Abstract
Acute kidney injury (AKI) is a common condition associated with significant morbidity, mortality, and cost. Injured kidney tissue can regenerate after many forms of AKI. However, there are no treatments in routine clinical practice to encourage recovery. In part, this shortcoming is due to an incomplete understanding of the genetic mechanisms that orchestrate kidney recovery. The advent of high-throughput sequencing technologies and genetic mouse models has opened an unprecedented window into the transcriptional dynamics that accompany both successful and maladaptive repair. AKI recovery shares similar cell-state transformations with kidney development, which can suggest common mechanisms of gene regulation. Several powerful bioinformatic strategies have been developed to infer the activity of gene regulatory networks by combining multiple forms of sequencing data at single-cell resolution. These studies highlight not only shared stress responses but also key changes in gene regulatory networks controlling metabolism. Furthermore, chromatin immunoprecipitation studies in injured kidneys have revealed dynamic epigenetic modifications at enhancer elements near target genes. This review will highlight how these studies have enhanced our understanding of gene regulation in injury response and regeneration.
Keywords: acute kidney injury, ischemia, regeneration, transcription factors, Pax genes, failed repair
Acute kidney injury (AKI) is a common condition affecting up to 20% of hospitalized patients and contributes to increased morbidity, mortality, and cost (1, 2). Remarkably, the injured kidney can regenerate after many forms of AKI if the degree of injury is sublethal. Unfortunately, there are no treatments to stimulate or enhance regeneration other than supportive care in routine clinical practice, in part owing to an incomplete understanding of the molecular and genetic controls that limit injury or promote regeneration. During repair, genetic cell lineage tracing supports a model where surviving tubular epithelial cells dedifferentiate, proliferate, and eventually re-express specialized proximal tubule genes required for normal functions (reviewed in Ref. (3)). To accomplish this feat, cells must alter their stable patterns of gene expression in response to the high cellular stress induced by injury. Cells must activate a program of survival and repair while retaining the memory of their original identity. Finally, they must re-establish a stable differentiated expression pattern indicative of functional proximal tubules. The precise genetic and epigenetic mechanisms that regulate these transformations and re-entry into mitosis remain poorly defined. However, recent advances in single-cell molecular methods, tractable genetic mouse models, and integrated bioinformatic analyses have opened an unprecedented window into the genetic controls that regulate tubular regeneration. Defining these molecular and genetic mechanisms may identify novel kidney-specific targets to promote successful regeneration or limit the degree of injury for patients suffering from AKI. This review will highlight recent advances in our understanding of how transcription factors drive the injury response and impact regeneration.
Overview of ischemic injury
Kidney injury can occur from multiple hemodynamic, toxic, and obstructive insults. A common cause of severe kidney injury in hospitalized patients is transient ischemia leading to injury and death of tubular epithelial cells (reviewed in Refs. (4, 5)). Of the nephron segments, the proximal tubule is most sensitive to this type of injury, though injury to other segments also occurs (6). The proximal tubule uses highly specialized metabolism that relies on oxidation of fatty acids to fuel the energy-intensive resorption of fluid and electrolytes from the glomerular ultrafiltrate, which is predominantly performed by this nephron segment. Concurrently, the microvascular anatomy of the kidney produces a gradient of oxygen availability from the cortex to medulla (7). Thus, in the setting of kidney hypoperfusion, a mismatch between oxygen delivery and demand results in oxidative stress, injury, and cell death via several pathways. These processes occur predominantly in the S3 segment of the proximal tubule, which sits at the border of the cortex and medulla (8) (Fig. 1). Remarkably, the damaged tubular epithelium can regenerate, restoring kidney function for some patients. However, we now appreciate that this regeneration is rarely complete such that patients who develop AKI have a greater chance of developing chronic kidney disease and progressing to end-stage kidney disease (9, 10, 11).
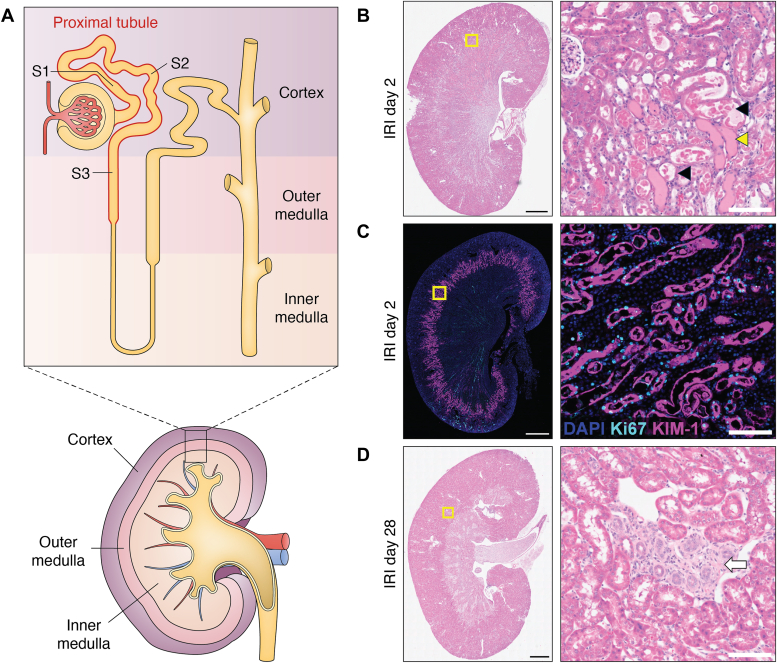
Figure 1.
Patterns of injury after ischemic kidney injury.A, the nephron segments of the proximal tubule (red outline) involved in acute kidney injury and their position in the gross structure of the kidney. For clarity, distal segments including the loop of Henle, distal tubule, and collecting ducts are not labeled. B, H&E-stained mouse kidney section 2 days after ischemia–reperfusion injury (IRI) showing injured tubules in the S3 segment. Note tubular dilation, simplification of the epithelium, and intratubular debris (examples: black arrowheads) and casts (example: yellow arrowhead). C, immunofluorescence staining 2 days after IRI for kidney injury marker 1 (KIM-1, purple) and Ki67 (light blue), which mark injured epithelium and proliferating cells, respectively. Note that KIM-1 staining is highly localized to the S3 segment at the border between cortex and medulla. D, H&E staining of a mouse kidney 28 days after IRI. Note the focal area of fibrosis and tubular atrophy with inflammatory infiltrate (white arrow) surrounded by normal tubules. Scale bars represent 1 mm (whole section) and 100 μm (regions of interest).
Over the last 2 decades, a more comprehensive understanding of proximal tubule epithelial cell regeneration has emerged. Much of this work has been performed in genetically modified mouse models of AKI. Among the most widely used injury models is ischemia—reperfusion in which the renal artery and vein are temporarily clamped and then released. Alternatively, toxin models include cisplatin, glycerol-induced rhabdomyolysis, aristolochic acid, folic acid, and lipopolysaccharides. The rodent ischemia–reperfusion clamp model most closely resembles the most common forms of human AKI and will be the focus of this review. While rodent and human AKI are not equivalent, work with human kidney organoids and human AKI samples from the Kidney Precision Medicine Project (6, 12, 13) confirms homology among many transcriptional programs. Thus, genetically engineered mice can yield important insights into the transcriptional responses to AKI in humans where such manipulations are not possible.
The studies addressing the origin of cells that repopulate the injured nephron have led to multiple interpretations, including designated populations of renal stem cells, bone marrow–derived mesenchymal cells, or resident surviving epithelia. However, sophisticated genetic lineage tracing now supports a model where mature proximal tubule cells that survive the initial injury can dedifferentiate and re-enter mitosis. Although some cells in the nephron may disproportionately contribute to repair (14), most mature tubular epithelia appear to retain regenerative potential. Thus, there is neither evidence of a fixed nephron progenitor pool in the mature kidney nor evidence of cells of nonkidney origin contributing to repair (15, 16, 17). Rather, irreversibly tagged proximal tubule cells can generate clones within regenerated tubules to strongly support the idea that most of any surviving epithelial cell can proliferate to repair a damaged tubule. Therefore, the widespread and robust activation of proliferation markers after injury (18, 19) likely represents intrinsic nephron repair (14, 15, 20). Furthermore, single-cell RNA-Seq datasets also identified a novel population of cells after AKI that reflect incomplete or failed repair and are associated with areas of fibrosis and ongoing inflammation (5, 21, 22, 23) (Fig. 1D).
Mechanisms controlling cell state in regeneration
Dedifferentiation, proliferation, and the expression of developmental regulatory genes and pathways all suggest similarities between nephron development and regeneration after AKI (24, 25). For a more complete discussion of renal development and the genes and pathways involved, see the study by Schnell et al., 2022 (26). After gastrulation, the kidney develops from a region of mesoderm called intermediate, as it is located between the axial mesoderm and the lateral plate (Fig. 2). Two simple epithelial tubes called the nephric or Wolffian ducts run bilaterally from the midthoracic region to the posterior cloaca. At the posterior end, the ureteric bud grows out of the nephric duct and invades the metanephric mesenchyme to initiate metanephric kidney development. The ureteric bud begins to branch, ultimately forming much of the collecting duct system, whereas the mesenchyme aggregates at the tips of the branching ureteric buds in response to WNT signaling and begins the process of mesenchyme-to-epithelial transition (MET) that will generate most of the epithelia cells of the nephron. Also termed cap mesenchyme, these progenitor cells express unique combinations of markers including Pax2, Six2, and Cited1.
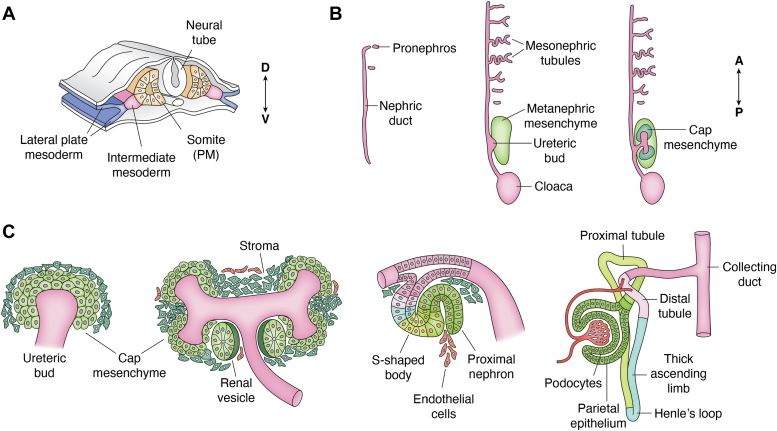
Figure 2.
Overview of kidney development.A, the kidney derives from a region of mesoderm termed intermediate, lying between the somites and lateral plate, as seen on a cross section of embryonic day 8.5 mouse or 3 weeks human. B, two bilateral epithelial ducts run longitudinally with a rudimentary pronephros at the most anterior end. In time (left-to-right), an outgrowth of the nephric duct, termed ureteric bud, invades surrounding mesenchyme and induces the adult or metanephric kidney. Cap mesenchyme (green) surrounds the ureteric bud tips in preparation for MET. C, the MET generates much of the nephron through a series of morphological stages as outlined from left to right. Continued branching of the ureteric bud induces more nephrons along the radial axis of the developing kidney. MET, mesenchyme-to-epithelial transition.
The cap mesenchyme aggregates and forms a primitive epithelial renal vesicle that first forms a comma-shaped and subsequently an s-shaped body patterned along the proximal–distal axis in part by Notch signaling (Fig. 2C). The distal end of the s-shaped body connects to the branching ureteric bud epithelia to form a continuous tubule from glomerular progenitor cells at the proximal end to collecting tubules at the distal end. Multiple transcription factors have been identified that drive the process of mesenchyme-to-epithelial conversion, proliferation, elongation, and renal epithelial cell–type specification. Among the most essential are Pax2/8, WT1, Six2, Eya1, HNF1b, HNF4a, and NOTCH intracellular domains.
Prior to induction by the ureteric bud, the metanephric mesenchyme already is predisposed toward the renal epithelial lineage fate and expresses a unique combination of transcription factors such as WT1 and Pax2, both of which are essential for responding to inductive signals and whose deletion results in complete renal agenesis (27, 28, 29). The related protein Pax8 has an identical DNA-binding domain to Pax2 but is only expressed in renal progenitor cells after induction and remains on in most all the adult renal epithelia, whereas Pax2 is downregulated in the nephron but remains expressed in the collecting duct system. Genetic studies suggest some redundancy in Pax2/8 functions, both in development and in the adult kidney (30, 31). Recent data suggest a Pax-dependent core transcriptional program driving the MET in cooperation with HNF1b (32). After induction, Six2 expression in the cap mesenchyme helps to maintain the undifferentiated state and allows for continued progenitor cell proliferation, as deletion of Six2 results in precocious differentiation and progenitor cell exhaustion (33). Inactivation of HNF1b in nephron progenitors results in rudimentary nephrons with mostly distal epithelial cell fates, suggesting a need for this protein in correct proximal–distal patterning (34). Proximal–distal nephron patterning is also affected by NOTCH ligands, which generate the DNA-binding Notch intracellular domain through proteolytic cleavage (35, 36). The Sry-box containing protein Sox9 is needed for branching of the ureteric bud epithelia and is also reactivated and necessary for regeneration after AKI (37, 38, 39). An HNF4a gene regulatory network has also been described in mice and human kidney organoids that appears essential for proximal tubule cell development (40, 41). While multiple developmental genes and pathways are re-expressed or upregulated in response to AKI (for more details, see the study by Little and Kairath, 2016 (24)), whether these are essential or are merely markers for a dedifferentiated state remains to be fully explored.
Analysis of transcriptional control
At the transcriptional level, gene expression is regulated by cis-acting DNA elements at promoters and enhancers and trans-acting factors that bind to such elements and recruit RNA polymerase and other cofactors necessary for initiation and elongation. Yet, enhancers and promoters function within the context of intact chromatin, which consists of DNA, associated histones, and other proteins. The histones within the nucleosome can be modified post-translationally by epigenetic imprints, including methylation or acetylation at lysine residues, which can determine the accessibility of chromatin to the transcription machinery. Indeed, cell- and tissue-specific enhancer modules can be identified solely by their specific patterns of histone methylation (42), which are imprinted by the histone methyltransferases of the Trithorax and Polycomb family of proteins. Originally identified in Drosophila as activators of embryonic gene expression, the Trithorax family of proteins includes the histone H3K4 methyltransferases, whereas the Polycomb family is thought to silence gene expression in part through methylation at H3K27 and H3K9. Thus, transcription factors that upregulate or downregulate specific genes generally act upon the accessible part of the genome, or epigenome, much of which is determined during embryonic development as individual cell fates are specified.
The ability to quantify the entire transcriptome of a cell has revolutionized how we think about cell identity, cell stress, and plasticity. Hybridization of RNA to microarrays was first used to quantify gene expression changes at genome-wide scale. Next-generation bulk RNA-Seq has refined such methods to enable quantification of mRNA and splice variants with precision and economy. Moreover, single-cell RNA-Seq has become the standard for identifying not only new cell types and gene expression changes but also for following cellular differentiation and lineage tracing in pseudotime. Building on decades of work that has mapped the targets of many key transcription factors associated with human physiology and disease, such advanced technologies and bioinformatics are beginning to deconvolve the activity of complex gene regulatory networks from such high-resolution transcriptional data (reviewed in detail in Ref. (43)).
In addition to gene expression measurements, the direct mapping of transcription factor localization to specific DNA sequences at genome-wide scale has progressed rapidly. Using specific antibodies against transcription factors coupled with chromatin immunoprecipitation and next-generation sequencing (ChIP-Seq), the binding profile of a transcription factor or the pattern of histone modifications can be mapped across the entire genome. Novel methods for mapping protein binding to chromatin, which require much less input material, include cleavage under targets and release using nuclease (CUT&RUN) or tagmentation (CUT&Tag). An extensive library of these data is publicly available via the ENCODE project (44) and is becoming available in murine (45) and human (13) injured kidney samples. These data complement experimental and computational approaches (46) to identify sequence motifs associated with transcription factor–binding sites that may regulate the response to renal injury.
Independently, high-throughput techniques to measure genome-wide chromatin accessibility have become widely available and include transposase-accessible chromatin with sequencing (ATAC-Seq) and DNAse sensitivity or hypersensitivity assays. These methods not only show which genes are accessible but more importantly, indicate which gene regulatory elements are open and accessible to other factors. Open gene regulatory regions that contain transcription factor–binding motifs can be combined with transcription factor expression and target gene activation to infer gene regulatory networks. As with RNA-Seq, these technologies can be scaled to single-cell resolution.
Several approaches have been deployed in the analysis of the transcriptional response to kidney injury. The first is to use deep sequencing technology to identify the differential expression of transcription factors themselves (47, 48, 49). However, transcription factor expression may be necessary but insufficient to infer activity. Post-translational modifications such as phosphorylation can impact transcription factor activity, stability, or nuclear localization to further regulate the response to any given factor. To overcome this limitation, several groups have used single-cell regulatory network inference and clustering (SCENIC) (50) to infer activity based on target gene expression and pathway activation. Briefly, this approach correlates sets of genes with transcription factor expression, then refines these sets by including only genes with nearby cis-regulatory elements (transcription factor motifs, Fig. 3, A–C). The activity of each transcription factor regulon is then measured in each transcriptome. Another strategy uses single-cell ATAC-Seq data (chromVAR (51)). Rather than pooling the expression of transcripts, the approach pools the differential accessibility of transcription factor–binding motifs. This allows for a direct readout of potential transcription factor activity in clusters of cells (Fig. 3, D and E). SCENIC and chromVAR were optimized for sparse single-cell datasets. Therefore, these approaches can be applied across thousands of single-cell transcriptomes to infer transitional cell states.
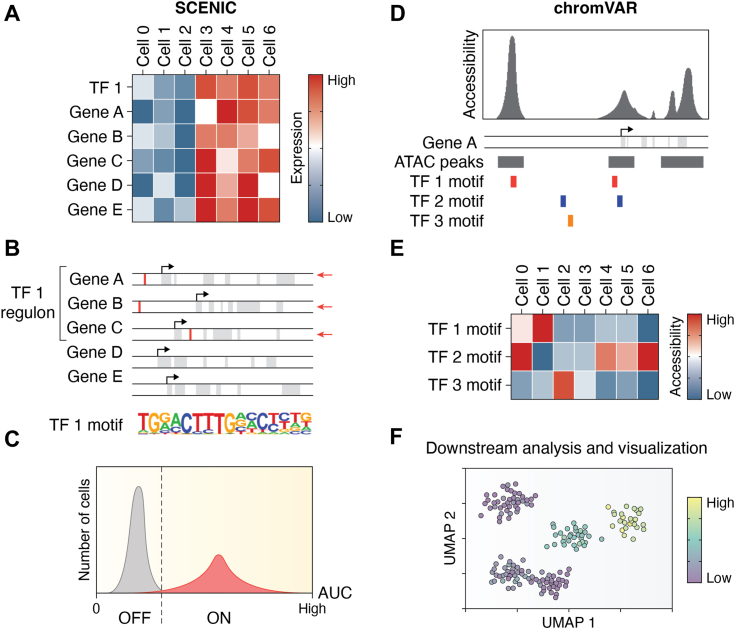
Figure 3.
Overview of gene regulatory network inference approaches in single cells.A, SCENIC identifies coexpression modules that are correlated with transcription factor (TF) expression. B, the subset of genes within these modules likely to be directly regulated by the TF (TF regulons) are defined by the presence of nearby TF-binding motifs. C, the activity of each regulon is determined by measuring the ranks of the regulon genes in each cell as an area under the curve (AUC) of the count-rank profile at a specified fraction of genes. The binary (ON/OFF) regulon activity for each cell can be inferred from the distribution of activities for the entire cell population. D, chromVAR uses differential chromatin accessibility to define TF activity. Accessibility (ATAC) peaks are mapped to TFs by the presence of a motif within the peak region. This yields a set of peaks associated with each TF motif. E, bias-corrected relative accessibility is estimated for the entire TF peak set, which helps mitigate the sparse reads obtained from single-cell ATAC data. F, either approach can be then fed into downstream visualization and analysis. Diagrams were based on those in the original publications describing their use (50, 51).
New strategies refine predictions of transcription factor activity using single-cell multiomic experiments that combine ATAC-Seq with single-cell RNA-Seq data from the same cell. Approaches used to study kidney injury include TRIPOD (13, 52) and RENIN (53). In particular, RENIN was developed and validated specifically with human kidney multiomic data. The regularized regression approach in RENIN uses two steps. First, differentially accessible regions defined by ATAC-Seq are correlated with gene expression in individual cells to identify putative cis-regulatory elements driving changes in expression. Second, target gene expression is linked to the expression of transcription factors with binding motifs in these cis-regulatory elements to model transcriptional networks. This approach enabled the prediction of several transcription factors driving a chronic injury state in proximal tubule cells that were validated experimentally (53).
However, these approaches are only accurate for transcription factors where binding motifs are known with high sensitivity and specificity. For many transcription factors, motif specificity is quite poor (54). Sequence motifs can have degeneracies leading to both high- and low-affinity sites. Indeed, recent studies on enhancer logic suggest that the highest affinity sites are not always the most biologically significant (55). Even with transcription factors whose motifs are tightly defined 6 to 8 bp sequences, only a fraction of sites on the genome are bound in vivo. Furthermore, actual bound sites, as detected with ChIP-Seq, may not represent functional transcription factor activity, since this technique only correlates protein location to specific loci (56). Also, binding targets can overlap considerably between transcription factors of the same family. Many transcription factors can heterodimerize with other family members to further increase the complexity and impact the specificity of binding motifs.
Despite limitations, interactive gene regulatory networks can be modeled based on transcription factor binding sites as determined by ChIP-Seq, the position of consensus motifs in promoters and enhancers, and gene expression changes in response to gain or loss of function. Yet constructing regulatory networks still requires simplified assumptions of transcription factor–target effects, which are often nonlinear and can vary among targets for the same transcription factors. Such regulatory models also do not account for the effects of post-translational modifications that can alter the essential function of many transcription factors. The potential for compounded errors because of each of these limitations increases with complexity, necessitating rigorous validation of predictions. However, validating the vast numbers of gene regulatory network predictions made by approaches such as SCENIC or chromVAR is challenging. The throughput of typical reductionist validation approaches is inadequate to match the vast number of predictions made. Furthermore, in the kidney, there are no tissue culture model systems available that can replicate the transcriptome of proximal tubule in vivo (57). As a result, only a few novel predictions have been validated experimentally (13, 47).
Early injury response
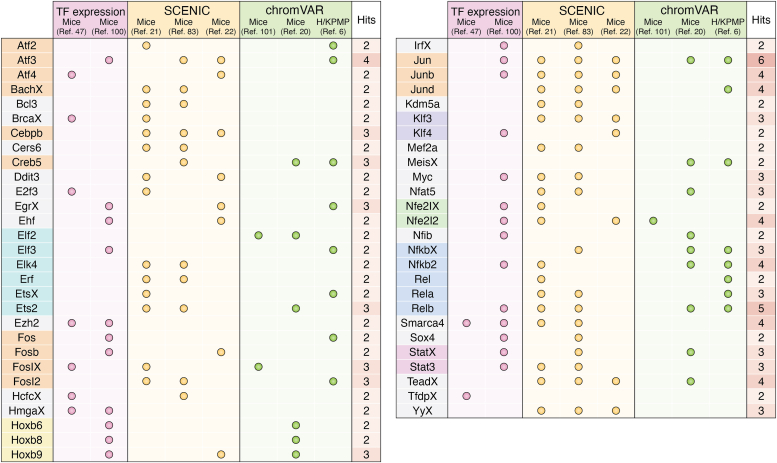
AKI leads to rapid changes in gene expression including many transcription factors that may be involved in regulating the adaptive and maladaptive responses to injury. Multiple studies have examined transcriptomic changes after injury in animal models of AKI (as listed in Table 1). Figure 4 summarizes transcription factors with increased expression and activity in proximal tubule cells early after ischemic injury, identified using the various strategies discussed previously. Only transcription factors identified in two or more studies were included. The strongest and most clearly defined signatures detected include the Fos/Jun and ATF/CREB basic leucine zipper families and the NF-κB family. These highly conserved transcriptional programs are not kidney specific; we will defer additional discussion here. While the results in Figure 4 validate computational approaches in a broad sense, these pathways represent generic cell responses to stress and are unlikely to reflect novel targets to promote kidney regeneration. Most transcription factors identified in these studies appear only once (143 of 198 across all studies, 72%). This inconsistency highlights the limitations of the computational approaches outlined previously as well as the sparse nature of single-cell RNA-Seq and ATAC-Seq data. Furthermore, several transcription factors with clear roles defined by genetic studies such as FOXM1 (47), KLF6 (58), and SOX9 (37, 38) (outlined later) are not robustly detected by these approaches.
Table 1.
Selected transcriptional profiling studies after kidney injury
| Study | Species | Source | Type of data | Cell-type specification | Transcription factor analysis |
|---|---|---|---|---|---|
| Liu et al. (49) | Mouse | Whole kidney | Microarray | TRAP for Six2 (nephron), Foxd1 (interstitial), Cdh5 (endothelial), and Lyz2 (myeloid) | Ingenuity canonical pathway analysis |
| Chang-Panesso et al., 2019 (47) | Mouse | Whole kidney | RNA-Seq | TRAP for Kim1 | Transcription factor expression |
| Kirita et al., 2020 (21) | Mouse | Whole kidney | snRNA-Seq | Clustering with annotation | SCENIC (50) |
| Legouis et al., 2020 (98) | Mouse | Whole kidney | snRNA-Seq | Clustering, Six2 enriched | PROGENy (99) |
| Rudman-Melnick et al., 2020 (100) | Mouse | Whole kidney | scRNA-Seq | Clustering with annotation | Transcription factor expression |
| Gerhardt et al., 2021 (83) | Mouse | Whole kidney | snRNA-Seq | Clustering, Krt20 and Six2 enriched | SCENIC (50) |
| Ide et al., 2021 (23) | Mouse | Whole kidney | scRNA-Seq | Clustering with annotation | Not specifically assessed |
| Balzer et al., 2022 (22) | Mouse | Whole kidney | scRNA-Seq | Clustering with annotation | SCENIC (50) |
| Ide et al., 2022 (97) | Mouse | Whole kidney | scRNA-Seq | Clustering with annotation | SCENIC (50) |
| Li et al., 2022 (101) | Mouse | Whole kidney | snRNA-Seq | Clustering with annotation | DoRothEA (102, 103) |
| Gerhardt et al., 2023 (20) | Mouse | Whole kidney | snRNA-Seq, snATAC-Seq | Clustering, Ki67 enriched | chromVAR (51) |
| Lake et al., 2023 (6) | Human | Research biopsies | sc/snRNA-Seq, SNARE-Seq, spatial transcriptomics | Clustering with annotation | chromVAR (51) |
| Li et al., 2024 (104) | Human | Kidney regions | snRNA-Seq, snATAC-Seq (SHARE-Seq) | Clustering with annotation | PROGENy (99), chromVAR (51) |
Abbreviation: TRAP, translating ribosome affinity purification.
Figure 4.
Transcription factors (TFs) and signatures identified in the early injury response by single-cell sequencing. TFs or gene regulatory networks identified in each study are summarized. About 198 unique TFs were identified across all studies. Only TFs that appear in two or more studies are shown (55 of 198, 27%). Only data available with the article were utilized (including figures, supporting figures, and supporting information) so identified pathways may not be comprehensive. Where statistics were available, only pathways with a Padj <0.05 were included. TFs are color coded by family. Columns are coded by the approach used to identify gene regulatory networks.
In contrast to the bioinformatic strategies, lineage-tagged transcriptional profiling has identified several transcription factors that have some specificity for epithelial injury response in the kidney (59). Among these is SOX9, a transcription factor that is activated during development in proximal and distal tubule epithelia but not in the glomeruli and collecting ducts (37, 38, 39). In the adult, some distal tubule cells express SOX9. However, after injury, there is widespread activation of SOX9 throughout the epithelia, colocalizing with markers of cell proliferation. Genetic loss of Sox9 function prior to injury impairs recovery (38, 60, 61), suggesting a critical need for Sox9 in the early epithelial response to AKI. Work by Pabla et al. has identified several critical mechanisms that regulate the function of SOX9. The early expression of SOX9 is induced by Zfp24, which is present in a phosphorylated state in uninjured kidney but is rapidly and robustly dephosphorylated in response to injury and enables binding to the SOX9 promoter (62). SOX9 function is further modulated by Cdkl5-mediated phosphorylation, which decreases SOX9 activity and stability (61, 63). While many SOX9 targets in other organs are well established (64), only some of the kidney-specific targets of SOX9 in AKI are defined, including VGF (65). Thus, SOX9 activity reflects the transcriptional and post-transcriptional mechanisms that can regulate the function of an essential transcription factor for recovery after AKI. These limitations are unaddressed by most computational strategies and may account for the lack of consistent signals in single-cell data despite robust evidence of its central importance. While these findings suggest that SOX9 helps to stabilize injured cells, allowing them to recover, the precise downstream genetic mechanisms remain to be determined.
Chang-Panesso et al. (47) recently used a kidney injury molecule 1 lineage tracing strategy with translating ribosome affinity purification to identify transcription factors during early and late repair. This approach identified FOXM1, which like SOX9 is important for regulating early proliferation. FOXM1 expression was localized predominantly to the S3 segment of the proximal tubule, consistent with the site of highest kidney injury molecule 1 expression and proliferation after injury. FOXM1 correlates with proliferation in a variety of cell types (66) and is upregulated by the EGF signaling pathway known to be critical for proximal tubule regeneration (67, 68). However, the genetic mechanisms specifically activating the expression of FOXM1 and the identities of its targets are still poorly defined.
In addition to the generalized stress response and proliferation, the early phase of recovery is also characterized by altered proximal tubule metabolism. This topic was recently reviewed in detail by Piret and Mallipattu (69) and will be summarized briefly here. Under physiologic conditions, the proximal tubule relies on highly efficient oxidation of fatty acids and amino acids to fuel energy-intensive transport. The transcriptional basis for this unique and specialized metabolism depends on a network of transcription factors, including peroxisome proliferator–activated receptor alpha (PPARA) (70) and PPARG (71), estrogen-related receptor alpha (ESRRα) (72), pregnane X receptor (73), farnesoid X receptor (71), HNF4a (41), Kruppel-like factor 15 (74), and PPARγ coactivator 1α (75). After injury, these pathways are all downregulated across a range of injury models. The mechanisms that define the interactions and relative importance of these factors and their impact on metabolism is an area of active investigation. In general, loss of any of these factors tends to reduce baseline expression of the genes needed for fatty acid metabolism, which, in turn, correlates with increased severity of injury to the proximal tubule. For example, metabolic switching after injury is regulated in part by a shift in Kruppel-like factor 15, which promotes expression of normal oxidative metabolism, to KLF6, which blocks the expression of genes required for the metabolism of branched-chain amino acids that contribute to oxidative metabolism (58, 74). Likewise, Dhillon et al. (72) showed that ESRRα motifs, together with HNF4A, HNF1B, and PPARA, are enriched in promoters of metabolic and transport genes lost after injury. ESRRα activity contributes to the oxidative capacity of proximal tubule cells. Importantly, the combined expression of ESSRα with other identified transcription factors (HNF4A, HNF1B, and PPARA) can have a synergistic effect on expression of transport genes. Similarly, Clark et al. (76) recently identified cooperative regulation by HNF4a and PPARγ coactivator 1α of QPRT, which is critical for NAD+ metabolism in proximal tubules. It is also notable that most of these factors also act as nuclear hormone receptors, which adds an additional ligand-dependent layer to their function (77, 78). Together, these data indicate that proximal tubule transcription factors function in an overlapping fashion to regulate metabolism and differentiation during recovery from injury. While available evidence suggests these metabolic factors are tightly coordinated, the precise mechanisms that govern how these programs are synchronously regulated remain an area of active investigation.
In addition to the mouse models, robust sets of cell-level transcriptional data from human AKI samples are now becoming available (6, 13). While there are many similarities (79), prior work has shown only partial overlap in the transcriptional AKI response between rodents and humans (80). At the level of transcriptional regulatory elements, there may be important cross-species differences between regenerating proximal tubules and the inflammatory responses that accompany injury and regeneration.
Late regeneration responses
Compared with the immediate-early response to AKI, far less is known about the gene regulatory networks that drive regeneration and re-establish normal proximal tubule function over time. This knowledge gap is a critical barrier to developing new therapies for AKI since this late phase of injury is when most patients are recognized clinically. However, we now know that recovery diverges to generate at least two distinct populations of cells, as determined by pseudotime trajectory and RNA velocity analyses of single nuclear/cell RNA-Seq data (21, 23). One population of cells reflects successful repair and re-establishment of proximal tubule cell fate. The second population develops chronic injury and has been termed failed repair proximal tubule cells (FR-PTCs) by some authors (20, 21).
Successful recovery is characterized by the re-establishment of a transcriptional profile akin to healthy proximal tubules (21). Genes and motifs associated with unique proximal tubule metabolism are enriched in this transition including HNF4A, HNF1B, PPARA, and ESRRA (21, 22, 72). While these recovered tubules appear normal by histology and overall transcriptomic signature, subtle differences in gene expression do emerge (20). The downregulated genes in recovered but previously injured cells are associated with transport and metabolic processes and have substantial overlap with genes altered by preinjury preconditioning regimens (81). Pathway and motif analyses revealed persistent activity of early injury pathways (FOS/JUN, ATF/CREB, and Stat) and suppression of HNF4a activity (20). This preconditioning phenotype is found in other injury or stress models beyond AKI and includes such phenomena as caloric restriction, hypoxic preconditioning, and sex-dependent resistance to AKI (82).
In contrast, the population of FR-PTCs exhibits increased expression of pathways seen during the early injury response (FOS/JUN) and a strong inflammatory signature associated with NF-kB (20, 21, 83). Newer multiomic methods have implicated NFAT5, HIVEP2, and CREB5 as key contributors as well (53). The failed repair population peaks several weeks after injury and gradually recedes (21). In the mouse, the evolution of this population appears to be segment specific. In the first week after injury, the S3 segment contains most FR-PTCs. By 4 weeks, few S3 FR-PTCs remain, while the cortical population is unchanged (83).
None of the transcriptional analyses to date, which generally pool all segments of the proximal tubule together, suggest a stable undifferentiated population of cells after injury. RNA velocity analysis suggests most cells progress toward either a normal or an inflammatory state (23). These data suggest that once established, FR-PTCs may develop a fixed transcriptional program that is difficult to reset. Therefore, the loss of the population over time is most likely because of cell death rather than recovery of normal function. Turnover appears to be highest in the S3 segment of the proximal tubule (83), but this has yet to be established experimentally.
While the transcriptional networks in both postinjury populations have been characterized by available profiling studies, the transcriptional mechanisms that induce and maintain these fate decisions remain unknown. In severe injury, cells arise that show incomplete or atypical cell-cycle progression (14, 84). Though these cells were identified prior to the advent of single-cell transcriptomics, they are likely a subpopulation of FR-PTCs. While persistent ongoing DNA damage after injury would delay cell cycle progression via well-established mechanisms (e.g., p53), how this damage induces the dedifferentiation of proximal tubule cells has only recently been explored. For example, the cyclin D1–cyclin dependent kinase 5 axis can promote cell cycle arrest but simultaneously promotes a FR-PTC-like phenotype independently of effects on cell cycle progression (85). Furthermore, IL-22 is secreted in response to DNA damage in proximal tubules and impacts the DNA-damage response in neighboring cells, which contributes to clustering of FR-PTCs (86). However, the genetic mechanisms by which these signaling pathways induce dedifferentiation remain to be elucidated.
Epigenetics and regeneration
As discussed briefly in an earlier section, cell fates during development are specified, at least in part, by epigenetic modifications of chromatin such that each lineage contains a unique epigenome, upon which transcription factors act. This epigenome is heritable in development and has inherent stability, also termed cellular memory. In the case of regeneration after injury, surviving dedifferentiated cells that re-enter mitosis must replicate that epigenome to maintain cellular fates. Yet, injury itself can induce epigenetic modifications and impact recovery to affect long-term outcomes after AKI. Global epigenetic changes after injury include histone modifications (87) and DNA methylation (88). The specific mechanisms and the impact on cell recovery are active areas of investigation, as recently reviewed in detail (89).
The dynamic changes in epigenetic marks and the need to re-establish a stable epigenome after AKI are starting to be addressed. Recently, Wilflingseder et al. comprehensively mapped the promoter- and enhancer-selective histone marks, H3K4me3 and H3K27ac, respectively, before and 2 days after ischemia–reperfusion injury (IRI) (45). Their data indicated that the H3K27ac mark was highly dynamic in the enhancer elements, relative to promoters, of differentially regulated genes. The DNA-binding motifs identified in differentially marked enhancer elements overlap strongly with those identified by snRNA-Seq and multimodal analyses outlined previously. Enhancers activated with IRI are enriched in AP-1 transcription factors, while those suppressed were associated with metabolic transcription factors, including RXR, ESRRA, and HNF4a, all of which had decreased enhancer binding after IRI. The preferential dynamics of H3K27ac/H3K4me1 marked enhancer regions relative to promoters are also suggested by human data (13). Many of the injury pathways associated with differential chromatin accessibility were common to mice and humans (6, 13). These studies use bulk chromatin samples, which pool a wide range of cell-specific chromatin patterns. This feature tends to attenuate the most dynamic signals and accounts for only modest differences observed in histone marks in most studies (13, 45).
While enhancer elements show dynamic activity after injury, there is also strong evidence that promoter histone modifications play a critical role in the transcriptional response to injury. For example, Ezh2 is a component of the Polycomb repressor complex that catalyzes H3K27me3 at sites of gene repression. Its expression is upregulated early after injury in repairing proximal tubules identified both in snRNA profiles (21) and Kim1 translating ribosome affinity purification (47). However, its depletion reduces the severity of injury in lipopolysaccharide-induced AKI (90). In contrast, PTIP is an essential component of the KMT2C/D histone methyltransferase complex that deposits H3K4me3 at active promoter regions (91). After folic acid injury, the regeneration of proximal tubules that lack PTIP is profoundly impaired because of an inability to re-establish the necessary histone marks (92).
Pax2 and Pax8 are closely related developmental transcription factors necessary for normal kidney development where they are critical for METs (93). Pax2/8 are also upregulated during normal recovery after AKI (15, 37, 94), which partly mimics aspects of development. Pax2/8 can provide locus specificity for both the Polycomb repressor complex and for the PTIP–KMT2C/D complex depending on post-translational modifications and cofactor interactions (95, 96). These observations suggested that Pax2/8 may be tubule-specific mediators of epigenetic modifications after injury. We recently investigated the role of Pax2/8 in the recovery from AKI using conditional alleles and a proximal tubule–specific driver (82). We anticipated that Pax2/8 mutant mice would not recover from ischemic AKI because of an inability to reset appropriate epigenetic marks during regeneration. Surprisingly, we discovered these mice were more resistant to ischemic injury (82). Uninjured Pax2/8 mutant proximal tubule cells in the S3 segment developed an expression pattern that overlapped with FR-PTCs described previously. We proposed that the loss of Pax2/8 mimics the cellular stresses observed in preconditioning or after injury and alters metabolism to make cells more resistant to ischemia. Interestingly, upregulation of Pax2/8 (21, 97) and increased accessibility of Pax-family binding motifs (20) are prominent in the FR-PTC population. With similar DNA-binding domains, Pax2 and Pax8 may be partially redundant in the proximal tubules. Whether Pax2 and Pax8 have unique or shared target genes in the adult kidney before or after injury remains to be determined.
Conclusions
The advent of high-throughput technologies for analyzing transcription at single-cell resolution as well as chromatin accessibility and transcription factor binding has resulted in an unprecedented wealth of new data to understand kidney regeneration. These data have firmly established the roles of stress-response genes both in early and late injury and the paramount importance of metabolic expression signatures with proximal tubule function and regeneration. The tools for integrating these massive and diverse datasets are powerful and everimproving. Yet, the logic of biological phenomena is not easily discerned from large datasets because of the complexity, redundancy, and noise inherent in most systems. While changes in gene expression are easily measured across the genome, it is not obvious whether any specific gene change directly impacts renal cell regeneration. We can measure genome-wide binding of many transcription factors; yet it is not always clear whether these binding sites are functional. Similarly, motif analyses often indicate that only a small percentage of sequence motifs are bound in vivo and do not account for post-translational modifications that can enhance or suppress activity. Yet models and predictions provide many hypotheses that can be tested in genetically tractable animal models and validated in human cohorts. Thus, as the complex transcriptional networks that regulate recovery from AKI become more clear, novel therapeutic approaches to treat this vexing clinical problem may be at hand.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
J. A. B., J. A. W., and G. R. D. conceptualization; J. A. B. and J. A. W. formal analysis; J. A. W. and G. R. D. resources; J. A. B. and J. A. W. data curation; J. A. B. and J. A. W. writing–original draft; J. A. B. and G. R. D. writing–review & editing; J. A. B. and G. R. D. supervision; and G. R. D. project administration.
Funding and additional information
This work was funded by the University of Michigan O’Brien Kidney Center (grant no.: DK-P30-081943, to J. A. B.); National Institutes of Health (NIH) grant K08 DK125776 (to J. A. B.); ASN-Kidney Cure Career Development Award (to J. A. W.); Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (grant no.: ES103361-01, to J. A. W.); and NIH (grant nos.: R01 DK054740 and R01 DK073722, to G. R. D.). A gift from Josh and Audrey Rumsey in memory of Isaiah. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Reviewed by members of the JBC Editorial Board. Edited by Ursula Jakob
References
- 1.Silver S.A., Long J., Zheng Y., Chertow G.M. Cost of acute kidney injury in hospitalized patients. J. Hosp. Med. 2017;12:70–76. doi: 10.12788/jhm.2683. [DOI] [PubMed] [Google Scholar]
- 2.Chertow G.M., Burdick E., Honour M., Bonventre J.V., Bates D.W. Acute kidney injury, mortality, Length of Stay, and costs in hospitalized patients. J. Am. Soc. Nephrol. 2005;16:3365–3370. doi: 10.1681/ASN.2004090740. [DOI] [PubMed] [Google Scholar]
- 3.Chang-Panesso M., Humphreys B.D. Cellular plasticity in kidney injury and repair. Nat. Rev. Nephrol. 2017;13:39–46. doi: 10.1038/nrneph.2016.169. [DOI] [PubMed] [Google Scholar]
- 4.Basile D.P., Anderson M.D., Sutton T.A. Pathophysiology of acute kidney injury. Compr. Physiol. 2012;2:1303–1353. doi: 10.1002/cphy.c110041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatachalam M.A., Weinberg J.M., Kriz W., Bidani A.K. Failed tubule recovery, AKI-CKD transition, and kidney disease progression. J. Am. Soc. Nephrol. 2015;26:1765–1776. doi: 10.1681/ASN.2015010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lake B.B., Menon R., Winfree S., Hu Q., Ferreira R.M., Kalhor K., et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature. 2023;619:585–594. doi: 10.1038/s41586-023-05769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad P.V., Li L.-P., Hack B., Leloudas N., Sprague S.M. Quantitative Blood Oxygenation level dependent Magnetic Resonance Imaging for estimating Intra-renal oxygen availability Demonstrates kidneys are Hypoxemic in human CKD. Kidney Int. Rep. 2023;8:1057–1067. doi: 10.1016/j.ekir.2023.02.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaub J.A., Venkatachalam M.A., Weinberg J.M. Proximal tubular oxidative metabolism in acute kidney injury and the transition to CKD. Kidney360. 2021;2:355–364. doi: 10.34067/KID.0004772020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatachalam M.A., Griffin K.A., Lan R., Geng H., Saikumar P., Bidani A.K. Acute kidney injury: a springboard for progression in chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2010;298:F1078–F1094. doi: 10.1152/ajprenal.00017.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffl H., Fischer R. Five-year outcomes of severe acute kidney injury requiring renal replacement therapy. Nephrol. Dial. Transplant. 2008;23:2235–2241. doi: 10.1093/ndt/gfn182. [DOI] [PubMed] [Google Scholar]
- 11.Schiffl H. Renal recovery from acute tubular necrosis requiring renal replacement therapy: a prospective study in critically ill patients. Nephrol. Dial. Transplant. 2006;21:1248–1252. doi: 10.1093/ndt/gfk069. [DOI] [PubMed] [Google Scholar]
- 12.Menon R., Bomback A.S., Lake B.B., Stutzke C., Grewenow S.M., Menez S., et al. Integrated single cell sequencing and histopathological analyses reveal diverse injury and repair responses in a participant with acute kidney injury: a clinical-molecular-pathologic correlation. Kidney Int. 2022;101:1116–1125. doi: 10.1016/j.kint.2022.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gisch D.L., Brennan M., Lake B.B., Basta J., Keller M.S., Ferreira R.M., et al. The chromatin landscape of healthy and injured cell types in the human kidney. Nat. Commun. 2024;15:433. doi: 10.1038/s41467-023-44467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazzeri E., Angelotti M.L., Peired A., Conte C., Marschner J.A., Maggi L., et al. Endocycle-related tubular cell hypertrophy and progenitor proliferation recover renal function after acute kidney injury. Nat. Commun. 2018;9:1344. doi: 10.1038/s41467-018-03753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusaba T., Lalli M., Kramann R., Kobayashi A., Humphreys B.D. Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1527–1532. doi: 10.1073/pnas.1310653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphreys B.D., Valerius T.M., Kobayashi A., Mugford J.W., Soeung S., Duffield J.S., et al. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2:284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Berger K., Bangen J.-M., Hammerich L., Liedtke C., Floege J., Smeets B., et al. Origin of regenerating tubular cells after acute kidney injury. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1533–1538. doi: 10.1073/pnas.1316177111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witzgall R., Brown D., Schwarz C., Bonventre J.V. Localization of proliferating cell nuclear antigen, vimentin, c-Fos, and clusterin in the postischemic kidney. Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J. Clin. Invest. 1994;93:2175–2188. doi: 10.1172/JCI117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphreys B.D., Czerniak S., DiRocco D.P., Hasnain W., Cheema R., Bonventre J.V. Repair of injured proximal tubule does not involve specialized progenitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9226–9231. doi: 10.1073/pnas.1100629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerhardt L.M.S., Koppitch K., Gestel J.v., Guo J., Cho S., Wu H., et al. Lineage tracing and single-Nucleus multiomics reveal novel features of adaptive and maladaptive repair after acute kidney injury. J. Am. Soc. Nephrol. 2023;34:554–571. doi: 10.1681/ASN.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirita Y., Wu H., Uchimura K., Wilson P.C., Humphreys B.D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. U. S. A. 2020;117:15874–15883. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balzer M.S., Doke T., Yang Y.-W., Aldridge D.L., Hu H., Mai H., et al. Single-cell analysis highlights differences in druggable pathways underlying adaptive or fibrotic kidney regeneration. Nat. Commun. 2022;13:4018. doi: 10.1038/s41467-022-31772-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ide S., Kobayashi Y., Ide K., Strausser S.A., Abe K., Herbek S., et al. Ferroptotic stress promotes the accumulation of pro-inflammatory proximal tubular cells in maladaptive renal repair. Elife. 2021;10 doi: 10.7554/eLife.68603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little M., Kairath P. Does renal repair Recapitulate kidney development? J. Am. Soc. Nephrol. 2016;28:34–46. doi: 10.1681/ASN.2016070748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiba T., Hukriede N., de Caestecker M.P. Kidney regeneration: lessons from development. Curr. Pathobiol. Rep. 2015;3:67–79. doi: 10.1007/s40139-015-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnell J., Achieng M., Lindström N.O. Principles of human and mouse nephron development. Nat. Rev. Nephrol. 2022;18:628–642. doi: 10.1038/s41581-022-00598-5. [DOI] [PubMed] [Google Scholar]
- 27.Kreidberg J.A., Sariola H., Loring J.M., Maeda M., Pelletier J., Housman D., et al. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 28.Torres M., Gomez-Pardo E., Dressler G.R., Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- 29.Brophy P.D., Ostrom L., Lang K.M., Dressler G.R. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development. 2001;128:4747–4756. doi: 10.1242/dev.128.23.4747. [DOI] [PubMed] [Google Scholar]
- 30.Narlis M., Grote D., Gaitan Y., Boualia S.K., Bouchard M. Pax2 and Pax8 regulate branching Morphogenesis and nephron differentiation in the developing kidney. J. Am. Soc. Nephrol. 2007;18:1121–1129. doi: 10.1681/ASN.2006070739. [DOI] [PubMed] [Google Scholar]
- 31.Laszczyk A.M., Higashi A.Y., Patel S.R., Johnson C.N., Soofi A., Abraham S., et al. Pax2 and Pax8 proteins regulate Urea Transporters and Aquaporins to control urine Concentration in the adult kidney. J. Am. Soc. Nephrol. 2020;31:1212–1225. doi: 10.1681/ASN.2019090962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng-Blichfeldt J.-P., Stewart B.J., Clatworthy M.R., Williams J.M., Röper K. Identification of a core transcriptional program driving the human renal mesenchymal-to-epithelial transition. Dev. Cell. 2024;59:595–612.e8. doi: 10.1016/j.devcel.2024.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Self M., Lagutin O.V., Bowling B., Hendrix J., Cai Y., Dressler G.R., et al. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heliot C., Desgrange A., Buisson I., Prunskaite-Hyyryläinen R., Shan J., Vainio S., et al. HNF1B controls proximal-intermediate nephron segment identity in vertebrates by regulating Notch signalling components and Irx1/2. Development. 2013;140:873–885. doi: 10.1242/dev.086538. [DOI] [PubMed] [Google Scholar]
- 35.Cheng H.-T., Kim M., Valerius M.T., Surendran K., Schuster-Gossler K., Gossler A., et al. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duvall K., Crist L., Perl A.J., Shakked N.P., Chaturvedi P., Kopan R. Revisiting the role of notch in nephron segmentation confirms a role for proximal fate selection during mouse and human nephrogenesis. Development. 2022;149 doi: 10.1242/dev.200446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang H.M., Huang S., Reidy K., Han S.H., Chinga F., Susztak K. Sox9-Positive progenitor cells play a key role in renal tubule epithelial regeneration in mice. Cell Rep. 2016;14:861–871. doi: 10.1016/j.celrep.2015.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S., Liu J., Pang P., Krautzberger M.A., Reginensi A., Akiyama H., et al. Sox9 activation highlights a cellular pathway of renal repair in the acutely injured mammalian kidney. Cell Rep. 2015;12:1325–1338. doi: 10.1016/j.celrep.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 39.Reginensi A., Clarkson M., Neirijnck Y., Lu B., Ohyama T., Groves A.K., et al. SOX9 controls epithelial branching by activating RET effector genes during kidney development. Hum. Mol. Genet. 2011;20:1143–1153. doi: 10.1093/hmg/ddq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura Y., Muto Y., Omachi K., Miner J.H., Humphreys B.D. Elucidating the proximal tubule HNF4A gene regulatory network in human kidney organoids. J. Am. Soc. Nephrol. 2023;34:1672–1686. doi: 10.1681/ASN.0000000000000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marable S.S., Chung E., Park J.-S. Hnf4a is required for the development of Cdh6-Expressing progenitors into proximal tubules in the mouse kidney. J. Am. Soc. Nephrol. 2020;31:2543–2558. doi: 10.1681/ASN.2020020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 43.Badia-i-Mompel P., Wessels L., Müller-Dott S., Trimbour R., Flores R.O.R., Argelaguet R., et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 2023;24:1–16. doi: 10.1038/s41576-023-00618-5. [DOI] [PubMed] [Google Scholar]
- 44.Luo Y., Hitz B.C., Gabdank I., Hilton J.A., Kagda M.S., Lam B., et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2019;48:D882–D889. doi: 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilflingseder J., Willi M., Lee H.K., Olauson H., Jankowski J., Ichimura T., et al. Enhancer and super-enhancer dynamics in repair after ischemic acute kidney injury. Nat. Commun. 2020;11:3383. doi: 10.1038/s41467-020-17205-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., et al. Determination and inference of Eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang-Panesso M., Kadyrov F.F., Lalli M., Wu H., Ikeda S., Kefaloyianni E., et al. FOXM1 drives proximal tubule proliferation during repair from acute ischemic kidney injury. J. Clin. Invest. 2019;129:5501–5517. doi: 10.1172/JCI125519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanamori M., Konno H., Osato N., Kawai J., Hayashizaki Y., Suzuki H. A genome-wide and nonredundant mouse transcription factor database. Biochem. Biophys. Res. Commun. 2004;322:787–793. doi: 10.1016/j.bbrc.2004.07.179. [DOI] [PubMed] [Google Scholar]
- 49.Liu J., Krautzberger M.A., Sui S.H., Hofmann O.M., Chen Y., Baetscher M., et al. Cell-specific translational profiling in acute kidney injury. J. Clin. Invest. 2014;124:1242–1254. doi: 10.1172/JCI72126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schep A.N., Wu B., Buenrostro J.D., Greenleaf W.J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods. 2017;14:975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang Y., Harigaya Y., Zhang Z., Zhang H., Zang C., Zhang N.R. Nonparametric single-cell multiomic characterization of trio relationships between transcription factors, target genes, and cis-regulatory regions. Cell Syst. 2022;13:737–751.e734. doi: 10.1016/j.cels.2022.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ledru N., Wilson P.C., Muto Y., Yoshimura Y., Wu H., Li D., et al. Predicting proximal tubule failed repair drivers through regularized regression analysis of single cell multiomic sequencing. Nat. Commun. 2024;15:1291. doi: 10.1038/s41467-024-45706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambrosini G., Vorontsov I., Penzar D., Groux R., Fornes O., Nikolaeva D.D., et al. Insights gained from a comprehensive all-against-all transcription factor binding motif benchmarking study. Genome Biol. 2020;21:114. doi: 10.1186/s13059-020-01996-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farley E.K., Olson K.M., Zhang W., Rokhsar D.S., Levine M.S. Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proc. Natl. Acad. Sci. U. S. A. 2016;113:6508–6513. doi: 10.1073/pnas.1605085113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunt R.W., Wasserman W.W. Non-targeted transcription factors motifs are a systemic component of ChIP-seq datasets. Genome Biol. 2014;15:412. doi: 10.1186/s13059-014-0412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khundmiri S.J., Chen L., Lederer E.D., Yang C.-R., Knepper M.A. Transcriptomes of Major proximal tubule cell culture models. J. Am. Soc. Nephrol. 2020;32:86–97. doi: 10.1681/ASN.2020010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piret S.E., Guo Y., Attallah A.A., Horne S.J., Zollman A., Owusu D., et al. Krüppel-like factor 6–mediated loss of BCAA catabolism contributes to kidney injury in mice and humans. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2024414118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piret S.E., Mallipattu S.K. Proximal tubular transcription factors in acute kidney injury: recent advances. Nephron. 2020;144:613–615. doi: 10.1159/000508856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang K.P., Lee J.E., Lee A.S., Jung Y.J., Kim D.A.L., Lee S.I.K., et al. Effect of gender differences on the regulation of renal ischemia-reperfusion-induced inflammation in mice. Mol. Med. Rep. 2014;9:2061–2068. doi: 10.3892/mmr.2014.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J.Y., Bai Y., Jayne L.A., Cianciolo R.E., Bajwa A., Pabla N.S. Involvement of the CDKL5-SOX9 signaling axis in rhabdomyolysis-associated acute kidney injury. Am. J. Physiol. Renal Physiol. 2020;319:F920–F929. doi: 10.1152/ajprenal.00429.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J.Y., Silvaroli J.A., Martinez G.V., Bisunke B., Ramirez A.V.L., Jayne L.A., et al. Zinc finger protein 24-dependent transcription factor SOX9 up-regulation protects tubular epithelial cells during acute kidney injury. Kidney Int. 2023;103:1093–1104. doi: 10.1016/j.kint.2023.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim J.Y., Bai Y., Jayne L.A., Hector R.D., Persaud A.K., Ong S.S., et al. A kinome-wide screen identifies a CDKL5-SOX9 regulatory axis in epithelial cell death and kidney injury. Nat. Commun. 2020;11:1924. doi: 10.1038/s41467-020-15638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ming Z., Vining B., Bagheri-Fam S., Harley V. SOX9 in organogenesis: shared and unique transcriptional functions. Cell. Mol. Life Sci. 2022;79:522. doi: 10.1007/s00018-022-04543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J.Y., Bai Y., Jayne L.A., Abdulkader F., Gandhi M., Perreau T., et al. SOX9 promotes stress-responsive transcription of VGF nerve growth factor inducible gene in renal tubular epithelial cells. J. Biol. Chem. 2020;295:16328–16341. doi: 10.1074/jbc.RA120.015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalinichenko V.V., Gusarova G.A., Tan Y., Wang I.C., Major M.L., Wang X., et al. Ubiquitous expression of the Forkhead box M1B Transgene Accelerates proliferation of distinct Pulmonary cell types following Lung injury. J. Biol. Chem. 2003;278:37888–37894. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- 67.Hallman M.A., Zhuang S., Schnellmann R.G. Regulation of dedifferentiation and Redifferentiation in renal proximal tubular cells by the Epidermal growth factor receptor. J. Pharmacol. Exp. Ther. 2008;325:520–528. doi: 10.1124/jpet.107.134031. [DOI] [PubMed] [Google Scholar]
- 68.Zeng F., Singh A.B., Harris R.C. The role of the EGF family of ligands and receptors in renal development, physiology and pathophysiology. Exp. Cell Res. 2009;315:602–610. doi: 10.1016/j.yexcr.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piret S.E., Mallipattu S.K. Transcriptional regulation of proximal tubular metabolism in acute kidney injury. Pediatr. Nephrol. 2022;38:1–12. doi: 10.1007/s00467-022-05748-2. [DOI] [PubMed] [Google Scholar]
- 70.Iwaki T., Bennion B.G., Stenson E.K., Lynn J.C., Otinga C., Djukovic D., et al. PPARα contributes to protection against metabolic and inflammatory derangements associated with acute kidney injury in experimental sepsis. Physiol. Rep. 2019;7 doi: 10.14814/phy2.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu S., Jia P., Fang Y., Jin J., Sun Z., Zhou W., et al. Nuclear farnesoid X receptor attenuates acute kidney injury through fatty acid oxidation. Kidney Int. 2022;101:987–1002. doi: 10.1016/j.kint.2022.01.029. [DOI] [PubMed] [Google Scholar]
- 72.Dhillon P., Park J., Pozo C.H.d., Li L., Doke T., Huang S., et al. The nuclear receptor ESRRA protects from kidney disease by coupling metabolism and differentiation. Cell Metab. 2021;33:379–394.e378. doi: 10.1016/j.cmet.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu X., Xu M., Meng X., Li S., Liu Q., Bai M., et al. Nuclear receptor PXR targets AKR1B7 to protect mitochondrial metabolism and renal function in AKI. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aay7591. [DOI] [PubMed] [Google Scholar]
- 74.Piret S.E., Attallah A.A., Gu X., Guo Y., Gujarati N.A., Henein J., et al. Loss of proximal tubular transcription factor Krüppel-like factor 15 exacerbates kidney injury through loss of fatty acid oxidation. Kidney Int. 2021;100:1250–1267. doi: 10.1016/j.kint.2021.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran M., Tam D., Bardia A., Bhasin M., Rowe G.C., Kher A., et al. PGC-1α promotes recovery after acute kidney injury during systemic inflammation in mice. J. Clin. Invest. 2011;121:4003–4014. doi: 10.1172/JCI58662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clark A.J., Saade M.C., Vemireddy V., Vu K.Q., Flores B.M., Etzrodt V., et al. Hepatocyte nuclear factor 4α mediated quinolinate phosphoribosylltransferase (QPRT) expression in the kidney facilitates resilience against acute kidney injury. Kidney Int. 2023;104:1150–1163. doi: 10.1016/j.kint.2023.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shulman A.I., Larson C., Mangelsdorf D.J., Ranganathan R. Structural Determinants of Allosteric ligand activation in RXR Heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 78.Monden T., Yamada M., Nihei Y., Kishi M., Tomaru T., Ishii S., et al. Unliganded RXR acts as an inhibitory factor on troglitazone-induced activation. Life Sci. 2004;76:731–741. doi: 10.1016/j.lfs.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 79.Gerhardt L.M.S., McMahon A.P. Identifying common molecular mechanisms in experimental and human acute kidney injury. Semin. Nephrol. 2022;42 doi: 10.1016/j.semnephrol.2022.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cippà P.E., Sun B., Liu J., Chen L., Naesens M., McMahon A.P. Transcriptional trajectories of human kidney injury progression. JCI Insight. 2018;3 doi: 10.1172/jci.insight.123151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnsen M., Kubacki T., Yeroslaviz A., Späth M.R., Mörsdorf J., Göbel H., et al. The integrated RNA landscape of renal preconditioning against ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2020;31:716–730. doi: 10.1681/ASN.2019050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beamish J.A., Telang A.C., McElliott M.C., Al-Suraimi A., Chowdhury M., Ference-Salo J.T., et al. Pax protein depletion in proximal tubules triggers conserved mechanisms of resistance to acute ischemic kidney injury preventing transition to chronic kidney disease. Kidney Int. 2024;105:312–327. doi: 10.1016/j.kint.2023.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gerhardt L.M.S., Liu J., Koppitch K., Cippà P.E., McMahon A.P. Single-nuclear transcriptomics reveals diversity of proximal tubule cell states in a dynamic response to acute kidney injury. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2026684118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang L., Besschetnova T.Y., Brooks C.R., Shah J.V., Bonventre J.V. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med. 2010;16:535–543. doi: 10.1038/nm.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taguchi K., Elias B.C., Sugahara S., Sant S., Freedman B.S., Waikar S.S., et al. Cyclin G1 induces maladaptive proximal tubule cell dedifferentiation and renal fibrosis through CDK5 activation. J. Clin. Invest. 2022;132 doi: 10.1172/JCI158096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taguchi K., Sugahara S., Elias B.C., Pabla N.S., Canaud G., Brooks C.R. IL-22 is secreted by proximal tubule cells and regulates DNA damage response and cell death in acute kidney injury. Kidney Int. 2023;105:99–114. doi: 10.1016/j.kint.2023.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zager R.A., Johnson A.C.M., Becker K. Acute unilateral ischemic renal injury induces progressive renal inflammation, lipid accumulation, histone modification, and “end-stage” kidney disease. Am. J. Physiol. Renal Physiol. 2011;301:F1334–F1345. doi: 10.1152/ajprenal.00431.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heylen L., Thienpont B., Naesens M., Busschaert P., Depreeuw J., Smeets D., et al. Ischemia-induced DNA Hypermethylation during kidney Transplant Predicts chronic Allograft injury. J. Am. Soc. Nephrol. 2018;29:1566–1576. doi: 10.1681/ASN.2017091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo C., Dong G., Liang X., Dong Z. Epigenetic regulation in AKI and kidney repair: mechanisms and therapeutic implications. Nat. Rev. Nephrol. 2019;15:220–239. doi: 10.1038/s41581-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li B., Xia Y., Mei S., Ye Z., Song B., Yan X., et al. Histone H3K27 methyltransferase EZH2 regulates apoptotic and inflammatory responses in sepsis-induced AKI. Theranostics. 2023;13:1860–1875. doi: 10.7150/thno.83353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Patel S.R., Kim D., Levitan I., Dressler G.R. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soofi A., Kutschat A.P., Azam M.H., Laszczyk A.M., Dressler G.R. Regeneration after acute kidney injury requires PTIP mediated epigenetic modifications. JCI Insight. 2020;5 doi: 10.1172/jci.insight.130204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rothenpieler U.W., Dressler G.R. Pax-2 is required for mesenchyme-to-epithelium conversion during kidney development. Development. 1993;119:711–720. doi: 10.1242/dev.119.3.711. [DOI] [PubMed] [Google Scholar]
- 94.Imgrund M., Gröne E., Gröne H.J., Kretzler M., Holzman L., Schlöndorff D., et al. Re-expression of the developmental gene Pax-2 during experimental acute tubular necrosis in mice1. Kidney Int. 1999;56:1423–1431. doi: 10.1046/j.1523-1755.1999.00663.x. [DOI] [PubMed] [Google Scholar]
- 95.Abraham S., Paknikar R., Bhumbra S., Luan D., Garg R., Dressler G.R., et al. The Groucho-associated Phosphatase PPM1B Displaces Pax Transactivation domain interacting protein (PTIP) to Switch the transcription factor Pax2 from a transcriptional activator to a repressor. J. Biol. Chem. 2015;290:7185–7194. doi: 10.1074/jbc.M114.607424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patel S.R., Ranghini E., Dressler G.R. Mechanisms of gene activation and repression by Pax proteins in the developing kidney. Pediatr. Nephrol. 2014;29:589–595. doi: 10.1007/s00467-013-2603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ide S., Ide K., Abe K., Kobayashi Y., Kitai H., McKey J., et al. Sex differences in resilience to ferroptosis underlie sexual dimorphism in kidney injury and repair. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Legouis D., Ricksten S.-E., Faivre A., Verissimo T., Gariani K., Verney C., et al. Altered proximal tubular cell glucose metabolism during acute kidney injury is associated with mortality. Nat. Metab. 2020;2:732–743. doi: 10.1038/s42255-020-0238-1. [DOI] [PubMed] [Google Scholar]
- 99.Schubert M., Klinger B., Klünemann M., Sieber A., Uhlitz F., Sauer S., et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018;9:20. doi: 10.1038/s41467-017-02391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rudman-Melnick V., Adam M., Potter A., Chokshi S.M., Ma Q., Drake K.A., et al. Single-cell profiling of aki in a murine model reveals novel transcriptional signatures, profibrotic phenotype, and epithelial-to-stromal crosstalk. J. Am. Soc. Nephrol. 2020;31:2793–2814. doi: 10.1681/ASN.2020010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H., Dixon E.E., Wu H., Humphreys B.D. Comprehensive single-cell transcriptional profiling defines shared and unique epithelial injury responses during kidney fibrosis. Cell Metab. 2022;34:1977–1998.e9. doi: 10.1016/j.cmet.2022.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garcia-Alonso L., Holland C.H., Ibrahim M.M., Turei D., Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019;29:1363–1375. doi: 10.1101/gr.240663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Holland C.H., Tanevski J., Perales-Patón J., Gleixner J., Kumar M.P., Mereu E., et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:36. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li H., Li D., Ledru N., Xuanyuan Q., Wu H., Asthana A., et al. Transcriptomic, epigenomic, and spatial metabolomic cell profiling redefines regional human kidney anatomy. Cell Metab. 2024;36:1105–1125.e10. doi: 10.1016/j.cmet.2024.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]