Abstract
Some inborn errors of metabolism due to deficiencies of soluble lysosomal enzymes cause global neurodegenerative disease. Representative examples include the infantile and late infantile forms of the ceroid lipofuscinoses (CLN1 or CLN2 deficiency, respectively) and mucopolysaccharidoses type VII (MPS VII), a deficiency of β-glucuronidase. Treatment of the central nervous system component of these disorders will require widespread protein or enzyme replacement, either through dissemination of the protein or through dissemination of a gene encoding it. We hypothesize that transduction of brain microcapillary endothelium (BME) with recombinant viral vectors, with secretion of enzyme product basolaterally, could allow for widespread enzyme dissemination. To achieve this, viruses should be modified to target the BME. This requires (i) identification of a BME-resident target receptor, (ii) identification of motifs targeted to that molecule, (iii) the construction of modified viruses to allow for binding to the target receptor, and (iv) demonstrated transduction of receptor-expressing cells. In proof of principal experiments, we chose the human transferrin receptor (hTfR), a molecule found at high density on human BME. A nonamer phage display library was panned for motifs which could bind hTfR. Forty-three clones were sequenced, most of which contained an AKxxK/R, KxKxPK/R, or KxK motif. Ten peptides representative of the three motifs were cloned into the HI loop of adenovirus type 5 fiber. All motifs tested retained their ability to trimerize and bind transferrin receptor, and seven allowed for recombinant adenovirus production. Importantly, the fiber-modified viruses facilitated increased gene transfer (2- to 34-fold) to hTfR expressing cell lines and human brain microcapillary endothelia expressing high levels of endogenous receptor. Our data indicate that adenoviruses can be modified in the HI loop for expanded tropism to the hTfR.
Many inherited metabolic disorders lead to central nervous system (CNS) deficits, either alone or in combination with systemic involvement (24). One approach to metabolic correction is by cellular transduction with virus vectors encoding a functional cDNA. For correction of the CNS component, therapies will likely require direct application to brain parenchyma, since closure of the blood-brain barrier (BBB) shortly after birth would restrict entry of the gene product or gene transfer vectors into the brain. In metabolic disorders due to deficiencies in soluble lysosomal proteins, genetic correction of all affected cells will not be required; secretion of overexpressed protein provides a pool of available enzyme for distribution to surrounding cells (cross-correction). Examples of such disorders include the ceroid lipofuscinoses I and II and mucopolysaccharidoses type VII (MPS VII). But even with cross-correction, spread of enzyme is limited. Thus, an important remaining problem for clinical application is how to affect global correction in these disorders.
Earlier studies using MPS VII mouse models (deficient in the lysosomal enzyme β-glucuronidase) have allowed testing of potential therapies for both the CNS and visceral components of this representative disease. Direct intraparenchymal gene transfer to mouse brain with adenovirus vectors expressing β-glucuronidase allowed for extensive distribution of enzyme and correction of the characteristic storage defect within the brains of β-glucuronidase-deficient mice (13, 29). The spread of enzyme beyond sites of transduction resulted from secretion of β-glucuronidase upon overexpression, with uptake and correction by nontransduced cells. Similar results were found with recombinant adeno-associated virus (25, 27, 31) and lentivirus (2) vectors expressing β-glucuronidase. Due to the larger size of a primate brain, however, focal gene delivery is unlikely to result in significant amounts of secreted enzyme reaching areas remote from the site of vector injection. An alternative to direct injection into the brain parenchyma for correction of global neurodegenerative disease would be to take advantage of the vasculature of the host. One approach could be to disrupt the tight junctions of the vascular endothelia for direct vector access to the underlying parenchyma. A second could be to transduce the vascular endothelium directly. For β-glucuronidase, which is capable of being secreted basolaterally from vascular endothelium (B.L.D., unpublished observations), distribution into the subpial and perivascular spaces (Virchow-Robin spaces) lining the penetrating blood vessels could allow access to the parenchyma since the pia does not form an impermeable barrier.
In earlier studies, we found that BBB disruption does not result in adequate vector access to parenchymal tissues. Our data showed that only several hundred cells could be transduced upon delivery of virus to mannitol-disrupted tight junctions (7). Rather than delivery of virus through disrupted tight junctions (7, 20), we propose to take advantage of the transferrin receptor (TfR) present on brain vascular endothelium. Human TfR (hTfR), a type II membrane protein, has been extensively characterized and consists of two identical 95-kDa subunits linked convalently by two disulfide bonds (30). In vitro, in vivo, and ex vivo studies by Pardridge and others showed that antibody or transferrin conjugates with specificity for the TfR allowed for delivery of substances to brain capillary endothelial cells (4, 12, 21, 26). We hypothesized that adenoviruses with motifs targeting the TfR could also allow for transduction of the brain microcapillary endothelium (BME).
Modification of the virus for targeting to the TfR could be accomplished through bifunctional antibodies or by genetically modifying the virus to display a specific TfR binding motif. Douglas et al. and others have reported the feasibility of using bifunctional antibodies for targeting in vitro (9, 16, 32, 34, 36). Experiments also showed that adenovirus capsids modified to contain a carboxyl-terminal polylysine tract allowed for improved gene transfer to many cell types via facilitated binding to cell surface heparan sulfate proteoglycans (3, 14, 28, 33, 35). Alteration of the HI loop of fiber to express an RGD motif, a sequence normally found in the penton base of adenovirus type 5 (Ad5), also improved binding to integrin-expressing cells (23).
Because of its relative simplicity we used phage display screening to identify epitopes specific to the TfR and then introduced the sequences encoding these peptides directly into the HI loop of Ad5 fiber. Modified recombinant adenoviruses were then tested for their ability to transduce TfR-expressing cell lines and human BME cells.
MATERIALS AND METHODS
Cell culture.
All media were supplemented with 10% fetal bovine serum (FBS) unless otherwise indicated. Human embryonic kidney cells (HEK 293) were maintained in Dulbecco's modified Eagle's medium (DMEM). Chinese hamster ovary cell (CHO) cells expressing human transferrin receptor (a kind gift from Martin Lawrence, Harvard University, Cambridge, Mass.), were maintained in F-12 Nutrient Mixture. HeLa cells (obtained from the American Type Culture Collection (ATCC), Rockville, Md.) were grown in minimal essential medium (MEM). The human prostate cancer cell line T24 was also from the ATCC and was maintained in 1640 medium. Human BME cells (kindly provided by Jay Nelson, Oregon Health Sciences University) were grown in 10% human AB serum (Sigma-Aldrich, St. Louis, Mo.) and in EBM (Clonetics, Walkersville, Md.).
Phage screening.
Ten microliters of amplified nonapeptide phage library (a generous gift from Al Jesaitis, Montana State University) was screened against the purified extracellular domain of hTfR (kindly provided by Martin Lawrence, Harvard University) coated on 96-well microtiter plates in 100 μl of 0.05 M carbonate buffer (pH 9.6). In the first panning, hTfR was coated at 100 μg/ml. Subsequent pannings were done with decreasing concentrations of hTfR for increased stringency (10 and 1 μg/ml for the second and third pannings, respectively). Bound phage were eluted with low-acid buffer (0.1 M glycine, pH 2.2) or ligand (iron-loaded human transferrin; Sigma-Aldrich) in TBS buffer (50 mM Tris-Cl, pH 7.5; 150 mM NaCl). After three successive rounds of panning and amplification, clones were picked and sequenced as described elsewhere (5). All sequencing was performed in the University of Iowa DNA sequencing facility.
Phage and peptide binding assays.
The extracellular domain of the hTfR was coated on 96-well microtiter well plates in 150 μl of 0.05 M carbonate buffer (pH 9.6) overnight (ON). The plates were then blocked (200 μl, 3% bovine serum albumin [BSA] for 2 h), washed, and incubated with purified phage (1010 phage) B1, B2, or a sequenced random clone from the library for 1 h at room temperature. Plates were washed and incubated with a rabbit anti-fd bacteriophage biotin conjugate (Sigma-Aldrich) directed against the M13 phage. Plates were developed using extravidin-peroxidase conjugates and diaminobenzidine (DAB) and then read at 490 nm using a microplate reader (Molecular Devices, Sunnyvale, Calif.). Data are presented as the mean of triplicates ± the standard error of the mean (SEM). The experiments were repeated three times.
B2 peptide binding was tested for specificity for hTfR in two separate assays. For both, hTfR was first coated onto 96-well plates. In one assay, plates were coated ON with 3.75 μl of hTfR in 150 μl, blocked, and washed, followed by the addition of 100 μl of peptide B2 conjugated to biotin (Genosys Biotechnologies, Inc., Woodlands, Tex.) at a concentration range of 500 to 62.5 μg/ml. In the second assay plates were coated with 150 μl of hTfR (range, 25 to 1.6 μg/ml). After overnight coating, wells were blocked and then incubated with B2-labeled biotin (25 μg). Plates were developed and read as described above. Data are presented as the means of triplicates ± the SEM. The experiments were repeated three times.
Construction of recombinant plasmids.
To facilitate the generation of HI loop-modified viruses, the Ad5 fiber gene was first cloned into the vaccinia virus expression vector pTM1 (a kind gift from Michael J. Welsh, University of Iowa) by PCR amplification. This plasmid was designated pTM1Ad5fiber. Unique restriction sites and the B2 sequence were then introduced into fiber. To accomplish this, two pairs of primers, F1 (5′-AGAAATGGAGATCTTACTGAAGGC-3′) and R1 (5′-CCCCTTCGGCCTCTTCACCTTATGACCAGTTGTGTCTCCTGTTTCCTGTGTACC-3′) and also F2 (5′-GGTCATAAGGTGAAGAGGCCGAAGGGGCCAAGTGCATACTCTATGTCATTTTCA-3′) and R2 (5′-AACCCCGGGACTAGTCTATTCTTGGGCAATGTATGAAAAAGTGTA-3′), were used to amplify a 210- and a 100-bp fragment of Ad5 fiber using purified virus genomic DNA as a template. The reaction products were gel purified and mixed, and contiguous sequences were generated by overlapping PCR using primers F1 and R2. The PCR amplification product contained a unique BglII site at the 5′ end and 3′ SpeI and SmaI sites. The restricted fragment was cloned into BglII- and SmaI-restricted pTM1Ad5 fiber. The resultant plasmid was named pTM1Ad5fiber/B2HI. All other motifs were similarly introduced using specific primer pairs. These plasmids were named pTM1Ad5fiber/B1HI, etc., and were used in in vitro expression systems to analyze the effects of the epitopes on fiber trimerization or binding to hTfR.
A shuttle was developed to allow insertion of modified HI loops into Ad5 fiber sequences. First, pTG1696 (obtained from Transgene S.A., Strasbourg, France) was cut by NotI and SpeI to remove approximately 8,000 bp of the first half of the plasmid. The plasmid was reclosed to generate pTGSN53 and contained adenovirus sequences from bp 29510 to 35935. pTM1Ad5fiber/B2HI was cut by SphI and SmaI, and the fragment was cloned into pTGSN53 to obtain pTGSS/B2HI plasmids. To facilitate homologous recombination in Escherichia coli, we next introduced more than 1.0 kb of adenovirus sequence at the 3′ end of the fiber sequence. The primers Fbs (5′-CCCACTAGTATCGTTTGTGTT-3′) and Rbs (5′-AAAGGATCCAGATCTGTTTGTCACGCCGCG-3′) were used to amplify a fragment containing SpeI and BamHI restriction sites (underlined) at the 5′ and 3′ ends of the fragment, respectively, using Ad5 genomic DNA as a template. The PCR product was cut by BamHI and SpeI and then cloned into pTGSS/B2HI. The resulting plasmid was designated pBS/B2HI (additional details and maps are available upon request). pBS/B2HI contains the hTfR-targeting peptide B2 in the HI loop of Ad5 fiber and the novel SpeI site at the end of fiber coding sequence. Moreover, this plasmid also has greater than 1 kb of flanking Ad5 DNA sequence on either side of the fiber. pBS/B2HI will be useful for the cloning of any identified motif into the HI loop or for the generation of chimeric fiber sequences.
For plasmids pBS/B1HI, pBS/B3HI, etc., overlapping PCR was used to generated fragments containing motifs B1 and B3 to B10. These fragments were cut with SpeI and SphI and cloned into similarly cut pBS/B2HI to generate pBS/BxHI. Sequences for overlapping PCR are available upon request.
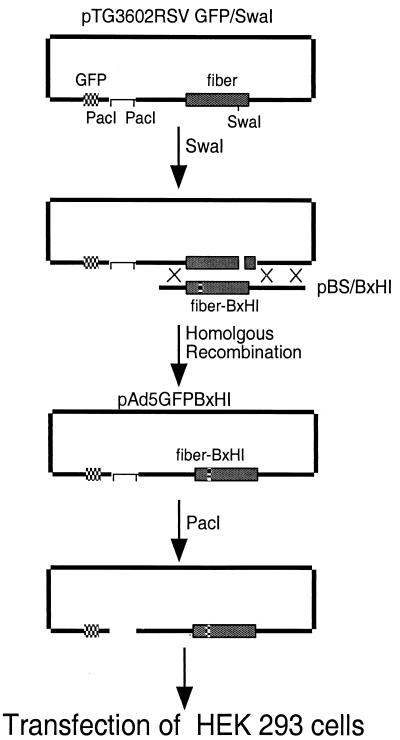
A full-length adenovirus backbone plasmid for recombination with pBS/BxHI was also generated. The plasmid pTG3602 (Transgene S.A.), which contains a wild-type fiber sequence, was modified to contain a unique SwaI site in fiber to facilitate homologous recombination. To accomplish this, pTG3602 was partially digested by NdeI and then ligated with an NdeI linker 5′-TACGCCCCATTTAAATGG-3′ containing an SwaI site (underlined). The plasmid was designated pTG3602/SwaI. pTG3602/SwaI was cut with ClaI and cotransformed into E. coli BJ5183 with ScaI-linearized pacAd5RSVGFP (1) to generate pTG3602RSVGFP/SwaI. Positive clones were screened by enzyme digestion and sequencing. A 4.6-kb BamHI and NotI restriction fragment was liberated from pBS/BxHI and cotransformed with SwaI-linearized pTG3602RSVGFP/SwaI into E. coli BJ5183 to generate pTG3602RSVGFP/BxHI (pAd5GFPBxHI; see Fig. 5).
FIG. 5.
Construction of recombinant adenoviruses with hTfR-targeting motifs in the HI loop. A series of HI loop-modified shuttle plasmids were generated (pBS/BxHI; see Materials and Methods). These shuttles were cut with BamHI and NotI, and the 4.6-kb fragment was cotransformed with SwaI-cut pTG3602RSVGFP/SwaI into E. coli BJ5183. Appropriate recombination resulted in full-length Ad5 genomes with a GFP expression cassette in the E1 region and B1, B2, etc., motifs in the HI loop. The recombinant plasmids were restricted with PacI and transfected into HEK 293 cells, and virus was harvested and propagated upon finding evidence of cytopathic effect.
Viruses.
Ad5GFP, with GFP under the control of Rous sarcoma virus (RSV) promoter, was from the Gene Transfer Vector Core, University of Iowa. hTfR-targeting viruses were generated by transfection of HEK 293 cells with PacI-digested peptide-modified virus vectors (6). pAd5GFPBxHI (10 to 15 μg) were digested with 16 U of PacI at 37°C for 2 h. The DNA was precipitated and transfected into HEK 293 cells using calcium phosphate (15). After 5 to 10 days, the lysates were harvested and further propagated on HEK 293 cells. Finally, viruses were purified by centrifugation in CsCl gradients according to standard protocols. Virus particle titers were determined spectrophotometrically (18). The viruses were named Ad5GFPBxHI, where “x” is the epitope number (B1, B2, etc.).
Fiber trimerization assays.
Fifty to 70% confluent HeLa cells in 150-mm plates were rinsed with serum-free MEM and then incubated with vaccinia virus VTF7-3 at a multiplicity of infection of 10 (a kind gift from Michael J. Welsh, University of Iowa) in 3 ml of MEM at 37°C for 1 h. Cells were washed and then transfected by pTM1Ad5fiber/BxHI plasmids or pTM1Ad5fiber (wild-type fiber) (10 μg) using Lipofectin (Gibco). After transfection, cells were rinsed and incubated in 30 ml of MEM–10% FBS at 37°C. The lysates were harvested 16 to 24 h later for trimerization assays. A 10-μl aliquot of lysate containing the recombinant proteins was subjected to reducing (31.25 mM Tris-Cl, pH 6.8; 1% sodium dodecyl sulfate [SDS]; 2.5% 2-mercaptoethanol [2-ME]; 10% glycerol) or nonreducing (the same except no 2-ME) conditions and fractionated by SDS–12% polyacrylamide gel electrophoresis (PAGE). The fractionated protein was transferred onto nitrocellulose membranes and probed by anti-fiber monoclonal antibody 4D2.5 (kindly provided by J. Engler, University of Alabama, Birmingham) (17). The film was developed using an ECL Kit (Amersham Pharmacia Biotech, Piscataway, N.J.) according to the manufacturer's recommendations.
TfR binding assays with Bx targeting motifs in context of fiber.
Protein G-conjugated agarose beads (60 μl; Pharmacia) were incubated with 15 μg of monoclonal antibody 128.1 directed against hTfR (generously provided by Ian Trowbridge, The Salk Institute, San Diego, Calif.). The 128.1-conjugated beads were resuspended in dilution buffer (10 mM Tris-Cl [pH 8.0], 140 mM NaCl, 0.1% Triton X-100, 0.1% BSA) and incubated with 15 μg of soluble hTfR for 1.5 h at 4°C. The complex was then incubated with 100 μl of vaccinia virus lysates containing wild-type or BxHI-modified fibers for 1.5 h at 4°C. The complexes were sequentially washed—twice with dilution buffer, twice with TSA buffer (10 mM Tris-Cl, pH 8.0; 140 mM NaCl), and once with 50 mM Tris-Cl (pH 6.8). The samples were denatured at 95°C for 3 min, followed by microcentrifugation. The disrupted complex was fractionated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed using 4D2.5 monoclonal antibody directed against an epitope in the Ad5 fiber tail (17). The experiments were repeated three times.
Transduction of TfR+ cells.
Experiments were carried out similarly to those described by Wickham et al. (33). hTfR+ CHO cells, human prostate cancer T24 cells, or human brain endothelial cells (105) were incubated with Ad5GFPBxHI or Ad5GFP (4 × 108 particles in 1 ml of DMEM–2% FBS) for 1 hour at 37°C. The cells were then washed three times with 2% FBS–DMEM, followed by incubation for 48 h at 37°C. The cells were then detached by trypsin and analyzed by using a fluorescence-activated cell sorter (FACS) at the University of Iowa FACS facility. In blocking experiments, hTfR+ CHO cells or T24 cells were incubated with human iron-loaded transferrin (40 μg/ml in 1 ml of DMEM–2% FBS) or soluble hTfR (15 μg/ml in 1 ml) for 30 min at 4°C before the addition of virus. Human brain endothelial cells were incubated with human iron-loaded transferrin (40 μg/ml in 1 ml) for 30 min at 4°C before the addition of virus. Data are presented as the means of triplicates ± the SEM. The experiments were repeated three times.
Immunohistochemistry.
T24 and BME cells were grown on 60-mm dishes, fixed in 2% paraformaldehyde, washed with PBS, and incubated with primary antibody (128.1 diluted 1:200 in PBS, 3% BSA, 0.3% Triton X-100, and 0.02% sodium azide) ON at 4°C. Plates were washed and incubated in rhodamine-conjugated goat anti-mouse (1:400) for 1 h at room temperature. Positive cells were visualized using an Olympus IX70 microscope, and images were captured using a SPOT RT digital camera.
RESULTS
hTfR targeting motifs.
The efficiency of adenovirus infection depends on the level of the coxackie adenovirus receptor (CAR) expression. However, the tropism of Ad5 can be modified using genetic methods (23, 33). As a first step toward extending the tropism of Ad5 to BME for use in CNS gene therapy, we screened a nonapeptide phage display library (5) against the extracellular domain of hTfR (30), a receptor found at high density on the BME. Enzyme-linked immunosorbent assay (ELISA) plates were coated with soluble hTfR, and bound phage were eluted by a low-pH buffer. Eluted phage were amplified in E. coli K91 and subsequently screened again for binding to the immobilized hTfR. In separate experiments, bound phage were eluted with purified transferrin holoenzyme. Three rounds of screening-amplification were done for each type of elution. From two independent experiments, a total of 43 clones were isolated, most from acid elution. Sequencing of isolated phage revealed the peptide motifs AKxxK/Rx, KxKxPK/R, or KxK in 31 of the clones (Table 1). Some clones were isolated more than once, arising from independent experiments and different elution parameters. GHKVKRPKG and IEAYAKKRK motifs were isolated 10 and 7 times, respectively. The peptide sequence KDKIKMDKK was present in four clones, while KNKIPKSPK was isolated twice. Only several peptides contained amino acid arrays identical to regions of human transferrin (Table 1), indicating that most motifs were likely conformational, rather than linear epitopes.
TABLE 1.
Displayed phage targeted to the TfRa
| Group | Sequence |
|---|---|
| 1 | LQAKKKRPK |
| VIAKIKKPK | |
| AIAKKHKWN | |
| VEAKGHKKK | |
| IEAYAKKRKc | |
| GHKAKGPRKb | |
| 2 | LQAKKKRPK |
| VIAKIKKPK | |
| KWKTPKVRV | |
| KNKIPKSPKc | |
| GHKAKGPRKb | |
| GHKVKRPKGc | |
| GKGPKWMRde | |
| 3 | VEAKGHKKK |
| AIAKKHKWN | |
| LQAKKKRPK | |
| VIAKIKKPK | |
| KWKTPKVRV | |
| KWKLHGHIK | |
| KNKIPKSPKc | |
| GHKAKGPRKb | |
| IEAYAKKRKc | |
| GHKVKRPKGc | |
| KDKIKMDKKbef |
Unless otherwise annotated, all epitopes were identified by acid elution. Underlined sequences indicate amino acids found in human transferrin.
Epitope identified using ligand elution parameters.
Epitope identified using both ligand and acid elution parameters.
This epitope contained only 8 amino acids.
Similar to KGKK in transferrin.
Similar to KDKSK in transferrin.
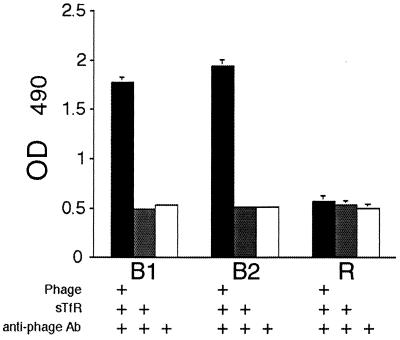
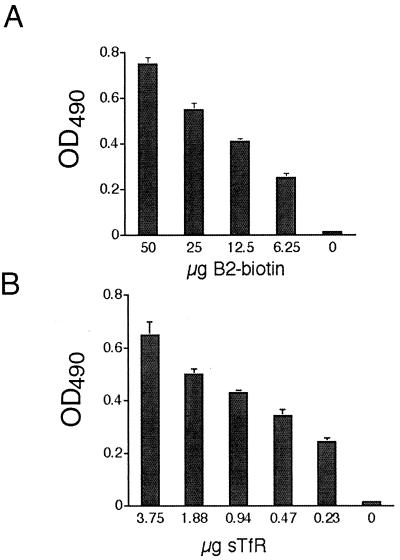
To test the specificity of peptide binding for human TfR, two motifs were randomly chosen: IEAYAKKRK (B1) and GHKVKRPKG (B2). First, phage expressing the B1 or B2 peptide and randomly isolated control phage (R) were subjected to large-scale amplification, followed by incubation with immobilized hTfR. Both B1 and B2 bound hTfR in contrast to control phage (Fig. 1). Binding of the B2 peptide motif to hTfR, independent of the phage sequences, was also tested. Biotinylated B2 was synthesized and incubated with immobilized hTfR using standard ELISA-based assays. The data in Fig. 2 show that peptide B2 alone bound to hTfR in a dose-dependent manner.
FIG. 1.
Phage expressing the hTfR-targeting motifs B1 or B2 bind immobilized hTfR. Amplified, purified phage containing B1, B2, or random peptide (R) were tested for their ability to bind hTfR on 96-well microtiter plates. Bound phage were detected using anti-fd antibodies as described in Materials and Methods. The data represent the means ± the SEM and are representative of three independent experiments.
FIG. 2.
Peptide motif B2 binds immobilized hTfR. B2 peptide was synthesized with an amino-terminal biotin moiety and tested for binding specificity to hTfR using an ELISA-based assay. (A) Soluble hTfR (3.75 μg) was coated onto ELISA plates, incubated with biotin-labeled B2 at various concentrations, and detected using extravidin as described in Materials and Methods. (B) Soluble hTfR at various concentrations was coated onto plates, followed by incubation with a constant concentration of B2-biotin (25 μg). Plates were developed using extravidin-horseradish peroxidase, followed by detection of the optical density at 490 nm as described in the text. The data represent the means ± the SEM from three independent experiments.
Characterization of recombinant fibers.
Motifs in phage may or may not be indicative of the same sequence in the context of Ad5 fiber. Moreover, the hTfR-targeting motifs could have deleterious effects on fiber trimerization inhibiting their assembly and in turn impairing virus capsid maturation (11, 17). To test if the identified nonapeptides retained their ability to bind hTfR in the context of fiber, immunoprecipitation experiments were performed. First, motifs were cloned into the HI loop and the resultant fibers were expressed in a mammalian expression system. Lysates containing the modified fibers were then incubated with hTfR, and the complex was immunoprecipitated with anti-hTfR antibodies. Western blot analysis for fiber using an antibody to the amino-terminal region indicated that the hTfR-targeting motifs, when placed in the HI loop of fiber, maintained their ability to bind to hTfR (Fig. 3, lanes B1 and B2). Wild-type fiber, though present at high levels in the expressed lysate (Fig. 3, lane WT*), was not pulled down with anti-hTfR antibodies (Fig. 3, lane WT), demonstrating specificity of the epitopes for the hTfR.
FIG. 3.
B2 or B1 epitopes in the HI loop of Ad5 fiber retain their ability to bind to hTfR. Wild-type or epitope-modified Ad5 fibers were expressed in HeLa cells using a vaccinia virus expression system as described in Materials and Methods. The expressed fibers were incubated with hTfR, and the complex was added to monoclonal antibody 128.1 (anti-hTfR antibody) previously adsorbed to protein G beads. The beads were isolated, and the complex was disassociated and fractionated by SDS-PAGE. Fibers pulled down by immunoprecipitation (lanes B1, B2, and WT) were detected using the 4D2.5 anti-fiber antibody, which recognizes an epitope in the tail region of the fiber (17). Lane WT* is vaccinia virus lysate expressing wild-type fiber that was not subjected to immunoprecipitation. The immunoblot is representative of three independent experiments.
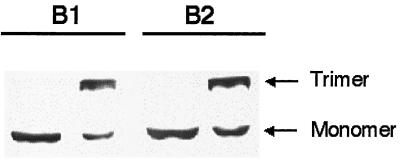
Lysates containing expressed, modified fibers were also used to determine the effects of the motifs on fiber trimerization. Other researchers have shown that trimerization can be disrupted by modifications at the carboxy terminus (17). Figure 4 shows that B1 and B2 motifs do not inhibit fiber trimerization. Similar results were seen for the other motifs (data not shown).
FIG. 4.
Effects of hTfR-targeting motifs on fiber trimerization. Epitope-modified Ad5 fibers were expressed in HeLa cells using a vaccinia virus expression system as described in Materials and Methods. The expressed fibers were subjected to reducing or nonreducing conditions prior to sample loading. Samples were fractionated by SDS-PAGE and blotted onto nitrocellulose. Expressed fibers were detected with antibody 4D2.5. The results are representative of the other hTfR-targeting motifs.
HI loop-modified viruses and gene transfer to hTfR-expressing cells.
The generation of multiple recombinant adenoviral vectors differing only in the HI loop could be best accomplished using the E. coli recombination system developed by Chartier et al. (6). For our purposes, two key plasmids were developed. The first was pBS/B2HI. This plasmid contained Ad5 fiber sequences with a unique SpeI site at the 3′ end of the fiber gene (see Materials and Methods). This site, in combination with an internal SphI site, allowed the subsequent introduction of all other HI modifications (pBS/B1HI, pBS/B3HI, etc.). It is important to note that pBS/B2HI will also allow the efficient introduction of any fiber modification. The fiber sequences contained more than 1.0 kb of flanking sequence for increased efficiency of recombination in E. coli.
A plasmid containing a full-length adenovirus genome with a green fluorescent protein (GFP) expression cassette in the E1 region and a unique SwaI site in fiber was also generated (see Materials and Methods). The SwaI site facilitated recombination with the pBS/BxHI plasmids (Fig. 5). The resultant plasmids were digested with PacI and transfected into HEK 293 cells. We noted considerable variability in the length of time from transfection to CPE and also our ability to further amplify the viruses once the initial lysates were harvested. Of the 10 peptides cloned into the fiber of Ad5, only 7 could be amplified and purified to concentrations adequate for further testing. These results indicate that amplification of our HI loop adenoviruses required retention of CAR binding for adequate growth in HEK 293 cells. Thus, the viruses have expanded rather than targeted tropism.
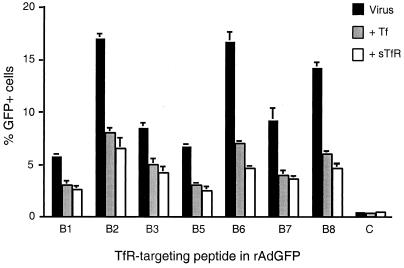
Peptide-modified recombinant adenoviruses were tested for their ability to transduce a CHO cell line previously transfected with recombinant hTfR. In this cell line, the endogenous CHO TfR has a 51-amino-acid deletion in the cytoplasmic domain leading to a nonfunctional receptor (22). Recombinant adenoviruses containing motifs B1 to B3 and B5 to B8 (Ad5GFPB1HI, Ad5GFPB2HI, etc.) facilitated gene transfer 11- to 34-fold over Ad5GFP (Fig. 6). In all cases, infection was inhibited by preincubating the cells with human transferrin, suggesting that gene transfer with TfR motif-modified adenoviruses occurred in part through specific binding to the hTfR. Preincubation of cells with soluble hTfR corroborated these results (Fig. 6).
FIG. 6.
Transduction of hTfR+ CHO cells by adenoviruses genetically modified to express hTfR-targeting epitopes. hTfR+ CHO cells (1 × 105) were infected by Ad5GFPB1HI, Ad5GFPB2HI, etc., or Ad5GFP (4 × 108 particles) for 1 h at 37°C, and GFP-positive cells were quantitated by FACS 48 h later. For blocking studies, cells were incubated with soluble hTfR (sTfR) or transferrin for 30 min at 4°C prior to the addition of virus. The various viruses are indicated by the TfR-targeting epitope (B1, B2, etc.). “C” corresponds to Ad5GFP. The data are the means ± the SEM from three independent experiments.
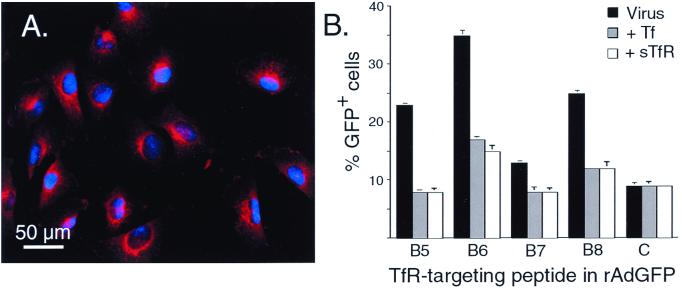
Ad5GFPB5HI through Ad5GFPB8HI were further tested on T24 cells, a cell line that has high endogenous levels of hTfR (Fig. 7A) and undetectable levels of CAR as assessed by reverse transcription-PCR and FACS analysis (Timothy Ratliff, unpublished observations). All motifs directed significant increases in gene transfer to T24 cells (Fig. 7B). Again, B6 and B8 epitopes were best. Ad5GFPB6HI and Ad5GFPB8HI allowed for transduction of 25 and 15% of cells (subtracting the background), respectively, a 3.9- or 2.8-fold increase over the control virus. Further, this increase could be abrogated by the prior addition of transferrin or purified soluble hTfR (Fig. 7B).
FIG. 7.
hTfR expression and transduction of T24 cells by hTfR-targeting epitope modified adenoviruses. (A) Immunostaining for hTfR expression. T24 cells were fixed and stained with monoclonal antibody 128.1, followed by rhodamine-conjugated goat anti-mouse antibodies. The photomicrograph is representative of the entire plate. (B) T24 cells (1 × 105) were transduced with Ad5GFPB5HI, Ad5GFPB6HI, etc., or Ad5GFP (4 × 108 particles), and GFP-positive cells were quantitated by FACS. For blocking experiments T24 cells, were incubated with human soluble hTfR or transferrin for 30 min at 4°C prior to the addition of virus. The various viruses are indicated by the hTfR-targeting epitope (B5, B6, etc.). “C” corresponds to Ad5GFP. The data are the means ± the SEM and are from three independent experiments.
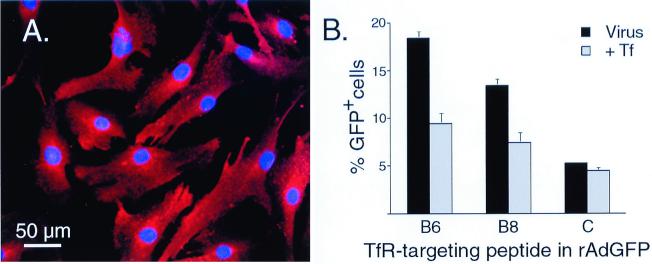
Human BME cells express a high level of the TfR (Fig. 8A). Initial studies showed that these cells are poorly transduced by recombinant adenoviruses expressing native Ad5 fiber sequences (Fig. 8B). However, when Ad5GFPB6HI and Ad5GFPB8HI were tested, gene transfer to human BME improved 3.5- and 2.5-fold, respectively. Again, transduction could be inhibited by preincubation with transferrin, indicating that the recombinant viruses were binding and entering BME cells via the targeted motif.
FIG. 8.
hTfR expression and Ad5GFPB6HI- or Ad5GFPB8HI-mediated transduction of human BME cells. (A) Expression of hTfR on human BME cells. Cells were fixed and stained as described in Fig. 7. (B) Human BME cells (1 × 105) were transduced with Ad5GFPB6HI, Ad5GFPB8HI, or Ad5GFP (4 × 108 particles), and GFP-positive cells were quantitated by FACS. For blocking experiments, cells were incubated with transferrin for 30 min at 4°C prior to the addition of virus. The data are the means ± the SEM and are from three independent experiments.
DISCUSSION
In this study we tested the feasibility of targeting recombinant viral vectors to the hTfR to transduce the BME. We took advantage of an available phage display library to identify motifs specific to the hTfR, and showed that these motifs did not inhibit fiber trimerization. In many cases fiber sequences modified to express the identified motifs in the HI loop allowed for the production of viable recombinant virus. Importantly, the sequences retained their ability to bind to hTfR and direct gene transfer to hTfR-expressing cells.
We found that the addition of peptide sequences into the HI loop of fiber was not uniformly well tolerated, even among sequences which were highly similar with regard to charge and side group. For instance, viruses containing the peptide B2 motif were capable of being amplified to some degree, while those containing the similar B6 motif (Table 2) had greatly improved growth properties. Similarly, the B4 sequence was not amenable to virus production, while the B3-containing virus grew quite well. Because we found that fiber trimerization was not affected, the block in virus production could result from steric hindrance of CAR binding. To overcome this problem for Ad5GFPB4HI, Ad5GFPB9HI, and Ad5GFPB10HI, amplification could be done using HEK 293 cells modified to express a surrogate receptor, such as previously described by others (8, 10).
TABLE 2.
Peptide motifs and their effects on recombinant adenovirus production
| Peptide | Epitope | Virus productiona |
|---|---|---|
| Control | ++++ | |
| IEAYAKKRK | B1 | ++ |
| GHKVKRPKG | B2 | ± |
| KDKIKMDKK | B3 | ++++ |
| G KNKIPKSPK LGSb | B4 | − |
| G GKGPKWMR LGS | B5 | +++ |
| G GHKAKGPRK LGS | B6 | ++++ |
| G VIAKIKKPK LGS | B7 | +++ |
| G KWKTPKVRV LGS | B8 | + |
| G LQAKKKRPK LGS | B9 | − |
| G VEAKGHKKK LGS | B10 | − |
“++++” indicates virus production characteristic of Ad5GFP. “−” indicates that the virus could not be amplified to reasonable titers.
Motifs B4 through B10 included an amino G and a carboxyl LGS linker.
Interestingly, earlier reports (8, 10) demonstrated the feasibility of using an epitope cloned into the HI loop or carboxy terminus of fiber, or penton base, to allow adenovirus binding and entry mediated by a membrane-bound ScFv directed to that epitope. Our data suggest that other ligand-receptor pairs may not be sufficient to support viral production when CAR-fiber interactions are impaired. HEK 293 cells express high levels of hTfR based on Western blot analyses and immunofluorescence microscopy (B.L.D., unpublished observations). However, production of B2 and B8 motif-modified adenoviruses was quite difficult even though binding to the hTfR was retained in the context of fiber and the intact virion.
The ability of a membrane-bound ScFv versus that of a recycling receptor-ligand pair (TfR-Tf) to serve as pseudoreceptor suggests that differences in recycling and internalization may be important. Data presented by Leopold and colleagues suggest that Ad5 escapes the endosome very early after internalization, probably before endosome-endosome fusion (19). It is possible that some HI loop-modified adenoviruses are impaired in their ability to direct early endosomal release, thereby allowing for some recycling and release of virus at the cell surface.
The B6 and B8 targeting motifs in recombinant adenovirus significantly improved gene transfer to hTfR+ CHO cells, T24 cells, and human BME cells. Also, both were more effective than the other motifs tested. The B2 epitope in Ad5GFPB2HI, which was only tested on hTfR+ CHO cell lines, has an amino acid sequence similar to that of B6. When cloned into adenovirus, both Ad5GFPB2HI and Ad5GFPB6HI resulted in an approximately 34-fold increase in gene transfer. The differences between the B2 and the B6 motifs are a valine in the +4 position (versus alanine), an arginine in the +6 position (versus glycine), a lysine in the +8 position (versus arginine), and a glycine in the +9 position (versus lysine). In cloning the B5-B8 epitopes into fiber, an amino-terminal glycine and a carboxyl-terminal LGS linker (relative to the nonapeptide sequence) was added. Thus, the linker did not appear to further improve or inhibit hTfR targeting. However, and importantly, the addition of the linker did facilitate virus growth as previously discussed.
At the outset of our studies, we expected that epitopes isolated by ligand elution would yield better results when cloned into the HI loop of fiber compared to those identified using acid wash. Our data show that the B3, B5, B7, and B8 sequences, identified by acid elution, facilitated adenovirus-mediated transduction to hTfR-expressing cells ca. 1.4-fold less effectively (on average) than did B6, B1, and B2. However, both B1 and B2 were identified by acid and transferrin elution, suggesting that there is only modest correlation between elution parameters and the ability of that motif to facilitate receptor targeting, at least for the hTfR.
Targeting recombinant virus vectors to a receptor expressed at high density on BME cells is a first step toward testing if the vascular system can be used to facilitate a global distribution of enzyme to the CNS for inhibition or reversal of neurodegeneration. If sufficient levels of transduction to the endothelia could be accomplished, basolateral secretion would provide a source of enzyme to an extensive area of the brain. In brains of larger animal models or in humans, such an approach would bypass the requirement of multiple parenchymal injections, which could result only in small, nonoverlapping spheres of correction.
In summary, our data suggest that several short motifs, when cloned into the HI loop of fiber, can support hTfR-targeted transduction with recombinant adenovirus vectors. It will be interesting to determine if these epitopes can also be used to improve gene transfer of other encapsidated viruses, such as adeno-associated virus.
ACKNOWLEDGMENTS
We thank Richard D. Anderson and Joseph Zabner for critical discussions and Christine McLennan for secretarial assistance.
This study was supported by the American Heart Association (H.X.), the State of Iowa Biosciences Initiative (Q.M.), and the National Institutes of Health (HD33531 and DK54759). B.L.D. is a fellow of the Roy J. Carver Trust.
REFERENCES
- 1.Anderson R D, Haskell R E, Xia H, Roessler B J, Davidson B L. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7:1034–1038. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- 2.Bosch A, Perret E, Desmaris N, Trono D, Heard J M. Reversal of pathology in the entire brain of mucopolysaccharidosis type VII mice after lentivirus-mediated gene transfer. Hum Gene Ther. 2000;11:1139–1150. doi: 10.1089/10430340050015194. [DOI] [PubMed] [Google Scholar]
- 3.Bouri K, Feero W G, Myerburg M M, Wickham T J, Kovesdi I, Hoffman E P, Clemens P R. Polylysine modification of adenoviral fiber protein enhances muscle cell transduction. Hum Gene Ther. 2000;10:1633–1640. doi: 10.1089/10430349950017635. [DOI] [PubMed] [Google Scholar]
- 4.Broadwell R D, Baker-Cairns M J, Friden P M, Oliver C, Villegas J C. Transcytosis of protein through the mammalian cerebral epithelium and endothelium. III. Receptor-mediated transcytosis through the blood-brain barrier of blood-borne transferrin and antibody against the transferrin receptor. Exp Neurol. 1996;142:47–65. doi: 10.1006/exnr.1996.0178. [DOI] [PubMed] [Google Scholar]
- 5.Burritt J B, Quinn M T, Jutila M A, Bond C W, Jesaitis A J. Topological mapping of neutrophil cytochrome b epitopes with phage-display libraries. J Biol Chem. 1995;270:16974–16980. doi: 10.1074/jbc.270.28.16974. [DOI] [PubMed] [Google Scholar]
- 6.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doran S E, Ren X D, Betz A L, Pagel M A, Neuwelt E A, Roessler B J, Davidson B L. Gene expression from recombinant viral vectors in the central nervous system after blood-brain barrier disruption. Neurosurgery. 1995;36:965–970. doi: 10.1227/00006123-199505000-00012. [DOI] [PubMed] [Google Scholar]
- 8.Douglas J T, Miller C R, Kim M, Dmitriev I, Mikheeva G, Krasnykh V, Curiel D T. A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat Biotechnol. 1999;17:470–475. doi: 10.1038/8647. [DOI] [PubMed] [Google Scholar]
- 9.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 10.Einfeld D A, Brough D E, Roelvink P W, Kovesdi I, Wickham T J. Construction of a pseudoreceptor that mediates transduction by adenoviruses expressing a ligand in fiber or penton base. J Virol. 1999;73:9130–9136. doi: 10.1128/jvi.73.11.9130-9136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falgout B, Ketner G. Characterization of adenovirus particles made by deletion mutants lacking the fiber gene. J Virol. 1988;62:622–625. doi: 10.1128/jvi.62.2.622-625.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friden P M, Olson T S, Obar R, Walus L R, Putney S D. Characterization, receptor mapping and blood-brain barrier transcytosis of antibodies to the human transferrin receptor. J Pharmacol Exp Ther. 1996;278:1491–1498. [PubMed] [Google Scholar]
- 13.Ghodsi A, Stein C, Derksen T, Yang G, Anderson R D, Davidson B L. Extensive β-glucuronidase activity in murine CNS after adenovirus mediated gene transfer to brain. Hum Gene Ther. 1998;9:2331–2340. doi: 10.1089/hum.1998.9.16-2331. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez R, Vereecque R, Wickham T J, Vanrumbeke M, Kovesdi I, Bauters F, Fenaux P, Quesnel B. Increased gene transfer in acute myeloid leukemic cells by an adenovirus vector containing a modified fiber protein. Gene Ther. 1999;6:314–320. doi: 10.1038/sj.gt.3300836. [DOI] [PubMed] [Google Scholar]
- 15.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 16.Harari O A, Wickham T J, Stocker C J, Kovesdi I, Segal D M, Huehns T Y, Sarraf C, Haskard D O. Targeting an adenoviral gene vector to cytokine-activated vascular endothelium via E-selectin. Gene Ther. 1999;6:801–807. doi: 10.1038/sj.gt.3300898. [DOI] [PubMed] [Google Scholar]
- 17.Hong J S, Engler J A. Domains required for assembly of adenovirus type 2 fiber trimers. J Virol. 1996;70:7071–7078. doi: 10.1128/jvi.70.10.7071-7078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishibashi M, Maizel J V., Jr The polypeptides of adenovirus. VI. Early and late glycopolypeptides. Virology. 1974;58:345–361. doi: 10.1016/0042-6822(74)90070-1. [DOI] [PubMed] [Google Scholar]
- 19.Leopold P L, Kreitzer G, Miyazawa N, Rempel S, Pfister K K, Rodriguez-Boulan E, Crystal R G. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 20.Nilaver G, Muldoon L L, Kroll R A, Pagel M A, Breakefield X O, Davidson B L, Neuwelt E A. Delivery of herpesvirus and adenovirus to nude rat intracerebral tumors after osmotic blood-brain barrier disruption. Proc Natl Acad Sci USA. 1995;92:9829–9833. doi: 10.1073/pnas.92.21.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pardridge W M, Buciak J L, Friden P M. Selective transport of an anti-transferrin receptor antibody through the blood-brain barrier in vivo. J Pharmacol Exp Ther. 1991;259:66–70. [PubMed] [Google Scholar]
- 22.Recht L D, Raso V, Davis R, Salmonsen R. Immunotoxin sensitivity of Chinese hamster ovary cells expressing human transferrin receptors with differing internalization rates. Cancer Immunol Immunother. 1996;42:357–361. doi: 10.1007/s002620050294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds P, Dmitriev I, Curiel D. Insertion of an RGD motif into the HI loop of adenovirus fiber protein alters the distribution of transgene expression of the systemically administered vector. Gene Ther. 1999;6:1336–1339. doi: 10.1038/sj.gt.3300941. [DOI] [PubMed] [Google Scholar]
- 24.Scriver C R, et al. The metabolic basis of inherited disease. New York, N.Y: McGraw-Hill Book Company; 1989. [Google Scholar]
- 25.Sferra T J, Qu G, McNeely D, Rennard R, Clark K R, Lo W D, Johnson P R. Recombinant adeno-associated virus-mediated correction of lysosomal storage within the central nervous system of the adult mucopolysaccharidosis type VII mouse. Hum Gene Ther. 2000;11:507–519. doi: 10.1089/10430340050015707. [DOI] [PubMed] [Google Scholar]
- 26.Shin S-U, Friden P, Moran M, Olson T, Kang Y-S, Pardridge W M, Morrison S L. Transferrin-antibody fusion proteins are effective in brain targeting. Proc Natl Acad Sci USA. 1995;92:2820–2824. doi: 10.1073/pnas.92.7.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skorupa A F, Fisher K J, Wilson J M, Parente M K, Wolfe J H. Sustained production of β-glucuronidase from localized sites after AAV vector gene transfer results in widespread distribution of enzyme and reversal of lysosomal storage lesions in a large volume of brain in mucopolysaccharidosis VII mice. Exp Neurol. 1999;160:17–27. doi: 10.1006/exnr.1999.7176. [DOI] [PubMed] [Google Scholar]
- 28.Staba M J, Wickham T J, Kovesdi I, Hallahan D E. Modifications of the fiber in adenovirus vectors increase tropism for malignant glioma models. Cancer Gene Ther. 2000;7:13–19. doi: 10.1038/sj.cgt.7700104. [DOI] [PubMed] [Google Scholar]
- 29.Stein C S, Ghodsi A, Derksen T, Davidson B L. Systemic and central nervous system correction of lysosomal storage in mucopolysaccharidosis type VII mice. J Virol. 1999;73:3424–3429. doi: 10.1128/jvi.73.4.3424-3429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testa U, Pelosi E, Peschle C. The transferrin receptor. Crit Rev Oncog. 1993;4:241–276. [PubMed] [Google Scholar]
- 31.Watson G L, Sayles J N, Chen C, Elliger S S, Elliger C A, Raju N R, Kurtzman G J, Podsakoff G M. Treatment of lysosomal storage disease in MPS VII mice using a recombinant adeno-associated virus. Gene Ther. 1998;5:1642–1649. doi: 10.1038/sj.gt.3300775. [DOI] [PubMed] [Google Scholar]
- 32.Wickham T J, Haskard D, Segal D, Kovesdi I. Targeting endothelium for gene therapy via receptors up-regulated during angiogenesis and inflammation. Cancer Immunol Immunother. 1997;45:149–151. doi: 10.1007/s002620050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 34.Wickham T J, Segal D M, Roelvink P W, Carrion M E, Lizonova A, Lee G M, Kovesdi I. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol. 1996;70:6831–6838. doi: 10.1128/jvi.70.10.6831-6838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickham T J, Tzeng E, Shears L L II, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon S-K, Mohr L, O'Riordan C R, Lachapelle A, Armentano D, Wands J R. Targeting a recombinant adenovirus vector to HCC cells using a bifunctional Fab-antibody conjugate. Biochem Biophys Res Commun. 2000;272:497–504. doi: 10.1006/bbrc.2000.2788. [DOI] [PubMed] [Google Scholar]