Abstract
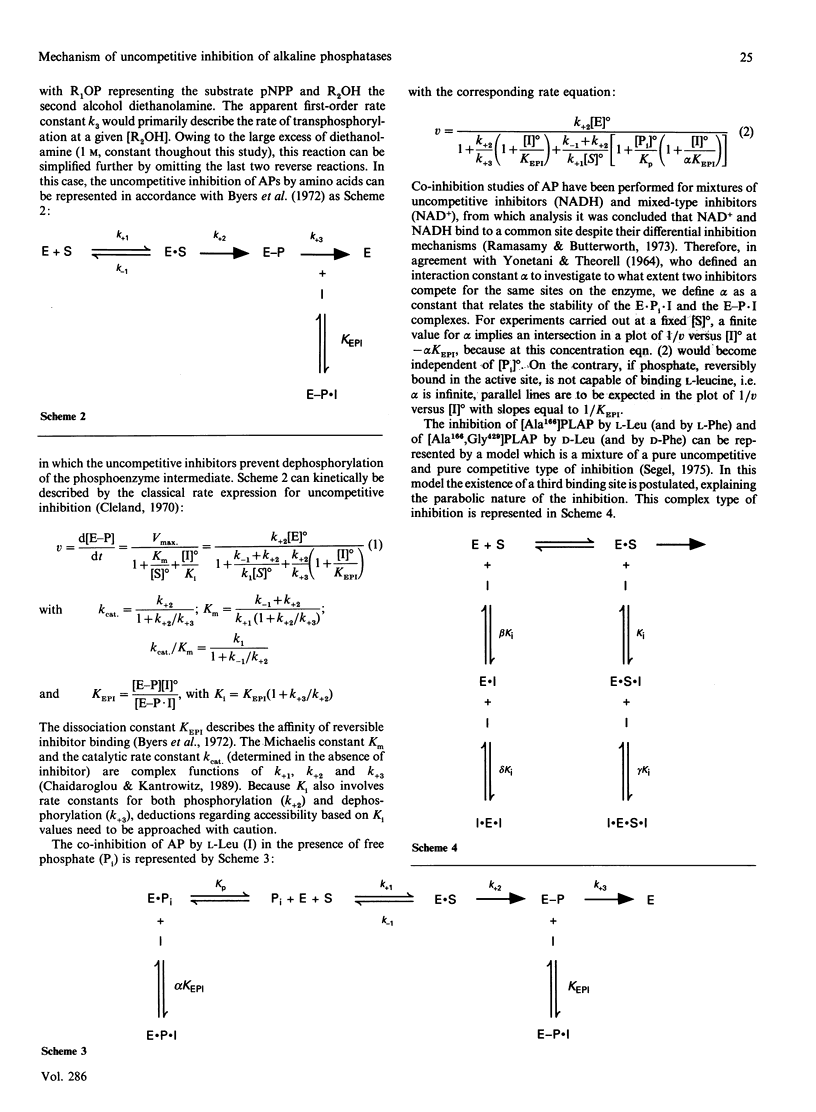
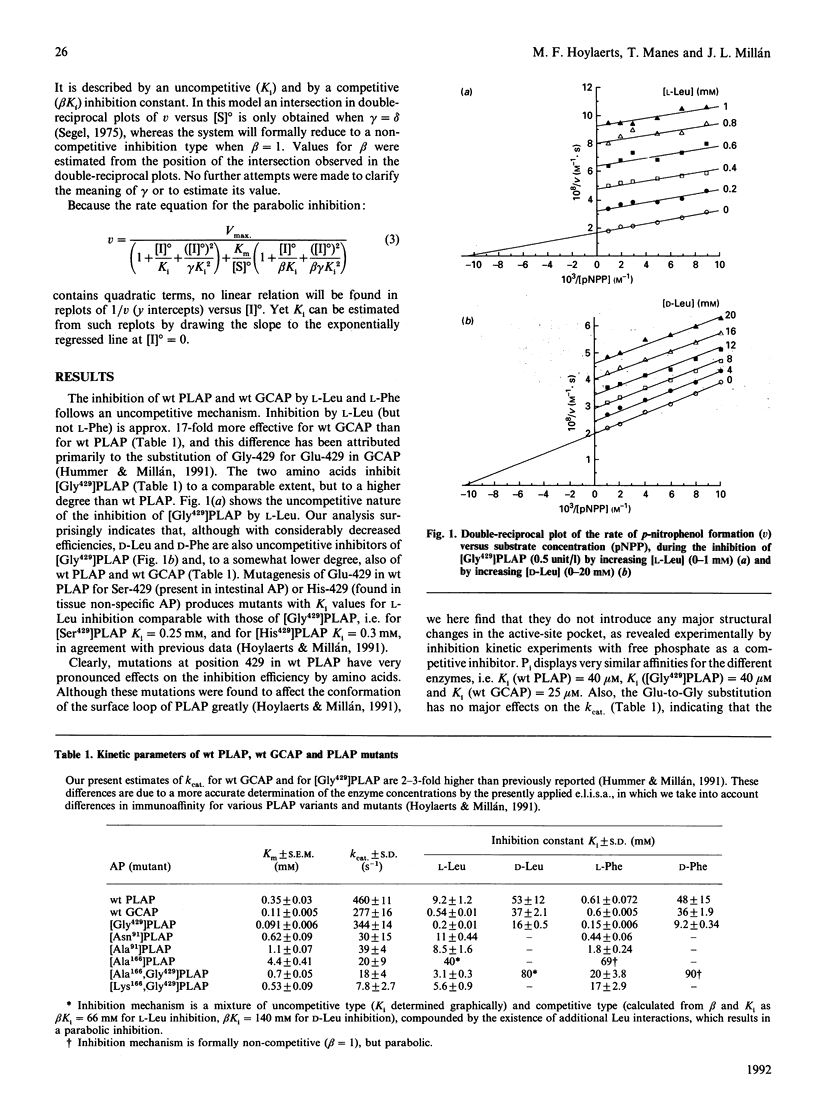
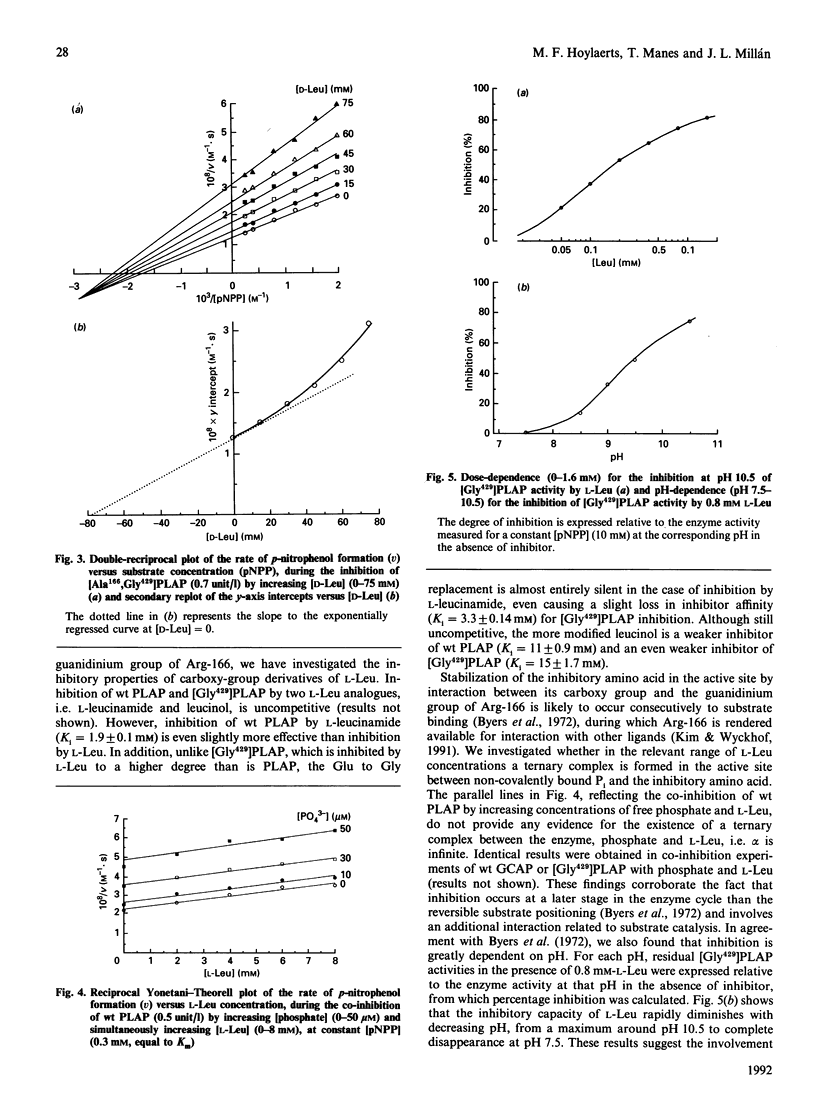
Placental (PLAP) and germ-cell (GCAP) alkaline phosphatases are inhibited uncompetitively by L-Leu and L-Phe. Whereas L-Phe inhibits PLAP and GCAP to the same extent, L-Leu inhibits GCAP 17-fold more strongly than it does PLAP. This difference has been attributed [Hummer & Millán (1991) Biochem. J 274, 91-95] to a Glu----Gly substitution at position 429 in GCAP. The D-Phe and D-Leu enantiomorphs are also inhibitory through an uncompetitive mechanism but with greatly decreased efficiencies. Replacement of the active-site residue Arg-166 by Ala-166 changes the inhibition mechanism of the resulting PLAP mutant to a more complex mixed-type inhibition, with decreased affinities for L-Leu and L-Phe. The uncompetitive mechanism is restored on the simultaneous introduction of Gly-429 in the Ala-166 mutant, but the inhibitions of [Ala166,Gly429]PLAP and even [Lys166,Gly429]PLAP by L-Leu and L-Phe are considerably decreased compared with that of [Gly429]PLAP. These findings point to the importance of Arg-166 during inhibition. Active-site binding of L-Leu requires the presence of covalently bound phosphate in the active-site pocket, and the inhibition of PLAP by L-Leu is pH-sensitive, gradually disappearing when the pH is decreased from 10.5 to 7.5. Our data are compatible with the following molecular model for the uncompetitive inhibition of PLAP and GCAP by L-Phe and L-Leu: after binding of a phosphorylated substrate to the active site, the guanidinium group of Arg-166 (normally involved in positioning phosphate) is redirected to the carboxy group of L-Leu (or L-Phe), thus stabilizing the inhibitor in the active site. Therefore leucinamide and leucinol are weaker inhibitors of [Gly429]PLAP than is L-Leu. During this Arg-166-regulated event, the amino acid side group is positioned in the loop containing Glu-429 or Gly-429, leading to further stabilization. Replacement of Glu-429 by Gly-429 eliminates steric constraints experienced by the bulky L-Leu side group during its positioning and also increases the active-site accessibility for the inhibitor, providing the basis for the 17-fold difference in inhibition efficiency between PLAP and GCAP. Finally, the inhibitor's unprotonated amino group co-ordinates with the active-site Zn2+ ion 1, interfering with the hydrolysis of the phosphoenzyme intermediate, a phenomenon that determines the uncompetitive nature of the inhibition.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird P., Gething M. J., Sambrook J. Translocation in yeast and mammalian cells: not all signal sequences are functionally equivalent. J Cell Biol. 1987 Dec;105(6 Pt 2):2905–2914. doi: 10.1083/jcb.105.6.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler-Ransohoff J. E., Kendall D. A., Kaiser E. T. Use of site-directed mutagenesis to elucidate the role of arginine-166 in the catalytic mechanism of alkaline phosphatase. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4276–4278. doi: 10.1073/pnas.85.12.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers D. A., Fernley H. N., Walker P. G. Studies on alkaline phosphatase. Inhibition of human-placental phosphoryl phosphatase by L-phenylalanine. Eur J Biochem. 1972 Sep 18;29(2):197–204. doi: 10.1111/j.1432-1033.1972.tb01975.x. [DOI] [PubMed] [Google Scholar]
- Chaidaroglou A., Brezinski D. J., Middleton S. A., Kantrowitz E. R. Function of arginine-166 in the active site of Escherichia coli alkaline phosphatase. Biochemistry. 1988 Nov 1;27(22):8338–8343. doi: 10.1021/bi00422a008. [DOI] [PubMed] [Google Scholar]
- Chaidaroglou A., Kantrowitz E. R. Alteration of aspartate 101 in the active site of Escherichia coli alkaline phosphatase enhances the catalytic activity. Protein Eng. 1989 Nov;3(2):127–132. doi: 10.1093/protein/3.2.127. [DOI] [PubMed] [Google Scholar]
- Doellgast G. J., Fishman W. H. L-leucine, a specific inhibitor of a rare human placental alkaline phosphatase phenotype. Nature. 1976 Jan 1;259(5538):49–51. doi: 10.1038/259049a0. [DOI] [PubMed] [Google Scholar]
- Fishman W. H. On the importance of being (stereo) specific. Clin Chim Acta. 1990 Jan 15;186(2):129–131. doi: 10.1016/0009-8981(90)90030-v. [DOI] [PubMed] [Google Scholar]
- Fishman W. H., Sie H. G. Organ-specific inhibition of human alkaline phosphatase isoenzymes of liver, bone, intestine and placenta; L-phenylalanine, L-tryptophan and L homoarginine. Enzymologia. 1971 Sep 30;41(3):141–167. [PubMed] [Google Scholar]
- Gasser K. W., Kirschner L. B. The inhibition and disposition of intestinal alkaline phosphatase. J Comp Physiol B. 1987;157(4):461–467. doi: 10.1007/BF00691830. [DOI] [PubMed] [Google Scholar]
- Ghosh N. K., Fishman W. H. On the mechanism of inhibition of intestinal alkaline phosphatase by L-phenylalanine. I. Kinetic studies. J Biol Chem. 1966 Jun 10;241(11):2516–2522. [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S., Anstiss C. L., Fishman W. H. Automated differential isoenzyme analysis. II. The fractionation of serum alkaline phosphatases into "liver", "intestinal" and "other" components. Enzymologia. 1971 Jul 31;41(1):9–26. [PubMed] [Google Scholar]
- Harris H. The human alkaline phosphatases: what we know and what we don't know. Clin Chim Acta. 1990 Jan 15;186(2):133–150. doi: 10.1016/0009-8981(90)90031-m. [DOI] [PubMed] [Google Scholar]
- Hoylaerts M. F., Millán J. L. Site-directed mutagenesis and epitope-mapped monoclonal antibodies define a catalytically important conformational difference between human placental and germ cell alkaline phosphatase. Eur J Biochem. 1991 Dec 5;202(2):605–616. doi: 10.1111/j.1432-1033.1991.tb16414.x. [DOI] [PubMed] [Google Scholar]
- Hummer C., Millán J. L. Gly429 is the major determinant of uncompetitive inhibition of human germ cell alkaline phosphatase by L-leucine. Biochem J. 1991 Feb 15;274(Pt 1):91–95. doi: 10.1042/bj2740091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson A., Wahren B., Brehmer-Andersson E., Silfverswärd C., Stigbrand T., Millán J. L. Eutopic expression of placental-like alkaline phosphatase in testicular tumors. Int J Cancer. 1984 Dec 15;34(6):757–761. doi: 10.1002/ijc.2910340604. [DOI] [PubMed] [Google Scholar]
- Kim E. E., Wyckoff H. W. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J Mol Biol. 1991 Mar 20;218(2):449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millán J. L., Manes T. Seminoma-derived Nagao isozyme is encoded by a germ-cell alkaline phosphatase gene. Proc Natl Acad Sci U S A. 1988 May;85(9):3024–3028. doi: 10.1073/pnas.85.9.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millán J. L. Molecular cloning and sequence analysis of human placental alkaline phosphatase. J Biol Chem. 1986 Mar 5;261(7):3112–3115. [PubMed] [Google Scholar]
- Millán J. L. Oncodevelopmental alkaline phosphatases: in search for a function. Prog Clin Biol Res. 1990;344:453–475. [PubMed] [Google Scholar]
- Millán J. L., Stigbrand T. Antigenic determinants of human placental and testicular placental-like alkaline phosphatases as mapped by monoclonal antibodies. Eur J Biochem. 1983 Oct 17;136(1):1–7. doi: 10.1111/j.1432-1033.1983.tb07697.x. [DOI] [PubMed] [Google Scholar]
- Mulivor R. A., Plotkin L. I., Harris H. Differential inhibition of the products of the human alkaline phosphatase loci. Ann Hum Genet. 1978 Jul;42(1):1–13. doi: 10.1111/j.1469-1809.1978.tb00927.x. [DOI] [PubMed] [Google Scholar]
- Paiva J., Damjanov I., Lange P. H., Harris H. Immunohistochemical localization of placental-like alkaline phosphatase in testis and germ-cell tumors using monoclonal antibodies. Am J Pathol. 1983 May;111(2):156–165. [PMC free article] [PubMed] [Google Scholar]
- Ramasamy I., Butterworth P. J. Nicotinamide-adenine dinucleotide inhibition of pig kidney alkaline phosphatase. Biochim Biophys Acta. 1975 Mar 28;384(1):146–158. doi: 10.1016/0005-2744(75)90104-7. [DOI] [PubMed] [Google Scholar]
- Ramasamy I., Butterworth P. J. The inhibition of pig kidney alkaline phosphatase by oxidized or reduced nicotinamide-adenine dinucleotide and related compounds. Biochem J. 1973 Feb;131(2):359–367. doi: 10.1042/bj1310359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowadski J. M., Handschumacher M. D., Murthy H. M., Foster B. A., Wyckoff H. W. Refined structure of alkaline phosphatase from Escherichia coli at 2.8 A resolution. J Mol Biol. 1985 Nov 20;186(2):417–433. doi: 10.1016/0022-2836(85)90115-9. [DOI] [PubMed] [Google Scholar]
- Trotman C. N., Greenwood C. Effects of zinc and other metal ions on the stability and activity of Escherichia coli alkaline phosphatase. Biochem J. 1971 Aug;124(1):25–30. doi: 10.1042/bj1240025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKINSON G. N. Statistical estimations in enzyme kinetics. Biochem J. 1961 Aug;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YONETANI T., THEORELL H. STUDIES ON LIVER ALCOHOL HYDROGENASE COMPLEXES. 3. MULTIPLE INHIBITION KINETICS IN THE PRESENCE OF TWO COMPETITIVE INHIBITORS. Arch Biochem Biophys. 1964 Jul 20;106:243–251. doi: 10.1016/0003-9861(64)90184-5. [DOI] [PubMed] [Google Scholar]


