Abstract
Osmotic stress significantly hampers plant growth and crop yields, emphasizing the need for a thorough comprehension of the underlying molecular responses. Previous research has demonstrated that osmotic stress rapidly induces calcium influx and signaling, along with the activation of a specific subset of protein kinases, notably the Raf-like protein (RAF)-sucrose nonfermenting-1–related protein kinase 2 (SnRK2) kinase cascades within minutes. However, the intricate interplay between calcium signaling and the activation of RAF-SnRK2 kinase cascades remains elusive. Here, in this study, we discovered that Raf-like protein (RAF) kinases undergo hyperphosphorylation in response to osmotic shocks. Intriguingly, treatment with the calcium chelator EGTA robustly activates RAF-SnRK2 cascades, mirroring the effects of osmotic treatment. Utilizing high-throughput data-independent acquisition–based phosphoproteomics, we unveiled the global impact of EGTA on protein phosphorylation. Beyond the activation of RAFs and SnRK2s, EGTA treatment also activates mitogen-activated protein kinase cascades, Calcium-dependent protein kinases, and receptor-like protein kinases, etc. Through overlapping assays, we identified potential roles of mitogen-activated protein kinase kinase kinase kinases and receptor-like protein kinases in the osmotic stress–induced activation of RAF-SnRK2 cascades. Our findings illuminate the regulation of phosphorylation and cellular events by Ca2+ signaling, offering insights into the (exocellular) Ca2+ deprivation during early hyperosmolality sensing and signaling.
Keywords: osmotic stress, calcium signaling, phosphorylation, RAF, SnRK2
Graphical Abstract
Highlights
-
•
Application of the calcium chelator EGTA activates RAF-SnRK2 cascades in Arabidopsis.
-
•
The phosphoproteomics profiles triggered by EGTA and mannitol largely overlap.
-
•
EGTA and mannitol activate MAP4Ks and RLKs independently of RAF-SnRK2 cascades.
In Brief
Osmotic stress impedes plant growth, necessitating understanding of molecular responses. Research shows calcium influx and RAF-SnRK2 kinase activation under osmotic stress. In this study, we found the calcium chelator EGTA activates RAF-SnRK2 cascades similarly to osmotic stress. High-throughput DIA phosphoproteomics revealed EGTA's broader impact on protein phosphorylation, activating MAPKs, CDPKs, and receptor-like protein kinases. Assays highlighted MAP4Ks and receptor-like kinases in osmotic stress–induced RAF-SnRK2 cascade activation, shedding light on Ca2+ signaling regulation and osmotic stress response.
Drought, cold, and salinity impose osmotic stress on plant cells, resulting in reduced water mobility and hindering plant growth, ultimately limiting crop yields (1, 2, 3, 4, 5). In response to osmotic stress, plants undergo a series of biochemical and physiological changes to adapt to these unfavorable conditions. These changes encompass rapid increases in cytosolic-free calcium concentration within seconds, activation of multiple protein kinases, phosphorylation of effector proteins, transcription of stress-responsive genes, biosynthesis of osmolytes, and reprogramming of growth and development. The elevation of cytosolic-free calcium concentration and activation of protein kinases are known to be the most rapid events in plant osmotic stress responses (6).
Previous studies have demonstrated that drought and salinity rapidly induce the accumulation of free calcium in the cytosol, with calcium peaks observed at 15 s and 1.5 min after osmotic shock (7). Utilizing an aequorin-based calcium imaging system, Yuan et al. (2014) identified a mutant, osca1 (reduced hyperosmolality induced [Ca2+]i increase 1), which displayed impaired osmotic calcium signaling in guard cells and root cells (8). OSCA1 encodes a hyperosmolality-gated calcium-permeable channel and is a homolog of the mammalian TMEM63 (9, 10, 11, 12). OSCA1.3 activity is regulated by botrytis-induced kinase 1 (BIK1), a receptor-associated protein kinase activated during immunity. Botrytis-induced kinase 1–mediated phosphorylation of the N-terminus increases the channel activity of OSCA1.3, which is crucial for stomatal closure during immune signaling (13). Similarly, using a similar approach, Chen et al. (2020) identified bonzai1 (bon1) as another mutant showing impaired calcium accumulation in response to osmotic stress (14). The bon1 and bon123 triple mutants exhibited reduced transcription of hyperosmolarity-responsive genes and increased sensitivity to osmotic stress. Although OSCA1 and BON1 have important roles in early osmotic stress signaling, the downstream effectors of OSCA1 and their exact roles in osmotic stress signaling, remain largely unknown.
Protein kinases and phosphorylation are also known to play central roles in osmotic sensing and early signaling (15). Osmotic stress rapidly triggers the activation of multiple protein kinases, including mitogen-activated protein kinases (MAPKs), calcium-dependent protein kinases, sucrose nonfermenting-1–related protein kinase 2s (SnRK2s), and Raf-like protein kinases (RAFs) (16, 17, 18, 19, 20, 21, 22, 23, 24, 25). Among these, RAFs and SnRK2s are activated within minutes by hyperosmotic shock and play central roles in osmotic stress signaling, which are the focus of this study. SnRK2s are plant-specific Ser/Thr protein kinases that play central roles in plant osmotic stress signaling (26, 27, 28, 29, 30). In the model plant Arabidopsis, there are ten members in the SnRK2 family, namely SnRK2.1 to SnRK2.10. Nine of these members, except SnRK2.9, are rapidly activated by hyperosmolarity (18, 26). Once activated, SnRK2s directly phosphorylate numerous target proteins, including transcription factors, chromatin modulators, ion channels, and others, to regulate a wide range of adaptation and developmental processes (27, 28, 29). The importance of SnRK2s in osmotic stress signaling is highlighted by the hypersensitivity to osmotic stress and inability to complete the life cycle observed in snrk2-decuple mutant lacking all ten SnRK2s (26).
In addition to their role in osmotic stress responses, three SnRK2s, SnRK2.6 (also known as open stomata 1, OST1), SnRK2.2, and SnRK2.3, act as positive regulators in the receptor-coupled signaling pathway of the phytohormone abscisic acid (ABA) (31, 32, 33, 34, 35). In the absence of ABA, A clade protein phosphatase 2C (PP2C) phosphatases bind to and dephosphorylate SnRK2s, and deactivate ABA signaling. Under multiple stresses such as cold and dehydration, ABA levels increase through de novo biosynthesis or release from conjugated forms like ABA-GE (36). ABA binds to ABA receptors, pyrabactin resistance/pyrabactin resistance-likes/regulation component of ABA receptors, forming a complex that inhibits PP2C proteins and releases SnRK2s (32, 37, 38). The SnRK2s are then phosphorylated and reactivated by a subgroup of Raf-like protein kinases (20, 21, 22, 39, 40, 41).
Raf-like protein kinases are named for their homologies of mammalian Raf kinases. In Arabidopsis, there are several Raf-like kinases divided into three groups: A-RAF (commonly referred to as mitogen-activated protein kinase kinase kinase or MAPKKK), B-RAF, and C-RAF (ZIK) (23, 42). The B-RAF group consists of 22 members classified into four subgroups: three B1 RAFs, six B2 RAFs, six B3 RAFs, and seven B4 RAFs. Recent studies have shown that B2, B3, and B4 RAFs are rapidly activated by osmotic stresses (20, 22, 41). Once activated, B-RAFs phosphorylate specific serine/threonine residues in the activation loop of SnRK2s, effectively reactivating the dephosphorylated inactive SnRK2s in vitro (20, 21, 22, 39, 40). In high-order mutants lacking B4 RAFs (OK130-null), osmotic stress–induced ABA-independent SnRK2 activation is greatly diminished, resulting in hypersensitivity to various osmotic stresses (20). B2 and B3 RAFs are required for the activation of ABA-dependent SnRK2s, and high-order mutants of B2/B3 RAFs show the ability to germinate and grow even under extremely high concentrations of ABA, resembling the phenotypes of snrk2.2/2.3/2.6 or high-order mutants of ABA receptors (39, 40, 43). Furthermore, a recent study reported that the guard cell–enriched B1RAF RAF15, along with B3 subgroup member RAF6, phosphorylates SnRK2.6 (SnRK2.6/OST1), which is essential for ABA-induced stomatal closure (44, 45). The B4 RAF kinases are also known as central mediators of rapid auxin responses across diverse plant species (24, 25, 46).
The relationship between Ca2+ signaling and RAF-SnRK2 cascades in osmotic stress sensing and early signaling is not yet fully understood (47). In vitro studies have shown that the kinase activity of recombinant SnRK2s is independent of Ca2+ (17). Furthermore, the application of EGTA, a calcium ion chelator, does not significantly affect the activation of SnRK2 (p42 and p44 in Figure 5B, Droillard et al., 2002) in response to hyperosmolarity in Arabidopsis cell suspensions (17). These findings suggest that the activation of SnRK2s may not directly depend on Ca2+ levels. Additionally, the activation of RAF-SnRK2 in response to mannitol-induced osmotic stress appears to be independent of OSCA1 and its homologs (20). This observation is supported by the identical activation of RAF-SnRK2 in the osca sep mutants, which are defective in OSCA1 and six homologs, compared to the WT (20). Interestingly, the activation of SnRK2 in response to mannitol is significantly enhanced in the bon123 triple mutants, which lack BON1, BON2, and BON3 proteins (14). Recently, our study revealed that SnRK2.6/OST1 phosphorylates cyclic nucleotide-gated channels and mediates Ca2+ in guard cells (48), suggesting that RAF-SnRK2 cascade may act upstream of Ca2+ signaling. Thus, the exact role of Ca2+ in the RAF-SnRK2 cascade activation and the involvement of Ca2+-dependent and independent phosphorylation events in the early osmotic response of plants require further investigation.
Fig. 5.
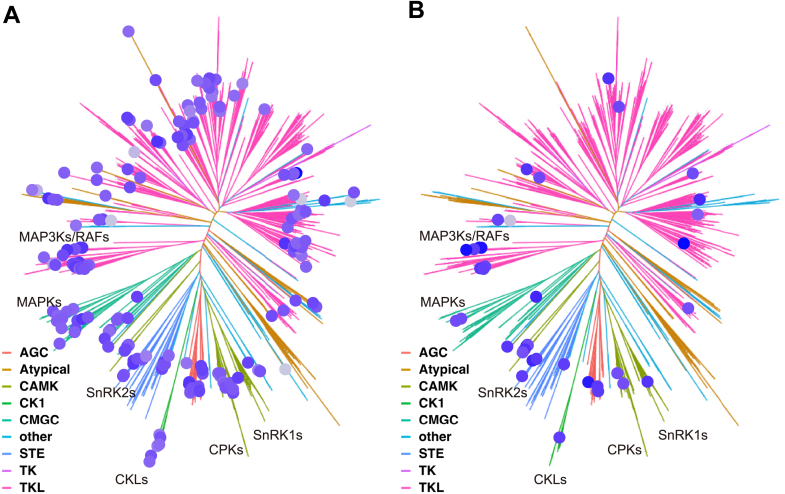
KinMap showing the EGTA and mannitol upregulated protein kinase.A, the protein kinase induced by EGTA treatment in our phosphoproteomics as shown in the KinMap. B, the protein kinase induced by mannitol treatment in our phosphoproteomics as shown in the KinMap.
In this study, we measured the RAF kinase activation in Arabidopsis seedlings upon application of Ca2+, Ca2+ chelator, and inhibitors or activators of the Ca2+ channel. We found that application EGTA, a Ca2+ chelator, quickly induced the activation of RAFs and SnRK2s. By data-independent acquisition (DIA)-based high-resolution phosphoproteomics, we in-depth profiled the EGTA- and mannitol-responsive phosphorylation events. We found that EGTA triggers a global alteration of protein phosphorylation at the proteomic level. In addition to induce the phosphorylation of RAFs, SnRK2, and a set of SnRK2 targets, EGTA treatment also activates MAPK cascades and promotes the phosphorylation of calcium-dependent protein kinases, receptor-like protein kinases. Our results reveal a potential role of (exocellular) Ca2+ deprivation in early hyperosmolality sensing and signaling and shed new light on Ca2+ signaling regulated phosphorylation and cellular events.
Experimental Procedures
Experimental Design and Statistical Rationale
To analyze the phosphoproteomic changes induced by mannitol and EGTA, three biological replicates were collected from 10-day-old Arabidopsis seedlings (see Plant materials and treatment). This results in three conditions, comprising nine samples. The DIA label-free approach was employed to quantify the perturbation in protein phosphorylation following treatment with EGTA and mannitol. The untreated seedlings were designated as the control condition. Each individual sample was analyzed through a single DIA run, leading to a total of nine DIA raw files. To generate the spectral library, phosphopeptides enriched from the three conditions were pooled and subsequently separated into eight fractions. Each fraction was analyzed twice, yielding a total of 16 data-dependent acquisition (DDA) runs. A hybrid library was generated in Spectronaut software (https://biognosys.com/software/spectronaut/) by combining 16 DDA runs from fractions with nine DIA runs from samples. The statistical analysis is described in the Data Analysis section.
Plant Materials and Treatment
Seeds were surface-sterilized with 75% ethanol and then grown vertically on the 1/2 Murashige-Skoog medium containing 0.75% agar in a growth chamber set to 22 °C, 60% humidity, and a 16 h light/8 h dark photoperiod. Ten-day-old seedlings were treated with either 800 mM mannitol for 5 min or 20 mM EGTA at pH 8.0 for 120 min. For phosphoproteomics analysis, the seedlings underwent one of three treatments for 30 min: EGTA, mannitol, or a mock treatment.
Generation of RAF24 Antibody
Rabbit was immunized with the 1 to 175 amino acids segment of the Arabidopsis RAF24 protein (ABclonal corporation). Then, the anti-RAF24 antibody, which was affinity purified, was validated by immunoblotting. The samples tested included these from the WT, raf24 (Wiscdslox430D04), raf42 (Salk_147328), OK130-weak, and the RAF24 KO mutant (OK130-null) (20) (Supplemental Fig. S1).
Immunoblotting
Approximately, 20 mg of 10-day-old seedlings, subjected to specified treatments, were ground into fine powders in liquid nitrogen. Total plant proteins were extracted using 60 μl of protein extraction buffer containing protease and phosphatase inhibitors (100 mM Hepes, pH 7.5, 5 mM EDTA, 5 mM EGTA, 10 mM DTT, 10 mM Na3VO4, 10 mM NaF, 50 mM β-glycerophosphate, 1 mM PMSF, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml aprotinin, 5% glycerol). The supernatant was collected by centrifugation at 12,000g at 4 °C for 20 min. For dephosphorylation, 20 μg of total protein was incubated with lambda protein phosphatase in protein metallo phosphatases buffer, supplemented with 7 mM MnCl2 (New England Biolabs, P0573L), for 30 min at 30 °C. Proteins (20 μg), both with and without dephosphorylation treatment, were separated by 6% or 10% SDS-PAGE and transferred onto a polyvinylidene fluoride membrane (Millipore, IPVH00010, immobilon-P polyvinylidene fluoride membrane, 0.45 μm pore size). The membrane was incubated overnight at 4 °C with antibody against RAF24 (ABclonal, customize, 0.2 μg/ml), p-S175-SnRK2.6 (49), p-S171-SnRK2.6 (39), pERK1/2 (Cell Signaling Technology, 9102S, 1:5000), MPK3 (Sigma-Aldrich, M8318, 1 μg/ml), and MPK6 (Sigma-Aldrich, A7104, 0.2 μg/ml) in Tris Buffered Saline with Tween 20 with 1% nonfat milk or BSA. The immunoblots were probed with an anti-actin antibody (ABclonal, AC009, 1:10,000) as the loading control. Primary antibodies were detected using either anti-rabbit (BioRad, 1706515, 0.05 μg/ml) or anti-mouse (Bio-Rad, 1706516, 0.05 μg/ml) secondary antibodies conjugated with horseradish peroxidase and visualized using enhanced chemiluminescence reagent (Shenger).
In-Gel Kinase Assay
For in-gel kinase assays, 20 μg of extracted proteins were used for SDS-PAGE, which was embedded with 0.5 mg/ml histone as the kinase substrate. After electrophoresis, the gel was washed at washing buffer (25 mM Tris–HCl, pH 7.5, 0.5 mM DTT, 10 mM Na3VO4, 5 mM NaF, 0.5 mg/ml BSA, and 0.1% Triton X-100) and subsequently incubated at 4 °C overnight with three changes of renaturing buffer (25 mM Tris–HCl, pH 7.5, 1 mM DTT, 0.1 mM Na3VO4, and 5 mM NaF). The gel was then incubated in 30 ml of reaction buffer (25 mM Tris–HCl, pH 7.5, 2 mM EGTA, 12 mM MgCl2, 1 mM DTT, and 0.1 mM Na3VO4) with 200 nM ATP plus 50 μCi of [γ-32P] ATP for 90 min. The reaction was stopped by transferring the gel into a solution of 5% (w/v) trichloroacetic acid and 1% (w/v) sodium pyrophosphate. The gel was washed for 5 h with five changes of the wash solution. Radioactivity was detected using a Typhoon 9500 scanner.
Extraction of Protein for Proteomics
Arabidopsis seedlings were ground to fine powders in liquid nitrogen in a mortar. Protein digestion was carried out using a suspension trapping (S-Trap) micro column (ProtiFi), following a previous report method (50). Briefly, total proteins were lysed by 5% (v/v) SDS in 50 mM triethylamine bicarbonate (TEAB). The protein concentration was determined by bicinchoninic acid assay (Thermo Fisher Scientific, 23227). One hundred micrograms of protein was reduced and alkylated using 10 mM tris(2-carboxyethyl)phosphine hydrochloride and 40 mM 2-chloroacetamide at 45 °C for 15 min. Subsequently, a final concentration of 5.5% (v/v) phosphoric acid was added, followed by a 6-fold volume of binding buffer (90%, v/v, methanol in 100 mM TEAB) to the protein solution. After gentle vortexing, the solution was loaded into an S-Trap microcolumn. The solution was removed by spinning the column at 4000g for 1 min. The column was washed three times with 150 μl binding buffer. Finally, 20 μl of digestion solution containing 1 unit Lys-C (Wako Chemicals, 10 AU/vial) and 1 μg trypsin (Promega, 0.5 μg/μl) in 50 mM TEAB was added to the column, and the mixture was incubated at 47 °C for 2 h. Each digested peptide was eluted using 120 μl of 1% (v/v) TFA in 80% (v/v) acetonitrile (ACN) and directly loaded into a Fe-immobilized metal ion affinity chromatography (IMAC) tip.
Phosphopeptide Enrichment
Phosphopeptides were enriched using a modified Fe-IMAC tip protocol (51). Briefly, an in-house IMAC tip was made by plugging a 20-μm polypropylene frit disk into the tip end and packed with 10 mg of nickel-nitrilotriacetic acid silica resin (Qiagen). The packed IMAC tip was inserted into a 2-ml Eppendorf tube. Ni2+ ions were first removed by adding 100 mM EDTA (200g, 1 min). Next, the tip was activated with 100 mM FeCl3 and equilibrated with 1% (v/v) acetic acid at pH 3.0 before loading the sample. The tryptic peptides were dissolved in 1% (v/v) TFA and 80% (v/v) ACN and loaded onto the IMAC tip. Washing steps were performed using 1% (v/v) TFA, 80% (v/v) ACN (200g, 1 min), and 1% (v/v) acetic acid (pH 3.0). Subsequently, the Fe-IMAC tip was inserted into an activated desalting SDB-XC (3M) StageTip. The bound phosphopeptides were eluted onto the activated desalting SDB-XC StageTip using 200 mM NH4H2PO4 and directly eluted into sample vials for LC-MS/MS analysis. The eluted peptides were dried under vacuum.
Basic pH Reverse Fractionation
The procedures followed the previously described method (52). Initially, 2 mg of Magic C18-AQ beads (5 μm particle size) were suspended in 100 μl of methanol and loaded into a 200 μl StageTip equipped with a 20 μm polypropylene frit. The C18 StageTips were activated with 100 μl of 40 mM NH4HCO2 at pH 10 in an 80% ACN solution and then equilibrated with 100 μl of 200 mM NH4HCO2 at pH 10. The isolated phosphopeptides were resuspended in 200 mM NH4HCO2 at pH 10, while the C18 StageTips were washed with 100 μl of 200 mM NH4HCO2 at pH 10. Subsequently, the bound phosphopeptides were fractionated from the StageTip using 50 μl of eight buffers containing varying ACN concentrations of ACN: 5%, 8%, 11%, 14%, 17%, 20%, 23%, and 80% ACN, all in a 200 mM NH4HCO2 solution at pH 10. The eluted phosphopeptides were dried and stored at −20 °C for further mass spectrometry (MS) analysis.
LC-MS/MS Analysis
The indexed retention time peptides (Biognosys, KI-3002-2) were dissolved in 50 μl of 0.1% (v/v) formic acid (FA) with 3% (v/v) ACN to achieve a 10× stock solution. The stock solution was then diluted 10-fold. Dried peptides were reconstituted in 10 μl of 0.1% (v/v) FA with 3% (v/v) ACN containing 1× indexed retention time peptides, and 4 μl of the sample was injected into an Ultimate 3000 UHPLC system coupled with a Q-Exactive mass spectrometer (Thermo Fisher Scientific). The mobile phases consisted of 0.1% FA (buffer A) and 0.1% FA in 80% ACN (buffer B). Peptides were separated on a 25-cm Thermo Acclaim PepMap RSLC column (75 μm × 25 cm, 2 μm, 100 Å) at a flow rate of 450 nl/min. The gradient settings for peptide separation were as follows: 3 to 5% buffer B in 5 min, 5 to 25% buffer B over 85 min, 25 to 85% buffer B in 2 min, maintained at 85% buffer B for 5 min, 85 to 3% buffer B in 1 min, and held at 3% buffer B for 22 min. For DDA analysis, the acquisition cycle of the MS data was performed in DDA mode with a full survey MS scan, followed by 10 tandem mass spectrometry (MS/MS) targeting of the top ten precursor ions from the MS scan. The MS1 scan was executed with a resolving power of 70,000 across a m/z range of 350 to 1,600, with an automatic gain control (AGC) target of 3e6. Dynamic exclusion was enabled. The MS/MS acquisitions utilized a 2 m/z isolation window, 27% normalized collision energy (NCE), an AGC target of 1e5, and a resolving power of 17,500. For DIA analysis, the MS1 scan mass range was set from 350 to 950 m/z, with a resolution of 70,000, an injection time of 30 ms, an AGC target of 3e6, and an NCE of 27%. For the MS2 scan, 28 windows of 20 m/z width each, with an overlap of 1 Da, covered the range from 400 to 906 m/z. The MS2 resolution was set to 17,500, injection time to 50 ms, AGC target to 1e6, and NCE to 27%.
Data Analysis
MS raw files were processed using Spectronaut software (version 18.4, Biognosys) (53). Hybrid libraries were generated within Spectronant by combining the DDA runs of fractionated samples with the DIA runs from the experiments. Araport 11 fasta file (27,533 entries; version date: 2022-09-15) was downloaded from the TAIR website for use. In phosphoproteomics analysis, serine/threonine/tyrosine phosphorylation, protein N-terminal acetylation, and methionine oxidation were set as variable modifications, while cysteine carbamidomethylation was set as a fixed modification. Trypsin was selected as the digestion enzyme following a maximum of two missed cleavages. The post-translational modification localization cut-off was set to 0. The false discovery rate (FDR) was set to 1% at both peptide and protein levels, with precursor and peptide q value cut-offs set to 0.2 and 0.01, respectively. The Protein q value experiment and run wide cut-offs were set to 0.01 and 0.05, respectively. Results were filtered for the best N fragments per peptide ranging between three and six. The MS1 and MS2 mass tolerance strategy was set to system default. The number of unique peptides assigned to each protein and the percentage of protein sequence coverage for each run are shown in Supplemental Table S1.
Further data and statistical analyses were conducted using Perseus software (version 2.0.5.0, https://www.maxquant.org/perseus/) (54). The phosphopeptides reports from Spectronaut were analyzed using the peptide collapse plug-in tool (version 1.4.4), with the localization cut-off set at 0.75 for class I sites (55). Summed phosphorylation site intensities were log2-transformed. Phosphorylation sites quantified in all three replicates in at least one condition were selected for cluster analysis. Missing values were imputed using the “replace missing value from normal distribution” function in Perseus software, with a width 0.2 and a down shift 2.0. A multiple-sample test (ANOVA, permutation-based FDR = 0.05) identified significantly changed phosphorylation sites, which were then z-scored before hierarchical clustering analysis. Principal component analysis was performed using the phosphopeptides identified across all nine DIA runs. For selecting significantly changed phosphorylation sites upon EGTA or mannitol treatment, statistical analysis was applied to phosphorylation sites identified in at least two replicates within at least one sample group. The missing value of abundance was replaced by the average abundance of phosphorylation sites in the group. Significantly upregulated phosphorylation sites were filtered by a fold change (FC) > 2, and a p < 0.05 (Student’s t test).
GO Analysis
Gene ontology (GO) analysis of EGTA-induced phosphoproteins and five different cluster proteins was conducted using AgriGO V2.0 (56) (http://systemsbiology.cau.edu.cn/agriGOv2/index.php). The singular enrichment analysis tool was utilized, selecting Arabidopsis thaliana as the species for this analysis. Other parameters in AgriGO were set as follows: Fisher’s exact tests, the statistical test method, Yekutieli (FDR under dependency), the significance level of 0.05. A minimum of five mapping entries was required, and the complete GO type was analyzed. Original GO enrichment results were filtered and refined using REVIGO to remove semantic redundancy (57). The kyoto encyclopedia of genes and genomes (KEGG) pathway analysis was performed using DAVID (58) (version 6.8) (https://david.ncifcrf.gov/summary.jsp), with results visualized using GraphPad Prism (version 10, https://www.graphpad-prism.cn) to plot. Supplemental Table S6 displays the significant GO terms and KEGG pathway results.
Arabidopsis kinMap
A phylogenetic tree of 1078 Arabidopsis kinases was calculated using FastTree (59) and visualized with R package ggtree (60). The sequence information for the kinases was downloaded from the iEKPD database (61) (http://iekpd.biocuckoo.org/download.php). The interactive Arabidopsis kinome MAP (AtKinMap) was constructed utilizing the R package shiny and is hosted on the shinyapps.io platform (https://little-ant.shinyapps.io/ath_kinases/#kinmap).
Results
EGTA Triggers the Phosphorylation of RAF-SnRK2 Cascades
In our previous studies, we observed that B2/B3 and B4 RAF kinases are quickly activated by hyperosmolarity using the in-gel kinase assay (20). However, this assay is challenging, requiring a large amount of radioactive ATP and meticulous attention to protein denaturation, renaturation, and activity assay procedures over 3 days. To overcome these challenges, we generated an antibody specifically recognizing RAF24 (AT2G35050) (Supplemental Fig. S1, A and B, which exhibited specific bands in Col-0 WT, raf42 mutant (20), OK130-weak (raf16;raf40/hcr1;RAF24Δ30;raf18;raf20;raf35;RAF42Δ18), a mutant allele with a 30 bp deletion in RAF24, but not in raf24 mutant (20) and OK130-null (raf16;raf40/hcr1;raf24;raf18;raf20;raf35;raf42) (20), a high-order mutant carrying knockout mutations in all seven B4 RAFs. Using this antibody, we discovered that mannitol treatment rapidly induced a size shift of RAF24 (Fig. 1A, lane 1–5). Dephosphorylation by lambda protein phosphatase completely abolished the mannitol-induced size shift of RAF24 (Fig. 1A, lane 6–10). Thus, osmotic stress quickly triggers the phosphorylation and size shift of RAF24, which could be used to indicate quick RAF activation in response to hyperosmolarity or other stimuli.
Fig. 1.
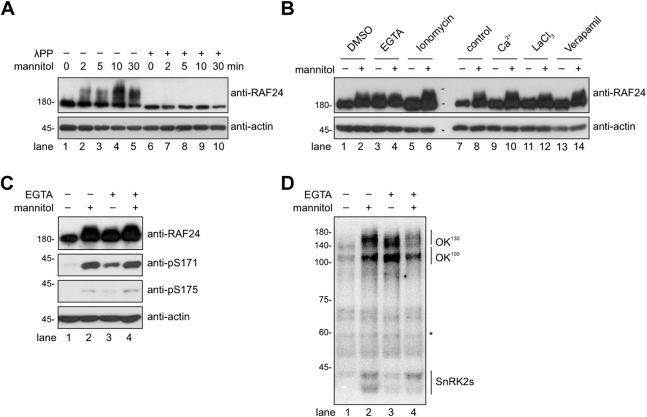
EGTA triggers activation of RAF-SnRK2 cascades.A, the anti-RAF24 immunoblot showing RAF24 hyperphosphorylation after the indicated time of mannitol treatment. The λPP treatment was performed to remove all phosphorylation in the samples. B, the anti-RAF24 immunoblot showing the RAF24 phosphorylation after the treatment of indicated chemicals. C, the immunoblot showing the phosphorylation of RAF24 and conserved serines in SnRK2s corresponding to Ser175 in SnRK2.6. D, in-gel kinase assay showing the SnRK2, OK100 (B2/3 RAF), and OK130 (B4 RAF) activities after 5 min treatment of 800 mM mannitol or 120 min treatment of 20 mM EGTA. Anti-actin immunoblot or the nonspecific band indicated by an asterisk are used as loading controls. The images shown are representative of three independent experiments. λPP, lambda protein phosphatase; SnRK2, sucrose nonfermenting-1–related protein kinase 2.
To investigate the role of Ca2+ in osmotic stress-triggered RAF and SnRK2 activation, we pretreated the Arabidopsis seedling with different chemicals inhibiting or inducing Ca2+ influx and measured the mannitol-induced RAF24 phosphorylation in these seedlings. These chemicals we used are as follows: lanthanum(III) chloride and verapamil (Ca2+ channel blockers), ionomycin (a calcium ionophore that allows free entry of calcium), and EGTA (a calcium chelator). As shown in Figure 1B, preincubation with CaCl2, lanthanum(III) chloride, Verapamil, or ionomycin for 120 min did not induce RAF24 phosphorylation or affect mannitol-induced RAF24 phosphorylation. Interestingly, the application of EGTA alone was sufficient to trigger RAF24 phosphorylation and size shift (Fig. 1B, lane 3), resembling the treatment of mannitol.
We then verified whether applying EGTA could induce the phosphorylation of SnRK2s using antibodies recognizing the phosphorylated serine residues corresponding to Ser171 and Ser175 in SnRK2.6 (49, 62, 63). The results showed that the application of EGTA also led to an increase in the phosphorylation of these two serine residues (Fig. 1C), known to occur upon SnRK2 activation (62, 64). Consistent with the immunoblot results, the in-gel kinase assay demonstrated that EGTA treatment significantly activated RAFs (OK100 and OK130) and SnRK2s, as indicated by their ability to phosphorylate histone proteins embedded in the gel matrix (Fig. 1D). Taken together, our findings indicate that the application of exogenous EGTA, mimicking mannitol treatment, can trigger the activation of both RAFs and SnRK2s.
Profiling of Phosphoproteomic Changes Upon Mannitol and EGTA Treatments
Inspired by the EGTA-induced RAF activation results, we employed the DIA phosphoproteomics approach to profile the phosphorylation-mediated signaling trigged by EGTA in Arabidopsis. In parallel, we utilized mannitol treatment, a known strong activator of the RAF and SnRK2 cascade, and no treatment as controls. To this end, we collected Arabidopsis seedlings from EGTA-treated, mannitol-treated, and no treatment conditions in triplicates. Digested phosphopeptides were enriched using the S-Trap-IMAC approach, as detailed in the Experimental Procedures section. Subsequently, we measured the enriched phosphopeptides using Q-Exactive with a 120-min DIA run.
As a result, we identified a total of 22,991 phosphorylated precursors and 21,209 unique phosphorylated peptides, corresponding to 6515 phosphorylation sites on 1790 proteins, using Spectronaut analysis of the triplicate of control, EGTA, and mannitol-treated seedlings. Among these, 19,231 phosphorylated precursors were quantified in at least two replicates of at least one experimental condition, indicating the depth of the Arabidopsis phosphoproteome (Supplemental Table S2). Notably, 5067 sites were identified as class I phosphorylation sites (Supplemental Table S3). We then evaluated the sample quality and reproducibility of each condition by conducting principal component analysis based on the identified phosphopeptides across nine replicates. The result showed that the three conditions were distinct from each other, but the biological replicates of each condition were clustered together (Supplemental Fig. S2). This suggests that the three conditions displayed good reproducibility and unique phosphoproteomic profiles.
EGTA Treatment Triggers Dramatic Changes in Global Phosphorylation
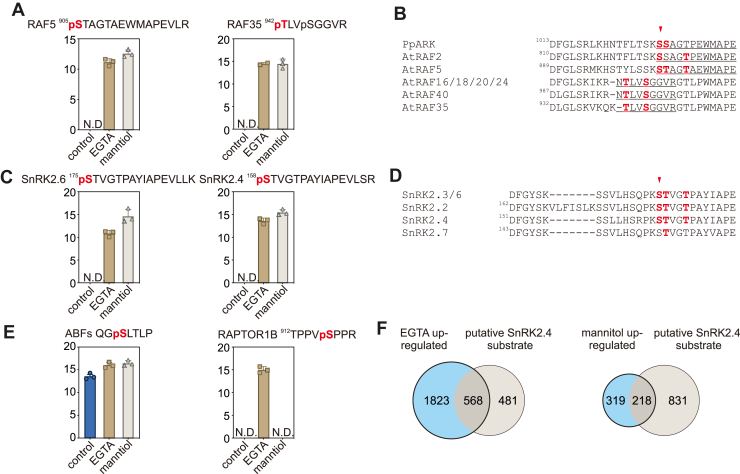
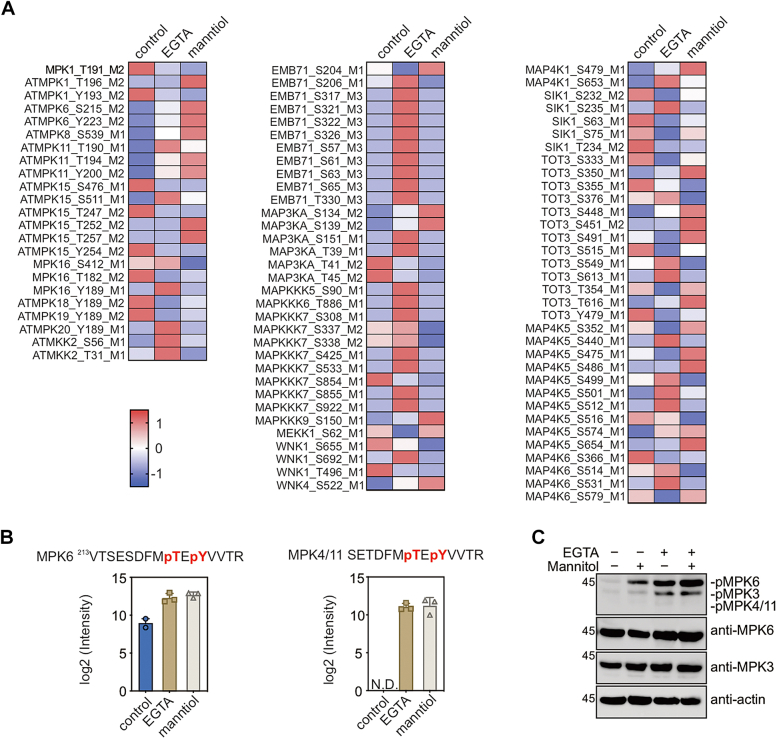
We observed that the EGTA treatment significantly impacted the Arabidopsis phosphoproteome. It induced the abundance of 1803 phosphopeptides while decreasing 964 phosphopeptides (Supplemental Table S4). Notably, an additional 8292 phosphopeptides were exclusively identified following the EGTA treatment, indicating a widespread rewiring of the phosphoproteome. Consistent with the observed activation of RAFs and SnRK2s upon EGTA treatment (Fig. 1), we noted a marked increase in the phosphorylation of more than 65 phosphosites in RAFs and 15 in SnRK2s (FC > 2, p < 0.05, Student’s t test) after EGTA treatment (Supplemental Table S4). For example, the phosphorylation of several conserved serine/threonine residues in the activation loop of RAF2/enhanced disease susceptibility 1(EDR1), RAF5/sugar insensitive 8(SIS8), and RAF35, which was induced by EGTA treatment or exclusively detected in EGTA-treated samples (Fig. 2A). The phosphorylation of Ser1029 in ABA- and abiotic stress-responsive Raf-like kinase (PpARK) (Fig. 2B, indicated by red arrow), a homology of Arabidopsis RAFs, is induced by either ABA or osmotic stress treatment (19). Importantly, Ser1029Ala mutation at this residue in PpARK, abolishing phosphorylation, resulted in the dysfunction of PpARK (65). Similarly, the conserved serine residues corresponding to Ser175 in SnRK2.6/OST1 (Fig. 2, C and D, indicated by red arrow)—a known dominant RAF target site—exhibited increased phosphorylation following EGTA treatment (Fig. 1C). In addition to RAF and SnRK2 subgroup protein kinases, we also noticed a set of downstream effector proteins of RAF-SnRK2 cascade, like ABA-responsive kinase substrate 1/2/3 (40, 66), BRAHMA (BRM) (67), regulatory associated protein of TOR (68, 69), VARICOSE (70), VARICOSE-related (71), SWEET11/12 (72), ABA-responsive element (ABRE)-binding protein/ABRE-binding factors (73), NADPH oxidase respiratory oxidase homolog D (74), At inhibitor-2 (75), RAF36 (76), five domain protein required for endosomal sorting 1 (77), SnRK2-substrate 1 (27), are presented in the list of EGTA upregulated phosphoproteins. The representative phosphosites in ABRE-binding factors and regulatory associated protein of TOR are shown in Figure 2E. Furthermore, our analysis revealed a high percentage overlap of EGTA- and mannitol-induced phosphoproteins with putative SnRK2.4 substrates identified by KALIP 2.0 (29). As shown in Figure 2F, 568 (54.1%) out of 1,049 putative SnRK2.4 substrates are overlapped with the 2,391 EGTA-induced phosphoproteins (FC > 2, p < 0.05, Students’s t test, Supplemental Table S5). Additionally, 218 (40.6%) out of 537 mannitol-induced phosphoproteins (FC > 2, p < 0.05, Students’s t test) were found in the list of putative SnRK2.4 substrates (Fig. 2F, right panel). These findings strongly supported that EGTA treatment activates RAF-SnRK2 cascades, characterized by the phosphorylation/activation of RAF and SnRK2 subfamily kinases, and phosphorylation of SnRK2 downstream substrates.
Fig. 2.
EGTA induces phosphorylation of RAFs and SnRK2s.A, the relative intensity of the phosphopeptides from RAF5 (B3 subgroup) and RAF35 (B4 subgroup) in seedlings without or with EGTA or mannitol treatment (n = 3 biological replicates). N.D., not detected. B, sequence alignment showing the conserved phosphosites in the Arabidopsis RAFs and PpARK. The phosphosites in Arabidopsis RAFs identified by this study and in PpARK/PpCTR1 are highlighted in red, and an arrow indicates the conserved serine residue corresponding to Ser1029 in PpARK/PpCTR1. C, the relative intensity of the conserved phosphosite in SnRK2.6 and SnRK2.4 in seedlings without or with EGTA or mannitol treatment (n = 3 biological replicates). N.D., not detected. D, sequence alignment showing the conserved phosphosites in SnRK2s. The conserved phosphosites are highlighted in red, and the conserved serine residue corresponding to Ser175 in SnRK2.6 is indicated by an arrow. E, the phosphorylation of conserved phosphorylation site in ABFs (left) and Raptor1B in seedlings without or with EGTA or mannitol treatment (n = 3 biological replicates). N.D., not detected. F, venn diagrams showing the overlaps of putative SnRK2.4 substrates (Wang et al., 2020) and EGTA (left) or mannitol (right) upregulated phosphoproteins. ABF, ABRE-binding factor; PpARK, ABA and abiotic stress-responsive Raf-like kinase; SnRK2, sucrose nonfermenting-1–related protein kinase 2.
Moreover, to activate the RAF-SnRK2 cascades, GO and KEGG pathway enrichment analysis (Fisher’s exact test, p < 0.05) revealed EGTA treatment also induced the phosphorylation of proteins involved in MAPK signaling pathway, mRNA processing, spliceosome, and photosynthesis (Supplemental Fig. S3, Supplemental Table S6). These comprehensive findings reveal the multifaceted roles of EGTA in the osmotic stress signaling mechanisms in Arabidopsis.
Comparison of EGTA- and Mannitol-Responsive Phosphoproteomics
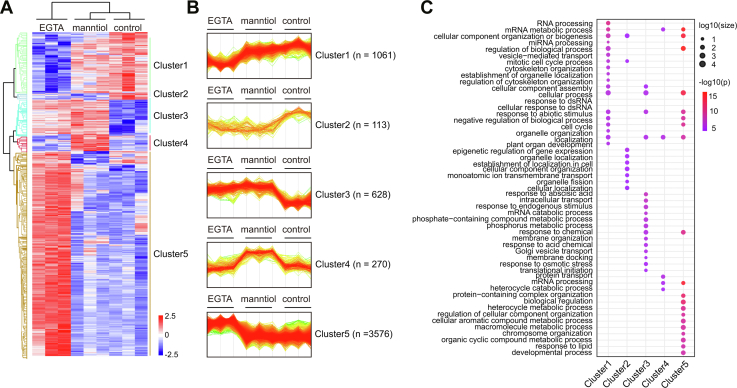
To gain further insights into the role of EGTA in hyperosmolarity signaling, we performed hierarchical clustering and GO analyses on the EGTA- and mannitol-responsive phosphosites and phosphoproteins. Our approach included an ANOVA test with a controlled FDR less than 0.05, followed by a hierarchical clustering analysis based on the z-scored levels of each phosphosite (Fig. 3A). This analysis allowed us to categorize the 5648 EGTA phosphosites responsive to EGTA and mannitol, representing 1658 phosphoproteins could be classified into five clusters (Fig. 3, A and B, Supplemental Table S7). Cluster 1 comprises 1061 phosphosites and represents 468 unique protein groups whose phosphorylation is exclusively repressed by EGTA treatment. GO term enrichment analysis of cluster 1 revealed that these proteins are primarily involved in RNA pressing, vesicle-mediated transport, and cytoskeleton organization (Supplemental Table S6). Cluster 2 contains 113 phosphosites, representing 69 proteins whose phosphorylation is repressed by both EGTA and mannitol treatments. The enriched GO terms in cluster 2 relate to plant organ development, cellular component organization, and organelle fission. Six hundred twenty-eight phosphosites in 249 proteins upregulated by EGTA and mannitol treatments are classified to cluster 3. GO term enrichment for cluster 3 highlights their roles in response to ABA and osmotic stress, membrane organization and docking, and golgi vesicle transport. Cluster 4 consists of 270 phosphosites in 131 phosphoproteins, exclusively upregulated by mannitol treatment, suggesting their specific response to this stimulu. Cluster 5 contains 3576 phosphosites in 1262 phosphoproteins that are exclusively upregulated by EGTA treatment. The phosphoproteins in cluster 5 are predominantly associated with functions related to lipid response and chromatin organization. In summary, our analysis not only sheds light on the extensive regulation of upregulated phosphoproteins by mannitol treatment but also reveals the broad influence of EGTA treatment on the phosphorylation of other proteins involved in development, metabolism, photoperiodism, cell cycle process, and chromatin organization.
Fig. 3.
Comparison of EGTA- and mannitol-responsive phosphoproteomics.A, heat map showing the result of the hierarchy clustering analysis of z-score intensities of phosphorylation sites in control, EGTA, or mannitol treatment. B, the profile plot of the five different clusters. C, the bubble matrix plot showing the GO terms enriched in each cluster. GO, gene ontology.
MAPKs and CPKs Upregulated by EGTA Application
In addition to the activation of RAFs and SnRK2s, we noticed the MAPK signaling pathway is enriched in the KEGG analysis results (Supplemental Fig. S3 and Supplemental Table S6). A classic MAPK cascade typically comprises a series of three serine/threonine kinases that act sequentially. In this cascade, the terminal components, known as MAPKs, are activated by MAPK kinases (MAPKKs or MEKs) through dual phosphorylation of conserved threonine and tyrosine residues within the T x Y motif located in the activation loop (T-loop). MAPKKs, in turn, are activated by MAPKK kinases (MAPKKKs or MEKKs) through the phosphorylation of conserved serine and/or threonine residues within their T-loop (23).
Our results unveiled that the canonical T x Y motif in MPK6, MPK15, and MPK4 exhibited mannitol- and EGTA-induced phosphorylation, suggesting that the potential activation of these MAPKs by mannitol and EGTA (Fig. 4A, left panel, and 4B, Supplemental Table S8). Furthermore, two MAPKKs, MKK2 and MKK5, displayed strong phosphorylation induced by EGTA. Of note, the MPK4, MKK2, and MEKK1 cascade plays a crucial role in basal resistance, and its disruption leads to dwarf and autoimmune phenotypes (78, 79). MKK5 is a well-known upstream activator of MPK3/6 and is involved in various processes such as stomatal development, abscission, inflorescence architecture, and responses to both abiotic and biotic stresses (80, 81, 82, 83).
Fig. 4.
MAPK cascade activated by EGTA and mannitol treatment.A, heat map showing the relative abundance of the phosphopeptides in MAPKs, MAPKKs, MAP3Ks, and MAP4Ks phosphoproteomics. The color intensity indicates the z-score intensities. B, the relative intensity of the phosphopeptides from MPK6 and MPK4/11 in seedlings without or with EGTA or mannitol treatment (n = 3 biological replicates). C, the immunoblot showing the EGTA- and mannitol-induced phosphorylation of MAPKs. The protein extract from 10-day-old seedlings with indicated treatments was subjected to immunoblots with anti-pERK1/2, anti-MPK3, and anti-MPK6 antibodies. The anti-actin immunoblot is shown as a protein loading control. The images shown are representative of two independent experiments. MAPK, mitogen-activated protein kinase; MAPKKK, mitogen-activated protein kinase kinase kinase.
Additionally, our observations extended to the hyperphosphorylation of several protein kinases belonging to the MAP4K family upon EGTA and mannitol treatments. There are ten putative MAP4Ks in Arabidopsis (84). Five out of them, MAP4K1, MAP4K3/salt inducible kinases 1 (SIK1s), MAP4K4/target of temperature 3 (TOT3), MAP4K5, and MPK4K6, show EGTA-induced phosphorylation in our result (Fig. 4A, right panel, Supplemental Table S8).
To validate our proteomics data, we performed an immunoblot to assay the MAPK activity using an antibody that recognizes the phosphorylated T x Y motif. As shown in Figure 4C, the phosphorylation of MPK3 and MPK6 is quickly induced by EGTA and mannitol treatments. We cannot detect the MPK4/11 band in immunoblot results, likely due to their relative low abundance. Taking the phosphoproteomics and immunoblot results together, multiple components in the MAPK cascade are activated by applying of EGTA. We also noticed that EGTA treatment triggers the activation of a group of calcium-dependent protein kinases (CPKs), including CPK6, CPK13, CPK19, CPK21, and CPK32. In the meantime, mannitol treatment induces the phosphorylation of several serine/threonine residues in CPK6, CPK9, and CPK21 (Supplemental Fig. S4).
KinMap analysis of EGTA and mannitol-regulated protein kinases
KinMap is a powerful tool to facilitate the phylogenetic information of human protein kinases (85). Unfortunately, such an analysis tool is lacking in plants. Here, we adopt the KinMap with 1068 annotated protein kinases in Arabidopsis and generate an AtKinMap tool (https://little-ant.shinyapps.io/ath_kinases/#kinmap). Using this online tool, the mannitol and EGTA-triggered phosphorylation of protein kinases are visualized (Fig. 5). Compared to mannitol treatment that mainly activates the phosphorylation of CPKs, SNF1-related kinase 1, SnRK2s, MAPKs, and MAP3Ks (Fig. 5B), the EGTA treatment has broader effects on induction of protein kinase phosphorylation (Fig. 5A). The 38 protein kinases whose phosphorylation are induced by both EGTA and mannitol treatments are showing in Supplemental Fig. S5 (see also Supplemental Table S9).
Discussion
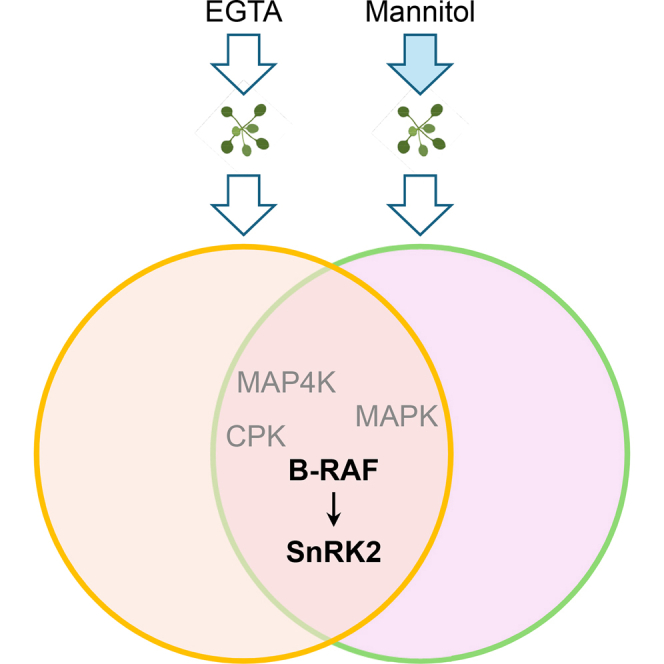
The RAF and SnRK2 cascades are quickly activated by hyperosmolarity in minutes and play a pivotal role in orchestrating a wide array of downstream stress adaptation processes (20, 25, 41). However, the mechanisms underlying the rapid activation of RAF-SnRK2 cascades during osmotic stress have remained poorly understood. This study shows that the Ca2+ chelator EGTA, but not other modulators on Ca2+ fluxes, can strongly activate both RAFs and SnRK2s. The EGTA treatment quickly induces the phosphorylation of RAF and SnRK2 protein kinases, including several key phosphosites in the activation loops of RAFs and SnRK2s required for their activation. Similar to mannitol treatment, the application of EGTA triggers the activation of RAFs and SnRK2s in the context of the capability to strongly phosphorylate histone proteins embodied in the sodium dodecyl (lauryl) sulfate-polyacrylamide gel matrix (Fig. 1D). To our knowledge, it is the first report of exocellular Ca2+ ions depletion, leading to the activation of the RAF-SnRK2 kinase cascades in plants.
It has been well documented that the exocellular Ca2+ serves dual roles in plants, contributing to both structural integrity and crucial signal functions. Calcium ion determines the rigidity of the cell wall by cross-linking acidic pectin residues and stabilizes cell membranes through the interaction with phospholipids (86). Application of EGTA on seedlings deprives the exocellular Ca2+, reducing the mechanical strength of cell walls (87). This loss of exocellular Ca2+ can influence membrane permeability, leading to ions and metabolite leakage. Furthermore, EGTA also triggers an elevation of cellular Ca2+ concertation, partially achieved by the release of Ca2+ from intracellular Ca2+ stores in tobacco cells (88). Consequently, the application of EGTA induces many disturbances, affecting cell wall integrity, membrane properties, and intracellular Ca2+ dynamics. This, in turn, results in a comprehensive reshaping of the protein phosphorylation landscape. Hyperosmolarity, induced by conditions such as drought or high salinity, triggers a series of transient biophysical changes within plant cells. These include the reduction of turgor pressure, shrinkage of the plasma membrane, and physical alterations of the cell wall. Osmotic stress also rapidly instigates a cytosolic Ca2+ signal, a process likely mediated through the mechanosensitive ion channel OSCA and Ca2+-responsive phospholipid-binding protein BONs (8, 14). Consequently, the application of both EGTA and mannitol, though differing in their modes of action, induces similar biophysical perturbation in the cell wall, membrane, and cytosolic Ca2+ signaling, resulting in a substantial overlap in downstream protein phosphorylation profiles. However, the distinctive feature of EGTA is its ability to induce more extensive changes in plant phosphoproteome compared to mannitol (Fig. 5), suggesting that EGTA exerts a more severe stimulus on plant cells. In alignment with this notion, a significant number of proteins related to membrane and cytoskeleton organization appear exclusively upregulated in response to EGTA treatment. Furthermore, the effects of EGTA treatment can be likened to those triggered by pathogen attacks or wounding, as evidenced by the phosphorylation of proteins involved in plant immunity, such as LysM-containing receptor-like kinase 3, enhanced disease resistance 2, pattern-triggered immunity compromised receptor-like cytoplasmic kinase 2. Notably, GO terms related to cell cycle and vegetative to reproductive phase transition of meristem are also enriched in EGTA exclusively upregulated phosphoproteins. For instance, during the G1 to S phase transition, cyclin-dependent kinase A;1/cell division control 2 phosphorylates retinoblastoma-related protein 1, releasing it from the E2F repression and activating the transcription of S-phase genes (89, 90). Both cyclin-dependent kinase A;1 and retinoblastoma-related protein 1 show EGTA upregulated phosphorylation in our result. This data may explain how EGTA and Ca2+ signals regulate the cell cycle observed 40 years ago (91, 92). Thus, identifying hundreds of phosphoproteins regulated by EGTA provides a valuable resource for comprehending the downstream events associated with the depletion of exocellular Ca2+ in plants.
As previously mentioned, the primary objective of this study was to investigate the phosphorylation events preceding the activation of RAF-SnRK2 signaling cascades in response to hyperosmolarity. Employing a similar strategy to that used in the quantitative comparison of mannitol-responsive phosphoproteomics between WT and snrk2-dec mutant plants, we successfully identified the involvement of B-subgroup RAFs in SnRK2 activation (20). In this study, we noticed that some protein kinases, like LysM RLK1-interacting kinase 1, MAP4K4/TOT3, MAP4K5, PBS1-like kinase 17 (PBL17), BS1-like kinase 27 (PBL27), and BR-signaling kinases, also rapidly activated by both mannitol and EGTA treatments. B-RAFs are classified into the canonical MAPKKKs family, which is highly conserved across yeast, animals, and plants (23). These MAPKKKs are not only activated directly by some receptor-like kinase but also, as seen in yeast and animals, can sometimes be phosphorylated by MAP4K. For example, in yeast under hypertonic stress, the yeast Ste20 acts as a MAP4K that directly phosphorylates Ste11, a MAP3K, to induce glycerol synthesis and initiate a cellular osmoregulatory response (93). The mammalian Ste20 homologs Pak1 and Pak2 are also activated under hyperosmotic stress and subsequently activate the MAP3K Raf-1 by direct phosphorylation (94, 95). In Arabidopsis, there are ten putative MAP4Ks, and three of which, MAP4K1, MAP4K4/TOT3, and MAP4K5, exhibit induced phosphorylation in response to both mannitol and EGTA treatments. Additionally, the phosphorylation of the other two members, MAP4K3/SIK1 and MAP4K6, is also induced by EGTA treatment. SIK1 can complement the ste20 yeast mutant phenotypes with respect to bud site selection and mitotic exit. The sik1 mutant displays dwarfism with reduced cell numbers, endoreduplication, and cell expansion (96). Besides MAP4K, two PBS1-like (PBL) kinases, PBL17 and PBL27, exhibited up-regulated phosphorylation in response to mannitol and EGTA treatments. PBL17 and PBL27 are crucial components of plant resistosomes, responsible for recognizing the different effectors produced by pathogens (97, 98). Specifically, PBL27 interacts ZED1-related kinase 3 to form complex with hopz-activated resistance 1 (ZAR1), which recognizes effectors HopF1r and small molecular Zaractin (99, 100). Upon activation, PBL27 has the capacity to phosphorylate MPKKK5, further activating the MAPK cascade (97, 101). While there is currently no direct evidence elucidating the role of MAP4Ks and resistosome/PBLs in osmotic stress signaling in plants, the identification of MAP4Ks/PBLs in our results suggests their potential roles in phosphorylating and activating the RAF-SnRK2 cascades. This discovery opens the door to future investigations to better understand the relationship between MAP4Ks, immune-related protein kinases, and their roles in osmotic sensing and early signaling in plants.
It is worth noting that the activation of RAF-SnRK2 cascades might not be contingent on calcium signaling. Several pioneering works have demonstrated that hyperosmolality-induced SnRK2 activation remains unaffected by the exogenous application of calcium ion chelators (17, 18). Genetic evidence further supports this observation; the malfunction of seven Arabidopsis OSCA homologs in osca-sep mutants does not impair RAF-SnRK2 activation upon mannitol treatment (20). Surprisingly, the bon1/2/3 triple mutant, which exhibits a substantial reduction in mannitol-induced cytosolic calcium accumulation, demonstrates stronger SnRK2 activation than the WT (14). However, it was reported that the C-terminal domain (domain II) of SnRK2s rich in acidic amino acids potentially involved in Ca2+ binding (102) and a possible existence of a calcium-dependent and ABA-activated SnRK2 in wheat (103). In addition, the SnRK2s are inhibited by the SnRK2-interacting calcium sensor in tobacco (104). The A clade PP2Cs, ABI2, AHG3/PP2CA, HAB1 could be inhibited by Ca2+ (105). In addition, SnRK2.6/OST1 phosphorylates cyclic nucleotide-gated channels in guard cells in ABA-mediated Ca2+ signaling (48). Taking these together, the activation of the RAF-SnRK2 cascades might be triggered by factors related to the cell wall, membrane, or other cellular perturbations rather than the cytosolic calcium signal induced by treatments with EGTA and mannitol. Consequently, the precise relationship between hyperosmolarity-induced calcium signaling and the RAF-SnRK2 cascade necessitates further in-depth investigation and clarification.
In this investigation, we leveraged DIA-based phosphoproteomics to explore the intricacies of the phosphoproteome in response to treatments with EGTA and mannitol. Compared to our previous study, which used a similar sample preparation procedure and the DDA method, we have found that DIA phosphoproteomics offers significantly enhanced phosphoproteome coverage and analytical throughput. This advantage of DIA phosphoproteomics has also been observed in a few recent studies in the field of plant biology, as reported by Zhou et al. (2021) (106), Tan et al. (2021) (107), and Qu et al. (2023) (108). In general, the DIA method allows the detection of more than 20,000 phosphopeptides, even when utilizing a 10-year-old Q-Exactive MS, reducing the gap between results generated from advanced and costly mass spectrometers. This capability makes it a valuable MS method for identifying low-abundance phosphopeptides, which are typically inaccessible by traditional DDA approaches. As a result, our findings yield more accurate and nuanced information about the dynamic plant phosphoproteome.
Data Availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (109) with the dataset identifier PXD046802.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We are grateful to Drs Zhen-Ming Pei of Duke University and Rainer Hedrich of Universität Würzburg for helpful discussion.
Funding and additional information
This work was supported by the National Key Research and Development Program of China, Grant 2021YFA1300402 to P. W., 110-2311-B-001-043-MY2 and 112-2311-B-001-024-MY3 from National Science and Technology Council to C.-C. H., and National Natural Science Foundation of China, Grant 32300261 to Z. L.
Author contributions
T. S., Y. D., P.-Y. L., C.-C. H., and P. W. data curation; T. S., C.-W. C., and Z. L. investigation; T. S., C.-W. C., Z. L.,Y. M., and M. H. methodology; T. S., Y. M., and Z. L. software; T. S. and Z. L. validation; T. S., Z. L., Y. D., W. A. T., C.-C. H., and P. W. writing–original draft; T. S., J.-K. Z., C.-C. H., and P. W. writing–review and editing; Z. L., C.-C. H., and P. W. funding acquisition; C.-C. H. and P. W. supervision; P, W. project administration; P. W. conceptualization.
Contributor Information
Chuan-Chih Hsu, Email: cchsu@gate.sinica.edu.tw.
Pengcheng Wang, Email: wangpc@sustech.edu.cn.
Supplemental Data
References
- 1.Gorgues L., Li X., Maurel C., Martinière A., Nacry P. Root osmotic sensing from local perception to systemic responses. Stress Biol. 2022;2:36. doi: 10.1007/s44154-022-00054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waadt R., Seller C.A., Hsu P.K., Takahashi Y., Munemasa S., Schroeder J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022;23:680–694. doi: 10.1038/s41580-022-00479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf S. Cell wall signaling in plant development and defense. Annu. Rev. Plant Biol. 2022;73:323–353. doi: 10.1146/annurev-arplant-102820-095312. [DOI] [PubMed] [Google Scholar]
- 4.Dong Q., Wallrad L., Almutairi B.O., Kudla J. Ca2+ signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 2022;64:287–300. doi: 10.1111/jipb.13228. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J.K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K., Isobe M., Muto S. An increase in cytosolic calcium ion concentration precedes hypoosmotic shock-induced activation of protein kinases in tobacco suspension culture cells. FEBS Lett. 1997;401:202–206. doi: 10.1016/s0014-5793(96)01472-x. [DOI] [PubMed] [Google Scholar]
- 7.Knight H., Trewavas A.J., Knight M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 2003;12:1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 8.Yuan F., Yang H., Xue Y., Kong D., Ye R., Li C., et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 2014;514:367–371. doi: 10.1038/nature13593. [DOI] [PubMed] [Google Scholar]
- 9.Maity K., Heumann J.M., McGrath A.P., Kopcho N.J., Hsu P.-K., Lee C.-W., et al. Cryo-EM structure of OSCA1.2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating. Proc. Natl. Acad. Sci. U. S. A. 2019;116:14309–14318. doi: 10.1073/pnas.1900774116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Wang J., Sun L. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 2018;9:5060. doi: 10.1038/s41467-018-07564-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murthy S.E., Dubin A.E., Whitwam T., Jojoa-Cruz S., Cahalan S.M., Mousavi S.A.R., et al. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. Elife. 2018;7 doi: 10.7554/eLife.41844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jojoa-Cruz S., Saotome K., Murthy S.E., Tsui C.C.A., Sansom M.S., Patapoutian A., et al. Cryo-EM structure of the mechanically activated ion channel OSCA1.2. Elife. 2018;7 doi: 10.7554/eLife.41845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thor K., Jiang S., Michard E., George J., Scherzer S., Huang S., et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature. 2020;585:569–573. doi: 10.1038/s41586-020-2702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K., Gao J., Sun S., Zhang Z., Yu B., Li J., et al. BONZAI proteins control global osmotic stress responses in plants. Curr. Biol. 2020;30:4815–4825.e4814. doi: 10.1016/j.cub.2020.09.016. [DOI] [PubMed] [Google Scholar]
- 15.Chen X., Ding Y., Yang Y., Song C., Wang B., Yang S., et al. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021;63:53–78. doi: 10.1111/jipb.13061. [DOI] [PubMed] [Google Scholar]
- 16.Hoyos M.E., Z S. Calcium-independent activation of salicylic acid-induced protein kinase and a 40-kilodalton protein kinase by hyperosmotic stress. Plant Physiol. 2000;122:1355–1363. doi: 10.1104/pp.122.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Droillard M.,B.M., Barbier-Brygoo H., Laurière C. Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett. 2002;527:43–50. doi: 10.1016/s0014-5793(02)03162-9. [DOI] [PubMed] [Google Scholar]
- 18.Boudsocq M., Barbier-Brygoo H., Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 19.Saruhashi M.,K.G.T., Arai K., Ishizaki Y., Hagiwara K., Komatsu K., Shiwa Y., et al. Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E6388–E6396. doi: 10.1073/pnas.1511238112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Z., Li Y., Zhang Z., Liu X., Hsu C.C., Du Y., et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 2020;11:613. doi: 10.1038/s41467-020-14477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsuta S., Masuda G., Bak H., Shinozawa A., Kamiyama Y., Umezawa T., et al. Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J. 2020;103:634–644. doi: 10.1111/tpj.14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soma F., Takahashi F., Suzuki T., Shinozaki K., Yamaguchi-Shinozaki K. Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 2020;11:1373. doi: 10.1038/s41467-020-15239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Group MAPK. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn A., Roosjen M., Mutte S., Dubey S.M., Carrillo Carrasco V.P., Boeren S., et al. RAF-like protein kinases mediate a deeply conserved, rapid auxin response. Cell. 2024;187:130–148.e17. doi: 10.1016/j.cell.2023.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P. Emerging multiple function of B-RAFs in plants. Trends Plant Sci. 2024;29 doi: 10.1016/j.tplants.2024.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Fujii H., Verslues P.E., Zhu J.K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umezawa T., S N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., et al. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013;6 doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- 28.Wang P., Xue L., Batelli G., Lee S., Hou Y.J., Van Oosten M.J., et al. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P., Hsu C.C., Du Y., Zhu P., Zhao C., Fu X., et al. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. U. S. A. 2020;117:3270–3280. doi: 10.1073/pnas.1919901117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao X., Li Y., Rehman S.U., Miao L., Zhang Y., Chen X., et al. The Sucrose Non-Fermenting 1-Related Protein Kinase 2 (SnRK2) Genes are multifaceted players in plant growth, development and response to environmental stimuli. Plant Cell Physiol. 2020;61:225–242. doi: 10.1093/pcp/pcz230. [DOI] [PubMed] [Google Scholar]
- 31.Fujii H., Zhu J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulik A., Wawer I., Krzywińska E., Bucholc M., Dobrowolska G. SnRK2 protein kinases—key regulators of plant response to abiotic stresses. OMICS. 2011;15:859–872. doi: 10.1089/omi.2011.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii H., Verslues P.E., Zhu J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burla B., Pfrunder S., Nagy R., Francisco R.M., Lee Y., Martinoia E. Vacuolar transport of abscisic acid glucosyl ester is mediated by ATP-binding cassette and proton-antiport mechanisms in Arabidopsis. Plant Physiol. 2013;163:1446–1458. doi: 10.1104/pp.113.222547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y.,S.I., Korte A., Moes D., Yang Y., Christmann A., Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 38.Park SY F.P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Z., Li Y., Wang Y., Liu X., Ma L., Zhang Z., et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021;12:2456. doi: 10.1038/s41467-021-22812-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi Y., Zhang J., Hsu P.K., Ceciliato P.H.O., Zhang L., Dubeaux G., et al. MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 2020;11:12. doi: 10.1038/s41467-019-13875-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabregas N., Yoshida T., Fernie A.R. Role of Raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Commun. 2020;11:6184. doi: 10.1038/s41467-020-19977-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao K.P., Richa T., Kumar K., Raghuram B., Sinha A.K. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 2010;17:139–153. doi: 10.1093/dnares/dsq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu P.K., Dubeaux G., Takahashi Y., Schroeder J.I. Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 2020;105:307–321. doi: 10.1111/tpj.15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H., Wang Y., Sang T., Lin Z., Li R., Ren W., et al. Cell type-specific proteomics uncovers a RAF15-SnRK2.6/OST1 kinase cascade in guard cells. J. Integr. Plant Biol. 2023;65:2122–2137. doi: 10.1111/jipb.13536. [DOI] [PubMed] [Google Scholar]
- 45.Hsu P.K., Takahashi Y., Merilo E., Costa A., Zhang L., Kernig K., et al. Raf-like kinases and receptor-like (pseudo)kinase GHR1 are required for stomatal vapor pressure difference response. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2107280118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P. Plant physiology: RAF kinases claim a conserved role in rapid auxin responses. Curr. Biol. 2024;34:R204–R206. doi: 10.1016/j.cub.2024.01.024. [DOI] [PubMed] [Google Scholar]
- 47.Maszkowska J., Szymanska K.P., Kasztelan A., Krzywinska E., Sztatelman O., Dobrowolska G. The multifaceted regulation of SnRK2 kinases. Cells. 2021;10:2180. doi: 10.3390/cells10092180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Tan Y.Q., Wang X., Li J.J., Du B.Y., Zhu M., et al. OPEN STOMATA1 phosphorylates CYCLIC NUCLEOTIDE-GATED CHANNELs to trigger Ca2+ signaling for ABA-induced stomatal closure in Arabidopsis. Plant Cell. 2024;36:2328–2358. doi: 10.1093/plcell/koae073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y., Zhang Z., Gao J., Wang P., Hu T., Wang Z., et al. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-Independent SnRK2 activity. Cell Rep. 2018;23:3340–3351.e3345. doi: 10.1016/j.celrep.2018.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C.W., Tsai C.F., Lin M.H., Lin S.Y., Hsu C.C. Suspension trapping-based sample preparation workflow for In-Depth plant phosphoproteomics. Anal. Chem. 2023;95:12232–12239. doi: 10.1021/acs.analchem.3c00786. [DOI] [PubMed] [Google Scholar]
- 51.Tsai C.F., Hsu C.C., Hung J.N., Wang Y.T., Choong W.K., Zeng M.Y., et al. Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Anal. Chem. 2014;86:685–693. doi: 10.1021/ac4031175. [DOI] [PubMed] [Google Scholar]
- 52.Hsu C.C., Zhu Y., Arrington J.V., Paez J.S., Wang P., Zhu P., et al. Universal plant phosphoproteomics workflow and its application to tomato signaling in response to cold stress. Mol. Cell. Proteomics. 2018;17:2068–2080. doi: 10.1074/mcp.TIR118.000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruderer R., Bernhardt O.M., Gandhi T., Miladinovic S.M., Cheng L.Y., Messner S., et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 55.Bekker-Jensen D.B., Bernhardt O.M., Hogrebe A., Martinez-Val A., Verbeke L., Gandhi T., et al. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 2020;11:787. doi: 10.1038/s41467-020-14609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibas C., Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price M.N.,D.P., Arkin A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.Y., McInerny G. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2016;8:28–36. [Google Scholar]
- 61.Guo Y., Peng D., Zhou J., Lin S., Wang C., Ning W., et al. iEKPD 2.0: an update with rich annotations for eukaryotic protein kinases, protein phosphatases and proteins containing phosphoprotein-binding domains. Nucleic Acids Res. 2019;47:D344–D350. doi: 10.1093/nar/gky1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vlad F., Droillard M.J., Valot B., Khafif M., Rodrigues A., Brault M., et al. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 2010;63:778–790. doi: 10.1111/j.1365-313X.2010.04281.x. [DOI] [PubMed] [Google Scholar]
- 63.Boudsocq M., Lauriere C. Osmotic signaling in plants: multiple pathways mediated by emerging kinase families. Plant Physiol. 2005;138:1185–1194. doi: 10.1104/pp.105.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soon FF N.L., Zhou X.E., West G.M., Kovach A., Tan M.H., Suino-Powell K.M., et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335:85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Islam M., Inoue T., Hiraide M., Khatun N., Jahan A., Kuwata K., et al. Activation of SnRK2 by Raf-like kinase ARK represents a primary mechanism of ABA and abiotic stress responses. Plant Physiol. 2021;185:533–546. doi: 10.1093/plphys/kiaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi Y., Ebisu Y., Shimazaki K.I. Reconstitution of abscisic acid signaling from the receptor to DNA via bHLH transcription factors. Plant Physiol. 2017;174:815–822. doi: 10.1104/pp.16.01825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peirats-Llobet M., Han S.K., Gonzalez-Guzman M., Jeong C.W., Rodriguez L., Belda-Palazon B., et al. A direct Link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol. Plant. 2016;9:136–147. doi: 10.1016/j.molp.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Wang P., Zhao Y., Li Z., Hsu C.C., Liu X., Fu L., et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell. 2018;69:100–112.e106. doi: 10.1016/j.molcel.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu C., Fu L., Xiong Y., Wang P. How long should a kiss last between a kinase and its substrate? J. Integr. Plant Biol. 2022;64:789–791. doi: 10.1111/jipb.13240. [DOI] [PubMed] [Google Scholar]
- 70.Soma F., Mogami J., Yoshida T., Abekura M., Takahashi F., Kidokoro S., et al. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants. 2017;3 doi: 10.1038/nplants.2016.204. [DOI] [PubMed] [Google Scholar]
- 71.Kawa D., Meyer A.J., Dekker H.L., Abd-El-Haliem A.M., Gevaert K., Van De Slijke E., et al. SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 2020;182:361–377. doi: 10.1104/pp.19.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Q., Hu T., Li X., Song C.P., Zhu J.K., Chen L., et al. Phosphorylation of SWEET sucrose transporters regulates plant root:shoot ratio under drought. Nat. Plants. 2022;8:68–77. doi: 10.1038/s41477-021-01040-7. [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 74.Sirichandra C., Gu D., Hu H.C., Davanture M., Lee S., Djaoui M., et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 75.Hou Y.J., Zhu Y., Wang P., Zhao Y., Xie S., Batelli G., et al. Type One Protein Phosphatase 1 and its regulatory protein inhibitor 2 negatively regulate ABA signaling. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamiyama Y., Hirotani M., Ishikawa S., Minegishi F., Katagiri S., Rogan C.J., et al. Arabidopsis group C Raf-like protein kinases negatively regulate abscisic acid signaling and are direct substrates of SnRK2. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2100073118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H., Li T., Li Y., Bai H., Dai Y., Liao Y., et al. The plant FYVE domain-containing protein FREE1 associates with microprocessor components to repress miRNA biogenesis. EMBO Rep. 2023;24 doi: 10.15252/embr.202255037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., et al. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- 79.Pitzschke A., Djamei A., Bitton F., Hirt H. A major role of the MEKK1–MKK1/2–MPK4 pathway in ROS signalling. Mol. Plant. 2009;2:120–137. doi: 10.1093/mp/ssn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang M., Wu H., Su J., Wang H., Zhu Q., Liu Y., et al. Maternal control of embryogenesis by MPK6 and its upstream MKK4/MKK5 in Arabidopsis. Plant J. 2017;92:1005–1019. doi: 10.1111/tpj.13737. [DOI] [PubMed] [Google Scholar]
- 81.Zhang M., Su J., Zhang Y., Xu J., Zhang S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018;45:1–10. doi: 10.1016/j.pbi.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 82.Lampard GR M.C., Bergmann D.C. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science. 2008;322:1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- 83.Bergmann D.C.,L.W., Somerville C.R. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 84.Pan L., De Smet I. Expanding the mitogen-activated protein kinase (MAPK) universe: an update on MAP4Ks. Front. Plant Sci. 2020;11:1220. doi: 10.3389/fpls.2020.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eid S., Turk S., Volkamer A., Rippmann F., Fulle S. KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics. 2017;18:16. doi: 10.1186/s12859-016-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hepler P.K. Calcium: a central regulator of plant growth and development. Plant Cell. 2005;17:2142–2155. doi: 10.1105/tpc.105.032508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang Y., Zhao D., Meng J., Tao J. EGTA reduces the inflorescence stem mechanical strength of herbaceous peony by modifying secondary wall biosynthesis. Hortic. Res. 2019;6:36. doi: 10.1038/s41438-019-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cessna S.G., Low P.S. An apoplastic Ca2+ sensor regulates internal Ca2+ release in aequorin-transformed tobacco cells. J. Biol. Chem. 2001;276:10655–10662. doi: 10.1074/jbc.M006989200. [DOI] [PubMed] [Google Scholar]
- 89.Nowack M.K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A.K., et al. Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell. 2012;22:1030–1040. doi: 10.1016/j.devcel.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 90.Harashima H., Sugimoto K. Integration of developmental and environmental signals into cell proliferation and differentiation through RETINOBLASTOMA-RELATED 1. Curr. Opin. Plant Biol. 2016;29:95–103. doi: 10.1016/j.pbi.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 91.Saunders M.J., H P. Calcium antagonists and calmodulin inhibitors block cytokinin-induced bud formation in Funaria. Dev. Biol. 1983;99:41–49. doi: 10.1016/0012-1606(83)90252-x. [DOI] [PubMed] [Google Scholar]
- 92.Zhang C., Shi W., Ma K., Li H., Zhang F. EGTA, a calcium chelator, affects cell cycle and increases DNA methylation in root tips of Triticum aestivum L. Acta Soc. Bot. Pol. 2016;85:3502. [Google Scholar]
- 93.Drogen F., O R S., Stucke V.M., Jaquenoud M., Neiman A.M., Peter M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- 94.Dan I.,W.N., Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 95.Zang M., Hayne C., Luo Z. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J. Biol. Chem. 2002;277:4395–4405. doi: 10.1074/jbc.M110000200. [DOI] [PubMed] [Google Scholar]
- 96.Xiong J., Cui X., Yuan X., Yu X., Sun J., Gong Q. The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot. 2016;67:1461–1475. doi: 10.1093/jxb/erv538. [DOI] [PubMed] [Google Scholar]
- 97.Yamada K., Yamaguchi K., Shirakawa T., Nakagami H., Mine A., Ishikawa K., et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016;35:2468–2483. doi: 10.15252/embj.201694248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang X., Bao Y., Zhang M., Du D., Rao S., Li Y., et al. A Phytophthora capsici RXLR effector targets and inhibits the central immune kinases to suppress plant immunity. New Phytol. 2021;232:264–278. doi: 10.1111/nph.17573. [DOI] [PubMed] [Google Scholar]
- 99.Seto D., Khan M., Bastedo D.P., Martel A., Vo T., Guttman D., et al. The small molecule Zaractin activates ZAR1-mediated immunity in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2116570118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seto D., Koulena N., Lo T., Menna A., Guttman D.S., Desveaux D. Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat. Plants. 2017;3 doi: 10.1038/nplants.2017.27. [DOI] [PubMed] [Google Scholar]
- 101.Shinya T., Yamaguchi K., Desaki Y., Yamada K., Narisawa T., Kobayashi Y., et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014;79:56–66. doi: 10.1111/tpj.12535. [DOI] [PubMed] [Google Scholar]
- 102.Harmon A.C. Calcium-regulated protein kinases of plants. Gravit. Space Biol. Bull. 2003;16:83–91. [PubMed] [Google Scholar]
- 103.Coello P., Hirano E., Hey S.J., Muttucumaru N., Martinez-Barajas E., Parry M.A., et al. Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 2012;63:913–924. doi: 10.1093/jxb/err320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bucholc M., Ciesielski A., Goch G., Anielska-Mazur A., Kulik A., Krzywinska E., et al. SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J. Biol. Chem. 2011;286:3429–3441. doi: 10.1074/jbc.M110.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maheshwari P., Du H., Sheen J., Assmann S.M., Albert R. Model-driven discovery of calcium-related protein-phosphatase inhibition in plant guard cell signaling. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1007429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou L., Wang Z.M., Wen S., Li J., Xiong L.L., Zhu Z.Q. Application prospects of cerebrospinal fluid DIA techniques in identifying key regulatory molecules for mental disorders in encephalitis. Ibrain. 2021;7:44–51. doi: 10.1002/j.2769-2795.2021.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan J.,Z.Z., Feng H., Xing J., Niu Y., Deng Z. Data-independent acquisition-based proteome and phosphoproteome profiling reveals early protein phosphorylation and dephosphorylation events in Arabidopsis seedlings upon cold exposure. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222312856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qu L., Liu M., Zheng L., Wang X., Xue H. Data-independent acquisition-based global phosphoproteomics reveal the diverse roles of casein kinase 1 in plant development. Sci. Bull. (Beijing) 2023;68:2077–2093. doi: 10.1016/j.scib.2023.08.017. [DOI] [PubMed] [Google Scholar]
- 109.Perez-Riverol Y., Bai J., Bandla C., Garcia-Seisdedos D., Hewapathirana S., Kamatchinathan S., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (109) with the dataset identifier PXD046802.