzymology of quinoproteins. Adv Enzymol Relat Areas Mol Biol. 1987;59:169–212. doi: 10.1002/9780470123058.ch4. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Westerling J. Purification and properties of methanol dehydrogenase from Hyphomicrobium x. Biochim Biophys Acta. 1978 Jun 9;524(2):277–287. doi: 10.1016/0005-2744(78)90164-x. [DOI] [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- Frank J., Jr, Dijkstra M., Duine J. A., Balny C. Kinetic and spectral studies on the redox forms of methanol dehydrogenase from Hyphomicrobium X. Eur J Biochem. 1988 Jun 1;174(2):331–338. doi: 10.1111/j.1432-1033.1988.tb14102.x. [DOI] [PubMed] [Google Scholar]
- Frank J., Jr, van Krimpen S. H., Verwiel P. E., Jongejan J. A., Mulder A. C., Duine J. A. On the mechanism of inhibition of methanol dehydrogenase by cyclopropane-derived inhibitors. Eur J Biochem. 1989 Sep 1;184(1):187–195. doi: 10.1111/j.1432-1033.1989.tb15006.x. [DOI] [PubMed] [Google Scholar]
- Groen B., Frank J., Jr, Duine J. A. Quinoprotein alcohol dehydrogenase from ethanol-grown Pseudomonas aeruginosa. Biochem J. 1984 Nov 1;223(3):921–924. doi: 10.1042/bj2230921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordling M., Young S., Karlsson B. G., Lundberg L. G. The structural gene for cytochrome c551 from Pseudomonas aeruginosa. The nucleotide sequence shows a location downstream of the nitrite reductase gene. FEBS Lett. 1990 Jan 1;259(2):230–232. doi: 10.1016/0014-5793(90)80015-b. [DOI] [PubMed] [Google Scholar]
- Nunn D. N., Day D., Anthony C. The second subunit of methanol dehydrogenase of Methylobacterium extorquens AM1. Biochem J. 1989 Jun 15;260(3):857–862. doi: 10.1042/bj2600857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp M., Görisch H. Purification, crystallisation and characterization of quinoprotein ethanol dehydrogenase from Pseudomonas aeruginosa. Biol Chem Hoppe Seyler. 1988 Jun;369(6):431–439. doi: 10.1515/bchm3.1988.369.1.431. [DOI] [PubMed] [Google Scholar]
- Singh J., Wharton D. C. Cytochrome c-556, a di-heme protein from Pseudomonas aeruginosa. Biochim Biophys Acta. 1973 Feb 22;292(2):391–401. doi: 10.1016/0005-2728(73)90045-5. [DOI] [PubMed] [Google Scholar]
- Thomas P. E., Ryan D., Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976 Sep;75(1):168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- Van Wielink J. E., Oltmann L. F., Leeuwerik F. J., De Hollander J. A., Stouthamer A. H. A method for in situ characterization of b- and c-type cytochromes in Escherichia coli and in complex III from beef heart mitochondria by combined spectrum deconvolution and potentiometric analysis. Biochim Biophys Acta. 1982 Aug 20;681(2):177–190. doi: 10.1016/0005-2728(82)90021-4. [DOI] [PubMed] [Google Scholar]
- van Iersel J., Jzn J. F., Duine J. A. Determination of absorption coefficients of purified proteins by conventional ultraviolet spectrophotometry and chromatography combined with multiwavelength detection. Anal Biochem. 1985 Nov 15;151(1):196–204. doi: 10.1016/0003-2697(85)90072-7. [DOI] [PubMed] [Google Scholar]


