Abstract
Chronic stress–induced epinephrine (EPI) accelerates breast cancer progression and metastasis, but the molecular mechanisms remain unclear. Herein, we found a strong positive correlation between circulating EPI levels and the tumoral expression of ubiquitin-specific peptidase 22 (USP22) in patients with breast cancer. USP22 facilitated EPI-induced breast cancer progression and metastasis by enhancing adipose triglyceride lipase (ATGL)–mediated lipolysis. Targeted USP22 deletion decreased ATGL expression and lipolysis, subsequently inhibiting EPI-mediated breast cancer lung metastasis. USP22 acts as a bona fide deubiquitinase for the Atgl gene transcription factor FOXO1, and EPI architects a lipolysis signaling pathway to stabilize USP22 through AKT-mediated phosphorylation. Notably, USP22 phosphorylation levels are positively associated with EPI and with downstream pathways involving both FOXO1 and ATGL in breast cancers. Pharmacological USP22 inhibition synergized with β-blockers in treating preclinical xenograft breast cancer models. This study reveals a molecular pathway behind EPI’s tumor-promoting effects and provides a strong rationale for combining USP22 inhibition with β-blockers to treat aggressive breast cancer.
Psychological stress promotes cancer metastasis through enhancing lipid metabolism.
INTRODUCTION
Breast cancer is the most common cancer in women worldwide. While patients with early-stage, nonmetastatic disease often experience positive outcomes, the mortality rate for those with metastatic disease to distant organs remains high (1). Patients with breast cancer frequently endure various forms of chronic emotional stress, including depression, anxiety, and fear (2), leading to increased production of stress hormones such as epinephrine (EPI) and norepinephrine. Chronic stress–induced neuroendocrine activity, particularly EPI, has been shown to promote the progression and metastasis of breast cancer through binding to β-adrenergic receptors (3–7). β-Blockers, which inhibit EPI-induced signaling, can suppress proliferation, invasiveness, and angiogenesis in preclinical breast cancer models. Retrospective studies suggest that β-blockers may improve the prognosis of patients with breast cancer (8). However, evidence from multiple clinical trials indicates that β-blockers are ineffective in treating more advanced breast cancers or human epidermal growth factor receptor 2 (HER2)-positive cancers. The molecular mechanisms by which EPI contributes to breast cancer pathogenesis and the resistance of late-stage cancers to β-blockers remain unclear.
The ubiquitin-specific peptidase 22 (USP22) was initially identified as 1 of the 11 death-from-cancer signature genes that operate a crucial role in regulating tumor cell growth and death (9). Further analysis of clinical samples has shown that USP22 is up-regulated in nearly all human cancer types including breast cancers, and its expression is further elevated with the progress of breast cancers, which has been correlated with poor prognosis (9–11). We and others have defined USP22 as an oncogene through c-Myc activation, antagonism of the tumor suppressive function of p53, and stabilization of cyclins including B1 and D1 (12–15). USP22 also restricts antitumor immunity through potentiating regulatory T cell (Treg) immune suppressive activity (16) and Treg adaptation in the tumor microenvironment (17). More recently, we have found that USP22 controls breast cancer cell stemness and metastasis through selectively up-regulating a group of integrin family members on cancer cells (18). However, while accumulated evidence suggests that USP22 is involved in metabolic regulation in cancer cells, the molecular pathways that links the USP22 oncogenic function to metabolic regulation such as during chronic stress have not been identified.
In this investigation, we uncovered a strong positive association between circulating EPI levels and USP22 protein expression in patients with breast cancer. Intriguingly, EPI signaling was found to stabilize USP22 protein, thereby facilitating the transcription of adipose triglyceride lipase (ATGL), an enzyme pivotal in triacylglycerol (TAG) breakdown into diacylglycerol (DAG) and free fatty acids (FFAs), which subsequently promotes breast cancer metastasis (19, 20). Furthermore, under EPI stimulation, USP22 functions as a deubiquitinase for the ATGL transcription factor, the forehead box O1 (FOXO1), a crucial regulator of lipid and glucose metabolism (21–27). This study underscores the role of stress-induced EPI in augmenting ATGL-mediated lipolysis, consequently propelling breast cancer metastasis through an AKT-USP22-FOXO1-ATGL axis. Notably, pharmacological inhibition of USP22, in combination with β-blockers, exhibits synergistic effects in breast cancer therapy.
RESULTS
High serum of EPI is associated with elevated tumoral USP22 expression in patients with breast cancer
To investigate the potential involvement of USP22 in EPI-induced breast cancer progression, we collected paired para-cancer and cancer tissues, along with serum samples, from 65 patients with breast cancer (Table 1). Serum EPI concentrations were quantified using enzyme-linked immunosorbent assay ELISA as previously reported (28), and tumoral USP22 expression was assessed via immunohistochemistry (IHC) staining. Consistent with previous findings (11), USP22 protein levels were notably elevated in cancer tissues compared to their adjacent para-cancerous tissues (Fig. 1, A and B). Across the patient cohort, the average serum EPI concentration was 30.4 ± 39.5 pg/ml, with a median of 13.03 pg/ml (Table 1). Notably, serum EPI levels showed no associations with tumor stage, volume, or other clinical-pathological characteristics (fig. S1 and table S1).
Table 1. All information from clinical patients.
| No. | Age | Gender | EPI concentration | Tumor stage | ER | HER | PR |
|---|---|---|---|---|---|---|---|
| 1 | 59 | Female | 30.08944917 | pT2N2M0 | + | − | + |
| 2 | 44 | Female | 19.76304268 | pT1cN1M0 | + | − | + |
| 3 | 52 | Female | 20.30978666 | pT1aN0M0 | + | + | + |
| 4 | 48 | Female | 16.4201555 | pT2N1M0 | + | − | + |
| 5 | 43 | Female | 16.91745943 | pT1N2M0 | − | + | − |
| 6 | 84 | Female | 17.03529099 | pT1cN0M0 | + | − | + |
| 7 | 65 | Female | 18.58534115 | pT2N3M0 | − | − | − |
| 8 | 65 | Female | 19.21640434 | pT2N2M0 | + | + | + |
| 9 | 58 | Female | 13.75522267 | pT4N2aM0 | + | − | + |
| 10 | 71 | Female | 15.59517372 | pT2N0M0 | + | − | + |
| 11 | 52 | Female | 20.91037779 | pT2N0M0 | − | − | − |
| 12 | 38 | Female | 22.22811182 | pT3N1M0 | + | − | + |
| 13 | 69 | Female | 22.98317942 | pT2N1M0 | + | − | + |
| 14 | 58 | Female | 24.97132521 | pT1cN0M0 | + | + | + |
| 15 | 61 | Female | 26.56093979 | pT1cN0M0 | + | − | + |
| 16 | 47 | Female | 123.7253926 | pT2N0M0 | − | + | − |
| 17 | 55 | Female | 28.32988565 | pT1cN1M0 | + | − | + |
| 18 | 53 | Female | 146.4413857 | pT1cN0M0 | + | − | + |
| 19 | 38 | Female | 37.55151171 | pT2N0M0 | − | − | − |
| 20 | 52 | Female | 39.99299396 | pT1cN0M0 | + | − | − |
| 21 | 60 | Female | 142.6477335 | pT2N1M0 | + | − | + |
| 22 | 77 | Female | 51.27230258 | pT1cN0M0 | + | − | + |
| 23 | 66 | Female | 62.83211285 | pT2N0M0 | + | − | + |
| 24 | 58 | Female | 74.56981374 | pT1cN0M0 | + | − | + |
| 25 | 48 | Female | 87.33969051 | pT2N1M0 | + | − | + |
| 26 | 33 | Female | 95.32837132 | pT2N0M0 | + | − | + |
| 27 | 57 | Female | 103.0518796 | pT1bN0M0 | + | − | + |
| 28 | 64 | Female | 111.7721427 | pT1cN0M0 | − | + | + |
| 29 | 55 | Female | 27.54935444 | pT2N0M0 | + | − | + |
| 30 | 52 | Female | 125.5680438 | pTis(DCIs)N0M0 | + | + | + |
| 31 | 82 | Female | 140.6945155 | pT1cN0M0 | − | + | − |
| 32 | 68 | Female | 49.59331707 | pT2N0M0 | + | − | + |
| 33 | 63 | Female | 13.031525244 | pT3N1M0 | − | + | − |
| 34 | 64 | Female | 1.53453364 | pT2N0M0 | + | + | + |
| 35 | 64 | Female | 2.653448814 | pT2N3M0 | + | + | − |
| 36 | 49 | Female | 2.973902716 | pT1bN0M0 | − | − | − |
| 37 | 57 | Female | 3.197128007 | pT1cN0M0 | + | − | + |
| 38 | 47 | Female | 11.929401027 | pT1cN0M0 | + | − | + |
| 39 | 39 | Female | 3.851752002 | pT2N1M0 | + | − | + |
| 40 | 64 | Female | 3.869871156 | pT2N1M0 | + | − | + |
| 41 | 53 | Female | 4.046442175 | pT2N0M0 | + | − | − |
| 42 | 61 | Female | 4.171561597 | pT2N1M0 | + | + | + |
| 43 | 62 | Female | 4.352005244 | pT2N3M0 | + | − | + |
| 44 | 54 | Female | 4.418306235 | ypT3N3M0 | + | − | + |
| 45 | 61 | Female | 4.742769948 | pT2N1M0 | + | − | + |
| 46 | 50 | Female | 5.362233546 | ypT3N2M0 | + | − | + |
| 47 | 49 | Female | 5.680236149 | pT1N0M0 | + | − | + |
| 48 | 66 | Female | 5.893910998 | pT1N1M0 | + | − | + |
| 49 | 66 | Female | 6.841821665 | pT2N0M0 | − | − | − |
| 50 | 52 | Female | 6.991946894 | pT2N0M0 | + | − | + |
| 51 | 58 | Female | 12.606259832 | pT1N0M0 | + | − | + |
| 52 | 34 | Female | 7.311725729 | pT2N0M0 | + | − | + |
| 53 | 77 | Female | 7.618189254 | pT3N1M0 | + | − | + |
| 54 | 72 | Female | 7.790290593 | pT2N0M0 | + | − | + |
| 55 | 65 | Female | 8.008178739 | pT1N0M0 | + | − | + |
| 56 | 69 | Female | 8.134525005 | pT2N0M0 | + | − | + |
| 57 | 76 | Female | 8.201616335 | pT1N0M0 | + | − | + |
| 58 | 66 | Female | 8.440578721 | pT1N1M0 | + | − | − |
| 59 | 55 | Female | 9.349572877 | pT2N0M0 | + | + | − |
| 60 | 76 | Female | 9.377327706 | pT1N0M0 | + | − | + |
| 61 | 77 | Female | 9.582287895 | pT1N0M0 | + | − | + |
| 62 | 63 | Female | 10.069014381 | pT1N0M0 | + | − | + |
| 63 | 60 | Female | 10.468748695 | pT2N0M0 | − | − | − |
| 64 | 74 | Female | 3.84711759 | pT1N1M0 | + | − | + |
| 65 | 81 | Female | 7.242336207 | pT2N1M0 | + | − | + |
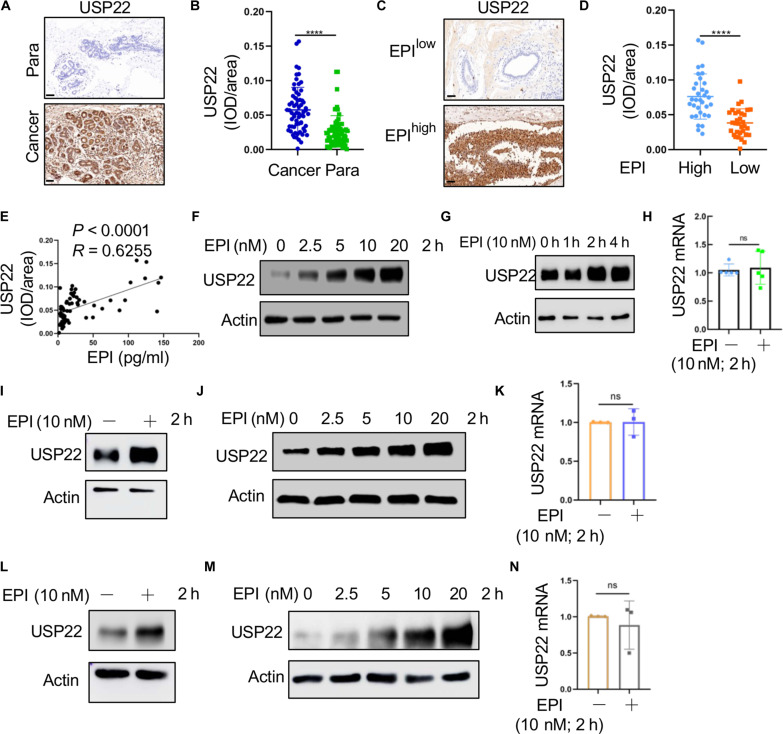
Fig. 1. High EPI in patients with breast cancer is accompanied with the increasing expression of USP22.
(A) Representative IHC photomicrographs of tissues stained with USP22 antibodies in patients with breast cancer in clinical breast cancer tissues and para-cancer tissues; Scale bars, 50 μm. (B) Quantification of USP22 expression in breast cancer tissues and para-cancer tissues (n = 65). (C and D) Representative images of USP22 staining in patients with breast cancer (EPIlow, n = 33; EPIhigh, n = 32); Scale bars, 50 μm (C). Analysis of USP22 expression in human breast cancer tissues in patients with high and low levels of EPI (D). (E) Relationship between serum EPI levels and USP22 from the IHC staining score in human breast cancer tissues. Pearson correlation coefficient was used as a measure of association. (F to N) Analysis of USP22 protein expression in MDA-MB-231 [(F) to (H)], MCF-7 [(I) to (K)], and SK-BR-3 [(L) to (N)] breast cancer cells treated with indicated concentrations of EPI [(F), (J), and (M)] or time course [(G), (I), and (L)]. Expression of mRNA of USP22 was measured by quantitative RT-PCR [(H), (K), and (N)]; Pearson correlation coefficient was used as a measure of association. Data are expressed as means ± SD of three or more independent experiments. Statistical significance was concluded by unpaired Student’s t test. ****P < 0.0001; ns, no statistical significance.
Patients were stratified into two groups based on EPI levels: EPIhigh with EPI levels higher than the median of 13.03 pg/ml and EPIlow (EPI ≤ 13.03). Cancer tissues from the EPIhigh group exhibited markedly elevated USP22 protein expression compared to those from the EPIlow group (Fig. 1, C and D). The serum EPI concentrations were positively correlating with tumoral USP22 protein expression levels (Fig. 1E). These findings suggest a potential mechanism whereby EPI may contribute to tumorigenesis, at least in part, through the up-regulation of USP22.
We sought to investigate if EPI stimulates USP22 expression in breast cancer cells. Treatment of human triple-negative breast cancer (TNBC) MDA-MB-231 cells with EPI resulted in a dose-dependent increase in USP22 protein expression (Fig. 1F). Intriguingly, the induction of USP22 protein expression by EPI reached a plateau as early as 2 hours after treatment (Fig. 1G). However, real-time reverse transcription polymerase chain reaction (RT-PCR) analysis revealed no alterations in USP22 mRNA levels following EPI treatment in MDA-MB-231 cells (Fig. 1H). Consistent with these findings, the treatment of MCF-7 and SK-BR-2 human breast cancer cells with EPI also markedly induced USP22 protein expression within 2 hours in a dose-dependent manner (Fig. 1, I, J, L, and M). In contrast, the mRNA levels of USP22 remained unchanged by EPI treatment (Fig. 1, K and N). These observations suggest the possibility that EPI induces USP22 protein expression posttranslationally in human breast cancers.
USP22 inhibition abolished EPI-induced breast cancer metastasis
To further investigate USP22’s involvement in EPI-mediated tumorigenesis, we generated USP22-null MDA-MB-231 and 4T1 cells using CRISPR technology. Western blotting analysis confirmed the complete deletion of USP22 (Fig. 2A). As expected, the genetic deletion of USP22 notably inhibited MDA-MB-231 xenograft tumor growth in immunodeficient NYG mice, as evidenced by reduced tumor volume (Fig. 2B), luminol fluorescence activity (Fig. 2, C to E), and tumor weight (Fig. 2, F and G). Notably, the treatment of mice with EPI at 2 mg/kg, as previously reported (29), markedly promoted the growth of wild-type (WT) MDA-MB-231 tumors but not USP22-null tumors (Fig. 2, B to D, F, and G), indicating that EPI drives breast cancer progression in a USP22-dependent manner. In addition, luminol fluorescence activity was observed in the lung area of mice with WT tumors but not in mice with USP22 knockout (KO) tumors, and EPI treatment increased this activity (Fig. 2, C and E). Histological analysis further confirmed EPI-induced breast cancer lung metastasis and the inhibitory effect of USP22 ablation (Fig. 2, H and I), underscoring the essential role of USP22 in EPI-induced breast cancer metastasis.
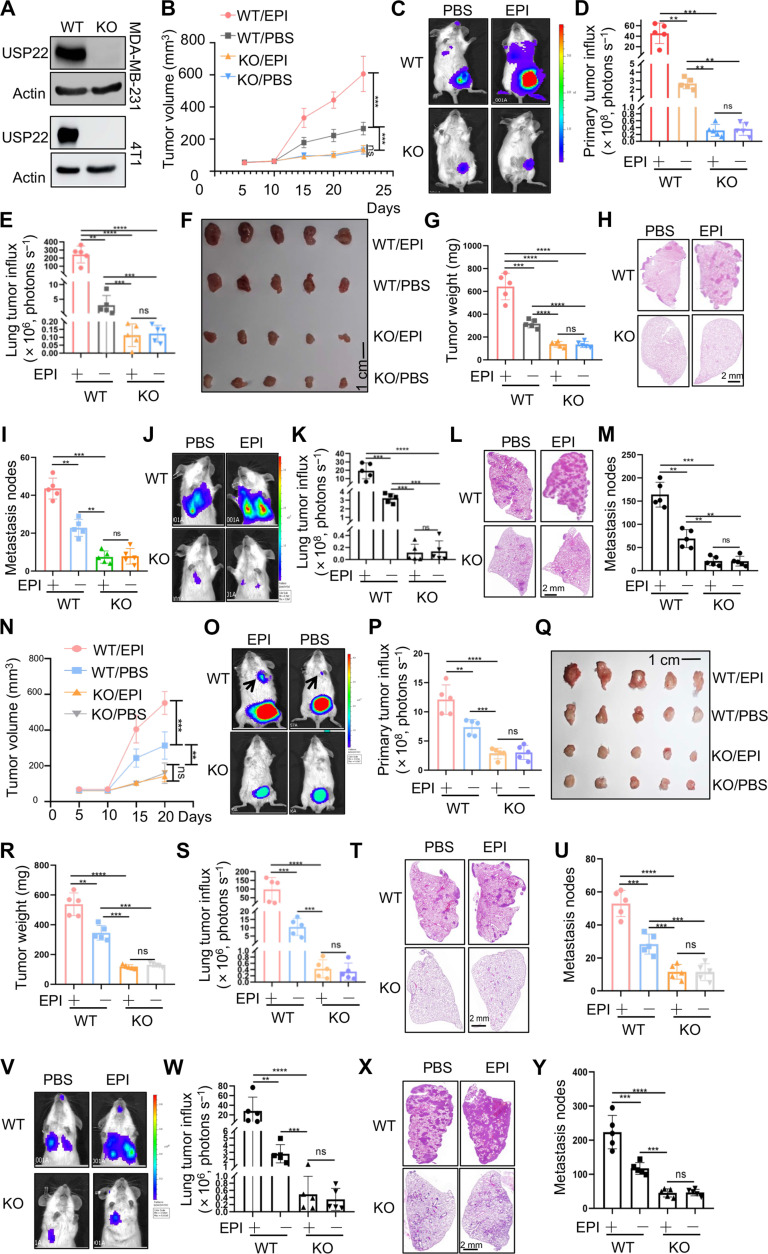
Fig. 2. EPI increases the expression of ATGL to induce the lipolysis of breast cancer through USP22.
(A) Validation of USP22 protein expression in WT and USP22 KO MDA-MB-231 cells (top) or 4T1 cells (bottom). (B to I) A total of 1 × 106 WT or USP22 KO MDA-MB-231 cells were injected into the mammary fat pad of immune compromised NYG mice and treated with PBS or EPI (2 mg/kg) per day for 7 days. Tumor volume (B), bioluminescence activity [(C) and (D)], and tumor weight [(F) and (G)] were measured. Lung metastases were measured by luminol fluorescence [(C) and (E)] and H&E staining of their lung sections [(H) and (I)]. N = 5 each group. (J to M) A total of 0.5 × 106 WT or USP22 KO MDA-MB-231 cells were injected via the tail vein into NYG mice. Lung metastases were measured by luminol fluorescence [(J) and (K)] and H&E staining [(L) and (M)]. (N to U) A total of 1 × 105 WT or USP22 KO 4T1 cells were injected into the mammary fat pad of BALB/c mice and treated with PBS or EPI (2 mg/kg) per day for 7 days. Tumor volume (N), bioluminescence activity [(O) and (P)], and tumor weight [(Q) and (R)] were measured. Lung metastases were measured by luminol fluorescence [(O) and (S)] and H&E staining of their lung sections [(T) and (U)]. N = 5 each group. (V to Y) A total of 0.5 × 105 WT or USP22 KO 4T1 cells were injected via the tail vein into BALB/c mice. Lung metastases were measured by luminol fluorescence [(V) and (W)] and H&E staining [(X) and (Y)]. Statistical significance was determined by one-way ANOVA test or unpaired Student’s t test. Data are expressed as means ± SD of five independent experiments. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, no statistical significance.
Further supporting this conclusion, EPI treatment markedly increased the number of lung tumor nodules upon intravenous injection of WT MDA-MB-231 cells into immune compromised NYG mice, whereas targeted deletion of USP22 completely prevented EPI-induced breast cancer lung metastasis as analyzed by luminol fluorescence activity (Fig. 2, J and K) and hematoxylin and eosin (H&E) staining of the lung tissue sections (Fig. 2, L and M). The serum EPI concentration was measured to be 335.53 ± 41.21 pg/ml in mice treated with EPI, nearly twice as high as in the control phosphate-buffered saline (PBS) group (175.75 ± 35.14 pg/ml) (fig. S2). These findings highlight the role of elevated EPI levels in both mice and humans in promoting breast cancer metastasis.
Subsequently, we validated our findings using an orthotopic model of 4T1 TNBC in BALB/c mice. As expected, USP22 ablation markedly impeded the growth of 4T1 breast cancer and markedly mitigated the tumor-promoting effects of EPI, as demonstrated by reductions in tumor volume, weights, and luminol fluorescence (Fig. 2, N to R). Moreover, USP22 ablation completely prevented lung metastasis, even in mice with tumors treated with EPI, because the lung luminol fluorescence activity and tumor nodules were both markedly decreased by USP22 deletion (Fig. 2, S to U). Similarly, intravenous injection of 4T1 cells showed that suppressing USP22 entirely nullified the EPI-induced enhancement in TNBC metastasis (Fig. 2, V to Y). Correspondingly, in vitro EPI treatment resulted in a statistical significance but modest increase in 4T1 cell growth (fig. S3A), while the migration of 4T1 cells was increased for more than threefold by EPI treatment (fig. S3, B and C). However, targeted deletion of USP22 completely abolished EPI-induced 4T1 cell growth and migration (fig. S3). These results reinforce our initial conclusion that USP22 plays a critical role in EPI-mediated breast cancer progression and metastasis.
USP22 is critical for EPI-induced lipolysis in breast cancer cells through up-regulating ATGL transcription
It has been established that EPI stimulates the expression of ATGL in adipocytes, promoting lipolysis, a process involved in cancer progression and metastasis (20, 30–33). Coupled with our discovery that targeted USP22 inhibition completely negated EPI-induced breast cancer progression and metastasis, we hypothesized that USP22 may play a role in regulating EPI-induced lipolysis in breast cancer. When comparing para-tumoral normal tissues to breast cancer tissues, there were lower levels of TAG but with notably higher levels of DAG and FFAs in the tumor tissues (fig. S4, A to C). This pattern aligns with the crucial roles of EPI in lipolysis as breast cancer tissues from patients in the EPIhigh group show reduced TAG levels accompanied by increased DAG and FFAs when compared with those from patients in the EPIlow group (fig. S4, D to F). IHC staining revealed that the expression levels of ATGL protein were elevated in human breast cancers compared to para-tumor control tissues (Fig. 3, A and B). ATGL protein expression was also higher in the breast cancer tissues from patients in the EPIhigh group comparing to that in the EPIlow group (Fig. 3, C and D), and it positively correlated with both serum EPI concentration and USP22 expression (Fig. 3, E and F). These findings suggest that EPI enhances lipolysis in patients with breast cancer, potentially through the up-regulation of ATGL expression.
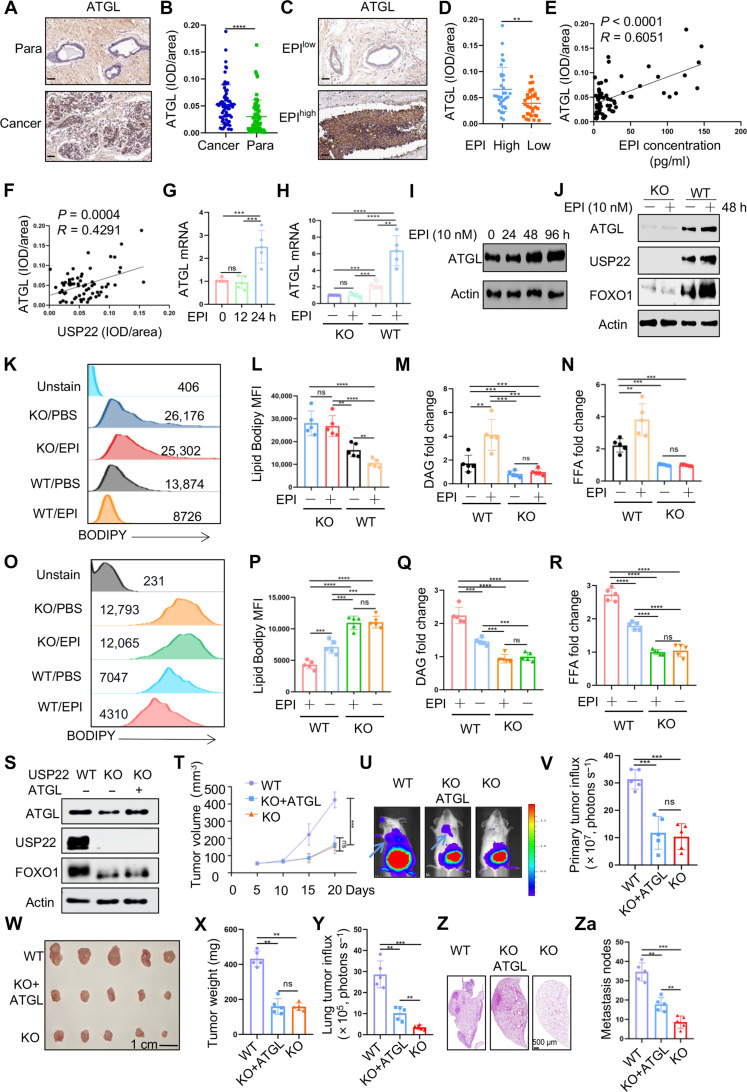
Fig. 3. USP22 promotes ATGL transcription and lipolysis in breast cancer cells.
(A to F) ATGL expression in breast cancer tissues by IHC staining. Representative images from para-tumor normal tissue and cancer sections (A) and quantification from 65 patients (B) are shown. Scale bars, 50 μm. The data were further analyzed by EPI levels [(C) and (D)]. The correlation of ATGL with EPI (E) and USP22 (F) was analyzed. (G to J) The mRNA [(G) and (H)] and protein [(I) and (J)] levels of ATGL in MDA-MB-231 cells treated with EPI (10 nM) were determined by real-time PCR and Western blotting, respectively. (K to M) Intracellular lipid analysis, including BODIPY [(K) and (L)], DAG (M), and FFAs (N) in WT and USP22-null MDA-MB-231 cells treated with EPI in vitro. (O to R) Intracellular lipid analysis in WT and USP22-null MDA-MB-231 cells isolated from the xenograft tumors as shown in Fig. 2E. (S) The protein levels of USP22, FOXO1, and ATGL in the WT 4T1 cells, USP22 KO 4T1 cells, and the USP22 KO 4T1 cells transduced with ATGL were measured. (T to Za) A total of 1 × 105 WT, USP22 KO 4T1 cells, or USP22 KO 4T1 cells transduced with ATGL were injected into the mammary fat pad of BALB/c mice. Tumor volume (T), bioluminescence activity [(U) and (V)], and tumor weight [(W) and (X)] were measured. Lung metastases were measured by luminol fluorescence [(U) and (Y)] and H&E staining of their lung sections [(Z) and (Za)]. N = 5 each group. Pearson correlation coefficient was used as a measure of association. Statistical significance was determined by one-way ANOVA test or unpaired Student’s t test. Data are expressed as means ± SD of five independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no statistical significance.
Both the mRNA and protein expression levels of ATGL were substantially up-regulated in MDA-MB-231 cells after treatment with EPI, indicating that EPI induces ATGL expression at the transcriptional level (Fig. 3, G and I). USP22 ablation led to a substantial reduction in ATGL expression, even in the presence of EPI treatment (Fig. 3, H and J). Consequently, USP22-targeted deletion resulted in a marked increase in lipid accumulation in breast cancer cells, regardless of EPI treatment (Fig. 3, K and L). Further analysis of intracellular DAG and FFA levels confirmed that USP22 inhibition largely diminished the EPI-induced DAG and FFAs increases in the in vitro cultured MDA-MB-231 cancer cells (Fig. 3, M and N). Similarly, analysis of the cancer cells isolated from MDA-MB-231 xenograft tumors from EPI-treated mice as shown in Fig. 2B demonstrated that USP22 suppression markedly reduced EPI-induced lipolysis (Fig. 3, O and P) and diminished EPI-induced DAG and FFA accumulation (Fig. 3, Q and R). A similar result was further confirmed in mouse 4T1 TNBCs (fig. S5). In addition, treating 4T1 TNBC cells with an ATGL-specific inhibitor, atglistatin, completely inhibited EPI-induced lipolysis in USP22 WT 4T1 cells but had no additional inhibitory effect on the lipolysis of USP22 KO cells (fig. S6), clearly indicating that ATGL is required for EPI-induced lipolysis in breast cancer cells. Unexpectedly, reconstitution of ATGL expression markedly, albeit not entirely, restored the migration but was ineffective in restoring the proliferation of USP22-null 4T1 cells (fig. S7). Collectively, our data demonstrate that USP22 plays a critical role in EPI-induced lipolysis by promoting Atgl gene transcription for TNBC migration.
An interesting question arises: Can stable ectopic expression of ATGL reverse the progression and metastasis of breast cancer? Unexpectedly, while ATGL expression did not restore the growth of 4T1 TNBC at the primary orthotopic sites, which is consistent with our in vitro analysis that ATGL failed to reconstitute USP22-null cell growth (fig. S7), it did partially mitigate their lung metastasis (Fig. 3, S to Za). Likewise, upon intravenous injection of both WT and USP22 KO 4T1 breast cancer cells, the quantity of lung tumor nodules markedly decreased by USP22 deletion, which was partially reinstated by stable ATGL expression (fig. S8). These studies suggest that USP22-mediated ATGL expression plays a key role in breast cancer metastasis in both MDA-MB-231 xenograft and 4T1 syngeneic tumor models.
USP22 promotes EPI-induced lipolysis through stabilizing the ATGL transcription factor FOXO1
Because USP22 promotes EPI-induced lipolysis through up-regulating Atgl mRNA transcription, together with the fact that EPI induces lipolysis of the transcription factor FOXO1-mediated ATGL expression (33, 34), we then investigated if USP22 regulates the transcription factor FOXO1 expression. When treated with EPI, FOXO1 protein levels increased without a corresponding increase in its mRNA expression (Fig. 4, A, B, and E), suggesting that EPI influences FOXO1 expression at a posttranscriptional level. Further support for this hypothesis, we detected that FOXO1 protein accumulated to similar levels in MDA-MB-231 cells treated with the proteasome inhibitor MG132, regardless of EPI stimulation (Fig. 4, C and D). Conversely, USP22 gene deletion resulted in a marked decrease in FOXO1 protein levels without affecting its mRNA expression and largely prevented EPI-induced FOXO1 up-regulation (Fig. 4, A, B, and E). This suggests that USP22 plays a role in promoting FOXO1 protein expression at the posttranslational level in breast cancer cells. Further IHC analysis confirmed that EPI treatment markedly increased the protein expression of USP22, FOXO1, and ATGL in xenograft MDA-MB-231 breast cancer tissues, while USP22 inhibition largely suppressed EPI-induced up-regulation of both FOXO1 and ATGL (Fig. 4, F to I). Similarly, EPI-induced USP22, FOXO1, and ATGL protein expression was also observed in 4T1 syngeneic breast cancer tissues by IHC staining. Targeted USP22 ablation completely suppressed EPI-induced FOXO1 and ATGL protein expression (fig. S9). In addition to mouse and human TNBC cells, we further demonstrated that treatment with EPI in vitro notably increased the protein expression levels of USP22, FOXO1, and ATGL in both ER+HR+HER2− MCF-7 and HER2+ SK-BR-3 cells (fig. S10, A and B). Consistent with our findings showing that targeted deletion of USP22 inhibited lipolysis in TNBC cells, pharmacological inhibition of USP22 completely prevented EPI-induced lipolysis in both MCF-7 and SK-BR-3 breast cancer cells (fig. S10, C to J). These findings indicate that USP22 plays a crucial role in EPI-induced lipolysis across a wide range of human breast cancers, irrespective of their HER expression status.
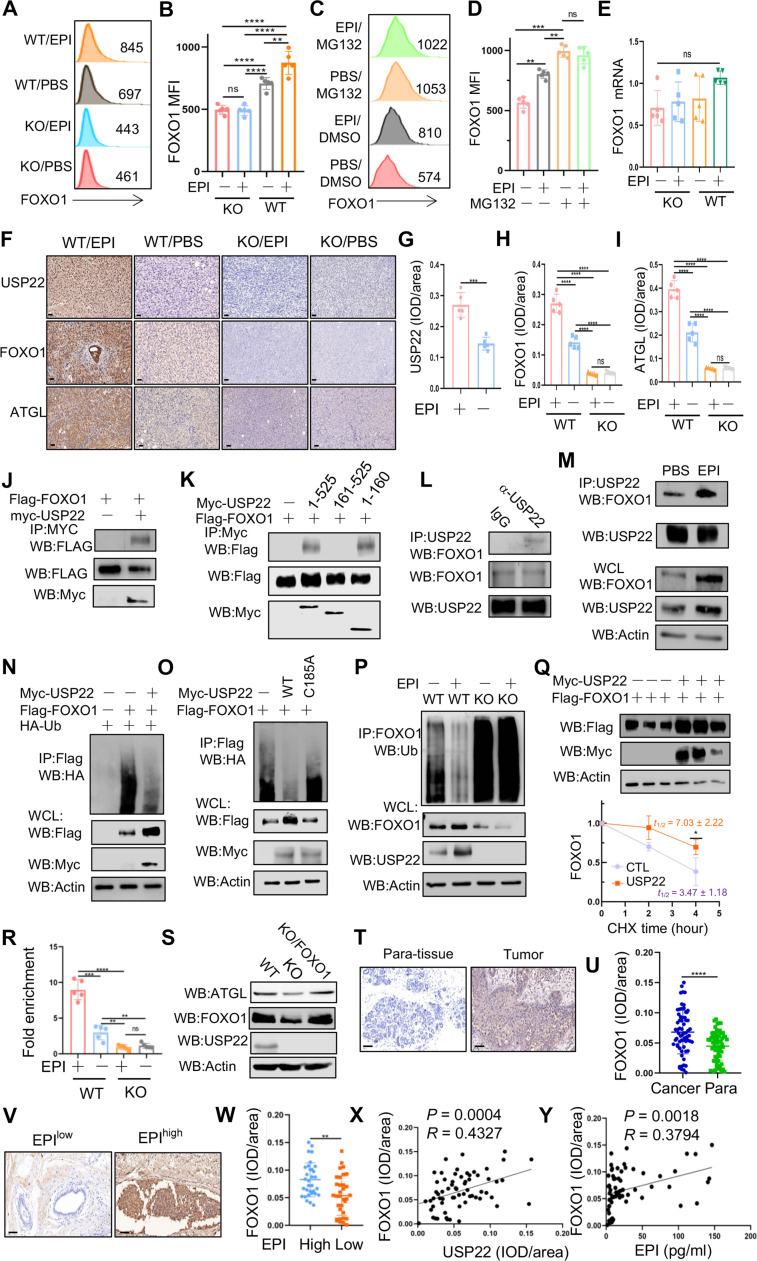
Fig. 4. USP22 is a deubiquitinase for Atgl transcription factor FOXO1.
(A to E) Effect of EPI on FOXO1 protein expression in WT and USP22-null MDA-MB-231 cells without [(A) and (B)] or with MG132 treatment [(C) and (D)]. The mRNA levels of FOXO1 were analyzed by real-time RT-PCR (E). (F to I) Analysis of USP22, FOXO1, and ATGL expression levels by IHC in syngeneic tumor tissues as shown in Fig. 2D. Representative images (F) (scale bars, 50 μm) and quantification data [(G) to (I)] are shown. (J to M) FOXO1 interaction in HEK239T cells transiently transfected with FOXO1 and USP22 (J) or USP22 truncated mutants (K) expressing plasmids and in MDA-MB-231 cells (L) or further with EPI treatment (M). WB, Western blotting. WCL, whole cell lysate. (N to P) Analysis of FOXO1 ubiquitination in HEK293T cells coexpressed with USP22 (N) and its mutants (O) and in USP22 WT and KO MDA-MB-231 cells (P). (Q) Immunoblot analysis of FOXO1 protein degradation in HEK293T cells transfected with or without USP22 (n = 3). (R) Analysis of FOXO1 binding to Atgl promoter by ChIP in WT and USP22 KO MDA-MB-231 cells (n = 5). (S) Reconstitution of FOXO1 expression in USP22-null MDA-MB-231 cells restored ATGL expression. (T to Y) Analysis of FOXO1 expression in human breast cancer tissues [(T) and (U) with para-tumor tissues as controls or (V) and (W) in EPIhigh and EPIlow groups] as well as its correlation with USP22 (X) and EPI (Y). Scale bars, 50 μm. Pearson correlation coefficient was used as a measure of association. Statistical significance was determined by one-way ANOVA test or unpaired Student’s t test. Data are expressed as means ± SD of three or more independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, no statistical significance.
To define USP22 functions as a possible FOXO1-specific deubiquitinating enzyme, we first determined if USP22 interacts with FOXO1. Western blotting analysis detected the Flag-tagged FOXO1 in anti-Myc immunoprecipitants of human embryonic kidney (HEK) 293T cells when myc-USP22 but not empty vector was cotransfected (Fig. 4J). The endogenous USP22 interaction with FOXO1 was further confirmed in MDA-MB-231 cells (Fig. 4L). Further analysis of FOXO1 interactions with USP22 truncated mutants revealed that the zinc finger–containing N terminus of USP22 mediates its interaction with FOXO1 (Fig. 4K). Stimulation with EPI markedly increased USP22 interaction with FOXO1 in breast cancer cells (Fig. 4M). These results indicate that USP22 is an interacting partner of FOXO1. As a consequence, coexpression of USP22 largely inhibited the polyubiquitin of FOXO1 (Fig. 4N). In contrast, the deubiquitinase catalytic-inactive USP22/C185A mutant failed to suppress FOXO1 ubiquitination (Fig. 4O). EPI stimulation, which induced USP22 protein expression and promoted USP22-FOXO1 interaction, markedly inhibited FOXO1 polyubiquitination in USP22 WT cancer cells, and targeted USP22 deletion resulted in a significant increase in FOXO1 ubiquitination and reduced FOXO1 protein expression even with EPI stimulation (Fig. 4P). These results indicate that USP22 is an EPI-induced FOXO1-specific deubiquitinase in breast cancer cells. Therefore, overexpression of USP22 protected FOXO1 protein from ubiquitination-mediated degradation and prolonged FOXO1 half-life (Fig. 4Q).
FOXO1 has been shown to regulate lipolysis through ATGL expression (34). Consistently, our chromatin immunoprecipitation (ChIP) analysis detected the binding of FOXO1 to ATGL promoter in breast cancer cells, and the treatment of cells with EPI markedly increased FOXO1 binding to ATGL promoter. CRISPR deletion of USP22 markedly reduced FOXO1 binding to ATGL promoter (Fig. 4R), indicating that USP22 facilitates breast cancer lipolysis through stabilizing FOXO1 to enhance Atgl gene transcription. To support this conclusion, we further detected a substantial reduction in ATGL protein expression by USP22-targeted deletion, and expression of FOXO1 fully rescued ATGL expression in USP22-null breast cancer cells (Fig. 4S). Therefore, USP22 achieves its oncogenic function through stabilizing FOXO1 protein to promote ATGL transcription. Similar to both USP22 and ATGL, IHC staining detected a statistically increased FOXO1 expression in cancer cells comparing to that in adjacent normal control tissues (Fig. 4, T and U), and FOXO1 protein was higher in the breast cancers from the EPIhigh group when compared to that in the EPIlow group (Fig. 4, V and W). The FOXO1 protein expression is positively associated with both USP22 protein expression and serum EPI concentration in human breast cancers (Fig. 4, X and Y). Therefore, our data suggest that EPI promotes USP22-mediated FOXO1 protein stabilization for ATGL up-regulation in breast cancer cells.
EPI stabilizes USP22 through AKT-mediated phosphorylation in breast cancer cells
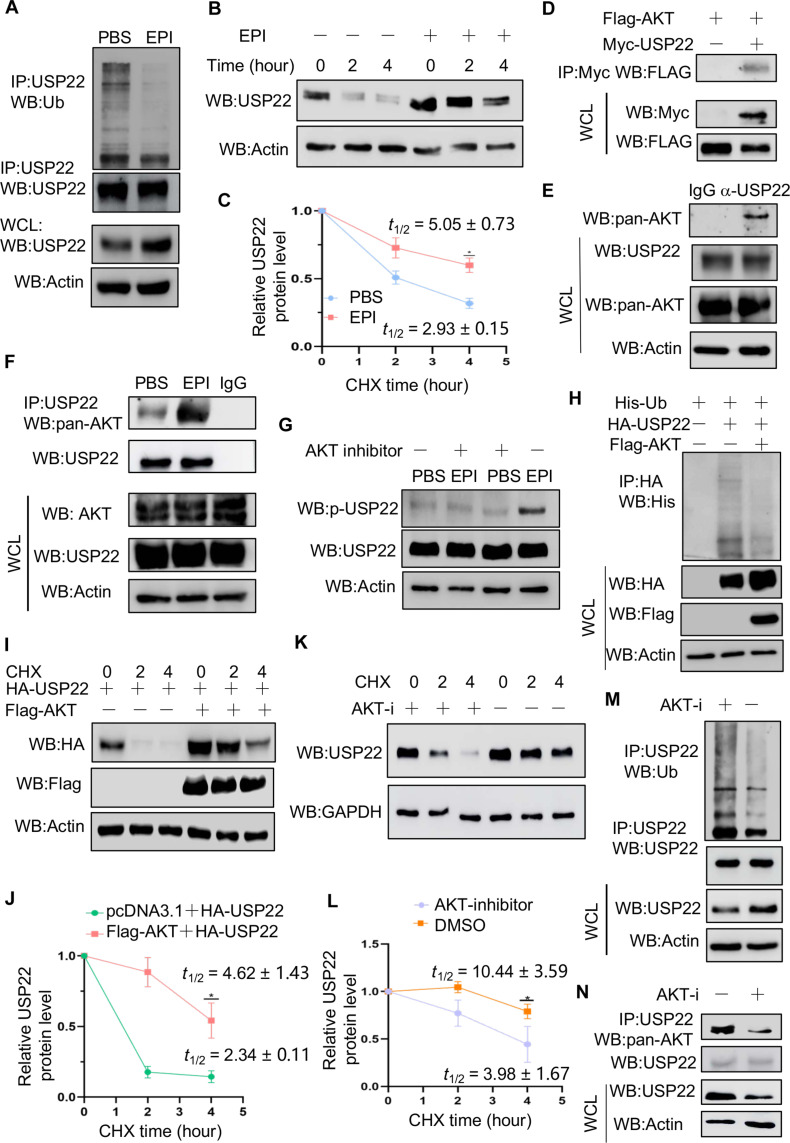
Because EPI induces USP22 protein expression without affecting its mRNA levels, we tested if EPI-mediated signaling controls USP22 protein expression at posttranslational levels. Stimulation with EPI markedly reduced USP22 ubiquitination levels (Fig. 5A), supporting our speculation that EPI protects USP22 from ubiquitination-mediated degradation. As expected, the half-life of USP22 was prolonged over 4 hours by EPI treatment (Fig. 5, B and C). It has been shown that EPI signaling is involved in protein stabilization through AKT-mediated phosphorylation (35–37). We then determined the potential cross-talk of AKT with USP22 in breast cancer cells. The interaction of AKT with USP22 was detected in transiently transfected HEK293T cells as well as in human MDA-MB-231 breast cancer cells (Fig. 5, D and E). Stimulation of MDA-MB-231 breast cancer cells with EPI markedly enhanced both AKT and USP22 interaction (Fig. 5F) as well as their phosphorylation (fig. S11, A and B). This p-USP22-T147 antibody is highly specific because it detected a band in USP22 KO cells when expressed with WT but not USP22/T147A mutant (fig. S11C). Further treatment with the AKT-specific inhibitor largely diminished EPI-induced USP22 phosphorylation (Fig. 5G), indicating that AKT is a kinase for USP22 phosphorylation in breast cancer cells upon EPI stimulation. As a consequence, overexpression of AKT markedly inhibited USP22 ubiquitination and prolonged its half-life (Fig. 5, H to J). Conversely, treatment with the AKT-specific inhibitor facilitated USP22 protein degradation (Fig. 5, K and L), increased the USP22 ubiquitination (Fig. 5M), and inhibited the AKT and USP22 interaction in breast cancer cells (Fig. 5N). These results indicate that EPI stabilizes USP22 through AKT-mediated USP22 phosphorylation.
Fig. 5. EPI promotes the phosphorylation of USP22 by AKT to enhance the stability of USP22.
(A to C) The effect of EPI on USP22 ubiquitination (A) and protein stability [(B) and (C)] in MDA-MB-231 cells were determined (n = 3). (D and E) Co-IP and immunoblot analysis of USP22 interaction with AKT in transiently transfected HEK293T cells (D) and MDA-MB-231 cells (E). (F and G) Effect of EPI on USP22 interaction with AKT and USP22 phosphorylation in MDA-MB-231 cells. (H to J) Effect of AKT overexpression on USP22 ubiquitination (H) and protein stability [(I) and (J)] (n = 3). (K to N) Effect of AKT inhibition on USP22 protein degradation [(K) and (L)] (n = 3), ubiquitination (M), and interaction with AKT (N) in MDA-MB-231 cells. AKT-i, AKT inhibitor. Statistical significance was determined by unpaired Student’s t test. Data are expressed as means ± SD of three independent experiments. *P < 0.05.
AKT-mediated phosphorylation links USP22 protein stability with its oncogenicity in breast cancer
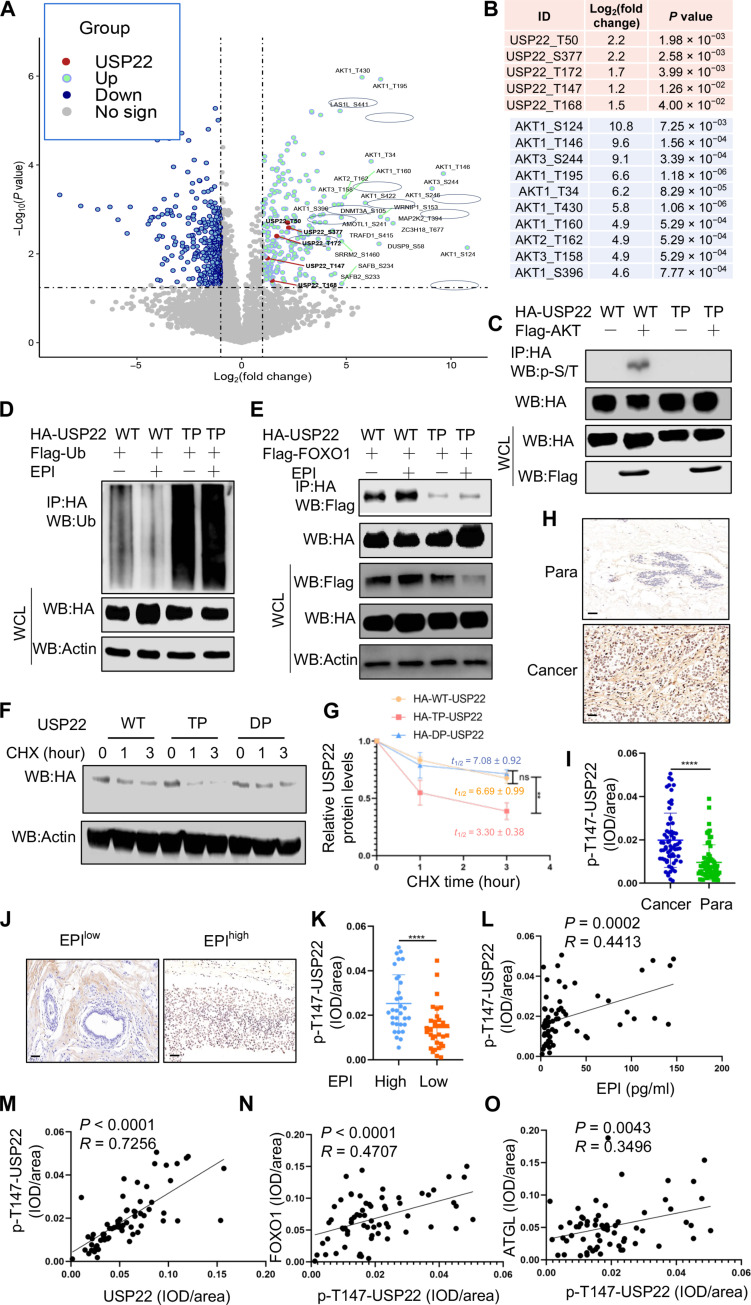
We then used an unbiased proteomic approach to identify the potential phosphorylation sites of USP22 by AKT. As shown in Fig. 6 (A and B), USP22 peptides carrying each of the five phosphorylated amino acid residues including T50, T147, T168, T173, and S355 were markedly enriched in cells when AKT was overexpressed (Fig. 6, A and B), indicating that AKT phosphorylates USP22 through multiple sites. Of note, mass spectrometry (MS) analysis also detected 10 AKT phosphorylated peptides in the anti-USP22 immunoprecipitants (Fig. 6, A and B), confirming our initial discovery of AKT-USP22 interaction and suggesting that USP22 interacts with the phosphorylated AKT upon EPI stimulation in breast cancer cells. We then generated a USP22 mutant by replacing all five identified S/T residues with alanine (A) (USP22-TP) and confirmed that the mutation totally abolished AKT-mediated USP22 phosphorylation (Fig. 6C). Loss of AKT-mediated phosphorylation markedly increased USP22 ubiquitination and abolished EPI-induced inhibitory efficacy (Fig. 6D). Therefore, mutation of the phosphorylation sites in USP22 promoted its degradation. Conversely, the phosphorylation mimicking mutation to aspartic acid at all five residues (USP22-DP) resulted in a marked improvement in USP22 stabilization (Fig. 6, F and G). These results indicate that AKT-mediated phosphorylation is critical for USP22 stabilization during EPI-induced tumorigenesis. To further support this conclusion, expression of WT USP22, but not its TP mutant, resulted in the increased FOXO1 and ATGL expression (fig. S12, A to C). As previously reported (38), the AKT inhibitor markedly inhibited tumor growth, even in mice treated with EPI, suggesting that EPI-induced tumor growth largely occurs through an AKT-dependent mechanism (fig. S13, A to C). Consistent with our in vitro findings (Fig. 5G), EPI treatment augmented USP22 phosphorylation and protein expression in the tumor tissues (fig. S13, D to F). Treatment of mice with the AKT inhibitor completely abolished EPI-induced USP22 phosphorylation in 4T1 tumors (fig. S13, D and F). These results indicate that EPI induces USP22 phosphorylation both in vitro and in tumor tissues in an AKT-dependent manner.
Fig. 6. AKT-mediated USP22 phosphorylation controls its protein stability.
(A and B) Proteomic analysis of USP22 phosphorylation from HEK293T cells transfected with or without Flag-AKT. The spectra of peptides carrying phosphorylated peptides (A) and the identified phosphorylation residues of USP22 (top) and AKT (bottom) (B) are shown. (C and D) Analysis of phosphorylation (C) and ubiquitination (D) with USP22 and its phosphorylation mutant. (E) Analysis of FOXO1 interaction with USP22 and its phosphorylation mutant. (F and G) Analysis of protein stability of USP22 and its phosphorylation mutant (n = 3). (H to O) Analysis of USP22 phosphorylation in human breast cancer by IHC. Representative images in tumor and para-controls (H) or in EPIhigh and EPIlow groups (J) as well as the quantification from 65 patients [(I) and (K)] are shown. Scale bars, 50 μm. Statistical analyses of the correlation p-USP22-Thr147 levels with EPI (L), total USP22 (M), FOXO1 (N), and ATGL (O) are shown. Pearson correlation coefficient was used as a measure of association. Statistical significance was determined by unpaired Student’s t test. Data are expressed as means ± SD of three or more independent experiments. **P < 0.01; ****P < 0.0001; ns, no statistical significance.
We then analyzed the phospho-USP22 (p-USP22) levels by IHC staining using specific antibodies to one of the USP22 phosphorylation sites, T147, that is catalyzed by AKT (Fig. 6, A and B). The levels of p-USP22 were elevated, as expected, in breast cancers comparing to that in adjacent normal controls (Fig. 6, H and I). Consistent with our conclusion that EPI induces USP22 phosphorylation through AKT activation, we observed a statistically substantial increase in p-USP22 levels in breast cancers from EPIhigh versus EPIlow groups (Fig. 6, J and K). Further analysis revealed a strong positive correlation between p-USP22 with serum EPI levels in patients with cancer (Fig. 6L) as well as with both FOXO1 and ATGL levels (Fig. 6, M to O). Unexpectedly, neither the expression levels of USP22, FOXO1, and ATGL nor USP22 phosphorylation show any statistically substantial increase in para-tumoral normal tissues (fig. S14).
Moreover, unbiased analysis of a dataset (GSM5740291) from a study on the treatment of a human medulloblastoma cell line, ONS-76 cells with the β-blocker propranolol, revealed a marked reduction (over 50%) in Atgl mRNA levels, while USP22 and FoxO1 mRNA levels remained unchanged (fig. S15). This aligns with our study, which suggests that EPI up-regulates USP22 and FoxO1 at posttranslational levels to stimulate Atgl mRNA transcription. Collectively, our study revealed a previously unappreciated molecular mechanism underlying how USP22 controls EPI-induced cancer progression and metastasis through potentiating FOXO1-medited ATGL expression and lipolysis.
Pharmacological USP22 inhibition synergizes with β-blockers in breast cancer treatment
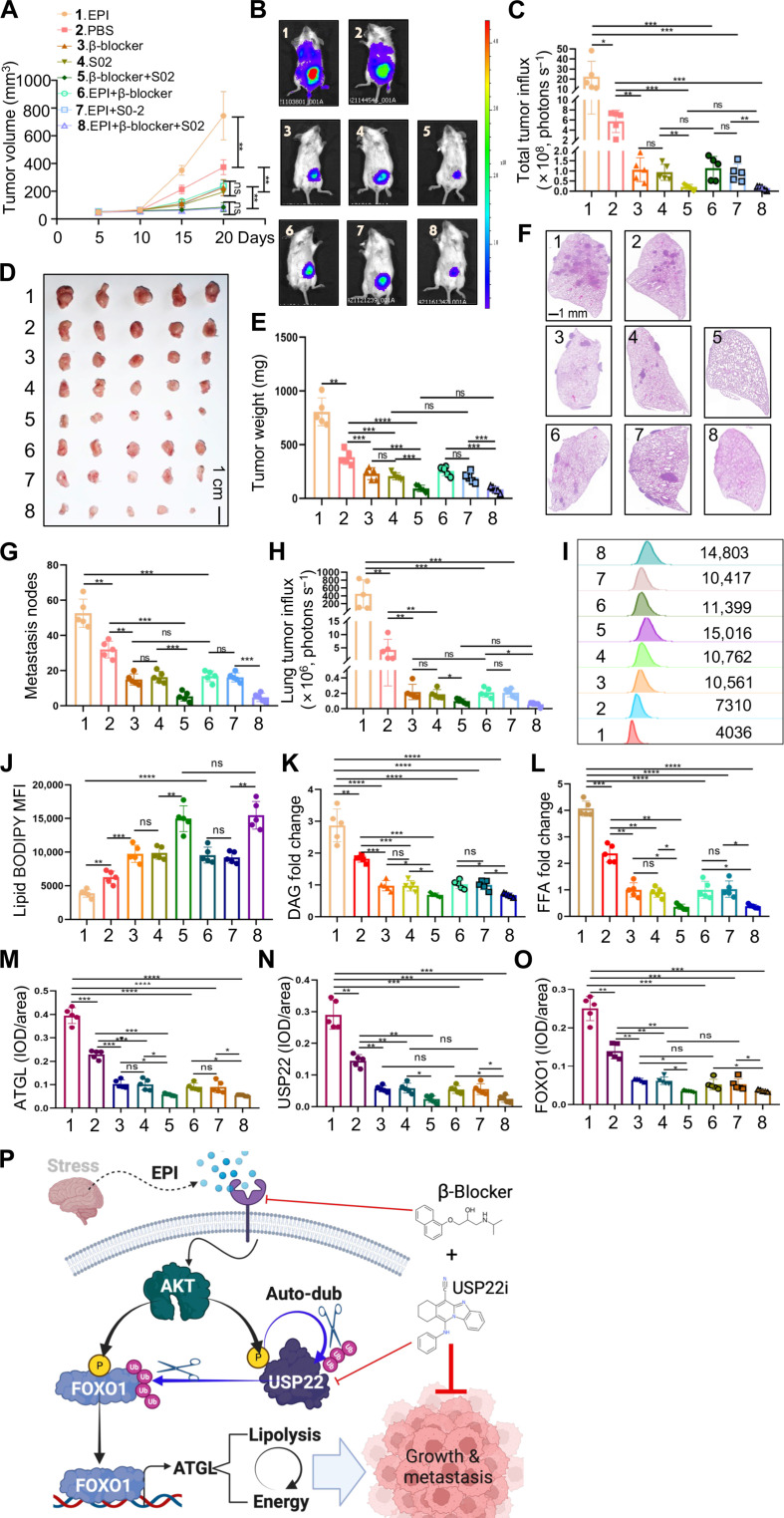
Our discovery that EPI promotes breast cancer growth and metastasis through an AKT-USP22-FOXO1-ATGL lipolysis axis provides a rationale for a combined synergistic treatment by USP22 inhibition with β-blockers. We then examined the effect of combination treatment with USP22 inhibitor (USP22i-S02) and β receptor inhibitor propranolol on tumor growth and metastasis using an orthotopic breast cancer model. Daily monitoring the tumor volume detected that USP22i-S02 treatment inhibited the tumor growth to a similar efficacy as that by β-blocker treatment, and the combined treatment leads to a near-complete resection of the tumor growth (Fig. 7A). Consistently, either USP22i-S02 or β-blocker treatment resulted in a significant reduction in the tumoral luminol fluorescence activity at 20 days after treatment, and the combined treatment largely diminished the luminol fluorescence activity, even in mice with EPI administration (Fig. 7, B and C), which was confirmed by tumor weight at 20 days after treatment (Fig. 7, D and E).
Fig. 7. Synergistic effects of USP22 and β receptor inhibitors on tumors.
(A to E) 4T1 TNBC cells were injected into the mammary fat pad of BALB/c mice (N = 5 each group). Tumor volume (A), bioluminescence activity [(B) and (C)], and tumor weight [(D) and (E)] were measured. (F to H) The metastases of the lung were measured by luminol fluorescence [(B) and (H)] and H&E staining [(F) and (G)]. Lung tissue sections were analyzed by H&E staining [(F) and (G)]. Representative lung metastasis (F) and quantifications of five mice each group (G) are shown. (I to L) Lipid analysis of syngeneic tumors from (D) by lipid BODIPY [(I) and (J)], DAG (K), and FFAs (L). MFI, mean fluorescence intensity. (M to O) IHC analysis of USP22, FOXO1, and ATGL expression levels in syngeneic tumor tissue sections from (D). Quantification data of all five mice each group [(M) to (O)] are shown. (P) Model of targeting chronic stress–mediated cancer metastasis and lipolysis by USP22. Statistical significance was determined by one-way ANOVA test or unpaired Student’s t test. Data are expressed as means ± SD of five independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, no statistical significance.
Treatment of mice with USP22i-S02 and β-blocker in treating 4T1 largely diminished the breast cancer lung metastasis as analyzed through histological analysis of metastatic nodules in lung tissue sections (Fig. 7, F and G) and luminol fluorescence activity (Fig. 7H). These results clearly indicate that targeting USP22 synergizes with β-blocker, which target the two steps in the EPI/AKT/USP22/FOXO1/ATGL lipolysis pathway in the treatment of the aggressive breast cancer. To support this, our flow cytometry analysis detected a marked reduction in lipid BODIPY, as well as DAG and FFAs, in breast cancer cells from syngeneic tumors treated with USP22i-S02 and β-blocker (Fig. 7, I to L). We have previously reported that USP22i-S02 inhibitor treatment resulted in a reduction of USP22 proteins in cancer cells (17). IHC staining detected that the protein expression levels of USP22, and its downstream FOXO1 and ATGL, were decreased by both USP22i-S02 and β-blocker treatment in syngeneic tumor tissues (Fig. 7, M to O, and fig. S16). Collectively, our study revealed a previously unknown tumorigenic metabolic pathway, EPI/AKT/USP22/FOXO1/ATGL lipolysis in breast cancer progression and metastasis, and USP22 inhibition synergizes with β-blocker to inhibit breast cancer growth and lung metastasis (Fig. 7P).
DISCUSSION
This study has defined a USP22-mediated lipolysis circuit that links the chronic stress–induced EPI production to breast cancer pathogenesis through the AKT-USP22-FOXO1-ATGL pathway. This conclusion is documented by the following observations: First, the increase in the death-from-cancer signature gene USP22 expression is positively associated with the circulating blood EPI levels in patients with breast cancer, and EPI promotes TNBC progression and metastasis in a USP22-dependent manner; second, EPI stabilizes USP22 protein through AKT-mediated phosphorylation, which consequently inhibits ubiquitination-induced USP22 degradation; third, USP22 functions as a bona fide deubiquitinase of the transcription factor FOXO1 to promote the expression of ATGL, an enzyme critical for cancer cell lipolysis; fourth, both genetic and pharmacological USP22 inhibition totally abolished EPI-induced lipolysis in breast cancer cells; and last, USP22 suppression synergizes with EPI blocking in the treatment of breast cancer metastasis.
It is well established that stress-induced activation of sympathetic nervous system in patients with cancer, especially adrenergic signaling, operates the important role in the cancer cell proliferation and metastasis (4, 6, 39–42). Chronic stress–induced EPI promoted breast cancer stem–like properties via promoting lactate dehydrogenase A expression and allowing breast cancer cells using lactate (29). In liver cancer, EPI has been shown to promote hypoxia-inducible factor 1α–dependent glucose metabolism through suppressing autophagy activity (35). In addition to lactate and glucose metabolism, we show here that EPI promotes breast cancer metastasis through a USP22-mediated lipolysis circuit. Molecularly, USP22 functions as an endogenous deubiquitinase of FOXO1, a transcription factor known for ATGL expression leading to the enhancement of lipolysis (33). It has been reported that Gα12 ablation exacerbates liver steatosis and obesity by suppressing USP22/SIRT1(Sirtuin 1)-regulated mitochondrial respiration (43). USP22 was also found to regulate lipogenesis through PPARγ (peroxisome proliferator–activated receptor γ) in hepatocellular carcinoma tumorigenesis (44). In addition, mice lacking Usp22 in all hematopoietic cells display profound systemic emergency hematopoiesis (45), and USP22 supports the aggressive behavior of basal-like breast cancer by stimulating cellular respiration (46). Therefore, chronic stress–induced EPI executes its tumor-promoting functions through metabolic reprogramming. It will be interesting to analyze the association between psychological stress levels and EPI concentrations in the patients as well as their correlation with lipid metabolism. Future studies integrated with a psychologist team are needed to address this critical question.
Our study revealed that USP22-mediated lipolysis circuit links the chronic stress–induced EPI production to breast cancer pathogenesis through the AKT-USP22-FOXO1-ATGL pathway across a wide range of human breast cancers, irrespective of their HER expression status. In addition, because the stress-induced EPI promotes the development and progression in a variety of human cancer types (47), the EPI-AKT-USP22-FOXO1-ATGL pathway may be a common molecular mechanism in tumorigenesis.
We acknowledge that, while reintroducing ATGL expression into USP22-null breast cancer cells fully restored their lung metastasis, it only partially rescued the growth of USP22-null tumors at the primary site. To support this, we further found that ATGL expression partially rescued USP22-null 4T1 cell transwell migration but not proliferation in vitro. This suggests that USP22 may have an ATGL-independent role in EPI-induced tumorigenesis. USP22 has been known to achieve its oncogenic functions through inhibiting cell apoptosis and promoting cell cycle progression through targeting cyclins, c-Myc, and SIRT1, which controls p53 expression (12, 14, 15, 48–50).
Our study identifies FOXO1, an evolutionarily conserved transcription factor of longevity and stress response (51, 52), as a bona fide substrate of USP22 in breast cancer cells. USP22 stabilization of FOXO1 is regulated by EPI-induced activation of AKT, which phosphorylates FOXO1, such as at the Ser256, Thr24, and Ser319 residues (53), to provide the docking sites for USP22 binding. Consequently, the elevated FOXO1 transcribe ATGL for promoting the lipolysis during breast cancer cell proliferation and metastasis. In addition to ATGL-mediated lipolysis, FOXO1 has been shown to promote the expression of SOX2 to facilitate the stem cells of breast cancer (54). Also, FOXO1 is shown to enhance transcription of the matrix metallopeptidase 1, leading to the enhanced breast cancer cell metastasis possibly through easing cell migration simply by breaking down matrix barriers (55). Therefore, USP22-mediated stabilization of FOXO1 may promote breast cancer metastasis possibly through multiple cellular and molecular mechanisms. In addition to USP22, many other downstream factors, such as breast cancer resistance protein of AKT, are involved in drug resistance of breast cancers (56); therefore, future works are needed to dissect each substrate of phosphatidylinositol 3-kinase–AKT pathways in breast cancer tumorigenesis.
One unexpected observation is the EPI-induced posttranscriptional stabilization of USP22, the cancer stem cell gene with increased expression in breast cancer stem cells (18). While USP22 has been known to regulate the stability of a variety of proteins at both physiological and pathological contact (15, 18, 57), the molecular mechanisms underlying how USP22 regulates its own protein stability remain undefined. Our data suggest that ATK-mediated phosphorylation of USP22 is critical for its stabilization because mutation of the phosphorylation residues increased USP22 ubiquitination and facilitated its proteasomal degradation. In addition, the deubiquitinase-inactive mutation completely abolished AKT-mediated USP22 stabilization. Therefore, ATK-mediated phosphorylation upon EPI stimulation promotes USP22 autodeubiquitination for self-stabilization. Our study does not only defines a molecular pathway of lipolysis in breast cancer metastasis but also provides a rationale for the combined therapy of β-blocker, which has been clinically tested to be beneficial in chronic stress–mediated tumorigenesis treatment (58), and USP22 inhibitor in the treatment of breast cancer metastasis. Our data show a synergy between propranolol and USP22-specific inhibitor S02 in the treatment of the xenograft TNBCs.
MATERIALS AND METHODS
Cell lines and culture conditions
The human breast cancer cell line MDA-MB-231 and the mouse breast cancer cell line 4T1 were obtained from Haixing Biology. The HEK293T cell line was generously provided by F. Wang from Dalian Medical University, Dalian, China. The MCF-7 and SK-BR-3 cell lines were kindly provided by W. Guo from Dalian Medical University, Dalian, China. MDA-MB-231, HEK293T, MCF-7, and SK-BR-3 cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS). The 4T1 cell line was maintained in RPMI 1640 medium also supplemented with 10% FBS. All cell lines were incubated at 37°C in a humidified incubator with 5% CO2.
Animal studies
Six- to 8-week-old female NYG and BALB/c mice were purchased from the Animal Center of Dalian Medical University (Dalian, China) for use in the study. In the orthotopic breast cancer model, MDA-MB-231-luc cells (1 × 106) suspended in a 1:1 mixture of PBS/Matrigel (Biocoat, 354234) were injected into the right mammary fat pad of NYG mice. In addition, 4T1-luc cells (1 × 105) suspended in PBS/Matrigel (1:1) were injected into the mammary fat pad of BALB/c mice. For the experimental metastasis model, MDA-MB-231-luc cells (5 × 105) suspended in 150 μl of PBS were injected into 4- to 6-week-old female NYG mice via the tail vein. Similarly, 4T1-luc cells (5 × 104) suspended in 150 μl of PBS were injected into female BALB/c mice via the tail vein. All cell lines were stably labeled with a luciferase-expressing vector. Monitoring of primary tumor growth and the occurrence of lung metastasis was conducted using bioluminescence imaging after intraperitoneal injection of d-luciferin sodium (150 mg/kg; MedChemExpress HY-12591). Bioluminescence signals were measured using an IVIS Spectrum (PerkinElmer). The mice were randomized into four groups and received intraperitoneal injections of either vehicle or EPI. Mice underwent a 7-day pretreatment of EPI acclimation to habituate them to the study conditions. The EPI group received EPI (2 mg/kg per day, subcutaneous) for 7 days before tumor cell injection to mimic the chronic stress conditions as previously reported (29).
After tumor implantation, primary tumor sizes in NYG mice were measured every 5 days, and lung metastasis was assessed at 25 days. In BALB/c mice, primary tumor sizes were also measured every 5 days, with lung metastasis assessed at 20 days. Following tail vein injection, lung metastasis in NYG mice was measured at 20 days, while in BALB/c mice, it was measured at 15 days. Tumor sizes were measured using calipers, estimating volumes using the formula (a2 × b)/2, where a is the shorter of the two dimensions and b is the longer one. At protocol-defined end points, mice were euthanized and tissues were collected and fixed in 4% paraformaldehyde (PFA). Paraffin-embedded lung samples were subjected to H&E staining. All animal studies were approved by the Dalian Medical University Institutional Animal Care and Use Committee (AEE22017).
Human sample
Patient tissue samples and serum were collected from The First Affiliated Hospital of Dalian Medical University (Dalian, China). The patients with breast cancer included in this study met the following criteria: (i) complete pathological data were available; (ii) tissue samples for immunohistochemical staining were provided; and (iii) no patient had received adjuvant chemotherapy, immunotherapy, or radiotherapy prior to surgery. Patients with metastatic disease at initial presentation, recurrent tumors at the study’s onset, or those who had undergone prior neoadjuvant treatment were excluded from the study. As part of standard clinical practice, estrogen or progesterone, and HER2 were routinely stained for all patients with breast cancer. Clinical and pathological data were obtained from medical charts and pathology reports. Tumor staging was based on the American Joint Committee on Cancer pathologic tumor-node-metastasis classification (eighth edition). Tumor size was measured in centimeters by a pathologist. The study received approval from the Ethical Review Committee of The First Affiliated Hospital of Dalian Medical University (certificate number PJ-KS-KY-2022-278). Informed consent was obtained from all participants included in the study. The study protocol was approved by the Ethical Review Committee of The First Affiliated Hospital of Dalian Medical University, and all procedures were performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Measurement of EPI concentration
Peripheral blood was collected from patients with clinical breast cancer using a syringe treated with EDTA to prevent clotting. The blood was centrifuged for 15 min at 3000g, and the serum layer was transferred to separate vials and stored at −80°C. In mouse studies, 12 hours after administration of EPI treatment on the fifth day, blood was drawn from the mice via the tail vein using an EDTA-treated syringe to prevent clotting. The blood samples were then centrifuged for 15 min at 3000g, and the serum layer was carefully transferred to separate vials and stored at −80°C. EPI levels were measured using an Adrenaline ELISA kit (Abcam, ab287788) following the manufacturer’s protocols.
Plasmids and other reagents
Hemagglutinin (HA)-ubiquitin, Flag-ubiquitin, HA-USP22, His-USP22, myc-USP22, myc-185a-USP22, HA-TP-USP22, HA-DP-USP22, Flag-AKT, Flag-FOXO1, Lenti-luciferase-P2A-Neo, and pLV3-CMV-Pnpla2(mouse)-Blast plasmids were purchased from Miaoling Biology. Plasmid pcDNA.3.1 was purchased from GenePharma Biology, HA-147a-USP22 plasmids were used as previously reported (57, 59), and USP22 mutations were generated by PCR.
Transfection, generating stable cell line, and cell treatment
Transfections were carried out using Lipofectamine 3000 (Invitrogen, L3000150) according to the manufacturer’s instructions. Cells were harvested 48 hours after transfection and subjected to various assays. CRISPR-Cas9–mediated ablation of the USP22 gene was achieved using CRISPR-Cas9 RNP provided by Haixing Bioscience, containing expression cassettes for hSpCas9 and chimeric guide RNA. To knock out the USP22 gene, two guide RNA sequences (listed in table S2) were selected via the http://crispr-era.stanford.edu website. The plasmid containing the guide RNA sequence was electrotransfected into cells using the Neon transfection system (Thermo Fisher Scientific) as per the manufacturer’s instructions. After 2 days, single colonies were transferred to 96-well plates. To detect insertions or deletions (indels) in USP22-targeted clones, genomic DNA was isolated using a Quick-DNA Miniprep kit (Zymo Research), and PCR amplification was performed using a 2× Taq Master Mix (Dye Plus; Vazyme, P112) with primers flanking the exon (primer sequences listed in table S3). Plasmids were isolated from 8 to 10 single colonies and sequenced by Sanger sequencing (GENEWIZ, China). Clones with mutations in both alleles were chosen for downstream studies.
To establish a luciferase-stable cell line, Lenti-luciferase-P2A-Neo plasmids were transfected into cells using Lipofectamine 3000 according to the manufacturer’s instructions. After 48 hours of transfection, cells were selected with G418 (500 μg/ml; MedChemExpress, HY-17561) for at least 2 days to generate stable cell lines. To overexpress ATGL in USP22 KO 4T1 cells, pLV3-CMV-Pnpla2(mouse)-Blast plasmids were transfected into USP22 KO 4T1 cells. After 48 hours, cells were selected with blasticidin (2 μg/ml; MedChemExpress, HY-103401) for at least 2 days to generate stable cell lines.
For cell degradation experiments, transfected HEK293T or MDA-MB-231 cells were treated with cycloheximide (CHX) (200 μg/ml; Cell Signaling Technology, 2112) for various times. Cells were treated with 10 nM EPI (Selleck, S2521) and 0.5 μM AKT inhibitor (MedChemExpress, HY-15431) for the indicated time.
Pharmacological studies
The concentrations of all drugs were based on effective activation or inhibition observed in prior studies: EPI (10 nM), AKT inhibitor (0.5 μM), CHX (200 μg/ml), atglistatin (Selleck, S7364, 50 μM), and USP22i-S02 (10 μM). The mice were pretreated with a dose of 2 mg/kg via subcutaneous injection once a day before tumor cell injection to mimic chronic stress conditions, as previously reported (29). Following tumor cell implantation, the mice received the same dose (2 mg/kg) via subcutaneous injection once a day for 25 days (NYG mice) or 20 days (BALB/c mice). USP22i-S02 was injected intraperitoneally at a dose of 10 mg/kg per day in 100 μl of corona oil, once daily, starting on the first day of tumor cell injection for a duration of 20 days. In pharmacological β-blocker experiments, propranolol was administered intraperitoneally at a dose of 2 mg/kg per day, once daily, starting from the initial day of tumor cell injection, and continued for 20 days. AKT inhibitor [100 mg/kg per day in 2.5% dimethyl sulfoxide (DMSO) and 25% β-cyclodextrin] was orally administered once daily, starting on the first day of tumor cell injection, and continued for 15 days.
Flow cytometry
To stain intracellular FOXO1, cells were initially treated with a viability dye (Invitrogen, L34955) at a 1:1000 dilution in fluorescence-activated cell sorting (FACS) Buffer (2% FBS in PBS) for 30 min at 4°C. After washing the cells three times in FACS Buffer, the cells were fixed, permeabilized, and stained for transcription factors using the Foxp3 Transcription Factor Staining Buffer Set (eBioscience, 00-5523-00), following the manufacturer’s instructions. The cells were then stained with specific antibodies against PE-FoxO1 (14262, Cell Signaling Technology) for 1 hour at 4°C. After washing the cells three times in FACS Buffer, they were analyzed by flow cytometry, and the data were evaluated using FlowJo software.
For lipid staining, cells were initially treated with a viability dye (Invitrogen, L34955) at a 1:1000 dilution in FACS Buffer (2% FBS in PBS) for 30 min at 4°C. After washing the cells three times in FACS Buffer, the cells were fixed with 4% formaldehyde for 15 min and stained with BODIPY 493/503 (Invitrogen, D3922; stock concentration, 1 mg/ml; working solution, 1:1000 dilution) for 15 min at room temperature. After washing the cells three times in FACS Buffer, they were analyzed by flow cytometry, and the data were evaluated using FlowJo software as we recently reported (60).
Chromatin immunoprecipitation
ChIP was performed in MDA-MB-231 cells using the Simple Chip Enzymatic Chromatin IP kit (9002, Cell Signaling Technology, United States), as previously reported (16). Cells were fixed with 1% formaldehyde for 10 min at room temperature, and the fixation was halted by adding glycine and incubating for an additional 5 min at room temperature. Afterward, the cells were scraped, pelleted, and lysed in 1 ml of Buffer A with a protease inhibitor cocktail and dithiothreitol (DTT) for 10 min on ice. Following centrifugation at 2000g for 5 min at 4°C, the pelleted nuclei were resuspended in 1 ml of Buffer B with DTT and incubated for 10 min at 4°C. After repeating the centrifugation step, the pelleted nuclei were resuspended in 100 μl of Buffer B, mixed with 0.5 μl of micrococcal nuclease, and incubated for 20 min at 37°C with frequent mixing to digest the DNA into ~150– to 900–base pair (bp) fragments. Digestion was stopped by adding 10 μl of 0.5 M EDTA. After centrifugation at 16,000g for 1 min at 4°C, the pelleted nuclei were resuspended in 500 μl of ChIP buffer and sonicated for 20 min. After centrifugation at 9400g for 10 min at 4°C, the supernatant, which contained cross-linked chromatin, was collected. For optimal ChIP results, 5 to 10 μg of digested, cross-linked chromatin were used per immunoprecipitation (IP) reaction.
For each IP reaction, immunoprecipitating antibodies were added to 500 μl of diluted chromatin. IP samples were incubated for 4 hours to overnight at 4°C with rotation, followed by the addition of ChIP-grade protein G agarose beads to each IP reaction. After a 2-hour incubation at 4°C with rotation, the beads were washed three times with 1 ml of low-salt wash buffer and once with 1 ml of high-salt wash buffer. DNA was eluted in elution buffer, and cross-links were reversed by incubating overnight at 65°C. RNA and protein were digested using ribonuclease A and proteinase K, and the DNA was purified by phenol-chloroform extraction and isopropanol precipitation. Target DNA abundance in ChIP eluates was assayed by quantitative PCR with primers designed to produce 100- to 200-bp products. An isotype-matched immunoglobulin G (IgG) antibody was used as a control. The antibodies used for ChIP assays were FOXO1 antibody (2880, Cell Signaling Technology) and a corresponding IgG control (Cell Signaling Technology, 2729). Sequences of the primers used for ChIP assays are listed in table S4.
Immunohistochemistry
The samples were fixed with 4% PFA and then embedded in paraffin. The paraffin-embedded human tissues were deparaffinized and rehydrated. Following heat-induced antigen retrieval, the samples were blocked with goat serum blocking solution at room temperature for 30 min and subsequently incubated with primary antibodies overnight at 4°C. The next day, the sections were washed and incubated with biotin-conjugated secondary antibodies for 1 hour at room temperature, followed by a 3,3′-diaminobenzidine chromogenic reaction. Nuclei were counterstained using hematoxylin staining solution.
Images were captured using a Nikon microscope and analyzed with Aipathwell software. Antibodies used in this study included USP22 (1:100), ATGL (1:200), FOXO1 (1:200), and p-USP22-Thr147 (1:50). Depending on the antibody instructions, either EDTA or citrate solution was used for antigen retrieval. Integrated optical density per area (IOD/area) was calculated to assess the staining intensity.
Co-IP and Western blotting
Cells were washed with ice-cold PBS and then lysed in radioimmunoprecipitation assay (RIPA) lysis buffer containing protease and phosphatase inhibitors. The lysates were incubated on ice for 15 min, followed by centrifugation at 15,000g for 15 min. The supernatants were precleaned three times with protein G Sepharose (GE Healthcare, 17-0618-02), each time for 15 min. The precleaned supernatants were then subjected to IP with the indicated antibodies, followed by a 2-hour incubation on ice. Next, 50 μl of protein G Sepharose beads was added and incubated for an additional 2 hours. The beads were washed five times and then boiled in 50 μl of 2× loading buffer for 5 min. Proteins were separated on 8 to 10% SDS–polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. The membranes were blocked in 5% fat-free dried milk in tris-buffered saline with 0.5% Tween 20 (TBST) for 1 hour. The membranes were then incubated with appropriate primary antibodies overnight at 4°C. After washing with TBST, the membranes were incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies (CST, HRP conjugate; goat, HRP conjugate) for 1 hour. The membranes were washed again with TBST, and signals were visualized using an enhanced chemiluminescence substrate (Abbkine, BMU102-CN) and quantified using the Bio-Rad Image software. If necessary, membranes were stripped using stripping buffer (Solarbio, SW3020) and reincubated with corresponding antibodies. Primary antibodies used are listed in table S5.
Quantitative RT-PCR
Total RNA was extracted using the Total RNA kit (Omega, R6834-01), followed by reverse transcription using the iScript cDNA Synthesis Kit (Bio-Rad, 1708891). Real-time RT-PCR was conducted with the SuperReal PreMix Plus Kit (Tiangen, FP205-02) following the manufacturer’s protocol. The mRNA levels were calculated using the ΔCt method and normalized to β-actin. Primers for human genes and β-actin were obtained from Thermo Fisher Scientific, while USP22, ATGL, and FOXO1 primers were sourced from GenePharma Biology. All primers used in this study are listed in table S6.
Measurement of TAG, DAG, and FFA contents
Lipid analysis was performed as previously reported (61). Briefly, TAG contents were determined using the Triglyceride Assay Kit (Abcam, ab65336). Tissue samples (10 mg) were washed with cold PBS and then resuspended and homogenized in a 5% NP-40/ddH2O solution using a Dounce homogenizer or pestle with 10 to 15 passes. The samples were slowly heated to 80° to 100°C in a water bath until the NP-40 solution became cloudy and then cooled to room temperature. This heating and cooling process was repeated to solubilize all triglycerides. Following the kit’s requirements, the operation continued, and the output was measured on a microplate reader at an optical density (OD) of 570 nm for colorimetric assay.
DAG contents were determined using the DAG ELISA Kit (BlueGene Biotech, E01D0010). Tissue homogenate preparation varied depending on the tissue type. For this assay, tissues were rinsed in ice-cold PBS to remove excess blood thoroughly and weighed before homogenization. Tissues (10 mg) were minced into small pieces and homogenized in a certain amount of PBS with a glass homogenizer on ice. The resulting suspension was subjected to ultrasonication or two freeze-thaw cycles to further break the cell membranes. Afterward, the homogenates were centrifuged for 15 min at 5000 rpm. The supernatant was removed, and the assay was performed immediately or aliquoted and stored at −20°C. Following the kit’s requirements, the operation proceeded, and the output was measured on a microplate reader at an OD of 450 nm for colorimetric assay.
FFA contents were determined using the Free Fatty Acid Quantitation Kit (Sigma-Aldrich, MAK044). The tissue (10 mg) was homogenized in 200 μl of a 1% (w/v) Triton X-100 in chloroform solution. The samples were then centrifuged at 13,000g for 10 min to remove the insoluble material. The organic phases (lower phase) were collected and air-dried at 50°C to remove chloroform. Vacuum drying was performed for 30 min to remove trace chloroform. The dried lipids were dissolved in 200 μl of Fatty Acid Assay Buffer by vertexing extensively for 5 min. Following the kit’s requirements, the operation continued, and the output was measured on a microplate reader at an OD of 570 nm for colorimetric assay.
Proteomic analysis of the phosphorylation sites in USP22 and AKT
Cells were harvested 24 hours after transfection, washed twice with ice-cold PBS, resuspended in RIPA lysis buffer, and sonicated. The protein concentrations were determined using a bicinchoninic acid assay. A 1-mg protein was incubated with 1 μl of anti-Myc tag monoclonal antibody (2 mg/ml) in a rotating mixer for ~6 hours at 4°C. Then, 30 μl of protein A/G agarose (Santa Cruz) was added to the mixture and rotated overnight. The beads were spun down at 700g for 2 min, and the pellets were resuspended in 100 μl of 50 mM Triethylammonium bicarbonate (TEABC) and 2 M urea. The samples were incubated with 10 mM DTT for 60 min, followed by alkylation with 20 mM indole-3-acetic acid in the dark for 30 min, and then 100 μl of 50 mM TEABC and 4 μg of trypsin were added to digest the enriched proteins on the beads at 37°C overnight. The phosphopeptides from the desalted peptides were enriched using the Phosphopeptide Enrichment Kit (Thermo Fisher Scientific, no. A32992) according to the provided instructions.
For MS, the enriched phosphopeptides were analyzed by a Q-Exactive HF-X coupled to an UltiMat 3000 RSLCnano system (Thermo Fisher Scientific, MA). Phosphopeptides were reconstituted in 0.1% formic acid (FA) and loaded onto a 360-μm outer diameter × 150-μm inner diameter silica-based column packed with ReproSil-Pur 120 C18. An 88-min liquid chromatography–tandem MS (MS/MS) method was used to analyze the sample with a flow rate of 300 nl/min. The solvent A (0.1% FA) and solvent B (80% acetonitrile/0.1% FA) were used as a loading pump solvent. The elution program was as follows: 0 to 10 min, 6% B; 15 min, 10% B; 70 min, 30% B; 80 min, 40% B; 80 to 85 min, 95% B; and 85 to 88 min, 6% B. The top 25 most intense ions from the MS1 scan spectrum [mass range from mass/charge ratio (m/z), 350 to 1550; resolution, 120,000; and Automatic Gain Control (AGC) target, 3 × 106] were measured to MS2 (mass range from m/z 200 to 2000; resolution, 15,000; AGC target, 2 × 104) with a normalized collision energy of 27; the dynamic exclusion time was set to 15 s.
Cell proliferation assay
The proliferation of indicated WT-4T1 cell was measured by Cell Counting Kit-8 (CCK-8) (MedChemExpress, HY-K0301) assay as previously reported (62). Briefly, cells were seeded into 96-well plates with 3 × 103 cells per well and incubated for the cell attachment. Then, each well was added with 10 μl of CCK-8 solution and incubated at 37°C for 2 hours. The absorption values were measured at 450 nm using a microplate reader.
In vitro migration assays
4T1 cells were pretreated with EPI for 5 days. Subsequently, cells (5 × 104) resuspended in serum-free medium were placed into uncoated membranes in the upper chamber (24-well insert, 8 μm, Corning Costar, China). The lower chamber was filled with growth medium supplemented with 10% FBS as an attractant. After 24 hours of incubation, cells that migrated through the membrane were fixed with 4% paraformaldehyde (Santa Cruz, United States) and stained with 0.1% crystal violet solution (Shanghai Sangon Company, China). Microscopic images of the stained cells were captured using an Olympus microscope (Japan), and counts were performed on five random fields at 10× magnification.
Data analysis
Raw MS files from the entire study were searched by MaxQuant (63) (version 2.0.1.0) against a nonredundant UniProt human database (containing 20,387 sequences and downloaded from the website www.uniprot.org). Carbamidomethyl cysteine was set as a fixed modification, and phosphorylation of serine, threonine, and tyrosine and oxidation of methionine were allowed as variable modifications. Trypsin was specified as the proteolytic enzyme with up to two missed cleavage sites allowed. Mass tolerances were 20 parts per million for precursor ions and 0.05 Da for Ion Trap MS, MS/MS ions. The false discovery rate was controlled to <0.01 at both the peptide and protein level. The other settings were the same as the conventional search. The reverse hits and potential contaminant hits were removed for further analysis. The identified peptides showed in this work were screened by Score ≥ 40. The obtained phosphorylated sites were further filtered by Localization Probability ≥ 0.75 and Score Difference ≥ 5, which means a high confidence level of phosphorylated sites. And all these parameters are commonly used in phosphoproteomics studies (64, 65). The MS data and MaxQuant output files have been deposited onto the iProX database (https://iprox.cn/) with an accession code of IPX0003657000.
The differential phosphorylated sites were processed by R package Prostar (66). Phosphorylation sites in which the number of missing data was observed in less than two replicates were kept in at least one condition. Then, the intensity data were logarithmic transformed and normalized by median centering within condition. The missing values were imputed with 1 quantile of each replicate. Differentially expressed phosphorylation sites were detected using a Welch’s t test. Phosphorylation sites were regarded as markedly differentially expressed between two groups with a P value of less than 0.05 and a fold change of <−2 or >2.
Statistical analyses
Statistical analyses were performed with SPSS software (version 21.0) or GraphPad Prism 8.0 (GraphPad Software Inc.). Shapiro-Wilk test to exam the data normality was used prior to the t test, one-way analysis of variance (ANOVA), and Pearson correlation. One-way ANOVA was used to compare the means of more than two groups. Student’s t test was used to compare the mean value of two groups. χ2 test was used to analyze the association of the plasma EPI level with clinical and pathological characteristics of patients with breast cancer. Pearson correlation analysis was performed to determine the correlation between two variables. Results are presented as means ± SD of three or more independent experiments. *P < 0.05; ** P < 0.01; *** P < 0.001.
Acknowledgments
We thank the Northwestern Lurie Cancer Center flow cytometry core and genomic sequencing core for the service support.
Funding: This work was supported by the National Institutes of Health (NIH) grants R01DK126908, R01DK120330, R01CA257520, and CA232347 to D.F. and the National Natural Science Foundation of China (no. 82073768) and Dalian High-level Talent Innovation Support Program (no. 2019RD03) to Z.S.
Author contributions: Y. Zhou, P.C., Y.W., N.L., Q.G., S.W., G.X., Y. Zhao, H.J., J. Song, Y.P., H. Zhu, J. Sun, S.M., C.S., B.H., Z.Z., H. Zhang, J.L., and J.W. performed the studies and analyzed the data. Y. Zhao and H.W. provided patient tissue samples. H.W., Z.S., and D.F. designed the study and wrote the manuscript.
Competing interests: D.F. is the inventor of a USP22-specific inhibitor and holds the US patent entitled “Development of USP22-specific inhibitor for treatment of cancer and other human diseases” (US63/201,330) and the cofounder of ExoMira Medicine Inc. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S16
Tables S1 to S6
REFERENCES AND NOTES
- 1.Weigelt B., Peterse J. L., van’t Veer L. J., Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 5, 591–602 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Spiegel D., Bloom J. R., Kraemer H. C., Gottheil E., Effect of psychosocial treatment on survival of patients with metastatic breast cancer. Lancet 2, 888–891 (1989). [DOI] [PubMed] [Google Scholar]
- 3.Borgi M., Collacchi B., Ortona E., Cirulli F., Stress and coping in women with breast cancer: Unravelling the mechanisms to improve resilience. Neurosci. Biobehav. Rev. 119, 406–421 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Zhu Z., Yu R., Yang C., Li D., Wang J., Yang W., Ji Y., Wang L., Wang Y., Jiang F., Stress-related hormone reduces autophagy through the regulation of phosphatidylethanolamine in breast cancer cells. Ann. Transl. Med. 9, 149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang A., Botteri E., Gillis R. D., Lofling L., Le C. P., Ziegler A. I., Chung N. C., Rowe M. C., Fabb S. A., Hartley B. J., Nowell C. J., Kurozumi S., Gandini S., Munzone E., Montagna E., Eikelis N., Phillips S. E., Honda C., Masuda K., Katayama A., Oyama T., Cole S. W., Lambert G. W., Walker A. K., Sloan E. K., Beta-blockade enhances anthracycline control of metastasis in triple-negative breast cancer. Sci. Transl. Med. 15, eadf1147 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Conceicao F., Sousa D. M., Paredes J., Lamghari M., Sympathetic activity in breast cancer and metastasis: Partners in crime. Bone Res. 9, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He B., Gao R., Lv S., Chen A., Huang J., Wang L., Feng Y., Feng J., Liu B., Lei J., Deng B., He B., Cui B., Peng F., Yan M., Wang Z., Lam E. W., Jin B., Shao Z., Li Y., Jiao J., Wang X., Liu Q., Cancer cell employs a microenvironmental neural signal trans-activating nucleus-mitochondria coordination to acquire stemness. Signal Transduct. Target. Ther. 8, 275 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lofling L. L., Stoer N. C., Sloan E. K., Chang A., Gandini S., Ursin G., Botteri E., β-blockers and breast cancer survival by molecular subtypes: A population-based cohort study and meta-analysis. Br. J. Cancer 127, 1086–1096 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glinsky G. V., Berezovska O., Glinskii A. B., Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115, 1503–1521 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahad J. P., Mills G. B., Coombes K. R., Stem cell-ness: A “magic marker” for cancer. J. Clin. Invest. 115, 1463–1467 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Yao L., Zhang X., Ji H., Wang L., Sun S., Pang D., Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J. Cancer Res. Clin. Oncol. 137, 1245–1253 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Lin Z., Tan C., Qiu Q., Kong S., Yang H., Zhao F., Liu Z., Li J., Kong Q., Gao B., Barrett T., Yang G. Y., Zhang J., Fang D., Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov. 1, 15028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atanassov B. S., Dent S. Y., USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 12, 924–930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atanassov B. S., Evrard Y. A., Multani A. S., Zhang Z., Tora L., Devys D., Chang S., Dent S. Y., Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell 35, 352–364 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Z., Yang H., Kong Q., Li J., Lee S. M., Gao B., Dong H., Wei J., Song J., Zhang D. D., Fang D., USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46, 484–494 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Cortez J. T., Montauti E., Shifrut E., Gatchalian J., Zhang Y., Shaked O., Xu Y., Roth T. L., Simeonov D. R., Zhang Y., Chen S., Li Z., Woo J. M., Ho J., Vogel I. A., Prator G. Y., Zhang B., Lee Y., Sun Z., Ifergan I., Van Gool F., Hargreaves D. C., Bluestone J. A., Marson A., Fang D., CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582, 416–420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montauti E., Weinberg S. E., Chu P., Chaudhuri S., Mani N. L., Iyer R., Zhou Y., Zhang Y., Liu C., Xin C., Gregory S., Wei J., Zhang Y., Chen W., Sun Z., Yan M., Fang D., A deubiquitination module essential for Treg fitness in the tumor microenvironment. Sci. Adv. 8, eabo4116 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu K., Gao Q., Jia Y., Wei J., Chaudhuri S., Wang S., Tang A., Mani N., Iyer R., Cheng Y., Gao B., Lu W., Sun Z., Liu H., Fang D., Ubiquitin-specific peptidase 22 controls integrin-dependent cancer cell stemness and metastasis. Res. Sq. 2023, 10.21203/rs.3.rs-2922367/v1 (2023). [DOI] [Google Scholar]
- 19.Lafontan M., Langin D., Lipolysis and lipid mobilization in human adipose tissue. Prog. Lipid Res. 48, 275–297 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Wang Y. Y., Attane C., Milhas D., Dirat B., Dauvillier S., Guerard A., Gilhodes J., Lazar I., Alet N., Laurent V., Le Gonidec S., Biard D., Herve C., Bost F., Ren G. S., Bono F., Escourrou G., Prentki M., Nieto L., Valet P., Muller C., Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight 2, e87489 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ow J. R., Caldez M. J., Zafer G., Foo J. C., Li H. Y., Ghosh S., Wollmann H., Cazenave-Gassiot A., Ong C. B., Wenk M. R., Han W., Choi H., Kaldis P., Remodeling of whole-body lipid metabolism and a diabetic-like phenotype caused by Loss of CDK1 and hepatocyte division. eLife 9, e63835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., Ma M., Li Y., Liu J., Sun C., Liu S., Ma Y., Yan Y., Tang Z., Shen S., Yu J., Wu Y., Jiang J., Wang L., Jin Z. B., Ying H., Li Y., miR-183 and miR-96 orchestrate both glucose and fat utilization in skeletal muscle. EMBO Rep. 22, e52247 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W., Bu S. Y., Mashek M. T., O-Sullivan I. S., Sibai Z., Khan S. A., Ilkayeva O., Newgard C. B., Mashek D. G., Unterman T. G., Integrated regulation of hepatic lipid and glucose metabolism by adipose triacylglycerol lipase and FoxO proteins. Cell Rep. 15, 349–359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Ma Z., Jiang S., Hu W., Li T., Di S., Wang D., Yang Y., A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog. Lipid Res. 66, 42–49 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Mihaylova M. M., Vasquez D. S., Ravnskjaer K., Denechaud P. D., Yu R. T., Alvarez J. G., Downes M., Evans R. M., Montminy M., Shaw R. J., Class Iia histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu Y., Rosso L. G., Huang P., Wang Z., Xu Y., Yao X., Bao M., Yan J., Song H., Wang G., Liver Med23 ablation improves glucose and lipid metabolism through modulating FOXO1 activity. Cell Res. 24, 1250–1265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langlet F., Haeusler R. A., Linden D., Ericson E., Norris T., Johansson A., Cook J. R., Aizawa K., Wang L., Buettner C., Accili D., Selective inhibition of FOXO1 activator/repressor balance modulates hepatic glucose handling. Cell 171, 824–835.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy J. F., Davies D. H., Smith C. J., The development of enzyme-linked immunosorbent assays (ELISA) for the catecholamines adrenalin and noradrenalin. J. Immunol. Methods 154, 89–98 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Cui B., Luo Y., Tian P., Peng F., Lu J., Yang Y., Su Q., Liu B., Yu J., Luo X., Yin L., Cheng W., An F., He B., Liang D., Wu S., Chu P., Song L., Liu X., Luo H., Xu J., Pan Y., Wang Y., Li D., Huang P., Yang Q., Zhang L., Zhou B. P., Liu S., Xu G., Lam E. W., Kelley K. W., Liu Q., Stress-induced epinephrine enhances lactate dehydrogenase a and promotes breast cancer stem-like cells. J. Clin. Invest. 129, 1030–1046 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Liang Y., Song R., Yang G., Han J., Lan Y., Pan S., Zhu M., Liu Y., Wang Y., Meng F., Cui Y., Wang J., Zhang B., Song X., Lu Z., Zheng T., Liu L., Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol. Cancer 17, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu B., Li S., Li M., Wang S., Mu G., Chen K., Wang M., Zhu W. G., Wang W., Wang J., Li Z., Yang J., Yang Y., KAT8 acetylation-controlled lipolysis affects the invasive and migratory potential of colorectal cancer cells. Cell Death Dis. 14, 164 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das S. K., Eder S., Schauer S., Diwoky C., Temmel H., Guertl B., Gorkiewicz G., Tamilarasan K. P., Kumari P., Trauner M., Zimmermann R., Vesely P., Haemmerle G., Zechner R., Hoefler G., Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 333, 233–238 (2011). [DOI] [PubMed] [Google Scholar]
- 33.El-Merahbi R., Viera J. T., Valdes A. L., Kolczynska K., Reuter S., Loffler M. C., Erk M., Ade C. P., Karwen T., Mayer A. E., Eilers M., Sumara G., The adrenergic-induced ERK3 pathway drives lipolysis and suppresses energy dissipation. Genes Dev. 34, 495–510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi T., Zamponi R., Chirico M., Pisanu M. E., Iorio E., Torricelli F., Gugnoni M., Ciarrocchi A., Pistoni M., BETi enhance ATGL expression and its lipase activity to exert their antitumoral effects in triple-negative breast cancer (TNBC) cells. J. Exp. Clin. Cancer Res. 42, 7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu F. Q., Fang T., Yu L. X., Lv G. S., Lv H. W., Liang D., Li T., Wang C. Z., Tan Y. X., Ding J., Chen Y., Tang L., Guo L. N., Tang S. H., Yang W., Wang H. Y., ADRB2 signaling promotes hcc progression and sorafenib resistance by inhibiting autophagic degradation of HIF1α. J. Hepatol. 65, 314–324 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Li J., Yang X. M., Wang Y. H., Feng M. X., Liu X. J., Zhang Y. L., Huang S., Wu Z., Xue F., Qin W. X., Gu J. R., Xia Q., Zhang Z. G., Monoamine oxidase a suppresses hepatocellular carcinoma metastasis by inhibiting the adrenergic system and its transactivation of EGFR signaling. J. Hepatol. 60, 1225–1234 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Hara M. R., Kovacs J. J., Whalen E. J., Rajagopal S., Strachan R. T., Grant W., Towers A. J., Williams B., Lam C. M., Xiao K., Shenoy S. K., Gregory S. G., Ahn S., Duckett D. R., Lefkowitz R. J., A stress response pathway regulates DNA damage through β2-adrenoreceptors and β-arrestin-1. Nature 477, 349–353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan S., Pullikuth A., Nelson K. C., Flores A., Karpova Y., Baiz D., Zhu S., Sui G., Huang Y., Choi Y. A., D’Agostino R. Jr., Hemal A., von Holzen U., Debinski W., Kulik G., β2-adrenoreceptor signaling increases therapy resistance in prostate cancer by upregulating MCL1. Mol. Cancer Res. 18, 1839–1848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole S. W., Sood A. K., Molecular pathways: Beta-adrenergic signaling in cancer. Clin. Cancer Res. 18, 1201–1206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassan S., Karpova Y., Baiz D., Yancey D., Pullikuth A., Flores A., Register T., Cline J. M., D’Agostino R. Jr., Danial N., Datta S. R., Kulik G., Behavioral stress accelerates prostate cancer development in mice. J. Clin. Invest. 123, 874–886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagaraja A. S., Sadaoui N. C., Dorniak P. L., Lutgendorf S. K., Sood A. K., SnapShot: Stress and disease. Cell Metab. 23, 388–388.e1 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Zhang Y., He Z., Yin K., Li B., Zhang L., Xu Z., Chronic stress promotes gastric cancer progression and metastasis: An essential role for ADRB2. Cell Death Dis. 10, 788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim T. H., Yang Y. M., Han C. Y., Koo J. H., Oh H., Kim S. S., You B. H., Choi Y. H., Park T. S., Lee C. H., Kurose H., Noureddin M., Seki E., Wan Y. Y., Choi C. S., Kim S. G., Gα12 ablation exacerbates liver steatosis and obesity by suppressing USP22/SIRT1-regulated mitochondrial respiration. J. Clin. Invest. 128, 5587–5602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ning Z., Guo X., Liu X., Lu C., Wang A., Wang X., Wang W., Chen H., Qin W., Liu X., Zhou L., Ma C., Du J., Lin Z., Luo H., Otkur W., Qi H., Chen D., Xia T., Liu J., Tan G., Xu G., Piao H. L., USP22 regulates lipidome accumulation by stabilizing Pparγ in hepatocellular carcinoma. Nat. Commun. 13, 2187 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dietlein N., Wang X., Metz J., Disson O., Shang F., Beyersdorffer C., Rodriguez Correa E., Lipka D. B., Begus-Nahrmann Y., Kosinsky R. L., Johnsen S. A., Lecuit M., Hofer T., Rodewald H. R., Usp22 is an intracellular regulator of systemic emergency hematopoiesis. Sci. Immunol. 7, eabq2061 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Prokakis E., Bamahmoud H., Jansari S., Fritsche L., Dietz A., Boshnakovska A., Rehling P., Johnsen S. A., Gallwas J., Wegwitz F., USP22 supports the aggressive behavior of basal-like breast cancer by stimulating cellular respiration. Cell Commun. Signal 22, 120 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai S., Mo Y., Wang Y., Xiang B., Liao Q., Zhou M., Li X., Li Y., Xiong W., Li G., Guo C., Zeng Z., Chronic stress promotes cancer development. Front. Oncol. 10, 1492 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H., Liu N., Zhao Y., Zhu X., Wang C., Liu Q., Gao C., Zhao X., Li J., Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging 11, 9643–9660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gennaro V. J., Stanek T. J., Peck A. R., Sun Y., Wang F., Qie S., Knudsen K. E., Rui H., Butt T., Diehl J. A., McMahon S. B., Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 115, E9298–E9307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X. Y., Varthi M., Sykes S. M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A. W., Berger S. L., McMahon S. B., The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29, 102–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X., Yalcin S., Lee D. F., Yeh T. Y., Lee S. M., Su J., Mungamuri S. K., Rimmelé P., Kennedy M., Sellers R., Landthaler M., Tuschl T., Chi N. W., Lemischka I., Keller G., Ghaffari S., FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat. Cell Biol. 13, 1092–1099 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia Whitlock A. E., Sostre-Colon J., Gavin M., Martin N. D., Baur J. A., Sims C. A., Titchenell P. M., Loss of FOXO transcription factors in the liver mitigates stress-induced hyperglycemia. Mol. Metab. 51, 101246 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arden K. C., FoxO: Linking new signaling pathways. Mol. Cell 14, 416–418 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Yu J. M., Sun W., Wang Z. H., Liang X., Hua F., Li K., Lv X. X., Zhang X. W., Liu Y. Y., Yu J. J., Liu S. S., Shang S., Wang F., Yang Z. N., Zhao C. X., Hou X. Y., Li P. P., Huang B., Cui B., Hu Z. W., TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat. Commun. 10, 5720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng X., Wu Z., Wu Y., Hankey W., Prior T. W., Li L., Ganju R. K., Shen R., Zou X., Cdc25A regulates matrix metalloprotease 1 through Foxo1 and mediates metastasis of breast cancer cells. Mol. Cell. Biol. 31, 3457–3471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Li Y., Wang Q., Chen Z., Li X., Wu Z., Hu C., Liao D., Zhang W., Chen Z. S., The PI3K subunits, P110α and P110β are potential targets for overcoming P-gp and BCRP-mediated MDR in cancer. Mol. Cancer 19, 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y., Wang Y., Gao B., Sun Y., Cao L., Genardi S. M., Wang C. R., Li H., Sun Z., Yang Y., Fang D., USP22 controls iNKT immunity through MED1 suppression of histone H2A monoubiquitination. J. Exp. Med. 217, e20182218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blaes A. H., Domingo-Musibay E., Kalinsky K., Propranolol: What is blocking its clinical investigation in breast cancer? Clin. Cancer Res. 26, 1781–1783 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Melo-Cardenas J., Xu Y., Wei J., Tan C., Kong S., Gao B., Montauti E., Kirsammer G., Licht J. D., Yu J., Ji P., Crispino J. D., Fang D., USP22 deficiency leads to myeloid leukemia upon oncogenic kras activation through a PU.1-dependent mechanism. Blood 132, 423–434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Gui M., Wang Y., Mani N., Chaudhuri S., Gao B., Li H., Kanwar Y. S., Lewis S. A., Dumas S. N., Ntambi J. M., Zhang K., Fang D., Inositol-requiring enzyme 1α-mediated synthesis of monounsaturated fatty acids as a driver of B cell differentiation and lupus-like autoimmune disease. Arthritis Rheumatol. 73, 2314–2326 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei J., Yuan Y., Chen L., Xu Y., Zhang Y., Wang Y., Yang Y., Peek C. B., Diebold L., Yang Y., Gao B., Jin C., Melo-Cardenas J., Chandel N. S., Zhang D. D., Pan H., Zhang K., Wang J., He F., Fang D., ER-associated ubiquitin ligase HRD1 programs liver metabolism by targeting multiple metabolic enzymes. Nat. Commun. 9, 3659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Z., Yang H., Tan C., Li J., Liu Z., Quan Q., Kong S., Ye J., Gao B., Fang D., USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 5, 1639–1649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Sharma K., D’Souza R. C. J., Tyanova S., Schaab C., Wiśniewski J. R., Cox J., Mann M., Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M., Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Wieczorek S., Combes F., Lazar C., Giai Gianetto Q., Gatto L., Dorffer A., Hesse A. M., Coute Y., Ferro M., Bruley C., Burger T., DAPAR & ProStaR: Software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics 33, 135–136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Tables S1 to S6